Abstract
By virtue of its control over major histocompatibility complex class II (MHC-II) gene expression, CIITA represents a key molecule in the regulation of adaptive immune responses. It was first identified as a factor that is defective in MHC-II deficiency, a hereditary disease characterized by the absence of MHC-II expression. CIITA is a highly regulated transactivator that governs all spatial, temporal, and quantitative aspects of MHC-II expression. It has been proposed to act as a non-DNA-binding transcriptional coactivator, but evidence that it actually functions at the level of MHC-II promoters was lacking. By means of chromatin immunoprecipitation assays, we show here for the first time that CIITA is physically associated with MHC-II, as well as HLA–DM, Ii, MHC-I, and β2m promoters in vivo. To dissect the mechanism by which CIITA is recruited to the promoter, we have developed a DNA-dependent coimmunoprecipitation assay and a pull-down assay using immobilized promoter templates. We demonstrate that CIITA recruitment depends on multiple, synergistic protein–protein interactions with DNA-bound factors constituting the MHC-II enhanceosome. CIITA therefore represents a paradigm for a novel type of regulatory and gene-specific transcriptional cofactor.
Keywords: MHC class II, CIITA, enhanceosome, coactivator, transcription, gene regulation
Major histocompatibility complex class II (MHC-II) molecules are heterodimeric (α chain/β chain) cell-surface glycoproteins specialized in the presentation of peptide antigens to CD4+ T helper cells. These molecules play central roles in the initiation and propagation of CD4+ T-cell-mediated immune responses, as well as in the development and homeostasis of the CD4+ T-cell population. Tightly regulated MHC-II expression therefore represents a key parameter in the control of the immune response. This is exemplified by the fact that a lack of MHC-II expression results in a severe immunodeficiency disease, whereas inappropriate expression is associated with autoimmune diseases. Elucidation of the mechanisms that regulate MHC-II expression therefore represents a major challenge in molecular immunology and immunopathology.
The genes encoding the α and β chains of all MHC-II isotypes (HLA–DR, HLA–DQ, and HLA–DP) are in general coregulated. In addition, the genes encoding the invariant chain (Ii) and HLA–DM are coexpressed with MHC-II genes. The latter play important accessory roles for antigen presentation by MHC-II molecules. Two modes of MHC-II, Ii, and HLA–DM expression, constitutive and inducible, are generally recognized (Benoist and Mathis 1990; Glimcher and Kara 1992; Ting and Baldwin 1993; Mach et al. 1996; Boss 1997). Constitutive expression is largely restricted to specialized cells of the immune system, including thymic epithelial cells and professional APC such as B cells, macrophages, and dendritic cells. The majority of other cell types lack MHC-II molecules, but can be induced to express them by exposure to various inducing agents, of which the most potent and well known is interferon-γ. Both modes of expression are controlled primarily at the level of transcription.
The major transcriptional control element is a short 150-bp regulatory module that has been conserved in the promoter proximal regions of all MHC-II, Ii, and HLA–DM genes (Benoist and Mathis 1990; Glimcher and Kara 1992; Ting and Baldwin 1993; Mach et al. 1996; Boss 1997). This regulatory module consists of four cis-acting sequences known as the W, X, X2, and Y boxes, which are conserved in their sequence, orientation, and spacing with respect to each other. The W-X-X2-Y region behaves as a single functional unit in which all four elements contribute synergistically to optimal promoter activity (Vilen et al. 1991, 1992; Reith et al. 1994b). Interestingly, a conserved homologous region has also been recently identified in promoters of the β2-microglobulin (β2m) and MHC-I genes (van den Elsen et al. 1998). MHC-I and β2m form heterodimers that are exposed at the cell surface and present antigens to CD8+ T lymphocytes. Contrary to MHC-II, MHC-I and β2m genes are constitutively expressed in virtually all nucleated cells, and their transcription is predominantly driven by cis-acting sequences situated outside of the W-X-X2-Y region (David-Watine et al. 1990; Singer and Maguire 1990; Ting and Baldwin 1993). Not surprisingly, the homology between MHC-I and MHC-II promoters has long escaped attention.
A great deal of our current understanding of the molecular machinery that regulates transcription of MHC-II genes has come from studies of the defects underlying MHC-II deficiency (also called the Bare Lymphocyte syndrome), which is a hereditary disease of MHC-II gene regulation (de Preval et al. 1985; Mach et al. 1996). Elucidation of the molecular basis for MHC-II deficiency has led to the identification of CIITA and RFX, two essential transactivators of MHC-II expression (Mach et al. 1996). RFX is a ubiquitously expressed X box-binding protein consisting of three subunits called RFX5, RFXAP, and RFXANK (RFX-B) (Reith et al. 1988; Steimle et al. 1995; Durand et al. 1997; Masternak et al. 1998; Nagarajan et al. 1999). RFX binds in concert with two other transcription factors, NF-Y and X2BP, which recognize, respectively, the Y and X2 boxes of MHC-II, Ii, and HLA–DM promoters. NF-Y (CBF, CP1) is a well characterized, ubiquitous CCAAT-binding protein that associates with a wide variety of eukaryotic RNA Pol II promoters (Maity and De Crombrugghe 1998; Mantovani 1999). The X2-binding protein (X2BP) belongs to a large family of bZip transcription factors. A number of these factors are capable of binding to the X2 boxes of MHC-II promoters in vitro (Benoist and Mathis 1990; Mach et al. 1996; Boss 1997) and the precise identity of X2BP has for long remained unclear. At least one member of the bZip family, the cAMP responsive element-binding protein 1 (CREB-1), may be considered as a bona fide candidate for X2BP, because chromatin immunoprecipitation experiments demonstrated that it associates with MHC-II promoters in vivo (Moreno et al. 1999). CREB-1 also activated transcription of a reporter gene in an X2-dependent manner (Moreno et al. 1999).
Binding of RFX, X2BP, and NF-Y to the promoter DNA is highly cooperative and results in the formation of a remarkably stable higher-order nucleoprotein complex (Reith et al. 1994a,b; Louis-Plence et al. 1997) that can be regarded as the MHC-II enhanceosome (Thanos and Maniatis 1995). In the absence of RFX, the enhanceosome cannot form such that MHC-II promoters remain unoccupied in vivo (Kara and Glimcher 1991, 1993). Enhanceosome assembly is essential but not sufficient for MHC-II expression, which ultimately depends on CIITA (Steimle et al. 1993), a highly regulated master control factor that determines the level, cell type specificity, inducibility, and extinction of MHC-II expression (Steimle et al. 1993, 1994; Silacci et al. 1994; Mach et al. 1996; Boss 1997; Otten et al. 1998). The complex control of CIITA transcription is in its turn achieved by the differential activation of multiple alternative promoters (Muhlethaler-Mottet et al. 1997).
The biological role of CIITA as a master controller of MHC-II genes has now been firmly established. On the other hand, despite >6 years of research, relatively little has been learned about its mode of action. CIITA does not bind to DNA (Steimle et al. 1993). Instead, it is believed to function as a transcriptional coactivator that is recruited to MHC-II promoters by interactions with promoter-bound factors (Riley et al. 1995; Zhou and Glimcher 1995; Scholl et al. 1997; Brown et al. 1998). However, data supporting this hypothetical working model has remained disappointingly indirect. Evidence consistent with the model was provided by studies suggesting that CIITA contains amino-terminal acidic and proline/serine/threonine-rich transcription activation domains that can contact general transcription factors and coactivators (Riley et al. 1995; Zhou and Glimcher 1995; Bontron et al. 1997b; Chin et al. 1997; Mahanta et al. 1997; Brown et al. 1998; Kretsovali et al. 1998; Wright et al. 1998; Fontes et al. 1999). On the other hand, no convincing proof for a direct physical interaction between CIITA and MHC-II promoter-binding factors has yet been provided. Here we demonstrate that CIITA is associated with MHC-II and related promoters in living cells. CIITA recruitment is mediated by multiple weak interactions between its carboxy-terminal moiety and several different components of the MHC-II enhanceosome. RFX, NF-Y, X2BP, and W box-binding proteins all participate in the generation of the CIITA interaction surface.
Results
CIITA is recruited to MHC-II and related promoters in vivo
To determine whether CIITA is physically associated with MHC-II promoters in living cells, we performed chromatin immunoprecipitation assays (Orlando et al. 1997). Briefly, formaldehyde cross-linked chromatin fragments were isolated from B cells, immunoprecipitated with antibodies directed against CIITA, and analyzed by PCR for the presence of specific DNA sequences corresponding to various promoters. The experiments were performed in parallel with two B-cell lines, RJ2.2.5 and RJ6.4. RJ2.2.5 is a mutant MHC-II-negative cell line lacking CIITA (Steimle et al. 1993). RJ6.4 was generated by stable complementation of RJ2.2.5 with a CIITA expression vector and is MHC-II positive. The promoters of three MHC-II genes (HLA–DRA, HLA–DRB1, and HLA–DPB1) were analyzed. All MHC-II promoters were immunoprecipitated with anti-CIITA antibodies in experiments performed with RJ6.4 cells, but not with RJ2.2.5 cells (Fig. 1). This demonstrates for the first time that CIITA is recruited to MHC-II promoters in vivo. Additionally, CIITA was found to be physically associated with other promoters containing the W-X-X2-Y homology region, such as the promoters of the class II-related genes HLA–DMB and Ii, the MHC-I genes HLA-A and HLA-B, and the β2m gene (Fig. 1).
Figure 1.
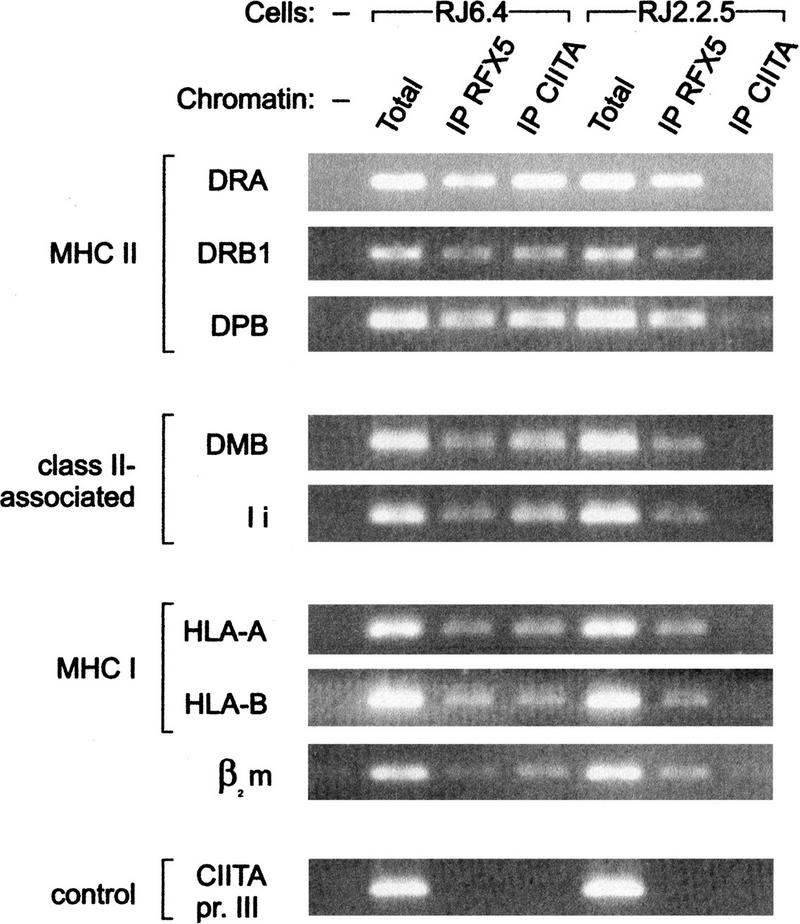
CIITA is associated with MHC-II and related promoters in vivo. Cross-linked chromatin isolated from CIITA-positive RJ6.4 cells and CIITA-deficient RJ2.2.5 cells was immunoprecipitated with antibodies against CIITA and RFX5. Immunoprecipitates were analyzed by PCR for the presence of promoter sequences of the MHC-II genes HLA–DRA, HLA–DRB1, and HLA–DPB1, class II-associated genes HLA–DMB and Ii, MHC-I genes HLA-A and HLA-B, and the β2m gene. Input chromatin (Total) and a sample without chromatin (−) were included as PCR controls. PCR amplification of promoter III of the MHC2TA gene (CIITA pr. III) was used as a control for the specificity of the immunoprecipitation.
In parallel, we also performed chromatin immunoprecipitation assays to analyze the binding of RFX, a well-established component of the MHC-II enhanceosome. As expected, all of the MHC-II and related promoters were immunoprecipitated with anti-RFX5 antibodies. In contrast to CIITA, however, the association of RFX with these promoters was equivalent in RJ6.4 and RJ2.2.5 cells (Fig. 1). This is consistent with previous in vivo footprinting studies demonstrating that the enhanceosome is a stable feature of MHC-II promoters in B cells, and that its assembly in these cells is independent of the presence of CIITA (Kara and Glimcher 1991). The B-cell-specific promoter III of the MHC2TA gene (Muhlethaler-Mottet et al. 1997; Ghosh et al. 1999) is not activated by RFX or CIITA and was not immunoprecipitated with either the CIITA or RFX5 antibodies (Fig. 1). This confirms the specificity of the assay.
CIITA binds to the assembled MHC-II enhanceosome
Classical protein–protein interaction studies have, to a large extent, failed to demonstrate direct contacts between CIITA and individual MHC-II promoter-binding proteins. There is, in fact, only one report suggesting the existence of an interaction between CIITA and RFX5. However, this study was not conclusive because the interaction was observed with a severely truncated nonfunctional version of RFX5, but not with the full-length wild-type RFX5 protein (Scholl et al. 1997). We hypothesized that the failure to demonstrate interactions between CIITA and isolated MHC-II promoter-binding factors reflects the weakness of individual interactions. Efficient recruitment of CIITA may require the synergistic contribution of several weak contacts with multiple components of the MHC-II enhanceosome. To test this possibility, we set out to analyze the interaction of CIITA with the entire enhanceosome, rather than with isolated factors. Enhanceosomes were allowed to assemble with crude B-cell extracts and DNA templates encompassing the W-X-X2-Y region of the HLA–DRA promoter (Fig. 2A). After enhanceosome formation, proteins were immunoprecipitated with anti-CIITA antibodies (Fig. 2B, lane 1), with antibodies directed against the RFXAP subunit of the RFX complex (lane 3), or with preimmune serum (lane 5). In negative control reactions, HLA–DRA promoter fragments were replaced by random DNA (Fig. 2B, lanes 2,4,6). Immunoprecipitates were analyzed for the presence of CIITA, RFX, and NF-Y. The latter two factors are well-defined components of the enhanceosome and are both stable heterotrimeric complexes. To detect RFX and NF-Y by immunoblot, we therefore used antibodies specific for the RFX5 and NF-YB subunits. Efficient coimmunoprecipitation of RFX, NF-Y, and CIITA is observed only when the promoter DNA template was included (Fig. 2B, lanes 1,3). It is not observed in control reactions lacking the promoter template (Fig. 2B, lanes 2,4). The trace amounts of CIITA apparent in the RFXAP immunoprecipitation done without promoter template (lane 4) do not reflect a specific RFX–CIITA interaction because they are also seen with preimmune serum (Fig. 2B, lanes 5,6). In contrast, the small amount of NF-Y that copurified with RFX in the absence of DNA template (lane 4) is not observed with preimmune serum (Fig. 2B, lanes 5,6). This suggests that a weak interaction between these two proteins occurs in solution, which is in line with previous observations (Wright et al. 1994).
Figure 2.
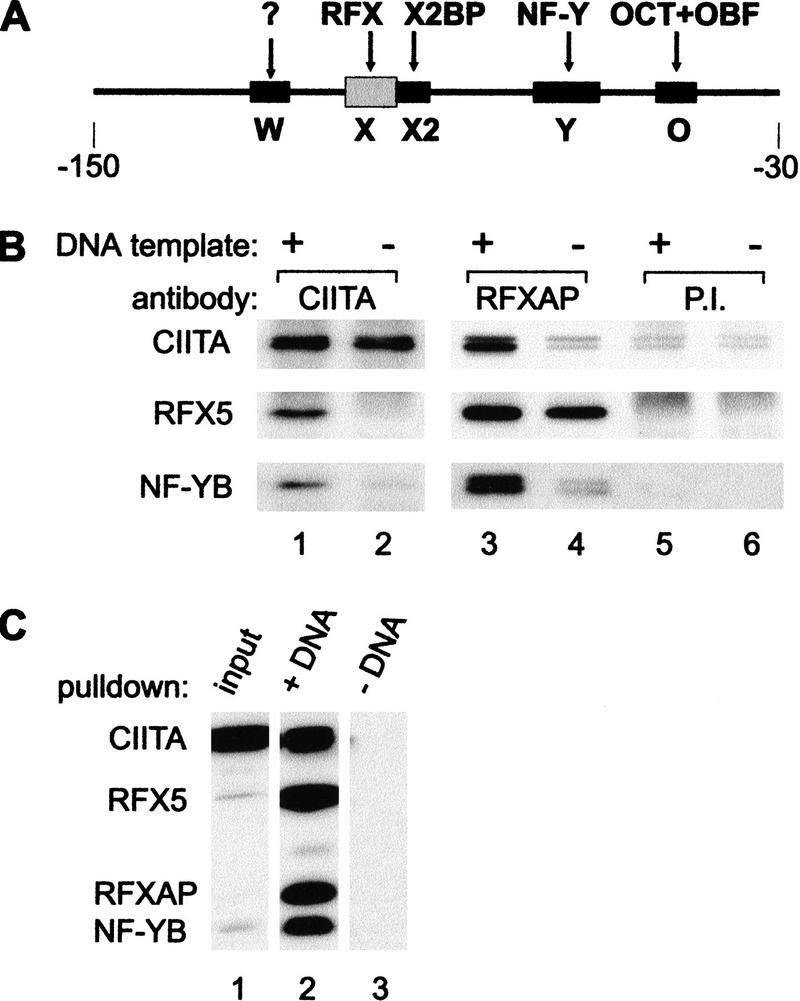
CIITA interacts with the MHC-II enhanceosome. (A) The −150 to −30 HLA–DRA promoter fragment used for enhanceosome assembly contains the W, X, X2, Y, and octamer (O) sequences. Factors (RFX, X2BP, NF-Y, OCT, OBF-1) binding to these sequences are shown. The W-binding factor remains undefined. (B) DNA-dependent coimmunoprecipitation of enhanceosome components and CIITA. RJ6.4 extract was incubated with the HLA–DRA promoter fragment shown above (A). Protein complexes assembled on promoter DNA templates (+) or in the presence of nonspecific DNA (−) were immunoprecipitated with antibodies against CIITA, the RFXAP subunit of RFX, or preimmune serum (P.I.). Immunoprecipitates were analyzed by immunoblotting with antibodies against CIITA, RFX5, and the B subunit of NF-Y. (C) Recruitment of CIITA to HLA–DRA promoter fragments immobilized on magnetic beads. RJ6.4 extract (Input) was subjected to a pull-down assay with immobilized promoter DNA templates (+DNA) or with empty magnetic beads (−DNA). Proteins purified on magnetic beads were analyzed by immunoblotting for the presence of CIITA, RFX5, RFXAP, and NF-YB. A total of 1% of the input extract was analyzed in parallel to visualize the enrichment obtained.
The DNA-dependent coimmunoprecipitation experiments demonstrated formally that at physiological conditions found in cell extracts, CIITA interacts physically with the assembled enhanceosome rather than with isolated components such as RFX or NF-Y. To confirm the interactions observed in DNA-dependent coimmunoprecipitation experiments, we developed a pull-down assay using HLA–DRA promoter templates immobilized on magnetic beads (Fig. 2C). Recruitment of CIITA to the enhanceosome is demonstrated by copurification of CIITA with RFX and NF-Y from a B-cell extract that was incubated with the immobilized templates (Fig. 2C, lane 2). CIITA recruitment occurred also in extracts from MHC-II-negative fibroblasts transfected with CIITA (data not shown), indicating that the enhanceosome components required for recruitment are not B-cell-specific. This is consistent with the finding that transfection of CIITA is sufficient to activate MHC-II expression in various MHC-II-negative cells (Silacci et al. 1994; Mach et al. 1996; Boss 1997; Piskurich et al. 1999; Villard et al. 1999). The interactions observed in the pull-down assay between promoter DNA, enhanceosome components, and CIITA are specific, as no purification of any of the factors was observed when empty magnetic beads were used (Fig. 2C, lane 3) or when a nonspecific DNA was used as immobilized template (data not shown). The specificity of the assay for CIITA and enhanceosome components was further demonstrated by the fact that different unrelated factors, such as the transcription factor JunB, subunits of TFIID (TBP, TAF250), or the coactivator CBP, were not purified by this procedure (data not shown).
Recruitment of CIITA to the promoter requires multiple enhanceosome components
To identify sequence elements critical for CIITA recruitment, mutated HLA–DRA promoter templates were immobilized on magnetic beads and tested in pull-down assays. Mutations of the W, X2, and Y boxes all strongly reduced CIITA recruitment (Fig. 3A, lanes 3,5,6 and B, lane 4). The Y mutation specifically eliminated binding of NF-Y, indicating that this protein is crucial. The drastic effect of the W and X2 mutations also demonstrates the importance of X2BP and W-binding factors for CIITA recruitment. These factors are likely to provide direct contacts with CIITA because the X2 and W mutations did not interfere with binding of RFX or NF-Y. The W-binding factor could not be assayed because its identity remains unknown. Concerning X2BP, a recent report has proposed that it contains CREB-1 (Moreno et al. 1999). We observed some X2 box-dependent binding of CREB-1 to the HLA–DRA promoter templates in the pull-down assay (Fig. 3B, cf. lanes 2,3 with 4). However, in contrast to RFX and NF-Y, CREB-1 was not enriched in the pull-down assay (Fig. 3B, cf. lanes 1 and 2). Moreover, the efficiency of CIITA recruitment was not affected in CREB-1-depleted extracts (data not shown), which suggests that CREB-1 is not obligatory for enhanceosome formation and CIITA recruitment. Perhaps X2BP function can also be provided by other members of the CREB/ATF family of transcription factors.
Figure 3.
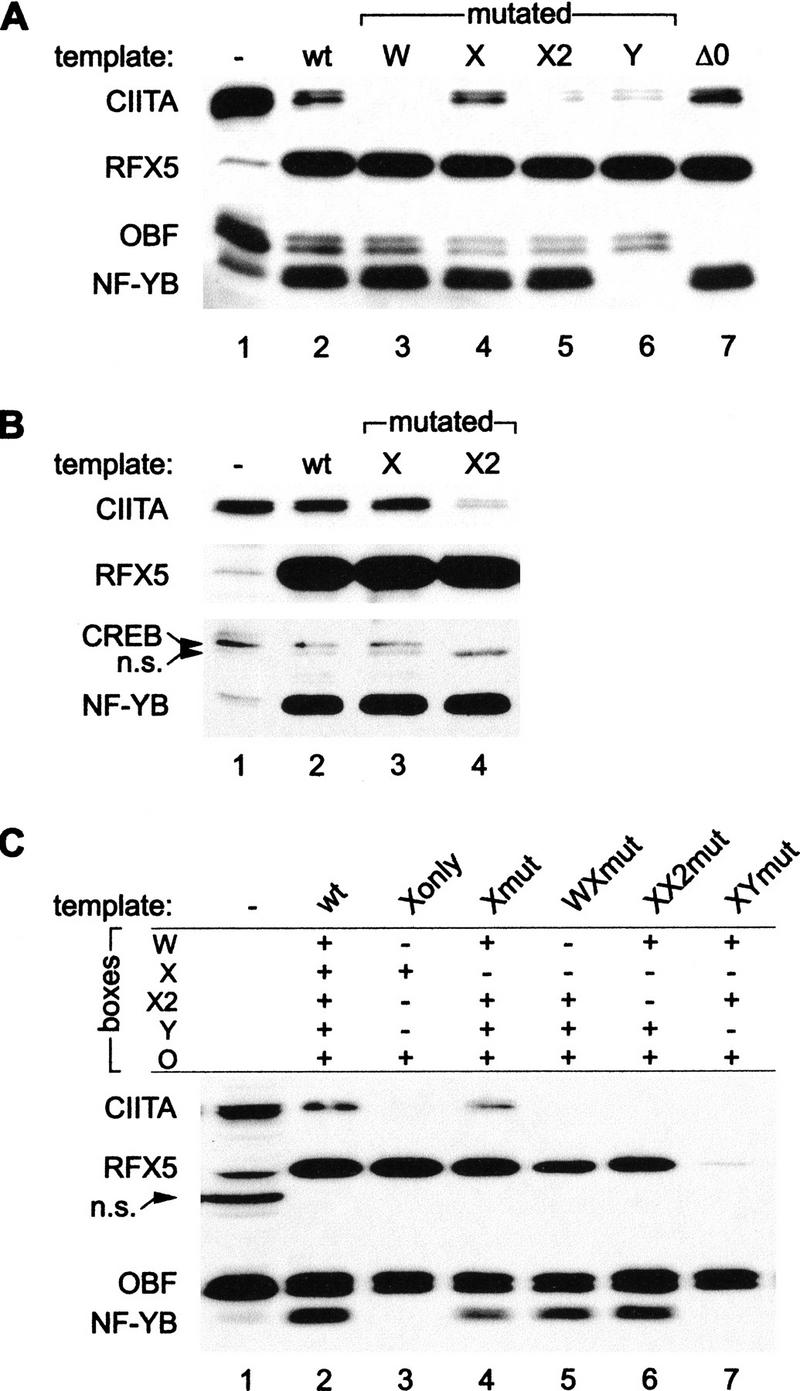
Recruitment of CIITA requires multiple promoter binding factors. (A,B) Mutations in the W, X, X2, or Y boxes, and a deletion of the octamer site (ΔO), were tested for their effect on CIITA recruitment in a promoter pull-down assay. Mutated or wild-type (wt) promoter fragments immobilized on magnetic beads were incubated with RJ6.4 extract and the purified proteins were analyzed by immunoblotting with CIITA, RFX5, NF-YB, and OBF-1 antibodies (A) or with CIITA, RFX5, NF-YB, and CREB-1 antibodies (B); (n.s.) nonspecific signal. A total of 3% (A) or 1% (B) of the input extract were run in parallel (lane 1). (C) Mutations in the W, X2, or Y boxes were tested for their effect on the cooperative retention of RFX on a HLA–DRA promoter template containing a mutated X box. Wild-type (wt) HLA–DRA promoter fragments, or promoter fragments carrying mutations in selected sequences (boxes), were tested in a pull-down assay as in A. Five percent of the input extract was loaded in lane 1.
The octamer-binding site of the HLA–DRA promoter is required for maximal expression in B cells (Sherman et al. 1989; Tsang et al. 1990) and has been proposed to bind the lymphoid-specific factor Oct-2 (Abdulkadir et al. 1995) together with the coactivator OBF-1, also referred to as Bob-1 or OCA-B (Gstaiger et al. 1995; Luo and Roeder 1995; Strubin et al. 1995; Fontes et al. 1996). The fact that OBF-1 is retained on the HLA–DRA promoter fragment in our pull-down assay is consistent with this hypothesis (Fig. 3A, lane 2). Binding of OBF-1 to the promoter does not require CIITA, because the loss of CIITA recruitment does not affect binding of OBF-1 (Fig. 3A, lanes 3,5,6). To study the potential role of octamer-binding proteins in CIITA recruitment, we performed pull-down assays with a promoter fragment lacking the octamer site. Removal of the octamer site abolished retention of OBF-1, but had no detectable effect on binding of CIITA (Fig. 3A, lane 7). We conclude that the octamer site and its cognate activator proteins are not required for tethering of CIITA to the promoter. This is not surprising considering the fact that the HLA–DRA promoter is the only MHC-II promoter that contains an octamer site.
Unexpectedly, the X mutation had no effect on CIITA recruitment. However, this was not informative with respect to the role of RFX in CIITA recruitment, because the X mutation did not eliminate binding of RFX (Fig. 3A, lane 4 and B, lane 3). The lack of effect of the X mutation on binding of RFX in the pull-down assay was unanticipated, because the same mutation is known to abolish binding of RFX in gel-retardation experiments (Reith et al. 1994b). It seemed likely that the explanation for this discrepancy resides in the strong cooperative binding interactions that RFX entertains with other enhanceosome components (Reith et al. 1994a,b; Louis-Plence et al. 1997). These interactions could be sufficient to retain RFX on the promoter despite the mutated X box. Interactions with NF-Y and X2BP were demonstrated previously to stabilize binding of RFX to severely mutated X boxes as well as to the natural low-affinity X boxes present in many MHC-II promoters, such as those of the HLA–DQA, HLA–DPB, and HLA–DRB genes (Reith et al. 1994a; Louis-Plence et al. 1997). We have actually shown here that binding of RFX to such low-affinity target sites occurs readily in vivo (see Fig. 1). To establish the contribution of particular enhanceosome components to the retention of RFX on the mutated X box, we introduced additional mutations into the HLA–DRA promoter and analyzed these multiple-mutated templates in a pull-down experiment (Fig. 3C). Binding of RFX to an X box-mutated template (Fig. 3C, lane 4) was nearly abolished by a concomitant Y-box mutation (lane 7). This is not surprising, as RFX binding in vitro and in vivo is known to be stabilized by NF-Y (Reith et al. 1994b; Wright et al. 1994). RFX binding was also reduced by the W-box mutation (lane 5), whereas the effect of the X2-box mutation was less apparent (lane 6). As anticipated from previous studies (Kobr et al. 1990), the wild-type X box of the HLA–DRA promoter was sufficient for RFX binding independently of the W, X2, and Y boxes (Fig. 3C, lane 3). Unsurprisingly, none of the multiple-mutated templates was able to sustain CIITA recruitment (Fig. 3, lanes 3,5–7), whereas OBF binding remained unaffected by the mutations in the W, X, X2, and Y boxes.
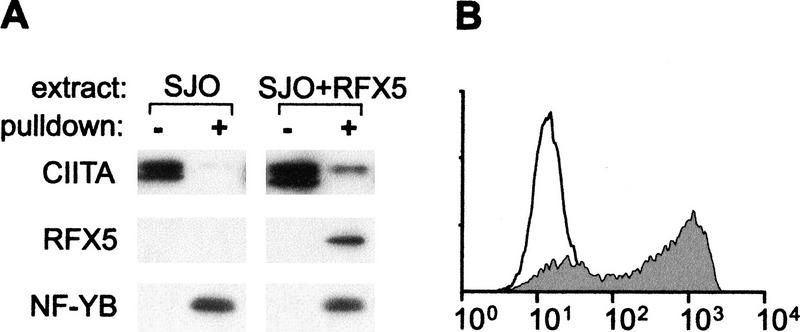
To determine whether the RFX complex is actually required for CIITA recruitment, we used cell lines derived from MHC-II deficiency patients having mutations in the genes encoding its three subunits (Steimle et al. 1995; Durand et al. 1997; Masternak et al. 1998; Nagarajan et al. 1999). Binding of RFX and recruitment of CIITA were not observed in pull-down assays performed with extracts from the RFX5-deficient cell line SJO (Steimle et al. 1995). Similar results were obtained with cell lines lacking RFXAP or RFXANK (data not shown). Binding of RFX, recruitment of CIITA, and MHC-II expression were restored in SJO cells complemented with RFX5 (Fig. 4A,B). This confirms that incorporation of RFX into the enhanceosome is essential for recruitment of CIITA and promoter activation.
Figure 4.
RFX is essential for CIITA recruitment and MHC-II expression. (A) Pull-down assays were performed with the wild-type promoter and extracts from SJO cells or SJO cells transfected with RFX5. A total of 2% of the input extract (−) and proteins purified by the pull-down assay (+) are shown. (B) MHC-II expression was analyzed by flow cytometry in SJO cells (open profile) and complemented SJO cells (gray profile).
Interactions of CIITA mutants with the enhanceosome
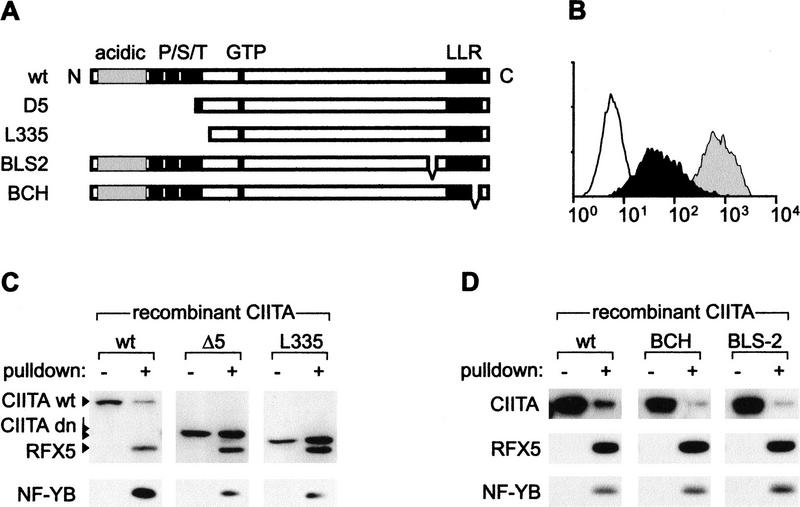
To determine the molecular basis of the phenotypes exhibited by known CIITA mutants, we performed promoter pull-down assays with a CIITA-deficient cell extract (RJ2.2.5) that was supplemented with recombinant wild-type or mutant CIITA proteins expressed in a Vaccinia virus-T7 RNA polymerase system (Fuerst et al. 1986). Two dominant-negative mutants (Bontron et al. 1997b) lacking the amino-terminal transcription activation domains were tested (Fig. 5A). Transfection of MHC-II-positive cells with NLS-L335 and NLS-D5 leads to a tenfold reduction in MHC-II expression (Bontron et al. 1997b). NLS-D5 and NLS-L335 retained the ability to bind to the enhanceosome, indicating that the carboxy-terminal moiety of CIITA is sufficient for this function (Fig. 5C). Remarkably, recruitment of NLS-D5 and NLS-L335 was considerably more efficient than that of wild-type CIITA, indicating that their dominant-negative phenotype can be explained by an increased affinity for the enhanceosome. This would lead to competitive inhibition of wild-type CIITA recruitment in transfected cells, which typically express the mutant proteins at levels greater than that of the endogenous protein (Bontron et al. 1997b).
Figure 5.
Mutations of CIITA affecting recruitment. (A) Schematic representation of wild-type (wt) and mutant CIITA proteins. The acidic and proline/serine/threonine rich activation domains, GTP-binding cassette, and leucine-rich repeat region (LRR) are indicated. (B) MHC-II expression was analyzed by flow cytometry in RJ2.2.5 cells (open profile) and Raji cells transfected with empty expression vector (gray profile) or NLS-L335 (black profile). (C,D) Pull-down assays were performed with RJ2.2.5 extracts supplemented with the indicated recombinant CIITA proteins. A total of 1% of the input extract (−) and proteins purified by the pull down assay (+) were analyzed by immunoblotting. In C, a carboxy-terminal-specific CIITA antibody was used for immunoblotting.
Two loss-of-function mutants isolated from MHC-II deficiency patients BLS-2 and BCH (Steimle et al. 1993; Bontron et al. 1997a) were also tested. They contain small in-frame deletions situated adjacent to or within a putative protein–protein interaction domain consisting of leucine-rich repeats. The BCH and BLS-2 mutants were recruited less efficiently than wild-type CIITA (Fig. 5D). Deletion of sequences involved in recruitment thus accounts, at least in part, for their loss-of-function phenotype. However, additional functions of CIITA could also be affected by these mutations. For instance, a recent report has indicated that the BLS-2 mutation affects nuclear import (Cressman et al. 1999). The finding that the BLS-2 and BCH mutations inhibit recruitment only partially suggests that CIITA contains more than one region involved in binding to the enhanceosome. This is in agreement with the fact that multiple DNA-binding proteins form the landing pad for CIITA (Figs. 3,4).
Discussion
The data presented in this study demonstrate that, at physiological concentrations, CIITA recruitment to the MHC-II enhanceosome requires multiple protein–protein interactions with the DNA-bound activators (Figs. 3,4). Factors binding to the W, X, X2, and Y sequences are all involved in creating the CIITA docking interface (Fig. 6). The contribution of these proteins is synergistic because elimination of any one of them leads to a dramatic loss of recruitment. This explains previously published data demonstrating that CIITA exerts its function via all of these four promoter elements (Riley et al. 1995; Zhou and Glimcher 1995; Brown et al. 1998).
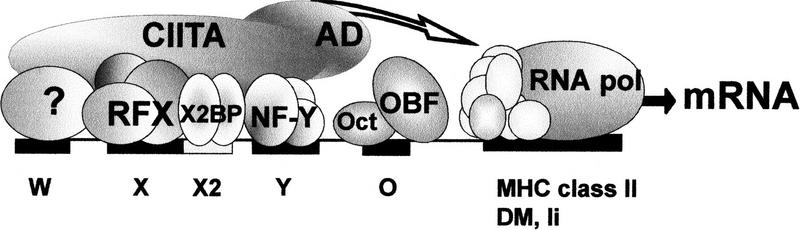
Figure 6.
Recruitment of CIITA to MHC-II promoters. A prototypical MHC-II promoter (HLA–DRA) is represented schematically with the W, X, X2, and Y sequences conserved in all MHC-II, Ii, and HLA–DM promoters. RFX, X2BP, NF-Y, and an as yet undefined W-binding protein bind cooperatively to these sequences and assemble into a stable higher order nucleoprotein complex referred to here as the MHC-II enhanceosome. CIITA is tethered to the enhanceosome via multiple weak protein–protein interactions with the W, X, X2, and Y-binding factors. The octamer site found in the HLA–DRA promoter (O), and its cognate activators (Oct and OBF-1) are not required for recruitment of CIITA. CIITA is proposed to activate transcription (arrow) via its amino-terminal activation domains (AD), which contact the RNA polymerase II basal transcription machinery.
Each individual interaction between CIITA and one of the isolated enhanceosome components is too weak on its own to permit recruitment in vivo. Interactions between CIITA and RFX in solution, or between CIITA and NF-Y, are not observed in coimmunoprecipitation experiments performed with cell extracts containing physiological concentrations of these proteins (Fig. 2). To detect these interactions by coimmunoprecipitation, it is necessary to work with extracts in which the interaction partners are strongly overexpressed (S. Hake and V. Steimle, in prep.). When overexpressed, we found that CIITA can interact with two RFX subunits (RFX5 and RFXANK) and two NF-Y subunits (NF-YB and NF-YC), thereby confirming the existence of multiple CIITA-enhanceosome contacts (S. Hake and V. Steimle, in prep.). Although the W and X2 boxes are essential for recruitment, physical interactions between CIITA and the W- and X2-binding factors have not been assayed.
The carboxy-terminal region of CIITA is required for recruitment (Fig. 5D), a finding that is consistent with previous experiments demonstrating that this region of CIITA mediates specificity for MHC-II promoters (Zhou and Glimcher 1995). We show that at least part of the CIITA-enhanceosome interactions must involve the leucine-rich repeat (LRR) region of CIITA, which is a known protein–protein interaction motif. Mutations within or near this region reduce the efficiency of recruitment (Fig. 5D). The latter result is in agreement with the fact that the LRR motif is essential for the function of CIITA (Bontron et al. 1997a). However, as pointed out earlier, an inhibition of CIITA recruitment may not be the sole consequence of mutations affecting the LRR region. A recent report has demonstrated that the LRR region is also involved in nuclear import (Cressman et al. 1999).
Multiple CIITA-enhanceosome interactions are expected to exert a reciprocal stabilization effect. They would not only enhance binding of CIITA, but also contribute to promoter occupancy by stabilizing interactions between the components of the enhanceosome. Stabilization of the enhanceosome by CIITA could underlie a number of unexplained observations. First, in certain cell types, in vivo occupation of MHC-II promoters requires expression of CIITA (Silacci et al. 1994; Wright et al. 1998; Villard et al. 1999). Second, in some RFX-deficient cells, overexpression of CIITA leads to a partial rescue of MHC-II expression (van den Elsen et al. 1998). Finally, Rfx5−/− mice exhibit residual MHC-II expression in such cell types as activated B cells and dendritic cells, which are likely to express high levels of CIITA (Clausen et al. 1998).
Under the conditions used here for the promoter pull-down assays, typically <1% of the CIITA was recruited to the wild-type HLA–DRA template. This modest efficiency reflects the low affinity of the CIITA-enhanceosome interaction. The concentration of CIITA was found to be the limiting factor for the interaction (data not shown). This is consistent with previous findings indicating that the levels of CIITA and MHC-II expression show a nearly linear correlation in vivo (Otten et al. 1998). The low affinity of the CIITA-enhanceosome interaction ensures that the level of MHC-II expression is proportional to the CIITA concentration. In fact, a high-affinity interaction between CIITA and the enhanceosome would not have been expected because this would not permit quantitative control of MHC-II expression.
The approaches we have developed here can now be exploited to address a number of subsequent unresolved issues concerning the mode of action of CIITA. These include which additional known or unknown factors it brings to the MHC-II promoter, which general transcription factors it is capable of recruiting, and how it collaborates with the other enhanceosome components to activate transcription. They will also contribute to the identification of the functionally relevant W-binding factor. The data presented here demonstrate that the ability to collaborate with RFX, X2BP, and NF-Y in tethering CIITA to MHC-II promoters must represent a key characteristic of a candidate W-binding factor.
Synergistic recruitment by DNA-bound activators is characteristic for the RNA Pol II general transcription factor complex (Carey 1998). This mechanism has also been described recently for recruitment of the RNA Pol II holoenzyme-associated general coactivators CBP/p300 to the interferon-β enhanceosome (Merika et al. 1998). CIITA however, is a gene-specific regulatory factor and is not known to be associated with the general transcription machinery. The type of recruitment described here for CIITA has not been documented previously for other regulatory factors functioning as transcriptional coactivators.
Coactivators are usually ubiquitously expressed proteins that are pleiotropic in their function, and are recruited to many unrelated promoters by interactions with a diverse array of DNA-bound activators (Roeder 1998; Wu and Hampsey 1999; Xu et al. 1999). In general, coactivators simply serve as relays or effectors mediating chromatin remodeling or transcription activation (Roeder 1998; Xu et al. 1999); they usually do not play a regulatory role. Regulation is instead almost invariably achieved by the combinatorial control exerted by multiple DNA-binding activators assembling on promoters and/or enhancers (Carey 1998). The situation is strikingly different in the MHC-II system, in which transcriptional regulation relies entirely upon a coactivator. The MHC-II enhanceosome consists of constitutively expressed DNA-binding proteins. In contrast, CIITA is expressed in a complex and highly regulated pattern that governs all spatial, temporal, and quantitative aspects of MHC-II expression (Steimle et al. 1994; Mach et al. 1996; Boss 1997; Otten et al. 1998). CIITA is thus a paradigm for a novel type of coactivator that acts both as a master regulator exerting a tight qualitative and quantitative control, and as a specificity factor recognizing the promoter via the macromolecular surface of the enhanceosome. This finding raises a number of intriguing and unresolved questions. Why has the mechanism controlling MHC-II expression evolved such a strict dependence on a coactivator rather than relying on DNA-binding activators as is the most common situation in other systems? Is this situation unique, or have cofactors with similar properties not yet been identified in other systems? Examples of other coactivators that share some of the unique features of CIITA are rare. These include tissue-specific cofactors, such as OBF-1 (Bob-1, OCA-B) in B cells (Gstaiger et al. 1995; Luo and Roeder 1995; Strubin et al. 1995), ACT in testis (Fimia et al. 1999), and PGC-1 in brown adipose cells (Puigserver et al. 1998). Like CIITA, these coactivators exhibit a restricted pattern of expression and show selectivity with respect to the DNA-bound activators with which they interact.
We show here that CIITA is recruited in vivo to the promoters of the coregulated MHC-II, Ii, and HLA–DM genes (Fig. 1). However, CIITA and RFX were also found to be associated with the promoters of the β2m and MHC-I genes, which share the W-X-X2-Y homology region, but are not coregulated with MHC-II genes. MHC-I transcription depends, to a large extent, on cis-acting sequence elements that lie outside of the W-X-X2-Y region (David-Watine et al. 1990; Singer and Maguire 1990; Ting and Baldwin 1993). Functional evidence for the involvement of CIITA in the regulation of MHC-I expression is conflicting. On one hand, CIITA up-regulates MHC-I expression in cell lines that express MHC-I at low levels, and has been shown to activate MHC-I promoter-driven reporter gene expression in transient cotransfection assays (Gobin et al. 1997; Martin et al. 1997). On the other hand, MHC-I expression is affected neither in CIITA knockout mice (Chang et al. 1996; Williams et al. 1998; Itoh-Lindstrom et al. 1999), nor in cell lines derived from MHC-II deficiency patients in group A (Mach et al. 1996). Thus, the effect of CIITA on MHC-I expression may vary, depending on the species, genetic background, and cell type, confirming that the regulatory pathway implicating CIITA plays only an accessory role in the system. Further studies will be needed to determine the actual importance of this pathway for MHC-I expression in vivo. Apart from its fundamental role in MHC-II expression, and a possible involvement in fine tuning of MHC-I expression, CIITA may be implicated in the regulation of other genes as well. A recent report suggests that CIITA down-regulates the expression of Interleukin 4 and possibly other cytokines during T-cell differentiation (Gourley et al. 1999).
Via its decisive control over MHC-II expression, CIITA plays a key role in the regulation of adaptive immune responses. We demonstrated here that CIITA interacts with a macromolecular complex assembled on MHC-II promoters. In the future, defining the protein–protein contacts that contribute to CIITA recruitment will be of major medical relevance. This is anticipated to lead to the development of novel immunosuppressive drugs that function by interfering with these protein–protein interactions.
Materials and methods
Cell lines, transfections, and flow cytometry
The B-cell lines Raji, RJ2.2.5, SJO, and SJO transfected stably with RFX5 have been described (Accolla 1983; Steimle et al. 1993, 1995; Villard et al. 2000). The RJ6.4 cell line was produced by stable transfection of RJ2.2.5 with CIITA tagged at its amino terminus with a haemagglutinin epitope. Cell culture, transfections, selection with hygromycin, and flow cytometry using HLA–DR antibodies were done as described (Steimle et al. 1993; Durand et al. 1997; Bontron et al. 1997b).
Chromatin immunoprecipitation assay
Chromatin was prepared as described (Orlando et al. 1997) with several modifications. B cells (4 × 108) were treated with 1% formaldehyde for 10 min at room temperature. Glycine was added to 0.125 m to stop the cross-linking. The cells were washed once with cold PBS and lysed in TE buffer (10 mm Tris-HCl at pH 8.0, 1 mm EDTA) containing protease inhibitors and 0.5% NP-40. Nuclei were pelleted and lysed in TE buffer containing 0.5 m NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.5% sarcosyl. Cross-linked chromatin was resuspended in TEN buffer (10 mm Tris-HCl at pH 8.0, 1 mm EDTA, 100 mm NaCl), sheared by sonication, and cleared by 2 cycles of centrifugation at 15,000g for 15 min. The resulting chromatin supernatant was supplemented with 0.1% sarcosyl and stored at −70°C. Immunoprecipitations were done at room temperature. Chromatin supernatant (∼2.5 μg, corresponding to 3 million cells) was diluted 10 times with IP buffer (20 mm Tris-HCl at pH 8.0, 200 mm NaCl, 2 mm EDTA, 0.1% sodium deoxycholate, 0.1% SDS, 1 mm PMSF) containing 50 μg/ml salmon sperm DNA, 100 μg/ml Escherichia coli tRNA, and 1 mg/ml BSA. After preincubation with protein A–Sepharose beads (Pharmacia) for 30 min on a rotator, supernatant was cleared by centrifugation and supplemented with 5 μl of anti-CIITA or anti-RFX5 serum. Immune complexes were allowed to form for 3 hr, protein A–Sepharose beads were added, and incubation was continued for 1 hr. Beads were washed twice with IP buffer, twice with IP buffer with 500 mm NaCl, twice with 20 mm Tris-HCl (pH 8.0), 0.25 m LiCl, 2 mm EDTA, 0.5% NP-40, 0.5% sodium deoxycholate, and once in TE containing 0.1% NP-40. Immune complexes were eluted with 100 mm Tris-HCl (pH 8.0), 1% SDS for 10 min at 65°C, diluted twofold with 400 mm NaCl, 200 μg/ml proteinase K, digested for 1 hr at 37°C, and incubated O/N at 65°C to reverse the cross-links. After extraction with phenol/chloroform and chloroform, DNA was precipitated with isopropanol in the presence of 5 μg of glycogen, washed in 80% ethanol, and resuspended in TE. Immunoprecipitated DNA (corresponding to 100 ng of input chromatin supernatant) and input chromatin (10 ng) were analyzed by PCR using promoter-specific primer pairs. The following primers were used: HLA–DRA, 5′-GTTGTCCTGTTTGTTTAAGAAC-3′ sense and 5′-GCTCTTTTGGGAGTCAG-3′ antisense; HLA–DRB1*0301, 5′-CAACTGGTTCAAACCTTTCAAGTAC-3′ sense and 5′-AGTCTCACTCAGGGAGAACTATGAAC-3′ antisense; HLA–DPB1, 5′-ATGAGTATCACTGTCTTTCCTCCG-3′ sense and 5′-GAGACCATGAACCCAAGTAGTCTTC-3′ antisense; HLA–DMB, 5′-CTACGGAAATCTACTGGTTGTTCTG-3′ sense and 5′-ATTAAATCTGTTCCTTCCAGCTCAC-3′ antisense; Ii, 5′-GGTGTCTTCTGTTTCAAAGTGCTTTC-3′ sense and 5′-CACTCCGCCCACTTGGTAGAT-3′ antisense; HLA-A, 5′-TCCGCAGTTTCTTTTCTCCC-3′ sense and 5′-GGAGAATCTGAGTCCCGGTGG-3′ antisense; HLA-B, 5′-TCTCAGGGTCTCAGGCTCCGAG-3′ sense and 5′-TGCGTGGGGACTTTAGAACTGG-3′ antisense; β2m, 5′-CATGCCTTCTTAAACATCACGAGAC-3′ sense and 5′-CAGGCCAGAAAGAGAGAGTAGCG-3′ antisense; MHC2TA promoter III, 5′-TAACCAGTCACCAGTTGGGAGCCCG-3′ sense and 5′-CTGGCACCAGTGCGGTTCCATTGTG-3′ antisense.
Extracts and recombinant CIITA expression
Cells were resuspended in two packed cell volumes of a buffer containing 50 mm HEPES at pH 7.9, 400 mm KCl, 1 mm EDTA, 1 mm EGTA, 2 mm DTT, 5 μg/ml leupeptin, 1 mm PMSF, 0.5 mm NaF, 0.5 mm Na3VO4, 0.01% NP-40, 20% glycerol, and a cocktail of antiproteases (Complete, Roche Diagnostics). Whole cell extracts were obtained by three freeze-thaw cycles, cleared by centrifugation, and stored at −80°C. Recombinant CIITA proteins were expressed in HeLa cells using a Vaccinia-T7 system (Fuerst et al. 1986), and extracts from these cells were prepared as above.
HLA–DRA promoter templates
Wild-type and mutated HLA–DRA promoter fragments were constructed by PCR on a DRsyn template (Tsang et al. 1990). The W-box sequence GGACCCTTTGCAAG was mutated to TACATAGCGTACGT. The X2-box sequence TGCGTCA was mutated to GACAAGT. The mutations in the X and Y boxes were described previously (Durand et al. 1994; Reith et al. 1994). The ΔOct template (−150 to −56) was obtained by digestion of the wild-type DRsyn fragment with BglII.
DNA-dependent immunoprecipitation and promoter pull-down assays
All steps were done at 4°C. For immunoprecipitation experiments, extracts (15 μl, 0.5–0.6 mg) were diluted twofold with a buffer containing 20 mm HEPES at pH 7.9, 9 mm MgCl2, 1 mm DTT, 20% glycerol, 0.01% NP-40, and a cocktail of antiproteases (Complete, Roche Diagnostics). Diluted extracts were cleared by centrifugation and supplemented with 0.15 mg/ml single-stranded E. coli DNA and 0.15 mg/ml poly[d(I-C)]. HLA–DRA promoter fragments (2.5 pmoles), or an equivalent amount of salmon sperm DNA (0.2 μg), were added and protein–DNA complexes were assembled for 2 hr. Before immunoprecipitation, extracts were preincubated with protein A–Sepharose beads for 30 min and cleared by centrifugation. Supernatants were then incubated for 1 hr with anti-RFXAP or anti-CIITA-N antibodies coupled to protein A–Sepharose. The beads were washed three times with buffer D (20 mm HEPES at pH 7.9, 100 mm KCl, 6 mm MgCl2, 1 mm DTT, 20% glycerol, 0.01% NP-40) containing 1 mg/ml BSA, and proteins were eluted with SDS-PAGE sample buffer. For pull-down assays, protein–DNA complexes were assembled, washed, and eluted as described above, except that the promoter templates biotinylated at the 5′ end of the upper strand and coupled to streptavidin-coated magnetic beads (Promega) were used. In experiments with recombinant CIITA, extracts from CIITA-negative RJ2.2.5 B cells and from HeLa cells expressing the recombinant proteins were mixed before adding the remaining reaction components. Concentrations of recombinant CIITA added to the assembly reactions were comparable with the endogenous CIITA concentration in B-cell extracts. Conditions of the pull-down assay were set up such that retrieval of RFX and NF-Y from crude cell extracts was, respectively, 100% and 20%–50%. Under these conditions, typically <1% of the CIITA was recruited to the wild-type HLA–DRA template. Experiments shown in Figures 2, 3, and 4 were performed with extracts from RJ6.4 cells expressing two to three times more CIITA than our model B-cell line Raji. This led to a proportional increase in CIITA recruitment, yielding neater results with a higher signal-to-noise ratio. Notwithstanding, the results obtained with Raji cells were essentially identical. Extending the promoter template upstream to position −196 or downstream to +51 did not affect enhanceosome assembly or recruitment of CIITA.
Antibodies and immunoblotting
Polyclonal anti-CIITA and anti-RFX5 sera, and the immunoaffinity-purified RFXAP antibodies have been described (Steimle et al. 1995; Bontron et al. 1997b; Masternak et al. 1998). Antibodies specific for the amino terminus of CIITA (anti-CIITA-N, used in Figs. 2, 3, 4, and 5D) were obtained by affinity purification of the anti-CIITA serum on an amino-terminal His6-tagged CIITA fragment (amino acids 25–300) covalently coupled to Sepharose beads. Antibodies specific for the carboxyl terminus of CIITA (anti-CIITA-C, used in Fig. 5C) were retrieved from the unbound fraction by a second affinity purification step using full-length recombinant CIITA. The NF-YB antibody was a gift from Roberto Mantovani (Universitá di Milano, Italy). The TBP antibody was a gift from Pierre Chambon (IGBMC, Strasbourg, France). The other antibodies were purchased from Santa Cruz Biotechnology (α-OBF-1, sc-955; α-CREB-1, sc-271; α-CREB-1, sc-186; α-CREB-1, sc-58; α-JunB, sc-46; α-CBP, sc-583; α-hTAFII p250, sc-735). Proteins were analyzed by immunoblotting according to standard protocols. In immunoblots done with B-cell extracts (Figs. 2 and 3), CIITA is detected as a double band, probably due to the use of alternative initiation codons.
Acknowledgments
We thank R. Mantovani, P. Chambon, S. Clarkson, and V. Ducas for antibodies, S. Bontron for plasmids, C. Ucla for expert technical assistance, and M. Strubin and D. Trono for helpful discussion. This work was supported by the Louis-Jeantet Foundation, the Swiss National Science Foundation, and Novartis Stifttung. A.M. was supported by the Marie Heim-Vögtlin Foundation. We are indebted to B. Mach who provided the scientific environment in which this work was first initiated.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Walter.Reith@medecine.unige.ch; FAX 41-22-7025702.
References
- Abdulkadir SA, Krishna S, Thanos D, Maniatis T, Strominger JL, Ono SJ. Functional roles of the transcription factor Oct-2A and the high mobility group protein I/Y in HLA–DRA gene expression. J Exp Med. 1995;182:487–500. doi: 10.1084/jem.182.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accolla RS. Human B cell variants immunoselected against a single Ia antigen subset have lost expression in several Ia antigen subsets. J Exp Med. 1983;157:1053–1058. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoist C, Mathis D. Regulation of major histocompatibility complex class II genes: X, Y and other letters of the alphabet. Annu Rev Immunol. 1990;8:681–715. doi: 10.1146/annurev.iy.08.040190.003341. [DOI] [PubMed] [Google Scholar]
- Bontron S, Steimle V, Ucla C, Mach B. Two novel mutations in the MHC class II transactivator CIITA in a second patient from MHC class II deficiency complementation group A. Hum Genet. 1997a;99:541–546. doi: 10.1007/s004390050403. [DOI] [PubMed] [Google Scholar]
- Bontron S, Ucla C, Mach B, Steimle V. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol Cell Biol. 1997b;17:4249–4258. doi: 10.1128/mcb.17.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss JM. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- Brown JA, Rogers EM, Boss JM. The MHC class II transactivator (CIITA) requires conserved leucine charged domains for interactions with the conserved W box promoter element. Nucleic Acids Res. 1998;26:4128–4136. doi: 10.1093/nar/26.18.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Chang CH, Guerder S, Hong SC, van Ewijk W, Flavell RA. Mice lacking the MHC class II transactivator (CIITA) show tissue-specific impairment of MHC class II expression. Immunity. 1996;4:167–178. doi: 10.1016/s1074-7613(00)80681-0. [DOI] [PubMed] [Google Scholar]
- Chin KC, Li GG, Ting JP. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: Generation of transdominant-negative mutants. Proc Natl Acad Sci. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen BE, Waldburger JM, Schwenk F, Barras E, Mach B, Rajewsky K, Forster I, Reith W. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998;8:143–155. doi: 10.1016/s1074-7613(00)80467-7. [DOI] [PubMed] [Google Scholar]
- Cressman DE, Chin KC, Taxman DJ, Ting JP. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- David-Watine B, Israel A, Kourilsky P. The regulation and expression of MHC class I genes. Immunol Today. 1990;11:286–292. doi: 10.1016/0167-5699(90)90114-o. [DOI] [PubMed] [Google Scholar]
- de Preval C, Lisowska-Grospierre B, Loche M, Griscelli C, Mach B. A trans-acting class II regulatory gene unlinked to the MHC controls expression of HLA class II genes. Nature. 1985;318:291–293. doi: 10.1038/318291a0. [DOI] [PubMed] [Google Scholar]
- Durand B, Kobr M, Reith W, Mach B. Functional complementation of MHC class II regulatory mutants by the purified X box binding protein RFX. Mol Cell Biol. 1994;14:6839–6847. doi: 10.1128/mcb.14.10.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, De Cesare D, Sassone-Corsi P. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- Fontes JD, Jabrane-Ferrat N, Toth CR, Peterlin BM. Binding and cooperative interactions between two B cell-specific transcriptional coactivators. J Exp Med. 1996;183:2517–2521. doi: 10.1084/jem.183.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes JD, Kanazawa S, Jean D, Peterlin BM. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh N, Piskurich JF, Wright G, Hassani K, Ting JP, Wright KL. A novel element and a TEF-2-like element activate the major histocompatibility complex class II transactivator in B-lymphocytes. J Biol Chem. 1999;274:32342–32350. doi: 10.1074/jbc.274.45.32342. [DOI] [PubMed] [Google Scholar]
- Glimcher LH, Kara CJ. Sequences and factors: A guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- Gobin SJ, Peijnenburg A, Keijsers V, van den Elsen PJ. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: The ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601–611. doi: 10.1016/s1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- Gourley T, Roys S, Lukacs NW, Kunkel SL, Flavell RA, Chang CH. A novel role for the major histocompatibility complex class II transactivator CIITA in the repression of IL-4 production. Immunity. 1999;10:377–386. doi: 10.1016/s1074-7613(00)80037-0. [DOI] [PubMed] [Google Scholar]
- Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens CM. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- Itoh-Lindstrom Y, Piskurich JF, Felix NJ, Wang Y, Brickey WJ, Platt JL, Koller BH, Ting JP. Reduced IL-4-, lipopolysaccharide-, and IFN-gamma-induced MHC class II expression in mice lacking class II transactivator due to targeted deletion of the GTP-binding domain. J Immunol. 1999;163:2425–2431. [PubMed] [Google Scholar]
- Kara CJ, Glimcher LH. In vivo footprinting of MHC class II genes: Bare promoters in the bare lymphocyte syndrome. Science. 1991;252:709–712. doi: 10.1126/science.1902592. [DOI] [PubMed] [Google Scholar]
- ————— Three in vivo promoter phenotypes in MHC class II deficient combined immunodeficiency. Immunogenetics. 1993;37:227–230. doi: 10.1007/BF00191890. [DOI] [PubMed] [Google Scholar]
- Kobr M, Reith W, Herrero Sanchez C, Mach B. Two DNA-binding proteins discriminate between the promoters of different members of the major histocompatibility complex class II multigene family. Mol Cell Biol. 1990;10:965–971. doi: 10.1128/mcb.10.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis-Plence P, Moreno CS, Boss JM. Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J Immunol. 1997;159:3899–3909. [PubMed] [Google Scholar]
- Luo Y, Roeder RG. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: Lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- Mahanta SK, Scholl T, Yang FC, Strominger JL. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity SN, De Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Martin BK, Chin KC, Olsen JC, Skinner CA, Dey A, Ozato K, Ting JP. Induction of MHC class I expression by the MHC class II transactivator CIITA. Immunity. 1997;6:591–600. doi: 10.1016/s1074-7613(00)80347-7. [DOI] [PubMed] [Google Scholar]
- Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez JC, Hochstrasser DF, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- Merika M, Williams AJ, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- Moreno CS, Beresford GW, Louis-Plence P, Morris AC, Boss JM. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan UM, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss JM. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Otten LA, Steimle V, Bontron S, Mach B. Quantitative control of MHC class II expression by the transactivator CIITA. Eur J Immunol. 1998;28:473–478. doi: 10.1002/(SICI)1521-4141(199802)28:02<473::AID-IMMU473>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Piskurich JF, Linhoff MW, Wang Y, Ting JP. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor beta. Mol Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Reith W, Satola S, Herrero Sanchez C, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam MR, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- Reith W, Kobr M, Emery P, Durand B, Siegrist CA, Mach B. Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J Biol Chem. 1994a;269:20020–20025. [PubMed] [Google Scholar]
- Reith W, Siegrist CA, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci. 1994b;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JL, Westerheide SD, Price JA, Brown JA, Boss JM. Activation of class II MHC genes requires both the X box and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- Roeder RG. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb Symp Quant Biol. 1998;63:201–218. doi: 10.1101/sqb.1998.63.201. [DOI] [PubMed] [Google Scholar]
- Scholl T, Mahanta SK, Strominger JL. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman PA, Basta PV, Heguy A, Wloch MK, Roeder RG, Ting JP. The octamer motif is a B-lymphocyte-specific regulatory element of the HLA-DR alpha gene promoter. Proc Natl Acad Sci. 1989;86:6739–6743. doi: 10.1073/pnas.86.17.6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silacci P, Mottet A, Steimle V, Reith W, Mach B. Developmental extinction of major histocompatibility complex class II gene expression in plasmocytes is mediated by silencing of the transactivator gene CIITA. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer DS, Maguire JE. Regulation of the expression of class I MHC genes. Crit Rev Immunol. 1990;10:235–257. [PubMed] [Google Scholar]
- Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- Steimle V, Siegrist C-A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by Interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- Steimle V, Durand B, Barras E, Zufferey M, Hadam MR, Mach B, Reith W. A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (Bare Lymphocyte Syndrome) Genes & Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with Octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- Ting JP, Baldwin AS. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- Tsang SY, Nakanishi M, Peterlin BM. Mutational analysis of the DRA promoter: Cis-acting sequences and trans-acting factors. Mol Cell Biol. 1990;10:711–719. doi: 10.1128/mcb.10.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elsen PJ, Peijnenburg A, van Eggermond MC, Gobin SJ. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol Today. 1998;19:308–312. doi: 10.1016/s0167-5699(98)01287-0. [DOI] [PubMed] [Google Scholar]
- Vilen BJ, Cogswell JP, Ting JP. Stereospecific alignment of the X and Y elements is required for major histocompatibility complex class II DRA promoter function. Mol Cell Biol. 1991;11:2406–2415. doi: 10.1128/mcb.11.5.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilen BJ, Penta JF, Ting JP-Y. Structural constraints within a trimeric transcriptional regulatory region: Constitutive and Interferon-g inducible expression of the HLA–DRA gene. J Biol Chem. 1992;267:23728–23734. [PubMed] [Google Scholar]
- Villard J, Muhlethaler-Mottet A, Bontron S, Mach B, Reith W. CIITA-induced occupation of MHC class II promoters is independent of the cooperative stabilization of the promoter-bound multi-protein complexes. Int Immunol. 1999;11:461–469. doi: 10.1093/intimm/11.3.461. [DOI] [PubMed] [Google Scholar]
- Villard, J., M. Peretti, K. Masternak, E. Barras, G. Caretti, R. Mantovani, and W. Reith. 2000. A functionally essential domain of RFX5 mediates activation of MHC class II promoters by promoting cooperative binding between RFX and NF-Y. Mol. Cell. Biol. (In press). [DOI] [PMC free article] [PubMed]
- Williams GS, Malin M, Vremec D, Chang CH, Boyd R, Benoist C, Mathis D. Mice lacking the transcription factor CIITA–a second look. Int Immunol. 1998;10:1957–1967. doi: 10.1093/intimm/10.12.1957. [DOI] [PubMed] [Google Scholar]
- Wright KL, Vilen BJ, Itoh Lindstrom Y, Moore TL, Li G, Criscitiello M, Cogswell P, Clarke JB, Ting JP. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright KL, Chin KC, Linhoff M, Skinner C, Brown JA, Boss JM, Stark GR, Ting JP. CIITA stimulation of transcription factor binding to major histocompatibility complex class II and associated promoters in vivo. Proc Natl Acad Sci. 1998;95:6267–6272. doi: 10.1073/pnas.95.11.6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WH, Hampsey M. Common cofactors and cooperative recruitment. Curr Biol. 1999;9:R606–R609. doi: 10.1016/s0960-9822(99)80385-6. [DOI] [PubMed] [Google Scholar]
- Xu L, Glass CK, Rosenfeld MG. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Zhou H, Glimcher LH. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]