Abstract
Malignant transformation usually inhibits terminal cell differentiation but the precise mechanisms involved are not understood. PU.1 is a hematopoietic-specific Ets family transcription factor that is required for development of some lymphoid and myeloid lineages. PU.1 can also act as an oncoprotein as activation of its expression in erythroid precursors by proviral insertion or transgenesis causes erythroleukemias in mice. Restoration of terminal differentiation in the mouse erythroleukemia (MEL) cells requires a decline in the level of PU.1, indicating that PU.1 can block erythroid differentiation. Here we investigate the mechanism by which PU.1 interferes with erythroid differentiation. We find that PU.1 interacts directly with GATA-1, a zinc finger transcription factor required for erythroid differentiation. Interaction between PU.1 and GATA-1 requires intact DNA-binding domains in both proteins. PU.1 represses GATA-1-mediated transcriptional activation. Both the DNA binding and transactivation domains of PU.1 are required for repression and both domains are also needed to block terminal differentiation in MEL cells. We also show that ectopic expression of PU.1 in Xenopus embryos is sufficient to block erythropoiesis during normal development. Furthermore, introduction of exogenous GATA-1 in both MEL cells and Xenopus embryos and explants relieves the block to erythroid differentiation imposed by PU.1. Our results indicate that the stoichiometry of directly interacting but opposing transcription factors may be a crucial determinant governing processes of normal differentiation and malignant transformation.
Keywords: PU.1, GATA-1, erythropoiesis, murine erythroleukemia, differentiation
The hallmark features of many cancers include both a loss of normal proliferation controls and a block to differentiation (Hunter 1997). The study of oncogenic viruses and the genetic lesions present in animal and human cancers has led to the identification of oncoproteins and tumor suppressor proteins and an increasingly deep understanding of how these proteins regulate cell proliferation. On the other hand, we know less about how malignant transformation affects the genes and gene products that regulate cell differentiation. Very frequently, leukemogenesis in humans and mice is associated with alterations in the structure or regulation of transcription factors that participate in normal blood cell development (Look 1997; Tenen et al. 1997). Such events often exhibit a very high degree of specificity for a particular hematopoietic lineage and stage of differentiation, but the molecular mechanisms leading to specific differentiation blockades are not understood.
Friend virus-induced erythroleukemias of mice have served as an important model for studying the multistep process of leukemia development (Ben-David and Bernstein 1991). Likewise the in vitro terminal differentiation that can be induced in the permanent murine erythroleukemia (MEL) cell lines established from such mice (Marks et al. 1987) has stimulated interest in approaches to cancer therapy based on forced differentiation of tumor cells. Friend virus consists of a complex of two retroviruses: the replication-defective spleen-focus-forming virus (SFFV) and a replication-competent, helper Friend murine leukemia virus (F-MuLV). There are at least three events that contribute to generation of malignant erythroid cells in Friend virus-infected mice (for review, see Ben-David and Bernstein 1991). The early stage of the disease is characterized by polyclonal proliferation of nonleukemic erythroid progenitors caused by interaction of the SFFV-encoded 55-kd fusion glycoprotein (gp 55) with the erythropoietin receptor, triggering constitutive activation of mitogenic signaling. The subsequent emergence of clonal or oligoclonal malignant cells is associated with two additional genetic events: inactivation of p53 and SFFV proviral insertions at the Spi-1 locus that encodes the PU.1 transcription factor (Moreau-Gachelin et al. 1988; Paul et al. 1989; Goebl 1990). Integration of SFFV does not disrupt PU.1 gene expression; rather it is thought to activate or deregulate PU.1 synthesis in erythroid precursors (Moreau-Gachelin et al. 1989). Proof that PU.1 plays a causative role in erythroleukemogenesis came from the observation that transgenic mice expressing PU.1 in erythroid cells develop erythroleukemias at a high rate (Moreau-Gachelin et al. 1996). Its role in transformation of erythroid cells is further supported by finding that a PU.1-encoding retrovirus can immortalize erythroblasts efficiently from infected long-term bone marrow cultures (Schuetze et al. 1993). PU.1 is a member of the Ets family of transcription factors. Interestingly, two other ets family genes, fli-1 and v-ets have also been implicated in erythroleukemic transformation (Ben-David and Bernstein 1991).
PU.1 is expressed specifically in hematopoietic tissues, with high levels of expression in cells of the monocytic, granulocytic and B lymphoid lineages (Galson et al. 1993; Hromas et al. 1993). A large number of presumptive PU.1 target genes have been identified (Fisher and Scott 1998). Many of these genes are expressed in myeloid and B cells, including some that are thought to have important roles in differentiation and survival of cells within these lineages. Consistent with this view, inactivation of the PU.1 gene in mice causes defects in the development of multiple hematopoietic lineages, including B and T lymphocytes, monocytes, and granulocytes (Scott et al. 1994; McKercher et al. 1996). PU.1 probably plays important roles at several stages of the differentiation process, and there is evidence that it is active at an early stage, mediating commitment of multipotent progenitors to the myeloid lineage (Nerlov and Graf 1998). However, PU.1 does not appear to be needed for production of megakaryocytes or erythrocytes.
The in vitro differentiation that can be induced in permanent MEL cell lines established from Friend virus-infected mice provides an opportunity to analyze events occurring when the leukemic phenotype is reversed and the cells reenter their erythroid differentiation program (Marks et al. 1987). MEL cells are transformed erythroid precursors that are blocked at about the proerythroblast stage of differentiation. Treatment of the cells with a variety of agents causes them to undergo differentiation over several days and culminates in terminal cell division and accumulation of hemoglobin and other erythrocyte-specific proteins. One of the early events occurring during this process is a marked decline in levels of PU.1 (Schuetze et al. 1992; Galson et al. 1993; Rao et al. 1997), suggesting that high levels of PU.1 may block erythroid differentiation. By transfecting MEL cells with expression vectors encoding PU.1 we (Rao et al. 1997) and other investigators (Yamada et al. 1997) have shown that PU.1 can indeed block erythroid differentiation of MEL cells. The effect of PU.1 on erythroid differentiation was also seen in other cell culture models (Quang et al. 1995; Delgado et al. 1998). An intriguing aspect of the block to differentiation in MEL cells is the fact that the cells contain substantial amounts of several transcription factors that are involved in erythroid differentiation, including GATA-1, a zinc finger protein that is essential for erythropoiesis and expression of most erythroid-specific genes (Shivdasasani and Orkin 1996). We show here that PU.1 binds directly to GATA-1 and specifically inhibits its ability to activate transcription of reporter genes. Moreover we show that the stoichiometry of PU.1 and GATA-1 affects erythroid differentiation capacity in MEL cells and Xenopus embryos and explants. We propose that PU.1 blocks differentiation in erythroleukemia cells and contributes to leukemogenesis by binding to and inhibiting the ability of GATA-1 to promote terminal erythroid differentiation. We also discuss the possibility that the opposing actions of PU.1 and GATA-1 or other GATA factors contributes to lineage determination during normal hematopoiesis.
Results
PU.1 and GATA-1 interact in vitro and in vivo
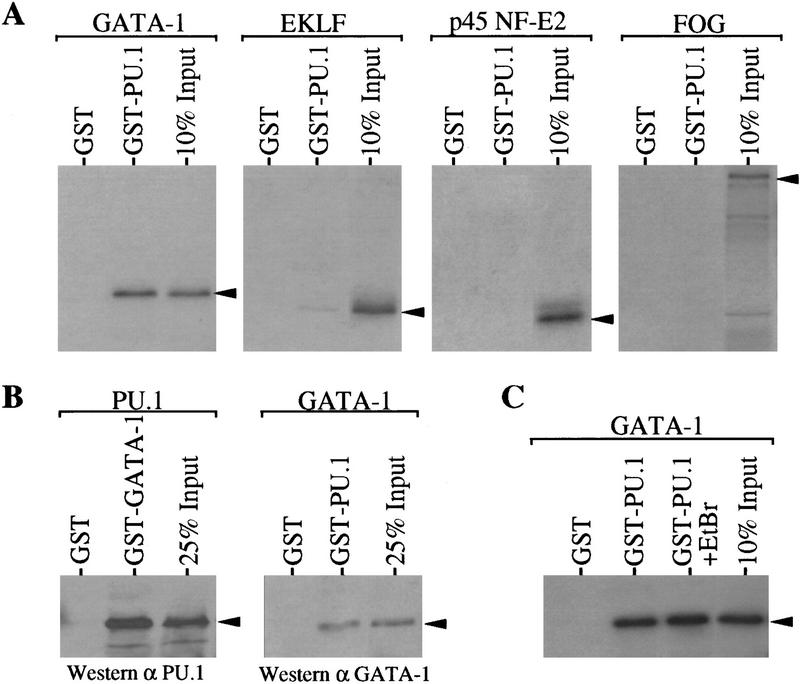
As mentioned above, several transcription factors known to be involved in erythroid differentiation are present in MEL cells. These factors include GATA-1 (Tsai et al. 1989; Leonard et al. 1993), erythroid Krüppel-like factor (EKLF; Miller and Bieker 1993), p45 NF-E2 (Lu et al. 1994), and Friend of GATA-1 (FOG; Tsang et al. 1997). Restoration of terminal differentiation in MEL cells requires a large decrease in the amount of PU.1. Therefore, we wondered whether PU.1 might interact directly with one or more of these transcription factors and inhibit their activities in promoting differentiation. We examined potential in vitro interactions between PU.1 and these factors by using GST–PU.1 immobilized on glutathione–Sepharose beads and in vitro synthesized 35S-labeled proteins. About 15% of input GATA–1 bound to GST–PU.1, whereas GATA-1 did not bind to immobilized GST (Fig. 1A). On the other hand, very little EKLF was bound to GST–PU.1 and binding of p45 NF-E2 or FOG to GST–PU.1 was not detected (Fig. 1A). A strong interaction was also evident when GST–GATA-1 and 35S-labeled PU.1 were used, whereas PU.1 did not bind to GST nor to another GST fusion protein, GST–GUK (see Fig. 3A). The in vitro interaction of PU.1 and GATA-1 occurs even in the presence of 1% NP-40 (not shown). Because the 35S-labeled proteins used in binding reactions were synthesized in rabbit reticulocyte lysates we sought to determine whether the PU.1–GATA-1 interaction is direct or dependent on a bridging molecule that might be present in the lysate. Interaction is direct because PU.1–GATA-1 complexes were formed using both molecules purified from bacteria (Fig.1B). Furthermore, formation of PU.1–GATA-1 complexes was unaffected by ethidium bromide, indicating that the interaction is not mediated by contaminating DNA (Fig. 1C).
Figure 1.
PU.1 and GATA-1 interact in vitro. (A) 35S-Labeled proteins, indicated above each panel, were prepared and tested for binding to GST or GST–PU.1 by a ‘pull-down’ assay using proteins immobilized on glutathione–Sepharose beads as described in Materials and Methods. Bound proteins were released and analyzed by SDS–PAGE and autoradiography. An autoradiogram of 10% of each 35S-labeled protein added to the binding reactions is shown in each panel (10% input). Arrowheads indicate the position of the 35S-labeled proteins on the gel. (B) PU.1 or GATA-1, as indicated above each panel, were prepared from the respective GST fusion proteins by thrombin cleavage as described in Materials and methods. The purified proteins were tested for binding to GST, GST–GATA-1 or GST–PU.1 by a pull-down assay. Bound proteins were analyzed by SDS–PAGE and immunoblotting with the indicated antibodies. An immunoblot of 25% of the purified PU.1 or GATA-1 added to the binding reactions is shown in each panel (25% Input). (C) As in A except that where indicated 100 μg/ml of ethidium bromide (EtBr) was added to the reaction.
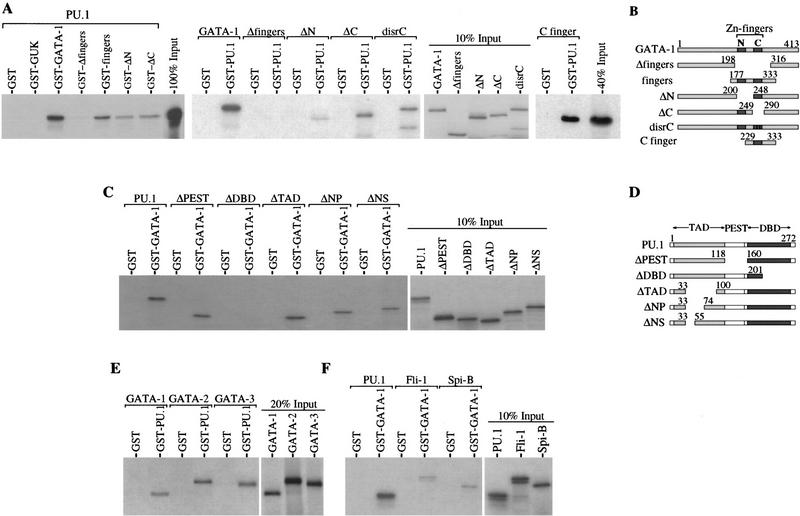
Figure 3.
The PU.1–GATA-1 interaction is mediated by the zinc finger region of GATA-1 and the Ets region of PU.1. Binding of 35S-labeled PU.1 to wild-type and mutant GST–GATA-1 fusion proteins (A, left), or 35S-labeled wild-type and mutant GATA-1 proteins to GST–PU.1 (A, middle and far right), or 35S-labeled wild-type and mutant PU.1 proteins to GST–GATA-1 (C) was tested as described in Fig.1. GST–GUK is a GST fusion protein of the human LIN2A guanylate kinase. (B,D) Schematic diagram of the mutant proteins used for the binding studies in A and C, respectively. The positions of amino acid residues at the termini of regions present in the mutant proteins are shown above the diagrams. (TAD) transactivation domain; (PEST) region rich in PEST residues; (DBD) DNA-binding domain. Binding of 35S-labeled GATA-1, GATA-2, and GATA-3 to GST or GST–PU.1 (E) or 35S-labeled PU.1, Fli-1, and Spi-B to GST or GST–GATA-1 (F) was tested as described in Fig. 1.
To test whether PU.1 and GATA-1 complexes form in vivo, we transfected expression vectors encoding PU.1 and GATA-1 into 293 cells and analyzed nuclear extracts for PU.1–GATA-1 complexes by immunoprecipitating with an anti-GATA-1 antibody and immunoblotting for PU.1. PU.1–GATA-1 complexes were detected in the transfected cell extracts and the presence of PU.1 in the anti-GATA-1 immunoprecipitate was dependent on cotransfection of both expression vectors (Fig. 2A). MEL cell extracts were also analyzed by coimmunoprecipitation experiments, using both anti-GATA-1 and anti-PU.1 antibodies to determine whether endogenous PU.1 and GATA-1 are associated in these cells. PU.1–GATA-1 complexes were readily detected in MEL cell nuclear extracts using either antibody, whereas the association was not seen when control antibodies were used (Fig. 2B). We estimate that a large fraction of GATA-1 is bound to PU.1 in the tumor cells, suggesting that the interaction may be important in the PU.1-mediated block to differentiation of the cells.
Figure 2.
PU.1 and GATA-1 interact in vivo. (A) Nuclear extracts from 293 cells transfected with expression vectors encoding either PU.1 or both PU.1 and GATA-1 were prepared and immunopreciptated with an anti-GATA-1 antibody as described in Materials and Methods. Immunoprecipitates were analyzed by SDS–PAGE and immunoblotting with the indicated antibodies. An immunoblot of 15% of the amount of extract that was immunoprecipitated is shown (Input). Arrowheads indicate the positions of the immunodetected proteins. The asterisked (*) arrowhead indicates the position of Ig chains, which are used in immunoprecipitation. (B) Nuclear extracts of MEL cells were prepared and immunoprecipitated with either anti-GATA-1 (α-GATA-1) and anti-p16 (α-control) antibodies or anti-PU.1 (α-PU.1) and anti-cyclin E (α-control) antibodies as described in Materials and Methods. Input lanes are shown as in A, except that 3% of the extract was analyzed for the immunoprecipitation of PU.1 and Western of PU.1 lane. Analysis of immunoprecipitates and other details were as in A. (C, D) Analysis of the effect of mutations in GATA-1 (C) and PU.1 (D) on the GATA-1–PU.1 interaction in vivo. Whole cell extracts from 293 cells transfected with expression vectors encoding the indicated GATA-1 and PU.1 proteins (see Fig. 3B,D) were immunoprecipitated and analyzed as in A. Input lanes are shown as in A.
PU.1 targets the zinc finger region of GATA-1
To define the regions within PU.1 and GATA-1 that are required for interaction of the two proteins, we tested a set of deletion mutants of each protein for their abilities to interact in vitro. Structure-function studies of both proteins have delineated specific regions or domains that contribute to their functions as transcription factors as well as their interactions with other proteins.
GATA-1 binding to DNA is mediated by a domain comprised of two zinc fingers. The carboxy-terminal finger is absolutely required for DNA binding, whereas the amino-terminal finger is thought to stabilize binding (Martin and Orkin 1990). Both fingers have been found to interact with other proteins (Tsang et al. 1997 and references therein). Transfection assays have defined several regions of GATA-1 contributing to transcriptional transactivation, the most potent of which lies near the amino-terminal end of the protein (Martin and Orkin 1990). We found that the finger region was both necessary and sufficient for interaction with PU.1 (Fig. 3A,B). Deletion of the region encoding both fingers of GATA-1 abolished completely the interaction, and a GST fusion protein comprising only the finger region of GATA-1 bound PU.1 quite well. Other regions in GATA-1 may also contribute to the interaction as the extent of binding of PU.1 to the GST fingers fusion protein was less than to full-length GST–GATA-1. Within the GATA-1 finger region both the amino-terminal finger and the carboxy-terminal finger appear to contribute to binding to PU.1 as GATA-1 proteins containing a deletion of either finger or a substitution of two cysteines that disrupt the carboxyl finger (disrC) bind less well to PU.1 than full-length GATA-1 (Fig. 3A). The contribution of both GATA-1 fingers to binding to PU.1 was confirmed by coimmunoprecipitaion experiments in transfected 293 cells. Consistent with the in vitro results, deletion of both fingers abolished the interaction, whereas GATA-1 proteins that are deleted for either finger bind PU.1 in vivo but with an apparent reduced affinity (Fig. 2C). Furthermore, a GATA-1 fragment comprising only the carboxy-terminal finger is able to bind to GST–PU.1 (Fig. 3A).
DNA binding by PU.1 is directed by the carboxy-terminal ets homology region (Klemsz et al. 1990). In addition to an intact DNA-binding domain, transactivation of reporter constructs containing PU.1-binding sites requires an amino-terminal region consisting of acidic and glutamine-rich subdomains (Klemsz and Maki 1996 and references therein). There is also a central region rich in proline, glutamic acid, serine, and threonine (PEST) residues (Moreau-Gachelin 1994). Certain interactions of PU.1 with other proteins are mediated by the transactivation domain (Hagemeir et al. 1993), whereas other interactions occur with the Ets domain (Nagulapalli et al. 1995; Petrovick et al. 1998). Deletion of the PEST region or deletions encompassing portions of the amino-terminal transactivation domain had no significant effect on PU.1–GATA-1 interaction in vitro (Fig. 3C,D). These deletions also did not affect interaction of PU.1 and GATA-1 in vivo (Fig. 2D). In contrast, deletion of the carboxy-terminal region of the DNA-binding domain of PU.1 abolished completely its interaction with GATA-1 (Fig. 3C). Thus, the two proteins appear to interact through their respective DNA-binding regions.
GATA-1 is highly related in its DNA-binding zinc finger region to GATA-2 and GATA-3, two other GATA factors involved in hematopoiesis (Shivdasani and Orkin 1996). The three proteins share little similarity outside of the finger region. We found that GATA-2 and GATA-3 bound well to GST–PU.1, with in vitro affinities approximately the same as that of GATA-1 (Fig. 3E).
PU.1 is a member of the Ets family of transcription factors. The most closely related Ets protein is Spi-B, a B cell-specific factor. Spi-B is most similar to PU.1 in the Ets domain; the two proteins are much less similar in other regions (Ray et al. 1992). Fli-1 is related more distantly to PU.1, but like PU.1 it has been implicated in mouse erythroleukemia (Ben-David et al. 1991). Both Ets proteins bound to GST–GATA-1, although the interaction was much weaker than with PU.1 (Fig. 3F). The weak interaction of Spi-B with GATA-1 points to a high degree of specificity in the interaction between PU.1 and GATA-1. The observed interactions of GATA and Ets family members with PU.1 and GATA-1, respectively, are consistent with the mapping of the interaction sites on PU.1 and GATA-1 to their respective DNA-binding regions. The significance of such interactions remains to be determined (see Discussion).
PU.1 inhibits GATA-1-dependent transcriptional activity
On the basis of the well-established function of GATA-1 as a transcription factor that promotes expression of many erythroid-expressed genes (Shivdasani and Orkin 1996), and our earlier finding that PU.1 can inhibit MEL cell erythroid differentiation (Rao et al. 1997), the observation that PU.1 and GATA-1 interact could suggest that PU.1 is able to inhibit GATA-1-mediated transcriptional activation. To test this possibility we performed cotransfection assays in NIH-3T3 cells and HeLa cells with three different reporter constructs containing either synthetic (M1α, Fig. 4A,B and αD3; Fig. 4C) or natural (p45 NF-E2; Fig. 4D) GATA-1-dependent promoters. The p45 NF-E2 promoter requires the synergistic action of GATA-1 and FOG as a cofactor (Tsang et al. 1997). Coexpression of PU.1 along with GATA-1 led to a marked reduction in GATA-1-mediated transactivation of all three reporters. Inhibition by PU.1 was dose dependent. With the natural p45 NF-E2 promoter, the largest amount of PU.1 expression plasmid used led to a reduction in the stimulation by GATA-1 plus FOG to the basal activitiy of the promoter (Fig. 4D). The effect of PU.1 is specific for GATA-1-mediated transactivation as coexpression of PU.1 did not affect another reporter that is activated by a GAL4–Sp1 fusion protein (Fig. 4E); PU.1 also did not affect the basal activity of each reporter. Furthermore, inhibition was not seen in cotransfections with the PU.1 cDNA in the antisense configuration (PU.1-AS; Fig. 4A). Immunoblotting of extracts from transfected cells showed that coexpression of PU.1 did not reduce the level of GATA-1 protein produced in the transfected cells (Fig. 4A,C insets). Coexpression of the carboxy-terminally deleted PU.1 protein that does not interact with GATA-1 failed to inhibit GATA-1-mediated transactivation. Deletion of the PU.1 PEST region, which does not affect interaction with GATA-1, also does not affect the ability of PU.1 to repress GATA-1-mediated transactivation. Interestingly, deletion of a portion of the PU.1 amino-terminal transactivation domain resulted in a loss in the ability to repress GATA-1-mediated transactivation. In fact, we observed a consistent, approximately twofold stimulation of GATA-1-mediated transactivation when this PU.1 mutant was coexpressed with GATA-1 (Fig. 4B,C). The mutant PU.1 proteins were produced at levels comparable to or greater than that of full-length PU.1 as determined by anti-HA immunoblotting of HA-tagged proteins (Fig. 4B inset). Furthermore, like wild-type PU.1, the three types of mutant PU.1 molecules are able to localize in the nucleus, as determined by immunoblotting of nuclear and cytoplasmic extracts of transfected cells (not shown). Thus, repression of GATA-1 by PU.1 requires both the carboxy-terminal DNA-binding region and the amino-terminal transactivation domain of PU.1.
Figure 4.
PU.1 inhibits GATA-1-dependent transcriptional activity with synthetic and natural promoters. M1α–GH reporter (15 ng) with and without either 150 ng (A) or 60 ng (B) of pXM–GATA-1 were cotransfected with pEBB or the indicated amounts (nanograms) of pEBB expression constructs encoding PU.1, reverse transcripts of PU.1 (AS–PU.1), or the indicated PU.1 mutants (see Fig. 3D) into NIH 3T3-cells as described in Materials and Methods. (C) αD3-LUC reporter (15 ng) with and without 30 ng of pXM–GATA-1 were cotransfected with pEBB or the indicated amounts of plasmids as in B into HeLa cells. (D) p45 NF-E2-GH reporter (10 ng) with and without 100 ng of pXM–GATA-1 and 100 ng pMT2–FOG were cotransfected with pEBB or the indicated amounts pEBB–PU.1 into NIH-3T3 cells as in A. (E) UAS–LUC reporter (100 ng) were cotransfected into NIH-3T3 with and without 100 ng of SP6–GAL4–Sp1 and 180 ng of pEBB–PU.1. The levels of growth hormone (A,B,D) or luciferase activity (C,E) were determined 48 hr after transfection as described in Materials and Methods. (Insets in A,B,C) The immunoblots with anti-GATA-1 antibody (A,C) and anti-HA antibody (B) of cell extracts corresponding to the indicated transfected samples. Arrowheads indicate the positions of GATA-1 and HA-tagged full-length and mutant PU.1 proteins; the asterisked (*) arrowhead indicates a nonspecific band detected with anti-HA antibody in 293 extracts. Numbers represent the average of at least two independent transfections. Error bars represent the s.e.m.. Each type of experiment was performed between three and seven times with two or three different DNA preparations and produced similar results.
Both the DNA-binding and transactivation domains of PU.1 are required for blocking erythroid differentiation
Treatment of MEL cells with several chemical agents that cause the cells to reinitiate terminal erythroid differentiation leads to a very rapid decline in PU.1 levels. Previously, we showed that transfection of the cells with an expression construct that constitutively produces PU.1, even in the presence of differentiation inducers, causes a block to differentiation (Rao et al. 1997). Having determined that the interaction between PU.1 and GATA-1 requires an intact PU.1 DNA-binding domain, and that repression of GATA-1 requires both that domain and the transactivation domain, we sought to determine whether these regions of PU.1 are also needed for its ability to block MEL cell differentiation. We prepared stable MEL cell transfectants expressing either an epitope-tagged (HA) PU.1 protein or tagged, mutant PU.1 proteins containing deletions in the DNA-binding domain or the transactivation domain. Stable MEL cell transfectants expressing wild-type and mutant proteins were identified by immunoblotting with an anti-HA antibody (Fig. 5). Transfectants expressing amounts of the mutant proteins similar to or greater than that of wild-type PU.1 were then tested for their ability to undergo differentiation and form benzidine-positive, hemoglobinized cells in response to hexamethylene bisacetamide (HMBA), an inducer of differentiation. Transfectants that did not express any detectable wild-type or mutant-tagged PU.1 were used as controls; these control transfectants differentiated well. Transfectants expressing wild-type PU.1 were blocked completely for differentiation, whereas clones expressing the mutant proteins were fully capable of differentiating (Table 1). On the other hand, deletion of the PEST region did not affect the ability of PU.1 to block MEL cell differentiation (F. Radparvar and A.I. Skoultchi, unpubl.). These results indicate that both regions of PU.1 required for repressing GATA-1-mediated transcription are also necessary for the activity of PU.1 in blocking differentiation.
Figure 5.
Detection of wild-type and mutant epitope-tagged PU.1 proteins in MEL cell transfectants. MEL cells were transfected with expression constructs encoding both resistance to puromycin and an influenza hemagglutinin (HA)-epitope tagged wild-type or mutant PU.1 proteins as described in Materials and Methods. Puromycin resistant clones were isolated and screened for expression of PU.1 proteins by immunoblotting of cell extracts with anti-HA antibody. The levels of PU.1 proteins (arrowhead) in two representative clones of each type of PU.1 transfectant are shown. PU.1-ΔDBD–HA is deleted for residues 201–272 and PU.1–ΔTAD–HA is deleted for residues 33–100 (see Fig. 3D).
Table 1.
Response of MEL cell transfectants to HMBA treatment
|
Clonea
|
Percent benzidinepositive cellsb
|
|
|---|---|---|
|
day 3
|
day 5
|
|
| PU.1–HA expressors | ||
| 35 | 0 | 0 |
| 39 | 0 | 0 |
| 51 | 0 | 2 |
| 64 | 0 | 1 |
| PU.1–HA nonexpressors | ||
| 12 | 74 | 100 |
| 15 | 74 | 93 |
| PU.1–ΔDBD–HA expressors | ||
| 29 | 90 | 91 |
| 44 | 76 | 90 |
| 49 | 95 | N.D. |
| 51 | 74 | 90 |
| PU.1–ΔDBD–HA nonexpressors | ||
| 25 | 90 | 92 |
| 43 | 62 | 90 |
| PU.1–ΔTAD–HA expressors | ||
| 37 | 91 | N.D. |
| 40 | 93 | 100 |
| 41 | 92 | 100 |
| 50 | 81 | N.D. |
| PU.1–ΔTAD–HA nonexpressors | ||
| 54 | 100 | N.D. |
| 60 | 95 | N.D. |
| MEL | 84 | 93 |
(N.D.) Not determined.
Expression levels of wild-type and mutant PU.1 proteins in representative transfectants are shown in Fig. 5. Clones indicated as nonexpressors did not produce any detectable epitope-tagged protein on immunoblots like that shown in Fig. 5. Similar results were obtained with 6–10 additional clones in each transfection.
Cells were treated with 5 mm HMBA for the indicated times, and the percent hemoglobinized, benzidine-positive cells was determined as described in Materials and Methods.
GATA-1 relieves a block to erythroid differentiation imposed by PU.1
As mentioned above, during chemically induced reinitiation of MEL cell differentiation PU.1 levels decline rapidly. We described previously (Rao et al. 1997) and in the preceding section, MEL cell transfectants that express PU.1 constitutively even in the presence of differentiation inducers. These cells are blocked from differentiating in response to inducers, attributable to the presence of exogenous PU.1. We wondered whether the differentiation block specifically imposed by PU.1 in these transfectants could be relieved by providing the cells with additional GATA-1. We transfected one such PU.1-transfectant (PU.1 clone 605; Rao et al. 1997) a second time with a construct expressing a conditionally active form of GATA-1, achieved by fusing GATA-1 coding sequences to the ligand-binding domain of the human estrogen receptor (ER). The GATA-1/ER fusion protein is activated for GATA-1 dependent transcription in the presence of estrogen (Tsang et al. 1997). Supertransfectants of the clone 605 expressing the GATA-1/ER fusion protein were identified by immunoblotting. These transfectants were studied for their ability to differentiate in response to various treatments. Like the parental clone 605 cells, the supertransfectants exhibited a block to differentiation in response to HMBA, producing <10% benzidine-positive, hemoglobinized cells when cultured for 5 days in HMBA (Fig. 6A–C). However, when these cells were treated with HMBA in the presence of estrogen they produced large numbers of differentiated cells, whereas the combination of HMBA and estrogen did not increase the percentage of differentiated cells in the parental clone 605 line above that observed with HMBA alone. The capacity of the 605 line to differentiate in response to HMBA was restored by GATA-1/ER in the presence of estrogen to levels comparable to individual clones of MEL cells. Moreover, we found that the presence of GATA-1/ER in these cells rendered them able to differentiate with estrogen treatment alone, without the need for HMBA (Fig. 6B,C). Thus, relief of the block to differentiation in the erythroleukemia cells appears to require only sufficient levels of active GATA-1. We hypothesize that the combination of HMBA and estrogen may induce more of the cells to differentiate than estrogen alone because HMBA may cause a reduction in expression of the endogenous PU.1 gene, effectively increasing the amount of active GATA-1. Whereas treatment of the clone 605 line with HMBA and estrogen led to only minimal accumulation of globin mRNA, differentiation induced in the GATA-1/ER supertransfected lines by HMBA plus estrogen led to a marked increase in globin mRNA levels (Fig. 6D), comparable to that induced in MEL cells by HMBA and other agents.
Figure 6.
Restoration of hemoglobinization and terminal growth arrest in PU.1-transfected MEL cells by a GATA-1/ER fusion protein. MEL cells (broken line) and PU.1-transfected MEL cells, clone 605 (A) and two representative GATA-1/ER supertransfectants of clone 605, clones 12 (B) and 13 (C) (solid lines), were grown without (♦) or with 5 mm HMBA (▴), 5 mm HMBA with 10−7m β-estradiol (●) or 10−7 m β-estradiol (█). At the indicated times, the percentage of hemoglobinized cells was determined by benzidine staining as described in Material and Methods. Similar results were obtained with four other GATA-1/ER supertransfectants. (D) Clone 605 and clone 12 were treated for 72 hr with 10−7 m β-estradiol, 5 mm HMBA, or both as indicated above the panel. β-Globin mRNA levels were determined by Northern blot hybridization of total cellular RNA with 32P-labeled β major globin DNA. The lower panel shows the ethidium bromide-stained gel before transfer. MEL cells (broken line) and cells of clones 605 (E) and clone 13 (F) (solid lines) were cultured as in A for the indicated times and plated in plasma clots with β-estradiol as described in Materials and Methods. After 7 days clots were removed, fixed, and stained with hematoxylin and the percentage of colonies with <50 cells was determined by microscopy. At least 200 colonies were counted for each determination.
An important aspect of the differentiation program induced in MEL cells by HMBA and other treatments is that, in addition to expression of many erythroid-specific markers, the cells undergo terminal cell division (Marks et al. 1987). This property can be studied by observing the proliferation of individual cells in semisolid media such as in plasma clots. Cells undergoing differentiation exhibit limited proliferative capacity and give rise to small colonies consisting of a maximum of 32–64 cells, whereas undifferentiated cells proliferate extensively and produce very large colonies consisting of hundreds or thousands of cells. Previously, we showed that PU.1 blocks HMBA-induced terminal cell division (Rao et al. 1997), allowing PU.1 transfectants like clone 605 to form large colonies in plasma clots after HMBA treatment (Fig. 6E). We tested the proliferation of GATA-1/ER transfectants treated with HMBA, estrogen, or both agents by plasma clot assays. As shown in Fig. 6F treatment with estrogen alone or estrogen and HMBA caused the cells to lose their proliferative capacity and form many small colonies, whereas when treated with HMBA alone they retained their ability to divide extensively and produced only very large colonies. On the other hand, treatment of the 605 parental line with estrogen or estrogen and HMBA did not lead to production of many small colonies showing that the proliferation capacity of the cells is not affected by estrogen in the absence of GATA-1/ER (Fig. 6E). We conclude that the differentiation program restored in the PU.1-blocked line by GATA-1/ER includes terminal cell division.
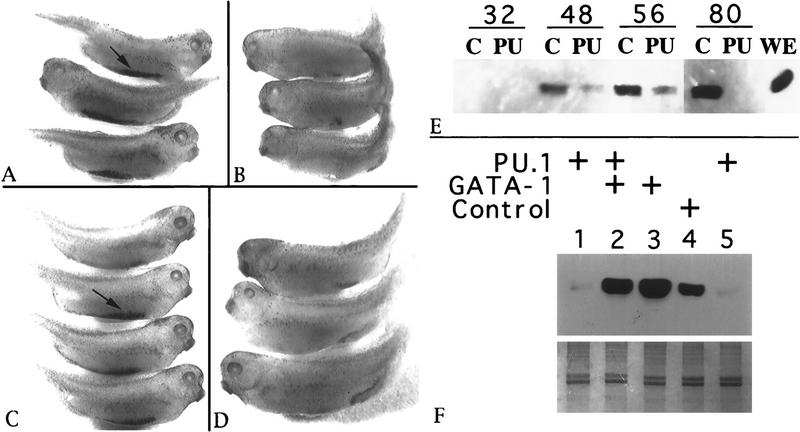
The foregoing results suggest that the relative levels of PU.1 and GATA-1 determine whether or not the erythroleukemia cells undergo terminal differentiation. To determine whether this is also true during normal erythropoiesis we studied the effects of PU.1 and GATA-1 on erythropoiesis in Xenopus embryos. In Xenopus, blood is derived from blastomeres that contribute to ventral mesoderm, and the first erythrocytes differentiate in the ‘blood island’ located along the ventral midline of 2-day-old embryos. The ventral blastomeres can be distinguished by the four-cell stage; the dorsal/ventral axis is established at fertilization after cortical rotation leading to the differential localization of maternal dorsal determinants (and pigment) that are separated physically by the second cleavage plane. Therefore, proteins can be expressed ectopically during blood development by injecting fertilized eggs or specific cleavage stage ventral blastomeres with mRNA or DNA expression plasmids. RNA encoding PU.1 was synthesized in vitro and injected into developing Xenopus embryos. Embryos developed normally after injecting both blastomeres of a two-cell embryo with 125 pg of reverse-strand (control) PU.1 RNA, as shown by typical benzidine staining of blood island regions in such embryos (arrow in Fig. 7A). In contrast, injection of 125 pg of PU.1 RNA led to a substantial block in primitive erythropoiesis, as shown by a lack of benzidine staining regions, except for occasional small patches of stained cells (Fig. 7B). These embryos also displayed defects unrelated to hematopoiesis in the developing dorsal axis, seen first as an uneven neural plate (not shown) and resulting in abnormally shaped tails. To test whether PU.1 expressed specifically in the blood island region was responsible for the defect in erythropoiesis, PU.1 RNA or control RNAs were injected into both ventral blastomeres of four-cell embryos. Injection of as much as 1 ng of reverse strand PU.1 RNA into ventral blastomeres had no effect on hemoglobin production (Fig. 7C). Embryos injected with 125 pg of PU.1 RNA displayed a lack of differentiated erythrocytes, but appeared otherwise normal (Fig. 7D).
Figure 7.
PU.1 inhibits and GATA-1 restores erythroid differentiation in Xenopus embryos and explants. (A–D) Shown are representative 2.5-day Xenopus embryos following benzidine staining to visualize primitive erythropoiesis in the ventral blood island region (indicated by arrows in A and C). The embryos were injected with 125 pg reverse-strand control RNA (A) or mRNA encoding PU.1 (B) in both blastomeres at the two-cell stage. Embryos shown in C (1 ng) and D (125 pg) had been injected similarly, except that the mRNA was targeted into the two ventral blastomeres at the four-cell stage. (E,F) Globin expression in VMZ explants was analyzed by Western blotting using a monoclonal antibody to embryonic α-globin. (E) Explants were cultured for the times (in hr) indicated above the lanes. Lysates were derived from embryos injected into both blastomeres at the two-cell stage with 25 pg of either a control (C) CMV expression plasmid, or the same plasmid containing the PU.1 cDNA (PU). The lysate in lane WE was from an uninjected 80-hr whole embryo. Each sample contains proteins from six VMZ explants, derived from two independent experiments. (F) For each sample lysates were prepared from 10 VMZ explants isolated from embryos previously injected at the four-cell stage with RNA into both ventral blastomeres. Embryos were injected with various combinations of PU.1, GATA-1, reverse-strand PU.1 (PU.1rs), or a frameshift mutant form of GATA-1 that encodes a nonfunctional protein (G1fs). To control for any nonspecific effects of RNA injection, a total of 350 pg mRNA was injected into each blastomere, supplemented with PU.1rs or G1fs as required. (Lane 1) 250 pg PU.1 + 100 pg G1fs; (lane 2) 250 pg PU.1 + 100 pg GATA-1; (lane 3): 250 pg GATA-1 + 100 pg G1fs; (lane 4): 250 pg PU.1rs + 100 pg G1fs; (lane 5) the same as lane 1 from an independent experiment. (Lower) Coomassie Blue staining of the gel demonstrates equal loading of total protein for each lane.
An advantage of the Xenopus system is that erythropoiesis also can be analyzed by culturing embryo explants isolated from the presumptive ventral margin zone (VMZ) that contains the blood progenitors. To test whether ectopic PU.1 expression can inhibit erythropoiesis in isolated explants, 25 pg of a plasmid encoding PU.1 or the empty vector were injected into both blastomeres of two-cell embryos. Injected plasmids are not transcribed until the midblastula transition. At the beginning of gastrulation, VMZ explants containing primitive erythroid progenitors were isolated and allowed to develop in culture. At various times, explants were harvested and lysates were analyzed for globin by immunoblotting (Fig. 7E). By 48 hr, when globin was detected in control explants, there was significantly less globin in the explants injected with the PU.1 expression plasmid. By 80 hr, the control explants expressed abundant globin, whereas those expressing ectopic PU.1 were negative for globin. PU.1 appears to inhibit transcription of the globin gene as determined by RT–PCR experiments for globin mRNA (not shown). Globin production also was blocked in isolated VMZ explants after injecting 250 pg of PU.1 RNA into the two ventral blastomeres at the four-cell stage (Fig. 7F, lanes 1,5). In this case, coinjection of 100 pg of RNA encoding Xenopus GATA-1 is sufficient to rescue erythropoiesis, assayed by globin production (Fig. 7F, lane 2). Rescue of blood island development in whole embryos coinjected with PU.1 and GATA-1 RNA into ventral blastomeres also was demonstrated by benzidine staining (data not shown). These results indicate that the relative levels of PU.1 and GATA-1 can determine the outcome of erythroid differentiation during normal development.
Discussion
Previous studies with MEL cells and other cell culture models showed that PU.1 can block terminal erythroid differentiation. To understand the mechanism by which PU.1 interferes with erythroid differentiation, we investigated potential interactions between PU.1 and several erythroid-specific transcription factors. Among the factors tested, GATA-1 showed very strong binding with PU.1. Our results show that PU.1 and GATA-1 interact directly both in vitro and in vivo and that the interaction leads to repression of GATA-1-mediated transcriptional activation. Furthermore, we show that PU.1 and GATA-1 oppose each other’s biological activities in both normal and leukemic erythroid cells. Our results suggest that PU.1 and GATA-1 can antagonize each other’s actions by binding to one another and altering some aspect of their functions as transcription factors.
Mechanism of PU.1-mediated repression of GATA-1
The results reported here show that PU.1 binds to the zinc finger region of GATA-1. This region of GATA-1 has been shown to bind DNA (Martin and Orkin 1990). It also participates in interactions with several other proteins. Most of these proteins, including FOG (Tsang et al. 1997), CREB-binding protein (CBP; Blobel et al. 1998), EKLF, and Sp1 stimulate GATA-1 transcriptional activation. However, two examples of repression of GATA factors have been described that involve interactions with the finger region. Inhibition of a Drosophila GATA homolog, Pannier, by U-shaped occurs through interaction with the amino finger (Haenlin et al. 1997). Inhibition of GATA-1 by the estrogen receptor appears to involve both the zinc finger region and another region of GATA-1 (Blobel et al. 1995). However, the mechanisms underlying stimulation or repression of GATA-1 activity by binding of these various proteins to the zinc finger region are not known.
Several different mechanisms can be envisaged for how binding of PU.1 to the finger region inhibits GATA-1 transcriptional activity. Any proposed mechanism must account for our finding that the transactivation domain of PU.1 is required for its ability to inhibit GATA-1-dependent transcription, as well as for blocking MEL cell differentiation, although not for binding to GATA-1. This finding makes it unlikely that PU.1 represses GATA-1 activity by simply preventing its binding to DNA or required cofactors that also bind to the GATA-1 zinc finger region. Consistent with this view, GATA-1 has been reported to be associated with target DNA sites (Strauss et al. 1992; Reddy and Shen 1993) and several cofactors (Osada et al. 1995; Tsang et al. 1997; Blobel et al. 1998) in MEL cells despite the presence of high levels of PU.1. Furthermore, we found that CBP and FOG binding to GATA-1 was not diminished in the presence of excess PU.1 in vitro (N. Rekhtman and A.I. Skoultchi, unpubl.). Taken together, these results suggest that GATA-1 may be able to assemble with associated proteins on DNA even in the presence of PU.1.
We suggest that a more likely mechanism involves association of PU.1 with the assembled GATA-1 protein complex on DNA and repression of its activity. In this model binding to the GATA-1 finger region is mediated by the PU.1 Ets domain, whereas the PU.1 transactivation domain would be needed to recruit other protein factors or to inhibit activities necessary for transcriptional activation. In this context, it may be relevant that the transactivation domain of PU.1 has been reported to interact with both the retinoblastoma protein (RB) and TATA-binding protein (TBP) (Hagemeier et al. 1993). Tethering of RB to promoters through protein–protein interactions has been implicated in transcriptional repression (Weintraub et al. 1995). Alternatively, or in addition, the transactivation domain of PU.1 might contact TBP and cause inhibition of transcription initiation by GATA-1. Although PU.1 has most often been shown to stimulate transcription of target genes containing PU.1-binding sites, a few instances have been reported in which PU.1 represses transcription (Borras et al. 1995; Lopez-Rodriguez and Corbi 1997).
Because the reporter constructs used in the current work do not contain PU.1 consensus-binding sequences in the vicinity of the GATA-1-binding sites, our results indicate that repression of GATA-1 activity by PU.1 can occur without PU.1 binding to DNA. It seems likely that in these experiments PU.1 is tethered through its strong interaction with GATA-1. However, it is quite possible that in some cases repression of GATA-1 activity by PU.1 occurs on genes containing binding sites for both proteins. Consistent with this notion, GATA-1 and PU.1 sites have been found in close proximity in the transcriptional regulatory regions of two erythoid-specific genes (Galson et al. 1993; Nemoto et al. 1996).
Mechanism of PU.1-mediated block to differentiation in erythroleukemia
The results reported here show that PU.1 has the capacity to block erythroid differentiation in both MEL cells and Xenopus embryos, and that in both situations increasing the amount of GATA-1 relative to PU.1 relieves the block and restores the differentiation process. These results suggest that GATA-1 is the principal target of the PU.1-mediated block to differentiation in the erythroleukemic cells. However, our results do not exclude other actions of PU.1 in the erythroleukemia.
A recently identified PU.1 target gene that is clearly relevant to erythroleukemia is another ets family member gene fli-1. The fli-1 locus has been shown to be the preferred viral integration site in SFFV-independent erythroleukemias induced in newborn mice by F-MuLV, the helper component of Friend virus (Ben-David et al. 1991). Fli-1 transcription is stimulated by PU.1 and Fli-1 can also block MEL cell differentiation (Starck et al. 1999). Therefore, fli-1 may be an effector gene, downstream of PU.1. Interestingly, the fli-1 promoter contains, in addition to multiple PU.1-binding sites, a GATA-1 site (Starck et al. 1999). Thus, Fli-1 expression could be modulated by GATA-1, although the role of the GATA-1 site in the fli-1 promoter is not clear at present. It is also noteworthy that we found that Fli-1 and GATA-1 interact in vitro, albeit weakly compared to PU.1 and GATA-1. Fli-1 binding to GATA-1 might amplify the affect of PU.1 on GATA-1.
Another possible mechanism of the PU.1-mediated block to erythroid differentiation might involve the binding of PU.1 to other erythroid transcription factors. Results presented here show that in vitro PU.1 can also bind to GATA-2 and EKLF. EKLF is an erythroid-specific factor that is required for expression of β-globin (Shivdasani and Orkin 1996) and possibly other genes in erythroid cells. Binding of PU.1 to EKLF, although much weaker than to GATA-1, could have similar consequences in inhibiting erythroid gene expression. However, we think that GATA-2 is unlikely to be involved in the PU.1-mediated block to differentiation in MEL cells because these cells have only very low levels of GATA-2 mRNA (Leonard et al. 1993). Other proposed targets of interaction with PU.1 in MEL cells include an RNA-binding protein p54nrb (Hallier et al. 1996), a potential splicing factor TLS (Hallier et al. 1998), and steroid hormone and vitamin receptors (Gauthier et al. 1993), but the contributions of such interactions to the differentiation block remain to be evaluated.
The results reported here also show with PU.1-blocked MEL cell transfectants, that simply shifting the balance in favor of GATA-1 by supertransfecting with a gene encoding a conditionally active GATA-1 not only restores hemoglobin synthesis but also the ability to undergo terminal cell division. We have performed similar experiments in MEL cells containing the GATA-1/ER fusion protein but without an exogenous PU.1 gene and found that estrogen treatment alone can induce 80–90% of the cells to undergo differentiation and terminal cell division. (F. Radparvar and A.I. Skoultchi, unpubl.). These findings suggest that it may be possible to reverse uncontrolled proliferation in tumor cells by introducing into them a single gene encoding the transcription factor that normally regulates their differentiation process.
Do PU.1 and GATA factors interact during normal development?
Although the results reported here clearly show that PU.1 can interact with several GATA factors in vitro and that PU.1 and GATA-1 interact in vivo in erythroleukemia cells, we do not know whether these interactions occur during normal development. The question as to whether PU.1 and GATA factors interact during normal development may be basic to our understanding of lineage commitment in the hematopoietic system.
Current views of lineage commitment processes suggest that such decisions require both increased expression of lineage-specific genes as well as decreased expression of other genes that are not needed in or are not compatible with individual differentiation programs (Cross and Enver 1997). The results described here using the Xenopus embryo system show that PU.1 and GATA-1 can antagonize each others actions in normal erythropoiesis. Thus, if PU.1 and GATA-1 are normally expressed in early hematopoietic progenitors, their relative levels could regulate lineage commitment decisions. It seems likely that PU.1 and GATA-1 are coexpressed in certain hematopoietic cells as their mRNAs have been detected in early erythroid precursors and also in multipotent progenitors (Cheng et al. 1996). The two proteins were also detected in mature mast cells (Henkel and Brolon 1994). An even more likely possibility for interaction during normal development may be between PU.1 and GATA-2. GATA-2 is expressed more widely than GATA-1 and it is thought that GATA-2 functions very early in hematopoietic cell development and proliferation (Shivdasani and Orkin 1996). Studies of GATA-2 null mice and embryonic stem (ES) cells support this view, indicating that GATA-2 may be needed for generation of many, if not all, hematopoietic progenitors (Tsai et al. 1994). Thus a functional interaction could occur between PU.1 and GATA-2 in erythroid, myeloid, or lymphoid lineage determination.
Finally, it should be noted that the consequences of PU.1–GATA factor interactions need not always result in inhibition of target promoter function. In at least one instance, PU.1 and GATA factors were shown to collaborate to stimulate transcription of a target gene containing adjacent PU.1 and GATA binding sites (Henkel and Brown 1991). Both PU.1 and GATA-2 have the capacity to block terminal erythroid differentiation. This could suggest that these two factors interact and collaborate to stimulate cell division in early erythroid progenitors. The association of PU.1 and GATA factors in hematopoietic cells and the consequences of such interactions remains to be determined.
Materials and methods
In vitro protein interaction studies
Glutathione S-transferase fusion proteins were prepared by standard procedures. Purity of the GST fusion proteins was analyzed by boiling proteins for 5 min in Laemmli sample buffer [60 mm Tris (pH 6.8), 2% SDS, 100 mm DTT, 10% glycerol, 0.001% bromphenol blue], separating by SDS-PAGE and Coomassie Blue staining. The concentration of each purified GST fusion protein was estimated by comparison to several amounts of bovine serum albumin also present on the gels. The GST fusion protein constructs included GATA-1 and GATA-1 mutants, PU.1 kindly provided by Y.K. Fung (USC, Los Angeles) and GUK a gift from R.A. DePinho (Dana Farber Cancer Institute, Boston, MA).
[35S]Methionine-labeled proteins were prepared by in vitro transcription/translation using a coupled TnT reticulocyte lysate system (Promega). Plasmids used included pGEM-PU.1 (Rao et al. 1997) for full-length PU.1, PU.1(ΔDBD) constructed by cloning the KpnI fragment from pGEM–PU.1 into the KpnI site of pBluescript (pBSK, Stratagene), PU.1(ΔPEST) (Klemsz and Maki 1996) subcloned in EcoRI site of pBSK, pBKS–PU.1(ΔNN) called here ΔTAD, (ΔNP), (ΔNS) (Klemsz and Maki 1996) kindly provided by M.J. Klemsz, pBSK-GATA-1(ΔN) (Blobel et al. 1998) a gift from G.A. Blobel, GATA-1(ΔC) and GATA-1(Δfingers) generated by subcloning the respective coding sequences derived from pXM–GATA-1(ΔC) (Martin and Orkin 1990) and pGEX-2T–GATA-1–Δfingers (Merika and Orkin 1995) into pBSK, p45 NF-E2 (Andrews et al. 1993) and FOG (Tsang et al. 1997) generated by subcloning the respective coding sequences into the pBSK vector, pL28-Spi-B, kindly provided by D.G. Tenen (Beth Israel Deaconness Medical Center, Boston, MA); pBKS-Fli-1, a kind gift of A. Bernstein (Mount Sinai Hospital, Toronto, Canada); pPET8C–GATA-1, GATA-2, and GATA-3 and pBSK–EKLF generously provided by M. Merika.
GST fusion proteins (2 μg) immobilized on beads and amounts of 35S-labeled proteins quantitated by fluorography were mixed in 400 μl of EBC buffer [120 mm NaCl, 1 m Tris-HCl (pH 8), 0.5% NP-40, 1 mm ZnCl2, protease inhibitors] and incubated for 2 hr at 4°C. The total amount of glutathione Sepharose used in each binding reaction was kept constant by adding an appropriate amount of beads without bound proteins. Beads were washed five times with EBC buffer. The concentration of NaCl in the final wash was increased to 500 mm. The bound proteins were released from the beads by boiling for 5 min in 40 μl of sample buffer and analyzed by SDS-PAGE followed by fluorography.
To generate proteins free of the GST moiety, 10 μg of purified GST fusion proteins immobilized on beads were treated with 0.1 μg of thrombin (Sigma) for 30 min at room temperature. Beads with uncleaved proteins were removed by centrifugation and the supernatants were incubated with fresh beads to remove any contaminating uncleaved proteins.
In vivo protein interaction studies
Clone DS19 MEL cells and 293 cells were grown as described previously (Rao et al. 1997). Nuclei were isolated as described by Andrews and Faller (1991) and nuclear extracts were prepared by lysis of nuclei by sonication in 50 mm HEPES (pH 7.5), 150 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1 mm DTT, 0.1% Tween 20, 10% glycerol, protease inhibitors. Whole cell extracts were prepared by treating cells with EBC buffer. 293 cells were transfected by the calcium phosphate precipitation method with pEBB expression vectors (Rao et al. 1997) encoding PU.1 and PU.1 mutants and pXM expression vectors encoding GATA-1 (Tsai et al. 1989) and GATA-1 mutants (Martin and Orkin 1990). GATA-1(Δfingers) was subcloned into the pEBB vector. Whole cell or nuclear extracts were prepared after 48 hr as above. Protein concentrations in the extracts were measured by the Bradford assay (BioRad). MEL cell extracts (600 μg) and 293 cell extracts (400 μg) were used in the immunoprecipitations.
For immunoprecipitation of GATA-1, 50 μl of Protein A–Sepharose beads (Pharmacia) were preincubated with 8 μl of anti-GATA-1 antibody N6 (Santa Cruz) and 2 μl of monoclonal anti-rat antibody (Sigma) overnight. As a control, 8 μl of mouse monoclonal anti-p16 antibody (DCS-50.1/A7; NeoMarkers) and 2 μl of anti-rat antibody were used. For immunoprecipitation of PU.1, 50 μl of protein A–Sepharose beads and 15 μl of anti-PU.1 antibody T21 (Santa Cruz) or 15 μl of a control anti-cyclin E antibody M-20 (Santa Cruz) were used. Antibody-saturated beads were collected and incubated with nuclear extracts for 2 hr. Beads were washed five times and bound proteins were released and analyzed by SDS-PAGE and immunoblotting with the appropriate antibodies.
Transactivation assays
NIH-3T3 and HeLa cells were cultured as above. Twenty-four hours before transfection 7 × 104 cells were plated in each well of 12-well plates. Cells were transfected by the Lipofectamine Plus method (GIBCO BRL) as recommended by the manufacturer in the presence of 5% serum. The total amount of DNA was maintained at 450 ng/well by adding the appropriated amount of empty pEBB vector DNA. Growth hormone production was measured by radioimmunoassay (Nichols Institute Diagnostics) on 100 μl (10%) of the culture medium. Luciferase production was measured on equal amounts of protein from whole cell extracts with the Promega Luciferase Assay system.
Constructs used in the reporter assays included M1αGH (Martin and Orkin 1990), p45 NF-E2-GH, pMT2-FOG (Tsang et al. 1997), αD3-LUC (Evans and Felsenfeld 1991), (UAS)5E1BTATA–LUC (Watanabe et al. 1996), pEBB–PU.1, pEBB–AS–PU.1 (Rao et al. 1997), pXM–GATA-1 and SP6–GAL4-Sp1 (Sif and Gilmore 1994).
Preparation and analysis of MEL cell stable transfectants
MEL cells (clone DS19) or cells of the PU.1–MEL transfectant (EF1α-PU.1 clone 605) were transfected as described previously (Rao et al. 1997). Selection with 5 μg/ml puromycin (MEL) or 5 μg/ml puromycin and 1 mg/ml G418 (clone 605) was initiated after 24 hr. Antibiotic-resistant clones were expanded and cell extracts were analyzed by immunoblotting. Benzidine staining, Northern blot hybridization for globin mRNA with [α-32P]dCTP-labeled pCA11, provided by W. Zimmer (University of South Alabama, Mobile), and plasma clot assays were performed as described previously (Rao et al. 1997).
Expression constructs for HA epitope-tagged PU.1 proteins were constructed by subcloning PU.1 cDNA fragments into a modified pEBB vector. The vector pEBB-HA-puro was generated by blunt-end ligation of the SalI fragment of pPGK-Puro into HindIII-digested pEBB-HA, kindly provided by G. Cheng (University of California, Los Angeles). PU.1–ΔTAD–HA and PU.1–ΔPEST–HA were prepared by substitution of the BamI–KpnI fragment of pEBB–PU.1–HA-puro with the mutated inserts from pBKS-ΔNN and pBSK–ΔPEST, respectively. PU.1–ΔDBD–HA was created by digestion of PU.1–HA with KpnI and NotI, filling in with Klenow and self-ligation. pEBB–GATA-1/ER-puro was constructed by subcloning GATA-1/ER into the pEBB-puro vector constructed as above.
Immunoblotting
Proteins separated by SDS-PAGE were transferred to nitrocellulose or PVDF membranes. Membranes were incubated with 1:500 dilution of either rat monoclonal anti-GATA-1 antibody N6 or 1:5000 dilution of mouse monoclonal anti-HA antibody 12CA5 (Boehringer Mannheim) or 1:300 dilution of rabbit polyclonal anti-PU.1 antibody T21 followed by incubation with either 1:8000 dilution of HRP-conjugated anti-mouse antibody (Sigma) or 1:3500 dilution of HRP-conjugated anti-rabbit antibody (Santa Cruz). HRP activity was detected using the enhanced chemiluminescence (ECL) system (Amersham).
Xenopus embryos and explants
Xenopus laevis eggs obtained from gonadotropin-induced females were fertilized in vitro. Dejellied embryos were placed into 0.1× MBS/5% Ficoll solution and injected with 4.5 pL of RNA or plasmid solution into blastomeres at the two-cell stage. Alternatively, four-cell stage embryos with an obvious pigment shift and precisely oriented cleavage planes were chosen to target injections into the two presumptive ventral blastomeres. Embryos were allowed to recover for several hours and then were cultured at room temperature in 0.1× MBS. Globin production was analyzed in whole embryos by benzidine staining. In some cases, when the dorsal lip was first visible at stage 10 1/4, VMZ explants were cut and incubated in 1× MMR for 56 hr or as indicated. Protein lysates were made by homogenizing explants in 10 μl of SDS sample buffer/explant and equal explant equivalents were electrophoresed in 17.5% acrylamide/3m urea SDS gels before blotting. The l4-26 mAb specific for embryonic α-globin chains was a kind gift of Dr. C. Katagiri. HRP-conjugated secondary antibodies were detected using ECL (Amersham).
The cDNAs encoding murine PU.1 or Xenopus GATA-1 were cloned into the SP64T transcription vector and mRNA was transcribed in vitro using SP6 polymerase. Control RNAs were transcribed from a plasmid containing the PU.1 cDNA in the reverse orientation (PU.1rs) or a frameshift mutant form of the GATA-1 cDNA that encoded a non-DNA-binding truncated isoform (Glfs). RNA was quantified by UV spectrometry and quality was analyzed before injection by gel electrophoresis and in vitro translation.
Acknowledgments
We thank M.J. Klemsz, S.H. Orkin, M. Merika, and G.A. Blobel for generously providing plasmids used in this study. T.E. thanks T. Torregroza for excellent technical help. A.I.S. receives support from National Cancer Institute Cancer Center grant 2P30CA13330, T.E. was supported by grants from the National Institutes of Health (NIH) (HL56132) and the American Cancer Society (ACS), F.R. was supported by NIH 5T32AG00194 and N.R. by NIH 5T32GM07491. This work was supported by NIH grant 5R37CA16368.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL skoultch@aecom.yu.edu; FAX (718) 430-8574.
References
- Andrews NC, Faller DV. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews NC, Kotkow KJ, Ney PA, Erdjument-Bromage H, Tempst P, Orkin SH. The ubiquitous subunit of erythroid transcription factor NF-E2 is a small basic-leucine zipper protein related to the v-maf oncogene. Proc Natl Acad Sci. 1993;90:11488–11492. doi: 10.1073/pnas.90.24.11488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David Y, Bernstein A. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell. 1991;66:831–834. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- Ben-David Y, Giddens EB, Letwin K, Bernstein A. Erythroleukemia induction by Friend murine leukemia virus: Insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes & Dev. 1991;5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Sieff CA, Orkin SH. Ligand-dependent repression of the erythroid transcription factor GATA-1 by the estrogen receptor. Mol Cell Biol. 1995;15:3147–3153. doi: 10.1128/mcb.15.6.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borras FE, Lloberas J, Maki RA, Celada A. Repression of I-A beta gene expression by the transcription factor PU.1. J Biol Chem. 1995;270:24385–24391. doi: 10.1074/jbc.270.41.24385. [DOI] [PubMed] [Google Scholar]
- Briegel K, Lim KC, Plank C, Beug H, Engel JD, Zenke M. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes & Dev. 1993;7:1097–1109. doi: 10.1101/gad.7.6.1097. [DOI] [PubMed] [Google Scholar]
- Cheng T, Shen H, Giokas D, Gere J, Tenen DG, Scadden DT. Temporal mapping of gene expression levels during the differentiation of individual primary hematopoietic cells. Proc Natl Acad Sci. 1996;93:13158–13163. doi: 10.1073/pnas.93.23.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross MA, Enver T. The lineage commitment of haemopoietic progenitor cells. Curr Opin Genet Dev. 1997;7:609–613. doi: 10.1016/s0959-437x(97)80007-x. [DOI] [PubMed] [Google Scholar]
- Delgado MD, Gutierrez P, Richard C, Cuadrado MA, Moreau-Gachelin F, Leon J. Spi-1/PU.1 proto-oncogene induces opposite effects on monocytic and erythroid differentiation of K562 cells. Biochem Biophys Res Comm. 1998;252:383–391. doi: 10.1006/bbrc.1998.9587. [DOI] [PubMed] [Google Scholar]
- Evans T, Felsenfeld G. Trans-Activation of a globin promoter in nonerythroid cells. Mol Cell Biol. 1991;11:843–853. doi: 10.1128/mcb.11.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RC, Scott EW. Role of PU.1 in hematopoiesis. Stem Cells. 1998;16:25–37. doi: 10.1002/stem.160025. [DOI] [PubMed] [Google Scholar]
- Galson DL, Hensold JO, Bishop TR, Schalling M, D’Andrea AD, Jones C, Auron PE, Housman DE. Mouse beta-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier JM, Bourachot B, Doucas V, Yaniv M, Moreau-Gachelin F. Functional interference between the Spi-1/PU.1 oncoprotein and steroid hormone or vitamin receptors. EMBO J. 1993;12:5089–5096. doi: 10.1002/j.1460-2075.1993.tb06203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl MK. The PU.1 transcription factor is the product of the putative oncogene Spi-1. Cell. 1990;61:1165–1166. doi: 10.1016/0092-8674(90)90676-6. [DOI] [PubMed] [Google Scholar]
- Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Transcriptional activity of Pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes & Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier C, Bannister AJ, Cook A, Kouzarides T. The activation domain of transcription factor PU.1 binds the retinoblastoma (RB) protein and the transcription factor TFIID in vitro: RB shows sequence similarity to TFIID and TFIIB. Proc Natl Acad Sci. 1993;90:1580–1584. doi: 10.1073/pnas.90.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallier M, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 binds RNA and interferes with the RNA-binding protein p54nrb. J Biol Chem. 1996;271:11177–11181. doi: 10.1074/jbc.271.19.11177. [DOI] [PubMed] [Google Scholar]
- Hallier M, Lerga A, Barnache S, Tavitian A, Moreau-Gachelin F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J Biol Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- Henkel G, Brown MA. PU.1 and GATA: Components of a mast cell-specific interleukin 4 intronic enhancer. Proc Natl Acad Sci. 1994;91:7737–7741. doi: 10.1073/pnas.91.16.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromas R, Orazi A, Neiman RS, Maki R, Van Beveran C, Moore J, Klemsz M. Hematopoietic lineage- and stage-restricted expression of the ETS oncogene family member PU.1. Blood. 1993;82:2998–3004. [PubMed] [Google Scholar]
- Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- Klemsz MJ, Maki RA. Activation of transcription by PU.1 requires both acidic and glutamine domains. Mol Cell Biol. 1996;16:390–397. doi: 10.1128/mcb.16.1.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemsz MJ, McKercher SR, Celada A, Van Beveren C, Maki RA. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- Krieg PA, Melton DA. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Leonard MW, Lim KC, Engel JD. Expression of the chicken GATA factor family during early erythroid development and differentiation. Development. 1993;119:519–531. doi: 10.1242/dev.119.2.519. [DOI] [PubMed] [Google Scholar]
- Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Corbi AL. PU.1 negatively regulates the CD11c integrin gene promoter through recognition of the major transcriptional start site. Eur J Immunol. 1997;27:1843–1847. doi: 10.1002/eji.1830270804. [DOI] [PubMed] [Google Scholar]
- Lu SJ, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: Evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Sheffery M, Rifkind RA. Induction of transformed cells to terminal differentiation and the modulation of gene expression. Cancer Res. 1987;47:659–666. [PubMed] [Google Scholar]
- Martin DI, Orkin SH. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes & Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau-Gachelin F. Spi-1/PU.1: An oncogene of the Ets family. Biochem Biophys Acta. 1994;1198:149–163. doi: 10.1016/0304-419x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- Moreau-Gachelin F, Ray D, Mattei MG, Tambourin P, Tavitian A. The putative oncogene Spi-1: Murine chromosomal localization and transcriptional activation in murine acute erythroleukemias. Oncogene. 1989;4:1449–1456. [PubMed] [Google Scholar]
- Moreau-Gachelin F, Wendling F, Molina T, Denis N, Titeux M, Grimber G, Briand P, Vainchenker W, Tavitian A. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagulapalli S, Pongubala JM, Atchison ML. Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3) J Immunol. 1995;155:4330–4338. [PubMed] [Google Scholar]
- Nemoto Y, Terajima M, Shoji W, Obinata M. Regulatory function of delta/YY-1 on the locus control region-like sequence of mouse glycophorin gene in erythroleukemia cells. J Biol Chem. 1996;271:13542–13548. doi: 10.1074/jbc.271.23.13542. [DOI] [PubMed] [Google Scholar]
- Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes & Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada H, Grutz G, Axelson H, Forster A, Rabbitts TH. Association of erythroid transcription factors: Complexes involving the LIM protein RBTN2 and the zinc finger protein GATA1. Proc Natl Acad Sci. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul R, Schuetze S, Kozak SL, Kabat D. A common site for immortalizing proviral integrations in Friend erythroleukemia: Molecular cloning and characterization. J Virol. 1989;63:4958–4961. doi: 10.1128/jvi.63.11.4958-4961.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovick MS, Hiebert SW, Friedman AD, Hetherington CJ, Tenen DG, Zhang DE. Multiple functional domains of AML1: PU.1 and C/EBPalpha synergize with different regions of AML1. Mol Cell Biol. 1998;18:3915–3925. doi: 10.1128/mcb.18.7.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quang CT, Pironin M, von Lindern M, Beug H, Ghysdael J. Spi-1 and mutant p53 regulate different aspects of the proliferation and differentiation control of primary erythroid progenitors. Oncogene. 1995;11:1229–1239. [PubMed] [Google Scholar]
- Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14:123–131. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- Ray D, Bosselut R, Ghysdael J, Mattei MG, Tavitian A, Moreau-Gachelin F. Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol Cell Biol. 1992;12:4297–4304. doi: 10.1128/mcb.12.10.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy PM, Shen CK. Erythroid differentiation of mouse erythroleukemia cells results in reorganization of protein–DNA complexes in the mouse beta major globin promoter but not its distal enhancer. Mol Cell Biol. 1993;13:1093–1103. doi: 10.1128/mcb.13.2.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetze S, Paul R, Gliniak BC, Kabat D. Role of the PU.1 transcription factor in controlling differentiation of Friend erythroleukemia cells. Mol Cell Biol. 1992;12:2967–2975. doi: 10.1128/mcb.12.7.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetze S, Stenberg PE, Kabat D. The Ets-related transcription factor PU.1 immortalizes erythroblasts. Mol Cell Biol. 1993;13:5670–5678. doi: 10.1128/mcb.13.9.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Shivdasani RA, Orkin SH. The transcriptional control of hematopoiesis. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- Sif S, Gilmore TD. Interaction of the v-Rel oncoprotein with cellular transcription factor Sp1. J Virol. 1994;68:7131–7138. doi: 10.1128/jvi.68.11.7131-7138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck J, Doubeikovski A, Sarrazin S, Gonnet C, Rao G, Skoultchi A, Godet J, Dusanter-Fourt I, Morle F. Spi-1/PU.1 is a positive regulator of the fli-1 gene involved in inhibition of erythroid differentiation in friend erythroleukemic cell lines. Mol Cell Biol. 1999;19:121–135. doi: 10.1128/mcb.19.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss EC, Andrews NC, Higgs DR, Orkin SH. In vivo footprinting of the human alpha-globin locus upstream regulatory element by guanine and adenine ligation-mediated polymerase chain reaction. Mol Cell Biol. 1992;12:2135–2142. doi: 10.1128/mcb.12.5.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- Tsai FY, Keller G, Kuo FC, Weiss M, Chen J, Rosenblatt M, Alt FW, Orkin SH. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- Tsai SF, Martin DI, Zon LI, D’Andrea AD, Wong GG, Orkin SH. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Watanabe G, Howe A, Lee RJ, Albanese C, Shu IW, Karnezis AN, Zon L, Kyriakis J, Rundell K, Pestell RG. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- Yamada T, Kondoh N, Matsumoto M, Yoshida M, Maekawa A, Oikawa T. Overexpression of PU.1 induces growth and differentiation inhibition and apoptotic cell death in murine erythroleukemia cells. Blood. 1997;89:1383–1393. [PubMed] [Google Scholar]