Abstract
A sequence element located immediately upstream of the TATA element, and having the consensus sequence 5′-G/C-G/C-G/A-C-G-C-C-3′, affects the ability of transcription factor IIB to enter transcription complexes and support transcription initiation. The sequence element is recognized directly by the transcription factor IIB. Recognition involves α-helices 4′ and 5′ of IIB, which comprise a helix–turn–helix DNA-binding motif. These observations establish that transcription initiation involves a fourth core promoter element, the IIB recognition element (BRE), in addition to the TATA element, the initiator element, and the downstream promoter element, and involves a second sequence-specific general transcription factor, IIB, in addition to transcription factor IID.
Keywords: IIB recognition element, TATA element, transcription factor IIB, helix–turn–helix, DNA binding
Efficient transcription initiation at a human protein-encoding gene requires assembly on promoter DNA of a multiprotein complex containing RNA polymerase II (Pol II) and six general transcription factors, IIA, IIB, IID, IIE, IIF, and IIH (for review, see Orphanides et al. 1996; Roeder 1996; Nikolov and Burley 1997). Previous work establishes that assembly of the complex involves three core promoter elements: (1) the TATA element, located near position −30, (2) the initiator element, located near position −1, and (3) the downstream promoter element, located near position +30 (Burke and Kadonaga 1996; Orphanides et al. 1996; Roeder 1996). Transcription factor IID is responsible for recognition of at least two of these elements. One subunit of IID, TATA-binding protein (TBP), is responsible for recognition of the TATA element; one or more of the remaining subunits of IID, TBP-associated factors (TAFs), is responsible for recognition of the downstream promoter element. It is not clear which factor is responsible for recognition of the initiator element.
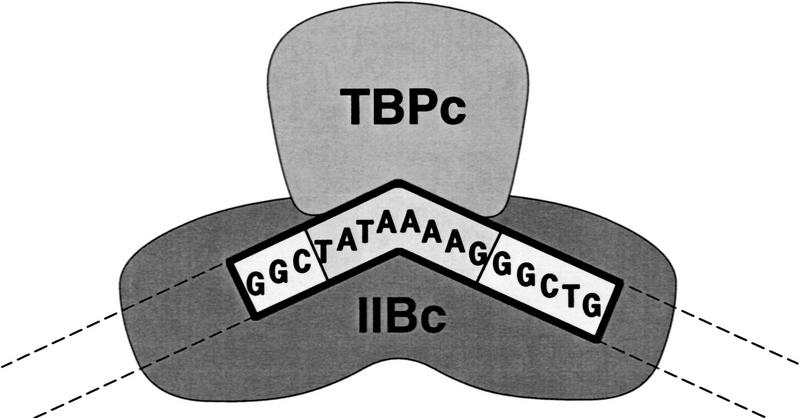
The crystallographic structure of a ternary complex of transcription factor IIB core domain (IIBc), TBP core domain (TBPc), and a 16-bp DNA fragment containing the TATA element shows that IIBc interacts with both TBPc and DNA, interacting with the DNA major groove immediately upstream of the TATA element and the DNA minor groove immediately downstream of the TATA element (Fig. 1; Nikolov et al. 1995). In the crystallographic structure, details of the interaction between IIBc and the DNA major groove upstream of the TATA element are incomplete, since the structure was determined using a DNA fragment containing only three nucleotide pairs upstream of the TATA element (Nikolov et al. 1995). However, DNA-binding and site-specific protein–DNA photocross-linking experiments confirm that interaction between IIB and the DNA major groove upstream of the TATA element occurs and indicate that the interaction is extensive, spanning up to 7–9 nucleotide pairs (Lee and Hahn 1995; Lagrange et al. 1996). The observation that IIB makes extensive interactions with the DNA major groove upstream of the TATA element raises the possibility that the ability of IIB to enter into transcription complexes and, thus, the ability of IIB to support transcription initiation, may be affected by the DNA sequence upstream of the TATA element (Lagrange et al. 1996).
Figure 1.
Structure of the IIB–TBP–TATA complex. Schematic representation of the crystallographic structure of a ternary complex of transcription factor IIB core domain (IIBc), TBP core domain (TBPc), and a 16-bp DNA fragment containing the TATA element (Nikolov et al. 1995). (TATAAAAG) TATA element; (GGC and GGCTG) DNA segments upstream and downstream of the TATA element in the 16-bp DNA fragment; (dashed lines) projected paths of distal upstream and downstream DNA segments.
In this study, we show that the seven nucleotide pairs immediately upstream of the TATA element constitute a new core promoter element, and we show that IIB directly recognizes this element.
Results
New core promoter element in Pol II-dependent transcription initiation
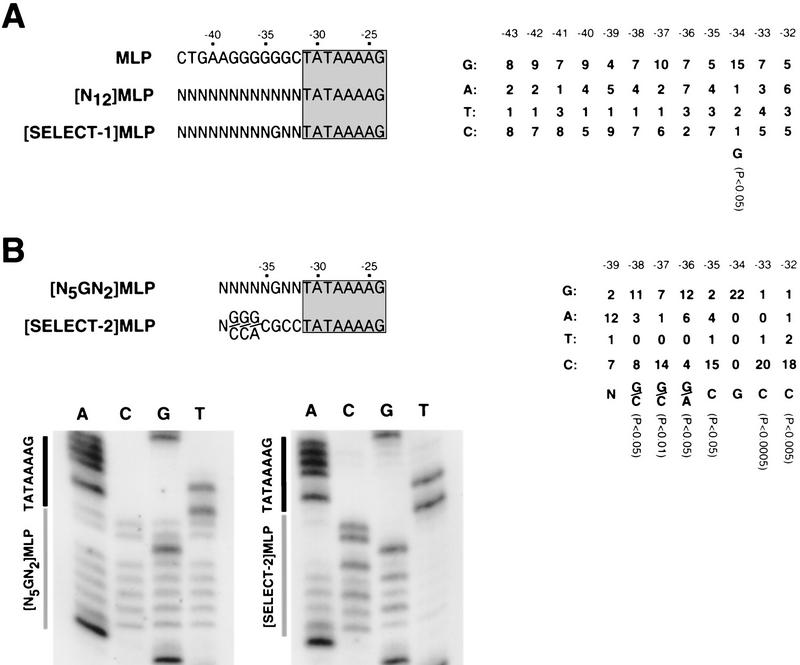
As a first step in testing the possibility that the ability of IIB to enter into transcription complexes may be affected by the DNA sequence immediately upstream of the TATA element, we performed binding-site selection (Oliphant et al. 1989; Blackwell and Weintraub 1990; Blackwell et al. 1990; Mavrothalassitis et al. 1990; Thiesen and Bach 1990). We constructed a library of DNA fragments having the TATA element of the adenovirus major late promoter preceded by 12 randomized nucleotide pairs (1.7 × 107 DNA fragments), formed IIB–TBP–DNA complexes using saturating TBP and limiting IIB, isolated IIB–TBP–DNA complexes by nondenaturing PAGE, and amplified DNA fragments in complexes by PCR. After four cycles of selection and amplification, electrophoretic mobility shift experiments indicated that the pooled selected DNA fragments exhibited an apparent fivefold higher affinity for IIB in IIB–TBPc–DNA complex formation than the unselected randomized DNA fragments, indicating that the procedure in fact had selected DNA fragments having above-average affinities for IIB (data not shown). Sequences of 19 cloned selected DNA fragments revealed a statistically significant (P < 0.002) preference for G three nucleotides upstream of the TATA element (Fig. 2A). The wild-type adenovirus major late promoter—a standard test promoter for analysis of Pol II-dependent transcription, chosen for its exceptional strength (Corden et al. 1980)—contains G at this position (i.e., position −34) (Fig. 2A, left). We suggest that G at this position is part of a sequence element correlated with high affinity for IIB in IIB–TBP–DNA complex formation.
Figure 2.
New sequence element. (A) Binding site selection, round 1. (Left) Positions −43 to −24 of the wild-type, N12-randomized, and consensus round-1-selected adenovirus major late promoter sequences (TATA element boxed and shaded). (Right) Summary of 19 round-1-selected sequences. (B) Binding site selection, round 2. (Left) Positions −39 to −24 of the N5GN2- randomized and consensus round-2-selected adenovirus major late promoter sequences (TATA element boxed and shaded). (Right) Summary of 22 round-2-selected sequences. (Bottom) Sequences of pooled N5GN2-randomized and round-2-selected DNA fragments. Probabilities in A and B are from χ2 analysis (Statistix v. 2.1).
To define further the putative sequence element, we performed a second round of binding site selection (“round-2”), in this round, using a library of DNA fragments having the TATA element of the adenovirus major late promoter preceded by the sequence 5′-N5GN2-3′, in which the nucleotides corresponding to positions −39 to −35 and −33 to −32 were randomized and the nucleotide corresponding to position −34 was constrained as G (1.6 × 104 DNA fragments). After seven cycles of selection and amplification, sequences of the pooled round-2-selected DNA fragments revealed strong nucleotide preferences at the seventh, sixth, fifth, fourth, second, and first positions upstream of the TATA element (Fig. 2B, bottom). Sequences of 22 cloned round-2-selected DNA fragments confirmed strong, statistically significant, nucleotide preferences at these positions (Fig. 2B, right). Taken together, the results of the first and second rounds of binding site selection yield the consensus sequence 5′-G/C-G/C-G/A-C-G-C-C-3′ (Fig. 2B, left).
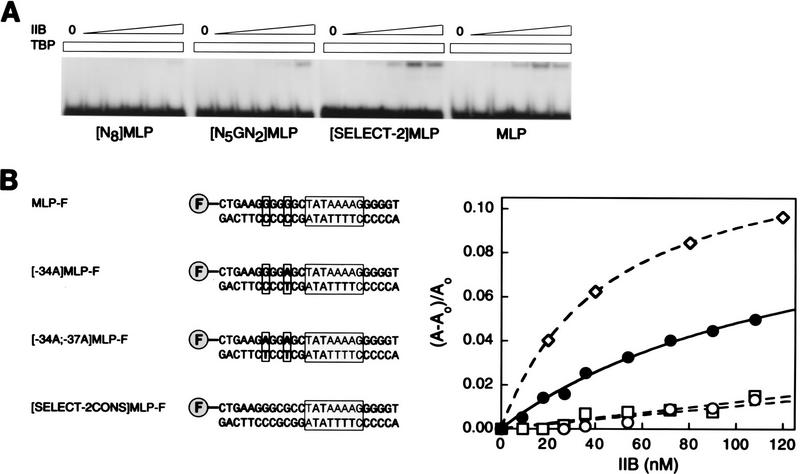
Results of electrophoretic mobility shift DNA-binding experiments indicate that the pooled round-2-selected DNA fragments exhibit an apparent affinity for IIB in IIB–TBP–DNA complex formation that is ∼30-fold higher than that of unselected N8-randomized DNA fragments, ∼8-fold higher than that of unselected N5GN2-randomized DNA fragments, and slightly higher than that of the wild-type adenovirus major late promoter (Fig. 3A). The electrophoretic mobility shift results confirm that the identity of the nucleotide corresponding to position −34 of the adenovirus major late promoter affects affinity for IIB (Fig. 3A, panel 1 vs. panel 2), confirm that the identities of the nucleotides corresponding to positions −39 to −35 and −33 to −32 of the adenovirus major late promoter affect affinity for IIB (Fig. 3A, panel 2 vs. panel 3), and indicate that the wild-type adenovirus major late promoter, which contains a 5-of-7 match to the consensus sequence (Fig. 2), has high, but not optimal, affinity for IIB (Fig. 3A, panel 3 vs. panel 4).
Figure 3.
New sequence element is important for TBP–IIB–DNA complex formation. (A) Electrophoretic mobility shift analysis of IIB–TBP–DNA complex formation. Experiments were performed with DNA fragments containing N8-randomized, N5GN2-randomized, pooled round-2-selected, and wild-type adenovirus major late promoter sequences. TBP was at 30 nm; IIB was at 0, 0.0625, 0.25, 1, 4, and 16 nm. The upper and lower bands correspond to the IIB–TBP–DNA complex and free DNA, respectively; under the conditions used, the TBP–DNA complex is not stable to electrophoresis (see Maldonado et al. 1990). (B) Fluorescence anisotropy analysis of IIB–TBPc–DNA complex formation. (Left) DNA fragments used. The DNA fragments contain fluorescein and wild-type, −34A, −37A;−34A, and consensus round-2-selected adenovirus major late promoter sequences. Fluorescein is indicated as “F”; positions −37 and −34 are boxed; the TATA element is boxed and shaded. (Right) Data. (•) DNA fragment MLP-F; (○) DNA fragment [−34A]MLP-F; (□) DNA fragment [−37A;−34A]MLP-F; (⋄) DNA fragment [SELECT-2CONS]MLP-F.
To confirm the electrophoretic mobility shift DNA-binding results in a true equilibrium assay—an assay not requiring separation of bound and free components—we performed fluorescence anisotropy DNA-binding experiments (for review, see Heyduk et al. 1996; Lundblad et al. 1996). Fluorescence anisotropy DNA-binding experiments are based on the fact that successive additions of proteins P1 and P2 to a fluorochrome-labeled DNA fragment normally result in successive decreases in molecular rotation of the DNA fragment, and therefore normally result in successive increases in fluorescence anisotropy. We started with preformed binary complexes of TBPc and a fluorochrome-labeled DNA fragment (Heyduk et al. 1996), titrated with increasing concentrations of IIB, and quantified IIB–TBPc–DNA complex formation by measurement of increases in fluorescence anisotropy. We constructed and analyzed four fluorochrome-labeled DNA fragments: one DNA fragment containing the consensus round-2-selected sequence, 5′-GGGCGCC-3′, and three DNA fragments containing the wild-type, −34A, and −37A;−34A adenovirus major late promoter sequences (Fig. 3B, left). The results establish that the consensus round-2-selected sequence has an affinity for IIB in IIB–TBPc–DNA complex formation threefold higher than that of the wild-type adenovirus major late promoter (KB = 2.2 ± 0.1 × 107/m vs. KB = 7.0 ± 1 × 106/m; Fig. 3B). The results further establish that the −34A single substitution and the −37A;−34A double substitution result in six- to sevenfold defects in affinity (KB = 9.5 ± 1 × 105/m and 1.1 ± 0.1 × 106/m; Fig. 3B). We conclude that the identities of the seven nucleotides immediately upstream of the TATA element affect affinity for IIB in IIB–TBP–DNA complex formation, and that G at the position corresponding to position −34 of the adenovirus major late promoter is an especially critical determinant.
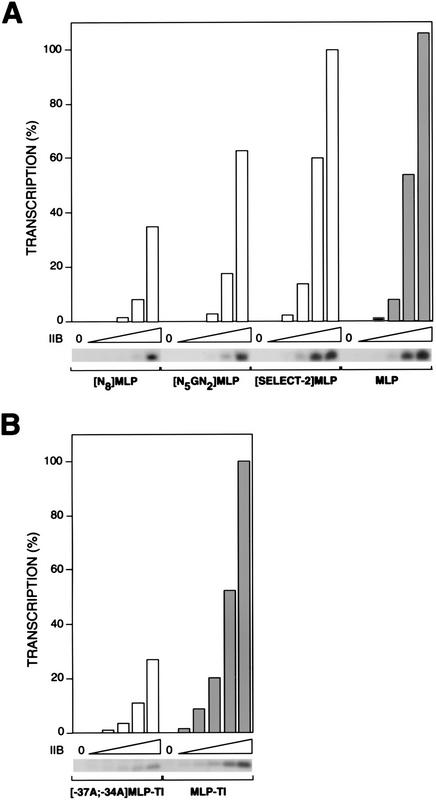
To determine whether the sequence element functions in transcription, we performed abortive initiation in vitro transcription experiments with adenovirus major late promoter derivatives containing the round-2-selected, N8-randomized, N5GN2-randomized, and wild-type sequences, using fixed concentrations of promoter DNA, TBP, IIE, IIF, IIH, and Pol II, and variable concentrations of IIB. The results establish that the IIB concentration dependence for transcription initiation at the promoter containing the round-2-selected sequence is about one-eighth that of the promoter containing the N8-randomized sequence (Fig. 4A, panels 1 and 3), about one-quarter that of the promoter containing the N5GN2- randomized sequence (Fig. 4A, panels 2 and 3), and slightly less than that of the wild-type adenovirus major late promoter (Fig. 4A, panels 3 and 4; see especially data for lower IIB concentrations). At each tested IIB concentration, the promoter containing the round-2-selected sequence is substantially more proficient in transcription initiation than the promoters containing the N8-randomized and N5GN2-randomized sequences, and, at four of five tested IIB concentrations, the promoter containing the round-2-selected sequences is more proficient in transcription initiation than the wild-type adenovirus major late promoter (Fig. 4A). Results of analogous experiments with adenovirus major late promoter derivatives containing the wild-type and −37A;−34A sequences indicate that the IIB concentration dependence for transcription initiation at the promoter containing the wild-type sequence is about one-third that of at the promoter containing the −37A;−34A double substitution (Fig. 4B).
Figure 4.
New sequence element is important for transcription initiation. (A) Transcription initiation at the N8-randomized, N5GN2-randomized, pooled round-2-selected, and wild-type adenovirus major late promoters. IIB was at 0, 15, 60, 240, 960, and 3800 pm. (B) Transcription initiation at −37A;−34A and wild-type adenovirus major late promoter derivatives. IIB was at 0, 20, 40, 80, 160, and 320 pm.
We conclude that the seven nucleotide pairs upstream of the TATA element constitute a bona fide core promoter element—i.e., a sequence element affecting basal preinitiation complex assembly and basal transcription initiation.
IIB recognizes the new core promoter element
The simplest interpretation of the results in Figures 2, 3, 4 is that IIB recognizes the new promoter element directly. However, because the experiments in Figures 2, 3, 4 were performed in the presence of TBP, and because IIB interacts with TBP (Nikolov et al. 1995; Tang et al. 1996), an alternative possible interpretation is that IIB recognizes the element indirectly (e.g., recognizing an element-dependent TBP conformation).
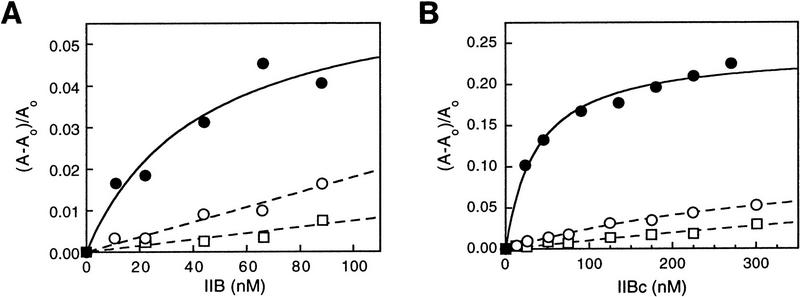
To determine whether IIB recognizes the new promoter element directly, we performed fluorescence anisotropy DNA-binding experiments (Heyduk et al. 1996; Lundblad et al. 1996) investigating whether IIB is able to bind to DNA in the absence of TBP and, if so, whether binding is affected by the element. The results in Figure 5A establish that IIB interacts with a fluorochrome-labeled DNA fragment containing the wild-type adenovirus major late promoter sequence to form a IIB–DNA complex with an equilibrium binding constant of 2.3 ± 1 × 107/m. The interaction is sequence specific; thus, the interaction is observable in the presence of a 400-fold mass excess of competitor DNA (see Materials and Methods). The interaction requires the new promoter element; thus, affinity is reduced ∼7-fold by the −34A substitution within the adenovirus major late promoter sequence and ∼20-fold by the −37A;−34A double substitution within the adenovirus major late promoter sequence (KB = 3.4 ± 0.3 × 106/m and 1.3 ± 0.4 × 106/m; Fig. 5A). Essentially identical results are obtained using IIB (Fig. 5A) and IIBc (Fig. 5B), indicating that the determinants of IIB for the interaction are contained within IIBc. We conclude that IIB is a sequence-specific DNA-binding protein and is directly responsible for recognition of the new promoter element.
Figure 5.
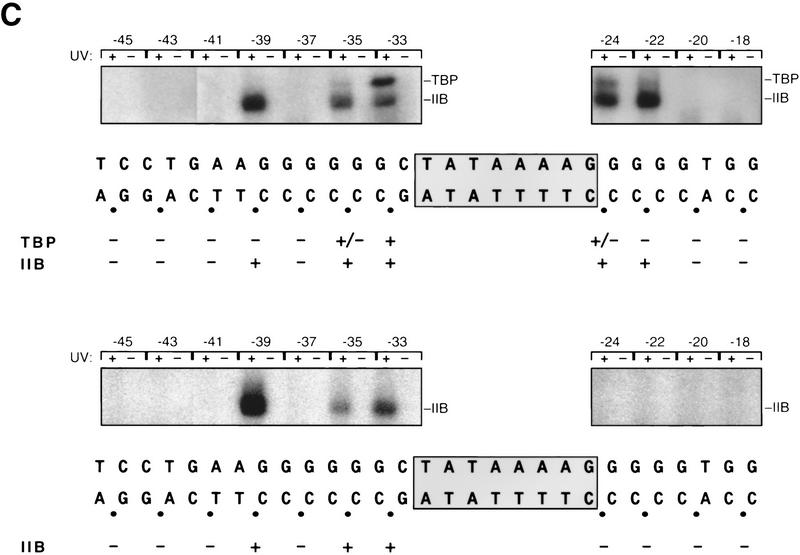
IIB recognizes the new sequence element. (A) Fluorescence anisotropy analysis of IIB–DNA complex formation. (•) DNA fragment MLP-F; (○) DNA fragment [−34A]MLP-F; (□) DNA fragment [−37A; −34A]MLP-F (sequences in Fig. 3B). (B) Fluorescence anisotropy analysis of IIBc–DNA complex formation (symbols as in A). (C) Site-specific protein–DNA photo-cross-linking analysis of IIB–TBP–DNA complex formation (top) and IIB–DNA complex formation (bottom). The TATA element is boxed and shaded. (•) Phosphates at which a phenyl-azide photoactivatible cross-linking agent was incorporated; (+, ±) sites at which strong and weak cross-linking, respectively, were observed. The autoradiographs present results of reactions with UV irradiation and control reactions without UV irradiation.
To confirm the ability of IIB to form IIB–DNA complexes, and to define the position of IIB relative to DNA in IIB–DNA complexes, we performed site-specific protein–DNA photo-cross-linking (Lagrange et al. 1996). We constructed a set of 11 site specifically derivatized adenovirus major late promoter DNA fragments, each having a phenyl-azide photoactivatible cross-linking agent and an adjacent radiolabel incorporated at a single, defined DNA phosphate (template strand phosphates −45, −43, −41, −39, −37, −35, and −33 upstream of the TATA element, and template strand phosphates −24, −22, −20, and −18 downstream of the TATA element; Fig. 5C). For each resulting DNA fragment, we equilibrated the DNA fragment with IIB and TBP or with IIB alone, UV-irradiated the reaction mixtures to initiate DNA → protein cross-linking, carried out nuclease digestion to eliminate uncross-linked DNA and to convert cross-linked DNA to a cross-linked, radiolabeled 3–5 nucleotide “tag,” and identified the “tagged” proteins.
Upstream of the TATA element—in the region corresponding to the new promoter element—the patterns of DNA → IIB cross-linking were identical in the reactions containing IIB and TBP (Fig. 5C, top) and the reactions containing IIB alone (Fig. 5C, bottom). In each case, DNA → IIB cross-linking occurred at template strand positions −39, −35, and −33. We conclude, in agreement with the conclusions from the fluorescence anisotropy DNA-binding experiments, that IIB is able to form IIB–DNA complexes, and we infer that IIB makes similar or identical interactions with DNA upstream of the TATA element in IIB–TBP–DNA and IIB–DNA complexes.
In contrast, downstream of the TATA element, DNA → IIB cross-linking occurred only in reactions containing both IIB and TBP (Fig. 5C). We infer that IIB does not, or does not strongly, interact with DNA immediately downstream of the TATA element in IIB–DNA complexes. We speculate that the difference in downstream interactions in IIB–TBP–DNA and IIB–DNA complexes is a consequence of the presence of the ∼80° TBP-induced DNA bend in the IIB–TBP–DNA complex (J. Kim et al. 1993; Y. Kim et al. 1993; Nikolov et al. 1995; see Fig. 1). The TBP-induced DNA bend in the IIB–TBP–DNA complex places the upstream and downstream DNA segments in proper spatial register for simultaneous IIB–DNA interactions with both DNA segments (Nikolov et al. 1995; see Fig. 1). The absence of a corresponding DNA bend in the IIB–DNA complex would preclude simultaneous IIB–DNA interactions with both DNA segments.
IIB recognizes the new core promoter element through a canonical helix–turn–helix motif
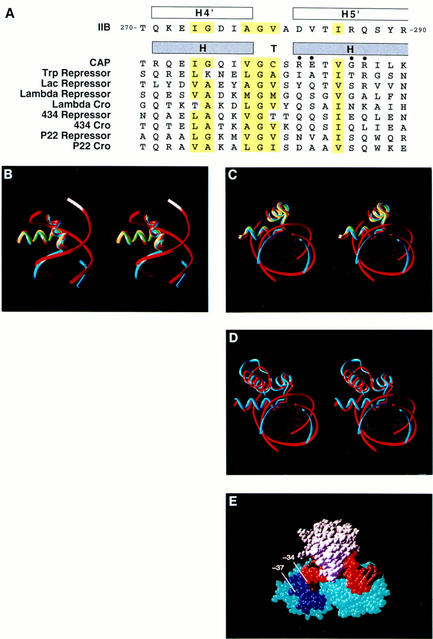
As a first step to define the structural basis for recognition of the new promoter element, we inspected the crystallographic structure of the IIBc–TBPc–TATA complex in the vicinity of the nucleotides corresponding to positions −34 to −32 of the adenovirus major late promoter and in the projected vicinity of the nucleotides corresponding to positions −38 to −35 of the adenovirus major late promoter (Figs. 1 and 6E). Inspection indicated that the interaction between IIB and the DNA major groove immediately upstream of the TATA element exhibits unambiguous, but previously unrecognized, structural similarity to the interaction between a helix–turn–helix (HTH) DNA-binding protein and DNA (for review of HTH, see Pabo and Sauer 1992) (Figs. 1 and 6). The amino acid sequence of helices 4′ and 5′ of IIBc matches the sequence profile for HTHs with a Dodd and Egan (1990) score of 4.6 SD (Fig. 6A), and the backbone atoms of helices 4′ and 5′ of IIBc superimpose on the backbone atoms of the HTHs of CAP, λ repressor, and Trp repressor with root-mean-square differences of 0.8, 0.8, and 0.9 Å, respectively (Fig. 6B,C). The orientation of helices 4′ and 5′ of IIBc relative to DNA (modeling DNA upstream of the TATA element as B-DNA) falls within the range of orientations observed with canonical HTHs and, in particular, is similar to the orientations observed with the HTHs of λ repressor and Trp repressor (Fig. 6B,C).
Figure 6.
IIB recognizes the new sequence element through a helix–turn–helix motif: structural evidence. (A) Amino acid sequences of helices 4′ and 5′ of IIB (residues 270–290 of IIB; Bagby et al. 1995; Nikolov et al. 1995) and known HTHs (residues −1–20 of each HTH; Pabo and Sauer 1992). Amino acids conserved among known HTHs are highlighted in yellow; positions of amino acids that contact DNA base pairs in known structures of HTH–DNA complex structures are marked with filled circles. (B,C) Superimposition of helices 4′ and 5′ of IIB (blue; Nikolov et al. 1995; coordinates kindly supplied by S. Burley, Rockefeller University, New York, NY) on the HTHs of CAP (yellow; Parkinson et al. 1996), λ repressor (green; Beamer and Pabo 1992; coordinates obtained from Brookhaven Protein Data Bank, accession code 1LMB), and Trp repressor (red; Otwinowski et al. 1988; coordinates obtained from Brookhaven Protein Data Bank, accession code 1TRO) (two orthogonal views). The 3 nucleotide pairs upstream of the TATA element present in the crystallographic structure of the IIBc–TBPc–TATA complex are shown in ribbon representation and colored blue; these nucleotide pairs correspond to positions −34 to −32 of the adenovirus major late promoter (position −32 at bottom in B). Nine nucleotide pairs in the Trp repressor–DNA complex are shown in ribbon representation and colored red. (D) Superimposition of helices 3′–5′ of IIB (blue) on helices C–E of Trp repressor (red). (E) Structure of the IIBc–TBPc–TATA complex (Nikolov et al. 1995; Fig. 1) showing helices 4′ and 5′ of IIB (dark blue), the nucleotide pair corresponding to position −34 of the adenovirus major late promoter (arrow), and the projected location of the nucleotide pair corresponding to position −37 of the adenovirus major late promoter (arrow). IIBc is blue, TBP is white, the DNA nontemplate strand is bright red, and the DNA template strand is dark red.
The structural similarity of IIBc–DNA interaction to Trp repressor–DNA interaction is especially striking. Three consecutive helices of IIBc (helices 3′–5′) and Trp repressor (helices C–E) can be superimposed with a root-mean-square difference of 1.6 Å, and the bound DNA segments simultaneously can be superimposed (Fig. 6D).
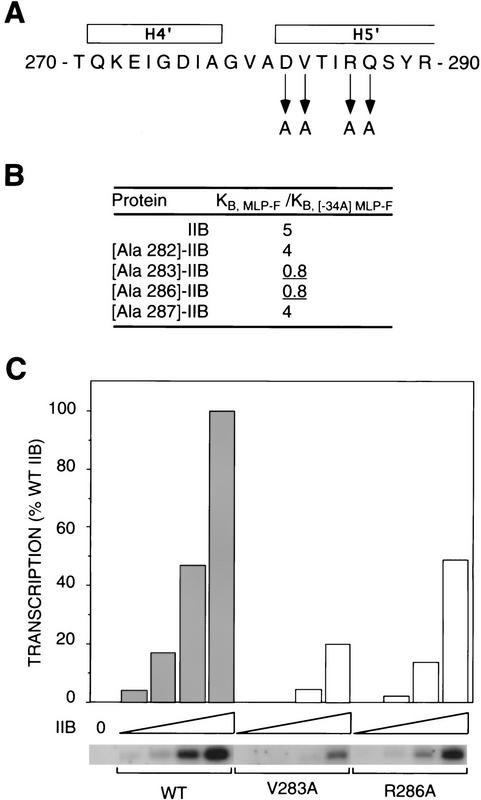
To test the functional significance of these structural similarities, we have constructed alanine substitutions at the amino acids of IIB corresponding to the amino acids that in HTH DNA-binding proteins make specificity-determining contacts with DNA base pairs (i.e., Asp-282, Val-283, Arg-286, and Gln-287 of IIBc) (Figs. 6A and 7A), and we have assessed the effects on specificity at position −34 of the adenovirus major late promoter (Fig. 7B). We find that the Val-283 → Ala and Arg-286 → Ala substitutions eliminate specificity at position −34; thus, whereas wild-type IIB exhibits a fivefold preference for G:C versus A:T at position −34 (Figs. 2, 3, and 7B), the substituted IIB derivatives exhibit no preference for G:C—or even a small preference for A:T (Fig. 7B). In addition, the Val-283 → Ala and Arg-286 → Ala substitutions result in two- to threefold decreased affinity for IIB–DNA complex formation with the wild-type adenovirus major late promoter sequence (data not shown) and two- to threefold decreased effectiveness in supporting transcription initiation at the wild-type adenovirus major late promoter (Fig. 7C). The Val-283 → Ala and Arg-286 → Ala substitutions exhibit the two defining characteristics of substitutions that permit identification of amino acids that make specificity-determining contacts with DNA base pairs: (1) elimination of specificity between consensus and nonconsensus base pairs at one position within the DNA site, and (2) reduction in affinity for the consensus DNA site (“loss-of-contact” substitutions; for review, see Ebright 1991). We conclude that Val-283 and Arg-286 of IIB functionally determine specificity for G:C versus A:T at position −34, and we propose that Val-283 and Arg-286 make direct contact with G:C at position −34.
Figure 7.
IIB recognizes the new sequence element through a HTH motif: functional evidence. (A) Substitutions of amino acids of IIB that correspond to base-pair-contacting amino acids of HTH DNA-binding proteins (see Fig. 6A). (B) Effects of substitutions on specificity between G:C and A:T at position −34 of the adenovirus major late promoter. Data are from fluorescence anisotropy assays of IIB–TBPc–DNA complex formation. (C) Effects on transcription initiation at the adenovirus major late promoter. Data are from run-off transcription assays. IIB and IIB derivatives were at 1, 2, 4, and 8 nm.
Details of the proposed interactions by Val-283 and Arg-286 of IIB must await determination of a crystallographic structure of a IIB–TBP–TATA complex with a DNA fragment sufficiently long, and having the correct sequence, to encompass the proposed interactions. However, model building suggests that Val-283 may be positioned to make van der Waals interactions with the cytosine C5 atom of G:C at position −34 and that Arg-286 may be positioned to interact with the guanine O6 and N7 atoms of G:C at position −34 and/or with the DNA–phosphate backbone at position −34.
Discussion
Our results establish the existence of a fourth core promoter element, the “IIB recognition element” (BRE), in addition to the TATA element, the initiator element, and the downstream promoter element, and a second sequence-specific general transcription factor, IIB, in addition to IID. Our results further establish that the degree of similarity between a promoter BRE and the consensus BRE determines the IIB concentration dependences for preinitiation complex assembly and transcription initiation at the promoter—at least in vitro. Determination of whether similarity between a promoter BRE and the consensus BRE has similar effects in vivo awaits further analysis. Nevertheless, on the basis of our results, it is possible that IIB–BRE interaction will play a role in determining the overall strength of a promoter, the order of preinitiation complex assembly at a promoter (e.g., initial binding by TBP, initial binding by IIB, or concerted binding by TBP and IIB), the rate-limiting step in transcription initiation at a promoter, and—given the observation that several transcriptional activators interact with IIB (Roberts et al. 1995 and references therein)—the responsiveness of a promoter to specific transcriptional activators. It also is possible that IIB–BRE interaction will play a role in determining the upstream → downstream directionality of preinitiation complex assembly and transcription initiation, supplementing the TBP–TATA interaction, which, because of its high degree of twofold symmetry (J. Kim et al. 1993; Y. Kim et al. 1993), is insufficient to determine directionality (Xu et al. 1991; J. Cox, M. Hayward, J. Sanchez, L. Gegnas, S. van der Zee, J. Dennis, P. Sigler, and A. Schepartz, in prep.). Finally, it is possible that IIB–BRE interaction will play a role, possibly a dominant role, in preinitiation complex assembly and transcription initiation at TATA-less promoters (see Weis and Reinberg 1997). The effects of the upstream element (UP element) in prokaryotic transcription provide an informative precedent for effects of a supplementary promoter element on multiple aspects of transcription initiation and transcriptional activation (for review, see Busby and Ebright 1994).
A search of the Eukaryotic Promoter Database (EPD) (Bucher 1996) reveals that, of Pol II-dependent vertebrate promoters that have a consensus TATA element at a consensus position (T1A2T3A4 with T1 at position −32, −31, −30, −29, or −28 relative to the principal transcription start; 315 promoters), 12% have a 5-of-7 or better match to the consensus BRE immediately upstream of the TATA element, 34% have a 4-of-7 or better match to the consensus BRE immediately upstream of the TATA element, and 40% have the consensus nucleotide pair G:C 3 nucleotide pairs upstream of the TATA element. Of Pol II-dependent vertebrate promoters that do not contain a consensus TATA element at a consensus position (550 promoters), 15% have a 6-of-7 or better match to the consensus BRE between positions −41 and −27, and 62% have a 5-of-7 or better match to the consensus BRE between positions −41 and −27. All of these frequencies are significantly higher than those expected by chance (0.94%, 1.7%, and 25%; 2.9% and 8.5%, respectively), consistent with the hypothesis that BRE is functionally significant in vivo. The observation that a substantial minority, rather than a majority, of Pol II-dependent promoters contains a consensus or near-consensus BRE accounts for the fact that the BRE was not identified through global sequence alignments of Pol II-dependent promoters.
The amino acid sequence of helices 4′ and 5′ of IIB—the region shown here to fold as an HTH (Fig. 6)—is conserved in IIB from all eukaryotes for which sequence data are available and in transcription factor B (TFB) from all archaea for which sequence data are available (9 IIB and 5 TFB sequences in SWISSPROT), suggesting that this region folds as an HTH in all cases. Val-283 and Arg-286 of IIB—the amino acids shown here to determine specificity at the G:C nucleotide pair corresponding to position −34 of the adenovirus major late promoter (Fig. 7)—are invariant in IIB from all eukaryotes, except plants and fungi, and in TFB from all archaea, suggesting that IIB from all eukaryotes, possibly excluding plants and fungi, and TFB from all archaea may have related DNA-binding specificities. Consistent with these suggestions, helices 4′ and 5′ of TFB from the archaeon Pyrococcus woesi fold as a canonical HTH (Kosa et al. 1997; coordinates obtained from Brookhaven Protein Data Bank, accession code 1AIS), and results obtained while this manuscript was in preparation suggest that TFB from the archaeon Sulfolobus shibatae recognizes a promoter element located immediately upstream of the TATA element and containing a critical G:C nucleotide pair (S. Qureshi and S. Jackson, pers. comm.).
The starting point for this work was the observation that IIB makes extensive interactions with the DNA major groove (Lee and Hahn 1995; Nikolov et al. 1995; Lagrange et al. 1996). Site-specific protein–DNA photocross-linking indicates that at least three other components of the human Pol II-dependent preinitiation complex make extensive interactions with the DNA major groove (i.e., IIA, IIF, and Pol II) (Lagrange et al. 1996; Kim et al. 1997). It will be important to investigate whether there exist promoter elements recognized by IIA, IIF, and Pol II. (Preliminary binding site selection results support the existence of a promoter element recognized by IIA; H. Tang, D. Reinberg, and R.H. Ebright, unpubl.). The possible existence of additional promoter elements is attractive in that it would enable the “encoding” in promoter DNA of a wide range of promoter strengths and promoter characteristics, including responsiveness to specific transcriptional activators and repressors (see also Shen and Green 1997). If documented, the existence of additional promoter elements would require revision of the current view of the promoter DNA as an information-poor scaffold for preinitiation complex assembly to a new view of promoter DNA as an information-rich participant in, and director of, preinitiation complex assembly.
Materials and methods
IIB derivatives
Plasmids encoding amino-terminally hexahistidine-tagged human IIB (20 nonnative amino acids followed by amino acids 1–316 of IIB; pNHisIIB) or amino-terminally hexahistidine-tagged human IIBc (23 nonnative amino acids followed by amino acids 112–316 of IIB; pNHisIIBc) under control of the bacteriophage T7 gene 10 promoter were constructed by replacement of the NheI–BamHI segment of pET28a(+) (Novagen) by the NheI–BamHI segments of phIIB and phIIBΔ4-111 (Ha et al. 1993), respectively. Substitutions were introduced into pNHisIIB by the method of Kunkel et al. (1991). Cultures of Escherichia coli strain BL21(DE3) (Novagen) transformed with pNHisIIB, pNHisIIB derivative, or pNHisIIBc were shaken at 37°C in 1 liter of LB (Miller 1972) plus 40 μg/ml of kanamycin until OD600 = 0.6, induced by addition of IPTG to 1 mm, and shaken for an additional 3 hr at 37°C. Cultures were harvested by centrifugation (4000g; 15 min at 4°C), cell pellets were resuspended in 20 ml of buffer A [20 mm Tris-HCl (pH 7.9), 500 mm NaCl, 5 mm imidazole, 0.1% NP-40], cells were lysed by sonication, and lysates were cleared by centrifugation (30,000g; 30 min at 4°C). Samples were adsorbed onto a 1.5-ml column of Ni2+–NTA agarose (Qiagen) in buffer A at 4°C, washed with 25 ml of buffer A, washed with 50 ml of buffer A plus 40 mm imidazole, eluted with 16 ml of buffer A plus 300 mm imidazole, desalted into or dialyzed against 10 mm Tris-HCl (pH 7.9), 500 mm KCl, 0.2 mm EDTA, 0.2 mm PMSF, and 20% glycerol, and stored in aliquots at −80°C. The yield was 5–10 mg and the purity was >98%.
TBP, IIE, IIF, IIH, RNA Pol II
Recombinant human TBP, recombinant human TBPc, recombinant human IIE, recombinant human IIF, human IIH (Mono-S and 0.5 m phenyl–Superose fractions), and human Pol II (DEAE–5PW fraction) were prepared as described (Lu et al. 1991; Peterson et al. 1991; Flores et al. 1992; Ha et al. 1993; Wang et al. 1993; Heyduk et al. 1996; Tang et al. 1996).
Binding site selection, round 1
Reaction mixtures for round 1, cycle 1, contained (20 μl): 1 nm 32P-end-labeled DNA fragment [N12]MLP [10 Bq/fmole; positions −43 to −12 of the adenovirus major late promoter, with −43 to −32 randomized, flanked by 16 bp of nonnative sequence upstream and 15 bp of nonnative sequence downstream; prepared by annealing of 5′-AGACGGATCCATTCGANNNNNNNNNNNNTATAAAAGGGGGTGGGGGCGCTGTAGGAATTCGGA-3′ and primer A (5′-TCCGAATTCCTACAG-3′) followed by extension with Sequenase (Amersham) and [α-32P]dNTPs], 10 nm TBPc, 0.5–20 nm IIB, 20 mm Tris-HCl (pH 7.9), 20 mm HEPES–NaOH (pH 7.9), 60 mm KCl, 10 mm MgCl2, 8 mm (NH4)2SO4, 0.5 mm DTT, 0.05 mm EDTA, 0.1 mm PMSF, 25 μg/ml poly[d(G-C)]:poly[d(G-C)] (average molecular mass, 800 kD; Pharmacia), 25 μg/ml of PEG (average molecular mass, 8 kD), and 5% glycerol. After reaction for 30 min at 30°C, reaction products were separated by nondenaturing electrophoresis as in Tang et al. (1996), and gels were dried, autoradiographed, and phosphorimaged. The reaction containing 2 nm IIB resulted in ∼2% saturation of IIB–TBPc–DNA complex formation. For this reaction, the band corresponding to the IIB–TBPc–DNA complex was excised, and the DNA in the band was eluted by boiling 10 min in 100 μl of water, extracted with phenol/chloroform/isoamyl alcohol (25:24:1) after addition of 5 μg of yeast tRNA as carrier, ethanol precipitated, amplified by PCR with primer A, primer B (5′-AGACGGATCCATTCGA-3′), Taq DNA polymerase, and [α-32P]dNTPs, and purified by 12% PAGE. The selected, amplified DNA was subjected to three additional cycles of selection and amplification, in each cycle selecting DNA from reactions with IIB concentrations that resulted in 2%–4% saturation of IIB–TBPc–DNA complex formation (0.5, 0.5, and 0.1 nm IIB in cycles 2, 3, and 4, respectively). Selected, amplified DNA fragments from cycle 4 were treated with EcoRI and BamHI, ligated into EcoRI/BamHI-digested plasmid pBluescript II SK(-) (Stratagene), cloned by transformation into strain XL1-Blue (Stratagene), and sequenced using the T7 promoter primer (Stratagene).
Binding site selection, round 2
Reaction mixtures for round 2, cycle 1, contained (20 μl): 50 pm 32P-end-labeled DNA fragment [N5GN2]MLP56 [150 Bq/fmole; positions −53 to +3 of the adenovirus major late promoter, with −39 to −35, −33, and −32 randomized; prepared by annealing of 5′-GATCGAATTCCGGAGCCGGGTGTTCCTGANNNNNGNNTATAAAAGGGGGTGGGGGCG-3′ and 5′-GATGGGATCCATTAGAGTGAGGACGAACGCGCCCCCACCCCCTTTTA-3′, extension with T4 DNA polymerase and dNTPs, PCR with primer C (5′-GATCGAATTCCGGAG-3′), primer D (5′-GATGGGATCCATTAG-3′), Taq DNA polymerase, and dNTPs, and radiophosphorylation with T4 polynucleotide kinase and [γ-32P]ATP], 10 nm TBP, 0.5–80 nm IIB, 5 mm Tris-HCl (pH 8.0), 60 mm KCl, 5 mm MgCl2, 1 mm DTT, 25 μg/ml poly[d(G-C)]:poly[d(G-C)] (average molecular mass, 800 kD; Pharmacia), 100 μg/ml of BSA, 0.1% Brij-58, and 4% glycerol. Reactions were performed, and DNA was isolated and amplified, essentially as in binding site selection, round 1, except that electrophoresis was performed at 4°C in gels containing 1 mm Mg(OAc)2 and not containing glycerol. Seven cycles of selection and amplification were performed, in the first cycle selecting DNA from reactions with IIB concentrations that resulted in ∼10% saturation of IIB–TBP–DNA complex formation, and in subsequent cycles selecting DNA from reactions with IIB concentrations that resulted in 2%–4% saturation (7, 1, 0.2, 0.06, 0.01, 0.02, and 0.05 nm IIB). After each cycle, DNA fragment pools were sequenced directly using primer C (Blackwell and Weintraub 1990). After the seventh cycle, DNA fragments were digested with EcoRI and BamHI, ligated into EcoRI/BamHI-digested plasmid pBluescript II KS(−) (Stratagene), cloned by transformation into strain XL1-Blue (Stratagene), and sequenced using primer C.
Electrophoretic mobility shift DNA-binding experiments
Electrophoretic mobility shift DNA-binding experiments were performed as in Maldonado et al. (1990), using 0.1 nm 32P-labeled DNA fragment [N12]MLP or [SELECT-1]MLP (10 Bq/fmole; positions −43 to −12 of the N12-randomized or pooled round-1-selected adenovirus major late promoter, flanked by 16 bp of nonnative sequence upstream and 15 bp of nonnative sequence downstream; see above) or 0.1 nm 32P-end-labeled DNA fragment MLP56, [N8]MLP56, [N5GN2]MLP56, or [SELECT-2]MLP56 (500 Bq/fmole; positions −53 to +3 of the wild-type, N8-randomized, N5GN2-randomized, or round-2-selected adenovirus major late promoter; prepared by PCR of the corresponding synthetic DNA fragments or pooled round-2-selected DNA fragments with 32P-end-labeled primer C and unlabeled primer D), 30 nm TBP, and 0.05–20 nm IIB.
Fluorescence anisotropy DNA binding experiments
Assay mixtures contained (1.2 ml in 12 × 75 mm borosilicate glass disposable culture tubes): 0.3–1 nm fluorescein-end-labeled DNA fragment MLP-F, [SELECT-2CONS]MLP-F, [−34A]MLP-F, or [−37A;−34A]MLP-F (positions −43 to −19 of the wild-type, consensus round-2-selected, −34A, or −37A;−34A adenovirus major late promoter; prepared as described for MLP24FL in Heyduk et al. 1996), 0 or 80 nm TBPc, 20 mm Tris-HCl (pH 8.0), 20 mm HEPES–NaOH (pH 8.0), 60 mm KCl, 10 mm MgCl2, 8 mm (NH4)2SO4, 0.05 mm EDTA, 0.5 mm DTT, 0.1 mm PMSF, 7 μg/ml of poly[d(G-C)]:poly[d(G-C)] (average molecular mass, 800 kD; Pharmacia), 25 μg/ml PEG (average molecular mass, 8 kD), 100 μg/ml of BSA, 0.1% NP-40, and 5% glycerol. Assay mixtures were incubated 25 min at 28°C (25°C for IIB–DNA and IIBc–DNA complex formation) and then titrated by successive addition of 0.5–2 μl aliquots of IIB or IIBc in the same buffer at 28°C (25°C for IIB–DNA and IIBc–DNA complex formation). Fluorescence anisotropy was determined using a PanVera Beacon fluorescence polarization instrument at the start of the titration and 4 min after each successive addition in the titration. Equilibrium-binding constants were calculated using nonlinear regression (Gunasekera et al. 1992).
Protein–DNA photo-cross-linking
Protein–DNA photo-cross-linking was performed as in Lagrange et al. (1996), using 0 or 6 nm TBP and 20 nm IIB or IIBc.
Transcription experiments
Reaction mixtures for abortive initiation experiments contained (20 μl): 1.0 nm DNA fragment MLP112, [N8]MLP112, [N5GN2]MLP112, [SELECT-2]MLP112 [positions −68 to +44 of the wild-type, N8-randomized, N5GN2-randomized, or round-2-selected adenovirus major late promoter; prepared by add-on PCR of the corresponding synthetic DNA fragments or pooled round-2-selected DNA fragments; concentration determined flurometrically using PicoGreen reagent (Molecular Probes)] or 1.0 nm DNA fragment MLP-TI or [−37A;−34A]MLP-TI (positions −45 to +9 of the wild-type or −37A;−34A adenovirus major late promoter flanked by nonnative sequence and with positions −23 to −6 replaced by nonnative sequence; prepared from corresponding plasmid clones), 20 nm TBP, 0.02–4 nm IIB, 20 nm IIF, 30 nm IIE, 10 nm IIH (Mono-S fraction), 20 nm RNA Pol II (DEAE-5PW fraction), 1 mm [α-32P]CTP (30 Bq/pmole), 1 mm ATP, 10 mm Tris-HCl (pH 7.8), 10 mm HEPES-KOH (pH 7.8), 50 mm KCl, 4 mm MgCl2, 4 mm (NH4)2SO4, 0.1 mm EDTA, 0.7 mm DTT, 400 U/ml of rRNasin (Promega), and 10% glycerol. Reactions were initiated by addition of [α-32P]CTP and ATP, were allowed to proceed for 30 min at 28°C, and were terminated and analyzed as in Goodrich and Tjian (1994). Reaction mixtures for run-off transcription experiments contained (20 μl): 2 nm plasmid pΔML, which carries the adenovirus major late promoter followed by a 210-nucleotide G-less cassette [derivative of pML(C2AT)Δ-50 (Sawadogo and Roeder 1985) with shortened G-less cassette], 20 mm TBP, 1–8 nm IIB or derivative, 20 nm IIF, 30 nm IIE, 50 nm IIH (0.5 m phenyl–Superose fraction), 50 nm Pol II (DEAE-5PW fraction), 0.15 mm [α-32P]UTP, 0.6 mm ATP, 0.6 mm CTP, 1000 U/ml of RNase T1, 20 mm HEPES–NaOH (pH 7.9), 50 mm KCl, 10 mm (NH4)2SO4, 10 mm β-mercaptoethanol, 20 mg/ml of PEG (average molecular mass, 8 kD), and 10% glycerol. Reactions were initiated by addition of [α-32P]UTP, ATP, and CTP, were allowed to proceed for 20 min at 28°C, and were terminated by addition of 80 μl of 100 mm sodium acetate (pH 5.5), 10 mm EDTA, 1 mg/ml of yeast tRNA, and 0.2% SDS. Reaction products were phenol/chloroform extracted, ethanol precipitated, and analyzed by 8 m urea–15% PAGE followed by phosphorimaging and autoradiography.
Acknowledgments
We thank S. Burley for atomic coordinates and L. Weis for database searches. This work was supported by National Institutes of Health (NIH) grant GM37120 and a Howard Hughes Medical Institute Investigatorship to D.R. and by NIH grant GM53665 to R.H.E. A.N.K. was supported in part by the State Scholarship Foundation, Greece, and the Hellenic Medical Society of New York.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL ebright@mbc1.rutgers.edu; FAX (732) 445-5735.
References
- Bagby S, Kim S, Maldonado E, Tonk K, Reinberg D, Ikura M. Solution structure of the carboxy-terminal core domain of human TFIIB: Similarity to cyclin A and interaction with TATA-binding protein. Cell. 1995;82:857–867. doi: 10.1016/0092-8674(95)90483-2. [DOI] [PubMed] [Google Scholar]
- Beamer L, Pabo C. Refined 1.8 Å crystal structure of the lambda repressor-operator complex. J Mol Biol. 1992;227:177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- Blackwell T, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Blackwell T, Kretzner L, Blackwood E, Eisenman R, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- Bucher P. The Eukaryotic Promoter Database EPD, EMBL, nucleotide sequence data library, release 48. Cambridge, UK: European Bioinformatics Institute; 1996. [Google Scholar]
- Burke T, Kadonaga J. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright R. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Corden J, Wasylyk B, Buchwalder A, Sassone-Corsi P, Kedinger C, Chambon P. Promoter sequences of eukaryotic protein-encoding genes. Science. 1980;209:1406–1414. doi: 10.1126/science.6251548. [DOI] [PubMed] [Google Scholar]
- Dodd B, Egan J. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. Identification of amino acid-base pair contacts by genetic methods. Methods Enzymol. 1991;208:620–640. doi: 10.1016/0076-6879(91)08032-d. [DOI] [PubMed] [Google Scholar]
- Flores O, Lu H, Reinberg D. Factors involved in specific transcription initiation by mammalian RNA polymerase II: Identification and characterization of factor IIH. J Biol Chem. 1992;267:2786–2793. [PubMed] [Google Scholar]
- Goodrich J, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Gunasekera A, Ebright Y, Ebright R. DNA-sequence determinants for binding of the Escherichia coli catabolite gene activator protein. J Biol Chem. 1992;267:14713–14720. [PubMed] [Google Scholar]
- Ha I, Roberts S, Maldonado E, Sun X, Kim L-U, Green M, Reinberg D. Multiple functional domains of human transcription factor IIB: Distinct interactions with two general transcription factors and RNA polymerase. Genes & Dev. 1993;7:1021–1032. doi: 10.1101/gad.7.6.1021. [DOI] [PubMed] [Google Scholar]
- Heyduk T, Ma Y, Tang H, Ebright R. Fluorescence anisotropy: Rapid, quantitative assay for protein-DNA and protein-protein interaction. Methods Enzymol. 1996;274:492–502. doi: 10.1016/s0076-6879(96)74039-9. [DOI] [PubMed] [Google Scholar]
- Kim J, Nikolov D, Burley S. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y, Geiger J, Hahn S, Sigler P. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Kim T-K, Lagrange T, Wang Y-H, Griffith J, Reinberg D, Ebright R. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosa P, Ghosh G, DeDecker B, Sigler P. The 2.1-Å crystal structure of an archaeal preinitiation complex: TATA-box-binding protein/transcription factor (II)B core/TATA-box. Proc Natl Acad Sci. 1997;94:6042–6047. doi: 10.1073/pnas.94.12.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–138. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- Lagrange T, Kim T-K, Orphanides G, Ebright Y, Ebright R, Reinberg D. High-resolution mapping of nucleoprotein complexes by site-specific protein-DNA photocross-linking: Organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci. 1996;93:10620–10625. doi: 10.1073/pnas.93.20.10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Hahn S. A model for TFIIB binding to the TBP-DNA complex. Nature. 1995;376:609–612. doi: 10.1038/376609a0. [DOI] [PubMed] [Google Scholar]
- Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad J, Laurance M, Goodman R. Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol. 1996;10:607–612. doi: 10.1210/mend.10.6.8776720. [DOI] [PubMed] [Google Scholar]
- Maldonado E, Ha I, Cortes P, Weis L, Reinberg D. Role of transcription factors TFIIA, TFIID, and TFIIB during formation of a transcription-competent complex. Mol Cell Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrothalassitis G, Beal G, Papas T. Defining target sequences of DNA-binding proteins by random selection and PCR: Determination of the GCN4 binding sequence repertoire. DNA. 1990;9:783–788. doi: 10.1089/dna.1990.9.783. [DOI] [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Nikolov D, Burley S. RNA polymerase II transcription initiation: A structural view. Proc Natl Acad Sci. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov D, Chen H, Halay E, Usheva A, Hisatake K, Lee D, Roeder R, Burley S. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Oliphant A, Brandl C, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: Analysis of yeast GCN4 protein. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Schevitz R, Zhang R-G, Lawson C, Joachimiak A, Marmorstein R, Luisi B, Sigler P. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- Pabo C, Sauer R. Transcription factors: Structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Parkinson G, Wilson C, Gunasekera A, Ebright Y, Ebright R, Berman H. Structure of the CAP-DNA complex at 2.5 resolution: A complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- Peterson M, Inostroza J, Maxon M, Flores O, Admon A, Reinberg D, Tjian R. Structure and functional properties of human general transcription factor IIE. Nature. 1991;354:369–373. doi: 10.1038/354369a0. [DOI] [PubMed] [Google Scholar]
- Roberts S, Choy B, Walker S, Lin Y-S, Green M. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- Roeder R. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Sawadogo M, Roeder R. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985;43:165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Shen W, Green M. Yeast TAFII145 functions as a core promoter selectivity factor, not a general coactivator. Cell. 1997;90:615–624. doi: 10.1016/s0092-8674(00)80523-1. [DOI] [PubMed] [Google Scholar]
- Tang H, Sun X, Reinberg D, Ebright R. Protein-protein interactions in eukaryotic transcription initiation: Structure of the pre-initiation complex. Proc Natl Acad Sci. 1996;93:1119–1124. doi: 10.1073/pnas.93.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiesen H-J, Bach C. Target Detection Assay (TDA): A versatile procedure to determine DNA binding sites as demonstrated on SP1 protein. Nucleic Acids Res. 1990;18:3203–3209. doi: 10.1093/nar/18.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BQ, Kostrun C, Finkelstein A, Burton Z. Production of human RAP30 and RAP74 in bacterial cells. Protein Express Purif. 1993;4:207–214. doi: 10.1006/prep.1993.1027. [DOI] [PubMed] [Google Scholar]
- Weis L, Reinberg D. Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding-protein-associated factors and initiator-binding proteins. Mol Cell Biol. 1997;17:2973–2984. doi: 10.1128/mcb.17.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Thali M, Schaffner W. Upstream box/TATA box order is the major determinant of the direction of transcription. Nucleic Acids Res. 1991;19:6699–6704. doi: 10.1093/nar/19.24.6699. [DOI] [PMC free article] [PubMed] [Google Scholar]