Abstract
The assembly and disassembly of protein complexes at replication origins play a crucial role in the regulation of chromosomal DNA replication. The sequential binding of the origin recognition complex (ORC), Cdc6, and the minichromosome maintenance (MCM/P1) proteins produces a licensed replication origin. Before the initiation of replication can occur, each licensed origin must be acted upon by S phase-inducing CDKs and the Cdc7 protein kinase. In the present report we describe the role of Xenopus Cdc7 (XCdc7) in DNA replication using cell-free extracts of Xenopus eggs. We show that XCdc7 binds to chromatin during G1 and S phase. XCdc7 associates with chromatin only once origins have been licensed, but this association does not require the continued presence of XORC or XCdc6 once they have fulfilled their essential role in licensing. Moreover, XCdc7 is required for the subsequent CDK-dependent loading of XCdc45 but is not required for the destabilization of origins that occurs once licensing is complete. Finally, we show that CDK activity is not necessary for XCdc7 to associate with chromatin, induce MCM/P1 phosphorylation, or perform its essential replicative function. From these results we suggest a simple model for the assembly of functional initiation complexes in the Xenopus system.
Keywords: DNA replication, XCdc7, XCdc45, licensing
The initiation of DNA replication forks occurs at replication origins distributed along chromosomes and must be strictly controlled to ensure that the DNA is replicated once and once only in each cell cycle (for review, see Diffley 1996; Stillman 1996; Donaldson and Blow 1999). The process of building an active replication origin can be divided into two phases. The first phase, occurring in late mitosis and early G1, comprises the sequential assembly onto replication origins of the origin recognition complex (ORC), the Cdc6 protein, and the RLF–M complex of Mcm2–7 proteins (also known as MCM/P1 proteins), which results in them becoming “licensed” for DNA replication in the subsequent S phase. The second phase involves the action of the Cdc7 and cyclin-dependent kinases (CDKs) on each origin to load the Cdc45 protein and to induce the initiation of a pair of replication forks. It is of considerable current interest to understand in more detail the sequence of events leading to the initiation of replication. In this paper we have concentrated on defining the precise stage in the process when the Cdc7 protein acts, using the biochemically tractable Xenopus cell-free DNA replication system.
Replication origins appear to be defined by binding the ORC (Bell and Stillman 1992). In yeast, ORC is bound to origins throughout the cell cycle (Diffley and Cocker 1992; Diffley et al. 1994), whereas in higher eukaryotes, ORC is probably displaced from the DNA during mitosis (Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1999). During late mitosis and early G1, Cdc6 is then assembled onto ORC-containing DNA (Coleman et al. 1996). Chromatin containing ORC and Cdc6 can then be licensed by loading the RLF-M complex of MCM/P1 proteins, a reaction also requiring the RLF-B component of the replication licensing system (Chong et al. 1995; Kubota et al. 1995, 1997; Thömmes et al. 1997; Tada et al. 1999; Prokhorova and Blow 2000). The complex of ORC, Cdc6, and MCM/P1 proteins appears to be responsible for the footprint of the prereplicative complex (pre-RC) observed in Saccharomyces cerevisiae on replication origins in late mitosis and early G1 (Diffley et al. 1994). Once licensing has occurred, ORC and Cdc6 become more loosely bound to DNA and have fulfilled their essential function in DNA replication (Hua and Newport 1998; Rowles et al. 1999).
For a licensed origin to initiate replication, two S phase-promoting protein kinases are then required: an S phase-promoting CDK and the Cdc7/Dbf4 protein kinase. Cdc7 is a serine threonine kinase conserved from yeast to humans that is required for the initiation of DNA replication (Hollingsworth et al. 1992; Jackson et al. 1993; Masai et al. 1995; Jiang and Hunter 1997; Sato et al. 1997; Hess et al. 1998). Although Cdc7 protein levels are approximately constant throughout the cell cycle, Cdc7 kinase activity peaks at the G1/S transition (Jackson et al. 1993; Yoon et al. 1993). This regulation is achieved in part by association with a regulatory subunit termed Dbf4 (Brown and Kelly 1999; Cheng et al. 1999; Jiang et al. 1999; Oshiro et al. 1999; Takeda et al. 1999). Instead of acting as a general initiator of S phase, Cdc7 is probably required to promote initiation at individual origins because, as well as being required for progression into early S phase, it is also required late in S phase to promote initiation at late-firing origins (Bousset and Diffley 1998; Donaldson et al. 1998a). Several lines of evidence suggest that the MCM/P1 proteins are the physiological substrate of Cdc7/Dbf4. In yeast, a mutant allele of Mcm5 (mcm5–bob1) can suppress a complete loss of Cdc7 or Dbf4 (Hardy et al. 1997). Conversely, a mutant of Dbf4 (dbf4-6) was isolated as an allele-specific suppressor of Mcm2 (Lei et al. 1997). Cdc7 and Dbf4 interact physically with MCM/P1 proteins (Lei et al. 1997; Roberts et al. 1999), and a number of reports have identified MCM/P1 proteins as excellent substrates of Cdc7/Dbf4 kinase in vitro (Lei et al. 1997; Sato et al. 1997). Moreover, certain phosphorylations of the MCM/P1 proteins (most notably of Mcm2) in vivo depend on Cdc7 (Lei et al. 1997; Jiang et al. 1999).
The ability of Cdc7 to phosphorylate the MCM/P1 proteins may be aided by its recruitment to replication origins because, in yeast, both Dbf4 and Cdc7 have been shown to interact with chromatin (Pasero et al. 1999; Weinreich and Stillman 1999), the association being dependent on ORC but not Cdc6 (Pasero et al. 1999). This interaction may be mediated by Dbf4, because a one-hybrid screen identified a domain of Dbf4 distinct from the Cdc7 interaction domain that recruited Dbf4 to replication origins (Dowell et al. 1994). Cdc7/Dbf4 could be a target of checkpoint kinases, because when yeast cells were treated with hydroxyurea to block progression through S phase, Dbf4 dissociated from the chromatin (Pasero et al. 1999). This treatment also induced the appearance of a phosphorylated form of Dbf4, which was dependent on the checkpoint kinase Rad53 (Brown and Kelly 1999; Cheng et al. 1999; Takeda et al. 1999; Weinreich and Stillman 1999).
CDKs are also required for the initiation of licensed replication origins. CDK activity leads to the assembly of the essential initiation protein Cdc45 onto origins, creating a “preinitiation complex” (Zou and Stillman 1998). Cdc45 interacts genetically with different components of the pre-RC including MCM/P1 proteins and ORC (Hopwood and Dalton 1996; Owens et al. 1997; Zou et al. 1997), and the association of Cdc45 with chromatin requires Cdc6 and Mcm2 (Zou and Stillman 1998). However, the binding of yeast Cdc45 to chromatin is independent of Cdc7 function, suggesting that CDKs and Cdc7 may act on parallel pathways to initiate replication (Zou and Stillman 1998). CDKs play an important role in executing the temporal program of origin activation during the course of S phase (Donaldson et al. 1998b). Like Cdc7, CDKs are probably required to promote initiation at individual origins, because the CDK-dependent loading of Cdc45 onto late-firing origins only occurs late in S phase (Aparicio et al. 1999).
In the present report, we have used cell-free extracts of Xenopus eggs to determine the precise stage in the process of origin activation at which Xenopus (X) Cdc7 acts. We show that XCdc7 binds to chromatin during G1 and S phase and that both the chromatin binding and the essential DNA replication function of XCdc7 are dependent on licensing but do not require the presence of XORC or XCdc6. We show that XCdc7 is required for the subsequent loading of XCdc45 onto chromatin by CDKs. Finally we show that XCdc7-dependent phosphorylation of XMcm2 and the essential function of XCdc7 can be performed in the absence of CDK activity. These results provide a simple model for the function of Cdc7 in the Xenopus system that appears to differ from that occurring in yeast.
Results
Association of XCdc7 with chromatin is dependent on licensing but independent of CDK activity
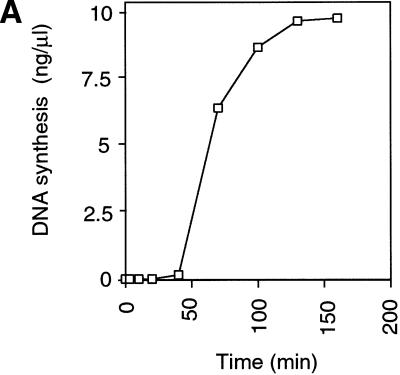
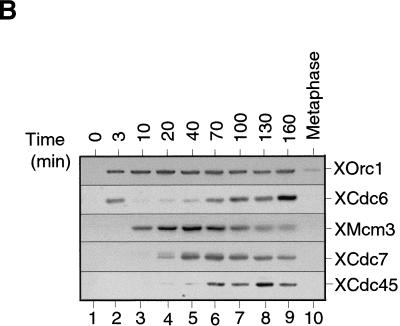
As a first step to understand the regulation of Xenopus Cdc7 in DNA replication, we investigated whether we could detect its association with chromatin during the course of the cell cycle. To get a good synchronization, we used metaphase-arrested extracts induced to enter interphase by the addition of calcium in the absence of new protein synthesis. During the first 40 min these extracts assembled sperm chromatin into interphase nuclei, and during the next 60 min the DNA was replicated precisely (Fig. 1A). These extracts then arrested in a G2-like state because of a lack of cyclin B synthesis. At the indicated times, chromatin was isolated from the extract and immunoblotted for XOrc1, XCdc6, XMcm3, XCdc7, and XCdc45 (Fig. 1B). Consistent with previous reports, XOrc1 and XCdc6 were first assembled onto the chromatin, followed by XMcm3 as the chromatin became licensed for a single round of replication (Chong et al. 1995; Kubota et al. 1995; Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1996). As a consequence of this, origins underwent “licensing-dependent origin inactivation”, and Cdc6 was removed (Hua and Newport 1998; Rowles et al. 1999). After licensing had occurred and before the time of replication initiation, XCdc7 became associated with chromatin. After XCdc7 association, XCdc45 bound to chromatin approximately at the time that DNA replication initiated (Mimura and Takisawa 1998). Then, as DNA was replicated during S phase, XMcm3 was removed. A similar, though slightly less extensive, removal of XCdc7 from chromatin was also observed during S phase. Consistent with previous reports, XCdc6 reassociated with chromatin during S-phase progression (Coleman et al. 1996). In a metaphase-arrested extract, only low levels of these proteins associated with chromatin (Rowles et al. 1999).
Figure 1.
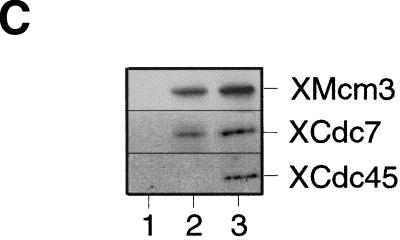
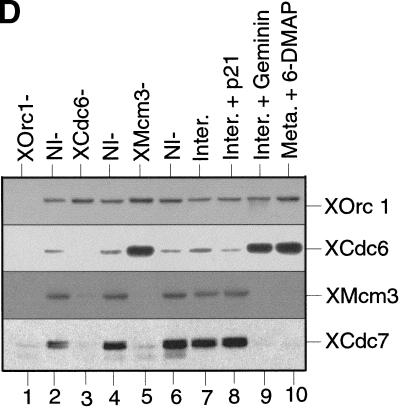
XCdc7 chromatin association is dependent on licensing. (A,B) Sperm nuclei were incubated at 10 ng/μl in interphase Xenopus egg extract. At the indicated times, (A) samples were assayed for DNA synthesis by [α-32P]dATP incorporation, or (B) chromatin was isolated and immunoblotted with antibodies specific for XOrc1, XCdc6, XMcm3, XCdc7, and XCdc45. (C) Sperm nuclei were incubated for 50 min in membrane-free extract (lane 2) or interphase extract (lane 3). Chromatin was isolated, subjected to SDS-PAGE, and blotted for XMcm3, XCdc7, and XCdc45. Lane 1 shows a control of membrane-free extract without added sperm. (D) Sperm nuclei were incubated for 40 min in interphase extracts immunodepleted previously with antibodies against XOrc1 (XOrc1−), XCdc6 (XCdc6−), and XMcm3 (XMcm3−), or with nonimmune antibodies (NI−), or in interphase extract (Inter.) minus or plus added p21 (Inter. + p21) or geminin (Inter. + geminin), or in metaphase arrested extract plus 3 mm 6-DMAP (Meta. + 6-DMAP). Chromatin was then isolated and immunoblotted for XOrc1, XCdc6, XMcm3, and XCdc7.
Because XCdc7 first associated with chromatin at about the same time that the nuclear envelope started to form in these extracts (Blow 1993), it was possible that XCdc7 loading depended on nuclear envelope assembly. To test this idea we compared the chromatin association of XCdc7 with sperm chromatin in whole interphase extract (Fig. 1C, lane 3) or in membrane-free extract that cannot support nuclear assembly (Fig. 1C, lane 2). Assembly of XCdc7 onto chromatin was identical in both extracts, as was the assembly of XMcm3. In contrast, XCdc45 was only loaded in whole extracts that could support nuclear assembly (Fig. 1C, lane 3). This result indicates that unlike XCdc45, the association of XCdc7 with chromatin does not require the prior assembly of DNA into an interphase nucleus.
Because XCdc7 loading took place after XMcm3 had been assembled onto chromatin and licensing had occurred, we investigated whether XCdc7 loading was dependent on licensing (Fig. 1D). Sperm nuclei were incubated in extracts that had been immunodepleted previously with antibodies specific for XOrc1 (which remove the entire ORC complex; Romanowski et al. 1996; Rowles et al. 1996), XCdc6, or XMcm3 (which remove the entire RLF-M complex; Kubota et al. 1997; Thömmes et al. 1997); chromatin was then isolated and immunoblotted for bound proteins. In XOrc1-depleted extract, neither XCdc6, XMcm3, nor XCdc7 associated with chromatin (Fig. 1D, lane 1). In XCdc6-depleted extract XOrc1 associated with chromatin, but XMcm3 or XCdc7 did not (Fig. 1D, lane 3). In XMcm3-depleted extract, XOrc1 and XCdc6 associated with chromatin, but XCdc7 did not (Fig. 1D, lane 5). This suggests that XCdc7's association with chromatin requires the chromatin to contain bound MCM/P1 proteins and to be functionally licensed, a process itself dependent on XORC and XCdc6 (Coleman et al. 1996; Romanowski et al. 1996; Rowles et al. 1996, 1999). To confirm the dependence of XCdc7 loading on licensing, we investigated the loading of XCdc7 onto chromatin in extracts that were defective in licensing. Addition of the kinase inhibitor 6-dimethylaminopurine (6-DMAP) to metaphase Xenopus egg extracts specifically blocks activation of the licensing system (Blow 1993; Mahbubani et al. 1997). In 6-DMAP-treated extracts, XOrc1 and XCdc6 were assembled onto sperm nuclei, but XMcm3 and XCdc7 were not (Fig. 1D, lane 10). A similar result was obtained in extract treated with geminin (Fig. 1D, lane 9), a replication inhibitor that blocks the assembly of MCM/P1 proteins onto chromatin (McGarry and Kirschner 1998; S. Tada and J.J. Blow, in prep.). In contrast, the CDK inhibitor p21Cip1, which inhibits XCdc45 loading and the initiation of replication but not licensing (Strausfeld et al. 1994; Chong et al. 1995; Mimura and Takisawa 1998), did not block the association of XCdc7 with chromatin (Fig. 1D, lane 8). These experiments demonstrate that licensing, but not CDK activity, is necessary for XCdc7 to be loaded onto chromatin.
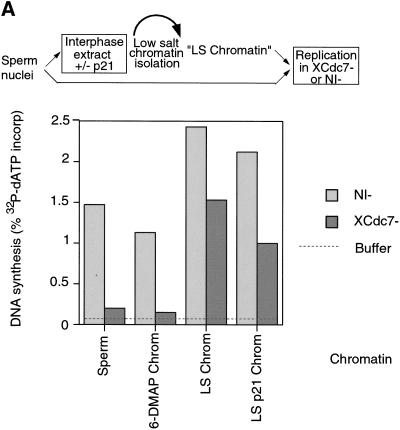
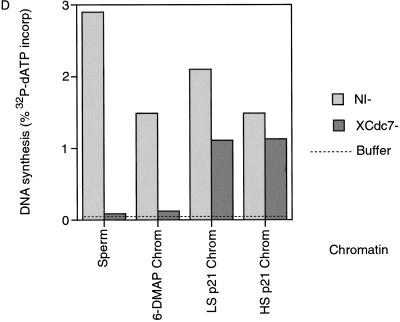
We then wanted to determine whether the association of XCdc7 with chromatin is functional. Extracts were immunodepleted with either antibodies to Cdc7 or with nonimmune antibodies. Figure 2A shows that in contrast to nonimmune-depleted extract, XCdc7-depleted extract was unable to replicate sperm nuclei (Fig. 2A, “Sperm”). This indicates that XCdc7 is required for DNA replication and is consistent with recent data showing inhibition of replication in a Xenopus extract by addition of anti-XCdc7 antibodies (Roberts et al. 1999). However, chromatin previously assembled in untreated extract that had bound XCdc7 (Fig. 1D) was able to replicate in the XCdc7-depleted extract (Fig. 2A, “LS Chrom”). Chromatin assembled in extract containing p21Cip1, which also has bound XCdc7 (Fig. 1D), was also able to replicate in the XCdc7-depleted extract (Fig. 2A, “LS p21 Chrom”). However, chromatin assembled in 6-DMAP-treated extract, which had bound neither XMcm3 nor XCdc7 (Fig. 1D), did not replicate in the XCdc7-depleted extract (Fig. 2A, “6-DMAP Chrom”). This suggests that the chromatin-bound XCdc7, whose presence is dependent on the chromatin having been licensed, is functional and capable of supporting the initiation of DNA replication.
Figure 2.
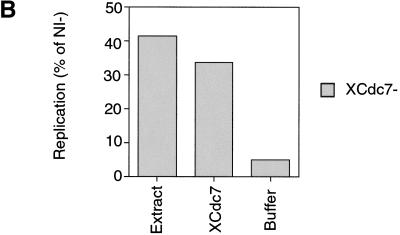
XCdc7 loaded onto chromatin is functional. Interphase Xenopus egg extracts were immunodepleted with antibodies against XCdc7 (XCdc7−) or with an equal quantity of nonimmune antibodies (NI−). Extract was supplemented with [α-32P]dATP and incubated for 3 hr with different DNA templates. DNA replication was assessed by the proportion of 32P incorporated into acid-insoluble material. (A) Replication in XCdc7-depleted and nonimmune-depleted extracts was measured using Xenopus sperm nuclei (Sperm) or various chromatin templates, prepared by incubating Xenopus sperm nuclei in interphase extract for 20 min minus (LS Chrom) or plus p21Cip1 (LS p21 Chrom) or for 15 min in 6-DMAP-treated extract (6-DMAP Chrom), followed by isolation in low salt buffer. (B) XCdc7-depleted extract was supplemented with 0.05 volume of different fractions, and its ability to replicate chromatin previously assembled in 6-DMAP-treated extract was assessed. Added fractions: (Extract) membrane-free egg extract; (XCdc7) partially purified XCdc7; (Buffer) LFB2/50. Replication is shown as the percentage obtained with the identical chromatin template incubated in nonimmune-depleted (NI−) extract.
To test the specificity of our XCdc7 depletion, we performed a partial chromatographic purification of XCdc7 from Xenopus egg extracts. The behavior of XCdc7 over four chromatographic separations was followed by immunoblotting (see Materials and Methods). This partially purified material was able to restore replicative capacity to XCdc7-depleted extracts (Fig. 2B). Comparison with unfractionated extract showed that the rescuing activity had been enriched ∼100-fold, supporting the idea that XCdc7 had been specifically depleted from the extract.
The loading of XCdc7 onto chromatin does not require the presence of XCdc6 and XORC
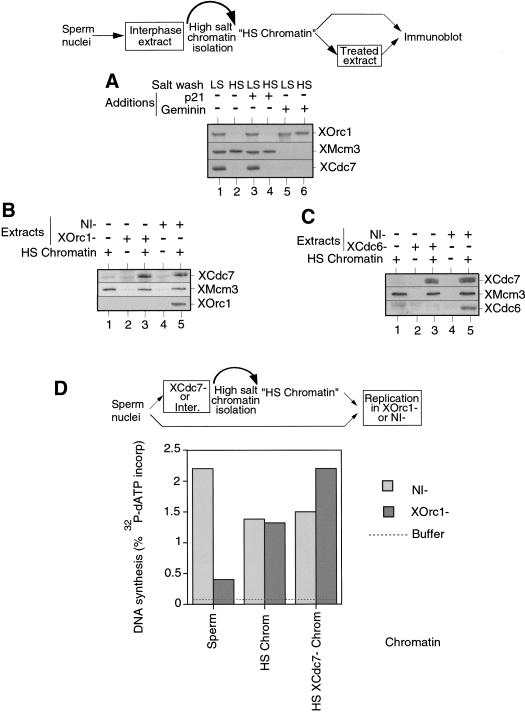
Although XORC and XCdc6 are required for loading the XMCM/P1 proteins onto chromatin, they are not required for the continued binding of XMCM/P1 proteins once they have been loaded. After licensing has occurred, XORC and XCdc6 can be removed from the chromatin by exposure to high CDK levels or by exposure to high salt, leaving functional XMCM/P1 proteins on the chromatin (Donovan et al. 1997; Hua and Newport 1998; Rowles et al. 1999). We therefore wanted to test whether the requirement for XORC and XCdc6 in the loading of XCdc7 onto chromatin (Fig. 1D) was because XCdc7 loading is also dependent on XORC and XCdc6 or whether it was simply because XCdc7 binding requires chromatin-bound XMCM/P1 proteins. Figure 3A shows that, consistent with previous reports (Rowles et al. 1999), XOrc1 was removed from licensed chromatin by treatment with high salt (∼250 mm) but was not removed from unlicensed chromatin by the same treatment (Fig. 3A, lanes 2,6). In contrast, XMcm3 remained bound to chromatin after high salt treatment (Fig. 3A, lanes 2, 4). High salt removal of XOrc1 also occurred on chromatin assembled in the presence of p21Cip1 (Fig. 3A, lane 4), indicating that CDK activity is not required for XOrc1 destabilization. XCdc7, like XOrc1, was completely removed from chromatin after high salt treatment, either in the presence or the absence of p21Cip1 (Fig. 3A, lanes 2,4).
Figure 3.
XCdc7 chromatin loading is independent of XOrc1 and XCdc6. (A) Xenopus sperm nuclei were incubated for 25 min in interphase Xenopus extract plus or minus p21Cip1 or geminin. Chromatin was isolated in low salt (LS) or high salt (HS) buffers and then immunoblotted for XOrc1, XMcm3, and XCdc7. (B,C). Sperm nuclei were incubated for 20 min in interphase extract, and chromatin was isolated in high salt buffer (HS Chromatin). An aliquot was used for immunoblot (lane 1) and the rest was reincubated in nonimmune-depleted (NI−) extracts or either XOrc1-depleted (XOrc1−) (B) or XCdc6-depleted (XCdc6−) (C) extracts for a further 50 min. Chromatin was re-isolated and blotted for XOrc1 (B) or XCdc6 (C), XMcm3, and XCdc7. Controls lacking added chromatin are also shown (lanes 2, and 4). A schematic representation of the experimental procedure for B and C is shown above. (D) Sperm nuclei were incubated for 20 min in XCdc7-depleted extract or untreated interphase extract, and chromatin was isolated in high salt buffer. Chromatin from the untreated extract (HS Chrom) or the XCdc7-depleted extract (HS XCdc7− Chrom) or untreated sperm nuclei (Sperm) were incubated for 3 hr in either XOrc1-depleted (XORC−) or in nonimmune-depleted extract (NI−) supplemented with [α-32P]dATP for 3 hr. DNA synthesis was assessed by the proportion of 32P incorporated into acid-insoluble material.
We then used this high salt treatment to test whether XORC and XCdc6 are required for XCdc7 loading beyond being required for licensing. Licensed chromatin was isolated in the presence of high salt to remove XORC, XCdc6, and XCdc7, but not MCM/P1, proteins (Fig. 3B,C, lane 1). This chromatin was then added to XOrc1- or XCdc6-depleted extracts and checked for XCdc7 reloading. Figure 3, B and C, shows that XCdc7 reloading occurred in both XOrc1- and XCdc6-depleted extracts (Fig. 3B,C, lane 3). These results indicate that neither XOrc1 nor XCdc6 are directly required for loading XCdc7 onto chromatin but are only indirectly required to license the DNA and load MCM/P1 proteins, which is in its turn required for XCdc7 loading. We confirmed this conclusion by showing that XCdc7 could be loaded onto licensed chromatin even after XORC and XCdc6 had been removed from the chromatin by the addition of cyclin A (data not shown).
We then wanted to determine whether the XCdc7 loaded onto chromatin in the absence of XORC is fully functional. Chromatin was assembled in whole extract or in XCdc7-depleted extract and was then washed in high salt to remove XORC (Fig. 3A; see also Fig. 4A). Consistent with previous results (Rowles et al. 1999), the washed chromatin from whole extract replicated efficiently in XOrc1-depleted extract (Fig. 3D, “HS Chrom”), showing that XORC had completed all its essential functions for DNA replication before isolation. When this protocol was repeated using chromatin assembled previously in XCdc7-depleted extract, the chromatin was again efficiently replicated in the XOrc1-depleted extract (Fig. 3D, “HS XCdc7− Chrom”). These results suggest that XCdc7 can load onto chromatin and perform all its essential functions for DNA replication in the absence of XORC. Because XCdc6 can only bind to chromatin in the presence of XORC, XCdc7 function is also likely be independent of XCdc6.
Figure 4.
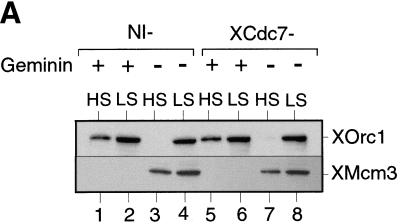
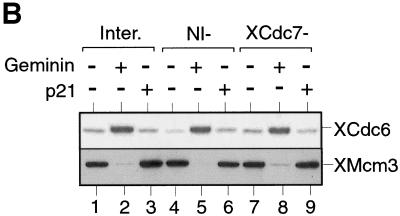
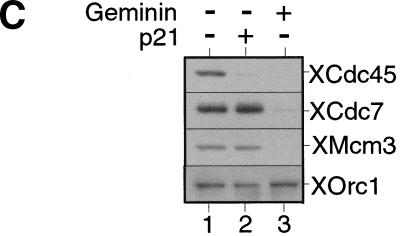
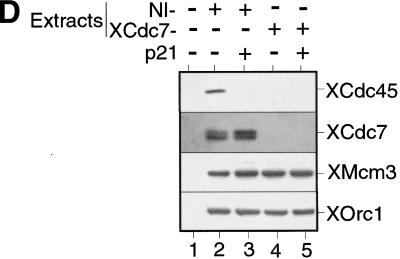
XCdc7 is not involved in licensing-dependent origin inactivation but is required for XCdc45 loading. (A) Sperm nuclei were incubated for 40 min in XCdc7-depleted extract (XCdc7−) or nonimmune-depleted extract (NI−) plus or minus geminin. Samples were split in two, and chromatin was isolated in low (LS) or high salt (HS) buffers and blotted for XOrc1 and XMcm3. (B) Sperm nuclei were incubated for 40 min in interphase extract (Inter.), XCdc7-depleted extract (XCdc7−) or nonimmune-depleted extract (NI−) plus or minus p21Cip1 or geminin. Chromatin was isolated in low salt buffer and immunoblotted for XCdc6 and XMcm3. (C) Sperm nuclei were incubated for 45 min in interphase extract plus or minus added p21Cip1 or geminin. Chromatin was isolated in low salt buffer and blotted for XCdc45, XCdc7, XMcm3, and XOrc1. (D) Sperm nuclei were incubated in XCdc7-depleted extract (XCdc7−) or nonimmune-depleted extract (NI−) plus or minus added p21Cip1 for 150 min (the long incubation is due to the slow rate that immunodepleted extracts assemble DNA into interphase nuclei). Complete nuclear envelope formation was verified by microscopy, and chromatin was then isolated in low salt buffer and immunoblotted for XCdc45, XCdc7, XMcm3, and XOrc1.
XCdc7 is not involved in licensing-dependent origin inactivation but is required for XCdc45 loading
The assembly of the pre-RC is a dynamic process in which the binding of some elements changes the stability of others. On unlicensed chromatin, XORC is stably bound to chromatin, resisting elution by high CDK activity and high salt; however, after licensing has occurred, XORC can be eluted by either of these treatments (Rowles et al. 1999). Similarly, XCdc6 binds avidly to unlicensed chromatin, but once licensing has occurred, its levels on chromatin drop significantly (Fig. 1B), and it can be fully eluted by exposure to CDKs. We have termed this effect licensing-dependent origin inactivation (Rowles et al. 1999). We then investigated whether XCdc7 is involved in this process (Fig. 4). Sperm nuclei were incubated in nonimmune-depleted or XCdc7-depleted extract plus or minus geminin (to inhibit licensing) or p21Cip1 (to inhibit CDKs). Chromatin was then isolated in high or low salt and then immunoblotted for XOrc1 (Fig. 4A) or XCdc6 (Fig. 4B). As expected, the elution of XOrc1 by high salt was inhibited when licensing was blocked by geminin, but this behavior was the same in nonimmune-depleted and XCdc7-depleted extracts (Fig. 4A). Similarly, high levels of XCdc6 were seen on chromatin only when licensing was inhibited by geminin (Fig. 4B, lane 2), but XCdc6 behavior was similar in nonimmune-depleted and XCdc7-depleted extracts (Fig. 4B, lanes 4–9). Blocking CDK activity by addition of p21Cip1 also had no significant effect on the level of XCdc6 bound to chromatin (Fig. 4B, lanes 3,6,9). These results suggest that neither XCdc7 nor CDKs are required for the destabilization of XORC and XCdc6 that occurs as a consequence of origin licensing.
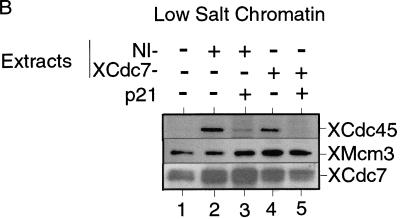
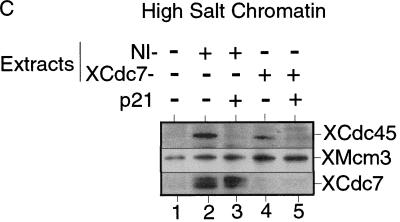
The loading of Cdc45 onto chromatin requires CDK activity, both in S. cerevisiae and in Xenopus (Mimura and Takisawa 1998; Zou and Stillman 1998). Moreover, in yeast it has been shown that this loading requires the assembly of a pre-RC (Zou and Stillman 1998). To test whether XCdc45 loading in Xenopus is dependent on the chromatin having been licensed, sperm nuclei were incubated in an interphase extract plus or minus added p21Cip1 (to inhibit CDKs) or geminin (to inhibit licensing). Consistent with a previous report (Mimura and Takisawa 1998), Figure 4C shows that XCdc45 loading was inhibited by p21Cip1 although licensing occurred normally (Fig. 4C, lanes 1,2). In contrast, XCdc45 loading did not occur in the presence of geminin (Fig. 4C, lane 3), suggesting that the binding of XCdc45 to chromatin is dependent on both XMCM/P1 proteins and CDK activity. Because XCdc7 loading is also dependent on the presence of MCM/P1 proteins, we then tested whether XCdc45 loading is dependent on XCdc7. Figure 4D shows that in nonimmune-depleted extract the association of XCdc45 with chromatin occurred normally and was inhibited by p21Cip1 (Fig. 4D, lanes 2,3). However, XCdc45 did not associate with chromatin in XCdc7-depleted extract despite the chromatin becoming fully licensed (Fig. 4D, lane 4). This suggests that both XCdc7 and CDKs are required for the loading of XCdc45 onto chromatin.
XCdc7-dependent phosphorylation of XMcm2 is independent of CDKs
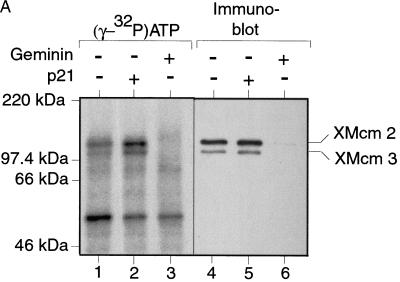
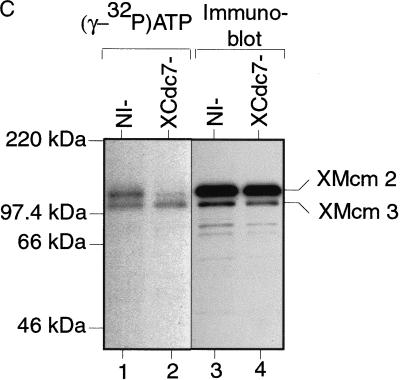
Experiments in a number of eukaryotic organisms have suggested that the major role of Cdc7 in DNA replication is to phosphorylate the MCM/P1 proteins (Hardy et al. 1997; Lei et al. 1997; Sato et al. 1997). We therefore wanted to see whether we could detect XCdc7-dependent MCM/P1 phosphorylation in the Xenopus system. Sperm nuclei were incubated in extracts containing [γ-32P]ATP, and chromatin was then isolated in a high salt buffer to remove most of the chromatin-bound proteins (such as XOrc1, XCdc6, and XCdc7) but retain the MCM/P1 proteins (Fig. 3A; Rowles et al. 1999). 32P-Labeled chromatin proteins were revealed by SDS-PAGE and autoradiography. Figure 5A shows that three main phosphorylated bands were detected, two of which comigrated with XMcm2 and XMcm3 (Fig. 5A, lanes 1,4). In extracts treated with geminin (to block the loading of MCM/P1 proteins onto DNA), these two phosphorylated bands were not detected (Fig. 5A, lanes 3,6). The identity of these two proteins was confirmed by their elution from chromatin in 1 m salt, which disrupts MCM/P1 complexes (Kubota et al. 1997; Prokhorova and Blow 2000), followed by immunoprecipitation with antibodies specific for XMcm2 and XMcm3 (Fig. 5B). Importantly, these two phosphorylated proteins were also detected at similar levels when the extract was treated with p21Cip1 (Fig. 5A, lanes 2,5), suggesting that these phosphorylations occur independently of CDK activity. To address whether these phosphorylations were dependent on XCdc7 activity, we compared labeling in control and XCdc7-depleted extracts (Fig. 5C). This showed that although XMcm3 phosphorylation was similar in the two extracts, XMcm2 phosphorylation was strongly reduced in the XCdc7-depleted extract (Fig. 5C, lane 2). This is consistent with the description of Cdc7-dependent Mcm2 phosphorylation in S. cerevisiae (Lei et al. 1997) and with the reported ability of Cdc7 to specifically phosphorylate Mcm2 when a complex of Mcm2-7 was used as substrate in Schizosaccharomyces pombe (Brown and Kelly 1998) and in HeLa cells (Jiang et al. 1999). Taken together, these results suggest that there is a significant level of XCdc7-dependent phosphorylation of XMcm2 that occurs in the absence of CDK activity and before DNA replication initiates.
Figure 5.
XCdc7-dependent XMcm2 phosphorylation is independent of CDK activity. (A,B) Sperm nuclei were incubated for 25 min in interphase extract containing [γ-32P]ATP plus or minus added p21Cip1 or geminin. Chromatin was then isolated in the presence of phosphatase inhibitors and 250 mm KCl. (A) Samples were subjected to SDS-PAGE and Western blotted; the filters were autoradiographed (lanes 1–3) and then probed with antibodies against XMcm2 and XMcm3 (lanes 4–6). (B) Isolated chromatin was treated with 1 m KCl to elute MCM/P1 proteins from chromatin. The supernatant was immunoprecipitated with XMcm2 and XMcm3 antibodies, and samples were run on SDS-PAGE and autoradiographed. (C) Sperm nuclei were incubated for 25 min in XCdc7-depleted extract (XCdc7−) or nonimmune-depleted extract (NI−) containing [γ-32P]ATP. Chromatin was isolated in the presence of phosphatase inhibitors and 250 mm KCl and subjected to SDS-PAGE and Western blotted; the filters were autoradiographed (lanes 1,2) and then probed with antibodies against XMcm2 and XMcm3 (lanes 3,4).
The replicative function of XCdc7 can occur independently of CDKs
Because MCM/P1 phosphorylation is expected to be one of the major functions of XCdc7 in the initiation of DNA replication and because we have detected an XCdc7-dependent phosphorylation of XMcm2 in the absence of CDK activity (Fig. 5), we wanted to determine whether the replicative function of XCdc7 could be fulfilled before the action of CDKs. We therefore prepared chromatin in extracts supplemented with p21Cip1 to block CDK activity, and isolated it in either low or high salt buffer (Fig. 6). Consistent with the results shown in Figure 3A, the chromatin isolated in low salt contained XMcm3 and XCdc7 (Fig. 6B, lane 1), but the chromatin isolated in high salt contained only XMcm3 (Fig. 6C, lane 1). XCdc45 was not present on either of the chromatin templates because of the added p21Cip1. Figure 6B shows that when the low salt chromatin (containing XCdc7 but lacking XCdc45) was incubated in an XCdc7-depleted extract, XCdc45 was loaded onto chromatin (Fig. 6B, lane 4). This loading was still inhibited by the addition of p21Cip1 to the XCdc7-depleted extract (Fig. 6B lane 5). Furthermore, this chromatin was able to replicate in XCdc7-depleted extract (Fig. 6D, “LS p21 Chrom”). This suggests that either the XCdc7 present on low salt chromatin was fully functional or that the essential function of XCdc7 had already been fulfilled on this chromatin template. The chromatin washed in high salt was used to distinguish these possibilities. Although the high salt washed chromatin did not contain bound XCdc7 (Fig. 6C, lane 1), it was still able to support XCdc45 loading when incubated in an XCdc7-depleted extract (Fig. 6C, lane 4). Again, this loading was inhibited by the addition of p21Cip1 to the XCdc7-depleted extract (Fig. 6C, lane 5). Furthermore, the high salt washed chromatin was able to replicate efficiently in XCdc7-depleted extract (Fig. 6D, “HS p21 Chrom”). Taken together, these results suggest that XCdc7 can phosphorylate XMcm2 (Fig. 5) and fulfil its essential function for XCdc45 loading and DNA replication in the absence of CDK activity (Fig. 6).
Figure 6.
XCdc7 can perform its essential function in the absence of CDK activity. (A) Schematic representation of the experimental procedure. Sperm nuclei were incubated for 25 min in interphase extract supplemented with p21Cip1. Chromatin was isolated in low or high salt buffer and reincubated for 150 min in XCdc7-depleted extract (XCdc7−) or nonimmune-depleted extract (NI−), plus or minus further added p21Cip1. DNA synthesis was assessed by incorporation of [α-32P]dATP; alternatively, chromatin was reisolated and immunoblotted for XCdc45, XMcm3, and XCdc7. (B) Low salt chromatin was blotted for bound XCdc45, XMcm3, and XCdc7 after reincubation in XCdc7- or nonimmune-depleted extract. (C) High salt chromatin was blotted for bound XCdc45, XMcm3, and XCdc7 after reincubation in XCdc7- or nonimmune-depleted extract. (D) DNA synthesis in XCdc7- or nonimmune-depleted extract. Chromatin templates were untreated sperm nuclei (Sperm), sperm nuclei incubated in 6-DMAP-treated extract and isolated under low salt conditions (6-DMAP Chrom), sperm nuclei incubated in p21Cip1-treated extract and isolated under low salt conditions as in B (LS p21 Chrom), or sperm nuclei incubated in p21Cip1-treated extract and isolated under high salt conditions as in C (HS p21 Chrom).
Discussion
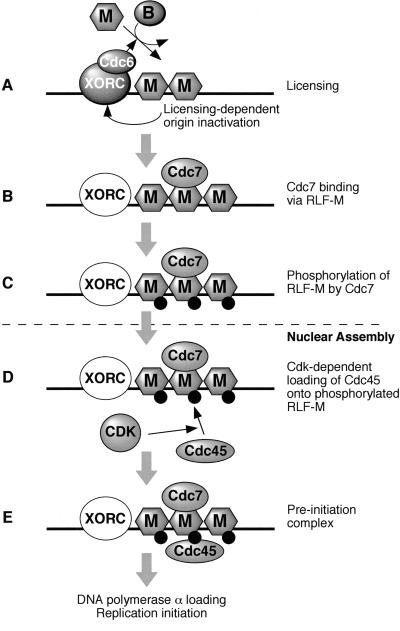
In this paper we have characterized the role of XCdc7 in DNA replication using the Xenopus cell-free system. This has allowed us to define the precise stage that XCdc7 functions in the assembly of an active replication origin. We have shown that XCdc7 associates with chromatin during the cell cycle and that this association requires the presence of the MCM/P1 proteins, but not XORC or XCdc6. We have shown that XCdc7 function is required for XMcm2 phosphorylation and for XCdc45 loading. Importantly, we show that XCdc7 can perform these functions in the absence of CDK activity. A model based on these results is shown in Figure 7.
Figure 7.
A model for XCdc7 function. (A) The loading of the RLF-M complex of XMCM/P1 proteins onto chromatin (licensing) is dependent on XORC and XCdc6 and RLF-B. As a consequence of licensing, XORC is destabilized and XCdc6 is removed, independent of XCdc7 function. (B) XCdc7 then binds to chromatin, plausibly by physical interaction with MCM/P1 proteins, and (C) phosphorylates them. After this phosphorylation, which can occur in the absence of CDK activity, XCdc7 is no longer required for DNA replication. (D) After nuclear assembly, the XCdc7-dependent phosphorylation of XMCM/P1 proteins is required for the CDK-dependent loading of XCdc45. (E) The preinitiation complex at origins, containing XORC, MCM/P1 proteins, XCdc7, and XCdc45 (though XORC and XCdc7 are no longer essential), promotes the loading of DNA polymerase α and the initiation of replication.
The association of XCdc7 with chromatin is dependent on licensing but not on the presence of XORC and XCdc6
Using Xenopus egg extracts that support cell cycle progression in vitro, we have shown that XCdc7 associates with chromatin from early interphase and is at least partially released during S-phase progression. The association of XCdc7 with chromatin in early interphase requires the chromatin to be functionally licensed for replication and contain bound MCM/P1 proteins (Fig. 7A,B). This mode of chromatin association provides a different picture from that obtained in S. cerevisiae. Although S. cerevisiae Dbf4 associates with chromatin during S and G2 phases, Cdc7 appears to associate with chromatin throughout the cell cycle (Weinreich and Stillman 1999). The binding of Dbf4 to a detergent-insoluble chromatin fraction has been shown to be dependent on ORC, but not on Cdc6 or MCM/P1 proteins (Pasero et al. 1999). The association of Xenopus Cdc7 with chromatin therefore differs from that seen in yeast for either Cdc7 or Dbf4.
Because MCM/P1 proteins are likely substrates for Cdc7 phosphorylation in vivo, the binding of XCdc7 to chromatin is probably to be by direct interaction with the MCM/P1 proteins (Fig. 7B). Although at present we have no direct proof of this, it would be consistent with the description of physical interaction between Cdc7 and MCM/P1 proteins (Lei et al. 1997; Roberts et al. 1999) and with our observations that XCdc7 is released from chromatin as the MCM/P1 proteins are released. Our demonstration that the association of XCdc7 with chromatin requires neither XORC nor XCdc6 once they have performed their essential function in origin licensing and requires neither nuclear assembly nor CDK activity is also consistent with a direct interaction between XCdc7 and the MCM/P1 proteins.
XCdc7-dependent phosphorylation of XMcm2
Our demonstration of an MCM/P1-dependent association of XCdc7 with chromatin and an XCdc7-dependent phosphorylation of XMcm2 is consistent with a number of observations suggesting that MCM/P1 proteins are the natural substrates for the Cdc7 kinase in vivo. In yeast, a mutant allele of Mcm5 (mcm5–bob1) is able to bypass the requirement for Cdc7 or Dbf4 (Hardy et al. 1997), and the phosphorylation of MCM/P1 proteins that normally occurs during S phase does not occur in cdc7 mutants (Lei et al. 1997). Furthermore, a number of reports have shown that the MCM/P1 proteins are excellent substrates of the Cdc7/Dbf4 kinase in vitro (Lei et al. 1997; Sato et al. 1997). The preferential phosphorylation of XMcm2 over the other MCM/P1 proteins that we see is also mirrored in other systems. In S. cerevisiae Mcm2p is preferentially phosphorylated in vivo in a Cdc7-dependent way (Lei et al. 1997). When human and S. pombe Cdc7 homologs are used to phosphorylate mixtures of MCM/P1 proteins in vitro, Mcm2 appears to be the preferred substrate (Brown and Kelly 1998; Jiang et al. 1999). Given that XMcm2 appears to be a good XCdc7 substrate, the XCdc7-dependent phosphorylation of XMcm2 that we observe is likely to be the result of direct phosphorylation by XCdc7 (Fig. 7C). However, our results do not rule out the possibility that XCdc7 also phosphorylates other MCM/P1 proteins as well.
XCdc7 is not required for licensing-dependent origin inactivation
After licensing has occurred in Xenopus, the association of XCdc6 and XORC with chromatin is destabilized, a phenomenon we have called licensing-dependent origin inactivation (Rowles et al. 1999). This is evidenced by a dramatic reduction in the quantity of chromatin-bound XCdc6 (Fig. 1B) and a switch of chromatin-bound XORC from a salt-resistant to a salt-sensitive form. Licensing-dependent origin inactivation may contribute to mechanisms preventing re-replication of DNA in a single cell cycle. In yeast and mammalian cells, there is evidence to show that Cdc6 function is regulated at least in part by CDKs (Jallepalli et al. 1997; Elsasser et al. 1999; Petersen et al. 1999). We provide evidence that in Xenopus, licensing-dependent origin inactivation is independent of XCdc7 or CDK activity (Fig. 7A). We could not reproduce the results of Hua and Newport (1998), who proposed that CDK activity is required for XCdc6 removal. We note, however, that in the Hua and Newport study, a comparison was not made between XCdc6 levels on chromatin before and after licensing, and so the full range of XCdc6 levels was never compared. Instead, our results suggest that origin destabilization is dependent only on licensing and that no other known events downstream of licensing are involved. When XORC and XCdc6 were removed from chromatin that had been licensed in the absence of XCdc7, the resultant chromatin was still able to replicate efficiently in extracts lacking XORC. This result supports the idea that the only function of XORC and XCdc6 is to support replication licensing, after which they have no further essential function in DNA replication (Hua and Newport 1998; Rowles et al. 1999).
XCdc45 chromatin loading requires XCdc7 but does not require physical interaction with XCdc7
Cdc45 is required for the initiation of DNA replication (Hopwood and Dalton 1996; Zou et al. 1997; Mimura and Takisawa 1998) and is assembled onto replication origins only in the presence of CDK activity (Mimura and Takisawa 1998; Zou and Stillman 1998). The Cdc45-containing protein complex found at replication origins in yeast has been termed the preinitiation complex (Zou and Stillman 1998). Cdc6 and Mcm2 are both required for Cdc45 to be loaded onto chromatin, suggesting that assembly of a licensed pre-RC is also necessary for Cdc45 loading (Zou and Stillman 1998). We show that this is also the case in Xenopus and that when licensing is blocked by geminin, XCdc45 is not loaded onto chromatin. We conclude that the MCM/P1 proteins are, therefore, essential components of the preinitiation complex in Xenopus.
We show here for the first time that the assembly of XCdc45 is dependent on XCdc7 function and does not occur in XCdc7-depleted extracts. However, the loading of XCdc45 does not require physical interaction with XCdc7, because once chromatin has interacted with XCdc7 and MCM/P1 phosphorylation has occurred, XCdc7 is no longer required for XCdc45 to bind to chromatin. This is most easily explained by a direct binding of XCdc45 to phosphorylated MCM/P1 proteins at licensed origins (Fig. 7D). Consistent with this interpretation, direct interactions between Cdc45 and different MCM/P1 proteins have been described in yeast (Hopwood and Dalton 1996; Zou and Stillman 1998). A requirement for interaction with XORC or XCdc6 can be ruled out in Xenopus because we also show that XCdc7 function can be performed in the complete absence of XORC, which is itself required for the binding of XCdc6 to chromatin. Thus, XORC and XCdc6 are not essential components of the preinitiation complex in Xenopus. These results are in apparent contrast with reports in yeast where it has been shown that the chromatin loading of Cdc45p occurs in a cdc7 mutant and that Cdc7 and Cdc45 functions are interdependent (Owens et al. 1997; Zou and Stillman 1998). This may represent a difference in the way that yeast and Xenopus regulate the assembly of replication origins (see below).
XCdc7 function can be executed in the absence of CDK activity
Although both Cdc7 and CDKs are required for the initiation of replication, it has been unclear whether they must act together or whether they act in a specific order. We show here that XCdc7 can fulfil all its known functions for the initiation of DNA replication in the absence of CDK activity. First, we show that the loading of XCdc7 onto chromatin is unaffected by the CDK inhibitor p21Cip1; similarly, XCdc7 can be loaded onto chromatin in the absence of nuclear assembly, whereas XCdc45, whose loading is CDK-dependent, cannot. Second, we show that the XCdc7-dependent phosphorylation of XMcm2 is unaffected by the presence of p21Cip1 at concentrations sufficient to block the initiation of replication and block XCdc45 loading. Third, we show that once XCdc7 has been allowed to interact with chromatin and phosphorylate MCM/P1 proteins in the presence of p21Cip1, XCdc7 can be washed off chromatin and is no longer required for subsequent XCdc45 loading and efficient DNA replication. These results strongly support the idea that the XCdc7 function can be completed in the absence of significant CDK activity. A simple model for Cdc7 function is suggested from these results. The MCM/P1 proteins would remain inactive until XCdc7 binds and phosphorylates them, events that are independent of nuclear assembly and CDK activity (Fig. 7B,C). This MCM/P1phosphorylation would produce a conformational change that is required for downstream events that are dependent on further phosphorylation by CDKs such as Cdc45 loading (Mimura and Takisawa 1998; Zou and Stillman 1998) (Fig. 7D,E). The association of Cdc45 with chromatin is then required for loading DNA polymerase α onto chromatin (Mimura and Takisawa 1998) and the initiation of replication. However, we cannot rule out at present the possibility that XCdc7 is required to phosphorylate other targets in the process of initiation.
Implications for the regulatory role of Cdc7
It is currently unclear what physiological role Cdc7 plays in the cell cycle. Because mcm5–bob1 mutant cells show essentially normal cell cycle timing in the absence of Cdc7 (Hardy et al. 1997), this suggests that Cdc7 is not primarily involved in ordering cell cycle events. Instead, there are indications that Cdc7 may be involved in checkpoint execution. In yeast, Cdc7 is required in late S phase to fire late replication origins (Bousset and Diffley 1998; Donaldson et al. 1998a), and XCdc45 is loaded onto late replication origins only late in S phase (Aparicio et al. 1999). However, if replication from early replication origins is blocked, then the firing of late origins is subjected to a Rad53-dependent checkpoint delay (Santocanale and Diffley 1998; Shirahige et al. 1998), which is associated with a phosphorylation of Dbf4 and its displacement from chromatin (Brown and Kelly 1999; Cheng et al. 1999; Pasero et al. 1999; Takeda et al. 1999; Weinreich and Stillman 1999). This suggests that the checkpoint delay might plausibly be mediated by Dbf4 and Cdc7 being displaced from chromatin to prevent it from phosphorylating MCM/P1 proteins. The very early execution of Xenopus Cdc7 function that we show here might at first sight seem to be incompatible with this S-phase checkpoint. However, there are several possible explanations for this apparent discrepancy. One possibility is that the Xenopus early embryo, which is known to have very reduced checkpoints compared with somatic cells (Hara et al. 1980; Kimelman et al. 1987), may have completely bypassed this S-phase checkpoint and so allowed XCdc7 to fulfil its function early in the cell cycle. An alternative possibility is that the S-phase checkpoint is active in the Xenopus system but that the replication we see when all XCdc7 function is provided in the absence of CDK activity derives from early firing, but not late-firing, replication origins. In this interpretation, the efficient replication that we observe could be explained by the passive replication of late replication origins by replication forks emanating from early firing origins. A role for XCdc7 in late S-phase to fire late replication origins would be consistent with our observation that XCdc7 remains associated with chromatin throughout S phase. However, further experimentation is required to investigate whether XCdc7 really does have a checkpoint function in the Xenopus early embryo.
Materials and methods
Preparation and use of egg extracts
Activated, metaphase-arrested, and licensing factor Xenopus egg extracts were prepared as described (Chong et al. 1997). Membrane-free extract is a 9% PEG precipitation of a licensing factor extract (Chong et al. 1997). For replication assays, the extracts were supplemented with 100 μg/ml cycloheximide, 25 mm phosphocreatine, 15 μg/ml creatine phosphokinase, 0.3 mm CaCl2 and [α-32P]dATP. 6-DMAP extracts were prepared in a similar way, except 3 mm 6-DMAP was added before CaCl2 addition (Blow 1993; Chong et al. 1997). Immunodepletion of interphase extracts with specific antibodies or with antibodies from nonimmune rabbit serum was performed as described (Chong et al. 1997). Immediately after immunodepletion extracts were snap-frozen in liquid nitrogen in 10-μl aliquots for future use. DNA synthesis was measured by incorporation of [α-32P]dATP into acid insoluble material as described (Chong et al. 1997). The final DNA concentration in these assays were kept at 1–10 ng of DNA/μl extract. All incubations were performed at 23°C.
Antibodies
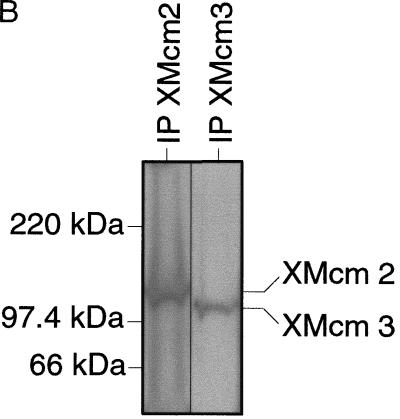
Antibodies raised against XOrc1 (Rowles et al. 1996), XCdc6 (Tada et al. 1999), and XMcm3 (Thömmes et al. 1997; Prokhorova and Blow 2000) were as described previously. Antiserum against XCdc45 was a kind gift of Dr. H. Takisawa (Mimura and Takisawa 1998). Rabbit polyclonal antibodies were raised against recombinant His-tagged full-length Xenopus Cdc7 (Sato et al. 1997). The XCdc7 antibody specifically immunoprecipitated a doublet of ∼55 kD from egg extract, as expected for full-length XCdc7 (53.5 kD; Sato et al. 1997), which comigrated with in vitro-translated XCdc7. A band of the same molecular weight was also specifically recognized by several affinity purified antibodies raised against XCdc7 (a kind gift of Dr. H. Masai; Sato et al. 1997). Although the antibody also cross-reacted in immunoblots of whole extracts with two other polypeptides, these polypeptides were not immunoprecipitated by the antibody. Furthermore, XCdc7 was the only polypeptide recognized on immunoblots of chromatin assembled in the Xenopus extracts.
Chromatin templates
Demembranated Xenopus sperm nuclei were prepared as described (Chong et al. 1997). They were assembled into chromatin by incubation at 20 ng/μl for appropriate times in treated extracts. In the case of immunodepleted extracts, sperm nuclei were incubated at 7 ng/μl, to compensate for the threefold dilution that occurs during depletion. Extract was then diluted 10- to 20-fold in nuclear isolation buffer (Chong et al. 1997) (NIB: 50 mm KCl, 50 mm HEPES–KOH at pH 7.6, 5 mm MgCl2, 2 mm DTT, 0.5 mm spermine–3HCl, 0.25 mm spermine–4HCl, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) supplemented with 0.25% Triton X-100 and then underlayered with the same buffer containing 15% sucrose. For high salt chromatin isolation additional KCl (to give 250 mm final) was added to the buffers. The chromatin was pelleted at 2100g in a swinging bucket centrifuge for 5 min at 4°C. The diluted extract and the top part of the cushion were carefully removed, and the chromatin pellet was resuspended in loading buffer and subjected to immunoblotting by standard techniques using 10% SDS-PAGE and ECL detection (Amersham). When isolated chromatin had to be reincubated in another extract, the isolation was performed in the same way but in the absence of Triton X-100, and a 10% sucrose cushion was used and underlayered with a cushion of treated extract (normally an extract depleted with specific antibodies). For reincubation of high salt chromatin, double 10% sucrose cushions were used, the buffer in the top cushion containing 250 mm KCl and the buffer in the bottom cushion containing 50 mm KCl. For the detection of XMCM/P1 phosphorylation [γ-32P]ATP was added to the extract. Chromatin isolation was performed as above but NIB and sucrose cushions were supplemented with a phosphatase inhibitor cocktail (0.1 mm sodium vanadate, 50 mm β-glycerophosphate, and 20 μm microcystin). After blotting, the membrane was revealed by autoradiography and ECL. Immunoprecipitation of XMCM/P1 proteins eluted from chromatin by 1 m KCl treatment was performed as described (Chong et al. 1997).
Partial purification of XCdc7
All chromatographic procedures were performed at 4°C, and fractions containing XCdc7 were identified by immunoblotting. A 4.5%–9% polyethylene glycol cut of “licensing factor extract” (Chong et al. 1997) was taken and resuspended at 1× in LFB1/100 (40 mm HEPES–KOH at pH 8.0, 20 mm K2HPO4/KH2PO4 at pH 8.0, 2 mm MgCl2, 1 mm EGTA, 2 mm DTT, 10% (wt/vol) sucrose, and 1 μg/ml each of leupeptin, pepstatin, and aprotinin, supplemented with 100 mm KCl). This fraction was precipitated with 36% saturated ammonium sulphate, resuspended in LFB1/100, and applied to a 1-ml Mono-S column (Amersham–Pharmacia Biotech) equilibrated in LFB1/100. Bound protein was eluted with a step to LFB1/500 and was then applied to a 100 μl Mono-Q column (Amersham–Pharmacia Biotech) equilibrated in LFB1/100. Bound proteins were eluted with a linear gradient to LFB1/500 over 10 column volumes, and 25-μl fractions were collected. Estimation of the protein content in the final XCdc7-containing fractions from the UV signal suggested that total protein content had been reduced by ∼705-fold, whereas the activity assay gave a yield of ∼15%.
Recombinant proteins
GST–p21Cip/Waf1 (Strausfeld et al. 1994), His–cyclin A (Brown et al. 1995), and GST–geminin (McGarry and Kirschner 1998) were prepared as described.
Acknowledgments
This work was supported by the Cancer Research Campaign [CRC] (program grant SP2385). P.J. is the recipient of a postdoctoral fellowship from Ministerio de Educacion y Cultura of Spain. We thank Jason Swedlow and Joost Zomerdijk for helpful comments on the manuscript and all the members of the lab, especially Peter Gillespie and Ben Hogdson, for useful discussions. We also thank Haruhiko Takisawa for providing the anti-XCdc45 antibody and Stephane Chevalier for making the anti-XCdc7 antibody.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
Zou et al. (Mol. Cell Biol. 20: 3086–3096) have recently reported that in S. cerevisiae, Cdc7/Dbf4 is required for the efficient loading of Cdc45 to origins. This is consistent with our data in Xenopus and reinforces the idea that Cdc45 loading is a key event controlled by Cdc7/Dbf4 and CDKs during origin activation.
Footnotes
E-MAIL j.j.blow@dundee.ac.uk; FAX 01382 345783.
References
- Aparicio OM, Stout AM, Bell SP. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci. 1999;96:9130–9135. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blow JJ. Preventing re-replication of DNA in a single cell cycle: Evidence for a replication licensing factor. J Cell Biol. 1993;122:993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes & Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- ————— Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc Natl Acad Sci. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Garman EF, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson LN. The crystal structure of cyclin A. Structure. 1995;3:1235–1247. doi: 10.1016/s0969-2126(01)00259-3. [DOI] [PubMed] [Google Scholar]
- Cheng L, Collyer T, Hardy CF. Cell cycle regulation of DNA replication initiator factor Dbf4p. Mol Cell Biol. 1999;19:4270–4278. doi: 10.1128/mcb.19.6.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JP, Mahbubani HM, Khoo CY, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Chong JP, Thömmes P, Rowles A, Mahbubani HM, Blow JJ. Characterization of the Xenopus replication licensing system. Methods Enzymol. 1997;283:549–564. doi: 10.1016/s0076-6879(97)83043-1. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes & Dev. 1996;10:2819–2830. doi: 10.1101/gad.10.22.2819. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Blow JJ. The regulation of replication origin activation. Curr Opin Genet Dev. 1999;9:62–68. doi: 10.1016/s0959-437x(99)80009-4. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes & Dev. 1998a;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998b;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JF. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Tydeman P, Kirschner M. A cytoplasmic clock with the same period as the division cycle in Xenopus eggs. Proc Natl Acad Sci. 1980;77:462–466. doi: 10.1073/pnas.77.1.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess GF, Drong RF, Weiland KL, Slightom JL, Sclafani RA, Hollingsworth RE. A human homolog of the yeast CDC7 gene is overexpressed in some tumors and transformed cell lines. Gene. 1998;211:133–140. doi: 10.1016/s0378-1119(98)00094-8. [DOI] [PubMed] [Google Scholar]
- Hollingsworth RE, Jr, Ostroff RM, Klein MB, Niswander LA, Sclafani RA. Molecular genetic studies of the Cdc7 protein kinase and induced mutagenesis in yeast. Genetics. 1992;132:53–62. doi: 10.1093/genetics/132.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood B, Dalton S. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc Natl Acad Sci. 1996;93:12309–12314. doi: 10.1073/pnas.93.22.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XH, Newport J. Identification of a preinitiation step in DNA replication that is independent of origin recognition complex and cdc6, but dependent on cdk2. J Cell Biol. 1998;140:271–281. doi: 10.1083/jcb.140.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Pahl PM, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Brown GW, Muzi Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes & Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, Scherson T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell. 1987;48:399–407. doi: 10.1016/0092-8674(87)90191-7. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of Xenopus DNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7–Dbf4 during the initiation of DNA synthesis. Genes & Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbubani HM, Chong JP, Chevalier S, Thömmes P, Blow JJ. Cell cycle regulation of the replication licensing system: Involvement of a Cdk-dependent inhibitor. J Cell Biol. 1997;136:125–135. doi: 10.1083/jcb.136.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiro G, Owens JC, Shellman Y, Sclafani RA, Li JJ. Cell cycle control of Cdc7p kinase activity through regulation of Dbf4p stability. Mol Cell Biol. 1999;19:4888–4896. doi: 10.1128/mcb.19.7.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JC, Detweiler CS, Li JJ. CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc Natl Acad Sci. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasero P, Duncker BP, Schwob E, Gasser SM. A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes & Dev. 1999;13:2159–2176. doi: 10.1101/gad.13.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by Cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhorova TA, Blow JJ. Sequential MCM/P1 subcomplex assembly is required to form a hetero-hexamer with replication licensing activity. J Biol Chem. 2000;275:2491–2504. doi: 10.1074/jbc.275.4.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BT, Ying CY, Gautier J, Maller JL. DNA replication in vertebrates requires a homolog of the Cdc7 protein kinase. Proc Natl Acad Sci. 1999;96:2800–2804. doi: 10.1073/pnas.96.6.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Rowles A, Tada S, Blow JJ. Changes in association of the Xenopus origin recognition complex with chromatin on licensing of replication origins. J Cell Sci. 1999;112:2011–2018. doi: 10.1242/jcs.112.12.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Sato N, Arai K, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: In vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Tada S, Chong JPJ, Mahbubani HM, Blow JJ. The RLF-B component of the replication licensing system is distinct from Cdc6 and functions after Cdc6 binds to chromatin. Curr Biol. 1999;9:211–214. doi: 10.1016/s0960-9822(99)80092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H. A fission yeast gene, him1(+)/dfp1(+), encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol Cell Biol. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thömmes P, Kubota Y, Takisawa H, Blow JJ. The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. EMBO J. 1997;16:3312–3319. doi: 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich M, Stillman B. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HJ, Loo S, Campbell JL. Regulation of Saccharomyces cerevisiae CDC7 function during the cell cycle. Mol Biol Cell. 1993;4:195–208. doi: 10.1091/mbc.4.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
- Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]