Abstract
Rule-based modeling provides a means to represent cell signaling systems in a way that captures site-specific details of molecular interactions. For rule-based models to be more widely understood and (re)used, conventions for model visualization and annotation are needed. We have developed the concepts of an extended contact map and a model guide for illustrating and annotating rule-based models. An extended contact map represents the scope of a model by providing an illustration of each molecule, molecular component, direct physical interaction, post-translational modification, and enzyme-substrate relationship considered in a model. A map can also illustrate allosteric effects, structural relationships among molecular components, and compartmental locations of molecules. A model guide associates elements of a contact map with annotation and elements of an underlying model, which may be fully or partially specified. A guide can also serve to document the biological knowledge upon which a model is based. We provide examples of a map and guide for a published rule-based model that characterizes early events in IgE receptor (FcεRI) signaling. We also provide examples of how to visualize a variety of processes that are common in cell signaling systems but not considered in the example model, such as ubiquitination. An extended contact map and an associated guide can document knowledge of a cell signaling system in a form that is visual as well as executable. As a tool for model annotation, a map and guide can communicate the content of a model clearly and with precision, even for large models.
Introduction
Cellular responses to environmental changes and signals are mediated by cell signaling systems. A cell signaling system is composed largely of a network of interacting proteins, which are responsible for information processing. A typical signaling protein contains multiple functional components. The components found in signaling proteins include catalytic domains1,2, modular protein interaction domains3, linear motifs4, and sites of post-translational modification5. Understanding the functional roles of protein components or sites is critical for a thorough understanding of cell signaling, because protein interactions generally depend on site-specific details. For example, many protein-protein interactions are modulated by tyrosine phosphorylation6. A large amount of information is available about the site-specific details of protein interactions. There is a need to be able to use this information to make predictions about system behaviors. In other words, we need mathematical/computational models to better understand cell signaling, which is complex7,8.
With recent developments in simulation methodology9–13, rule-based modeling14, discussed in detail below, now offers a viable approach for studying large numbers of protein interactions with consideration of site-specific details. Here, with the goal of making this modeling approach more accessible, we demonstrate how rule-based models can be better visualized and annotated, which is important for modeling efforts that aim to comprehensively capture the molecules and interactions involved in an entire cell signaling system or set of systems. A large, detailed model is of limited use unless it is presented in an understandable manner. The proteins and interactions included in a model, as well as the justification for modeling assumptions, should be communicated clearly and precisely if a model is to be understood, critically evaluated, and reused. To enable clear communication of rule-based models, we introduce the concept of an extended contact map, which serves to illustrate the scope of a rule-based model. We also introduce the concept of an associated model guide. A model guide attaches rules, which are formal representations of interactions, to arrows in an extended contact map. It also attaches molecule type definitions, which are formal representations of molecules, to boxes in a map. A map and a guide that annotates a complete model together provide a visual and executable means to document information about the site-specific details of molecular interactions in a cell signaling system. We expect that the concepts presented here should be useful for modelers as well as others interested in applying systems approaches to the study of cell signaling.
Background
Rule-based modeling
Rule-based modeling is a relatively new modeling approach in biology that is well-suited for capturing the dynamics of interactions among proteins14,15. The approach can be viewed as a particular type of agent-based modeling, in which agents (molecules) interact according to rules consistent with certain physicochemical principles.
A rule can be viewed as a coarse-grained representation of the kinetics of a class of (bio)chemical reactions. Each reaction within a class involves a common reaction center and transformation, which can take place in multiple contexts, but is characterized by a common rate law as an approximation. If the transformations that occur in a system can be assumed to be independent of most aspects of molecular context, then a modeler can use rules to concisely and comprehensively capture the consequences of the interactions and obtain model predictions consistent with a traditional physicochemical model that is defined implicitly. Thus, in the case of a well-mixed system, there exists a corresponding system of coupled ordinary differential equations (ODEs) that can in principle be derived from the set of rules15–17. The granularity of a rule can be refined by adjusting the necessary and sufficient conditions that are required of reactants (i.e., the molecular context that must be satisfied for a reaction to occur). A modeler is free to control the coarseness of model assumptions. At the finest level, a rule uniquely specifies a single chemical reaction. Thus, rule-based modeling can be viewed as a generalization of traditional modeling of (bio)chemical reaction kinetics.
The site-specific details of protein-protein interactions are difficult to capture in a conventional model, such as an ODE-based model, because of combinatorial complexity, an inherent feature of cell signaling systems18–21. On the other hand, such details can be naturally incorporated into a rule-based model. Rule-based modeling provides a needed capability for mechanistic modeling of cell signaling systems, and accordingly, it has been applied to model various aspects of various systems22–36. A number of software tools have been developed to enable rule-based modeling11–13,15,16,37–46. With the availability of these tools, we can expect to see more applications of the rule-based modeling approach. A discussion of how to visualize and annotate rule-based models seems timely.
BioNetGen language
Rule-based models can be encoded in the BioNetGen language (BNGL)15, which is used by a number of software tools11–13,15,16,41,42. This language is closely related to Kappa, which is used by yet other software tools44,47 (http://kappalanguage.org). In the graphical formalism upon which BNGL is based14,15,17,48,49, proteins and other molecules are represented using molecule type graphs, chemical species graphs and pattern graphs, which are called site graphs in Kappa47. The vertices of these graphs represent components, the functional parts of proteins (e.g., domains, linear motifs, and sites of post-translational modification). The vertices representing components may be associated with variable attributes, referred to as internal states. An internal state is often a useful abstraction, which can be used to represent the conformation, location, or post-translational modification status of a protein component. Protein-protein and other molecular interactions are represented using graph-rewriting rules, which designate what is required of molecules for an interaction to occur and how molecular components are affected by an interaction/transformation (see below). Rules are associated with rate laws (functions of properties of reactants, typically including the population levels of reactants), which are used to assign rates to transformations defined by rules.
A rule contains elements that are similar to those of a standard chemical reaction, as illustrated by the following BNGL-encoded rule:
| (1) |
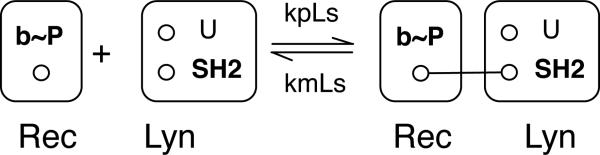
This rule, which is explained below, is visualized graphically in Fig. 1 in accordance with the conventions of Faeder et al.48. It is part of the model of Goldstein et al.22 and Faeder et al.23, which is given in the ESI (model.bngl). Note that the rule of Eq. (1) is identified as Rule 5 in the model-specification file. GetBonNie50 provides a tool, RuleBuilder Lite, for drawing rules and exporting BNGL code and for automatically visualizing BNGL code according to the conventions illustrated in Fig. 1. The graphs displayed in Fig. 1 are examples of pattern graphs or site graphs.
Fig. 1.
Visualization of the rule of Eq. (1). A molecule or part of a molecule is represented as a set of vertices, which represent molecular components. Vertices are labeled and may also be attributed. Here, the vertex b has the attribute P. Bonds are represented as edges. The reaction center defined by this rule is highlighted by bold vertex labels.
The rule of Eq. (1) specifies a reaction center, a set of components affected by a transformation. In the rule of Eq. (1) and in the rules of Appendix S1 (ESI), components in the reaction center are underlined. Components that are included in a rule but that are not part of the reaction center are contextual. In Eq. (1), the component U is contextual. The necessary and sufficient properties of reactants are specified on the left-hand side of Eq. (1), which indicates that FcεRI, the high-affinity receptor for IgE antibody (denoted Rec here), interacts reversibly with the Src-family protein tyrosine kinase Lyn (Lyn). The difference between the right- and left-hand sides of Eq. (1) indicates that the interaction results from binding of the tyrosine-phosphorylated β chain of the receptor (b~P) to the SH2 domain (SH2) of Lyn. The left-hand side of the rule indicates that the b component of Rec must be in the P internal state (i.e., it must be phosphorylated) to bind SH2. Furthermore, for a bond to form, the unique domain of Lyn (U) must be unbound, which is indicated by including U in the rule without associating this component with a bond label. If the unique domain had no impact on the interaction, it would be omitted from the rule. A bond label is preceded by a ‘!’ character. A bond, labeled ‘1,’ is identified on the right-hand side of Eq. (1). The ‘.’ character on the right-hand side of Eq. (1) is used to represent connectivity; here, it is redundant. The ‘~’ character precedes the name of an internal state of a component. Finally, the rule indicates that the interaction is characterized by certain on- and off-rate constants (kpLs, kmLs). By convention, it is understood that the rate law associated with this rule has the form of that for an elementary reaction. Non-elementary rate laws, such as the Michaelis-Menten rate law or a Hill function, can be specified if desired13,15.
An additional feature provided by compartmental BNGL (cBNGL), not demonstrated in Eq. 1, is the ability to explicitly represent compartments and trafficking of molecules between compartments51. For example, the following cBNGL-encoded rule represents translocation of the transcription factor NF-κB (denoted NFKB) from the cytoplasm (Cyt) to the nucleus (Nuc):
| (2) |
where the ‘@’ symbol is used to indicate a compartmental location. NF-κB translocates from the cytoplasm to the nucleus when it is not bound to an inhibitor, IκB, which interacts with the Rel homology domain (RHD) of NF-κB52,53. Note that inclusion of the component RHD (which denotes the RHD of NF-κB) in the rule of Eq. (2) represents a contextual constraint on translocation. As indicated in the rule, translocation requires that this component be free (of IκB).
As illustrated by the examples above, a rule provides a way of representing a molecular interaction with consideration of the site-specific details involved. Rules are executable14, meaning that they are formal elements of a model that can be simulated, and their precision makes them a useful way of summarizing information even if one does not intend to simulate a model.
A rule-based model for early events in FcεRI signaling
We will use the model of Goldstein et al.22 and Faeder et al.23, an early application of the rule-based modeling approach, to exemplify the basic conventions of an extended contact map and a model guide. Here, we provide an overview of this model, which we will refer to as the FcεRI model. A full specification of the model is provide in the ESI (model.bngl).
The FcεRI model22,23 is composed of 19 rules in total, and it captures early events in IgE receptor (FcεRI) signaling, which triggers allergic reactions. The receptor is composed of an α chain, a β chain, and a homodimer of two disulfide-linked γ chains. The extracellular portion of the α chain binds the Fc portion of IgE54; the interaction is long lived55. The β and γ chains each contain an immunoreceptor tyrosine-based activation motif (ITAM)56, a linear motif. Signaling is initiated when a multivalent antigen or other receptor crosslinking reagent bridges two receptors. In the model, receptor crosslinking is taken to be mediated by a chemically crosslinked dimer of IgE. Following receptor aggregation, the kinase Lyn, which constitutively interacts with the β chain, phosphorylates the β and γ ITAMs in neighboring receptors. As a result, the receptor can recruit Lyn and Syk, a second kinase involved in FcεRI signaling, through phosphorylation-dependent interactions. Syk is phosphorylated via two mechanisms: Lyn phosphorylates tyrosine residues in the linker region, and Syk trans-phosphorylates tyrosine residues in the activation loop of the kinase domain of a neighboring copy of Syk. In the model, all phosphorylation events are reversed by unspecified phosphatases, which are assumed to be available in excess. The model is based on several additional assumptions. For example, some tyrosine residues are treated as a single unit, i.e., lumped together as a virtual phosphorylation site.
Currently available methods for visualization of rule-based models
Models of biochemical processes, including rule-based models, are often easier to understand if they are visualized. Recently, efforts have been made to standardize visual representations of biochemical systems and models of biochemical systems. These efforts have culminated in Systems Biology Graphical Notation (SBGN)57. SBGN provides three sets of notational conventions, called languages, for various types of visualizations. Among these, the Process Description (PD) language can be used to visualize a biochemical reaction network or a model of such a network. Diagrams made using the PD language or the earlier related conventions of process diagrams58 are available that illustrate fairly large reaction networks59–61. For example, the diagram of Caron et al.61 accounts for 964 species and 777 reactions. Unfortunately, this network is small compared to some of the reaction networks underlying rule-based models14,62. Some rule-based models can be converted to conventional models and visualized using methods developed for such models, including SBGN, but there are many rule-based models that for all intents and purposes do not have conventional counterparts. It is especially for these cases that new visualization methods are needed. Below, we briefly review three visualization methods that have been used specifically for rule-based models and we discuss their limitations. Other methods are illustrated in Figs. S1 and S2 (ESI). Figure S1 illustrates a path25, a connected chain of reactions that each demonstrates an instance of a rule. The reactions in this path are visualized using the conventions of Faeder et al.48. Figure S2 illustrates an influence map62, which visualizes how execution of one rule influences the execution of other rules in a simulation.
Graphical representation of individual rules
A rule can be visualized via the graphical conventions of Faeder et al.48. These conventions are used in Fig. 1 to illustrate a rule in the FcεRI model22,23 that characterizes binding of Lyn to the phosphorylated β chain of FcεRI. The conventions of process diagrams58 may also be used to illustrate individual rules63. The approach of Fig. 1 is only adequate for illustrating one rule or a few rules. Individually illustrating every rule in a large model (i.e., a model composed of a large number of rules) will result in a diagram that is locally comprehensible but globally incomprehensible. Thus, illustration of individual rules is impractical for communicating the content of a large model.
Contact maps
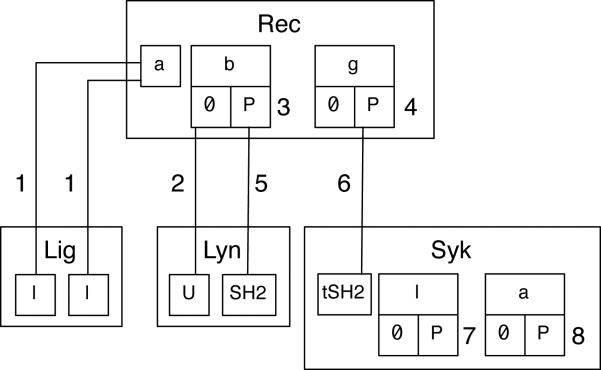
Danos et al.62 introduced contact maps, which facilitate static analysis of rules44,47. Contact maps are also useful for visualization purposes. (The term ‘contact map’ should not be confused with the term ‘protein contact map,’ which is used in structural biology64.) A contact map, which is a type of site graph, can be derived unambiguously from a rule-based model. A contact map identifies the molecules, the components of molecules, the possible internal states of components, and the possible bonds between components that are included in a model. Software tools are available for constructing a contact map automatically from a BNGL- or Kappa-encoded specification of a rule-based model50 (http://www.rulebase.org). A contact map for the FcεRI model22,23 is shown in Fig. 2.
Fig. 2.
A contact map for the FcεRI model22,23. A contact map shows the molecules, components, and direct binding interactions that are specified in a model. An outer box represents a molecule type. Inner boxes represent components. The possible internal states of components are also represented, by boxes below component boxes. Binding partners are connected by lines. This contact map indicates, for example, that the g component of Rec has two possible states (0 and P), and when in the P state, g may bind the tSH2 component of Syk. This component represents the two tandem SH2 domains of Syk. The labels 1, 2, 5 and 6 refer to sets of rules in the FcεRI model22,23 that characterize association/dissociation reactions. The labels 3, 4, 7 and 8 refer to sets of rules that characterize internal state change reactions, i.e., phosphorylation reactions in the FcεRI model22,23. All of these rule sets are described in Appendix S1 (ESI).
The contact map of Fig. 2 is derived directly from the FcεRI model22,23 (model.bngl, ESI). Thus, it reflects modeling assumptions, and fails to convey certain information about FcεRI signaling that was used in model specification. For example, the kinases responsible for phosphorylation events are not identified in Fig. 2. Typically, in a rule-based model, catalysts are not explicitly represented in rules, so contact maps generally will not reveal enzyme-substrate relationships. The graphical representation of molecules in Fig. 2 conforms to the underlying graphical formalism of BNGL49. In this formalism, only molecules and molecular components (i.e., only one layer of parent-child relationships) can be represented, even though molecular components can contain subcomponents. As a result, as discussed in detail by Lemons et al.65, structural relationships among the functional components and subcomponents of signaling proteins can be obscured. Explicit representation of enzyme-substrate relationships and structural relationships are generally not necessary for simulation purposes15, but omitting these types of details from an illustration of a model, such as that of Fig. 2, can hide the biological knowledge underlying a model specification.
Molecular interaction maps
Kohn et al.66 proposed conventions for representing a system marked by combinatorial complexity in the form of a molecular interaction map (MIM). Such a MIM can be used to visualize rules14. It should be noted that MIM-like diagrams can be specified using the Entity Relationship (ER) language of SBGN57. A MIM provides a visualization of a biological system by using boxes to represent molecules and a variety of symbols and lines/arrows to represent different types of interactions and influences. A MIM for the FcεRI model22,23 is shown in Fig. 3. Annotation of this MIM is provided in Appendix S2. The main purpose of Appendix S2 is to explain our use of MIM notation, i.e., why we used MIM notations as we did in our attempt to provide a MIM that accurately reflects the FcεRI model22,23. The conventions of a MIM call for the representation of a molecule only once so that all interactions involving a molecule can be traced to a common origin. This feature of a MIM, which is highly desirable as it avoids the need to represent every chemical species that can be populated, as in a conventional reaction scheme, is shared by a contact map. Unlike the situation for contact maps, software is not available for drawing MIMs automatically from model specifications. A MIM is a handcrafted illustration, although MIM construction is aided by a PathVisio67 plugin (http://discover.nci.nih.gov/mim).
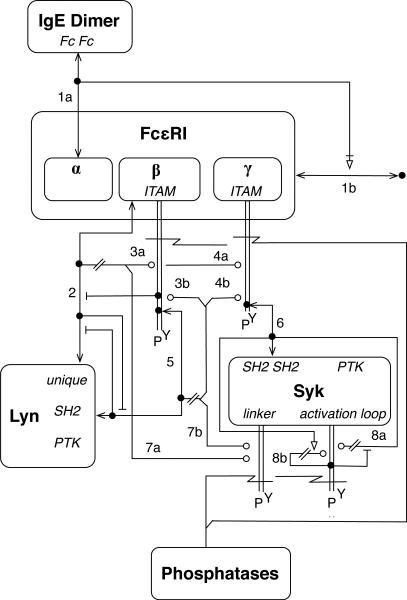
Fig. 3.
A molecular interaction map for the FcεRI model22,23. A MIM illustrates molecules, reactions, and contingencies. A line that begins and ends with an arrowhead represents noncovalent binding, whereas a pair of parallel lines with no arrowheads represents covalent binding. An open circle indicates enzymatic catalysis, an open triangle indicates stimulation, and a jagged line indicates cleavage of a covalent bond. For additional information about MIM conventions, see Kohn et al.66,68. The numbers 1–8 refer to sections of Appendix S1 (ESI). Annotation of this MIM is provided in Appendix S2. Note that this MIM is intended to be read using the combinatorial interpretation of MIM notations66.
Interactions illustrated in a MIM by lines/arrows fall into two categories: direct interactions, or reactions, and contingencies, which characterize how interactions/reactions affect one another. In other words, a MIM depicts molecular interactions as well as the way in which interactions are affected by the context in which they take place. For example, the MIM of Fig. 3 shows that the SH2 domain of Lyn interacts with the phosphorylated β ITAM of FcεRI (see the arrow labeled ‘5,’ which depicts a reaction), and it also shows that this interaction is mutually exclusive with binding of the unique domain of Lyn to unphosphorylated β (see the pair of inhibition arrows between the arrows labeled ‘2’ and ‘5’).
The conventions of Kohn et al.66 do not allow for the explicit representation of molecular substructures and site-specific details of molecular interactions. As shown in Fig. 3, boxes are used to represent molecules, and molecular components are represented using plain text inside molecule boxes. Components are not assigned their own boxes, and there is no provision for subcomponents. Thus, structural relationships can be difficult to visualize. For example, it is difficult to visually suggest that the ‘activation loop,’ identified as a site of phosphorylation in Fig. 3, is located within the PTK domain of Syk. Furthermore, interaction arrows and glyphs for post-translational modifications terminate at the edge of a molecule box, which makes it difficult to identify the components responsible for an interaction or the components affected by post-translational modifications. As demonstrated in Fig. 3, arrows can be positioned to suggest which components are responsible for an interaction, but nevertheless, with respect to representation of interactions at the level of molecular components, the conventions of Kohn et al.66 are somewhat imprecise and less precise than the conventions of Danos et al.62 (cf. Figs. 2 and 3). On the other hand, a MIM provides a clearer picture of the enzymes responsible for post-translational modifications than a contact map (cf. Figs. 2 and 3). The conventions used to draw the MIM of Fig. 3 date back to 2006. An update of these conventions recently became available (http://discover.nci.nih.gov/mim), which allows for better representation of molecular components and site-specific details of molecular interactions69. The updated conventions introduce ‘entity feature’ glyphs, which essentially allow boxes to be used to represent molecular components, not just whole molecules. These conventions differ from those that we will recommend below and do not specifically address visualization or annotation of rule-based models.
Results and Discussion
Having briefly reviewed the background material presented above, we are now prepared to introduce the concept of an extended contact map, which combines features of a plain, model-derived contact map (Fig. 2) with features of a MIM (Fig. 3). Our intention is to provide a means to visualize site-specific details of molecular interactions in cell signaling systems as well as to provide a means to illustrate and annotate rule-based models, which typically account for such details.
One can view the conventions proposed here as a tuning of the established MIM and contact map conventions of Kohn et al.66 and Danos et al.62 to make these conventions more useful for visualization of (large) rule-based models, protein substructures and site-specific details of protein interactions. Our notations are largely consistent with MIM conventions, but there are differences. For example, we introduce nesting of boxes to better represent protein substructures, and we propose the linking of maps to rules, and vice versa. Importantly, because rules are powerful tools for concisely and precisely representing contextual constraints on molecular interactions, we deemphasize the visualization of contextual aspects of interactions.
Below, we first provide an overview of the basic principles of an extended contact map and we then present several example visualizations. These examples serve to elaborate the concept of an extended contact map and to illustrate how various cell signaling processes can be visualized within the framework of an extended contact map. Finally, we discuss the concept of a map guide, which can be associated with an extended contact map to document additional information about the molecules and molecular interactions visualized in the map, particularly the contextual dependencies of the interactions. A map guide can also be used to specify and annotate an executable rule-based model encompassing the molecules and interactions visualized in a map. The model specification may be partial or complete. If a guide serves to annotate a model, it can be referred to as a model guide. The conventions presented here can be used to visualize and annotate an existing model or to depict a set of interactions before they are formalized as rules.
Basic features of an extended contact map
An extended contact map for early events in FcεRI signaling is shown in Fig. 4. In fact, this map illustrates the FcεRI model22,23. A guide for the map of Fig. 4 and the associated model is included in the ESI (Appendix S1) and will be discussed later. The guide lists and annotates proteins and interactions included in the model. Arrows in the map are numbered to correspond to sections in the guide. Each section includes a summary of available knowledge about an interaction and a set of rules. The rules in a set are related, in that they share a common reaction center. In other words, the rules in a set describe the same interaction, but in different contexts. In general, if an interaction depicted in a map occurs in more than one contextual setting, then a rule can be provided for each contextual setting of interest. Also included in the ESI is a version of Fig. 4 that is more aligned with the diagrammatic conventions of SBGN57 (Fig. S3). However, SBGN does not presently provide conventions for illustrating molecular substructures, site-specific details of molecular interactions, or rule-based models. Thus, Fig. S3 serves as a proposal for an extension of the conventions for ER diagrams in SBGN, which complements existing proposals for ER language development (http://sbgn.org/ER development).
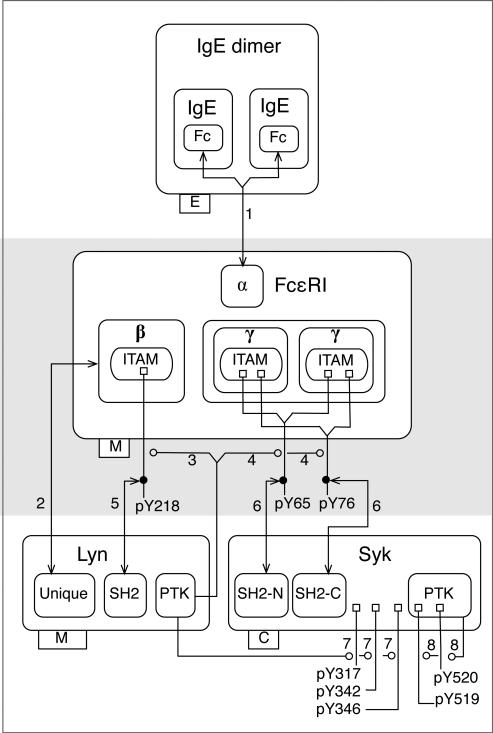
Fig. 4.
An extended contact map for the FcεRI model22,23. Molecules are represented with nested boxes. A direct physical interaction is represented by an arrow that begins and ends with an arrowhead. An enzyme-substrate relationship is represented by an arrow that begins at an enzyme or catalytic subunit box and terminates with an open circle, which identifies the substrate. Tags attached to lower left corners of molecule boxes identify compartmental locations: E stands for extracellular, M stands for plasma membrane (the subunits of FcεRI are transmembrane proteins and Lyn is anchored to the inner leaflet of the plasma membrane), and C stands for cytoplasmic (Syk is able to freely diffuse in the cytoplasm). Phosphatase activity is not depicted in this map, as phosphatases are considered only implicitly in the model22,23. Note that Arrow 5 corresponds to the rule of Eq. (1) and Fig. 1. The labels 1–8 next to arrows refer to sections of Appendix S1 (ESI). Each section includes a summary of available knowledge about an interaction and a set of rules that formally characterize the interaction. Note that the numbered arrows in this diagram correspond to the numbered lines/arrows in the diagrams of Figs. 2 and 3.
The map of Fig. 4 has three layers, which are indicated with shading. The concept of layers is based on the conventions of Kohn et al.70 The top layer includes a depiction of an IgE dimer, the receptor crosslinking reagent that initiates signaling in the FcεRI model22,23. The second layer contains FcεRI, which is the only molecule in the model to interact with IgE. The third layer contains the kinases Lyn and Syk, which interact with FcεRI. In general, the idea is to organize molecules in a layout to reflect the causality of events in cell signaling. A molecule or set of molecules is chosen as the starting point of the signaling process and is placed at a certain location in a map (e.g., at the top), which defines the first layer. The second layer contains molecules that interact with the molecules in the first layer, the third layer contains molecules that interact with molecules of the second layer, and so on. This layout is not strictly a representation of causality or information flow, which is better represented with a path25 (Fig. S1, ESI) or story62. A path or story (i.e., a minimal path) can be used to guide the numbering of arrows and the layering of an extended contact map. For example, Arrows 1, 2, and 3 in Fig. 4 correspond to Steps 1, 2 and 3 in the path of Fig. S1 (ESI).
As can be seen in Fig. 4, nested boxes are used to represent molecules (all proteins in this example) and their component and subcomponent parts. These nested boxes correspond to hierarchical graphs. Lemons et al.65 have recently proposed conventions that allow such graphs to be used to annotate rule-based models. (Incidentally, these conventions are consistent with the related representational formalism of Yang et al.71.) In Fig. 4, components of a protein are arranged linearly with the most N-terminal component at the left and the most C-terminal component at the right, a recommended convention consistent with many diagrammatic representations of proteins. The use of nested boxes allows for explicit representation of the structural relationships among the components and subcomponents of a molecule. For example, the β and γ chains of the receptor are shown to have multiple levels of internal structure: each contains an ITAM, which each contains a tyrosine residue that is a substrate of Lyn. We generally recommend that a protein be depicted in a map only once. A complex can be depicted if the complex is treated as an indivisible unit in a model. In Fig. 4, the γ chain is depicted twice, because the two γ chains are covalently coupled to each other by disulfide bonds and are constituent components of a multimeric protein (FcεRI), which is treated as an indivisible molecular entity in the FcεRI model22,23.
Two types of interactions are illustrated in the map of Fig. 4: direct physical interactions marked by reversible binding and enzyme-substrate interactions marked by covalent bond formation. A direct physical interaction is represented by a line that begins and ends with an arrowhead. The arrows labeled 1, 2, 5, and 6 in Fig. 4 represent direct physical interactions. For example, Arrow 2 indicates that the unique domain of Lyn interacts with the β chain of FcεRI. At the time that the FcεRI model22,23 was originally formulated it was unclear how the unique domain of Lyn interacts with the β chain specifically. Accordingly, the arrow from the unique domain is terminated at the border of the β chain instead of extending further. An enzyme-substrate interaction that results in formation of a covalent bond is represented by an arrow that begins at an enzyme or catalytic domain box and terminates with an open circle at a modification flag, which identifies the modification (i.e., the covalent bond formed) and the substrate. The arrows labeled 3, 4, 7, and 8 in Fig. 4 represent enzyme-substrate interactions. For example, Arrow 3 indicates that Lyn catalyzes phosphorylation of tyrosine 218 in the β ITAM of FcεRI.
As can be seen in Fig. 4, flags are attached to molecule boxes to indicate sites of post-translational modifications. A flag represents a covalent bond between a protein and a functional group (e.g., phosphate) or a small protein, such as ubiquitin. We later demonstrate how a similar notation can be used to represent covalent bonds in general. Post-translational modification flags have three parts: the ‘base’ of the flag is a small square that represents an amino acid residue in a polypeptide chain, the ‘pole’ is a line that represents a covalent bond, and the ‘flag’ itself is a text label. The text label of a modification flag (e.g., pY218) is used to identify the type of modification (e.g., ‘p’ represents phosphorylation) and the location of the modification (e.g., the single-letter amino acid code and number of a residue within a polypeptide chain). If a direct physical interaction depends on a post-translational modification, the arrow representing this interaction may originate/terminate at a modification flag, where a solid dot is placed as a point of origin/termination, in accordance with the conventions of Kohn et al.68. For example, the SH2 domain of Lyn interacts with phosphorylated tyrosine 218 in the β chain of FcεRI; Arrow 5 connects the SH2 domain box of Lyn to a dot on the pY218 modification flag. If an unmodified amino acid must be represented, it is simply drawn as a component, i.e., absence of a modification flag indicates absence of modification. If modification of an amino acid residue inhibits, rather than enables, an interaction, an inhibition arrow originating from a dot on the flag for this modification and terminating at the appropriate interaction arrow may be used to represent the negative effect of the modification on interaction. Flags in maps will tend to correspond to internal states of components of proteins included in a model, and flags will tend to be connected to arrows representing rules that define internal state changes.
It may be useful to point out how Fig. 4 differs from Figs. 2 and 3. Figure 4 contains information not shown in Fig. 2. This missing information in Fig. 2 is information that cannot be directly derived from the BNGL-encoded specification of the FcεRI model22,23, which is given in the ESI (model.bngl). As mentioned above, explicit representation of catalysts is usually missing in BNGL-encoded rules, and the list of rules included in model.bngl (ESI) is not an exception. Thus, enzyme-substrate relationships are not revealed in Fig. 2, whereas such relationships are revealed in Fig. 4. This is one reason why we refer to Fig. 4 as an extended contact map. Another example of information provided in Fig. 4 beyond that provided in Fig. 2 is identification of the individual sites of phosphorylation within the linker region and activation loop of the PTK domain of Syk. When an extended contact map is used to illustrate a model, we recommend that the map illustrate the biological knowledge underlying the model specification, i.e., the information available to the modeler and considered in model formulation. Comparison of an extended contact map and the corresponding model-derived contact map can then reveal how biological knowledge of a cell signaling system has been translated into a formal specification of a model for the system.
Visually, some of the differences between Fig. 3 (a MIM) and Fig. 4 (an extended contact map) may seem super-ficial. However, Fig. 4 introduces conventions that are essential for the consideration of molecular substructure and site-specific details of molecular interactions, most prominently nested boxes for the representation of structural relationships. Another key difference is that Fig. 3 contains information about the contextual dependencies of molecular interactions that is not represented in Fig. 4. For example, binding of an IgE dimer to the α chain of FcεRI is indicated to be a prerequisite for receptor dimerization in Fig. 3, but not in Fig. 4. In fact, Fig. 4 does not explicitly show that FcεRI dimerizes, although this can be inferred. Another example of context depicted in Fig. 3 but not in Fig. 4 is the case of the rightmost phosphorylation glyph attached to Syk. As explained in Appendix S2 (ESI), the various arrows terminating and originating at this glyph are intended to indicate that Syk trans-phosphorylates a second copy of Syk in a dimeric receptor complex and that the rate of phosphorylation is enhanced when the first copy of Syk is phosphorylated in its activation loop. A MIM tends to emphasize the contextual constraints on interactions rather than the component parts of molecules responsible for interactions. The opposite holds true for an extended contact map. We recommend a minimal representation of contextual information in an extended contact map because it is difficult to represent this type of information in the form of a diagram without sacrificing precision and/or readability. Thus, for example, avidity effects such as those considered in the model of Barua et al.27 would not be depicted in a map. In our experience, visualization of contextual dependencies tends to result in an overloaded diagram, especially in the case of large models. Our position is that a rule is usually the best way of capturing the contextual dependencies of an interaction. Therefore, we suggest that interaction arrows in an extended contact map be cross-referenced to a list of rules. As noted above, the interaction arrows of Fig. 4 are labeled 1–8 and these labels correspond to sections of the associated guide of Appendix S1 (ESI), where rules representing the interactions are listed and annotated.
A MIM can serve as a stand-alone summary of available biological knowledge. An extended contact map can also serve the same purpose. However, we recommend that a map always be accompanied by a guide containing rules for interactions. The guide need not fully specify a model. For example, a guide containing rules but omitting rate laws for rules, which are required for simulations, can still be useful, because rules are suitable for providing details that are not easily captured in a map. A MIM can be supplemented with annotation (for example, see Kohn et al.70). What is different here is that we are proposing that the annotation associated with an extended contact map include formal elements of an executable rule-based model, especially rules. It should be noted that rules, because they are formal representations of interactions, are more easily associated with arrows in a map, which also are representations of interactions, than the formal elements of a conventional model. In an ODE-based model, for example, multiple terms in multiple equations are typically required to capture the effects of a single interaction14,62. For an example of a MIM for which a corresponding conventional model is available, see Kim et al.72.
We have now introduced the basic features of an extended contact map by way of example. Below, we give additional guidance about the representation of molecules and molecular interactions before introducing several additional simple examples, which illustrate cell signaling processes and types of molecules that are not included in Fig. 4 but that are commonly found in cell signaling systems.
General guidelines for representation of molecules
As described above, proteins in an extended contact map are represented with nested boxes that correspond to hierarchical graphs, and sites of post-translational modifications are marked with modification flags. Recommended box and flag glyphs are summarized in Fig. 5. The components of a protein are ordered from N-terminal to C-terminal. When this type of ordering is not possible, as with separate polypeptide chains in a multimeric protein, individual polypeptides may be arranged in a way that reflects their physical organization. For example, in the case of a multimeric cell-surface receptor (e.g., FcεRI), a mostly extracellular subunit (e.g., FcεRIα) may be placed above other mostly cytoplasmic subunits (FcεRIβ and γ2).
Fig. 5.
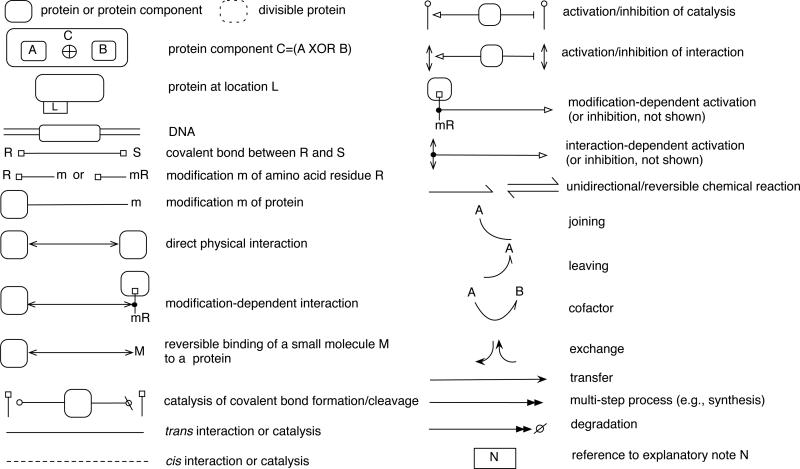
Boxes, flags, arrows and other symbols that can be used to draw an extended contact map. All of these glyphs are used in example diagrams that follow; see the main text for discussion. The label ‘m’ represents a post-translational modification. A separator, such as ‘:,’ can be inserted between the two parts of a modification flag label. Thus, ‘mR’ and ‘m:R’ are both acceptable forms of a modification flag label. For post-translational modifications commonly involved in cell signaling73, we recommend the following labels: Ac for acetylation, Me for methylation, OH for hydroxylation, p or P for phosphorylation, and Ub for ubiquitination. The ‘joining,’ ‘leaving,’ and ‘cofactor’ arrows are intended for use in combination with chemical reaction arrows. Note that the joining and leaving arrows together equal a cofactor arrow. In the ESI, the glyphs shown here are available electronically in the form of an OmniGraffle stencil package (Contact Maps.gstencil). OmniGraffle is a general-purpose drawing tool that is commercially available for the Mac platform (The Omni Group, Seattle, WA). The ESI also contains instructions for use of the stencil package (see README.txt). Updates of the stencil package will be made available at the BioNetGen web site (http://bionetgen.org).
To maintain compactness of a diagram, we recommend that only components of interest (e.g., domains, motifs, and amino acid residues that are included in a model) be shown in a map. For a map illustrating a BNGL-encoded rule-based model, the representation of molecules should reflect the BNGL molecule type definitions15 of the model. A more complete annotation of known molecular substructure can be included in a map/model guide if desired. In addition, a molecule is generally only shown once in an extended contact map, with the exception of molecules that are represented using plain text (see below) and molecules that are present in multiple copies in a complex (e.g., the γ chains of FcεRI). To avoid redundancy in the depiction of post-translational modifications, we recommend that the line segments (i.e., the poles) of modification flags attached to repeating component boxes in a map be consolidated so that they emerge from a molecule box as a single line. An example of this practice is shown in Fig. 4; see the pY65 and pY76 flags.
In addition to representing protein substructure, an extended contact map can provide other information about a protein, namely its location(s) and products of proteolytic cleavage. To indicate the possible compartmental locations of a protein, one can attach a compartment tab to a molecule box. Labels within the tab represent different compartments. A label need not be included for a compartmental location that can be inferred. For example, the compartment tab of the Syk molecule box in Fig. 4 contains the label ‘C’ (cytosolic) but not ‘M’ (plasma membrane). This is because membrane association of Syk can be inferred by the association of Syk with FcεRI, a membrane protein. If a model includes rules for translocation of proteins, such as the rule of Eq. (2), a tab can be associated with multiple labels to indicate all of the compartments in which a protein can be found, and the compartment tab can also be associated with a set of translocation rules, which can be listed and annotated in a model guide. To indicate that a protein is divisible (i.e., cleaved by the action of a protease into two or more smaller proteins), one can use a dotted molecule box to represent the protein. This also applies to the representation of divisible components. However, a dotted box should only be used when the protein fragments that result from proteolytic cleavage are relevant for understanding the system depicted in a map. One would not use a dotted molecule box to simply indicate that a protein is degraded.
Here, we emphasize visualization of proteins, but an extended contact map can also include other types of macromolecules, such as DNA, as well as small-molecule compounds, such as lipids, drugs, and metabolites. We recommend that boxes be reserved for macromolecules and we recommend that small-molecule compounds be represented using plain text.
General guidelines for representation of molecular interactions
Interactions among molecules are visualized with arrows in an extended contact map (Fig. 5). The same set of interactions can also generally be represented with rules, and thus an arrow in a map can be linked to one or more rules in a model. This connection is made through a model guide: arrows in a map are numbered, and rules and sections in a model guide are numbered to correspond with arrows. An arrow may correspond to more than one rule if a set of rules share a reaction center. A reaction center is defined as the set of vertices (components) that undergo modification in a graph-rewriting operation defined by a rule15. When a reaction center is common to multiple distinct rules, it means that the rules are representing a common interaction that takes place in multiple contexts. Rules that share a common reaction center can be mapped to a single interaction arrow in an extended contact map and the contextual differences need not be captured in the map, as these differences are accounted for in the rules themselves.
It is important to note that arrows are drawn as specifically as possible; in other words, they extend as many layers into the molecule as available knowledge allows, but not further. If an exact binding site is not known, an arrow is terminated at an outer layer and may even terminate at the outermost border of a molecule box. To accommodate space limitations in a map, arrows may branch. As seen in Fig. 4, a catalysis arrow from Lyn branches to show phosphorylation of the β and γ chains. When an arrow branches, a short diagonal segment or pair of diagonal segments can be introduced, which helps identify the box from which the arrow originates (see Arrows 1, 3 and 4 in Fig. 4). A catalytic arrow can be broken and extended to point to multiple modifications flags (see Arrows 4, 7 and 8 in Fig. 4). If an arrow crosses a modification flag that it does not affect, it may be drawn continuously or broken into segments; breaking of a line into segments is a stylistic option that does not affect the meaning of an arrow. Recommended arrows are summarized in Fig. 5. Unless otherwise noted, all arrows drawn with solid lines should be assumed to depict trans interactions; cis interactions are depicted with dotted lines. This convention can be reversed if convenient, e.g., in a case where most arrows in a map represent cis interactions. A reversal of the convention should be duly noted.
Example visualizations of common cell signaling processes
We now demonstrate how the conventions described above can be used to represent various biochemical processes found in cell signaling systems (Figs. 6–8). BNGL-encoded rules to accompany these diagrams are provided in Appendix S3, which serves as a primer on using rules to represent cell signaling processes. Other primers are available15,74,75.
Fig. 6.
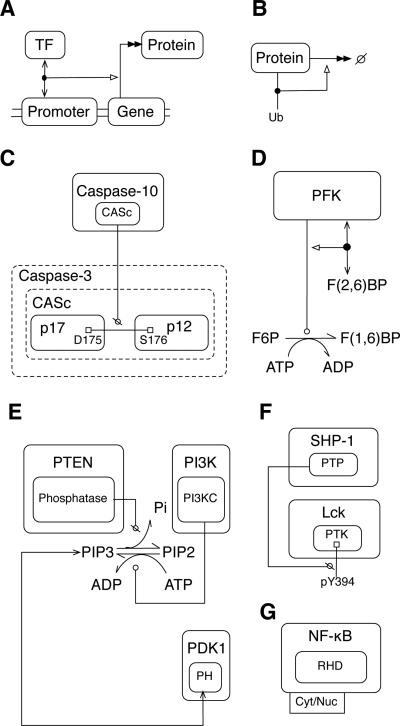
Visualization of various cell signaling processes. A) Stimulation of transcription and translation by transcription factor (TF) binding to DNA. B) Stimulation of protein degradation by ubiquitination. C) Proteolysis. Dotted box lines identify the parts of the caspase-3 polypeptide chain affected by the proteolytic action of caspase-10. Cleavage of the indicated peptide bond breaks the chain itself and the CASc segment. D) Allosteric regulation of a metabolic reaction, conversion of F6P (fructose 6-phosphate) to F(1,6)BP (fructose 1,6-bisphosphate). The effector is F(2,6)BP (fructose 2,6-bisphosphate). E) Lipid phosphorylation and dephosphorylation, and PH domain interaction with phosphatidylinositol (3,4,5)-trisphosphate (PIP3). The small-molecule metabolites ATP and ADP and inorganic phosphate (Pi) are shown to participate in the reactions between phosphatidylinositol (4,5)-bisphosphate (PIP2) and PIP3 to illustrate how such details can be represented in a map if desired. However, such details would normally be omitted for simplicity. F) Protein dephosphorylation. G) A protein (NF-κB) that traffics between the cytoplasm (Cyt) and nucleus (Nuc).
Fig. 8.
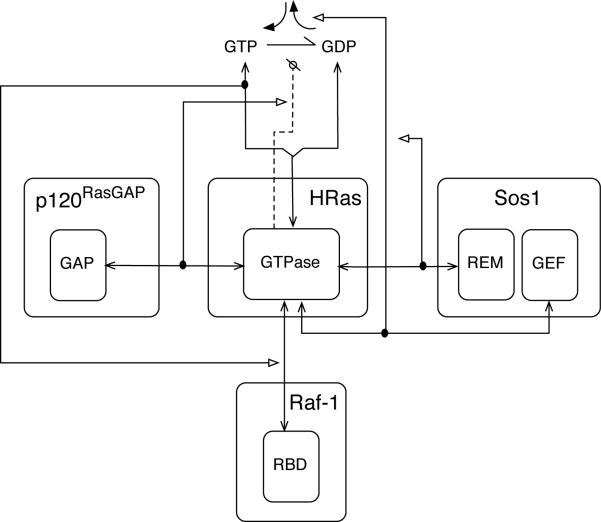
Visualization of Ras regulation. HRas, which has GTPase activity, is drawn with branched arrows pointing to GTP and GDP to indicate that HRas contains a single binding site for the guanine nucleotides. The reaction arrow between GTP and GDP represents the transition from one bound state of GTPase to another. A catalysis arrow is drawn from the GTPase domain of HRas to the GTP-to-GDP reaction arrow to represent the intrinsic catalytic activity of HRas. The arrow is dotted to indicate that it represents a cis interaction (i.e., the GTPase acts on a GTP molecule that is bound to itself). A stimulation arrow is drawn from the GTPase-GAP interaction arrow to the catalysis arrow to indicate GAP-mediated upregulation of GTPase activity. Exchange of GDP for GTP is represented with a pair of bent arrows, and an activation arrow indicates that Sos1 stimulates exchange. Sos1 is allosterically activated by HRas binding to the REM domain.
Protein synthesis and interaction of a transcription factor with a DNA binding site
According to the central dogma of molecular biology, protein synthesis consists of two basic steps: transcription of DNA into mRNA, and translation of mRNA into a polypeptide76. These steps may be regulated in many ways and additional steps may be involved in de novo protein synthesis; however, we are often only interested in the relationship between a gene and its protein product. In this case, one can use a shorthand notation to indicate synthesis of a protein encoded by a gene (Fig. 6A). A double-headed arrow points from a molecule box for a gene to a molecule box for a protein to represent the multistep process of transcription/translation. The double arrowhead is intended to suggest that steps are not shown. DNA is represented as a pair of parallel lines, and boxes for genes, promoters and other regulatory elements are embedded within these lines. This example also shows binding of a transcription factor (TF) to DNA and indicates that this interaction stimulates transcription/translation. A solid dot placed on the DNA-TF interaction arrow serves as a point of origin for an activation arrow. In general, a dot is placed on an arrow when it is necessary for another arrow to begin or end at that point. A similar combination of symbols could be used to represent other synthetic processes.
Proteolysis and protein degradation
Cells routinely degrade proteins: unnecessary or misfolded proteins are dismantled, and protein degradation is used to regulate the rates of biochemical reactions. Much protein degradation takes place in proteasomes76. In an extended contact map, degradation can be simply depicted as a double-headed arrow pointing from the degraded protein to a ‘null’ symbol (Fig. 6B). Proteases catalyze cleavage of peptide bonds between amino acids. This process has a role in protein degradation as well as in regulation of enzymatic activity. For example, caspase signaling involves caspase-catalyzed cleavage of caspase proteins, which liberates enzymatic subunits to assemble into active caspase enzymes77. The uncleaved form of a protein may be represented with a dotted border, indicating that it is divisible (Fig. 6C). The proteins that result from the cleavage event are represented within this box. They are connected by a solid line with squares at either end, representing a covalent bond. A ‘no’ arrowhead points from the catalytic domain of a protease to the covalent bond, indicating that the bond is cleaved. A more elaborate example of representation of a proteolytic cascade is provided in Fig. S4, which depicts proteolytic cleavage of complement component C3 to C3d78–80. This figure illustrates how a proteolytic cascade that results in cleavage of a protein at multiple sites can be represented in an extended contact map.
Allosteric regulation of a metabolic reaction
Allosteric regulation occurs when an effector molecule alters an enzyme's activity by binding to a site on the enzyme that is distinct from the active site. The result may be either an increase or decrease in catalytic activity. An example of an enzyme controlled by allosteric regulation is phosphofructokinase-1 (PFK-1). This enzyme catalyzes a key, irreversible step in the glycolysis pathway, and it is a central point of regulation. For example, PFK-1 is positively regulated by fructose-2,6-bisphosphate76. In an extended contact map, allosteric regulation of enzymatic activity by a small-molecule effector is represented as follows. A direct physical interaction arrow is drawn between the enzyme and effector. An activation or inhibition arrow then originates from the interaction arrow and points to the catalysis arrow between the enzyme and substrate (Fig. 6D). We generally discourage the use of activation and inhibition arrows because they tend to be ambiguous, but they are useful for representing allosteric regulation. In this example, plain text is used to represent metabolites, rather than boxes, to make a distinction between small molecules and macromolecules. If a material component considered in a model is not treated as a structured object (i.e., a graph) in a model, it and the reactions in which it participates can be represented using conventional means for representing biochemical reaction networks.
Dephosphorylation
Representation of phosphorylation is demonstrated in Fig. 4. The reverse process, dephosphorylation, is the enzyme-catalyzed removal of a phosphate group from an amino acid residue. Dephosphorylation can be just as important as phosphorylation in regulating protein interactions and catalytic activities. Unregulated basal dephosphorylation by unspecified phosphatases can be omitted from an extended contact map, as in Fig. 4, because it would necessitate an additional arrow for every phosphorylated residue, making the map less readable. However, it is sometimes significant that a specific phosphatase acts on a specific substrate. For example, dephosphorylation of the C-terminal regulatory tyrosine in the kinase Lck by SHP-1 prevents the formation of an intramolecular bond, which regulates Lck kinase activity81. As in the MIM of Fig. 3, Kohn and co-workers use a jagged line to represent dephosphorylation68. As an alternative that is more compact and more consistent with our notation for catalysis of covalent bond formation, we suggest depicting dephosphorylation (and more generally cleavage of a covalent bond) with a ‘no’ symbol (Fig. 6F). In the case of lipids (e.g., dephosphorylation of phosphatidylinositol (3,4,5)-trisphosphate by PTEN82), dephosphorylation can be represented as a standard chemical reaction with a catalysis arrow pointing from the enzyme to the reaction (Fig. 6E). We also use this example to demonstrate an interaction between a lipid and a protein: PIP3 binds the pleckstrin homology (PH) domain of PDK1, recruiting PDK1 to the plasma membrane83.
Transport
An extended contact map does not aim to illustrate transport or trafficking between compartments, but a map can be used to indicate compartmental locations of molecules. Compartments and transport between compartments can be represented explicitly using cBNGL51. The names of the compartments in which a molecule can be found can be included in an extended contact map in the form of a tag attached to a molecule box. In Fig. 6G, two location labels, ‘Cyt’ and ‘Nuc,’ are included within a single location tab attached to a molecule box for NF-κB. The tag indicates that NF-κB is considered to have two possible compartmental locations. A location tag can be associated with a rule, such as the rule of Eq. (2), to clarify details about trafficking between compartments. In the case of Fig. 4, molecules are considered that are found in three compartmental locations, and all the molecules are represented in the same map. In more complicated cases, it may be convenient to draw separate maps for separate compartments. Note that compartmental locations that can be inferred from interactions need not be included in a map. For example, the location tag attached to the Syk molecule box in Fig. 4 only indicates that Syk is cytoplasmic. It can be inferred that Syk is membrane associated when it interacts with FcεRI, so a membrane location label is not included in the Syk location tag.
Association
The extended contact map of Fig. 4 demonstrates how direct physical interactions between protein binding partners (see Arrows 1 and 2) and phosphorylation-dependent interactions (see Arrows 5 and 6) can be represented. Interactions that depend on other types of post-translational modifications can be represented in the same way as a phosphorylation-dependent interaction. A direct physical interaction between a protein and DNA can be represented as shown in Fig. 6A. A direct physical interaction between a protein and a small molecule can be represented as shown in panels D and E of Fig. 6. If two proteins are associated indirectly via an unknown linker, the boxes representing the proteins can be connected via a direct physical interaction arrow and the arrow can be attached to a note tag, a rectangle enclosing a reference to a note of explanation.
Conjugation and transfer: ubiquitin and ubiquitin-like proteins
Ubiquitin is a small protein that may be covalently coupled to copies of itself and to other proteins. Ubiquitination (Ub) tags proteins for degradation and serves various other functions84.
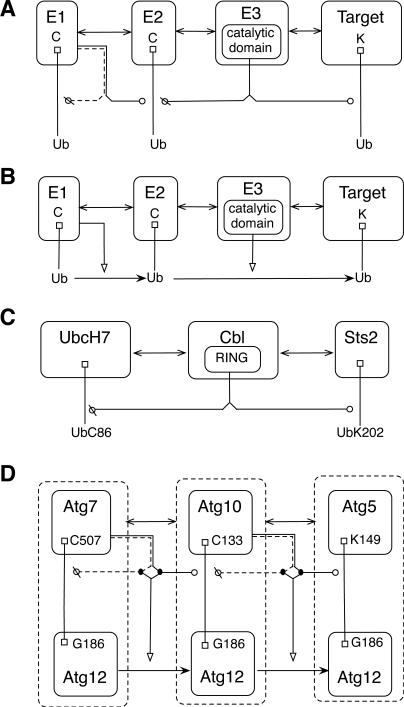
Representation of ubiquitination can be similar to representation of phosphorylation: a catalysis arrow can point from an enzyme to a substrate, where the type of modification (‘Ub’ for ubiquitination) and the location of the modification are specified. However, unlike phosphorylation, multiple enzymes are involved in the ubiquitination process: an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase. Ubiquitin is bound to a cysteine residue in the active site of E1, transferred to the active site of E2, and then bound to the target substrate in a reaction catalyzed by E386. Representation of ubiquitination in an extended contact map may vary. A detailed representation of ubiquitination includes all three enzymes and the target substrate. Arrowheads representing catalysis of covalent bond formation and cleavage can be used to implicitly represent transfer of Ub from one protein to the next (Fig. 7A). These reactions result in transfer of Ub, which can be alternatively represented with a transfer arrow, as depicted in Fig. 7B. Note that the arrowheads used for binding and transfer arrows are similar but distinct. See Fig. 5. Further note that Figs. 7A and 7B need not represent different models; the two diagrams could represent the same set of rules. In Fig. 7A, the dotted arrow from E1 indicates that an E1 enzyme removes ubiquitin from itself, rather than from a second E1 molecule. A more specific representation of ubiquitination in the style of Fig. 7A is shown in Fig. 7C. In some cases, specific residues in ubiquitin or ubiquitin-like proteins may be of interest. In the example of Fig. 7D, a specific glycine residue in the ubiquitin-like protein Atg12 is shown to form covalent bonds with specific residues in Atg7, Atg5, and Atg1087. In addition, activation arrows point from catalytic arrows to transfer arrows, which represent the sequential transfer of Atg12 from Atg7 to Atg10 to Atg5. The activation arrows, which emerge from dots on the catalytic arrows, are intended to indicate that enzyme-catalyzed cleavage and formation of the indicated covalent bonds serve to transfer Atg12. Dashed borders for the molecules containing Atg12 indicate that these entities are divisible. Note that Fig. 7D illustrates how the styles of Figs. 7A and 7B can be combined. Lastly, it is worth mentioning that monoubuiqitination can be distinguished from polyubiquitination (i.e., formation of a ubiquitin chain86) in the label of a modification flag. For example, the label ‘UbnK’ can be used to represent a chain of n ubiquitin molecules.
Fig. 7.
Visualization of ubiquitin and ubiquitin-like protein conjugation and transfer reactions. A) A general representation of ubiquitination that includes E1, E2, and E3 enzymes and a target substrate. E1 catalyzes cleavage of ubiquitin from itself and formation of a covalent bond between ubiquitin and E2. E3 catalyzes cleavage of ubiquitin from E2 and formation of a covalent bond between ubiquitin and the target. B) In an alternative representation, ubiquitination is represented by stimulated transfer of ubiquitin from one molecule to the next. C) A specific representation of ubiquitination showing only an E2 (UbcH7), an E3 (Cbl), and a target (Sts2), which is ubiquitinated at K20285. D) A specific glycine residue in the ubiquitin-like protein Atg12 is shown to form covalent bonds with Atg7, Atg5, and Atg10. Activation arrows point from catalytic arrows to transfer arrows. Dashed borders surrounding molecules containing Atg12 indicate that these entities are divisible.
Exchange: Ras
GTPases in the Ras family of proteins are hydrolase enzymes that bind and act on guanosine triphosphate (GTP) to yield guanosine diphosphate (GDP). In cell signaling, GTPases function as switches, being ‘on’ when bound to GTP (i.e., able to bind an effector) and ‘off’ when bound to GDP (i.e., unable to bind an effector). Transitions between these two states are mediated by GTPase activating proteins (GAPs), which stimulate a GTPase's intrinsic catalytic activity thereby accelerating the rate at which GTP is converted to GDP, and guanine nucleotide exchange factors (GEFs), which facilitate exchange of GDP for GTP by loosening the binding of a GTPase to both GTP and GDP. GTP is at a higher concentration than GDP in cells and is more likely to bind an empty binding site. HRas is a GTPase that is acted upon by p120RasGAP, a GAP, and by Sos1, a GEF88. In Fig. 8, HRas is drawn with a branched interaction arrow pointing to GTP and GDP. A unidirectional chemical reaction arrow from GTP to GDP represents the conversion of GTP to GDP. A cis (dashed) catalytic arrow from HRas to the reaction arrow indicates that HRas catalyzes the cleavage of a covalent bond and converts GTP to GDP. Exchange of GDP for GTP is represented with a special exchange glyph consisting of a pair of bent arrows. An activation arrow from the p120RasGAP-HRas interaction arrow indicates that RasGAP stimulates GTPase activity. An activation arrow from the HRas-Sos1 interaction arrow pointing to the exchange glyph indicates that Sos1 stimulates GTP/GDP exchange. As depicted in Fig. 8, interaction between HRas and the REM domain of Sos1 allosterically activates GEF activity89. The HRas molecule that allosterically activates Sos1 is distinct from the HRas molecule affected by the GEF activity of Sos1, and GDP- and GTP-loaded HRas have different allosteric effects, but these distinctions are not made in an extended contact map. Instead, rules in an associated model guide would clarify the mechanism depicted in the map. See Appendix S3. As depicted in Fig. 8, the GTP-bound form of HRas is able to bind Raf-190. The dependence of this interaction on GTP loading is indicated by the activation arrow extending from a solid dot on the GTP-HRas interaction arrow to the interaction arrow between HRas and Raf-1. The diagram of Fig. 8 contains a number of activation arrows. As mentioned earlier, we generally discourage the use of activation and inhibition arrows, but Fig. 8 provides an example of where these arrows are useful for representing allosteric regulation.
Example visualizations of miscellaneous molecule types
We will now demonstrate how various molecule types not yet considered may be represented (Fig. 9).
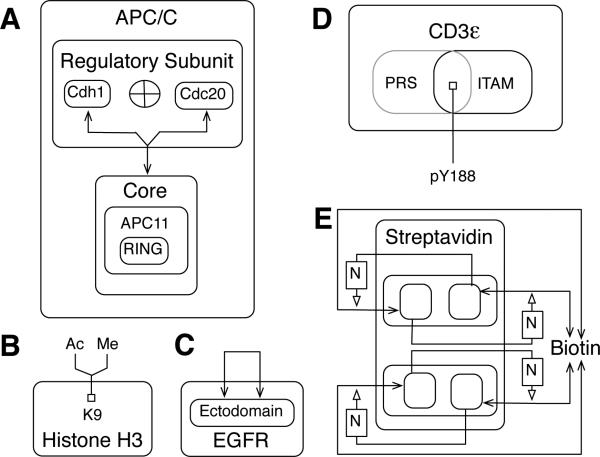
Fig. 9.
Visualization of various molecule types. A) A multimeric protein with two possible regulatory subunits. The two possible regulatory subunits of APC/C, Cdh1 and Cdc20, are shown as boxes within a ‘Regulatory Subunit’ box, separated with an ‘XOR’ (exclusive or) symbol to indicate that only one of these proteins may associate with the core at a time. B) A protein containing a site that can be modified in multiple ways. C) A protein that dimerizes. D) A protein with overlapping linear motifs. Boxes of overlapping components are distinguished by different shades (as shown) or colors. The PRS box is gray and the ITAM box is black. E) Discontinuous biotin binding sites of streptavidin. The use of activation arrows in this diagram can be considered nonstandard, so a note box is included to provide a note of explanation. The letter ‘N’ serves to label an explanatory note, ‘adjacent monomers form biotin binding sites.’ In general, we recommend that explanatory notes be labeled with letters, although longer labels are acceptable. Note that the monomeric subunits of streptavidin, a dimer of dimers, are not labeled, nor are the constituent dimers.
Divisible proteins
All proteins are divisible, i.e., their peptide bonds may be cleaved. However, in some models it is relevant to track the cleavage of a particular protein. In such cases, a special notation for divisible proteins is useful. A protein that may be cleaved is represented with a dotted molecule box, which encloses the fragments that result from cleavage. A divisible protein, caspase-3, is visualized in Fig. 6C. Caspase-3 is cleaved by the action of caspase-10, which allows the p17 and p12 components of the CASc domain of caspase-3 to assemble into an active caspase77. A representation of complement component C3 is given in Supplemental Figure S4 (ESI).
Alternate subunits: APC/C
Many enzymes are multimeric proteins. An example is APC/C, a cullin-RING domain E3 ubiquitin ligase, the specificity of which is determined by a regulatory subunit. The regulatory subunit can be either Cdh1 or Cdc2091. In Fig. 9A, a component box is introduced for a regulatory subunit in which the two possible components are included, separated by an XOR symbol, indicating that only one may be associated with core APC/C at a time.
Sites of multiple modifications: Histone H3
Histone modification regulates chromatin structure. As depicted in Fig. 9B, lysine 9 in histone H3 may be modified in two possible ways, by acetylation and by methylation. The balance between these two modifications may influence gene regulation over the course of the cell cycle92.
Homodimer: EGFR
Binding of epidermal growth factor (EGF) to the EGF receptor (EGFR) leads to formation of EGFR dimers. As depicted in Fig. 9C, receptors dimerize via ectodomain interactions93. Note that the arrow in Fig. 9C represent a trans interaction.
Overlapping linear motifs: CD3ε
The CD3ε chain of the T-cell receptor (TCR) contains a proline-rich sequence (PRS) and an ITAM that overlap. In the region of overlap there is a tyrosine residue (Y188), which is a substrate of kinases and phosphatases. As part of the ITAM, Y188 is phosphorylated during TCR signaling. Phosphorylation of Y188 inhibits binding of the PRS to SH3 domains in interaction partners, and binding of the PRS inhibits phosphorylation of Y18894. Thus, it is relevant to show that the PRS and ITAM overlap. In Fig. 9D, the PRS and ITAM are represented as overlapping boxes with Y188 located in the overlapping region. The two component boxes can be distinguished by using box lines that differ in shading (as shown) or color. In complicated cases, it may be necessary to explain overlaps in a note or map/model guide.
Discontinuous binding sites: biotin and streptavidin
Binding sites may be composed of parts of distinct components of a protein or protein complex, and there are various possibilities for how such binding sites and their interactions can be represented in an extended contact map. For example, the four biotin binding sites in a streptavidin tetramer are formed by residues of adjacent monomers that interact as functional dimers95. In Fig. 9E, the interaction of biotin with a streptavidin monomer is shown to be activated by a neighboring monomer. This diagram can be considered nonstandard. In such a case, a reference to an explanatory note can be included in a diagram. Here, ‘N’ is a label that refers to the explanatory note ‘adjacent monomers form biotin binding sites.’ In general, a rectangle enclosing a label can be introduced to clarify aspects of map by providing a reference to a note of explanation.
Basic features of a map/model guide
An extended contact map can be associated with a map guide or a model guide. A map guide complements an extended contact map by providing annotation about molecules and interactions visualized in a map. A model guide goes beyond a map guide by attaching formal elements of a rule-based model, molecule type definitions and rules, to boxes and arrows. An example of a model guide is provided in Appendix S1 (ESI). We recommend that a model guide be organized so that sections in the guide correspond to blocks of a BioNetGen input file15. A model guide essentially serves as a specification of a rule-based model, although the specification need not be complete. It can serve to annotate not only an extended contact map but also the underlying model illustrated by the map. We recommend that rules in a model guide be specified using BNGL15 because of the availability of various BNGL-compatible software tools11–13,15–17,41,42,45,50. However, any language for specifying rule-based models could be used.
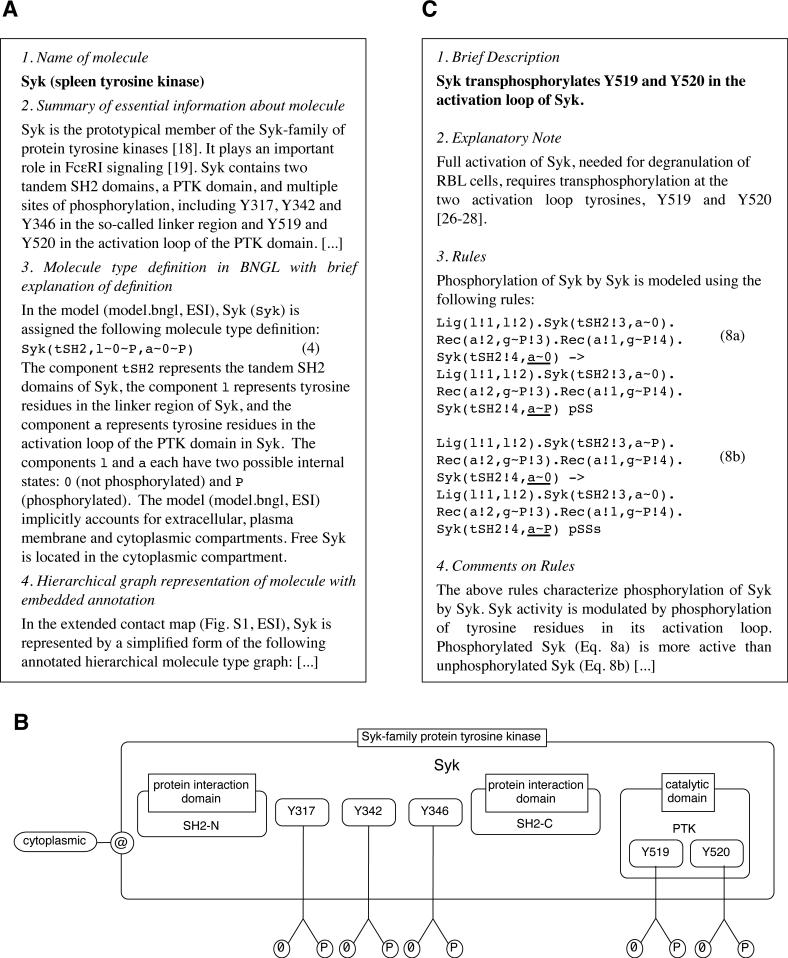
A guide may contain representations of molecules in the form of BNGL molecule type definitions15,49. A molecule type definition includes a list of internal states for all components that have internal states, as well as locations for components if one is using cBNGL51. A guide may also contain additional information that is not included in an extended contact map, such as links to online resources (e.g., UniProt96, Pfam97, and Phopho.ELM4), a narrative summary of available information about a protein, and estimates of protein copy numbers. A guide can include diagrams of complete domain structures of proteins in the form of domain graphs98 and/or diagrams that define the compartmental locations of molecules. Such diagrams can be included in a guide to provide a more complete picture of individual proteins. As discussed previously, only components of interest are included in protein representations in an extended contact map; a protein may contain other elements, but depicting all of them in a map is discouraged, in part because the practice would tend to make maps difficult to read. In the case of a map used to illustrate a model, protein representations should reflect the components considered in the formulation of the model. Consistent with the conventions of Kohn et al.66, a modification flag in a map only indicates the modified state of an amino acid residue, even though a residue may also have an unmodified state. An unmodified state may be specified in a guide if desired. In rule-based models, post-translational modifications are often represented using internal states, which are simply variable attributes associated with vertices of graphs. The value of an attribute associated with a particular modification state is arbitrary. Thus, it can be useful to specify a mapping of modification states of an amino acid residue (including an unmodified state) to the values of the corresponding internal state attribute in a model. Figure 10A shows annotation for Syk included in the example model guide (Appendix S1, ESI). In Fig. 10B, a diagram of Syk is shown with embedded annotation for the molecule and individual components (e.g., the SH2 domains of Syk are identified as protein interaction domains), possible internal states (‘0’ for unmodified and ‘P’ for phosphorylated), and compartmental location (‘cytoplasmic’).
Fig. 10.
Excerpts from the model guide of Appendix S1 (ESI). A) A protein annotation includes the name of a protein, a molecule type definition in BNGL, and a summary of relevant information from the literature. Other information may be included as well, such as a UniProt accession number (http://uniprot.org) and experimental data. B) Illustration of Syk with embedded annotation. This panel demonstrates how an ad hoc illustration of a molecule can be included in a guide to supplement a standardized representation of a molecule in a map. C) An interaction annotation includes a brief description of the interaction, a listing of all rules that characterize the interaction, and an explanation of modeling assumptions.
A map guide also serves to annotate the interactions represented by rules. Each interaction arrow in an extended contact map corresponds to either a rule or a set of rules in which all rules contain a common reaction center. An interaction annotation, such as that shown in Fig. 10C, has three parts: a summary of available information about an interaction, including citations from the primary literature; the rules used to model the interactions and/or to summarize the contextual dependencies of the interactions; and an explanation of the rules, including modeling assumptions. If a guide describes a fully specified model, rules will be associated with rate laws and estimates of parameters in the rate laws.
Typically, rules contain contextual information, but every interaction in an extended contact map can be trivially associated with a context-free rule. Thus, every extended contact map corresponds to a set of rules that comprise an executable model composed of context-free rules. A context-free rule is one in which all components are part of a reaction center. Consider the rules of Eqs. (1) and (2), which include contextual components: U and RHD, respectively. If these contextual components are omitted, the rules of Eqs. (1) and (2) become context-free rules.
An extended contact map (e.g., Fig. 4) and a model guide (e.g., Appendix S1, ESI) capture more details about a biological system than a BNGL-encoded specification of a model for the system (e.g., model.bngl, ESI) or a plain model-derived contact map (e.g., Fig. 2). As discussed previously, explicit representations of enzyme-substrate interactions are often omitted from rules, which is reflected in a model-derived contact map. In contrast, enzyme-substrate relationships are shown in an extended contact map. For example, Lyn-mediated phosphorylation of the linker region in Syk is shown in Fig. 4 but not in Fig. 2. The reason for extra details being included in an extended contact map is that these details are considered in the formulation of a model. If information is collected by a modeler and used to formulate a model, the information should not be lost or separated from a model specification simply because model simulations do not require the explicit incorporation of the information into the formal elements of a model. In addition, an extended contact map and model guide elucidate modeling assumptions. For example, the BNGL-encoded specification of the FcεRI model22,23 (model.bngl, ESI), contains a number of modeling assumptions, such as the lumping together of multiple tyrosine residues in the linker region of Syk as a single component, l. Accordingly, a l component appears in Fig. 2, without information about the tyrosine residues that are phosphorylated. In contrast, Fig. 4 identifies three tyrosine residues in the linker region that are phosphorylated during signaling. Fig. 4 also identifies specific tyrosine residues in the activation loop of the PTK domain of Syk and in the β and γ ITAMs of the receptor that are not shown in Fig. 2. As illustrated by these examples, an extended contact map and a model-derived contact map can be compared to reveal the assumptions of a model.
A guide can be used to specify and annotate a rule-based model, and an extended contact map can be used to illustrate the model. The map provides an extended description of the model, one that goes beyond that provided by the formal model specification. For example, Fig. 4 provides an extended description of the model specified in Appendix S1 (ESI), in that Fig. 4 is more detailed than Fig. 2, which is derived directly from the model and is therefore representative of the formal model specification. Although Fig. 4 is more detailed than Fig. 2, Fig. 4 is restricted in scope to the same molecules, molecular components, post-translational modifications, and interactions considered in the FcεRI model22,23. Consider dephosphorylation. Phosphatases play an important role in regulating FcεRI signaling99 but no specific phosphatases are included in the model. Instead, unspecified phosphatases are assumed to be available in excess. Accordingly, no phosphatase is shown in Fig. 4. Similarly, phosphorylation and dephosphorylation of a C-terminal tyrosine residue of Lyn is important for regulating Lyn activity and FcεRI signaling99, but this residue is not included in the model. Rather a certain fraction of total Lyn is assumed to be in active form, a form in which the C-terminal regulatory tyrosine is not phosphorylated. As a general guideline, we suggest that an extended contact map be drawn to reflect the biological knowledge that underlies the model being illustrated by the map.
Tools for drawing maps
The diagrams presented above were handcrafted using a general-purpose drawing tool, OmniGraffle (The Omni Group, Seattle, WA). The diagrams that appear in Figs. 4-10 are provided electronically in the templates.graffle file in the ESI. The ESI also contains an OmniGraffle stencil package, which provides access to the glyphs of Fig. 5 and should facilitate rapid construction of maps compliant with the guidelines recommended here. For instructions on using the stencil, see README.txt (ESI). OmniGraffle is only available for the Mac platform. Comparable software available on the Windows platform includes Microsoft Visio. Files can be exchanged between OmniGraffle and Microsoft Visio using the Microsoft Visio XML file format. We provide no software for automatically drawing an extended contact map for a given set of rules or for automatically writing context-free rules for a given map. The requirement for manual construction of a map should not be onerous but there are potential pitfalls. For example, a map could be drawn incorrectly so that it is not entirely consistent with an underlying model as intended, or during the process of model development, map and guide updates could fall significantly out-of-sync. However, our goal has been to present a set of standards that are easy to follow and, if followed, should facilitate the understanding and reuse of rule-based models.
To provide software for automatically drawing an extended contact map, we will first need to formalize the relationship between a model and a map and then extend one of the languages for specifying rule-based models (e.g., BNGL or Kappa). These languages do not currently provide a satisfactory means for encoding all of the information that one may wish to visualize in an extended contact map. For example, the catalyst responsible for a reaction represented by a rule is not usually discernible from the rule specification alone. An extension of BNGL could perhaps be introduced to allow for the identification of catalysts and enzyme-substrate interactions in the form of metadata attached to rules or to incorporate the hierarchical graphs of Lemons et al.65 for more natural representation of structural relationships. The development of software for drawing extended contact maps, such as the software available for drawing contact maps like that of Fig. 2, is beyond the intended scope of the work presented here, which is focused primarily on establishing guidelines for visualizing and annotating rule-based models.
Conclusions
Large rule-based models are on the horizon. The motivation to develop such models derives in part from the need for analysis tools, such as models, to interpret molecular properties of cancer cells and to guide the treatment of patients on the basis of molecular profiling data100. As models become larger, it will become increasingly important that models of cell signaling systems be documented and communicated in an understandable way. For the purpose of clear communication of complex information, diagrams have generally proven to be valuable. Readability is essential and weighs against diagrams overloaded with details.
The visualization and annotation guidelines recommended here for rule-based models are likely to aid modelers in three specific ways: 1) in specification of a model, 2) in communication and evaluation of a model, and 3) in reuse of models. As a starting point for modeling, an extended contact map can provide a way of summarizing and assembling information about interactions of interest before the formal elements of a model are specified. A map also provides an outline for organizing the elements of a model. In fact, a map can be used to organize the work of model specification and model annotation: sections in a guide corresponding to elements of a map can be completed one by one using appropriate parts of Fig. 10 (or Appendix S1, ESI) as templates. Model communication and evaluation are aided because a map and guide together provide documentation of the basis for a model. In the hands of a reviewer, a map should be especially useful. A map identifies what molecules and interactions are included in a model. The accompanying guide explains how these molecules and molecular interactions are modeled. If one is an expert on a particular molecule or is concerned about representation of a particular interaction, one can use a map and guide to quickly identify the parts of a model that should be scrutinized. Finally, model reuse is facilitated in part because biological knowledge and modeling assumptions are clearly delineated in a guide. Many parts of a guide, perhaps especially the parts related to biological knowledge, can likely be reused if a model is revised and/or extended, easing the burden of model specification and documentation for modelers who wish to build on the work of others. In fact, because a model specification is divided/organized into units (the sections of a model guide), new models can be quickly built through composition of these units. These benefits are perhaps meager for small models but they should be invaluable for large models and more apparent as more models become available.
We expect that the ideas presented here will be immediately useful for the visualization of (large) rule-based models, as well as for more general-purpose visualization of cell signaling systems when one is concerned about protein substructures and site-specific details of protein interactions. Models can be evaluated more efficiently when their contents can be visualized and their connections to biological knowledge can be identified. A map and associated guide provide an effective way of making these connections for rule-based models. We have attempted to anticipate the needs of those who wish to build large rule-based models of cell signaling systems, considering the visualization of an array of molecule types and molecular interactions found in cell signaling systems (see Fig. 4 and Figs. 6–9). Also, to help ensure serviceable recommendations, we have leveraged the notational conventions of Kohn and co-workers66,72. However, at present, the development of large models is not routine, and the guidelines presented here may require modification at some point. In the immediate future, we are dedicated to using and testing these guidelines in our modeling efforts.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants GM076570, GM085273, and GM035556 and DOE contract DE-AC52-06NA25396. We thank Dipak Barua, James A. Cahill, Matthew S. Creamer, Jérôme Feret, Justin S. Hogg, Sarah E. Huff, Garrit Jentsch, Kurt W. Kohn, Augustin Luna, Katie R. Martin, Michael I. Monine, John A. P. Sekar, Michael W. Sneddon, and Edward C. Stites for helpful discussions.
Footnotes
Published as part of a Molecular BioSystems themed issue on Computational Biology: Guest Editor Michael Blinov. Electronic Supplementary Information (ESI) available: [Figs. S1, S2, S3 and S4 (in a single .pdf file); Appendices S1, S2 and S3 (in separate .pdf files); model.bngl; templates.graffle; and Contact Maps.gstencil and README.txt]. See DOI: 10.1039/b000000x/
References
- 1.Bradshaw JM. Cell Signal. 2010;22:1175–1184. doi: 10.1016/j.cellsig.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Lemmon MA, Schlessinger J. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawson T, Nash P. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 4.Gould CM, Diella F, Via A, Puntervoll P, Gemünd C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, Seiler M, Davey NE, Haslam N, Weatheritt RJ, Budd A, Hughes T, Pas J, Rychlewski L, Travé G, Aasland R, Helmer-Citterich M, Linding R, Gibson TJ. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 6.Hunter T. Curr Opin Cell Biol. 2009;21:140–146. doi: 10.1016/j.ceb.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kholodenko BN. Nat Rev Mol Cell Biol. 2006;7:165–76. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kholodenko BN, Hancock JF, Kolch W. Nat Rev Mol Cell Biol. 2010;11:414–426. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danos V, Feret J, Fontana W, Krivine J. Lect Notes Comput Sci. 2007;4807:139–157. [Google Scholar]
- 10.Yang J, Monine MI, Faeder JR, Hlavacek WS. Phys Rev E. 2008;78:031910. doi: 10.1103/PhysRevE.78.031910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colvin J, Monine MI, Faeder JR, Hlavacek WS, Von Hoff DD, Posner RG. Bioinformatics. 2009;25:910–917. doi: 10.1093/bioinformatics/btp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colvin J, Monine MI, Gutenkunst RN, Hlavacek WS, Von Hoff DD, Posner RG. BMC Bioinformatics. 2010;11:404. doi: 10.1186/1471-2105-11-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sneddon MW, Faeder JR, Emonet T. Nat Methods. 2011;8:177–183. doi: 10.1038/nmeth.1546. [DOI] [PubMed] [Google Scholar]
- 14.Hlavacek WS, Faeder JR, Blinov ML, Posner RG, Hucka M, Fontana W. Sci STKE. 2006;2006:re6. doi: 10.1126/stke.3442006re6. [DOI] [PubMed] [Google Scholar]
- 15.Faeder JR, Blinov ML, Hlavacek WS. Methods Mol Biol. 2009;500:113–167. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 16.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. Bioinformatics. 2006;20:3289–3291. doi: 10.1093/bioinformatics/bth378. [DOI] [PubMed] [Google Scholar]
- 17.Faeder JR, Blinov ML, Goldstein B, Hlavacek WS. Complexity. 2005;10:22–41. doi: 10.1049/sb:20045031. [DOI] [PubMed] [Google Scholar]
- 18.Endy D, Brent R. Nature. 2001;409:391–395. doi: 10.1038/35053181. [DOI] [PubMed] [Google Scholar]
- 19.Hlavacek WS, Faeder JR, Blinov ML, Perelson AS, Goldstein B. Biotechnol Bioeng. 2003;84:783–94. doi: 10.1002/bit.10842. [DOI] [PubMed] [Google Scholar]
- 20.Kholodenko BN. Nat Rev Mol Cell Biol. 2006;7:165–176. doi: 10.1038/nrm1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayer BJ, Blinov ML, Loew LM. J Biol. 2009;8:81. doi: 10.1186/jbiol185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein B, Faeder JR, Hlavacek WS, Blinov ML, Redondo A, Wofsy C. Mol Immunol. 2002;38:1213–1219. doi: 10.1016/s0161-5890(02)00066-4. [DOI] [PubMed] [Google Scholar]
- 23.Faeder JR, Hlavacek WS, Reischl I, Blinov ML, Metzger H, Redondo A, Wofsy C, Goldstein B. J Immunol. 2003;170:3769–81. doi: 10.4049/jimmunol.170.7.3769. [DOI] [PubMed] [Google Scholar]
- 24.Blinov ML, Faeder JR, Goldstein B, Hlavacek WS. Biosystems. 2006;83:136–151. doi: 10.1016/j.biosystems.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Faeder JR, Blinov ML, Goldstein B, Hlavacek WS. Syst Biol. 2005;2:5–15. doi: 10.1049/sb:20045031. [DOI] [PubMed] [Google Scholar]
- 26.Barua D, Faeder JR, Haugh JM. Biophys J. 2007;92:2290–2300. doi: 10.1529/biophysj.106.093484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barua D, Faeder JR, Haugh JM. J Biol Chem. 2008;283:7338–7345. doi: 10.1074/jbc.M708359200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barua D, Faeder JR, Haugh JM. PLoS Comput Biol. 2009;5:e1000364. doi: 10.1371/journal.pcbi.1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heiser LM, Wang NJ, Talcott CL, Laderoute KR, Knapp M, Guan Y, Hu Z, Ziyad S, Weber BL, Laquerre S, Jackson JR, Wooster RF, Kuo WL, Gray JW, Spellman PT. Genome Biol. 2009;10:R31. doi: 10.1186/gb-2009-10-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nag A, Monine MI, Faeder JR, Goldstein B. Biophys J. 2009;96:2604–2623. doi: 10.1016/j.bpj.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nag A, Monine MI, Blinov ML, Goldstein B. J Immunol. 2010;185:3268–3276. doi: 10.4049/jimmunol.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nag A, Faeder JR, Goldstein B. IET Syst Biol. 2010;4:334–347. doi: 10.1049/iet-syb.2010.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malleshaiah MK, Shahrezaei V, Swain PS, Michnick SW. Nature. 2010;465:101–105. doi: 10.1038/nature08946. [DOI] [PubMed] [Google Scholar]
- 34.Monine MI, Posner RG, Savage PB, Faeder JR, Hlavacek WS. Biophys J. 2010;98:48–56. doi: 10.1016/j.bpj.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray JCJ, Igoshin OA. PLoS Comput Biol. 2010;6:e1000676. doi: 10.1371/journal.pcbi.1000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith GR, Shanley DP. PLoS One. 2010;5:e11092. doi: 10.1371/journal.pone.0011092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lok L, Brent R. Nat Biotechnol. 2005;23:131–136. doi: 10.1038/nbt1054. [DOI] [PubMed] [Google Scholar]
- 38.Andrews SS, Addy NJ, Brent R, Arkin AP. PLoS Comput Biol. 2010;6:e1000705. doi: 10.1371/journal.pcbi.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meier-Schellersheim M, Xu X, Angermann B, Kunkel EJ, Jin T, Germain RN. PLoS Comput Biol. 2006;2:e82. doi: 10.1371/journal.pcbi.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koschorreck M, Gilles ED. BMC Syst Biol. 2008;2:91. doi: 10.1186/1752-0509-2-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moraru II, Schaff JC, Slepchenko BM, L BM, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM. IET Syst Biol. 2008;2:352–362. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallavarapu A, Thomson M, Ullian B, Gunawardena J. J R Soc Interface. 2009;6:257–270. doi: 10.1098/rsif.2008.0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lis M, Artyomov MN, Devadas S, Chakraborty AK. Bioinformatics. 2009;25:2289–2291. doi: 10.1093/bioinformatics/btp387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feret J, Danos V, Krivine J, Harmer R, Fontana W. Proc Natl Acad Sci USA. 2009;106:6453–6458. doi: 10.1073/pnas.0809908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gruenert G, Ibrahim B, Lenser T, Lohel M, Hinze T, Dittrich P. BMC Bioinformatics. 2010;11:307. doi: 10.1186/1471-2105-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ollivier JF, Shahrezaei V, Swain PS. PLoS Comput Biol. 2010;6:e1000975. doi: 10.1371/journal.pcbi.1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harmer R, Danos V, Feret J, Krivine J, Fontana W. Chaos. 2010;20:037108. doi: 10.1063/1.3491100. [DOI] [PubMed] [Google Scholar]
- 48.Faeder JR, Blinov ML, Hlavacek WS. Proceedings of the 2005 ACM Symposium on Applied Computing; Santa Fe, NM. 13-17 March 2005.2005. pp. 133–140. [Google Scholar]
- 49.Blinov ML, Yang J, Faeder JR, Hlavacek WS. Lect Notes Comput Sci. 2006;4230:89–106. [Google Scholar]
- 50.Hu B, Fricke GM, Faeder JR, Posner RG, Hlavacek WS. Bioinformatics. 2009;25:1457–1460. doi: 10.1093/bioinformatics/btp173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harris LA, Hogg JS, Faeder JM. Proceedings of the 2009 Winter Simulation Conference.2009. pp. 908–919. [Google Scholar]
- 52.Jacobs MD, Harrison SC. Cell. 1998;11:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 53.O'Dea E, Hoffmann A. Cold Spring Harb Perspect Biol. 2010;2:a000216. doi: 10.1101/cshperspect.a000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garman SC, Wurzburg BA, Tarchevskaya SS, Kinet JP, Jardetzky TS. Nature. 2000;406:259–266. doi: 10.1038/35018500. [DOI] [PubMed] [Google Scholar]
- 55.Kulczycki A, Jr., Metzger H. J. Exp. Med. 1974;140:1676–1695. doi: 10.1084/jem.140.6.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cambier JC. J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 57.Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM, Bergman FT, Gauges R, Ghazal P, Kawaji H, Li L, Matsuoka Y, Villeger A, Boyd SE, Calzone L, Courtot M, Dogrusoz U, Freeman TC, Funahashi A, Ghosh S, Jouraku A, Kim S, Kolpakov F, Luna A, Sahle S, Schmidt E, Watterson S, Wu G, Goryanin I, Kell DB, Sander C, Sauro H, Snoep JL, Kohn K, Kitano H. Nat Biotechnol. 2009;27:735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- 58.Kitano H, Funahashi A, Matsuoka Y, Oda K. Nat Biotechnol. 2005;23:961–966. doi: 10.1038/nbt1111. [DOI] [PubMed] [Google Scholar]
- 59.Oda K, Matsuoka Y, Funahashi A, Kitano H. Mol Syst Biol. 2005;1:200, 0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oda K, Kitano H. Mol Syst Biol. 2006;2:2006, 0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H. Mol Syst Biol. 2010;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Danos V, Feret J, Fontana W, Harmer R, Krivine J. Lect Notes Comput Sci. 2007;4703:17–41. [Google Scholar]
- 63.Blinov ML, Yang J, Faeder JR, Hlavacek WS. Nat Biotechnol. 2006;24:137–138. doi: 10.1038/nbt0206-137. [DOI] [PubMed] [Google Scholar]
- 64.Holm L, Sander C. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 65.Lemons NW, Hu B, Hlavacek WS. BMC Bioinformatics. 2011;12:45. doi: 10.1186/1471-2105-12-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohn KW, Aladjem MI, Kim S, Weinstein JN, Pommier Y. Mol Syst Biol. 2006;2:51. doi: 10.1038/msb4100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Iersel ML, Kelder T, Pic AR, Hanspers K, Coort S, Conklin BR, Evelo C. BMC Bioinformatics. 2008;9:399. doi: 10.1186/1471-2105-9-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Mol Biol Cell. 2006;17:1–13. doi: 10.1091/mbc.E05-09-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luna A, Karac EI, Sunshine M, Chang L, Aladjem MI, Kohn KW. 2010. [DOI] [PMC free article] [PubMed]
- 70.Kohn KW, Aladjem MI, Weinstein JN, Pommier Y. Cell Cycle. 2009;8:2281–2299. doi: 10.4161/cc.8.14.9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Meng X, Hlavacek W. IET Syst Biol. 2010;4:453–466. doi: 10.1049/iet-syb.2010.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim S, Aladjem MI, McFadden GB, Kohn KW. PLoS Comput Biol. 2010;6:e1000665. doi: 10.1371/journal.pcbi.1000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deribe YL, Pawson T, Dikic I. Nat Struct Mol Biol. 2010;17:666–672. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 74.Krivine J, Danos V, Benecke A. Lect Notes Comput Sci. 2009;5643:17–32. [Google Scholar]
- 75.Sekar JAP, Faeder JR. Methods Mol Biol. doi: 10.1007/978-1-61779-833-7_9. in press. [DOI] [PubMed] [Google Scholar]
- 76.Nelson DL, Cox MM. In: Lehninger Principles of Biochemistry. 5th Edition Freeman WH, editor. 2008. [Google Scholar]
- 77.Taylor RC, Cullen SP, Martin SJ. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 78.Law SK, Dodds AW. Protein Sci. 1997;6:263–274. doi: 10.1002/pro.5560060201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sahu A, Lambris JD. Immunol Rev. 2001;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 80.Janssen BJ, Huizinga EG, Raaijmakers HC, Roos A, Daha MR, Nilsson-Ekdahl K, Nilsson B, Gros P. Nature. 2005;437:505–511. doi: 10.1038/nature04005. [DOI] [PubMed] [Google Scholar]
- 81.Chiang GG, Sefton BM. J Biol Chem. 2001;276:23173–23178. doi: 10.1074/jbc.M101219200. [DOI] [PubMed] [Google Scholar]
- 82.Vasquez F, Devreotes P. Cell Cycle. 2006;5:1523–1527. doi: 10.4161/cc.5.14.3005. [DOI] [PubMed] [Google Scholar]
- 83.Miao B, Skidan I, Yang J, Lugovsky A, Reibarkh M, Long K, Brazell T, Durugkar KA, Maki J, Ramana CV, Schaffhausen B, Wagner G, Torchilin V, Yuan J, Degterev A. Proc Natl Acad Sci USA. 2010;107:20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schnell JD, Hicke L. J Biol Chem. 2003;278:48–56. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 85.Hoeller D, Crosetto N, Blagoev B, Raiborg C, Tikkanen R, Wagner S, Kowanetz K, Breitling R, Mann M, Stenmark H, Dikic I. Nat Cell Biol. 2006;8:163–9. doi: 10.1038/ncb1354. [DOI] [PubMed] [Google Scholar]
- 86.Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 87.Yang Z, Klionsky DJ. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colicelli J. Sci STKE. 2004;2004:re13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gureasko J, Kuchment O, Makino DL, Sondermann H, Bar-Sagi D, Kuriyan J. Proc Natl Acad Sci USA. 2010;107:3430–5. doi: 10.1073/pnas.0913915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang XF, Settleman J, Kyrakis JM, Takeuchi-Suzuki E, Elledge SJ, Marshall MS, Bruder JT, Rapp UR, Avruch J. Nature. 1993;364:308–312. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 91.Pesin JA, Orr-Weaver TL. Annu Rev Cell Dev Biol. 2008;24:475–99. doi: 10.1146/annurev.cellbio.041408.115949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nicolas E, Roumillac C, Trouche D. Mol Cell Biol. 2003;23:1614–1622. doi: 10.1128/MCB.23.5.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. Mol. Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- 94.Kesti T, Ruppelt A, Wang JH, Liss M, Wagner R, Tasken K, Saksela K. J Immunol. 2007;179:878–885. doi: 10.4049/jimmunol.179.2.878. [DOI] [PubMed] [Google Scholar]
- 95.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 96.Jain E, Bairoch A, Duvaud S, Phan I, Redaschi N, Suzek BE, Martin MJ, McGarvey P, Gasteiger E. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. Cell Res. 2009;19:271–273. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
- 99.Rivera J, Fierro NA, Olivera A, Suzuk R. Adv Immunol. 2008;98:85–120. doi: 10.1016/S0065-2776(08)00403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kreeger PK, Lauffenburger DA. Carcinogenesis. 2010;31:2–8. doi: 10.1093/carcin/bgp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.