Abstract
Stress granules (SG) are cytoplasmic aggregates of stalled translation preinitiation complexes that form in cells exposed to various environmental stresses. Here, we show that stress granules assemble in cells infected with Theiler's murine encephalomyelitis virus (TMEV) mutants carrying alterations in the leader (L) protein, but not in cells infected with wild-type TMEV. Stress granules also formed in STAT1-deficient cells, suggesting that SG formation was not a consequence of increased type I interferon (IFN) production when cells were infected with the mutant virus. Ectopic expression of the wild-type L protein was sufficient to inhibit stress granule formation induced by sodium arsenite or thapsigargin treatment. In conclusion, TMEV infection induces stress granule assembly, but this process is inhibited by the L protein. Unlike poliovirus-induced stress granules, TMEV-induced stress granules did not contain the nuclear protein Sam68 but contained polypyrimidine tract binding protein (PTB), an internal ribosome entry site (IRES)-interacting protein. Moreover, G3BP was not degraded and was found in SG after TMEV infection, suggesting that SG content could be virus specific. Despite the colocalization of PTB with SG and the known interaction of PTB with viral RNA, in situ hybridization and immunofluorescence assays failed to detect viral RNA trapped in infection-induced SG. Recombinant Theiler's viruses expressing the L protein of Saffold virus 2 (SAFV-2), a closely related human theilovirus, or the L protein of mengovirus, an encephalomyocarditis virus (EMCV) strain, also inhibited infection-induced stress granule assembly, suggesting that stress granule antagonism is a common feature of cardiovirus L proteins.
INTRODUCTION
Theiler's murine encephalomyelitis virus (TMEV) belongs to the Cardiovirus genus, within the picornavirus family. Other cardioviruses are Saffold virus (SAFV), a recently described human virus closely related to TMEV, and encephalomyocarditis virus (EMCV). The genomes of these viruses are composed of nonsegmented positive-stranded RNA molecules of approximately 8 kb. During infection, these viruses produce a short protein cleaved from the amino-terminal end of the viral polyprotein and therefore called leader (L) protein (21).
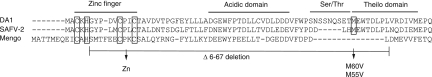
The DA strain of TMEV is responsible for persistent infection of the central nervous system of the mouse, leading to chronic demyelinating lesions reminiscent of those found in multiple sclerosis (5, 22). Disruption of the host immune response is critical for the establishment of viral persistence, and the L protein of the virus plays a crucial role in this process. Cardiovirus L proteins are closely related multifunctional proteins shown to interfere with critical cellular processes such as interferon (IFN) and chemokine production (16, 32), nucleocytoplasmic trafficking (4, 8, 17, 27, 30), apoptosis (12, 31), and mitogen-activated protein (MAP) kinase activity (28). Cardiovirus L proteins are very small proteins (about 70 amino acids) in which several domains have been described (Fig. 1): (i) an amino-terminal zinc finger, (ii) a glutamate/aspartate-rich domain, which confers a very acidic character to the protein (pI about 3.8), (iii) a serine/threonine-rich domain, and (iv) a carboxy-terminal domain, present in the L proteins of TMEV and SAFV (theilovirus species) but lacking in EMCV L and therefore called the Theilo domain. Mutations introduced either in the zinc finger or in the Theilo domain strongly inhibit all the known activities of the TMEV L protein (29).
Fig. 1.
Alignment of L protein sequences used in this work. Shown are the sequences of L proteins from TMEV (strain DA1), SAFV-2, and mengovirus. Protein domains are indicated. Cysteine and histidine residues forming the Zn finger, and the M60/M55 residue of the Theilo domain, are framed. L mutations are indicated under the alignment.
Polypyrimidine tract binding protein (PTB) is a RNA-binding protein involved in splicing regulation and 3′ end processing of RNA molecules. In TMEV-infected cells, PTB was found to relocalize from the nucleus to the cytoplasm (8), where it is believed to interact with the internal ribosome entry site (IRES) of viral RNA (24). Binding of PTB to the IRES is thought to stabilize the RNA structure, thereby promoting translation of viral proteins. Such translation-modulating activity of PTB has been confirmed for the GDVII TMEV strain (25).
Stress granules (SG) are cytoplasmic foci appearing in cells exposed to various environmental stresses, such as UV exposure, oxidative stress, hypoxia, and heat shock. They are mainly composed of stalled translation preinitiation complexes that aggregate in response to stress (3). The best-known event triggering SG assembly is the phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α). eIF2α phosphorylation can be mediated by four kinases activated in response to different stress signals: PKR, activated by cytosolic double-stranded RNA; PERK, activated by endoplasmic reticulum stress; GCN2, activated by amino acid starvation; and HRI, activated by oxidative stress or heme depletion (9). Phosphorylation of eIF2α leads to translational arrest and to the assembly of incomplete translation preinitiation complexes (lacking eIF2) on the caps of mRNA molecules. In this process, TIA-1, a nuclear prion-like protein, delocalizes from the nucleus to the cytoplasm and binds the eIF2-deficient preinitiation complexes, inducing their aggregation and thereby assembly of SG (13). SG are thought to play a role in translation inhibition during stress, through sequestration of cellular mRNA. When cells recover from stress, SG disperse and mRNA can be sent back to effective translation or targeted to processing bodies (P-bodies). P-bodies are cytosolic complexes containing enzymes for decapping and degradation of mRNA, as well as proteins involved in small interfering RNA (siRNA)-mediated repression of mRNA translation. Therefore, P-bodies are thought to be a place where mRNA is degraded (2). Factors regulating the fate of sequestered mRNA during recovery are incompletely known.
In recent years, several viruses have been shown to induce a stress granule response in host cells. Depending on the virus, the response can be pro- or antiviral. For example, respiratory syncytial virus (RSV) preferentially replicates in host cells forming stress granules (18). Conversely, poliovirus inhibits both P-body and SG formation (11, 33). Poliovirus 3C protease has been shown to block infection-induced SG formation by cleaving G3BP, a host cell protein involved in the SG assembly process. Expression of a noncleavable form of G3BP in host cells significantly lowered poliovirus replication (33).
Some cellular proteins, such as eIF3, TIA-1, and G3BP, are concentrated in SG and are thus used as ubiquitous SG markers. However, certain proteins are associated only with a particular type of SG. For example, Sam68, a nuclear factor involved in splicing regulation, was reported to be associated with poliovirus-induced SG, but not with oxidative stress or heat shock-induced SG (26).
We previously observed a stress granule-like cytoplasmic distribution of PTB in cells infected with TMEV expressing a mutated L protein, but not in cells infected with a wild-type (wt) virus. This observation led us to investigate the relationship between TMEV infection and the stress granule response.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
HeLa, U3A 2FTGH (19) (kindly provided by Ian Kerr), and L929 cells were maintained in Dulbecco's modified Eagle medium (Lonza) supplemented with 10% fetal calf serum (MP Biomedicals), 100 IU penicillin/ml, and 100 μg streptomycin/ml.
Viruses used in this study (Table 1) were derivatives of the DA1 molecular clone of the persistent Daniels (DA) TMEV strain (7, 20). L-mutant viruses derived from DA1 were TM564 carrying a deletion encompassing codons 6 to 67 of the L region (LΔ6–67), TM598 carrying mutations disrupting the L zinc finger (LZn) (32), and FB05 carrying a M60V substitution in the L Theilo domain (LM60V) (29).
Table 1.
Plasmids carrying full-length viral genomes
| Plasmid | Virus | L proteina | Virus background, characteristics |
|---|---|---|---|
| pTMDA1 | DA1 | Lwt | Molecular clone of TMEV DA strain |
| pTM564 | TM564 | LΔ6–67 | DA1 |
| pTM598 | TM598 | LZn | DA1 |
| pFB05 | FB05 | LM60V | DA1 |
| pKJ6 | KJ6 | Lwt | DA1, capsid adapted to infect L929 cells |
| pSB3 | SB3 | LΔ6–67 | KJ6, capsid adapted to infect L929 cells |
| pTM659 | TM659 | LZn | KJ6, capsid adapted to infect L929 cells |
| pFB09 | FB09 | LM60V | KJ6, capsid adapted to infect L929 cells |
| pFB26 | FB26 | SAFV-2 Lwt | KJ6, capsid adapted to infect L929 cells |
| pFB27 | FB27 | SAFV-2 LM55V | KJ6, capsid adapted to infect L929 cells |
| pSPA24 | SPA24 | Mengovirus Lwt | KJ6, capsid adapted to infect L929 cells |
| pSPA28 | SPA28 | Mengovirus LZn | KJ6, capsid adapted to infect L929 cells |
Unless specified, the leader protein was that of TMEV DA1.
For infection of L929 and U3A cells, we used virus KJ6 and L mutants thereof. KJ6 is a DA1 derivative carrying capsid mutations enabling the virus to infect L929 cells with high efficiency (15). This virus is further referred to as wild type in this work as it expresses a wild-type L protein (Lwt). L mutant KJ6 derivatives were SB3 (LΔ6–67), TM659 (LZn) (32), and FB09 (LM60V) (29). KJ6 derivatives expressing the wild-type L protein of mengovirus (SPA24) and its corresponding zinc finger mutant (SPA28) were described previously (23). FB26 and FB27 are KJ6 derivatives expressing the wild-type L protein and an M55V mutant of the L protein from Saffold virus 2 (SAFV-2; GenBank accession no. AM922293), respectively. These viruses were constructed as follows. The LSAFV-2 coding sequence was subcloned as a BamHI-XbaI restriction fragment from a synthetic plasmid (ordered to MrGene) in pBS-KS+, giving pFB16. This fragment was also cloned in pTM624, a pcDNA3 derivative containing the TMEV IRES followed by the enhanced green fluorescent protein (eGFP) coding sequence, giving pFB14. Plasmid pFB14 thus allows expression of a bicistonic LSAFV-2-IRES-eGFP construct from the cytomegalovirus (CMV) immediate-early promoter. The M55V mutation was introduced in LSAFV-2 by PCR amplification of the L region of pFB16 with primers TM952 (5′AAA GGA TCC GCC ACC ATG GCG TGC) and TM956 (5′ GGT AGA TCT GTC CAT TCC ACA TGG AGG TCA TCA GGA TAA). TM956 encodes the M55V mutation (A163TG→GTG). The resulting PCR product was used to replace the corresponding NcoI-BglII restriction fragment in pFB16 to yield pFB18. To construct TMEV cDNA clones expressing the SAFV-2 L protein, the mutated coding region of the LSAFV-2 gene was then transferred back from pFB18 to pFB14 as a BamHI-XbaI restriction fragment. The resulting plasmid was called pFB19. The wild-type and mutated LSAFV-2 gene coding sequences were then PCR amplified from pFB14 and pFB19 with primers TM952 and TM954 (5′ AAA CCT GAG GAC TGG GAG TTA CTC TTG TCA GAT GAA GAG GCG TTT CCT TGT GGT TCC ATT TCA ATG TC). The resulting PCR products were cloned into pTM565 (a subclone of pTMDA1) as NcoI-Bsu36I restriction fragments to yield pFB20 and pFB21. Finally, the XbaI-MscI fragment carrying the L gene coding region was extracted from these plasmids and used to replace the corresponding fragment in pKJ6. The resulting plasmids, pFB26 and pFB27, carry the full-length cDNA of KJ6 derivatives coding for the wild-type and M55V mutant SAFV-2 L proteins, respectively. Synthetic and PCR-amplified regions of all constructs were sequenced to check that no unexpected mutation occurred during the cloning steps. Plasmid constructs carrying full-length virus cDNA and the corresponding viruses are presented in Table 1.
All wild-type and mutant viruses were produced from the corresponding cDNA clones, as described previously (20). Viruses were collected 48 to 72 h after electroporation of BHK-21 cells with in vitro-transcribed viral RNA. Viruses were titrated in parallel by standard plaque assay in BHK-21 cells.
Bicistronic constructs expressing IRES-eGFP alone (pTM624), LTMEVwt-IRES-eGFP (pTM625), or LTMEVM60V-IRES-eGFP (pCER48) were described previously (30).
Immunostaining and in situ hybridization.
Immunostainings and in situ hybridizations were performed on cells cultivated on glass coverslips treated with poly-l-lysine and placed in 24-well plates. Prior to immunostaining, cells were fixed for 4 to 10 min in 300 μl of phosphate-buffered saline (PBS)–4% paraformaldehyde (PFA). Cells were then washed in 500 μl of PBS and permeabilized for 5 min at room temperature in 500 μl of PBS–0.1% Triton X-100. Blocking occurred for 1 h at room temperature in 300 μl of TNB blocking reagent (Perkin Elmer). Cells were next incubated with the primary antibody diluted in TNB at the following dilutions: PTB (mouse; Zymed 32-4800), 1/50; VP1 (mouse; F12B3 clone; kind gift from M. Brahic), 1/10; eIF3 (goat; Santa Cruz sc-16377), 1/200; TIA-1 (goat; Santa Cruz sc-1751 C20), 1/100; K1 or J2 (mouse, anti-double-stranded RNA [anti-dsRNA], English & Scientific Consulting Bt.), 1/200; Sam68 (rabbit; Santa Cruz sc-333; C20), 1/100; G3BP (mouse; BD Transduction Laboratories 611126), 1/500. After 1 h of incubation at room temperature, cells were washed 3 times for 5 min in 500 μl PBS–0.1% Tween 20. Secondary antibodies (Alexa Fluor 488- or 594-conjugated antibodies; Invitrogen) were incubated for 1 h at a 1/800 dilution in TNB. Finally, cells were washed 3 times in 500 μl PBS–0.1% Tween 20 and mounted with Mowiol for fluorescence microscopy.
Experiments involving in situ hybridization coupled to eIF3 immunostaining were performed according to a protocol adapted from that of Chakraborty et al. (6) using a 3′ biotinylated DNA probe (Eurogentec) complementary to the positive strand of viral RNA (5′ AGG GGT GCC TTT TCT TTC CAG GTG AGC CAT ATT CGG GAG AAA ATT). All reagents were prepared in 0.5% diethyl pyrocarbonate (DEPC)-treated water or PBS. Cells were fixed for 8 min in 500 μl of PBS–4% PFA prior to in situ hybridization. After 3 washes in 500 μl PBS, cells were permeabilized for 5 min at 4°C in 500 μl PBS–0.1% Triton X-100. Fifty microliters of prehybridization solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] containing 1 mg/ml of Escherichia coli tRNA [Roche 109 541], 10% dextran sulfate [Sigma D-6001], and 25% formamide) was then carefully pipetted over the cells on the coverslips, which were in a humidifying chamber. After incubations of 15 min at room temperature and 15 min at 42°C, the coverslips were drained on a piece of Tork paper and flipped over a 50-μl drop of hybridization solution (prehybridization solution containing 100 μg/ml of the biotinylated DNA probe) in the humidifying chamber at 42°C. After one night, the cells were washed two times in 2× SSC and one time in 0.5× SSC at 42°C for 15 min. Cells were then fixed for 8 min in PBS–4% PFA and washed 3 times in PBS. Coverslips were then turned on a drop of PBS containing Cy3-conjugated streptavidin (Sigma) diluted at 1/100 and anti-eIF3 antibody diluted at 1/200. After 1 h, the cells were washed two times in PBS–0.2% Triton X-100 and incubated with the secondary antibody (chicken anti-goat IgG–Alexa 488) diluted at 1/400 in PBS. Cells were washed twice for 15 min in PBS–0.2% Triton X-100 and twice for 15 min in PBS before being mounted with Mowiol for microscopy.
Fluorescence microscopy was performed with a DMIRB inverted microscope (Leica) equipped with a DC200 digital camera (Leica), a microscope (Zeiss) equipped for confocal microscopy (Bio-Rad; MRC-1024), or a spinning disk confocal microscope (Zeiss). Intensity, contrast, and color balance of images were equilibrated using ImageJ or Adobe Photoshop.
Plasmid transfection.
Transfection of plasmid DNA was performed on cells grown on poly-l-lysine-treated coverslips placed in 24-well plates. Cells were plated the day before transfection at a density of 6 × 104 cells per well. TransIT-LT1 (Mirus) was used as the transfection reagent, with a DNA/transfection reagent ratio of 1 μg/3 μl, according to the manufacturer's recommendations.
Immunoblotting.
Protein extracts were run on Tris-glycine-sodium dodecyl sulfate–8 or 10% polyacrylamide gels and transferred on polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore). Primary antibodies were anti-VP1 (F12B3 monoclonal antibody; kindly provided by M. Brahic) and anti-G3BP (reference no. 611126; BD Transduction Laboratories).
RESULTS
TMEV infection induces SG assembly, but the process is inhibited by the leader protein.
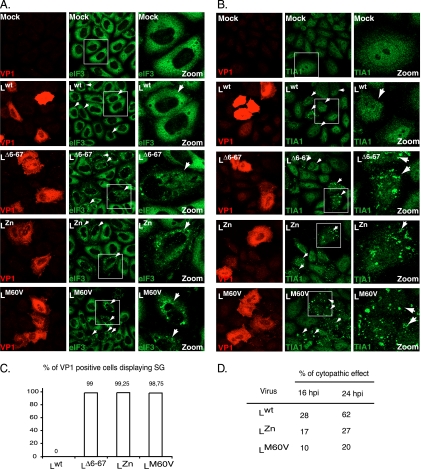
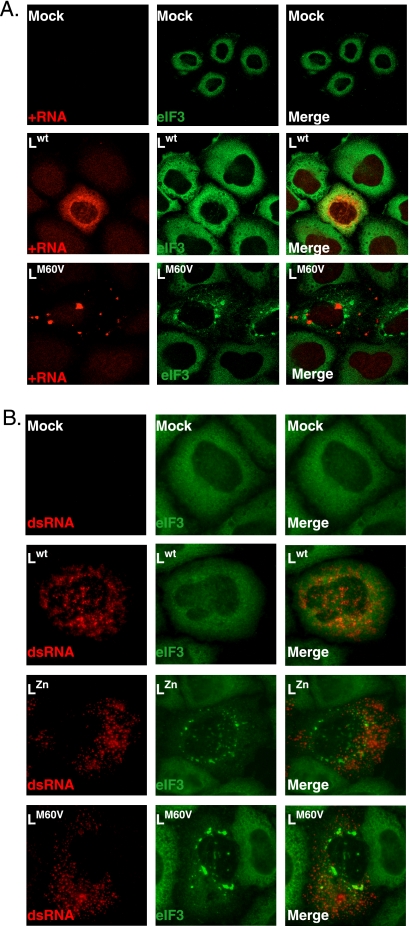
We analyzed whether stress granules (SG) are formed during infection of cells with TMEV. Therefore, HeLa cells were infected with the wild-type DA1 strain of TMEV or with L-mutant viruses carrying a deletion encompassing residues 6 to 67 of the L region (LΔ6–67), mutations in the Zn finger domain of L (LZn), or an M60V substitution in the Theilo domain of L (LM60V). Twelve and 16 h postinfection, cells were immunostained for viral capsid antigen VP1 and for eIF3, used as an SG marker. As shown in Fig. 2A and C, eIF3 was distributed homogeneously in the cytoplasm of mock-infected cells or of cells infected with the wild-type virus. In contrast, granular cytoplasmic aggregates of eIF3, evoking SG, were clearly visible in cells infected with the three L-mutant viruses. Identical results were obtained after immunostaining of TIA-1, another SG marker (Fig. 2B). In time course experiments, SG appeared from 8 h postinfection in HeLa cells infected with the L-mutant viruses and did not disappear before the development of cytopathic effect (CPE), around 24 h postinfection (Fig. 2D). In the case of the wild-type virus, SG were not detected in infected cells at any time point. In contrast to what was observed for poliovirus (26), we did not detect transient SG assembly early after infection with either the wild-type virus or mutant viruses (data not shown). The lack of SG in cells infected with the wild-type virus was not a consequence of faster CPE occurrence for this virus since SG were detected from 8 h postinfection, long before CPE development with either virus (Fig. 2D).
Fig. 2.
TMEV infection induces stress granule assembly. (A) HeLa cells were infected with 10 PFU/cell of wild-type TMEV (Lwt) or with LΔ6–67, LZn, or LM60V mutant virus. Twelve hours postinfection cells were fixed, coimmunostained for the VP1 capsid viral antigen (red) and eIF3 (green), and examined by confocal microscopy. Arrowheads point to infected cells (i.e., VP1 positive). The right column (zoom) shows a higher-magnification view of the region framed in the middle column. Note that some VP1-negative cells also display SG in the wells infected with the L-mutant viruses. This is probably due to the fact that VP1 had not reached a detectable level at the time when the cells were fixed. (B) Same experiment as in panel A but with immunostaining of VP1 (red) and TIA-1 (green). (C) Histogram showing the percentage (mean of two independent experiments) of VP1-positive cells displaying SG according to the virus used. Note that cells presenting CPE were excluded from the counts. (D) Percentages (means of two independent experiments) of cytopathic effect observed in HeLa cells infected with the wild-type and mutant viruses at 16 and 24 h postinfection.
Thus, our results suggest that TMEV infection triggers SG formation but that this process is inhibited by the L protein.
L protein blocks SG formation, independently of its inhibition of interferon production.
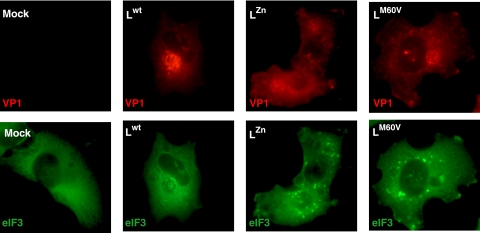
Due to L's antagonism of type I interferon (IFN) production, viruses carrying mutations in the Zn finger or in the Theilo domain of L were shown to trigger enhanced IFN production in infected cells, compared to wild-type virus. Since type I IFN can sensitize cells to translation blockade and apoptosis through PKR-mediated eIF2α phosphorylation, we tested whether SG formation in cells infected with L-mutant viruses was a consequence of the type I IFN response. To this end, SG formation was monitored in STAT-1-deficient U3A cells, which are unresponsive to type I IFN. As in other cell types, SG were detected in U3A cells after infection with the L-mutant viruses but not with the wt virus (Fig. 3). Kinetics of SG appearance did not differ from that observed in STAT-1+/+ parental 2ftgh cells (data not shown). In conclusion, type I IFN is not required for SG formation in TMEV-infected cells and L-mediated inhibition of SG formation is not a consequence of IFN antagonism by L.
Fig. 3.
Inhibition of SG assembly is independent of L inhibition of interferon production. U3A cells were infected with 5 PFU/cell of wild-type TMEV (Lwt) or with LZn and LM60V mutant viruses. At 8 hours postinfection, cells were processed for VP1 and eIF3 coimmunolabeling and examined by fluorescence microscopy. SG appear as bright spots of eIF3 staining in the cytoplasm.
L can block the assembly of SG induced by nonviral stresses.
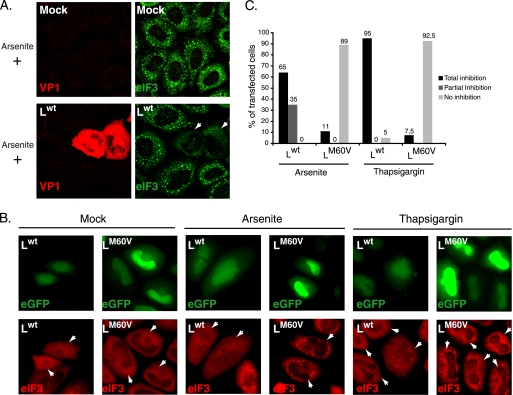
The absence of SG in cells infected with the wild-type virus can result either from a lack of SG induction, possibly due to a lack of virus detection by cell sensors, or from an “active” blockade of the SG formation process. To discriminate between these possibilities, we checked if the virus was able to inhibit SG assembly induced by stresses other than infection. To this end, HeLa cells infected for 12 h with wild-type and L-mutant viruses were treated with sodium arsenite, a strong SG inducer. eIF3 labeling was performed 45 min after sodium arsenite treatment. As shown in Fig. 4A, oxidative stress-induced SG formation was strongly inhibited in cells infected by the wild-type virus. This was due to the L protein because SG formation was not inhibited in cells infected with L-mutant viruses (not shown). These results show that inhibition of infection-induced SG results from an active process of TMEV L.
Fig. 4.
Ectopic expression of L is sufficient to inhibit arsenite- or thapsigargin-induced SG assembly. (A) HeLa cells were infected with a wild-type TMEV (Lwt) or with L-mutant viruses (not shown). Twelve hours postinfection, cells were treated with 0.5 mM sodium arsenite for 45 min, fixed, and processed for coimmunolabeling of VP1 and eIF3 (confocal microscopy images). (B) HeLa cells were transfected with bicistronic constructs expressing Lwt and eGFP or LM60V and eGFP. Sixteen hours posttransfection cells were mock treated or treated with 0.5 mM sodium arsenite for 45 min or with 15 μM thapsigargin for 50 min, fixed, and processed for eIF3 immunolabeling. White arrowheads indicate transfected cells. Note that cells transfected with the Lwt-IRES-eGFP construct had a much lower eGFP fluorescence level because Lwt expression represses eGFP expression (30). Moreover, eIF3 exhibited a partially nuclear localization in many cells expressing Lwt, in agreement with the reported effect of this protein on nucleocytoplasmic transport. (C) Histogram showing the percentages of transfected (eGFP-positive) cells displaying total or partial inhibition or no inhibition of arsenite- and thapsigargin-induced SG. In view of ectopically expressed L protein toxicity, only cells with unaltered morphology were taken into account.
We next asked whether the L protein inhibited SG assembly when expressed ectopically, in the absence of other virus components. HeLa cells were transfected with bicistronic constructs coexpressing eGFP and either Lwt or LM60V or with a control plasmid expressing eGFP alone. Sixteen hours posttransfection, cells were treated with sodium arsenite for 45 min, and the formation of SG was monitored by immunofluorescent staining of eIF3 (red) in transfected cells expressing eGFP (green) (Fig. 4B). Transfection of pTM624, expressing eGFP alone, did not induce SG formation in untreated cells and did not affect SG formation in cells treated with sodium arsenite (not shown). In contrast, expression of Lwt clearly inhibited sodium arsenite-induced SG assembly (Fig. 4B and C). This inhibition was not observed in cells expressing the mutated LM60V protein (Fig. 4B and C).
Thapsigargin treatment of the cells induced SG that were less conspicuous than those induced by sodium arsenite treatment. Again, assembly of these SG was inhibited after ectopic expression of the wild-type protein but not of the mutant LM60V protein (Fig. 4B and C). When SG assembly was induced by heat shock (50 min at 44°C), ectopic expression of Lwt triggered rapid apoptosis of most cells, preventing assessment of SG formation (not shown).
Taken together, these results show that TMEV infection triggers SG assembly but that the L protein produced by the virus inhibits this process, in a way that is independent of its antagonism of IFN production.
PTB but not Sam68 partially colocalizes with TMEV-induced SG.
To further characterize the composition of SG induced by mutant TMEVs, we performed different combinations of double immunofluorescent labeling in infected and, as a control, in sodium arsenite-treated HeLa cells. Granules induced by TMEV infection were much more heterogenous than sodium arsenite-induced SG (not shown). However, in both infected and arsenite-treated cells, we observed a perfect match between eIF3-G3BP and G3BP–TIA-1 localizations, suggesting that infection, like arsenite treatment, triggered the assembly of bona fide SG (Fig. 5A).
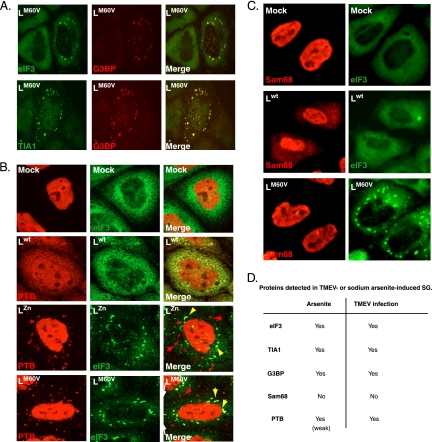
Fig. 5.
PTB, but not Sam68, partially colocalizes with TMEV infection-induced SG. (A) Confocal microscopy images showing coimmunostaining of eIF3 and G3BP or TIA-1 and G3BP in HeLa cells infected for 16 h (10 PFU/cell) with the LM60V mutant virus. (B) Confocal microscopy images showing coimmunostaining of eIF3 and PTB in HeLa cells infected for 12 h (10 PFU/cell) with wild-type TMEV or with LZn and LM60V mutant viruses. Yellow arrowheads indicate colocalization between PTB and stress granules. Red arrowheads indicate cytoplasmic PTB foci in which no eIF3 was detected. (C) Double fluorescence microscopy images showing representative U3A cells coimmunostained for Sam68 and eIF3, 8 h after infection with 5 PFU/cell of wild-type or L-mutant (LM60V) TMEV. (D) Detected stress granule markers associated with arsenite-induced SG or TMEV infection-induced SG.
We previously observed PTB aggregates in the cytoplasm of L929 and HeLa cells infected with L-mutant viruses. We thus explored whether these PTB aggregates colocalized with SG using confocal fluorescence microscopy. As shown in Fig. 5B, all infection-induced SG contained PTB. In contrast, some PTB aggregates did not colocalize with SG, as detected by eIF3 labeling. These additional granules are either not SG or SG with prominent PTB and low eIF3 content. Similar results were obtained after PTB/TIA-1 coimmunostaining. Interestingly, small amounts of PTB were also systematically detected in sodium arsenite-induced stress granules (not shown).
Sam68 is a nuclear protein that was shown to be incorporated into poliovirus-induced SG but not into oxidative stress-induced SG (26). We tested whether Sam68 incorporation into SG was a hallmark of virus-induced SG or whether the composition of SG varied according to the virus involved. As previously observed in the case of PTB, the virus expressing Lwt induced some diffusion of Sam68 out of the nucleus. However, unlike PTB, Sam68 failed to form visible spots in the cytoplasm of cells infected by L-mutant viruses and was therefore not visible in SG (Fig. 5C). Thus, stress granule composition appears to vary according to the inducing stimulus. Figure 5D sums up the detected SG markers, according to the stress stimulus used.
Viral RNA is not detected in stress granules.
Stress granules have been shown to trap cellular mRNA during stress (2). We hypothesized that infection-induced SG could sequester viral RNA and thereby negatively impact the viral cycle. The assumption that SG could sequester viral RNA was reinforced by the fact that PTB, which is reported to interact with TMEV RNA, was detected in TMEV-induced SG. To test whether viral RNA was trapped in SG, we used combined in situ hybridization for detection of positive-stranded viral RNA and immunofluorescence for detection of eIF3. In cells infected with the wild-type virus, positive-stranded viral RNA was detected in dense and large, sometimes focal perinuclear areas (Fig. 6A). Intriguingly, in cells infected with L-mutant viruses, positive-stranded viral RNA was detected in a punctated pattern. Yet the spots of viral RNA did not colocalize with SG.
Fig. 6.
Viral RNA is not detected in TMEV-induced SG. (A) In situ hybridization (ISH) to detect positive-stranded viral RNA coupled to immunolabeling of eIF3. HeLa cells were mock infected or infected with wild-type or LM60V TMEV at 10 PFU per cell. After 12 h of infection, cells were fixed and processed for ISH (red) and immunolabeling of eIF3 (green) (confocal microscopy images). (B) Coimmunolabeling of the viral double-stranded RNA (dsRNA; detected with the K1 antibody; red) and eIF3 (green). Experimental conditions were the same as in Fig. 2. (conventional microscopy images).
Viral double-stranded RNA, considered to be characteristic of replication complexes, was detected in infected cells using the J2 or K1 anti-dsRNA monoclonal antibodies. Here, double-stranded RNA was detected as a spotted pattern for the wild-type and mutant viruses. Again, this form of viral RNA was not detected in stress granules (Fig. 6B).
Inhibition of SG assembly is a common activity of cardiovirus L proteins.
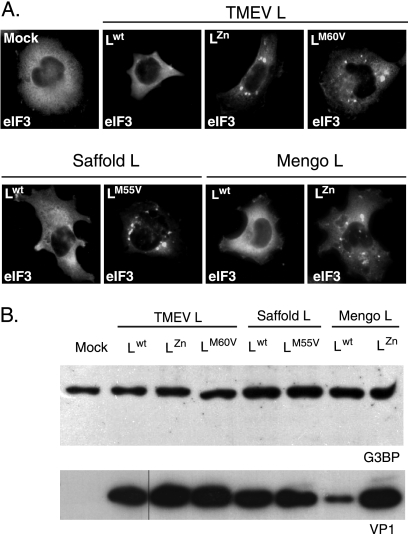
The L proteins of TMEV and EMCV share 35% identity. This percentage increases to 60% for identity between TMEV and Saffold virus L proteins (Fig. 1). We used recombinant TMEV derivatives to test whether EMCV L (mengovirus strain) or Saffold virus L (SAFV-2 strain) also antagonized SG formation. Therefore, U3A cells were infected in parallel with wild-type TMEV or with TMEV derivatives expressing TMEV LZn, TMEV LM60V, SAFV-2 Lwt, SAFV-2 LM55V, mengovirus Lwt, or mengovirus LZn. Coimmunolabeling of eIF3 and VP1 was performed 8 h postinfection. As shown in Fig. 7A, SG appeared in cells infected with the recombinants expressing mutated L proteins but not in cells infected with the recombinant viruses expressing the wt L protein of TMEV, SAFV-2, or EMCV. Thus, the L proteins of the different cardioviruses share the ability to inhibit infection-induced SG assembly. Western blot analysis of infected cell extracts failed to show G3BP degradation, suggesting that L proteins act in a different fashion than poliovirus 3C (Fig. 7B).
Fig. 7.
Inhibition of SG assembly is a common activity of cardiovirus L protein. (A) U3A cells were infected in parallel with a wild-type TMEV or with TMEV recombinants expressing TMEV LZn, TMEV LM60V, SAFV-2 Lwt, SAFV-2 LM55V, mengovirus Lwt, or mengovirus LZn. At 8 hours postinfection cells were fixed and processed for coimmunolabeling of VP1 (not shown) and eIF3. Each picture shows the distribution of eIF3 in a VP1-positive cell (not shown), except for the control, which was VP1 negative. (B) Western blot detection of G3BP in extracts from U3A cells infected as in panel A. No sign of G3BP cleavage or decrease in band intensity was observed when comparing wild-type and L-mutant viruses. Note that lower VP1 expression in SPA24 (Lwt mengovirus)-infected cells was noticed previously (23) and is thought to result from an altered synchronism between viral replication and L activity, which can indirectly impair viral replication.
DISCUSSION
The leader proteins of cardioviruses are very small proteins endowed with pleiotropic functions (1). They interfere with IFN and chemokine production, thereby slowing down innate immune responses against the virus; they promote hyperphosphorylation of nucleoporins that are critical components of the nuclear pore complex and perturb nucleocytoplasmic trafficking of mRNA and proteins; they also modulate the apoptotic response of the cell, either positively or negatively, according to the experimental conditions. Recently, EMCV L protein was found to trigger activation of extracellular signal-regulated kinase 1/2 (ERK1/2) and p38 kinase (28). Here, we report a new activity of cardiovirus L proteins: the inhibition of stress granule formation in infected cells. It is not clear how this new L activity relates to the previously identified L functions. We could exclude the possibility that SG formation inhibition was a mere consequence of IFN production antagonism by L. Indeed, SG formation was also promoted by L-mutant viruses in STAT-1-deficient cells, which are not responsive to IFN. The effect of L on SG formation might relate to nucleocytoplasmic trafficking alteration since factors involved in SG assembly, like TIA-1, are known to shuttle between nucleus and cytoplasm. In favor of a link between the different activities of L is the observation that mutations in either the Zn finger or in the Theilo domain abrogate all known activities of TMEV L (29), including SG formation inhibition (this work).
Poliovirus, which also belongs to the picornavirus family, was similarly reported to inhibit SG formation. However, in the case of poliovirus, SG formation inhibition involves the proteolytic activity of protein 3C, which was shown to cleave G3BP, a factor involved in the formation of SG (11, 33). In contrast, G3BP clearly accumulated in TMEV-induced SG. Moreover, in cells infected with TMEV derivatives expressing the various cardiovirus L proteins, there was no evidence of G3BP cleavage. This is in line with the fact that cardiovirus L does not possess protease activity. It is interesting to note the convergent evolution of cardiovirus L proteins and of poliovirus and rhinovirus proteases. Although these proteins appear to act by totally different mechanisms, they play very similar roles (10). Among the functions exerted by L proteins, IFN antagonism and nucleocytoplasmic trafficking perturbation are exerted by the poliovirus and rhinovirus 2A proteases while SG formation inhibition is exerted by poliovirus protease 3C.
A negative impact of SG assembly on viral replication has previously been demonstrated in the case of poliovirus (33). A mechanism by which SG could affect viral production would be the sequestration of viral RNA and the consequent inhibition of viral RNA translation. However, in situ hybridization experiments and immunolabeling experiments failed to detect any viral RNA in infection-induced SG. A similar observation was made in the case of poliovirus (26). One cannot, however, exclude the possibility that the sensitivity of the method used to detect viral RNA is not high enough to detect a minor pool of viral RNA trapped in SG. Another hypothesis to explain the negative impact of SG on viral replication would be that translation factors and other proteins required for the expression of the picornavirus genome (eIF3, PTB, etc.) would be made unavailable for virus genome translation when sequestered in SG.
An intriguing observation is the fact that positive-stranded TMEV RNA from wild-type and L-mutant strains yielded quite different patterns in infected cells. Although wild-type positive-stranded genomes were detected in large focal areas, generally perinuclear, genomes from L-mutant viruses were detected as small patches scattered into the cell cytoplasm. The reason for this is unknown.
Finally, it appears that SG formed in different experimental conditions might differ in their content: PTB was associated with TMEV-induced SG, while Sam68 was detected in poliovirus-induced but not in TMEV-induced SG. However, such differences might be more subtle than they appear since immunofluorescent labeling does not allow comparisons of the relative abundances of different proteins and since detection thresholds might vary according to the experimental set-up. For instance, conflicting data regarding Sam68 detection in oxidative stress-induced SG have been reported (14, 26).
In conclusion, it comes to light that more and more viruses have developed strategies to affect the stress granule response in host cells. However, the impact of SG formation on virus replication and spread and on cell resistance to viral infection is still not fully understood and warrants further studies.
ACKNOWLEDGMENTS
F.B. is the recipient of a fellowship from the Belgian FRIA. This work was supported by a DIANE convention of the Walloon region, by ARC (communauté française de Belgique), and by FRSM (Fonds national de la recherche Médicale convention 3.4576.08 and crédit aux chercheurs).
Footnotes
Published ahead of print on 13 July 2011.
REFERENCES
- 1. Agol V. I., Gmyl A. P. 2010. Viral security proteins: counteracting host defences. Nat. Rev. Microbiol. 8:867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson P., Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436 [DOI] [PubMed] [Google Scholar]
- 3. Anderson P., Kedersha N. 2009. Stress granules. Curr. Biol. 19:R397–R398 [DOI] [PubMed] [Google Scholar]
- 4. Bardina M. V., et al. 2009. Mengovirus-induced rearrangement of the nuclear pore complex: hijacking cellular phosphorylation machinery. J. Virol. 83:3150–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brahic M., Bureau J. F., Michiels T. 2005. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu. Rev. Microbiol. 59:279–298 [DOI] [PubMed] [Google Scholar]
- 6. Chakraborty P., Satterly N., Fontoura B. M. 2006. Nuclear export assays for poly(A) RNAs. Methods 39:363–369 [DOI] [PubMed] [Google Scholar]
- 7. Daniels J. B., Pappenheimer A. M., Richardson S. 1952. Observations on encephalomyelitis of mice (DA strain). J. Exp. Med. 96:517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Delhaye S., van Pesch V., Michiels T. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78:4357–4362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dever T. E. 2002. Gene-specific regulation by general translation factors. Cell 108:545–556 [DOI] [PubMed] [Google Scholar]
- 10. Dougherty J. D., Park N., Gustin K. E., Lloyd R. E. 2010. Interference with cellular gene expression, p. 165–180 In Ehrenfeld E., Domingo E., Roos R. P. (ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 11. Dougherty J. D., White J. P., Lloyd R. E. 2011. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 85:64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fan J., Son K. N., Arslan S. Y., Liang Z., Lipton H. L. 2009. Theiler's murine encephalomyelitis virus leader protein is the only nonstructural protein tested that induces apoptosis when transfected into mammalian cells. J. Virol. 83:6546–6553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gilks N., et al. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Henao-Mejia J., He J. J. 2009. Sam68 relocalization into stress granules in response to oxidative stress through complexing with TIA-1. Exp. Cell Res. 315:3381–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jnaoui K., Michiels T. 1998. Adaptation of Theiler's virus to L929 cells: mutations in the putative receptor binding site on the capsid map to neutralization sites and modulate viral persistence. Virology 244:397–404 [DOI] [PubMed] [Google Scholar]
- 16. Kong W. P., Ghadge G. D., Roos R. P. 1994. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc. Natl. Acad. Sci. U. S. A. 91:1796–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lidsky P. V., et al. 2006. Nucleocytoplasmic traffic disorder induced by cardioviruses. J. Virol. 80:2705–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindquist M. E., Lifland A. W., Utley T. J., Santangelo P. J., Crowe J. E., Jr 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKendry R., et al. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. U. S. A. 88:11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michiels T., Dejong V., Rodrigus R., Shaw-Jackson C. 1997. Protein 2A is not required for Theiler's virus replication. J. Virol. 71:9549–9556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Michiels T., Roos R. P. 2010. Theiler's virus central nervous system infection, p. 411–428 In Ehrenfeld E., Domingo E., Roos R. P. (ed.), The picornaviruses. ASM Press, Washington, DC [Google Scholar]
- 22. Oleszak E. L., Chang J. R., Friedman H., Katsetos C. D., Platsoucas C. D. 2004. Theiler's virus infection: a model for multiple sclerosis. Clin. Microbiol. Rev. 17:174–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Paul S., Michiels T. 2006. Cardiovirus leader proteins are functionally interchangeable and have evolved to adapt to virus replication fitness. J. Gen. Virol. 87:1237–1246 [DOI] [PubMed] [Google Scholar]
- 24. Pilipenko E. V., et al. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028–2045 [PMC free article] [PubMed] [Google Scholar]
- 25. Pilipenko E. V., Viktorova E. G., Guest S. T., Agol V. I., Roos R. P. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 20:6899–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Piotrowska J., et al. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84:3654–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Porter F. W., Bochkov Y. A., Albee A. J., Wiese C., Palmenberg A. C. 2006. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc. Natl. Acad. Sci. U. S. A. 103:12417–12422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Porter F. W., Brown B., Palmenberg A. C. 2010. Nucleoporin phosphorylation triggered by the encephalomyocarditis virus leader protein is mediated by mitogen-activated protein kinases. J. Virol. 84:12538–12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ricour C., et al. 2009. Random mutagenesis defines a domain of Theiler's virus leader protein which is essential for antagonism of nucleocytoplasmic trafficking and of cytokine gene expression. J. Virol. 83:11223–11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ricour C., et al. 2009. Inhibition of mRNA export and dimerization of interferon regulatory factor 3 by Theiler's virus leader protein. J. Gen. Virol. 90:177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romanova L. I., et al. 2009. Antiapoptotic activity of the cardiovirus leader protein, a viral “security” protein. J. Virol. 83:7273–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Pesch V., van Eyll O., Michiels T. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811–7817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. White J. P., Cardenas A. M., Marissen W. E., Lloyd R. E. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]