Abstract
The Gram-negative bacterium Francisella tularensis is the causative agent of tularemia, a disease intimately associated with the multiplication of the bacterium within host macrophages. This in turn requires the expression of Francisella pathogenicity island (FPI) genes, believed to encode a type VI secretion system. While the exact functions of many of the components have yet to be revealed, some have been found to contribute to the ability of Francisella to cause systemic infection in mice as well as to prevent phagolysosomal fusion and facilitate escape into the host cytosol. Upon reaching this compartment, the bacterium rapidly multiplies, inhibits activation of the inflammasome, and ultimately causes apoptosis of the host cell. In this study, we analyzed the contribution of the FPI-encoded proteins IglG, IglI, and PdpE to the aforementioned processes in F. tularensis LVS. The ΔpdpE mutant behaved similarly to the parental strain in all investigated assays. In contrast, ΔiglG and ΔiglI mutants, although they were efficiently replicating in J774A.1 cells, both exhibited delayed phagosomal escape, conferred a delayed activation of the inflammasome, and exhibited reduced cytopathogenicity as well as marked attenuation in the mouse model. Thus, IglG and IglI play key roles for modulation of the intracellular host response and also for the virulence of F. tularensis.
INTRODUCTION
Francisella tularensis is a facultative intracellular Gram-negative pathogen that causes the zoonotic disease tularemia. Human infections can occur through contact with infected mammals, especially rodents and lagomorphs, as well as from the bites of blood-feeding arthropods (39). The clinical manifestations of infection largely depend on infection route, of which inhalation resulting in pneumonia is potentially the most harmful (26, 56). The pathogenicity of F. tularensis is not completely understood, but the primary replication site in humans appears to be macrophages and involves escape from the phagosome prior to lysosomal fusion (19, 30), followed by cytosolic replication (68). Upon encounter with a host cell, rapid induction of a proinflammatory response which is completely or partly repressed upon bacterial internalization occurs (32, 60, 74). Moreover, F. tularensis actively suppresses the ability of both dendritic cells and macrophages to secrete cytokines in response to secondary stimuli (8, 21, 73). Thus, F. tularensis is clearly able to modulate many levels of the host immune response to facilitate its intracellular survival. After replication, bacterial egress is thought to occur via the induction of host cell death (41), which involves activation of both caspase-3- and caspase-1-dependent mechanisms and release of proinflammatory cytokines (52, 77). This Francisella-induced cell death has also been proposed to be an innate immune macrophage response to cytosolic bacteria aimed at restricting bacterial multiplication (52). Another host defense mechanism may involve the entry of Francisella into autophagosomes after cytosolic replication (14, 36).
Genes necessary for intracellular survival and virulence are found within the Francisella pathogenicity island (FPI), a 34-kb region which is duplicated in the highly virulent organisms F. tularensis subsp. tularensis and F. tularensis subsp. holarctica and which is under the positive control of at least five regulatory proteins: MglA, SspA, FevR (PigR), MigR, and PmrA (reviewed in reference 10). The iglABCD operon encodes gene products shown to be required for intramacrophage replication and virulence (9, 31, 46, 63), and at least IglA, IglB, and IglC are all essential for bacterial escape from the phagosome into the cytoplasm (7, 9, 46, 63). While the exact function(s) of this operon remains unknown, iglA and iglB have homologs in many bacterial species that encode type VI secretion systems (T6SSs) (5, 24), and their importance for substrate secretion has been experimentally demonstrated in some cases (6, 25, 61, 78). Bioinformatic analyses have identified additional FPI genes with limited homology to conserved T6SS components, including icmF, vgrG, clpV, dotU, and hcp (4, 24). Recently, Francisella tularensis subsp. novicida was shown to encode a functional T6SS that promoted VgrG (valine-glycine repeat protein G)- and IcmF-dependent translocation of IglI, a protein with no known homologs, into the macrophage cytosol during infection (4). This putative effector protein was shown to be required for phagosome escape, cytosolic replication, induction of inflammasome-dependent interleukin-1β (IL-1β) release, and virulence (4). Interestingly, FPI-independent secretion of VgrG within macrophages was also detected. In contrast, while the secretion of Hcp (hemolysin-coregulated protein) proteins is common to many bacterial pathogens (18, 54, 58, 66, 72, 78), secretion of Francisella Hcp/PdpE could not be detected (4).
In this study, we analyzed the roles of Hcp/PdpE and IglI in F. tularensis LVS. We also included IglG, a hitherto uncharacterized FPI protein, which, similar to IglI, lacks known homologs and therefore may represent a secreted substrate. While PdpE and IglG were shown to be outer membrane proteins, IglI localized to both soluble and membrane fractions. IglI, in contrast to IglG, was also shown to be secreted into the macrophage cytosol during infection. Despite the ability of the ΔiglG mutant to replicate in J774 cells, peritoneal exudate cells (PECs), and bone marrow-derived macrophages (BMDMs) and of the ΔiglI mutant to replicate in the J774 cells but not PECs or BMDMs, both mutants exhibited delayed phagosomal escape and inflammasome activation and displayed a diminished cytopathogenic response and marked attenuation for dissemination to and replication within internal organs upon intradermal infection of mice. In contrast, the ΔpdpE mutant showed essentially identical phenotypes to LVS in all investigated assays.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Escherichia coli strains were cultured in Luria-Bertani broth (LB) or on Luria agar plates at 37°C. F. tularensis was grown on modified GC agar base or in liquid Chamberlain's medium (13) at 37°C. When necessary, carbenicillin (Cb; 100 μg/ml), kanamycin (Km; 50 μg/ml for E. coli, 10 μg/ml for F. tularensis), or chloramphenicol (Cm; 25 μg/ml for E. coli, 2.5 μg/ml for F. tularensis) was used.
Cultivation and infection of macrophages.
J774A.1 (J774) macrophages and mouse peritoneal macrophages (PECs) or BMDMs were used in the cell infection assays. J774 macrophages were cultured and maintained in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Grand Island, NY) with 10% heat-inactivated fetal bovine serum (FBS; Gibco). PECs were isolated from 8- to 10-week-old C57BL/6 mice 3 days after intraperitoneal injection of 2 ml of 10% proteose peptone as previously described (47). BMDMs were generated by flushing bone marrow cells from the femurs and tibias of C57BL/6 mice. These cells were cultured for 4 days in DMEM containing 10% FBS, 5 μg/ml gentamicin, and 20% conditioned medium (CM) from L929 cells (ATCC no. CCL-1) overexpressing macrophage colony-stimulating factor, after which they were grown in medium lacking gentamicin. CM was replaced every 2 to 3 days.
On the day before infection, macrophages were seeded in tissue culture plates in DMEM with 10% FBS. Following incubation overnight, cells were washed, reconstituted with fresh culture medium, and allowed to recover for at least 30 min prior to infection. A multiplicity of infection (MOI) of 200 was used in all infection experiments, with the exception of the tumor necrosis factor alpha (TNF-α) secretion assay, where we used an MOI of 500 (73), and for the transmission electron microcopy (TEM) study, where an MOI of 1,000 was used.
Construction of pdpE, iglG, and iglI null mutants in F. tularensis LVS.
Primer combinations used to construct the pdpE, iglG, and iglI null mutants in LVS are listed in Table S2 in the supplemental material. Upstream and downstream flanking regions of ∼1,200 to 1,300 bp were amplified by PCR, cloned into the pCR4-TOPO TA cloning vector (Invitrogen AB, Stockholm, Sweden), and then sequenced by Eurofins MWG Operon (Ebersberg, Germany). Next, fragments were sequentially cloned into pBluescript SK+ (Stratagene, La Jolla, CA) using the XhoI/BamHI and BamHI/SacI sites, respectively (pdpE and iglG mutants), or the XhoI/EcoRV and EcoRV/SpeI sites, respectively (iglI mutant), thereby generating a fragment encoding PdpE with residues 4 to 188 deleted (PdpE Δ4-188), IglG Δ3-169, and IglI Δ4-361 with flanking regions joined by a BamHI or a EcoRV site. These fragments were cloned into XhoI/SacI or XhoI/SpeI-digested pDM4 (53), generating pJEB750 (pΔpdpE), pJEB751 (pΔiglI), and pJEB753 (pΔiglG). Conjugal mating experiments using S17-1λ pir as the donor strain allowed for the allelic exchange of the suicide plasmids within regions of complementary sequence on the LVS chromosome as described previously (31). To remove both copies of the pdpE, iglI, and iglG genes, the procedure was repeated, resulting in the null mutants, here designated ΔpdpE, ΔiglI, and ΔiglG mutants. In all cases, PCR screening was used to verify that the anticipated genetic event had occurred.
Construction of expression vectors.
Plasmids used in this study are listed in Table S1 in the supplemental material. All amplified fragments were first cloned into the pCR4-TOPO TA cloning vector (Invitrogen AB) to facilitate sequencing (Eurofins MWG Operon), before proceeding with the cloning. Plasmids used for trans-complementation studies were constructed as follows: PCR-amplified iglG 6×His tagged at the C terminus was introduced into plasmid pKK289Km, to allow constitutive expression from the groEL promoter (7). C-terminally glycogen synthase kinase (GSK)-tagged versions of pdpE and iglI were constructed as follows: a fragment corresponding to the HindIII to EcoRI region of the pUC19 MCS cassette with an added upstream NdeI site was PCR amplified using primers pUC19_MCS_F (5′-CAT ATG CTC GAG AAG CTT GCA TGC CTG GAG-3′; the NdeI region is in italics) and pUC19_MCS_R (5′-GAA TTC GAG CTC GGT ACC-3′; the EcoRI region is in italics). This fragment was introduced into NdeI/EcoRI-digested pKK289Km, generating pMOL42. A PCR fragment encoding GSK was amplified using primers GSK_F (5′-GGT ACC ATG TCA GGT AGA CCA AGA-3′; the KpnI region is in italics) and GSK_R (5′-GAA TTC TTA TGA TTC AGC AAA TGA AG-3′; the EcoRI region is in italics) and introduced into KpnI/EcoRI-digested pMOL42, resulting in pMOL52. Into this backbone, NdeI/KpnI fragments of pdpE or iglI were subsequently introduced. To generate the IglG-CyaA and IglI-CyaA expression constructs pJEB835 and pJEB851, respectively, a fragment carrying iglG or iglI was introduced into NcoI/NdeI-cut pKEK1012 (4), replacing the existing vgrG allele. The primer combinations and restriction sites used to generate the pdpE, iglG, and iglI alleles are listed in Table S2 in the supplemental material. Plasmids were transferred into F. tularensis by electroporation.
Fractionation of F. tularensis.
Francisella bacteria grown overnight at 37°C in 40 ml Chamberlain's medium with appropriate antibiotics were harvested and resuspended in 5 ml of ice-cold TE (Tris-EDTA) buffer. Cells were lysed by sonication, and unbroken cells were removed by 30 min of centrifugation (Multifuge 3 S-R, 75006445 swing-out rotor; Heraeus) at 3,452 × g and 4°C. The lysate was subjected to ultracentrifugation (Optima L-80 XP, rotor-type SW 41 Ti; Beckman) at 154,324 × g for 3 h at 4°C, upon which the supernatant (soluble protein fraction) was collected and recentrifuged for 1 h (154,324 × g, 4°C) to remove contaminants, while the membrane pellet was resuspended in 5 ml of 0.5% Sarkosyl (Sigma) and incubated for 90 min at 4°C while it was shaken. The Sarkosyl-soluble (inner membrane) and the Sarkosyl-insoluble (outer membrane) fractions were separated by a second ultracentrifugation step at 154,324 × g for 3 h at 4°C. Protein fractions were separated by SDS-PAGE and analyzed using standard Western blot techniques and an enhanced chemiluminescence (ECL) system (Amersham Biosciences, Uppsala, Sweden). PdpE, IglI, and IglG proteins were fused to the 6×His (IglG) or GSK (PdpE and IglI) tag at the C terminus and detected using antibodies recognizing 6×His (Qiagen, Solna, Sweden) and GSK (Cell Signaling Technology, Danvers, MA), respectively. Antisera against the inner membrane protein PdpB/IcmF (BEI Resources, Manassas, VA) and the outer membrane protein Tul4 were used to determine the purity of inner and outer membrane fractions, respectively, and to exclude membrane contamination of soluble fractions. For the latter purpose, antiserum against IglC (BEI Resources, Manassas, VA), suggested to be an exclusively soluble protein (24), was also used. Antimouse antibodies coupled to horseradish peroxidase (Santa Cruz Biotechnology, CA) were used as secondary antibodies in all cases, except for GSK, where antirabbit secondary antibodies coupled to horseradish peroxidase (GE Healthcare, United Kingdom) were used for detection. Due to low expression of IglG-6×His, 20 μg of the fraction was loaded to enable detection of this protein, while 5 μg of the fractions was used for detection of all other proteins. Protein concentrations were determined using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific, DE).
qPCR.
Protocols for isolation of bacterial RNA, cDNA synthesis, and quantitative real-time PCR (qPCR) have been described in detail elsewhere (9). Primers used will be provided upon request. For all samples, controls were made with either template or superscript omitted during cDNA synthesis. All reactions were performed in triplicate on three independent RNA preparations with a 7900HT sequence detection system (Applied Biosystems) using sequence detection system software. Samples were normalized against the F. tularensis 17-kDa housekeeping gene tul4 (FTL0421) and compared to the respective genes in LVS. Results were analyzed using the delta delta threshold cycle (CT) method of analysis and converted to a relative expression ratio (2−ΔΔCT) for statistical analysis (49). Paired two-tailed t tests were used to compare means.
Intracellular replication in macrophages.
To determine the ability of F. tularensis to grow within macrophages, we essentially followed our previously established protocols (31). In short, bacteria were added to each well at an MOI of 200, and bacterial uptake was allowed to occur for 2 h. The monolayers were washed three times and incubated in DMEM with FBS supplemented with 5 μg/ml of gentamicin. The time point after 30 min of gentamicin treatment was defined as time zero. At 0, 24, and 48 h, the macrophage monolayers were lysed in phosphate-buffered saline (PBS) with 0.1% deoxycholate, serially diluted in PBS, and plated on modified GC agar base plates for determination of viable counts. A two-sided t test with equal variance was used to determine whether the growth of a strain differed significantly from that of LVS.
Intracellular immunofluorescence assay.
To assess phagosomal escape, green fluorescent protein (GFP)-expressing F. tularensis was used in J774 cell infections as described previously (9), with the modification that, upon infection and subsequent washing, cells were incubated for up to 6 h to follow escape of mutants which showed a delayed phenotype. Cells were then stained for the lysosomal-associated membrane protein 1 (LAMP-1) glycoprotein as described previously (7). Colocalization of GFP-labeled F. tularensis and LAMP-1 was analyzed with an epifluorescence microscope (Axioskop2; Carl Zeiss MicroImaging GmbH, Germany) and a confocal microscope (Eclipse 90i; Nikon, Japan). From two separate experiments, each with a total number of 5 glass slides per strain, 50 bacteria/slide were scored. To verify that the colocalization level was significantly different from that of LVS, a Wilcoxon rank-sum test was used.
Transmission electron microscopy.
J774 cells seeded at a density of 1 × 106 cells/well in DMEM with FBS in 6-well tissue culture plates were infected at an MOI of 1,000 for 2 h, washed three times, and incubated in medium for 2 or 6 h. Monolayers were washed with DMEM before fixation in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Upon fixation, cells were washed in 0.1 M sodium cacodylate buffer, scraped from the dishes, and fixed for 1 h with 1% osmium tetroxide. After washing, the cell pellet was dissolved in 2% agarose and centrifuged in a warm centrifuge. The embedded pellet was cut into small cubes and stained with uranyl acetate (1% solution in methanol) overnight. Following dehydration using an ethanol series, the specimens were embedded in Spurr resin (Sigma-Aldrich, MO). Ultrathin sections (70 nm) were cut and stained with uranyl acetate and lead citrate before they were viewed with a JEM 1230 transmission electron microscope (Jeol Ltd., Tokyo, Japan). To examine membrane integrity, at least 100 bacteria from different sections were analyzed for each time point and categorized as having (i) an intact phagosomal membrane, (ii) a slightly damaged phagosomal membrane (<50% of membrane integrity affected), (iii) a highly damaged phagosomal membrane (>50% of membrane integrity affected), or (iv) no residual membrane.
Adenylate cyclase assay.
Determination of adenylate cyclase activity in J774 macrophages infected with F. tularensis expressing CyaA fusion proteins was performed as follows: macrophages seeded at a density of 1 × 105 cells/well in 96-well plates were infected at an MOI of 200 for 2 h. Media were aspirated and cells were incubated in the presence of 10 μg/ml gentamicin for 3 h, upon which measurement of cytosolic cyclic AMP (cAMP) accumulation was completed by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's instructions (cAMP Biotrak enzyme immunoassay system; Amersham Biosciences). To verify that the cAMP levels of the mutants were significantly different from the level of parental strain LVS, a 2-tailed t test was used.
TNF-α secretion assay.
Quantification of TNF-α secretion into the cell culture medium of lipopolysaccharide (LPS)-stimulated J774 macrophages infected with F. tularensis was determined using an OptEIA mouse TNF-α ELISA set (BD Biosciences) according to the manufacturer's instructions. Cells were infected at an MOI of 500 for 2 h, washed twice, and incubated in the presence of 5 μg/ml gentamicin and 50 ng/ml of LPS derived from E. coli O111:B4 (Sigma). After 1 or 2 h, culture supernatants were collected, centrifuged at 16,000 × g for 10 min at 4°C to remove any bacterial and cell contaminants, and used in the ELISA.
IL-1β secretion assay.
Peritoneal macrophages or BMDMs were infected at an MOI of 200 for 2 h in DMEM with FBS, washed twice, and incubated in the presence of 5 μg/ml gentamicin. When appropriate, this medium was supplemented with acetyl-YVAD-CMK (Ac-YVAD-CMK) caspase-1 inhibitor II (Calbiochem, La Jolla, CA) to a final concentration of 100 μM. Importantly, this concentration did not affect the extent of intracellular growth or lactate dehydrogenase (LDH) release by F. tularensis. After 0, 5, 8, or 24 h, culture supernatants were collected, centrifuged at 16,000 × g for 10 min at 4°C to remove any bacterial and cell contaminants, and used in the OptEIA mouse IL-1β ELISA set (BD Biosciences) according to the manufacturer's instructions.
LDH release assay.
J774 cells, PECs, or BMDMs were infected for 2 h (MOI, 200), washed twice, and incubated in the presence of 5 μg/ml gentamicin for 30 min (which corresponds to time zero). Supernatants were sampled at 0, 24, or 48 h and assayed for the presence of released LDH using a Cytotox 96 kit (Promega, Madison, WI) according to the manufacturer's instructions. Absorbance at 490 nm was determined using a microplate reader (Tecan Systems, San Jose, CA). Data are means ± standard deviations of three wells from one representative experiment of three. Uninfected J774 cells lysed in PBS with 0.1% deoxycholate served as a positive control, and the value for this control was arbitrarily considered 100% cell lysis. Sample absorbance was expressed as the percentage of the positive-control value.
Mouse infections.
For determination of the killing capacity of each strain, C57BL/6 female mice (n=5) were infected intradermally with approximately 7 × 106 or 2 × 108 CFU of F. tularensis. Aliquots of the diluted cultures were also plated on GC agar to determine the numbers of CFU injected, which were as follows: 7.8 × 106 or 1.9 × 108 for LVS, 7.6 × 106 or 1.9 × 108 for the ΔpdpE mutant, 7.1 × 106 or 1.8 × 108 for the ΔiglI mutant, 3.4 × 106 or 0.8 × 108 for the ΔiglI/pMOL59 mutant, 6.2 × 106 or 1.6 × 108 for the ΔiglG mutant, and 3.4 × 106 or 0.8 × 108 for the ΔiglG/pMOL103 mutant. In a follow-up experiment, we increased the doses of the ΔiglI and ΔiglG mutants further to 9.5 × 108 (ΔiglI mutant) and 6.5 × 108 (ΔiglG mutant) without any change in outcome. Mice were examined twice daily for signs of severe infection and euthanized by CO2 asphyxiation as soon as they displayed signs of irreversible morbidity. In our experience, such mice were at most 24 h from death, and the time to death of these animals was estimated on the basis of this premise. To measure the bacterial burden in skin, spleen, and liver, BALB/cJ female mice (n=5) were infected intradermally with LVS (2.7 × 105 CFU), the ΔiglG mutant (2.5 × 105), and the ΔiglI mutant (3.9 × 105 CFU). At days 3, 5, and 7 postinfection, mice were killed and serial dilutions of the homogenized organs were plated. All animal experiments were approved by the local Ethical Committee on Laboratory Animals, Umeå, Sweden (approval no. A113-08).
RESULTS
Construction of ΔpdpE, ΔiglG, and ΔiglI null mutants.
A hallmark of most T6SSs is the presence of Hcp and VgrG proteins in culture supernatants, which are components critical for the assembly and function of the secretion machineries (54, 57, 59). Indeed, the VgrG homolog of F. tularensis subsp. novicida was recently shown to be secreted during in vitro growth as well as during intracellular infection, while secretion of PdpE, the putative Hcp homolog, has yet to be demonstrated (4). Nevertheless, on the basis of the aforementioned critical role exhibited by Hcp proteins in T6S, we would predict that the loss of PdpE would have profound effects on the ability of Francisella to cause disease if it has an Hcp-like function. We therefore characterized the phenotype of a ΔpdpE mutant of LVS. We also included two additional mutants, the ΔiglI and ΔiglG mutants, since the corresponding gene products lack apparent homology to T6SS proteins of other bacterial systems and may therefore represent secretion substrates unique to Francisella. This was recently validated when IglI of F. tularensis subsp. novicida, which shares 97.9% identity to IglI from LVS, was shown to be secreted within macrophages (4). The iglG gene encodes a 24.6-kDa hypothetical protein, for which the function(s) has yet to be determined. Thus, we constructed in-frame deletion mutants, deleting both copies of each gene. The resulting null mutant strains, the ΔpdpE, ΔiglG, and ΔiglI mutants, were used in various biological assays. To verify the absence of gene expression in the mutants, real-time PCR was used to quantify levels of pdpE, iglG, and iglI transcripts in the ΔpdpE, ΔiglG, and ΔiglI mutants, respectively. In all cases, levels were below the detection limit of the assay (data not shown). Importantly, real-time PCR was also used to demonstrate the absence of polar effects on downstream genes in the in-frame deletion mutants. In fact, each mutant was found to produce wild-type levels of transcripts for all the other 16 genes of the pathogenicity island (from pdpA to pdpD) (see Table S3 in the supplemental material).
Localization of PdpE, IglG, and IglI in F. tularensis LVS.
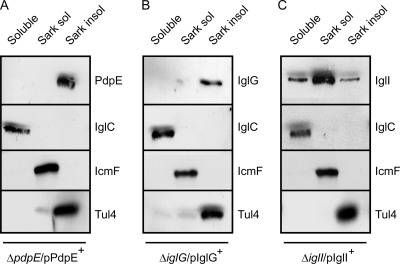
Knowledge of the subcellular localization of a protein is an important piece of information to infer its biological role. However, a bioinformatic analysis using the PSORTb tool (http://www.psort.org/psortb/) did not provide any useful information regarding the location of either PdpE, IglG, or IglI, with the exception that an N-terminal signal peptide was predicted for PdpE using the SignalP server (http://www.cbs.dtu.dk/services/SignalP/). Thus, to investigate the subcellular localization of these proteins, we fractionated LVS bacteria into soluble, inner membrane, and outer membrane fractions and determined the amounts of the proteins in each fraction by immunoblot analysis. To enable detection, the proteins were fused to either GSK or 6×His at the C terminus and expressed from the groEL promoter of pKK289Km (7) in the corresponding mutant background, resulting in ΔpdpE/pPdpE+(pMOL61), ΔiglG/pIglG+ (pMOL103), and ΔiglI/pIglI+(pMOL59) strains. Importantly, these small tags are not likely to impact the pattern of protein localization, since tagged PdpE, IglG, and IglI all behaved identically to their nontagged counterparts expressed from pKK289Km in the subsequent analyses (data not shown). The data from this fractionation experiment revealed that PdpE is exclusively an outer membrane protein, while IglI is found in all fractions, although it is enriched in the inner membrane (Fig. 1). IglG was predominantly found in the outer membrane fraction, although a small portion occasionally localized to the inner membrane (Fig. 1 and data not shown). As controls of the purity of the fractions, we detected IglC only in the soluble fraction, as reported previously (50), and the inner membrane protein IcmF/PdpB only in the Sarkosyl-soluble fraction (50), and the outer membrane protein Tul4 (40, 69), absent in the soluble fraction, was clearly enriched in the Sarkosyl-insoluble fraction, as expected (Fig. 1).
Fig. 1.
Subcellular localization of PdpE, IglG, and IglI in Francisella. C-terminal fusion proteins of PdpE and IglI (GSK tagged) and IglG (6×His tagged) were expressed from pKK289Km in the isogenic mutant background, resulting in ΔpdpE/pPdpE+(pMOL61) (A), ΔiglG/pIglG+(pMOL103) (B), and ΔiglI/pIglI+(pMOL59) (C) strains. Upon fractionation into soluble and membrane-associated fractions, Sarkosyl solubilization was used to further separate inner (Sarkosyl-soluble) and outer (Sarkosyl-insoluble) membranes. Protein fractions were separated by SDS-PAGE and analyzed using standard Western blot techniques and appropriate antisera. To detect PdpE and IglI, anti-GSK antiserum was used, while IglG was visualized using anti-His antibodies. Antibodies recognizing IglC, PdpB, or Tul4 were used as markers for soluble, inner membrane, and outer membrane fractions, respectively.
Requirement for PdpE, IglG, and IglI for phagosomal escape and in vitro growth in macrophages.
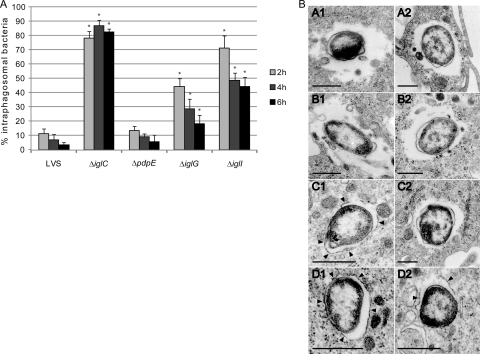
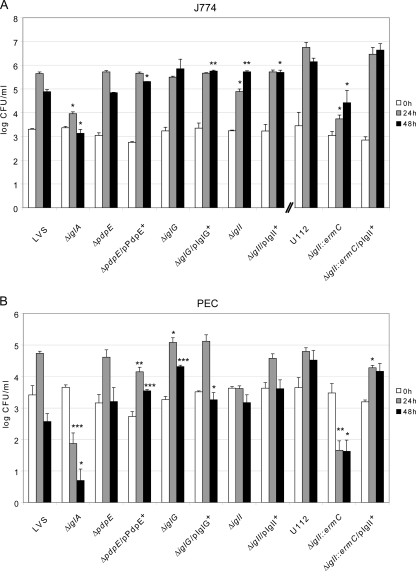
Many FPI mutants have been found to be defective for the escape from the phagosomal compartment. To determine if F. tularensis has an intraphagosomal localization, the most commonly used marker is LAMP-1 (reviewed in references 16 and 62), which is a late endosomal and lysosomal marker acquired within 30 min by the Francisella-containing phagosome (7, 14, 17, 19, 30, 64). Thus, we used microscopy to determine the percentage of LAMP-1 colocalization for ΔpdpE, ΔiglG, or ΔiglI mutant bacteria expressing GFP at 2, 4, and 6 h postinfection of J774 macrophages (Fig. 2A). As controls, LVS and the ΔiglC mutant were used. At 2 h postinfection, 11.2% ± 3.6% of the LVS strain and 13.2% ± 2.3% of the ΔpdpE mutant (P > 0.05 versus LVS) colocalized with LAMP-1. In contrast, 44.4% ± 5.6% of the ΔiglG mutant (P < 0.01 versus LVS), 71.2% ± 8.7% of the ΔiglI mutant (P < 0.01 versus LVS), and 78.0% ± 4.9% of the ΔiglC mutant (P < 0.01 versus LVS) colocalized with LAMP-1. At 6 h, only 3.2% ± 1.8% of LVS and 5.5% ± 1.9% of the ΔpdpE mutant (P > 0.05 versus LVS) associated with phagosomes, while the corresponding numbers for the mutants were 18.0% ± 6.2% for the ΔiglG mutant, 44.4% ± 6.5% for the ΔiglI mutant, and 82.4% ± 2.2% for the ΔiglC mutant (P < 0.01 versus LVS for all three mutants) (Fig. 2A). Altogether, these results clearly demonstrated that the ΔiglG mutant and, even more so, the ΔiglI mutant showed significantly delayed phagosomal escape. To corroborate the results from the confocal microscopy, we also performed transmission electron microscopy. J774 cells were infected with LVS or the ΔpdpE, ΔiglG, or ΔiglI mutant, and the percentage of bacterial escape was determined. Already after 2 h, the majority of LVS and ΔpdpE mutant bacteria were found free in the cytoplasm (69% and 70%, respectively) or were surrounded by highly damaged vacuolar membranes (19% and 20%, respectively), while only a minor fraction was found enclosed by intact or slightly damaged vacuolar membranes (12% and 11%, respectively) (Fig. 2B). At the same time point, only 33% of the ΔiglG mutant bacteria were found free in the cytoplasm, while the majority was surrounded by vacuolar membranes that were either highly damaged (<50% of membrane intact; 39%), slightly damaged (>50% of membrane intact; 30%), or intact (6%) (Fig. 2B), suggesting that the ΔiglG mutant exhibits a delayed escape from the phagosomes, analogous to what we observed in the LAMP-1 colocalization experiment. In support of this, similar numbers of ΔiglG (93%), ΔpdpE (94%), and LVS (92%) bacteria were found free in the cytosol when they were examined at the 6-h time point (Fig. 2B). Compared to the ΔiglG mutant, the ΔiglI mutant was even more defective for phagosomal escape: at 2 h, the majority of ΔiglI mutant bacteria was surrounded by intact (41%) or only slightly damaged (49%) bacterial membranes, while at 6 h, the majority was found to be either free in the cytosol (44%) or enclosed by highly damaged vacuolar membranes (41%) (Fig. 2B). Again, these results corroborate the findings from the confocal microscopy and LAMP-1 staining. Thus, in contrast to IglC, which is essential for phagolysosomal escape and subsequent replication in the cytosol (7, 31), PdpE, IglG, and IglI are not. To determine whether the delayed escape of the ΔiglG and ΔiglI mutants correlated with impaired growth in J774 cells, we performed viable counts at different time points postinfection. To our surprise, the ΔiglG and ΔiglI mutants replicated as efficiently as parental LVS and the ΔpdpE mutant over a time period of 48 h (Fig. 3 A). Compared to LVS, there were somewhat fewer ΔiglI bacteria at 24 h (P < 0.05), but at 48 h more bacteria were recovered (P < 0.01). These small differences were consistently found throughout all experiments (data not shown). Thus, the delayed escape observed for the ΔiglG and ΔiglI mutants was apparently not severe enough to greatly impact their ability to replicate in macrophages, at least not at the time points tested. To verify these results, we also investigated their ability to multiply within mouse PECs, which have a higher killing capacity than J774 cells, as well as resting murine BMDMs. In PECs, the ΔiglG mutant multiplied as efficiently as parental strain LVS and the ΔpdpE mutant over a time period of 48 h (Fig. 3B). In fact, compared to LVS, there were somewhat more ΔiglG bacteria at 24 h (P < 0.05) and at 48 h (P < 0.001). Efficient growth of the ΔiglG mutant was also seen in BMDMs (data not shown). The ΔiglI mutant, however, was unable to grow within PECs (P < 0.001) and BMDMs (Fig. 3B and data not shown). Growth could, however, be restored by supplying IglI in trans from pMOL59 (Fig. 3B and data not shown), suggesting that the growth defect observed is cell type specific. Still, in comparison with the nonreplicating ΔiglA mutant (9), ΔiglI mutant numbers did not decrease over time in PECs (Fig. 3B), indicating that the ΔiglI mutant is more resistant to macrophage killing than the ΔiglA mutant.
Fig. 2.
Phagosomal escape of F. tularensis. (A) GFP-expressing F. tularensis strains colocalized with LAMP-1. J774 cells were infected for 2 h with F. tularensis strains expressing GFP at an MOI of 200 and, after washing, incubated for 2, 4, or 6 h. Colocalization of GFP-labeled F. tularensis and LAMP-1 on fixed and labeled specimens was analyzed using an epifluorescence microscope (Axioskop2; Carl Zeiss MicroImaging GmbH, Germany) or a confocal microscope (Eclipse 90i; Nikon, Japan). Each bar represents the mean values, and the error bar indicates the standard deviation of one representative experiment of two. Asterisks denote that the percent colocalization is statistically different from that for LVS (*, P ≤ 0.05), as determined by a Wilcoxon rank-sum test. (B) Electron (TEM) micrographs of J774 cells infected with F. tularensis LVS (A1 and A2) or the ΔpdpE (B1 and B2), ΔiglG (C1 and C2), or ΔiglI (D1 and D2) mutant. Cells were infected for 2 h at an MOI of 1,000 and after washing were incubated for either 2 h (panels denoted with a 1 suffix) or 6 h (panels denoted with a 2 suffix). Electron micrographs were acquired with a JEM 1230 transmission electron microscope (Jeol Ltd., Tokyo, Japan). Black arrowheads indicate vacuolar membranes surrounding intracellular bacteria. Scale bars, 0.5 μm.
Fig. 3.
Intracellular growth of different strains of F. tularensis. J774 cells (A) or PECs (B) were infected by various strains of F. tularensis at an MOI of 200 for 2 h. Upon gentamicin treatment, cells were allowed to recover for 30 min, after which they were lysed immediately (which corresponds to 0 h) or after 24 h or 48 h with PBS-buffered 0.1% sodium deoxycholate solution and plated to determine the number of viable bacteria (log10). All infections were repeated two to three times with triplicate data sets. Representative experiments for strain LVS and derivatives thereof and F. tularensis subsp. novicida U112 and derivatives thereof are shown. Each bar represents the mean values, and the error bar indicates the standard deviation. The asterisks indicate that the log10 number of CFU was significantly different from that for the parental strain (LVS or U112), as determined by a 2-sided t test with equal variance (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Intriguingly, these results are in contrast to a previous report, where an ΔiglI::ermC mutant of F. tularensis subsp. novicida was found to be defective for intramacrophage growth in J774 cells (4). We verified these findings (P < 0.05 versus strain U112) (Fig. 3A) and extended these results to also include PECs (Fig. 3B). Similar to the ΔiglA mutant of strain LVS, the numbers of F. tularensis subsp. novicida ΔiglI::ermC cells were drastically reduced at 24 h (P < 0.01 versus U112) and 48 h (P < 0.05 versus U112) postinfection (Fig. 3B). This growth defect could be efficiently restored upon expression of IglI in trans (Fig. 3A and B). Thus, an F. tularensis subsp. novicida ΔiglI mutant appears to be more defective for intracellular growth and is more susceptible to macrophage killing than an ΔiglI mutant of LVS. This interesting finding led us to include the F. tularensis subsp. novicida mutant in several of the subsequent assays where the LVS mutants were characterized.
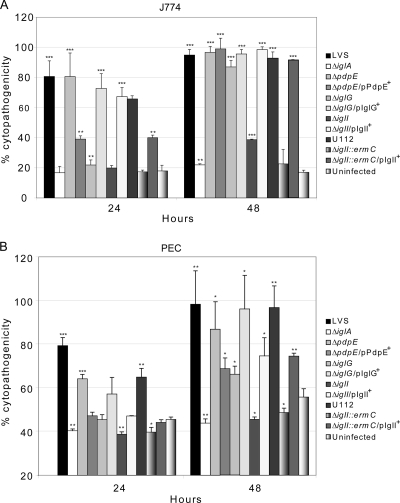
ΔiglG and ΔiglI mutants are unable to elicit an efficient cytopathogenic response.
Strain LVS causes a pronounced cytopathogenic response of infected macrophages, resulting in morphological changes such as membrane blebbing, cell detachment, LDH release, and an overall degenerate appearance of the cells (2, 41). Defects in phagosomal escape and cytosolic replication generally correlate with an inability to induce cytotoxicity (7, 11, 31, 51, 52). For this reason, we would expect the delayed phagosomal escape of ΔiglG and ΔiglI mutants to impair their ability to induce an efficient cytopathogenic response compared to LVS and the ΔpdpE mutant. To test this hypothesis, we infected J774 cells, PECs, as well as BMDMs with the null mutant strains and measured the release of LDH into the cell culture supernatants. In addition, the morphological effect on cells was investigated using phase-contrast microscopy. At 24 h postinfection, both LVS and the ΔpdpE mutant induced significant LDH release from the infected cells, which were also notably morphologically affected (Fig. 4 A and B and data not shown). Somewhat less LDH was released from PECs infected with the ΔpdpE mutant (P < 0.01) than PECs infected with LVS (Fig. 4B). We were, however, unable to complement this phenotype, since expression of PdpE in trans (pMOL61) was found to result in even more reduced levels of LDH release (Fig. 4B). The same was observed when the complemented strain was used to infect J774 cells (Fig. 4A). This may indicate that PdpE overexpression is toxic to Francisella; however, the complemented ΔpdpE mutant showed wild-type levels of intracellular growth (Fig. 3), excluding this possibility. At 24 h postinfection, LDH levels in culture supernatants and the morphological appearance of cells infected with either the ΔiglG or ΔiglI mutant did not differ much from those of uninfected cells or cells infected with the nonreplicating ΔiglA mutant (Fig. 4A and B and data not shown). However, at 48 h, the ΔiglG mutant promoted LDH release in J774 cells (87% of the positive lysis control), PECs (66% of the positive lysis control), as well as BMDMs (73% of the positive lysis control). These levels were significantly higher (P < 0.001, P < 0.05, and P < 0.01, respectively) than those for the uninfected controls (17%, 56%, and 37%, respectively) (Fig. 4A and B and data not shown). At the same time point, the ΔiglI mutant caused only minor LDH release in J774 cells (39%) (Fig. 4A). Still, at this time point, this was significantly higher (P < 0.001) than the levels of LDH release for the uninfected control (17%) or cells infected with the ΔiglA mutant (22%) or the F. tularensis subsp. novicida ΔiglI::ermC mutant (24%) (Fig. 4A). In contrast, the ΔiglI mutant, F. tularensis subsp. novicida ΔiglI::ermC, and the ΔiglA mutant all failed to induce LDH release in PECs (Fig. 4B), while at 48 h, low levels of LDH (43%) were released from ΔiglI mutant-infected BMDMs, and these levels were significantly higher than those of ΔiglA mutant-infected BMDMs (29%; P < 0.001) or the uninfected control (36%; P < 0.01) (data not shown). Importantly, expression of IglG (pMOL103) and IglI (pMOL59) in trans efficiently restored the cytopathogenic response of ΔiglG and ΔiglI mutants, respectively, in J774 cells and BMDMs but did so only partially in PECs (Fig. 4A and B and data not shown). Thus, IglG and, even more so, IglI play important but not essential roles in the ability of Francisella LVS to induce prominent cytopathogenicity, whereas PdpE is of minor importance for this process.
Fig. 4.
Cytopathogenicity of F. tularensis strains. Culture supernatants of J774 cells (A) or PECs (B) infected as described in Materials and Methods were assayed for LDH activity at 0, 24, and 48 h, and the activity was expressed as a percentage of the level for noninfected lysed cells (positive lysis control). At 0 h, infected as well as noninfected J774 cells showed ∼11% activity, which differed at most by 1% between strains (data not shown). For PECs, this activity was higher (∼39% activity of the infected as well as noninfected cells) but differed at most by ∼4% between strains (data not shown). Shown are means and standard deviations of triplicate wells from one representative experiment of two. The asterisks indicate that the cytopathogenicity levels were significantly higher than those of uninfected cells at a given time point, as determined by a 2-sided t test with equal variance (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Modulation of macrophage inflammatory responses.
Stimulation of macrophages with E. coli LPS leads to TNF-α production and secretion, an inflammasome-independent event, which is efficiently inhibited upon infection by F. tularensis LVS (73). In contrast, infection with an ΔiglC mutant or an ΔmglA mutant resulted in TNF-α production that was augmented when cells were activated by E. coli LPS (32, 73). Hence, to further characterize the role of the FPI in the inhibition of TNF-α production, we infected J774 cells with the ΔpdpE, ΔiglG, or ΔiglI mutant as well as the complemented mutant strains. After 1 or 2 h of stimulation with E. coli LPS, cell culture supernatants were collected and assayed for the presence of TNF-α. While the uninfected control resulted in significant TNF-α release, efficient inhibition of TNF-α production was observed for both LVS and the ΔpdpE mutant (Table 1). In contrast, the control ΔiglA strain was completely unable to inhibit TNF-α release (Table 1). In comparison to the ΔiglA mutant, the ΔiglG and ΔiglI mutants showed markedly stronger inhibition after 2 h of LPS stimulation (both P < 0.001), although it was not as strong as that for LVS (Table 1). Inhibition could, however, be fully restored by expressing IglG and IglI, respectively, in trans (Table 1). Thus, IglG and IglI, but not PdpE, are required for efficient inhibition of TNF-α production in infected macrophages. As seen before (52), F. tularensis subsp. novicida is completely unable to inhibit TNF-α release (Table 1).
Table 1.
TNF-α secretion of F. tularensis-infected J774 cells
| Strain | TNF-α secretion (pg/ml)a |
|
|---|---|---|
| 1 h | 2 h | |
| Noninfected | 156.2 ± 6.3*** | 168.5 ± 10.6*** |
| F. tularensis subsp. holarctica | ||
| LVS | 54.4 ± 3.3 | 40.9 ± 2.8 |
| ΔiglA | 228.1 ± 49.9* | 563.6 ± 14.3*** |
| ΔpdpE | 69.5 ± 5.7 | 37.2 ± 4.1 |
| ΔpdpE/pPdpE+ | 18.9 ± 0.8*** | 27.9 ± 0.9 |
| ΔiglG | 263.9 ± 16.6*** | 240.8 ± 47.4** |
| ΔiglG/pIglG+ | 39.9 ± 0.6*** | 35.0 ± 7.2** |
| ΔiglI | 226.0 ± 37.2** | 381.7 ± 33.9*** |
| ΔiglI/pIglI+ | 67.9 ± 18.7** | 37.5 ± 4.3*** |
| F. tularensis subsp. novicida | ||
| U112 | 225.0 ± 26.6 | 438.1 ± 40.1 |
| ΔiglI::ermC | 266.7 ± 20.0 | 645.9 ± 52.4* |
| ΔiglI::ermC/pIglI+ | 355.0 ± 42.3 | 625.7 ± 40.1 |
F. tularensis-infected (MOI=500) or noninfected J774 cells were incubated in the presence of E. coli-derived LPS (50 ng/ml) for 1 or 2 h. The average TNF-α secretion and standard errors from quadruple samples (n=4) from one out of two representative experiments are shown. In the absence of LPS, the cytokine levels were below the limit of detection for the assay (<15 pg/ml) (data not shown). A Student's 2-sided ttest was used to determine whether the TNF-α secretion induced by the mutants was significantly different from that of the parental strain (Pvalues indicated for each mutant) and whether the TNF-α secretion induced by the complemented strains was statistically different from that induced by their isogenic mutants (P values indicated for each complemented strain).
P < 0.05;
P < 0.01;
P < 0.001.
F. tularensis phagosomal escape into the macrophage cytosol is critical for the inflammasome-dependent induction of IL-1β secretion (4, 21, 28, 38, 52). Consequently, macrophages infected with strains with mutations in mglA, iglC, vgrG, or iglI have been shown to display abrogated IL-1β release (4, 21, 28, 38, 76). To determine whether PdpE, IglG, and IglI play a role in this process, we measured the concentration of IL-1β in culture supernatants of macrophages infected with the corresponding LVS mutants at 0, 5, 8, or 24 h postinfection. Mouse peritoneal exudate cells or BMDMs infected with LVS or the ΔpdpE mutant induced high levels of IL-1β release, although they were somewhat less for ΔpdpE mutant-infected PECs at 24 h (Table 2). This defect was, however, not statistically significant and could not be restored by expressing PdpE in trans. In fact, it became even more pronounced for PECs infected with the complemented strain at 24 h postinfection (P < 0.05 compared to LVS), while the same phenomenon did not occur when BMDMs were used (Table 2). Thus, PdpE overexpression diminishes IL-1β secretion (PECs only) as well as cytopathogenicity (Fig.4) but not intracellular growth (Fig. 3) or virulence (data not shown). The ΔiglG mutant showed an interesting phenotype with regard to IL-1β release: in contrast to LVS and the ΔpdpE mutant, PECs infected with the ΔiglG mutant produced intermediate levels of IL-1β secretion at 5 h (P < 0.05 compared to LVS) as well as 24 h (P < 0.01 compared to LVS), while ΔiglG mutant-infected BMDMs released 2 to 7 times more IL-1β than LVS-infected BMDMs in all four experiments (P < 0.01) (Table 2 and data not shown). We are currently lacking an explanation for this cell-specific defect in IL-1β secretion. The ΔiglI mutant infection resulted in only low levels of release at 24 h in PECs (below limit of detection at 5 h) and an intermediate level of release in BMDMs, while cells infected with F. tularensis subsp. novicida ΔiglI::ermC or the control ΔiglA strain showed completely abrogated release (PECs) or release similar to that for the noninfected control (BMDMs) (Table 2). Upon complementation in trans, IL-1β secretion was either partially or fully restored in the ΔiglG and ΔiglI mutants as well as F. tularensis subsp. novicida ΔiglI::ermC (Table 2). Thus, both IglG, to some extent, and, even more so, IglI are important for the IL-1β release by LVS, consistent with their delayed escape from the phagosomes. Thus, as suggested previously, there is a strong correlation between production of active IL-1β protein and a cytoplasmic location of the bacterium (4, 21, 28, 38).
Table 2.
IL-1β secretion of F. tularensis-infected PECs and BMDMs
| Strain | IL-1β secretion (pg/ml)a |
|||
|---|---|---|---|---|
| PECs |
BMDMs |
|||
| 5 h | 24 h | 8 h | 24 h | |
| Noninfected | BDL | BDL | BDL | 44.7 ± 4.7 |
| F. tularensis subsp. holarctica | ||||
| LVS | 93.6 ± 3.7 | 657.5 ± 51.9 | 103.0 ± 4.5 | 263.3 ± 28.4 |
| ΔiglA | BDL | BDL | 40.5 ± 2.4*** | 59.1 ± 6.9*** |
| ΔpdpE | 87.4 ± 15.4 | 492.6 ± 82.2 | 112.7 ± 9.0 | 295.67 ± 51.4 |
| ΔpdpE/pPdpE+ | 96.8 ± 15.6 | 374.4 ± 60.1 | 393.4 ± 39.4*** | 685.3 ± 67.8*** |
| ΔiglG | 75.5 ± 1.4* | 175.5 ± 27.3** | 156.8 ± 18.3** | 493.9 ± 52.4*** |
| ΔiglG/pIglG+ | 138.5 ± 7.5** | 543.7 ± 58.8** | 92.1 ± 9.6 | 278.7 ± 29.9* |
| ΔiglI | BDL | 47.0 ± 5.2** | 78.2 ± 4.1*** | 157.5 ± 27.3** |
| ΔiglI/pIglI+ | 118.2 ± 95.1 | 306.4 ± 55.4** | 126.8 ± 8.5*** | 406.2 ± 37.2*** |
| F. tularensis subsp. novicida | ||||
| U112 | NT | 2,415.0 ± 158.3 | 2,119.2 ± 64.1 | 2,286.7 ± 82.9 |
| ΔiglI::ermC | NT | BDL | 47.6 ± 5.5*** | 50.2 ± 13.7*** |
| ΔiglI::ermC/pIglI+ | NT | 440.4 ± 97.4 | 1,579.6 ± 150.6** | 2,124.8 ± 70.8 |
Francisella-infected (MOI=200) or noninfected PECs or BMDMs were incubated for 0, 5, 8, or 24 h after gentamicin treatment. The average IL-1β secretion and standard error of the mean from triplicate samples (n=3) from one out of two representative experiments are shown. BDL, cytokine levels were below the limit of detection for the assay (<31.25 pg/ml) (data not shown). At 0 h, the cytokine levels were below the limit of detection for all strains (data not shown). A Student's 2-sided t test was used to determine whether the IL-1β release induced by the mutants was significantly different from that by the parental strain (P values indicated for each mutant) and whether the IL-1β secretion induced by the complemented strains was statistically different from that of their isogenic mutants (P values indicated for each complemented strain.
P < 0.05;
P < 0.01;
P < 0.001; NT, not tested.
Combined work from several groups has demonstrated the importance of inflammasome activation for the induction of IL-1β secretion by F. tularensis subsp. novicida (reviewed in reference 29). More specifically, using caspase-1-deficient macrophages, the requirement of caspase-1 for production of active IL-1β has been established (12, 23, 38, 52, 75). In contrast, its role for production for LVS-dependent secretion is less thoroughly investigated (37, 52, 75) and has been established using cells initially primed with LPS (52, 75), a known trigger of inflammasome activation (34). To verify the dependency of caspase-1 for IL-1β secretion by F. tularensis-infected macrophages, we used Ac-YVAD-CMK caspase-1 inhibitor II. This inhibitor has been shown to inhibit caspase-1-dependent functions in pathogens like Salmonella, Shigella, and Yersinia spp. (33, 44, 55). Accordingly, in the presence of Ac-YVAD-CMK, PECs infected with F. tularensis subsp. novicida U112 displayed an ∼71% reduction in IL-1β secretion at 24 h postinfection (from 2,350 ± 366 pg/ml to 690 ± 161 pg/ml [standard error of the mean {SEM}]; P < 0.001, average of 2 experiments). In contrast, we observed no significant inhibition of the IL-1β release induced by macrophages infected with LVS or the ΔpdpE, ΔiglI, or ΔiglG mutant at 8 h or 24 h postinfection (data not shown). Thus, in infected PECs, the effects on IL-1β secretion of the caspase-1 inhibitor appeared to be distinct between the LVS and F. tularensis subsp. novicida infections.
Involvement of PdpE, IglG, and IglI in secretion of IglI and VgrG.
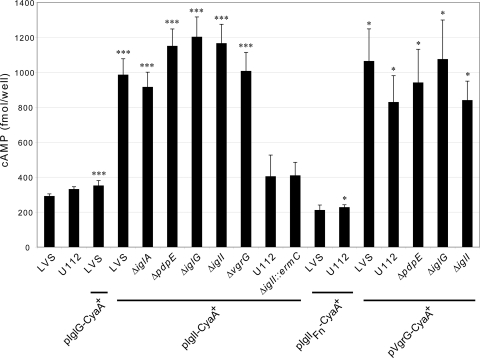
Recently, CyaA fusions of VgrG and IglI were shown to be secreted into the cytosol of F. tularensis subsp. novicida-infected macrophages, while PdpE-CyaA was not (4). To determine whether IglG is also a secreted substrate, we constructed a C-terminal IglG-CyaA fusion and expressed this protein from pJEB835 in LVS or the ΔiglG mutant. We also analyzed the contribution of PdpE, IglG, IglI, and IglA to the secretion of VgrG by infecting J774 cells with the LVS, ΔpdpE, ΔiglG, ΔiglI, ΔiglA, or ΔvgrG strain expressing VgrG-CyaA from pKEK1012 (4). Using this approach, we were unable to detect any considerable increases in cAMP levels indicative of IglG-CyaA secretion into the macrophage cytosol (Fig. 5 and data not shown). This was not due to a lack of protein expression, since the same construct was able to restore the cytopathogenic defect of an ΔiglG mutant to wild-type levels (data not shown). Interestingly, secretion of VgrG-CyaA occurred regardless of strain background, although to somewhat varying degrees (Fig. 5 and data not shown). This suggests that FPI-independent mechanisms are likely to operate to promote VgrG secretion in LVS, similar to what was recently reported for F. tularensis subsp. novicida (4). In the latter study, secretion of IglI-CyaA into macrophages was shown to be FPI-dependent, since it did not occur in a ΔvgrG or an ΔicmF mutant. Intriguingly, the very same mutants, as well as an ΔFPI mutant, were able to secrete IglI-FLAG in vitro, suggesting that IglI export may also occur by an FPI-independent mechanism in F. tularensis subsp. novicida (4). To analyze whether IglI-CyaA can be secreted by LVS during infection, we infected J774 cells with LVS or U112 expressing IglI-CyaA from pKEK1051 (4). In our hands, neither LVS nor U112 was able to secrete IglI-CyaA to any significant levels; in fact, most of the time secretion did not occur at all, irrespective of the MOI or time of infection (Fig. 5 and data not shown). We therefore constructed a new IglI-CyaA construct (pJEB851), based on the IglI derived from LVS (97.8% identical to F. tularensis subsp. novicida IglI). In contrast to pKEK1051, pJEB851 resulted in much higher levels of cAMP when it was expressed in LVS and somewhat elevated levels when it was expressed in U112. We therefore expressed this construct in the LVS, ΔiglA, ΔpdpE, ΔiglG, ΔvgrG, or ΔiglI strain to determine the contributions of IglA, PdpE, IglG, and VgrG to secretion of IglI. Surprisingly, in contrast to previous findings (4), IglI-CyaA was found to be secreted in all backgrounds tested, suggesting that in LVS, both VgrG and IglI secretion occurs by an FPI-independent mechanism.
Fig. 5.
Secretion of FPI protein-CyaA fusions into the cytosol of infected macrophages. J774 cells were infected with various Francisella strains expressing either IglG-CyaA, VgrG-CyaA, or IglI-CyaA. IglI was derived from either strain LVS or F. tularensis subsp. novicida (Fn), and intracellular cAMP levels were measured as described in Materials and Methods. The assay was performed in quadruplicate in four separate experiments, and a representative is shown. Shown are means and standard errors. The asterisks indicate that the cAMP levels were significantly different than those of LVS-infected cells (negative-control strain lacking CyaA construct), as determined by a 2-sided t test with equal variance (*, P ≤ 0.05; ***, P ≤ 0.001).
IglG and IglI are required for virulence.
The attenuated phenotypes observed for the ΔiglG and ΔiglI null mutants with respect to phagosomal escape, LDH release, and inhibition of TNF-α secretion may suggest that they are likely to be affected for virulence, while the ΔpdpE mutant is not. To test this, we infected C57BL/6 mice by the intradermal route. With an infection dose of 2 × 108 CFU (approximately 10 times the 50% lethal dose) (27), the LVS and ΔpdpE strains caused 100% mortality (mean times to death, 4.0 ± 0.0 days and 4.5 ± 1.3 days, respectively). In contrast, no mice died after infection with the ΔiglG or ΔiglI mutant (follow-up period of 20 days). Expression of IglG in trans resulted in 100% mortality (mean time to death, 4.2 ± 0.4 days), while 60% of the mice died after infection with the complemented ΔiglI mutant (mean time to death, 7 ± 2 days). A 4- to 8-fold increase in dose did not lead to killing of mice by the ΔiglG or ΔiglI mutant (data not shown), clearly demonstrating the requirement of IglG and IglI for the virulence of F. tularensis LVS.
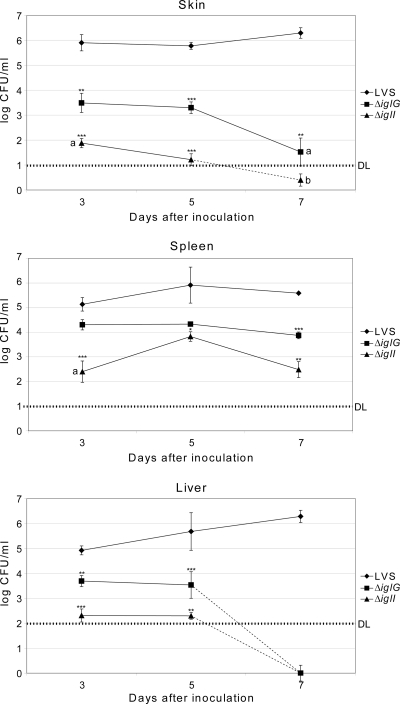
To get a better understanding of the in vivo phenotype of the ΔiglG or ΔiglI mutant, the bacterial numbers in skin, spleen, and liver after intradermal infection with 2 × 105 CFU of the LVS, ΔiglG, or ΔiglI strain were determined at days 3, 5, and 7 postinfection. For LVS-infected mice, the bacterial titers remained high in all three organs during the course of infection (Fig. 6). However, considerable killing of the ΔiglG mutant and even more killing of the ΔiglI mutant were observed in the skin, and as a consequence, the numbers of mutant bacteria recovered from spleen and liver were significantly lower (Fig. 6). As seen in previous studies (48, 70), comparable numbers of wild-type bacteria were recovered from spleen and liver at all time points (Fig. 6). Notably, however, the ΔiglG mutant was specifically killed within the liver, since counts on day 7 were >1,000-fold lower in liver than spleen. Thus, while the ΔiglG mutant efficiently persisted in visibly enlarged spleens up to day 7, it was detected in liver only up to day 5 (Fig. 6 and data not shown). Also, the ΔiglI mutant was able to colonize the spleen, but at much reduced numbers than the LVS and ΔiglG strains. Moreover, this organ showed no visible signs of infection, such as enlargement or discoloration (Fig. 6 and data not shown). ΔiglI mutant counts in liver were similar to those in spleen but barely above the detection limit (100 CFU) on days 3 and 5 and below the detection limit at day 7 postinfection (Fig. 6). Taken together, these results clearly demonstrate that both IglG and IglI are required for survival and hence optimal spread from the initial site of entry to cause systemic infections in mice.
Fig. 6.
Growth of F. tularensis strains in host tissues following intradermal infection. Mice inoculated intradermally with 2 × 105 CFU of the stated strain were killed at day 3, 5, or 7 postinfection, and bacterial burdens in skin spleen and liver were determined. The means ± SEMs for five mice per group and time point are shown. At day 7, only two out of five strain LVS-infected mice were alive and could be tested. A significant difference in the bacterial numbers of mutant strains versus LVS is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. At day 7, the numbers of bacteria in the liver of ΔiglG- or ΔiglI mutant-infected mice were below the detection level. a, bacteria were detected in only 4 out of 5 organs; b, bacteria were detected in only 2 out of 5 organs; DL, lower limit of detection.
DISCUSSION
The highly virulent bacterium F. tularensis is capable of intracellular growth within the cytosol of monocytic cells, which is preceded by its initial escape from the phagosome. The molecular mechanisms behind this intracellular lifestyle are most elusive but have been shown to require several genes of the Francisella pathogenicity island, most notably, the iglABCD operon genes (46, 52, 64). Evidence indicates that many of the FPI genes collectively constitute a T6SS; however, while such systems have been identified in nearly 100 different bacterial species to date, their homologies to the FPI system are weak, placing the F. tularensis T6SS in an evolutionarily distinct group (4, 5, 24). Two protein components, corresponding to IglA and IglB of F. tularensis, are conserved between all T6SSs described so far, and their importance for substrate secretion has been directly demonstrated for Vibrio cholerae, Pseudomonas aeruginosa, Edwardsiella tarda, and enteroaggregative E. coli (6, 25, 61, 78). Interestingly, of all the FPI proteins, IglA and IglB show the highest similarities to T6SS homologs of other bacteria. Although their exact functions are still unknown, we have shown that they physically interact and that this interaction involves a key α-helical region within IglA. Even single point mutations within this domain resulted in markedly impaired intramacrophage replication and loss of virulence in mice (9). A few other FPI gene products show modest homologies to conserved T6SS components, e.g., IcmF, VgrG, ClpV, DotU, and Hcp (4, 24); however, there is no evidence that this homology also extends to functional conservation. For example, IglF, the putative ClpV homolog, is missing the characteristic Walker A and B boxes, indicating that it lacks the ATPase activity essential for T6S in P. aeruginosa (54). Also, the Walker A box present in IcmF homologs is missing from PdpB (78). Additionally, the F. tularensis VgrG homolog is much smaller than cognate proteins in other species, e.g., V. cholerae, and the former also lacks the C-terminal enzymatic extensions found in many VgrG proteins (57). Thus, with the exception of IglA and IglB, there are no obvious deductions regarding the functions of the FPI proteins that can be drawn from other bacteria.
A hallmark of most T6SSs is the ability to secrete Hcp and VgrG, which also constitute essential components of the apparatus itself (59). However, there is yet no evidence that PdpE, the putative F. tularensis Hcp homolog, is secreted (4), nor has the effect of a pdpE in-frame deletion mutation on Francisella pathogenicity been determined to date. We therefore generated a ΔpdpE mutant of LVS. We also studied the ΔiglG and ΔiglI mutants, since the encoded gene products lack homology to known bacterial proteins and could therefore represent secreted substrates unique to Francisella. In none of the three mutants did we observe polar effects on downstream genes, nor did we observe any drastic alterations in the levels of transcription of other FPI genes. Thus, none of the three proteins appear to regulate expression of other FPI proteins. We found that PdpE, like IglG, localized to the outer membrane, while IglI was enriched in the inner membrane fraction but was also present in both the outer membrane and cytosolic fractions. Although the mechanism behind IglG and IglI export is elusive, an N-terminal secretion signal was predicted for PdpE, implying that it may be exported across the inner membrane by the Sec pathway. Importantly, supporting our finding of an outer membrane localization for PdpE, a dsbA mutation was recently shown to result in increased levels of PdpE in the membrane fraction of LVS (71).
We discovered that the ΔiglG mutant to some extent and more markedly the ΔiglI mutant showed delayed phagosomal escape in J774 cells. In particular, it was obvious at the 2-h time point postinfection, since at that time only 11% of LVS bacteria showed colocalization with LAMP-1, whereas the corresponding numbers for the ΔiglG and ΔiglI mutants were 44% and 71%, respectively. Using TEM, the percentages of bacteria contained within more or less intact phagosomes were at the same time point 12% (LVS), 36% (ΔiglG mutant), and 90% (ΔiglI mutant). Unexpectedly, this delay in phagosomal escape did not correlate with any marked deficiency in intracellular replication, at least not after 48 h. A previous study has shown that mutations within carA or carB render LVS unable to grow within primary macrophages, while it efficiently replicates in J774 cells (67). We therefore analyzed the ability of our mutants to grow within bone marrow-derived macrophages as well as peritoneal exudate cells, of which the latter exhibit a higher killing capacity than J774 (31). In none of these cells did the ΔiglI mutant show net growth. Of note, however, in PECs the ΔiglI mutant numbers stayed quite consistent over a time period of 48 h. This suggests that the ΔiglI mutant possesses some resistance to PEC killing, in contrast to the nonreplicating ΔiglA mutant, which showed a dramatic drop in numbers during the same time period. In contrast, the ΔiglG mutant readily replicated within both BMDMs and PECs. This appears to be the first demonstration of an FPI mutant with delayed phagosomal escape that does not lead to a marked defect in intracellular replication. The findings also imply that IglG and perhaps IglI, to some extent, play critical roles for the so far elusive mechanism of F. tularensis-mediated degradation of the phagosomal membrane.
Analogous to the delayed phagosomal escape, we also observed that the cytopathogenic response was very distinct between LVS- or ΔpdpE mutant-infected J774 cells, PECs, and BMDMs on one hand and ΔiglG mutant- and ΔiglI mutant-infected cells on the other hand, since the LVS and ΔpdpE mutant infections led to marked morphological effects on the cells and high levels of LDH release, while the ΔiglG mutant- and ΔiglI mutant-infected cells were essentially indistinguishable from uninfected cells at 24 h postinfection. However, over time, LDH release increased markedly for the ΔiglG mutant, while the ΔiglI mutant caused only minor release. Again, these findings demonstrate key roles for IglG and IglI in the interaction of F. tularensis with phagocytic cells. The loss of IglG or IglI also resulted in severe attenuation of LVS in mice, correlating with increased bacterial killing and diminished spread to internal organs. Intriguingly, the ΔiglG mutant was specifically susceptible to host killing when it was located within the liver, resulting in a rapid decrease of bacterial numbers within this organ compared to when it was located in the spleen. To our knowledge, this renders the ΔiglG mutant unique from any previously described Francisella mutants. This is also the first demonstration of an F. tularensis FPI mutant that behaves similarly to wild-type bacteria with regard to intramacrophage replication but still shows marked attenuation in vivo. Importantly, recent work on the role of PyrF in strains LVS and Schu S4 has suggested that replication in nonmacrophages could contribute to the pathogenesis of F. tularensis (35). We are currently pursuing the idea that IglG may possess a similar function in LVS.
Recently, it was demonstrated that VgrG and IglI of F. tularensis subsp. novicida were required for intramacrophage growth and virulence in mice and that CyaA fusion variants were secreted into the cytosol of macrophages (4). Though we failed to demonstrate secretion of IglG using the same CyaA-based approach, our results support the notion of an FPI-independent export of VgrG also in strain LVS, since it occurred in the absence of either PdpE, IglG, IglI, or IglA. We also demonstrated FPI-independent export of IglI during LVS infection, which contrasts to previous findings on IglI in F. tularensis subsp. novicida (4). Importantly, the latter study did show FPI-independent export of IglI under in vitro growth conditions, which might reconcile our different findings. So far, however, we have not been able to identify any condition under which FPI-dependent export of proteins occurs during in vitro growth of LVS. Thus, the real substrates of the putative T6SS of F. tularensis LVS still remain to be identified.
The lack of replication of the F. tularensis subsp. novicida ΔiglI::ermC mutant in J774 cells contrasted to our findings on the LVS ΔiglI mutant. Moreover, the F. tularensis subsp. novicida mutant was rapidly killed within PECs. This is intriguing and raises the issue of whether IglI possesses distinct functions in different subspecies of F. tularensis. Currently, there is no direct evidence for this, but the possibility cannot be dismissed, despite the very high degree of sequence conservation (97.9% identity), since it is possible that differences in gene regulation may exist between the subspecies. Importantly, we observed that LVS-derived IglI could restore the growth defect of the F. tularensis subsp. novicida mutant in macrophages, which indicates that the differences in growth behavior and resistance to macrophage killing displayed by the mutants must be due to fundamental differences in the two bacterial species and not due to the IglI protein itself. Contradicting this conclusion, however, is our observation that a CyaA fusion of LVS-derived IglI was more efficiently secreted than a fusion based on F. tularensis subsp. novicida IglI. A possible explanation for these differences may, however, be subtle differences in the cloning strategies that led to slightly different spacing between the Shine-Dalgarno sequence and the initiation codon (15) and possibly also iglI expression.
Inflammasomes are multiprotein molecular platforms that recruit and activate inflammatory caspase, such as caspase-1, upon stimulation. There are numerous studies demonstrating an absent in melanoma 2 (AIM2)-, apoptosis-associated speck-like protein (ASC)-, and pyrin-dependent activation of the inflammasome in F. tularensis subsp. novicida-infected cells, resulting in cleavage of caspase-1 and efficient IL-1β release (12, 22, 38, 52). Inefficient IL-1β secretion by the iglC mutant of LVS and F. tularensis subsp. novicida vgrG and iglI mutants has been established (4, 21). In accordance, we observed that PEC release of IL-1β was lower after ΔiglG or ΔiglI mutant infection than after strain LVS infection. Thus, there appears to be a strong correlation between phagosomal escape, intracellular replication, and inflammasome activation. It has been hypothesized that triggering of the AIM2 inflammasome occurs via release of DNA through spontaneous bacterial lysis (38). Indeed, a study on Listeria monocytogenes has provided direct evidence in support of this hypothesis (65). Moreover, recent publications on F. tularensis have demonstrated enhanced inflammatory properties of several mutants, many of which appear to have cell wall defects (42, 45, 75). Thus, a straightforward explanation for the correlation between phagosomal escape and inflammasome activation may be that an increased number of bacteria in the cytoplasm will lead to more lysed bacteria as well.
When the prerequisites for IL-1β secretion from F. tularensis subsp. novicida-infected cells have been investigated, it has consistently been found to be caspase-1 dependent. However, there are fewer data available on the mechanisms of strain LVS-induced IL-1β secretion, and in several instances it has been stated that secretion occurs only after priming with endotoxin (28, 75) and that the activation state of the infected cell likely affects the levels produced (37). Thus, there appear to be clear distinctions between LVS and F. tularensis subsp. novicida with regard to their ability to trigger IL-1β secretion. In agreement with a previous publication (21), we observed that LVS infection resulted in secretion of IL-1β from PECs, even in the absence of priming with endotoxin, although levels were much lower than those for F. tularensis subsp. novicida (28, 52). Also, the LVS and ΔpdpE strains induced higher levels in PECs than in BMDMs, whereas the opposite occurred after infection with the ΔiglG or ΔiglI mutant. Thus, the higher activation status of PECs did not lead to consistently higher levels of IL-1β secretion. Moreover, we observed efficient inhibition of F. tularensis subsp. novicida-induced IL-1β secretion upon treatment with a caspase-1 inhibitor, whereas the effect on the LVS-induced secretion was absent, again emphasizing the distinct characteristics of the two infections. Li et al. (43) and Huang et al. (37) demonstrated that a caspase-1 inhibitor conferred a reduction in LVS-induced IL-1β secretion; however, both studies employed human cells, so those results cannot be directly compared to our results.
Previously, we and others have demonstrated that an LVS infection renders the infected cells unable to respond to Toll-like receptor 2 (TLR2) or TLR4 stimuli, such as bacterial lipoprotein or E. coli LPS (1, 74). To this end, we investigated the effects of infection with the LVS mutants on the E. coli LPS-induced TNF-α secretion, an inflammasome-independent event. We found that there was a close relationship between the mitigation of the LPS-induced inflammatory response and the subcellular localization of F. tularensis. This indicates that the degree of perturbation of the phagosomal membrane is somehow linked to the capability of the infected cells to appropriately respond to TLR stimuli. It is possible that the escape mechanism is linked to manipulation of intracellular signaling pathways required for the normal inflammatory response. Indeed, recent studies by Cole et al. have shown that retention of bacteria in the phagosome, achieved by deletion of iglC, enhances TLR2-dependent gene expression, possibly by sustaining the interaction of Francisella with TLR2 from within the phagosome (20, 21). Altogether, the data show that a cytosolic localization of Francisella is a prerequisite for inducing release of IL-1β and mitigation of LPS-induced TNF-α secretion.
Although a ΔpdpE mutant infection of PECs resulted in a somewhat reduced cytopathogenic response compared to what was observed for macrophages infected with strain LVS, our data collectively indicate that PdpE has no direct function that affects the phagosomal escape, intramacrophage replication, or virulence of F. tularensis LVS. Further work will be required to determine if more subtle functions can be attributed to the protein. The results do not support any functional resemblance between PdpE and Hcp homologs, most likely due to the weak similarities between the proteins. Moreover, while the Hcp homolog of P. aeruginosa has been shown to assemble into tubular structures by the stacking of hexameric Hcp rings (3, 54), we have failed to detect homodimer formation indicative of multimerization for PdpE using a Saccharomyces cerevisiae two-hybrid system, while VgrG homodimerization readily occurred (M. Lavander, unpublished results).
On the other hand, our data indicate that both IglG and IglI play key roles in modulating the interaction between F. tularensis and the phagocytic cell. Both mutants showed unique phenotypes characterized by a minor cytopathogenic effect, delayed phagosomal escape, effective intramacrophage growth (at least for the ΔiglG mutant), impaired modulation of the macrophage inflammatory response, and marked attenuation in vivo. While IglG may perform its critical function at the surface of the bacterium, the scattered distribution of IglI could indicate multiple roles both within the bacterium and perhaps as a secreted substrate. Nevertheless, the elusive mechanisms behind FPI-independent export of IglI, as well as VgrG, clearly need more thorough investigation. Further elucidation of IglG and IglI functions will likely yield critical information to the understanding of the enigmatic mechanisms behind the intracellular lifestyle of F. tularensis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants 2006-3426 (to J.E.B.) and 2006-2877 and 2009-5026 (to A.S.) from the Swedish Research Council and a grant from the Medical Faculty, Umeå University, Umeå, Sweden. The work was performed in part at the Umeå Centre for Microbial Research (UCMR).
We are indebted to Karl Klose for the kind gift of the F. tularensis subsp. novicida iglI mutant as well as the VgrG-CyaA and IglI-Cya expression constructs. We also thank Rafael Björk and Monica Normark for valuable help with statistics, Lena Lindgren for technical assistance with the mouse infections, and Igor Golovliov and Nelson O. Gekara for kindly providing us with PECs and BMDMs. We are also indebted to Lenore Johansson for technical assistance with the transmission electron microscopy.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Present address: Livsmedelsverket, SE-751 26 Uppsala, Sweden.
Published ahead of print on 20 June 2011.
REFERENCES
- 1. Abplanalp A. L., Morris I. R., Parida B. K., Teale J. M., Berton M. T. 2009. TLR-dependent control of Francisella tularensis infection and host inflammatory responses. PLoS One 4: e7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anthony L. D., Burke R. D., Nano F. E. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59: 3291–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ballister E. R., Lai A. H., Zuckermann R. N., Cheng Y., Mougous J. D. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U. S. A. 105: 3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barker J. R., et al. 2009. The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74: 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bingle L. E., Bailey C. M., Pallen M. J. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11: 3–8 [DOI] [PubMed] [Google Scholar]
- 6. Bönemann G., Pietrosiuk A., Diemand A., Zentgraf H., Mogk A. 2009. Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28: 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bönquist L., Lindgren H., Golovliov I., Guina T., Sjöstedt A. 2008. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76: 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosio C. M., Dow S. W. 2005. Francisella tularensis induces aberrant activation of pulmonary dendritic cells. J. Immunol. 175: 6792–6801 [DOI] [PubMed] [Google Scholar]
- 9. Bröms J. E., Lavander M., Sjöstedt A. 2009. A conserved α-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191: 2431–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bröms J. E., Lavander M., Sjöstedt A. 2010. The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signalling. Frontiers Microbiol. 1: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brotcke A., et al. 2006. Identification of MglA-regulated genes reveals novel virulence factors in Francisella tularensis. Infect. Immun. 74: 6642–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broz P., von Moltke J., Jones J. W., Vance R. E., Monack D. M. 2010. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8: 471–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chamberlain R. E. 1965. Evaluation of live tularemia vaccine prepared in a chemically defined medium. Appl. Microbiol. 13: 232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Checroun C., Wehrly T. D., Fischer E. R., Hayes S. F., Celli J. 2006. Autophagy-mediated reentry of Francisella tularensis into the endocytic compartment after cytoplasmic replication. Proc. Natl. Acad. Sci. U. S. A. 103: 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen H., Bjerknes M., Kumar R., Jay E. 1994. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 22: 4953–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chong A., Celli J. 2010. The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Frontiers Microbiol. 1: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chong A., et al. 2008. The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect. Immun. 76: 5488–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chow J., Mazmanian S. K. 2010. A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7: 265–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clemens D. L., Lee B. Y., Horwitz M. A. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72: 3204–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cole L. E., et al. 2009. Phagosomal retention of Francisella tularensis results in TIRAP/Mal-independent TLR2 signaling. J. Leukoc. Biol. 87: 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole L. E., et al. 2008. Macrophage proinflammatory response to Francisella tularensis live vaccine strain requires coordination of multiple signaling pathways. J. Immunol. 180: 6885–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cremer T. J., Amer A., Tridandapani S., Butchar J. P. 2009. Francisella tularensis regulates autophagy-related host cell signaling pathways. Autophagy 5: 125–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cremer T. J., et al. 2009. MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response. PLoS One 4: e8508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Bruin O. M., Ludu J. S., Nano F. E. 2007. The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dudley E. G., Thomson N. R., Parkhill J., Morin N. P., Nataro J. P. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61: 1267–1282 [DOI] [PubMed] [Google Scholar]
- 26. Ellis J., Oyston P. C., Green M., Titball R. W. 2002. Tularemia. Clin. Microbiol. Rev. 15: 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Forslund A. L., et al. 2006. Direct repeat-mediated deletion of a type IV pilin gene results in major virulence attenuation of Francisella tularensis. Mol. Microbiol. 59: 1818–1830 [DOI] [PubMed] [Google Scholar]
- 28. Gavrilin M. A., et al. 2006. Internalization and phagosome escape required for Francisella to induce human monocyte IL-1beta processing and release. Proc. Natl. Acad. Sci. U. S. A. 103: 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gavrilin M. A., Wewers M. D. 2011. Francisella recognition by inflammasomes: differences between mice and men. Frontiers Microbiol. 2: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Golovliov I., Baranov V., Krocova Z., Kovarova H., Sjöstedt A. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71: 5940–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Golovliov I., Sjöstedt A., Mokrievich A., Pavlov V. 2003. A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222: 273–280 [DOI] [PubMed] [Google Scholar]
- 32. Hazlett K. R., et al. 2008. Adaptation of Francisella tularensis to the mammalian environment is governed by cues which can be mimicked in vitro. Infect. Immun. 76: 4479–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hilbi H., Chen Y., Thirumalai K., Zychlinsky A. 1997. The interleukin 1beta-converting enzyme, caspase 1, is activated during Shigella flexneri-induced apoptosis in human monocyte-derived macrophages. Infect. Immun. 65: 5165–5170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hornung V., Latz E. 2010. Critical functions of priming and lysosomal damage for NLRP3 activation. Eur. J. Immunol. 40: 620–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Horzempa J., O'Dee D. M., Shanks R. M., Nau G. J. 2010. Francisella tularensis DeltapyrF mutants show that replication in nonmacrophages is sufficient for pathogenesis in vivo. Infect. Immun. 78: 2607–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hrstka R., et al. 2007. Francisella tularensis strain LVS resides in MHC II-positive autophagic vacuoles in macrophages. Folia Microbiol. (Praha) 52: 631–636 [DOI] [PubMed] [Google Scholar]
- 37. Huang M. T., et al. 2010. Deletion of ripA alleviates suppression of the inflammasome and MAPK by Francisella tularensis. J. Immunol. 185: 5476–5485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jones J. W., et al. 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. U. S. A. 107: 9771–9776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keim P., Johansson A., Wagner D. M. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann. N. Y. Acad. Sci. 1105: 30–66 [DOI] [PubMed] [Google Scholar]
- 40. Khlebnikov V. S., et al. 1996. Outer membranes of a lipopolysaccharide-protein complex (LPS-17 kDa protein) as chemical tularemia vaccines. FEMS Immunol. Med. Microbiol. 13: 227–233 [DOI] [PubMed] [Google Scholar]
- 41. Lai X. H., Golovliov I., Sjöstedt A. 2001. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69: 4691–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lai X. H., et al. 2010. Mutations of Francisella novicida that alter the mechanism of its phagocytosis by murine macrophages. PLoS One 5: e11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H., Nookala S., Bina X. R., Bina J. E., Re F. 2006. Innate immune response to Francisella tularensis is mediated by TLR2 and caspase-1 activation. J. Leukoc. Biol. 80: 766–773 [DOI] [PubMed] [Google Scholar]
- 44. Lilo S., Zheng Y., Bliska J. B. 2008. Caspase-1 activation in macrophages infected with Yersinia pestis KIM requires the type III secretion system effector Yop J. Infect. Immun. 76: 3911–3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lindemann S. R., et al. 2011. Francisella tularensis Schu S4 O-antigen and capsule biosynthesis gene mutants induce early cell death in human macrophages. Infect. Immun. 79: 581–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lindgren H., et al. 2004. Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53: 953–958 [DOI] [PubMed] [Google Scholar]
- 47. Lindgren H., Stenman L., Tärnvik A., Sjöstedt A. 2005. The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7: 467–475 [DOI] [PubMed] [Google Scholar]
- 48. Lindgren H., Stenmark S., Chen W., Tärnvik A., Sjöstedt A. 2004. Distinct roles of reactive nitrogen and oxygen species to control infection with the facultative intracellular bacterium Francisella tularensis. Infect. Immun. 72: 7172–7182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- 50. Ludu J. S., et al. 2008. The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J. Bacteriol. 190: 4584–4595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maier T. M., et al. 2007. Identification of Francisella tularensis Himar1-based transposon mutants defective for replication in macrophages. Infect. Immun. 75: 5376–5389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mariathasan S., Weiss D. S., Dixit V. M., Monack D. M. 2005. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 202: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Milton D. L., O'Toole R., Hörstedt P., Wolf-Watz H. 1996. Flagellin A is essential for the virulence of Vibrio anguillarum. J. Bacteriol. 178: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mougous J. D., et al. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312: 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Obregon C., et al. 2003. Human alveolar macrophages infected by virulent bacteria expressing SipB are a major source of active interleukin-18. Infect. Immun. 71: 4382–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oyston P. C., Sjöstedt A., Titball R. W. 2004. Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2: 967–978 [DOI] [PubMed] [Google Scholar]
- 57. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104: 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pukatzki S., et al. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103: 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pukatzki S., McAuley S. B., Miyata S. T. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12: 11–17 [DOI] [PubMed] [Google Scholar]
- 60. Rajaram M. V., et al. 2006. Akt/protein kinase B modulates macrophage inflammatory response to Francisella infection and confers a survival advantage in mice. J. Immunol. 177: 6317–6324 [DOI] [PubMed] [Google Scholar]
- 61. Rao P. S., Yamada Y., Tan Y. P., Leung K. Y. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53: 573–586 [DOI] [PubMed] [Google Scholar]
- 62. Santic M., Al-Khodor S., Abu Kwaik Y. 2010. Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12: 129–139 [DOI] [PubMed] [Google Scholar]
- 63. Santic M., et al. 2007. A Francisella tularensis pathogenicity island protein essential for bacterial proliferation within the host cell cytosol. Cell. Microbiol. 9: 2391–2403 [DOI] [PubMed] [Google Scholar]
- 64. Santic M., Molmeret M., Klose K. E., Jones S., Kwaik Y. A. 2005. The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7: 969–979 [DOI] [PubMed] [Google Scholar]
- 65. Sauer J. D., et al. 2010. Listeria monocytogenes triggers AIM2-mediated pyroptosis upon infrequent bacteriolysis in the macrophage cytosol. Cell Host Microbe 7: 412–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schell M. A., et al. 2007. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol. Microbiol. 64: 1466–1485 [DOI] [PubMed] [Google Scholar]
- 67. Schulert G. S., et al. 2009. Francisella tularensis genes required for inhibition of the neutrophil respiratory burst and intramacrophage growth identified by random transposon mutagenesis of strain LVS. Infect. Immun. 77: 1324–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sjöstedt A. 2006. Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8: 561–567 [DOI] [PubMed] [Google Scholar]
- 69. Sjöstedt A., Tärnvik A., Sandström G. 1991. The T-cell-stimulating 17-kilodalton protein of Francisella tularensis LVS is a lipoprotein. Infect. Immun. 59: 3163–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stenmark S., Lindgren H., Tärnvik A., Sjöstedt A. 2003. Specific antibodies contribute to the host protection against strains of Francisella tularensis subspecies holarctica. Microb. Pathog. 35: 73–80 [DOI] [PubMed] [Google Scholar]
- 71. Straskova A., et al. 2009. Proteome analysis of an attenuated Francisella tularensis dsbA mutant: identification of potential DsbA substrate proteins. J. Proteome Res. 8: 5336–5346 [DOI] [PubMed] [Google Scholar]
- 72. Suarez G., et al. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44: 344–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Telepnev M., Golovliov I., Grundström T., Tärnvik A., Sjöstedt A. 2003. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 5: 41–51 [DOI] [PubMed] [Google Scholar]
- 74. Telepnev M., Golovliov I., Sjöstedt A. 2005. Francisella tularensis LVS initially activates but subsequently down-regulates intracellular signaling and cytokine secretion in mouse monocytic and human peripheral blood mononuclear cells. Microb. Pathog. 38: 239–247 [DOI] [PubMed] [Google Scholar]
- 75. Ulland T. K., et al. 2010. Cutting edge: mutation of Francisella tularensis mviN leads to increased macrophage absent in melanoma 2 inflammasome activation and a loss of virulence. J. Immunol. 185: 2670–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weiss D. S., et al. 2007. In vivo negative selection screen identifies genes required for Francisella virulence. Proc. Natl. Acad. Sci. U. S. A. 104: 6037–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wickstrum J. R., et al. 2009. Francisella tularensis induces extensive caspase-3 activation and apoptotic cell death in the tissues of infected mice. Infect. Immun. 77: 4827–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zheng J., Leung K. Y. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66: 1192–1206 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.