Abstract
Eukaryotic mRNA processing and export is mediated by various heterogeneous nuclear ribonucleoproteins (hnRNPs). Many of these hnRNPs are methylated on arginine residues. In the yeast, Saccharomyces cerevisiae, the predominant enzyme responsible for arginine methylation is Hmt1p. Hmt1p methylates both Npl3p and Hrp1p, which are shuttling hnRNPs involved in mRNA processing and export. Here, we employ an in vivo nuclear export assay to show that arginine methylation is important for the nuclear export of these hnRNPs. Both Npl3p and Hrp1p fail to exit the nucleus in cells lacking Hmt1p, and overexpression of Hmt1p enhances Npl3p export. The export of a novel hnRNP-like protein, Hrb1p, which does not bind poly(A)+ RNA, however, is not affected by the lack of methylation. Furthermore, we find a genetic relationship between Hmt1p and cap-binding protein 80 (CBP80). Together, these findings establish that one biological role for arginine methylation is in facilitating the export of certain hnRNPs out of the nucleus.
Keywords: HMT1, NPL3, HRP1, CBP80, heterogeneous ribonucleoprotein (hnRNP), arginine methylation, mRNA export
While in the nucleus, mRNA precursors, referred to as pre-mRNAs or heterogeneous nuclear RNAs (hnRNAs), undergo a series of processing events before traveling to the cytoplasm. These maturation events include capping at the 5′ end, splicing, and 3′-end cleavage followed by polyadenylation. From the time they leave the transcription complex, hnRNAs are associated with proteins, some of which have been proposed to be mediators of RNA export (for review, see Lee and Silver 1997). The set of proteins that bind hnRNAs, with the exception of small nuclear RNPs, are referred to as heterogeneous nuclear ribonucleoproteins (hnRNPs).
Among the most abundant proteins in the nucleus (Kiledjian et al. 1994), there are over 20 mammalian hnRNPs, proposed to function in nearly every step of mRNA maturation including export (Piñol-Roma 1997). One of the best-studied hnRNPs, hnRNP A1, travels back and forth between the nucleus and the cytoplasm in a process termed nucleocytoplasmic shuttling (Piñol-Roma and Dreyfuss 1992). In addition, an hnRNP A1-like protein in the insect Chironomus tentans can be seen by immunoelectron microscopy to be associated with pre-mRNA traveling through the nuclear pore to the cytoplasm (Visa et al. 1996a).
The discovery of a nuclear export signal (NES) in some hnRNPs has suggested further that these hnRNPs could play an active role in RNA export. A 38-amino-acid sequence within hnRNP A1 termed M9 has been found to be necessary and sufficient for export of the protein to the cytoplasm (Michael et al. 1995). Microinjection experiments have shown that saturating amounts of the M9 domain block mRNA export in Xenopus oocytes (Izaurralde et al. 1997). A model for mRNA export has been proposed in which the export signals on the shuttling hnRNPs are directly responsible for the translocation of bound mRNAs into the cytoplasm (Fischer et al. 1996; Nigg 1997). This model is best supported by studies of the HIV Rev protein. Through its leucine-rich NES, Rev facilitates the nuclear export of partially spliced and unspliced viral RNAs containing the Rev response element (Fischer et al. 1996).
Further evidence for the involvement of hnRNPs in mRNA export comes from studies in the yeast Saccharomyces cerevisiae. Npl3p is a yeast hnRNP (Bossie et al. 1992; Russell and Tollervey 1992) with structural features similar to those found in some mammalian hnRNPs, such as hnRNP A1. These include two RNP-type RNA-recognition motifs (RRMs), and a glycine-rich domain in the form of 15 arginine–glycine–glycine (RGG) repeats (Birney et al. 1993). Although Npl3p appears to localize to the nucleus in a distinct non-nucleolar pattern similar to human hnRNPs (Flach et al. 1994), it actually shuttles between the nucleus and the cytoplasm (Flach et al. 1994; Lee et al. 1996). Strains bearing mutant alleles of NPL3 accumulate poly(A)+ RNA in the nucleus (Russell and Tollervey 1992; Singleton et al. 1995; Henry et al. 1996; Lee et al. 1996). Export of these mutant Npl3p proteins from the nucleus is also defective, providing further evidence that Npl3p is a poly(A)+ RNA carrier (Lee et al. 1996). Moreover, Npl3p export depends on ongoing RNA polymerase II transcription, suggesting a tight association between the export of poly(A)+ RNA and Npl3p (Lee et al. 1996).
A second yeast hnRNP, HRP1, was identified in a screen for suppressors of the temperature-sensitive mutant, npl3-1 (Henry et al. 1996). This essential gene encodes a protein with two RNA-recognition motifs most similar to those found in human hnRNPs A1, A2/B1, and D/AUF1. In addition, the carboxyl terminus of the protein is arginine and glycine rich. Hrp1p has recently been shown to be required for proper 3′ end formation in S. cerevisiae (Kessler et al. 1997). As such, it binds mRNA at the UA-rich efficiency element, which is required for both cleavage and polyadenylation. Although Hrp1p localizes to the nucleus at steady state, it has also been found to efficiently exit the nucleus (Kessler et al. 1997). These data suggest a link between polyadenylation and mRNA export in yeast.
It has become increasingly apparent that hnRNPs play key regulatory roles at all levels of RNA processing (for review, see Dreyfuss et al. 1993). Such regulation may be mediated by post-translational modifications of the hnRNPs themselves. In fact, most hnRNPs do undergo such modifications including phosphorylation (e.g., Cobianchi et al. 1993; Piñol-Roma and Dreyfuss 1993), glycosylation (Soulard et al. 1993), and methylation (e.g., Liu and Dreyfuss 1995). Partially purified mammalian hnRNP complexes contain proteins with a high level of the unusually modified amino acid NG,NG-dimethylarginine (Beyer et al. 1977). In fact, hnRNPs contain about 65% of methylated arginines in the cell nucleus (Boffa et al. 1977), indicating that the methylation state of these proteins is likely to have an important effect on their functions. In addition, these methylated arginine residues are found in other non-hnRNP RNA-binding proteins, all of which appear to contain RGG repeats (Najbauer et al. 1993 and references therein). Despite speculation that methylation affects the RNA-binding activity of hnRNPs (Rajpurohit et al. 1994), or the interactions of hnRNPs with other proteins (Liu and Dreyfuss 1995), no in vivo function has been defined.
The recent identification of the enzymes responsible for arginine methylation provides an important gateway to further our understanding of its true biological role. Protein arginine methyltransferases specific for hnRNP A1 have been partially purified from several sources (Ghosh et al. 1988; Rawal et al. 1994; Liu and Dreyfuss 1995). The cDNAs encoding human and rat arginine methyltransferases have been identified (Lin et al. 1996; Nikawa et al. 1996; Katsanis et al. 1997). In S. cerevisiae, a homolog of the rat protein arginine methyltransferase (PRMT1) termed HMT1/RMT1 has been identified in a synthetic lethal screen with the temperature-sensitive mutant, npl3-1 (Henry and Silver 1996). HMT1/RMT1 was independently identified as a gene whose protein product represents the major arginine methyltransferase activity present in S. cerevisiae (Gary et al. 1996). Hmt1p has been shown to methylate Npl3p both in vitro and in vivo (Henry and Silver 1996; Siebel and Guthrie 1996), and to localize to the nucleus (Henry and Silver 1996). In a strain containing a null allele of HMT1, methylation of Npl3p is no longer detectable (Henry and Silver 1996). Taken together, these data suggest that Hmt1p may play a role in the regulation of Npl3p function.
In this investigation, we show that, like Npl3p, Hrp1p is methylated by Hmt1p. More importantly, we demonstrate that, in the absence of Hmt1p methylation, the ability of Npl3p and Hrp1p to exit the nucleus is impaired. This study is the first in vivo demonstration of a function for hnRNP arginine methylation and implicates this modification as part of the mechanism by which protein–RNA complexes are recognized for nuclear export.
Results
Effect of methylation on the nuclear export of mRNA-binding proteins
We have shown previously that both Npl3p and Hrp1p exit the nucleus (Flach et al. 1994; Lee et al. 1996; Kessler et al. 1997). To test whether methylation of Npl3p and Hrp1p has an effect on their export, we utilized a nuclear export assay (Lee et al. 1996). This assay exploits the properties of a temperature-sensitive mutant of the nucleoporin Nup49p (Doye et al. 1994). At the nonpermissive temperature of 36.5°C, import of proteins into the nucleus is blocked in the nup49-313 strain, whereas nuclear export is not appreciably affected. Because of this phenotype, shuttling proteins accumulate in the cytoplasm after exit from the nucleus. To monitor this export visually, the green fluorescent protein (GFP) is fused to the protein of interest and placed under a galactose inducible (GAL1) promoter. After induction, cells are either shifted to the nonpermissive temperature or maintained at the permissive temperature of 25°C.
The deletion of HMT1 alone results in no poly(A)+ RNA export defects or nuclear protein mislocalization (Henry and Silver 1996). To study the effect of methylation through the use of this export assay, a strain containing the nup49-313 mutation and a deletion of the HMT1 gene was constructed (see Materials and Methods).
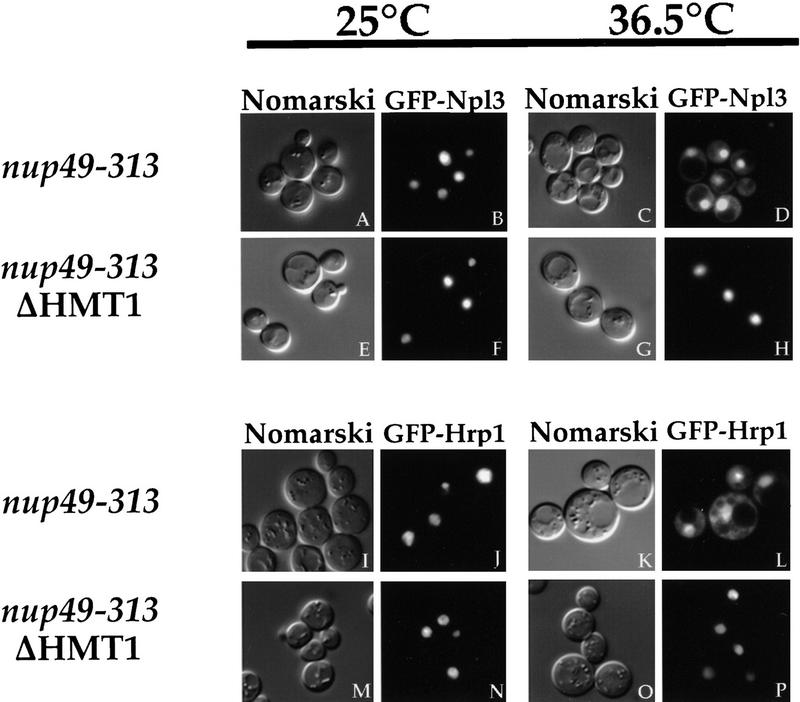
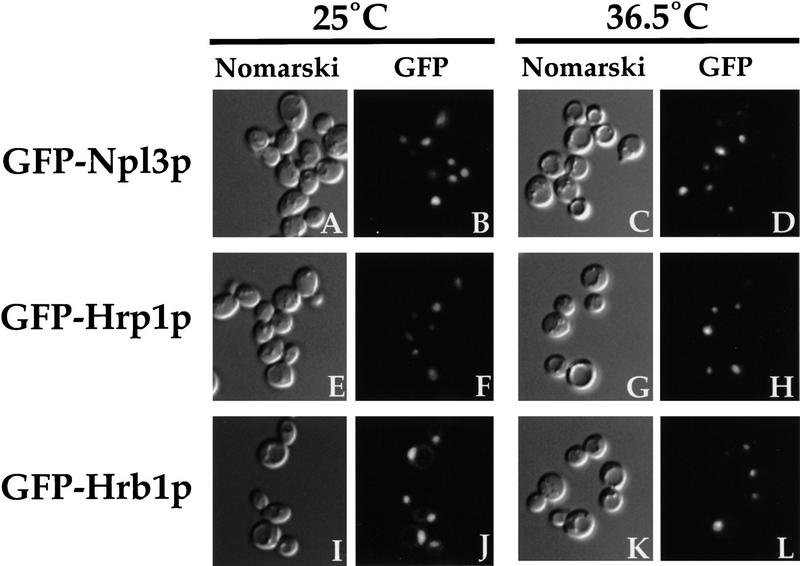
To monitor the localization of Npl3p, GFP was fused to its amino terminus, and the resulting fusion protein was placed under the GAL1 promoter (Lee et al. 1996). GFP–Npl3p accurately reflects the behavior of its endogenous counterpart (Lee et al. 1996) under all conditions examined. The nuclear export assay was performed in the nup49-313 ΔHMT1 strain, as well as the nup49-313 strain. At the permissive temperature, GFP–Npl3p localizes entirely in the nucleus in both strains (Fig. 1 B,F). Athough GFP–Npl3p accumulates in the cytoplasm of the nup49-313 cells at 36.5°C (Fig. 1D), the signal remains completely nuclear in the nup49-313 ΔHMT1 cells (Fig. 1H). As in the ΔHMT1 strain (Henry and Silver 1996), Npl3p is no longer methylated in the double mutant (data not shown). These results indicate that the export of Npl3p out of the nucleus is affected by methylation.
Figure 1.
Export of Npl3p and Hrp1p is blocked in the absence of Hmt1p. Either nup49-313 cells (A–D and I–L) or the nup49-313 strain in which HMT1 has been deleted (E–H and M–P) were incubated in the export assay. The reporter used was GFP–Npl3p (pPS811; A–H) or GFP–Hrp1p (pPS1358; I–P). Cells were incubated at 25°C (A,B,E,F,I,J,M,N) or at 36.5°C (C,D,G,H,K, L,O,P) for 5 hr. Cells were photographed by use of Nomarski optics (A,C,E,G,I,K,M,O) or for GFP fluorescence (B,D,F, H,J,L,N,P).
To determine whether this effect extended to another shuttling hnRNP, Hrp1p (Henry et al. 1996; Kessler et al. 1997), the nuclear export assay was performed using GFP–Hrp1p as the reporter protein. The localization patterns observed for GFP–Hrp1p paralleled those seen previously with myc–Hrp1p (Henry et al. 1996), and the fusion protein has been shown to be functional (Kessler et al. 1997). When the cells were incubated at 25°C, GFP–Hrp1p was seen in the nucleus in both strains (Fig. 1J,N). As shown in our previous study (Kessler et al. 1997), GFP–Hrp1p accumulated in the cytoplasm in nup49-313 cells (Fig. 1L) following shift to 36.5°C. Conversely, the fusion protein failed to leave the nucleus in the nup49-313 ΔHMT1 strain (Fig. 1P). These results demonstrate that nuclear export of Hrp1p, like that of Npl3p, is affected by methylation. The failure of GFP–Hrp1p and GFP–Npl3p to leave the nucleus at the restrictive temperature may be attributable to a slower rate of export in the strain lacking Hmt1p.
Hrp1p is methylated by Hmt1p
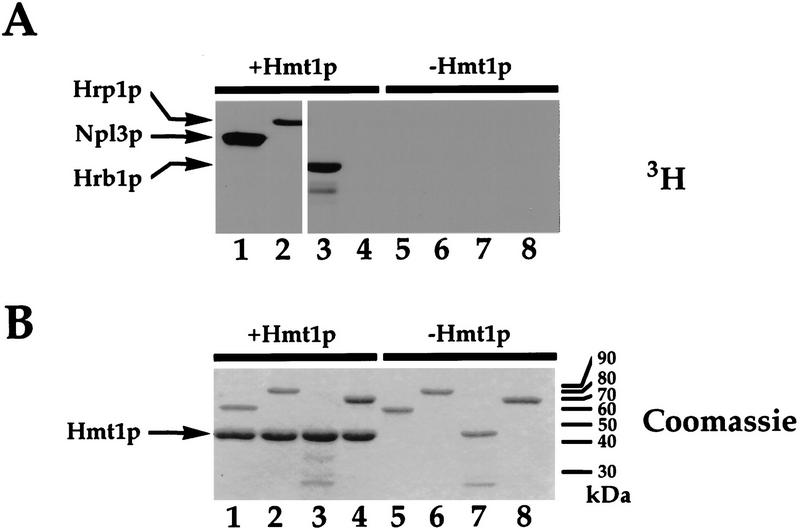
Previous work has shown that Hmt1p is required for Npl3p methylation and that this methylation occurs in the RGG-containing carboxyl terminus (Henry and Silver 1996; Siebel and Guthrie 1996). Because HRP1 was originally identified because of its genetic interactions with NPL3 and the encoded protein (Hrp1p) contains three RGGs, we tested whether Hmt1p was able to methylate Hrp1p as well. Recombinant Hmt1p and Hrp1p, both purified from Escherichia coli, were incubated with [methyl3H]-S-adenosylmethionine ([methyl3H]-SAM; see Materials and Methods). The proteins were resolved by SDS-PAGE and labeling was detected by fluorography. Under these conditions, Hrp1p is methylated by Hmt1p (Fig. 2A, lane 2). In addition, the assay was repeated with Npl3p purified from E. coli (Fig. 2A, lane 1). Quantitation of the incorporated label indicates that Npl3p is methylated approximately sixfold more than as Hrp1p. When the assay was performed in the absence of Hmt1p, no labeling of Npl3p or Hrp1p was detected (Fig. 2A, lanes 5,6).
Figure 2.
Npl3p, Hrp1p and Hrb1p are methylated by Hmt1p. Recombinant substrates Npl3p (lanes 1,5), Hrp1p (lanes 2,6), Hrb1p (lanes 3,7) were purified from E. coli and BSA was used as a negative control (lanes 4,8). Substrates were incubated with [methyl3H]-SAM in the presence (lanes 1–4) or absence (lanes 5–8) of Hmt1p purified from E. coli. Samples were resolved by SDS-PAGE. Proteins were detected by Coomassie staining (B) and labeling was detected by fluorography (A). Protein molecular weights are indicated in kilodaltons.
Methylation is not required for the export of all hnRNPs
The novel hnRNP, HRB1 (TOM34, YNL004w; Philippsen et al. 1997), was identified during the course of a screen for chromosomal mutations that are lethal in combination with a deletion of the nonessential nucleoporin NUP2. HRB1 was cloned as a very weak suppressor of the synthetically lethal combination of ΔNUP2 and a currently unknown mutant (M.S. Lee and P.A. Silver, unpubl.). The protein contains three RRMs, and an amino- terminal domain that contains two repeats of the tripeptide RGG. HRB1 is not essential for vegetative growth (see Materials and Methods).
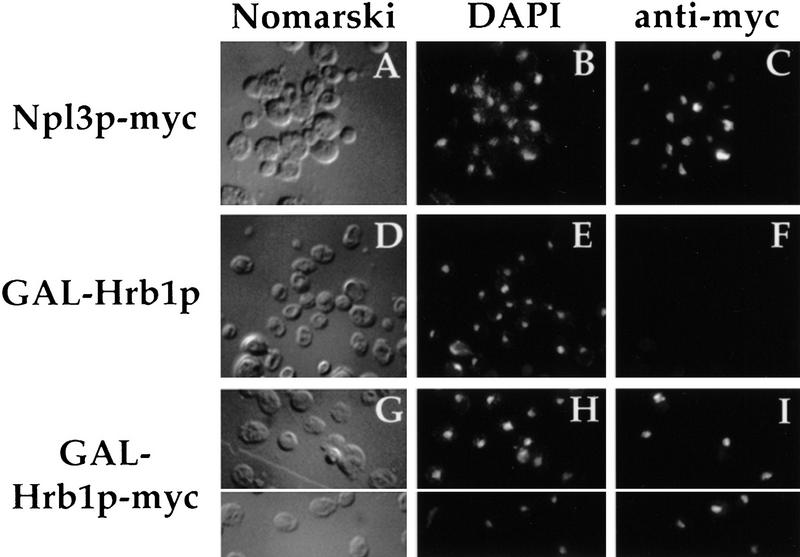
To examine the subcellular localization of Hrb1p, an epitope-tagged version of this protein was generated with a single myc epitope fused to the carboxyl terminus of the protein. Tagged and untagged versions of Hrb1p under the control of the inducible GAL1 promoter were constructed and harbored in a strain lacking the chromosomal copy of HRB1. Indirect immunofluorescence microscopy performed on these cells with the anti-Myc monoclonal 9E10 indicates that Hrb1p–Myc is located primarily in the nucleus (Fig. 3I). This is similar to the localization of Npl3–Myc (Fig. 3C). Virtually no staining is observed in cells containing Hrb1p lacking the epitope tag (Fig. 3F). Nuclear Hrb1p–Myc is also seen with a centromeric plasmid containing HRB1 under its own promoter, although the signal is weaker (data not shown).
Figure 3.
Hrb1p is a novel nuclear hnRNP. Cells harboring plasmids expressing Npl3p–Myc expressed from its endogenous promoter (A–C), or Hrb1p (D–F) and Hrb1p–Myc (G–I) under control of the inducible GAL1 promoter were subjected to immunofluorescence microscopy with the 9E10 monoclonal antibody. The Myc-tagged proteins were visualized with FITC labeled anti-mouse antibody (C,F,I). DNA was visualized using DAPI (B,E,H). Cells were photographed by use of Nomarski optics (A,D,G).
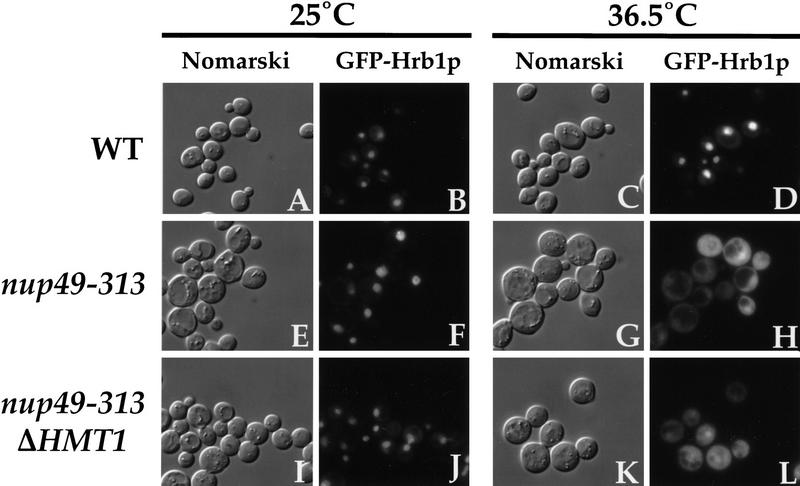
To examine whether Hrb1p exits the nucleus, the nuclear export assay was performed with a GFP–Hrb1p fusion (Fig. 4). The localization of GFP–Hrb1p was first examined in a wild-type strain. After induction of the fusion protein and incubation at either 25°C or 36.5°C, GFP–Hrb1p is found predominantly in the nucleus with some faint cytoplasmic signal (Fig. 4B,D). The same predominantly nuclear localization is seen when the nup49-313 strain containing this reporter is induced followed by incubation at 25°C (Fig. 4F). When nup49-313 cells are induced followed by a shift to 36.5°C, however, GFP–Hrb1p accumulates in the cytoplasm, indicating export from the nucleus (Fig. 4H). Furthermore, GFP–Hrb1p is still capable of export from the nucleus in a nup49-313 strain lacking HMT1 (Fig. 4L). These results indicate that although Hrb1p is located primarily in the nucleus, its localization represents a dynamic state within the cell. In addition, in contrast to the situation found with Hrp1p and Npl3p, the absence of functional Hmt1p has no effect on the ability of Hrb1p to exit the nucleus.
Figure 4.
Hrb1p export is unaffected by the absence of Hmt1p. Wild-type cells (A–D), nup49-313 cells (E–H), and nup49-313 ΔHMT1 double-mutant cells (I–L) containing the reporter plasmid pPS1341 expressing GFP–Hrb1p were subjected to the export assay. Cells were incubated at 25°C (A,B,E,F,I,J) or 36.5°C (C,D,G,H,K,L). Cells were photographed either using Nomarski optics (A,C,E,G,I,K) or for the GFP signal (B,D,F,H,J,L).
Hrb1p is methylated by Hmt1p
Because Hrb1p contains RGG repeats, we tested whether this protein is also methylated by Hmt1p. Recombinant Hrb1p was purified from E. coli and incubated with [methyl3H]-SAM in the presence or absence of bacterially produced Hmt1p (Fig. 2). Reaction products were separated by SDS-PAGE, and labeling was detected by fluorography. In the presence of Hmt1p, Hrb1p is methylated (Fig. 2A, lane 3). When Hmt1p is omitted from the reaction, no labeling of Hrb1p is seen (Fig. 2A, lane 7). Note that in the autoradiograph in Figure 2A, lanes 3–8 were exposed eight times longer than Figure 2A, lanes 1 and 2, although equal amounts of substrate protein were present in each reaction (Fig. 2B). Quantitation of the amount of radioactivity incorporated into the individual substrate in each reaction indicates that Npl3p contains 26 times as much label as Hrb1p. Similarly, Hrp1p contains 4.5 times as much label as Hrb1p.
Nuclear protein export and RNA synthesis
Because the nuclear export of Npl3p requires ongoing RNA synthesis, we tested whether the same is true for Hrp1p and Hrb1p. To accomplish this, we employed a temperature-sensitive allele of RNA Pol II termed rpb1-1 (Nonet et al. 1987). In the rpb1-1 strain, RNA Pol II transcription is inhibited at the nonpermissive temperature. The nuclear export assay was performed with a strain containing both the nup49-313 and rpb1-1 mutant alleles (Lee et al. 1996). Consistent with previous observations (Lee et al. 1996), GFP–Npl3p failed to exit the nucleus in this strain at the nonpermissive temperature (Fig. 5D). Furthermore, both GFP–Hrp1p and GFP–Hrb1p also failed to accumulate in the cytoplasm (Fig. 5, H and L, respectively) at the nonpermissive temperature. This result is not attributable to a lack of protein synthesis because, when the assay was performed in the presence or absence of the protein synthesis inhibitor cycloheximide, GFP–Hrp1p or GFP–Hrb1p accumulated in the cytoplasm at equal levels (data not shown). These results indicate that RNA Pol II transcription, but not new protein synthesis, is required for the export of Hrp1p and Hrb1p from the nucleus.
Figure 5.
Export is blocked in the absence of RNA Pol II transcription. Double-mutant nup49-313 rpb1-1 cells carrying reporter plasmids expressing GFP–Npl3p (A–D), GFP–Hrp1p (E–H), or GFP–Hrb1p (I–L) were subjected to the export assay. Cells were incubated at 25°C (A,B,E,F,I,J) or 36.5°C (C,D,G,H,K,L) for 5 hr. Living cells were photographed by Nomarski optics (A,C,E,G,I,K) or for the GFP signal (B,D,F,H,J,L).
Both Npl3p and Hrp1p bind poly(A)+ RNA (Wilson et al. 1994; Kessler et al. 1997). Hrb1p was also examined for its ability to bind to poly(A)+ RNA by irradiation of cells with ultraviolet light to crosslink RNA to proteins. Poly(A)+ RNA was then isolated from these cells and analyzed by Western blotting for the presence of Myc-tagged proteins. Hrb1p was not detected in the poly(A)+ RNA fraction when expressed either from its own promoter or from the high expression GAL1 promoter under conditions in which Npl3p was readily detectable (data not shown).
Suppression of export defects by HMT1
We previously generated and characterized a temperature-sensitive allele of NPL3, npl3-17 (Lee et al. 1996). This mutant displays a fully recessive phenotype that includes accumulation of poly(A)+ RNA in the nucleus at the nonpermissive temperature. This allele contains two amino-acid substitutions, one in the first RRM, phenylalanine 160 changed to leucine (F160L), and one in the second RRM, isoleucine 268 changed to threonine (I268T). Despite the presence of mutations in both RRMs, this protein can still be crosslinked to poly(A)+ RNA in vivo (data not shown).
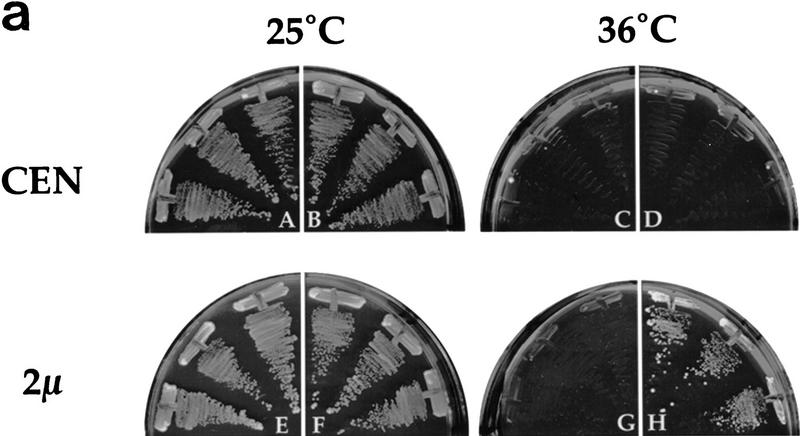
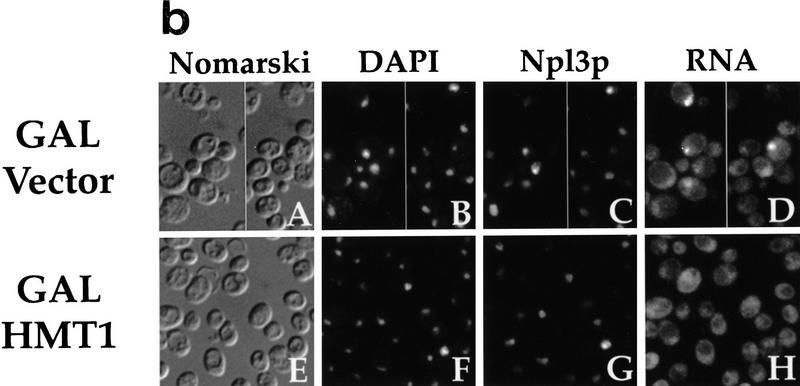
Overproduction of Hmt1p either on a high-copy plasmid (Fig. 6a), or by induction with the strong GAL1 promoter (data not shown) suppresses the temperature-sensitive growth defect of npl3-17 cells. The associated RNA export defect in npl3-17 is also suppressed by the overproduction of Hmt1p from a high-copy vector (data not shown) or with the GAL1 promoter (Fig. 6b, panel H), but not by the vector alone (Fig. 6b, panel D). We have shown previously that the single mutation in the first RRM, F160L, when present in the GFP–Npl3p reporter protein (GFP–F160L) results in a mutant protein that is defective in export from the nucleus (Lee et al. 1996). Taken together, these observations suggest the possibility that overproduction of Hmt1p rescues the temperature-sensitive growth and RNA export defects of npl3-17 by restoring the ability of this mutant to exit the nucleus at the nonpermissive temperature.
Figure 6.
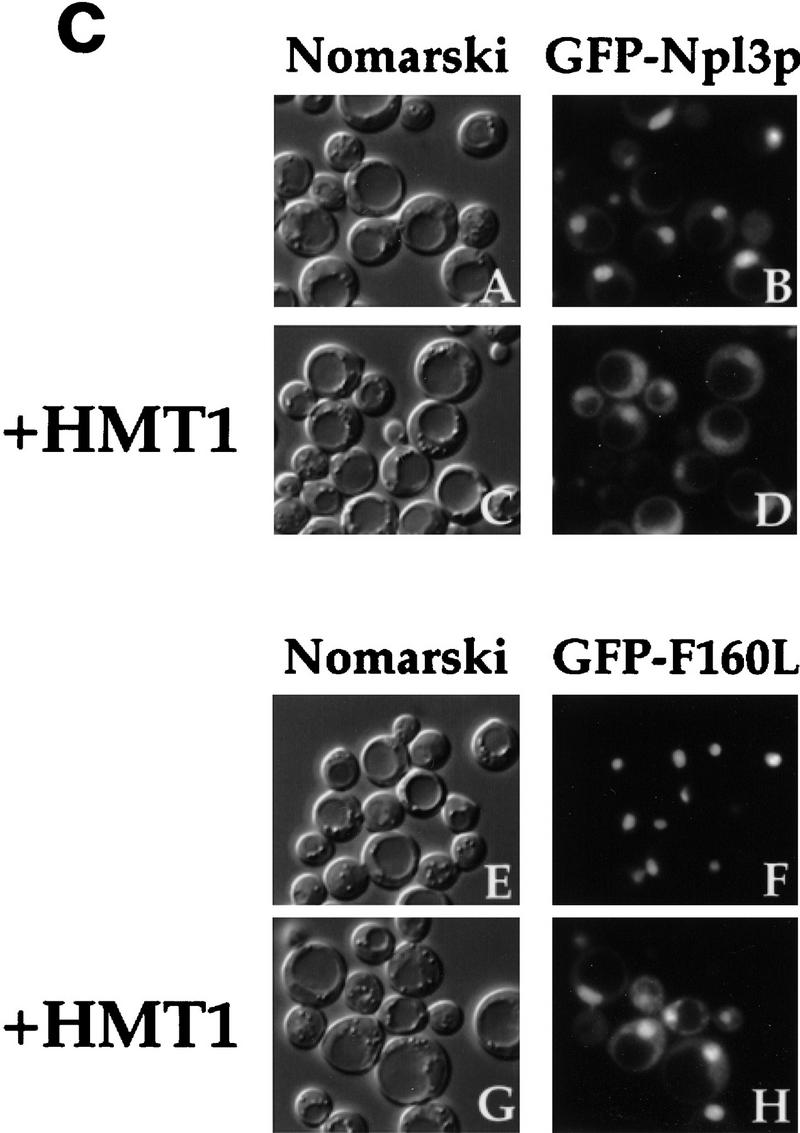
Overproduction of Hmt1p rescues the export defects of npl3-17. (a) A npl3-17 strain was transformed with the following plasmids and streaked on synthetic complete plates lacking leucine and containing glucose as the carbon source: CEN vector alone (pRS315; panels A,C), CEN HMT1 (pPS1305; panels B,D), 2μ vector alone (YEp351; panels E,G), 2μ HMT1 (pPS1308; panels F,H). Plates were incubated at either 25°C (panels A,B,E,F) or 36°C (panels C,D,G,H) for 3 days. (b) npl3-17 cells harboring GAL1 vector pPS311 (panels A–D) or GAL1 HMT1 pPS1141 (panels E–H). Transformants were induced for protein expression for 2 hr then incubated at 36°C for 4 hr, fixed with formaldehyde and processed for in situ hybridization and immunofluorescence microscopy to detect Npl3p (panels C,G) and poly(A)+ RNA (panels D,H). DNA was visualized by DAPI staining (panels B,F), and cells were photographed by use of Nomarski optics (panels A,E). (c) nup49-313 cells containing GFP–Npl3p (pPS811; panels A,B), GFP–Npl3p with HMT1 (pPS1348; panels C,D), GFP–F160L (pPS879; panels E,F), and GFP–F160L with HMT1 (pPS1349; panels G,H) were subjected to the export assay. Cells shifted to 36.5°C for 5 hr are shown. Cells were photographed by use of Nomarski optics (panels A,C,E,G) and for the GFP signal (panels B,D,F,H).
To address this possibility, we constructed high-copy plasmids that carry HMT1 under its own promoter in addition to the GFP–Npl3p or GFP–F160L fusion proteins under control of the GAL1 promoter. We then compared the export of GFP–Npl3p and GFP–F160L in the nuclear export assay in the presence or absence of this additional plasmid-borne copy of HMT1 (Fig. 6c). As we have reported previously, there is often a nuclear GFP–Npl3p signal still visible at the time that export is examined in the assay (Lee et al. 1996; Fig. 6c, panel B), indicating that not all the Npl3p was exported during the course of this experiment. Interestingly, when HMT1 is included on the reporter plasmid, the accumulation of GFP–Npl3p in the cytoplasm is enhanced and most of the nuclear signal is gone (Fig. 6c, panel D). This result suggests that the methylation of Npl3p is limiting in this assay, possibly because of the high overexpression of the GFP–Npl3p reporter in this system. Consistent with previous observations, the GFP–F160L reporter fails to accumulate in the cytoplasm after temperature shift in the export assay (Lee et al. 1996; Fig. 6c, panel F). The presence of HMT1 on the reporter plasmid, however, was able to rescue the defective export of GFP–F160L, resulting in accumulation of this reporter in the cytoplasm (Fig. 6c, panel H). These results indicate that overproduction of Hmt1p rescues the defective nuclear export of the npl3-17 temperature-sensitive mutant.
Synthetic lethality with HMT1
Given the role of Hmt1p in the export of Npl3p and Hrp1p from the nucleus, it is puzzling that HMT1 is not an essential gene. To identify genes that function in conjunction with Hmt1p in nuclear export, a synthetic lethal screen was conducted with a strain that contains a deletion of the HMT1 gene and a wild-type copy of HMT1 on a URA3 plasmid. This strain was mutagenized with ethyl methanesulfonate (EMS) to kill ∼50% of the cells. Cells containing mutations that are synthetically lethal with the HMT1 deletion were selected by the colony color assay (see Materials and Methods). Following mutagenesis, only one recessive mutant was found to require the wild-type HMT1 gene for viability. The corresponding wild-type gene was cloned from a yeast genomic library by screening for restoration of the sectoring phenotype. Subcloning of complementing plasmids localized the minimal region required to a 4.2-kb ClaI DNA fragment. DNA sequence analysis and BLASTn searches revealed a single complete ORF that encodes the yeast homolog of the human cap-binding protein 80 (CBP80) (Izaurralde et al. 1994).
To establish linkage to the synthetic lethality, wild-type CBP80 was marked on the chromosome with the LEU2 gene in a ΔHMT1 strain (see Materials and Methods). The resulting haploid was crossed to the synthetic lethal mutant strain. The diploid was sporulated, and tetrads were analyzed. Each spore carried the null allele of HMT1. Assuming linkage, the spores that carried the LEU2-marked wild-type copy of CBP80 would be able to lose the URA3 HMT1 plasmid. Those that carried the mutant form of CBP80 from the synthetic lethal strain, however, would require the plasmid to remain viable. To test for the ability to lose the plasmid, complete tetrads were streaked on plates containing 5-fluoro-orotic acid (5-FOA). The presence of 5-FOA in the plates selects against cells that carry the URA3 plasmid. Fourteen tetrads were analyzed, and all displayed the behavior predicted for linkage; two Leu+ FOAR: two leu− FOAS spores.
Given that a mutant form of CBP80 is synthetically lethal with the deletion of ΔHMT1, we also tested whether the null allele of CBP80 would give the same result. It had been shown previously that CBP80 is a nonessential gene and that the deletion of CBP80 results in a slow growth phenotype (Uemera and Jigami 1992). We observed the same growth characteristics when constructing a null allele in our strain background (see Materials and Methods). A haploid containing the null allele of CBP80 was crossed to the ΔHMT1 strain. The diploid was sporulated and tetrads were dissected. Of the 47 tetrads analyzed, no spore with the genotype of ΔCBP80 ΔHMT1 was found, indicating that the null alleles are also synthetically lethal. To verify this result, we introduced a copy of HMT1 on a URA3 plasmid into the ΔHMT1 strain and crossed it to the ΔCBP80. The diploid was sporulated, and tetrads were subjected to analysis. The presence of the plasmid rescued the synthetic lethality. In addition, when tetrads were streaked on FOA plates, the double mutant failed to grow, confirming our conclusion that ΔCBP80 is synthetically lethal with ΔHMT1 (Fig. 7).
Figure 7.
Deletion of both CBP80 and HMT1 is lethal. One tetrad from a cross of ΔHMT1 covered with wild-type HMT1 and ΔCBP80 is shown. Two spores are wildtype for both alleles (A,D), and two contain both deletion mutants (B,C). Spores were streaked on synthetic complete media lacking uracil (top) or containing 5-FOA (bottom). Plates were incubated at 25°C for 2 days.
Discussion
In this report we have investigated the effect of arginine methylation on yeast hnRNPs. Previous studies have shown that the essential shuttling hnRNP, Npl3p, is modified by the arginine methyltransferase, Hmt1p. We now demonstrate that Hmt1p is also able to methylate another essential shuttling hnRNP, Hrp1p. By use of a nuclear export assay, the ability of Npl3p and Hrp1p to exit the nucleus was shown to depend on their methylation by Hmt1p. The export of a novel shuttling protein, Hrb1p, however, is not impaired by lack of methylation. Taken together, these data indicate that methylation is critical for the export of certain proteins that bind mRNAs.
Our discovery of genetic interactions between HMT1 and NPL3, in combination with our data that Npl3p is a substrate of Hmt1p, provide the first insight into at least one biological role of arginine methylation. We have been able to study the effect of methylation on Npl3p export by using a nuclear export assay that effectively uncouples export from reimport. Our results suggest that the rate of export is decreased in a strain lacking HMT1. Under the conditions of our assay, Npl3p does not leave the nucleus when HMT1 is absent, thereby indicating a role for methylation in its export. Because HMT1 is not an essential gene (Henry and Silver 1996), however, RNA export must still be occurring in vivo. One possibility is that when NPL3 is not methylated, it does not exit the nucleus, and its role in export is provided by another protein that does not require arginine methylation by Hmt1p. In our assay, GFP–Npl3p needs to reach a threshold level in the cytoplasm before it can be visually detected. In a strain lacking HMT1, this threshold is not attained, suggesting that Npl3p is exiting the nucleus at a slower rate. Consistent with this idea, when the nuclear export assay was performed with cells overexpressing Hmt1p, there was an increase in the cytoplasmic accumulation of GFP–Npl3p at the nonpermissive temperature (Fig. 6c, panel D). This result argues in favor of a role for methylation in the rate of export.
Is methylation affecting Npl3p export directly or through its effects on another protein? We have shown previously that overexpression of Hmt1p can suppress the temperature-sensitive growth of certain npl3 mutant alleles (Henry and Silver 1996). In this study, we have found that overexpression of Hmt1p is able to suppress the temperature-sensitive growth and mRNA export defect associated with the npl3-17 mutant allele. Suppression of these defects correlates with the restoration of the ability of mutant Npl3 to exit the nucleus. It is possible that the npl3-17 protein product is undermethylated in comparison to wild-type Npl3p. Hypermethylation by overproduction of Hmt1p could be enhancing the activity of a partially defective Npl3 encoded by the mutant allele of NPL3. The suppression does not occur by bypassing the need for Npl3p because overexpression of Hmt1p is unable to suppress a null allele of NPL3 (P. Chui and P.A. Silver, unpubl.). In sum, these data support the idea that methylation directly affects the export of Npl3p.
Interestingly, Npl3p is not the only protein whose export is affected by methylation. The export of another methylated mRNA-binding protein known to shuttle between the nucleus and the cytoplasm, Hrp1p, is also impaired when HMT1 is missing. Hrp1p binds specifically to pre-mRNA and promotes its proper polyadenylation (Kessler et al. 1997). Furthermore, mutations in HRP1 can suppress mRNA export defects associated with certain npl3 temperature-sensitive alleles (Henry et al. 1996). It may be that, in addition to its role in polyadenylation, Hrp1p acts together with Npl3p to promote mRNA export. Methylation of Hrp1p is not required for reconstitution of the cleavage and polyadenylation reactions (Kessler et al. 1997), suggesting further that this modification functions specifically in nuclear export. Thus, the efficient export of at least two shuttling hnRNPs implicated in RNA processing and transport requires arginine methylation.
The idea that methylation by Hmt1p has a direct effect on the export of Npl3p and Hrp1p is supported by our studies of Hrb1p. This novel hnRNP-like protein maintains its ability to exit the nucleus in the presence or absence of Hmt1p. One possibility is that although Hrb1p is methylated in vitro, it may not be a bona fide substrate of Hmt1p in vivo. In addition, unlike Npl3p and Hrp1p, we do not detect Hrb1p bound to poly(A)+ RNA. Nevertheless, ongoing RNA Pol II transcription is required for nuclear export of Hrb1p. One explanation for these results is that Hrb1p binds a different Pol II transcript such as snRNAs. These results demonstrate that methylation does not affect the export of all shuttling proteins. Furthermore, the effect of methylation appears to be specific for at least two proteins that bind poly(A)+ RNA.
There are several possibilities by which methylation could mediate an effect on a substrate. First, the modification could regulate protein–protein interactions. Methylation could lead to the formation and stabilization of an interaction or could disrupt a pre-existing interaction. Second, the modification could be involved in protein–RNA interactions (Calnan et al. 1991; Rajpurohit et al. 1994). It has been proposed that arginine methylation could regulate the specificity of RNA binding (Calnan et al. 1991). Alternatively, arginine methylation could also play a role in protein stability. It has been shown that methylated hnRNP A1 is more susceptible to trypsin digestion than its unmethylated counterpart (Rajpurohit et al. 1994). The stability of Npl3p, however, is unaffected in the absence of methylation (M.F. Henry and P.A. Silver, unpubl.). Finally, the presence of the NG, NG-dimethylarginine could help determine the localization of the modified protein (Pintucci et al. 1996) as demonstrated here. For instance, it could be mediating interactions of hnRNPs with the nuclear export machinery (Ullman et al. 1997).
Given the effect of methylation by Hmt1p on the exit of Npl3p and Hrp1p from the nucleus, it is surprising that HMT1 is not an essential gene. However, we have discovered a situation where HMT1 is essential. In strains containing a mutant allele of the gene encoding CBP80, HMT1 is required for viability. Cbp80p and another protein termed Cbp20p form the cap-binding protein complex (CBC) (Izaurralde et al. 1994; Lewis et al. 1996). Cbp80p and Cbp20p have been isolated in yeast by both genetic and biochemical approaches (Uemera and Jigami 1992; Colot et al. 1996; Gorlich et al. 1996). Homologs of both proteins have been identified in other organisms, including amphibia and insects (Izaurralde et al. 1995; Visa et al. 1996b). Studies in Xenopus oocytes and HeLa cell extracts have shown that CBC mediates efficient pre-mRNA splicing, as well as snRNA export (for review, see Lewis and Izaurralde 1997). Studies of the association of Cbp20p with a specific hnRNP complex, the Balbiani ring particle in C. tentans, indicate that Cbp20p binds to nascent hnRNA very soon after transcription initiation and remains attached during translocation through the nuclear pore (Visa et al. 1996b). While the 5′-cap structure is not required for successful export, it has been shown to enhance the rate of mRNA export from the nucleus (Hamm and Mattaj 1990). Thus, the CBC is a good candidate for the role of the mediator of this effect.
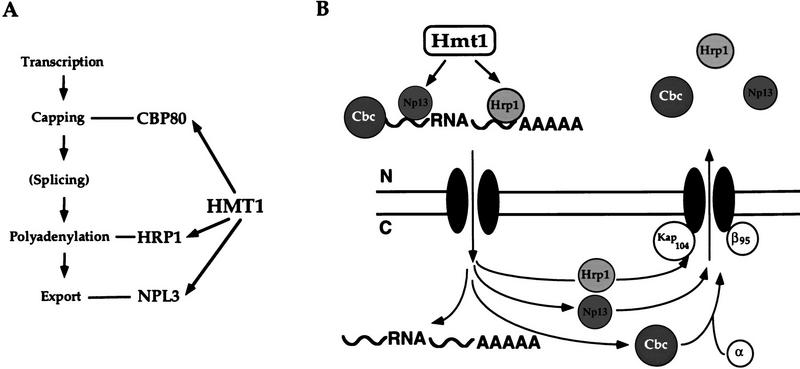
How do Cbp80p, Hrp1p, and Npl3p all affect the export of mRNA from the nucleus? From our genetic data, it is possible that Cbp80p, Hrp1p, and Npl3p all act in a linear pathway of export, with each step being affected by Hmt1p (Fig. 8A). As mentioned previously, a mutant allele of HRP1 is able to suppress a temperature-sensitive allele of NPL3. Suppression could occur by the ability of the mutant Hrp1p to compensate for the defective Npl3p. In this scheme, methylation would play a role at the different levels of mRNA metabolism including capping, polyadenylation, and export. First, Hmt1p has an effect on CBP80 function, as seen by the synthetic lethal interactions. Second, Hmt1p interacts biochemically to modify Hrp1p, a protein necessary for cleavage and polyadenylation. Finally, Hmt1p also modifies Npl3p, a potential carrier of mRNA out of the nucleus. Successful completion of each step, mediated by Hmt1p, could lead to efficient export of the mRNA.
Figure 8.
Models for HMT1 function in pre-mRNA metabolism, packaging, and export. (A) Genetic data suggest that HMT1 affects diverse functions in RNA metabolism including capping, polyadenylation, and export. (B) Hmt1p methylates at least two nuclear RNA binding proteins that play a role in the export of RNA, possibly providing a signal to facilitate association of properly packaged RNAs with the export machinery. Once in the cytoplasm, RNA binding proteins are released and re-enter the nucleus.
A molecular model supported by the data is that proteins such as Cbp80p, Npl3p, and Hrp1p package the RNA for export, and that methylation plays a role in the efficiency of this packaging (Fig. 8B). All three factors are known to exit the nucleus and to bind pre-mRNAs (Flach et al. 1994; Gorlich et al. 1996; Lee et al. 1996; Kessler et al. 1997), thus supporting the possibility that they could be actively involved in export. The presence of proteins associated with capping and polyadenylation could lead to more efficient packaging and export of the RNA to the cytoplasm. Consistent with this concept, we have found that a null allele of CBP80 is synthetically lethal with a temperature-sensitive allele of NPL3 (E.C. Shen and P.A. Silver, unpubl.). In this case, the presence of one defective packaging protein would cause the cell to be sick but still viable. In the presence of more than one defective packaging protein, however, RNA export would be so impaired as to result in cell death.
In this packaging model, Hmt1p could affect the functions of these proposed carriers of mRNA in several ways. One possibility is that methylation of Hrp1p and Npl3p could be affecting the interaction of these factors with each other. In addition, the modification could mediate interactions between these proteins and Cbp80p, or other hnRNPs that are presumably binding pre-mRNAs. Formation of the proper export complex would be crucial for efficient exit of mRNA out of the nucleus. Another possibility is that methylation mediates interactions of these factors with the export machinery such as the recently identified nuclear export receptor, Xpo1p/Crm1p (Adachi and Yanagida 1989; Fornerod et al. 1997; Neville et al. 1997; Ossareh-Nazari et al. 1997; Stade et al. 1997). Modification by Hmt1p could also be mediating the specificity of RNA binding by Hrp1p and Npl3p and hence formation of a functional transport unit. Lack of modification by Hmt1p could result in less efficient packaging of this transport unit, and lead to slower export of mRNA out of the nucleus.
Conservation of the players in the packaging model from yeast to mammals suggests that this model can be applied to higher eukaryotes. The knowledge that human arginine-methylated hnRNPs (Liu and Dreyfuss 1995) are involved in mRNA processing makes it likely that some are involved in the packaging of the RNA. Notably, some viruses exploit the cellular export machinery to facilitate the maturation of their RNAs. Finally, viral RNA-binding proteins such as the herpes simplex virus ICP27 protein are known to be arginine-methylated (Mears and Rice 1996). We should be able to extend our new understanding of the biological role of Hmt1p and arginine methylation to virally infected cells and the design of possible therapeutics.
Materials and methods
Plasmids and strains
Plasmids used in this study are listed in Table 1. Yeast strains used are listed in Table 2. The diploid JU4-2xJR26-19B (gift from Ed Hurt, Biochimie-Zentrum Heidelberg, Germany) was used to generate wild-type haploids PSY603 and PSY609. The npl3-17 allele was integrated by homologous recombination. To create the nup49-313 ΔHMT1 double mutant, nup49-313 was mated to PSY1122, a strain created exactly as described for PSY865 (Henry and Silver 1996). A PCR strategy (Baudin et al. 1993) was employed to disrupt HRB1 in the wild-type strain PSY615. After sporulation, all tetrads had four viable spores, indicating that HRB1 is not essential for vegetative growth. All other strain constructions are described under headings for individual experiments.
Table 1.
Plasmids used in this study
| Plasmid
|
Features
|
Source
|
|---|---|---|
| pETHisHrp1 | HisHRP1 in pET15b | M. Kessler and C. Moore (Tufts Medical School, Boston, MA) |
| pET9c–NPL3 | NPL3 in pET9c | A. Krainer and A. Mayeda (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) |
| pPS293 | yeast URA3 2μ GAL1 promoter expression vector | Lee et al. (1996) |
| pPS311 | yeast CEN URA3 GAL1 promoter expression vector | Flach et al. (1994) |
| pPS709 | yeast 2μ URA3 ADE3 | Koepp et al. (1996) |
| pPS811 | yeast 2μ URA3 GFP–Np13p under GAL1 promoter | Lee et al. (1996) |
| pPS877 | np13-17 allele in pRS306 | Lee et al. (1996) |
| pPS879 | yeast 2μ URA3 GFP–F160L under GAL1 promoter | Lee et al. (1996) |
| pPS892 | yeast 2μ URA3 GAL1 GST fusion expression vector | Koepp et al. (1996) |
| pPS1141 | HMT1 ORF in pPS311 | Henry and Silver (1996) |
| pPS1303 | yeast 2μ URA3 GAL1 sGFP fusion expression vector | this study |
| pPS1305 | 2.7-kb XbaI HMT1 fragment in pRS315 | Henry and Silver (1996) |
| pPS1307 | 2.7-kb XbaI fragment containing HMT1 in pRS316 | Henry and Silver (1996) |
| pPS1308 | 2.7-kb XbaI–HindIII HMTI fragment in YEp351 | Henry and Silver (1996) |
| pPS1319 | NdeI–BamHI His HMT1 PCR fragment in PET15b | this study |
| pPS1320 | 1.8-kb BamHI–AvrII HRB1 fragment in pPS892 | this study |
| pPS1321 | 1766-bp PCR product containing HRB1 in pBSK+ | this study |
| pPS1323 | 2.3-kb BalI–SalI GST–HRB1 fragment in pGEX-4T-1 | this study |
| pPS1324 | 12.5-kb genomic insert in YCp50 (original HRB1 clone) | this study |
| pPS1325 | 3.6-kb XhoI HRB1 fragment from pPS1324 in pRS315 | this study |
| pPS1327 | 2.6-kb SpeI–AvrII HRB1 fragment in PRS315 | this study |
| pPS1332 | 1.8-kb BamHI–AvrII HRB1 fragment in pPS293 | this study |
| pPS1333 | 1.3-kb HRB1–myc PCR product in pPS1332 | this study |
| pPS1340 | 1-kb BsaBI–SalI HRB1-myc fragment in pPS1327 | this study |
| pPS1341 | 1.1-kb HRB1 PCR product in pPS1303 (HRB1–sGFP) | this study |
| pPS1347 | 2.5-kb BamHI HMT1 fragment from pPS1307 in pPS709 | this study |
| pPS1348 | 2.7-kb XbaI HMT1 fragment from pPS1307 in pPS811 | this study |
| pPS1349 | 2.7-kb XbaI HMT1 fragment from pPS1307 in pPS879 | this study |
| pPS1352 | 4.2-kb ClaI fragment containing CBP80 in pRS316 | this study |
| pPS1353 | PstI–XhoI CBP80 fragment from pPS1352 in pRS305 | this study |
| pPS1356 | 3.5-kb ScaI NLP3–myc fragment in pRS315 | this study |
| pPS1357 | NPL3–myc in pET9c–Np13 | this study |
| pPS1358 | Hrp1 fused to the carboxyl terminus of GFP (pCGF–1C–HRP1) | Kessler et al. (1997) |
Table 2.
Yeast strains used in this study
| Strain
|
Genotype
|
Source
|
|---|---|---|
| FY23 | MATa ura3-52 leu2Δ1 trp1Δ63 | Fred Winston (Harvard Medical School, Boston, MA) |
| nup49-313 | MATα nup49∷TRP1 ura3 leu2 his3 ade2 ade3 plus pUN100–nup49ts–LEU2 | Doye et al. (1994) |
| CH1305 | MATa ura3 leu2 his3 ade2 ade3 can1 | Kranz and Holm (1990) |
| JU4-2 × JR26-19B | MATa/α ura3-52/−leu2-3/−his3/+ his4/+ trp1/+lys1-1/−ade2-1/− ade8/+ can1-100/− | Ed Hurt |
| PSY603 | MATα ura3-52 leu2-3 his4 trp1 lys1-1 ade2-1 ade8 can1-100 | this study |
| PSY609 | MATα ura3-52 leu2-3 his3 lys1-1 ade2-1 ade8 can1-100 | this study |
| PSY615 | MATa/α ura3-52/− leu2-3/−his3/− trp1/+ lys-1/− ade2-1/− ade8/− | Henry et al. (1996) |
| PSY814 | MATa npl3∷HIS3 ura3-52 leu2-3 his3 trp1 lys1-1 ade2-1 ade8 plus YCpNPL3-3 | Henry et al. (1996) |
| PSY865 | MATα hmt1∷HIS3 ura3 leu2 his3 lys1 ade2 ade8 | Henry and Silver (1996) |
| PSY902 | MATa/α ura3-52/− leu2Δ1/− his3Δ200/−trp1/+ lys2/+ade2/+ ade3/+ | this study |
| PSY992 | MATa/α npl3∷HIS3/npl3-17 ura3-52/− leu2-3/− his3/− trp1-1/+ lys1-1/− ade2-1/− ade8/−can1-100/− | this study |
| PSY1023 | MATa npl3-17 ura3-52 leu2-3 his3 lys1-1 ade2-1 ade8 can1-100 | this study |
| PSY1096 | MATa hmt1∷HIS3 nup49∷TRP ura3-52 leu2 lys− ade2 ade8 plus pUN100–nup49-313–LEU2 | this study |
| PSY1115 | MATa hmt1∷HIS3 ura3 leu2 lys− ade2 ade3 plus pPS1347 | this study |
| PSY1116 | MATa hmt1∷HIS3 cbp80-1 ura3 leu2 lys− ade2 ade3 plus pPS1347 | this study |
| PSY1117 | MATa hmt1∷HIS3 cbp80-1 ura3 leu2 lys− ade2 ade3 plus pPS1351 | this study |
| PSY1118 | MATα hmt1∷HIS3 CBP80:LEU2 ura3 leu2 his3 lys1 ade2 | this study |
| PSY1119 | MATa cbp80∷HIS3 ura3 leu2 his3 trp1 ade2 | this study |
| PSY1120 | MATα cbp80∷HIS3 ura3 leu2 lys1 ade2 ade8 | this study |
| PSY1121 | MATa/α npl3∷HIS3/NPL3 ura3-52/− leu2-3/− his3/− trp1-1/+ lys1-1/− ade2-1/− ade8/− can1-100/− | this study |
| PSY1122 | MATa hmt1∷HIS3 ura3 leu2 his3 lys1 ade2 ade8 | Henry and Silver (1996) |
| PSY1123 | MATa/α hrb1∷HIS3/HRB1 ura3/− leu2/− his3/− trp1-1/+ lys1/− ade2/− ade8/− can1-100/− | this study |
| PSY1124 | MATa hrb1∷HIS3 ura3 leu2 his3 trp1-1 lys1 ade2 ade8 can1-100 | this study |
| PSY1129 | MATa npl3∷HIS3 ura3-52 leu2-3 his3 trp1 lys1-1 ade2-1 ade8 can1-100 plus pPS1356 | this study |
Export assay
The nuclear export assay was performed exactly as described (Lee et al. 1996). Briefly, cells containing various reporter plasmids are induced for 2 hr in medium containing galactose to express the reporter followed by repression for 2 hr in medium containing glucose. Cells are then incubated at 25°C or 36.5°C for 5 hr and examined for GFP fluorescence.
Purification of recombinant proteins from E. coli
HisHmt1 was purified from E. coli by binding to a charged nickel column (Pharmacia). Bound proteins were eluted with 500 mm NaCl, 50 mm EDTA and dialyzed against buffer B (20 mm MOPS at pH 7.2, 50 mm NaCl, 2 mm EDTA, 5 mm DTT) containing 50 mm EDTA, then against buffer B. HisHrp1 was purified from E. coli like HisHmt1 except that eluted proteins were dialyzed against buffer C (20 mm potassium phosphate at pH 7.4, 50 mm KCl, 0.2 mm EDTA, 5 mm DTT) containing 50 mm EDTA and then against buffer C. HisHrp1 was then bound to a SP Sepharose column (Pharmacia) equilibrated with buffer C, and eluted with a step gradient of buffer C containing 250 mm, 500 mm, and then 750 mm KCl. Proteins were then dialyzed into buffer D (20 mm Tris at pH 8.0, 50 mm KCl, 0.2 mm EDTA, 5 mm DTT). GST–Hrb1p was purified from E. coli by affinity to glutathione–sepharose (Pharmacia) and released from GST by cleavage with thrombin (Sigma) in the presence of 20 mm glutathione. Free GST was removed by incubation with glutathione–agarose, and the remaining protein was dialyzed against methylation assay buffer (50 mm MOPS at pH 7.2, 2 mm EDTA, 300 mm NaCl). HisHmt1p, HisHrp1p, and Hrb1p were concentrated with a Centriprep-10 ultra centrifugation unit (Amicon). Npl3P–Myc was purified from E. coli by cesium chloride gradient fractionation as described (Krainer et al. 1991) by monitoring fractions with the mouse monoclonal anti-c-Myc 9E10 antibody. Fractions containing Npl3p–Myc were purified further by anion-exchange chromatography on a Mono-Q column (Pharmacia) and cation-exchange chromatography on a Mono-S column (Pharmacia).
Methylation assay
Methylation of recombinant proteins was performed as described (Henry and Silver 1996). Briefly, the 40-μl reaction contained 50 mm MOPS at pH 7.2, 2 mm EDTA, 300 mm NaCl, 30 μm SAM, 5.5 μCi [methyl3H]-SAM (NEN Dupont, 80 Ci/mmole), and 3 μg of substrate protein. Npl3p–Myc, HisHrp1p, and Hrb1p were prepared from E. coli, and the BSA control was purchased from Sigma. Reactions were initiated by the addition of 4 μg of HisHmt1p or methylation assay buffer, and incubated at 30°C for 40 min. Reactions were terminated by the addition of SDS-PAGE sample buffer and boiling for 5 min at 100°C. Proteins were resolved by SDS-PAGE followed by Coomassie staining and fluorography.
Indirect immunofluorescence
Indirect immunofluorescence microscopy was performed exactly as described (Lee et al. 1996). Affinity purified rabbit anti-Npl3p (Bossie et al. 1992) was used at 1:1000, and 9E10 mouse monoclonal anti-Myc (Department of Molecular Biology, Princeton University, NJ) was used at 1:2. In situ hybridization to poly(A)+ RNA was performed exactly as described (Lee et al. 1996).
CBP80 synthetic lethal screen
To create the parental strain for the synthetic lethal screen, PSY865 was mated with CH1305 (Kranz and Holm 1990), diploids were selected, tetrads were dissected, and one ΔHMT1::HIS3 ade2 ade3 spore was selected and transformed with pPS1347 to generate PSY1115. pPS1347, a 2μ URA3 ADE3 HMT1 plasmid, was generated by subcloning of a 2.5-kb BamHI fragment from pPS1307 (Henry and Silver 1996) into the BamHI site of pPS709 (Koepp et al. 1996).
Cells were mutagenized with EMS to 50% killing. Approximately 53,000 colonies were screened. Three hundred and sixteen non-sectoring colonies were streaked on fresh YEPD medium plates. One candidate satisfied all the criteria (Henry and Silver 1996; Koepp et al. 1996) for synthetic lethality with ΔHMT1. A 4.2-kb ClaI fragment containing one complete ORF in pRS316 (pPS1352) was the minimal region required to restore sectoring. Linkage of the synthetic lethality to CBP80 was checked by mating PSY1118 with PSY1116. The diploid was sporulated and tetrads analyzed for FOA sensitivity. PSY1118 contains a LEU2 marked copy of CBP80 generated by homologous recombination. To disrupt the CBP80 gene, a PCR strategy was performed (Baudin et al. 1993). To check synthetic lethality between ΔHMT1 and ΔCBP80, PSY1119 was outcrossed three times to the wild-type strain PSY609. The resulting ΔCBP80 strain, PSY1120, was crossed to PSY865, and tetrads were analyzed.
Acknowledgments
We gratefully acknowledge Adrian Krainer and Akila Mayeda for pET9c–NPL3 and protocols for purification of recombinant protein from E. coli; Clarie Moore and Marco Kessler for pET15b-HisHrp1; Patricia Chui for unpublished results; and Ed Hurt and F. Winston for strains. We also thank Michael Rosbash, Robert Kingston, and members of the Silver laboratory for critical reading of the manuscript and helpful comments and discussion. This work was funded by grants from the National Institutes of Health to P.A.S., The Medical Foundation Charles King Trust to M.S.L., a predoctoral fellowship from Sandoz Pharmaceuticals to E.C.S., and a National Science Foundation predoctoral fellowship to V.H.W. S.R.V. is on leave of absence from Faculdade de Ciencias Farmaceuticas de Araraquara–Universidade Estadual Paulista and is a recipient of a postdoctoral fellowship from Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pamela_silver@dfci.harvard.edu; FAX (617) 632-5103.
References
- Adachi Y, Yanagida M. Higher order chromosome structure is affected by cold-sensitive mutations in a Schizosaccharomyces pombe gene CRM1+, which encodes a 115-kD protein preferentially localized in the nucleus and its periphery. J Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulou O, Denouel A, Lacroute F, Cullin C. A simple and efficient method of direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Christensen ME, Walker BW, LeStourgeon WM. Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell. 1977;11:127–138. doi: 10.1016/0092-8674(77)90323-3. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: Conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffa LC, Karn J, Vidali G, Allfrey VG. Distribution of NG-NG-dimethylarginine in nuclear protein fractions. Biochem Biophys Res Commun. 1977;74:969–976. doi: 10.1016/0006-291x(77)91613-8. [DOI] [PubMed] [Google Scholar]
- Bossie MA, DeHoratius C, Barcelo G, Silver P. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol Biol Cell. 1992;3:875–893. doi: 10.1091/mbc.3.8.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Arginine-mediated RNA recognition: The arginine fork. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Cobianchi F, Calvio C, Stoppin M, Buvoli M, Riva S. Phosphorylation of human hnRNP protein A1 abrogates in vitro strand annealing activity. Nucleic Acids Res. 1993;21:949–955. doi: 10.1093/nar/21.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap binding complex. Genes & Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+ RNA transport and nuclear pore distribution. EMBO J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Fischer U, Michael M, Luhrmann R, Dreyfuss G. Signal-mediated nuclear export pathways of proteins and RNAs. Trends Cell Biol. 1996;6:290–293. doi: 10.1016/0962-8924(96)20030-3. [DOI] [PubMed] [Google Scholar]
- Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins DA, Silver PA. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Gary JD, Lin WJ, Yang MC, Herschman HR, Clarke S. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J Biol Chem. 1996;271:12585–12594. doi: 10.1074/jbc.271.21.12585. [DOI] [PubMed] [Google Scholar]
- Ghosh SK, Paik WK, Kim S. Purification and molecular identification of two protein methylases I from calf brain. Myelin basic protein and histone specific enzyme. J Biol Chem. 1988;263:19024–19033. [PubMed] [Google Scholar]
- Gorlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey RA, Mattaj IW, Izaurralde E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- Henry M, Borland CZ, Bossie M, Silver PA. Potential RNA binding proteins in Saccharomyces cerevisiae identified as suppressors of temperature-sensitive mutations in NPL3. Genetics. 1996;142:103–115. doi: 10.1093/genetics/142.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MF, Silver PA. A novel methyltransferase (Hmt1p) modifies poly(A)+ RNA binding proteins. Mol Cell Biol. 1996;16:3668–3678. doi: 10.1128/mcb.16.7.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. 1997;137:27–35. doi: 10.1083/jcb.137.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Yaspo ML, Fisher EM. Identification and mapping of a novel human gene, HRMT1L1, homologous to the rat protein arginine N-methyltransferase 1 (PRMT1) gene. Mammal Genome. 1997;8:526–529. doi: 10.1007/s003359900491. [DOI] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes & Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Burd CG, Portman DS, Dreyfuss G. Structure and function of hnRNP proteins. In: Nagai K, Mattaj IW, editors. RNA–protein interactions: Frontiers in molecular biology. Oxford, UK: IRL Press; 1994. pp. 127–149. [Google Scholar]
- Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: Homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Kranz JE, Holm C. Cloning by function: An alternative approach for identifying yeast homologs from other organisms. Proc Natl Acad Sci. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Silver PA. RNA movement between the nucleus and the cytoplasm. Curr Opin. 1997;7:212–219. doi: 10.1016/s0959-437x(97)80131-1. [DOI] [PubMed] [Google Scholar]
- Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes & Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Gorlich D, Mattaj IW. A yeast cap-binding protein complex (yCBC) acts as an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WJ, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. MolCell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears WE, Rice SA. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: A signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Najbauer J, Johnson BA, Young AL, Aswad DW. Peptides with sequences similar to glycine, arginine-rich motifs in proteins interacting with RNA are efficiently recognized by methyltransferase(s) modifying arginine in numerous proteins. J Biol Chem. 1993;268:10501–10509. [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Nigg E. Nucleocytoplasmic transport: Signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- Nikawa J, Nakano H, Ohi N. Structural and functional conservation of human and yeast HCP1 genes which can suppress the growth defect of the Saccharomyces cerevisiae ire15 mutant. Gene. 1996;171:107–111. doi: 10.1016/0378-1119(96)00073-x. [DOI] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. Eukaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Philippsen P, Kleine K, Pohlmann R, Dusterhoft A, Hamberg K, Hegemann JH, Obermaier B, Urrestarazu LA, Aert R, Albermann K, et al. The nucleotide sequence of Saccharomyces cerevisiae chromosome XIV and its evolutionary implications. Nature. 1997;387:93–98. [PubMed] [Google Scholar]
- Piñol-Roma S. HnRNP proteins and the nuclear export of mRNA. Sem Cell Dev Biol. 1997;8:57–63. doi: 10.1006/scdb.1996.0122. [DOI] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- ————— Cell cycle-regulated phosphorylation of the pre-mRNA binding (heterogeneous nuclear ribonucleoprotein) C proteins. Mol Cell Biol. 1993;13:5762–5770. doi: 10.1128/mcb.13.9.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintucci G, Quarto N, Rifkin DB. Methylation of high molecular weight fibroblast growth factor-2 determines post-translational increases in molecular weight and affects its intracellular distribution. Mol Biol Cell. 1996;7:1249–1258. doi: 10.1091/mbc.7.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpurohit R, Paik WK, Kim S. Effect of enzymic methylation of heterogeneous ribonucleoprotein particle A1 on its nucleic-acid binding and controlled proteolysis. Biochem J. 1994;304:903–909. doi: 10.1042/bj3040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal N, Rajpurohit R, Paik WK, Kim S. Purification and characterization of S-adenosylmethionine protein arginine N-methyltransferase from rat liver. Biochem J. 1994;300:483–489. doi: 10.1042/bj3000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell ID, Tollervey D. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J Cell Biol. 1992;119:737–747. doi: 10.1083/jcb.119.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebel CW, Guthrie C. The essential yeast RNA binding protein Npl3p is methylated. Proc Natl Acad Sci. 1996;93:13641–13646. doi: 10.1073/pnas.93.24.13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton DR, Chen S, Hitomi M, Kumagai C, Tartakoff AM. A yeast protein that bidirectionally affects nucleocytoplasmic transport. J Cell Sci. 1995;108:265–272. doi: 10.1242/jcs.108.1.265. [DOI] [PubMed] [Google Scholar]
- Soulard M, Veronique DV, Siomi MC, Piñol-Roma S, Codogno P, Bauvy C, Bellini M, Lacroix JC, Monod G, Dreyfuss G, et al. hnRNP G: Sequence and characterization of a glycosylated RNA-binding protein. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Uemera H, Jigami Y. GCR3 encodes an acidic protein that is required for expression of glycolytic genes in Saccharomyces cerevisiae. J Bacteriol. 1992;174:5526–5532. doi: 10.1128/jb.174.17.5526-5532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman KS, Powers MA, Forbes DJ. Nuclear export receptors: From importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- Visa N, Alzhanova-Ericsson AT, Sun X, Kiseleva E, Bjorkroth B, Wurtz T, Daneholt B. A pre-mRNA-binding protein accompanies the RNA from the gene through the nuclear pores and into polysomes. Cell. 1996a;84:253–264. doi: 10.1016/s0092-8674(00)80980-0. [DOI] [PubMed] [Google Scholar]
- Visa N, Izaurralde E, Ferreira J, Daneholt B, Mattaj IW. A nuclear cap-binding complex binds Balbiani ring pre-mRNA cotranscriptionally and accompanies the ribonucleoprotein particle during nuclear export. J Cell Biol. 1996b;133:5–14. doi: 10.1083/jcb.133.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SM, Datar KV, Paddy MR, Swedlow JR, Swanson M. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J Cell Biol. 1994;127:1173–1184. doi: 10.1083/jcb.127.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]