Abstract
Glucocorticoids repress NFκB-mediated activation of proinflammatory genes such as interleukin-8 (IL-8) and ICAM-1. Our experiments suggest that the glucocorticoid receptor (GR) confers this effect by associating through protein–protein interactions with NFκB bound at each of these genes. That is, we show that the GR zinc binding region (ZBR), which includes the DNA binding and dimerization functions of the receptor, binds directly to the dimerization domain of the RelA subunit of NFκB in vitro and that the ZBR is sufficient to associate with RelA bound at NFκB response elements in vivo. Moreover, we demonstrate in vivo and in vitro that GR does not disrupt DNA binding by NFκB. In transient transfections, we found that the GR ligand binding domain is essential for repression of NFκB but not for association with it and that GR can repress an NFκB derivative bearing a heterologous activation domain. We used chromatin immunoprecipitation assays in untransfected A549 cells to infer the mechanism by which the tethered GR represses NFκB-activated transcription. As expected, we found that the inflammatory signal TNFα stimulated preinitiation complex (PIC) assembly at the IL-8 and ICAM-1 promoters and that the largest subunit of RNA polymerase II (pol II) in those complexes became phosphorylated at serines 2 and 5 in its carboxy-terminal domain (CTD) heptapeptide repeats (YSPTSPS); these modifications are required for transcription initiation. Remarkably, GR did not inhibit PIC assembly under repressing conditions, but rather interfered with phosphorylation of serine 2 of the pol II CTD.
Keywords: Glucocorticoid receptor, transcriptional repression, intracellular receptor, RelA, chromatin, anti-inflammation
Glucocorticoids are the most commonly prescribed treatment for inflammatory diseases such as rheumatoid arthritis (Da Silva 1999; Elenkov et al. 1999), and it is well established that glucocorticoids downregulate the transcription of proinflammatory genes (van de Stolpe et al. 1993; Mukaida et al. 1994; Ray and Prefontaine 1994). Those genes are themselves activated by the NFκB transcriptional regulator, but the mechanism by which glucocorticoids preclude activation has been a matter of debate.
The central member of the mammalian rel gene family is NFκB (Baldwin 1996; Ghosh et al. 1998); a major form of NFκB is a heterodimer of the RelA and p50 family members. In the inactivated state, RelA/p50 heterodimers are sequestered in the cytoplasm by the inhibitor protein IκB (Baeuerle and Baltimore 1988a,b). Proinflammatory signals such as tumor necrosis factor alpha (TNFα) and interleukin-1 (IL-1) trigger IκB phosphorylation, ubiquitination, and proteosomal degradation, enabling NFκB nuclear translocation and binding to specific genomic response elements (Chen et al. 1995; Scherer et al. 1995; Mercurio et al. 1997; Woronicz et al. 1997). Once bound to DNA, NFκB activates the transcription of proinflammatory genes such as IL-8 and ICAM-1 (van de Stolpe et al. 1993; Kunsch et al. 1994; Mukaida et al. 1994). Importantly, the NFκB response elements are the necessary and sufficient promoter sequences both for induction and for glucocorticoid-mediated inhibition (Mukaida et al. 1994; van de Stolpe et al. 1994; Caldenhoven et al. 1995).
The glucocorticoid receptor (GR) is the founding member of the intracellular receptor (IR) gene family (Tsai and O'Malley 1994; Yamamoto 1995). The GR zinc binding region (ZBR) harbors the DNA binding and protein dimerization functions of GR and is essential for regulation of all known GR target genes (Schena et al. 1989; Luisi et al. 1991). In the absence of hormone, GR is localized to the cytoplasm in association with a molecular chaperone complex that interacts with the GR ligand binding domain (LBD) (Picard and Yamamoto 1987; Rusconi and Yamamoto 1987; Picard et al. 1988; Howard et al. 1990). On hormone binding, GR releases the chaperone complex and translocates to the cell nucleus. Truncation derivatives that lack the LBD, but retain a functional ZBR, are constitutively nuclear and transcriptionally active (Godowski et al. 1987).
Transcriptional regulatory factors adopt distinct activities depending on the sequence and context, cellular and physiological, of the particular response elements with which they associate (Miner and Yamamoto 1991; Lefstin and Yamamoto 1998). Regulatory factors bind and function at response elements in three different modes: simple, composite, and tethering (Yamamoto et al. 1998). A regulatory factor binds directly to a simple response element, and it is the sole DNA binding factor necessary for regulation from that element. At a composite response element, the regulatory factor similarly binds directly to DNA, but its activities are defined by the composition of heterotypic regulators that also bind to the element. In contrast, at a tethering response element, the regulatory factor does not bind DNA, but rather associates via protein–protein interaction with a heterotypic regulator that itself is specifically bound to DNA. For example, an AP-1 site near the collagenase gene serves both as a simple AP-1 response element and as a tethering glucocorticoid response element (GRE); from that site, AP-1 activates transcription, and GR represses without disrupting AP-1 binding, apparently by direct interaction with AP-1 (Konig et al. 1992).
The fact that GR represses proinflammatory genes via NFκB response elements suggested to us that those elements might also serve as tethering GREs. However, two prior reports had suggested that glucocorticoids inhibit NFκB action by inducing IκB gene transcription (Auphan et al. 1995; Scheinman et al. 1995a). On the other hand, others had reported that the glucocorticoid effects occur independent of new protein synthesis (van de Stolpe et al. 1993; Wissink et al. 1998), or showed directly that GR inhibition of NFκB could be observed in the absence of glucocorticoid-mediated IκBα or IκBβ gene induction (Brostjan et al. 1996; De Bosscher et al. 1997; Heck et al. 1997; Newton et al. 1998; Wissink et al. 1998; Adcock et al. 1999). These findings, together with reports that GR binds selectively to the RelA subunit of NFκB (Ray and Prefontaine 1994; Caldenhoven et al. 1995; Scheinman et al. 1995b; Wissink et al. 1997), encouraged us to reexamine the tethering model and to explore the mechanism by which GR inhibits NFκB-mediated activation. In principle, GR could affect NFκB DNA binding, NFκB activation domain function, RNA polymerase II (pol II) preinitiation complex (PIC) formation, pol II phosphorylation, PIC isomerization, or promoter clearance.
We performed our studies in vitro and in CV-1 and A549 cultured cells. We used an in vitro protein–protein interaction assay to map regions of GR and RelA that interact. Transient transfections of CV-1 cells, which lack endogenous GR and RelA, enabled characterization of GR and RelA mutations, truncations, and chimeras. In contrast, A549 cells express endogenous GR and NFκB. In these cells, the proinflammatory stimulus, TNFα (Newton et al. 1998) potentiates NFκB-mediated activation of the endogenous IL-8 and ICAM-1 promoters, and the synthetic glucocorticoid dexamethasone triggers GR-mediated repression. We used these experimental approaches to determine whether GR blocks NFκB DNA binding, whether NFκB response elements are tethering GREs, and whether GR inhibition is activation-domain specific. Finally, we probed the biochemical composition of the pol II complexes during inhibition at the endogenous IL-8, ICAM-1, and IκB gene promoters in vivo.
Results
Direct repression of TNFα induced IL-8 transcription by GR
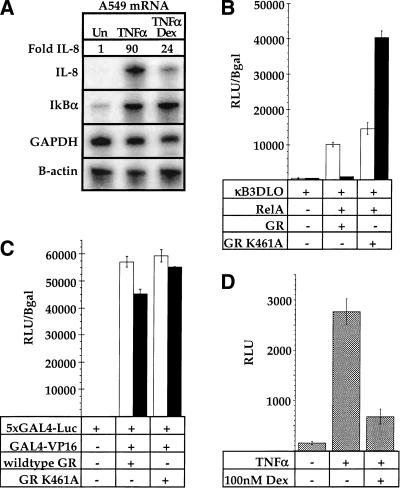
We focused on the IL-8 and ICAM-1 promoters in A549 cells because of their robust TNFα induction and glucocorticoid repression, strict dependence on NFκB response elements, and the physiological importance of IL-8 and ICAM-1 during inflammation (Mukaida et al. 1992; Harada et al. 1994; Mukaida et al. 1994; Caldenhoven et al. 1995). In these cells, which express endogenous GR, we found that IL-8 mRNA accumulation, measured by ribonuclease protection, was induced approximately 90-fold by TNFα, and that dexamethasone, a synthetic glucocorticoid, inhibited that induction approximately fourfold (Fig. 1A). As controls, we analyzed expression of the GAPDH and β-actin mRNAs, genes not regulated by either NFκB or GR, and did not detect significant alterations (Fig. 1A).
Figure 1.
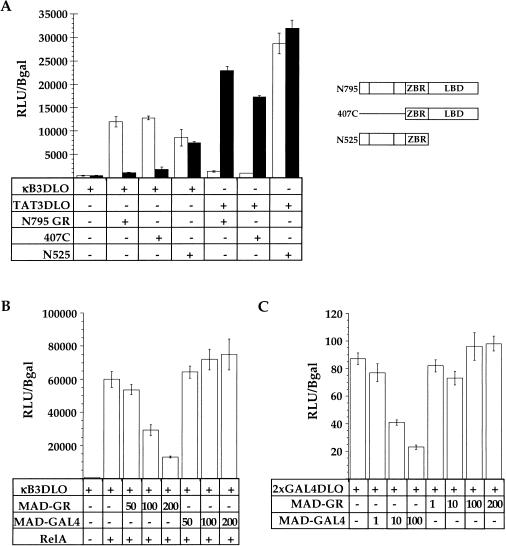
The specificity of glucocorticoid receptor (GR) inhibition of NFκB activity. (A) Regulation of the endogenous interleukin-8 (IL-8) and IκBα genes in A549-k9 cells. An RNAase protection assay was performed on total RNA isolated from untreated (Un) A549-k9 cells, or from cells treated for 2 hr with 2.5 ng/ml tumor necrosis factor alpha (TNFα), or cotreated for 2 hr with 2.5 ng/ml TNFα and 100 nM dexamethasone (TNFα/Dex). (B) Inhibition of NFκB-mediated activation by wild-type GR and further activation by GR mutant K461A. CV-1 cells were transiently transfected with NFκB response element reporter alone (κB3DLO), or cotransfected with expression vectors for RelA and GR (wild type or K461A, as indicated); reporter activity was measured in the presence of an ethanol vehicle (white bars) or 10 nM dexamethasone (black bars), expressed as relative light units (RLU) normalized to β-galactosidase activity from an internal control CMV–LacZ expression plasmid. (C) The effect on GAL4–VP16 activity by wild-type GR or GR mutant K461A. CV-1 cells were transiently transfected with GAL4 response element reporter alone (5xGAL4–e1b–Luc), or cotransfected with expression vectors for GAL4–VP16 and GR (wild type or K461A, as indicated); reporter activity was measured as in Fig. 1B. (D) Inhibition of NFκB-mediated activation by endogenous GR. A549-k9 cells, which contain a stable NFκB response element reporter transgene (κB3DLO), were treated for 8 hr with 2.5 ng/ml TNFα or cotreated with 2.5 ng/ml TNFα and 100 nM dexamethasone (TNFα/Dex) as indicated; reporter activity was expressed as RLU.
The GR–ZBR is required for both activation and repression of transcription (Schena et al. 1989), and response elements themselves can be allosteric effectors of GR activity (Lefstin et al. 1994; Lefstin and Yamamoto 1998). The response element signals appear to be detected or interpreted by rat GR residue K461 within the ZBR because GR mutant K461A activates transcription from composite and tethering response elements even under conditions in which the wild-type GR represses (Starr et al. 1996). Thus, if GR inhibits NFκB indirectly by inducing another factor such as IκBα, the K461A mutant should repress like wild-type GR. On the other hand, if NFκB response elements are tethering GREs, then the GR mutant K461A might enhance rather than repress RelA activity.
Transient transfections of CV-1 cells, which lack endogenous GR, revealed that the K461A mutant indeed activated transcription from the NFκB reporter (Fig. 1B). The effect was specific to RelA because the unrelated transcriptional regulator GAL4–VP16 (Sadowski et al. 1988) was unaffected by either wild-type or mutant GR (Fig. 1C). As expected, glucocorticoids had no effect in CV-1 cells on either RelA or GAL4–VP16 activity in the absence of a cotransfected GR expression plasmid (data not shown). Moreover, the reporter construct, as a stable transgene in A549 cells, was inducible with TNFα and repressed by the endogenous GR, demonstrating that the NFκB site confers both induction and repression, and that GR is a negative regulator of NFκB in these cells (Fig. 1D). Notably, dexamethasone treatment did not significantly increase IκBα mRNA levels in the A549 cells (Fig. 1A). These results are consistent with the view that the NFκB site is a tethering GRE.
NFκB expression and DNA binding are unaffected by GR
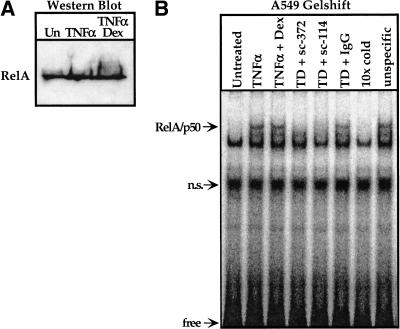
To begin to assess how GR affects RelA function, we first examined A549 whole cell extracts by immunoblotting and found that the glucocorticoids had no effect on RelA protein levels (Fig. 2A). Thus, GR must affect RelA activity and not its accumulation.
Figure 2.
The effect of glucocorticoid treatment on the expression level in A549-k9 cells and the DNA binding activity of the RelA protein in vitro. (A) An immunoblot was performed by using a RelA antibody (sc-372, Santa Cruz Biotechnology) on equal amounts of whole cell extract from A549-k9 cells treated as described in Fig. 1A. (B) An electrophoretic gel-mobility shift assay was performed by using a 32P end-labeled NFκB response element oligonucleotide derived from the interleukin-8 (IL-8) promoter and equal amounts of nuclear extract from A549 cells treated as described in Fig. 1A. An arrow indicates the RelA/p50 heterodimer, as determined by preincubating the TNFα and dexamethasone cotreated extract (TD) with the RelA antibody sc-372, or the p50 antibody (sc-114, Santa Cruz Biotechnology), blocking the specific bandshift (TD + sc-372, TD + sc-114). The normal rabbit IgG control did not affect the bandshift pattern (TD + IgG). To demonstrate the specificity of the bandshift pattern to the IL-8 probe, we added a tenfold molar excess of either unlabeled IL-8 probe (10× cold) or collagenase AP-1 site (unspecific) to TD extract. (n.s.) nonspecific.
To investigate whether glucocorticoids influence NFκB DNA binding in vitro, we analyzed A549 nuclear extracts by using an electrophoretic gel mobility-shift assay with the IL-8 NFκB binding site as a probe (Fig. 2B). Extracts from TNFα-treated A549 cells produced a readily detectable RelA/p50–DNA complex relative to extracts from untreated cells, and this induced signal was undiminished by dexamethasone (Fig. 2B).
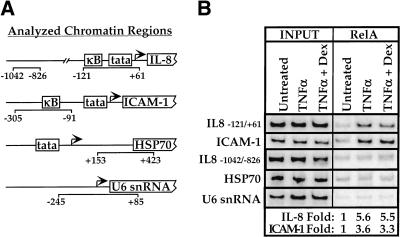
We used the chromatin immunoprecipitation assay (Braunstein et al. 1996; Orlando et al. 1997) to assess the effect of glucocorticoids on the occupancy in vivo of NFκB response elements associated with the IL-8 and ICAM-1 promoters (Fig. 3A). Normalized to the internal U6 snRNA control and relative to untreated cells, chromatin immunoprecipitation from TNFα treated cells with an antibody against RelA resulted in approximately fivefold enrichment of sequences containing the IL-8 NFκB response element (Fig. 3B). Cotreatment of A549 cells with TNFα and dexamethasone yielded a similar approximately fivefold enrichment (Fig. 3B). Thus, consistent with the in vitro assay, glucocorticoids do not inhibit NFκB binding to these sites in vivo. Similarly, ICAM-1 promoter sequences were enriched approximately threefold on gene activation, and this enrichment was unaffected by dexamethasone treatment (Fig. 3B). We monitored three additional chromatin regions as controls, none of which carries an NFκB binding site: a sequence 700 bp upstream of the IL-8 NFκB site, and the promoter regions from the HSP70 and U6 snRNA genes, which are transcribed by RNA polymerase II and III, respectively (Fig. 3A). Analyzing the immunoprecipitates for the IL-8 −1042/−826 control region demonstrated that TNFα dependent enrichment is promoter proximal (Fig. 3B). The HSP70 and U6 snRNA controls showed that enrichment is specific to NFκB regulated genes (Fig. 3B). These results lend further support to the tethering scheme, which requires site occupancy by NFκB during inhibition; it seems likely that glucocorticoids might similarly affect other proinflammatory genes.
Figure 3.
The effect of dexamethasone on NFκB response element occupancy in vivo. (A) Regions to be analyzed by chromatin immunoprecipitation. We chose the interleukin-8 (IL-8) promoter region -121/+61 and the ICAM-1 promoter region −305/−91, which contain functional NFκB binding sites as the experimental probes. For controls, we chose the IL-8 promoter 5′-region −1042/−826, the HSP70 gene region +153/+423, and the pol III transcribed U6 snRNA gene region −245/+85. (B) The effect of glucocorticoids on response element occupancy by RelA in vivo. A chromatin immunoprecipitation assay was performed by using a RelA antibody (sc-109, Santa Cruz Biotechnology) on A549-k9 cells treated as in Fig. 1A. The left panel (INPUT) shows that the starting chromatin extracts had equal amounts of the probed regions; the right panel (RelA) shows enrichment of the IL-8 and ICAM-1 experimental regions that contain NFκB response elements. The fold enrichment values for the experimental regions are determined by normalizing to the internal control U6 snRNA intensity.
The GR–ZBR directly binds the RelA dimerization domain
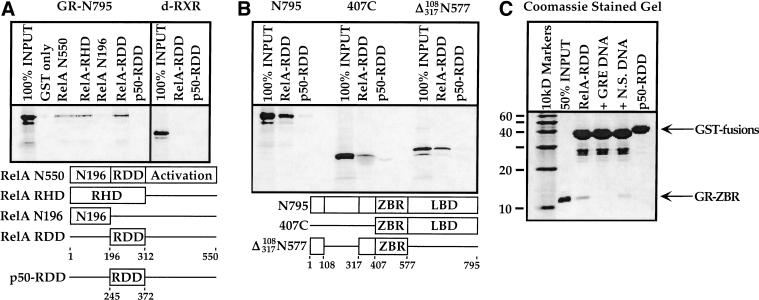
In view of these findings, we characterized in greater detail the physical interaction between GR and RelA (Ray and Prefontaine 1994; Caldenhoven et al. 1995; Scheinman et al. 1995b; Wissink et al. 1997). We constructed a series of GST fusions bearing full-length RelA, the rel homology domain (RHD), or its two subregions: the amino-terminal DNA interacting region (N196) and the rel dimerization domain (RDD) (Ghosh et al. 1995; Chen et al. 1998; Fig. 4A). We made a fourth GST fusion containing the RDD from the p50 protein for use as a negative control. In standard “pull down” assays (see Materials and Methods), we found that 35S-methionine-labeled GR bound to full-length RelA and to the RHD, but not to the RelA amino-terminal segment; moreover, the RelA dimerization domain (RelA-RDD) was necessary and sufficient for the interaction (Fig. 4A). In contrast, the p50-RDD, which is 50% identical to the RelA-RDD, supported no significant interaction (Fig. 4A). As an additional negative control, the Drosophila RXR homolog, ultraspiracle (Oro et al. 1990), failed to interact with either RDD fusion (Fig. 4A). Interestingly, the in vitro interaction was ligand-independent as measured in solution in these assays, and the GR mutant K461A displayed the same interaction profile as wild-type GR (data not shown).
Figure 4.
Protein domains required for physical interaction between RelA and glucocorticoid receptor (GR). (A) Protein–protein interaction domain mapping on the RelA protein. In vitro translated 35S-labeled full-length GR (N795) was tested on full-length RelA (RelA N550), the RelA–RHD, the amino-terminal 196 amino acids of RelA (RelA-N196), the RelA-RDD, or the p50-RDD. The distantly related intracellular receptor, d-RXR, included as a negative control, did not significantly interact with either the RelA-RDD or p50-RDD. (B) Protein–protein interaction domain mapping on the GR protein. In vitro translated 35S-labeled N795, 407C, and Δ(108–317)N577 constructs were tested on the RelA-RDD and p50-RDD proteins. (C) Interaction between the GR–ZBR and the RelA-RDD is direct. Recombinant, purified GR–ZBR (amino acids 407–525) and RelA-RDD were used in interaction assays as described earlier; where indicated, we included in the assay 200 nM DNA corresponding to either an idealized simple glucocorticoid response element or the unrelated collagenase AP-1 site. Coomassie stained SDS-polyacrylamide gel is shown; arrow indicates GR–ZBR protein.
We next tested two 35S-labeled GR deletion constructs for interaction with the RelA-RDD. We found that 407C, which lacks the amino-terminal 406 amino acids of GR but retains the ZBR and LBD, and Δ(108–317)N577, which lacks most of the amino terminus as well as the LBD (Fig. 4B), both remained competent for interaction with the RelA-RDD but not with the negative control p50-RDD. Because the two constructs have only the GR–ZBR in common, the simplest interpretation is that the GR–ZBR associates with the RelA-RDD (Fig. 4B).
To test whether this interaction is direct, we repeated these assays by using purified recombinant components visualized by Coomassie staining (Luisi et al. 1991). The GR–ZBR bound specifically to the RelA-RDD and not to the p50-RDD (Fig. 4C). Interestingly, an oligonucleotide bearing a simple GRE DNA binding site abrogated the RelA-RDD interaction, whereas a nonspecific oligonucleotide was less effective (Fig. 4C). We conclude that these regulatory proteins interact directly, and we suggest that GR binding to a simple GRE or to the RelA-RDD are mutually exclusive either because of overlapping interaction surfaces or because of an allosteric change elsewhere in the ZBR triggered by the response element.
The GR–LBD harbors repression activity
To determine which domains of GR are required for inhibition of RelA-mediated activation, we performed transient transfections of CV-1 cells to test GR deletion constructs for inhibition of RelA activity. The amino-terminal 406 residues of GR were dispensable for inhibition, whereas the LBD was essential (Fig. 5A). As a control, both receptor derivatives activated transcription from the simple GRE reporter, TAT3-DLO (Fig. 5A). Thus, we can distinguish the protein–protein interaction from the RelA repression functions within GR.
Figure 5.
Functional domain mapping on glucocorticoid receptor (GR) and functional test of a MAD–GR chimera. (A) Functional analysis of GR deletion constructs. CV-1 cells were transiently transfected with NFκB response element reporter alone (κB3DLO), or cotransfected with expression vectors for RelA and GR (N795, 407C, or N525, as indicated). Cells transfected with a simple glucocorticoid response element reporter (TAT3DLO) and GR N795, 407C or N525, are shown as indicated. Reporter activity was measured as in Fig. 1B. (B) Effect of MAD–GR chimera on RelA activity. CV-1 cells were transiently transfected with NFκB response element reporter alone (κB3DLO), or cotransfected with expression vectors for RelA and increasing amounts (in nanograms) of MAD–GR or Mad–GAL4, as indicated. Reporter activity was measured as described in Fig. 1B. (C) Effect of Mad–GAL4 chimera on basal transcription from the GAL4 response element reporter, 2xGAL4–DLO. CV-1 cells were transiently transfected with GAL4 response element reporter alone (2xGAL4–DLO), or cotransfected with increasing amounts (in nanograms) of MAD–GR or Mad–GAL4, as indicated. Reporter activity was measured as in Fig. 1B.
The GR–ZBR associates with RelA at NFκB response elements
The isolated GR–ZBR does not repress RelA activity (Fig. 5A; data not shown). If the protein–protein interaction observed in vitro occurs in vivo, then fusion of a heterologous repression domain, such as that from the Mad1 protein, to the GR–ZBR might restore repression of RelA activity. Mad1 is a member of the Mad/Myc/Max family of transcriptional regulators that functions by recruiting the N-CoR/mSin-3A/SMRT histone deacetylase (HDAC)-containing complex (Ayer et al. 1996; Alland et al. 1997; Hassig et al. 1997; Heinzel et al. 1997; Laherty et al. 1997; Zhang et al. 1997). We transiently transfected CV-1 cells with constructs expressing the Mad1 repression domain fused either to the GR–ZBR (MAD–GR) or to the unrelated GAL4–DBD (Mad–GAL4) (Fig. 5B,C). The MAD–GR construct inhibited RelA-mediated activation, but the control Mad–GAL4 construct did not (Fig. 5B). Both constructs were functional and specific because Mad–GAL4 inhibited basal transcription from a GAL4 site-driven reporter whereas MAD–GR had no effect (Fig. 5C). Notably, MAD–GR was less potent than wild-type GR, perhaps because of intrinsic differences between the Mad1 repression domain and GR–LBD. Regardless, the GR–ZBR, the minimal domain required for in vitro protein–protein interaction, is necessary and sufficient to associate with RelA in vivo.
The GR inhibitory function operates on a heterologous activation domain
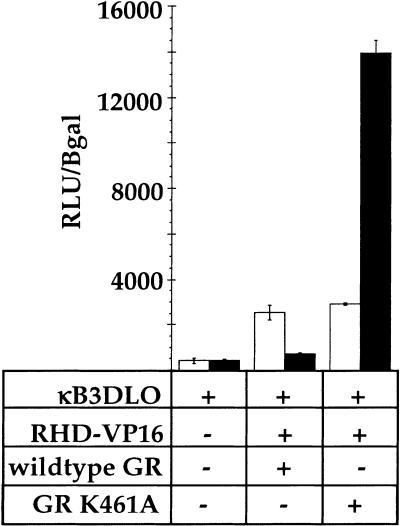
The yeast GAL80 protein negatively regulates galactose-inducible genes by selectively binding and inhibiting the yeast GAL4 transcriptional activation domain (Lue et al. 1987; Melcher and Johnston 1995; Grant et al. 1997; Yano and Fukasawa 1997; Ansari et al. 1998; Sil et al. 1999). To test whether the GR–LBD repression function is similarly limited to only the cognate activation domain of RelA, we constructed a fusion between the RelA–RHD and the activation domain from the herpesvirus VP16 protein (Triezenberg et al. 1988). The VP16 activation domain was insensitive to regulation by GR when fused to the GAL4–DBD (Fig. 1B). However, the same activation domain was inhibited by wild-type GR and activated by GR mutant K461A when expressed within the context of the RelA–RHD (Fig. 6). Thus, the inhibitory functions of GR are not dedicated to a single activation domain, suggesting that it functions downstream of the activators, perhaps affecting components of the basal transcription machinery.
Figure 6.
Effect of glucocorticoid receptor (GR) on the activity of a rel homology domain (RHD)–VP16 chimera. CV-1 cells were transiently transfected either with NFκB response element reporter alone (κB3DLO), or cotransfected with RHD–VP16 chimera and GR (wild type or K461A, as indicated). Reporter activity was measured as in Fig. 1B.
GR represses transcription after PIC assembly
The minimal PIC contains the general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIF, and pol II. The TFIID complex contains the TATA-binding protein (TBP) and several TBP-associated factors (TAFs), which together initiate PIC assembly by recognizing the TATA box, the initiator promoter elements, or both. TFIIB is incorporated second, followed by the TFIIF–pol II complex (Zawel and Reinberg 1995; Roeder 1996). In an alternative view, PICs might assemble by directly recruiting a pol II “holoenzyme” containing a subset of the GTFs (Koleske and Young 1994).
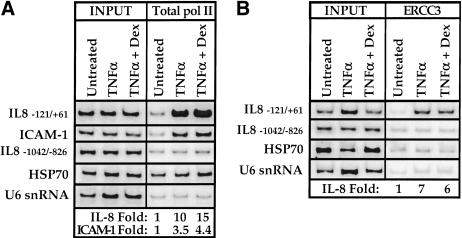
To determine whether GR inhibits NFκB-mediated activation by interfering with PIC assembly, we performed the chromatin immunoprecipitation assay by using an antibody against the amino terminus of the pol II rpb1 subunit (Fig. 7A). This antibody recognizes both the unphosphorylated (IIa) and the phosphorylated (IIo) forms of pol II, allowing determination of total pol II (IIa + IIo) recruitment. Treatment of A549 cells with TNFα stimulated pol II occupancy of the IL-8 and ICAM-1 promoter regions by approximately 10-fold and threefold, respectively (Fig. 7A).
Figure 7.
Effect of TNFα and glucocorticoids on promoter recruitment of pol II and the TFIIH helicase subunit ERCC3. Chromatin immunoprecipitation assays were performed on A549-k9 cells by using (A) a pol II antibody (sc-899, Santa Cruz Biotechnology) or (B) a TFIIH helicase subunit ERCC3 antibody (sc-293, Santa Cruz Biotechnology). Left panel (INPUT) shows that the starting chromatin extracts had equal amounts of the various chromatin regions; right panel (A, total pol II; B, ERCC3) shows enrichment of the interleukin-8 (IL-8) and ICAM-1 experimental regions; fold enrichment values for the experimental regions were determined by normalizing to the internal control U6 snRNA intensity.
Notably, under conditions of repression by dexamethasone, total pol II promoter occupancy was further increased to approximately 15-fold and fourfold, respectively (Fig. 7A). Regulated pol II recruitment was promoter region-specific because control sequences 700 bp upstream were not significantly enriched, and the inducing or repressing signals provoked no appreciable effects on pol II occupancy of the HSP70 or U6 snRNA genes (Fig. 7A). As expected, pol II was readily detectable at HSP70 (Fig. 7A) relative to the U6 snRNA gene (Birnstiel 1988; Fig. 7A) as a stalled, unphosphorylated pol II complex just downstream of this promoter in non-heat shocked cells (O'Brien et al. 1994). Thus, we conclude that GR represses NFκB-mediated activation by interfering with a step after PIC assembly, perhaps affecting initiation or promoter clearance.
PIC incorporation of TFIIH is unaffected by GR-mediated repression
In “ordered assembly” models of PIC formation, the complex is completed by recruitment of TFIIH, a nine-subunit GTF that contains multiple catalytic activities, including an ATP-dependent DNA helicase and a carboxy-terminal domain (CTD) kinase (Svejstrup et al. 1996). Promoter melting and clearance requires TFIIH, specifically, the helicase activity of the XPB/ERCC3 subunit (Guzman and Lis 1999; Moreland et al. 1999; Kim et al. 2000). Because PIC assembly in vivo is independent of at least one TFIIH subunit (Kuras and Struhl 1999), we tested the possibility that GR might repress transcription by blocking TFIIH recruitment. Treatment of A549 cells with TNFα induced approximately sevenfold recruitment of the XPB/ERCC3 DNA helicase subunit of TFIIH to the IL-8 promoter region, and this recruitment was unaffected by cotreatment with dexamethasone (Fig. 7B). As controls, XPB/ERCC3 recruitment was promoter specific because sequences 700 bp upstream of the IL-8 promoter were not enriched, and there was no appreciable effect on the HSP70 and U6 snRNA control chromatin regions (Fig. 7B). Thus, incorporation of XPB/ERCC3, and likely the whole nine-subunit TFIIH complex, into PICs is unaffected by glucocorticoid repression of NFκB-mediated activation.
Phosphorylation of CTD serine-5 is unaffected by GR-mediated repression
The rpb1 subunit of mammalian pol II includes a CTD consisting of 52 tandem repeats of a heptapeptide (YSPTSPS) that is essential for viability and is conserved among eukaryotes (Allison et al. 1985; West and Corden 1995). The CTD is unphosphorylated during PIC assembly, but initiation is accompanied by cooperative phosphorylation of the heptapeptide serine-2 and serine-5 residues (Nonet et al. 1987; Bartolomei et al. 1988; Cismowski et al. 1995; West and Corden 1995; Parada and Roeder 1996; Lee and Lis 1998; Trigon et al. 1998; Kuras and Struhl 1999). The cdk7 subunit of TFIIH selectively phosphorylates the CTD heptapeptide at serine-5 (Trigon et al. 1998). This phosphorylation event appears to be essential for transcription of most genes (Cismowski et al. 1995; Kuras and Struhl 1999).
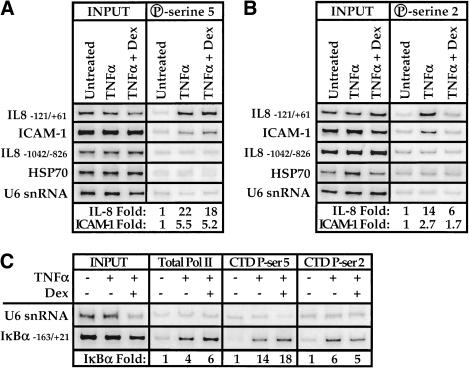
In chromatin immunoprecipitation assays in which we used a monoclonal antibody specific for the phosphoserine-5 heptapeptide repeat, we found that TNFα induction provoked an approximately 22-fold increase in binding of pol II bearing this modification at the IL-8 promoter and that dexamethasone repression had little effect on occupancy (Fig. 8A); parallel results were obtained at the ICAM-1 promoter, but to a lesser extent. Control regions of the HSP70 and U6 snRNA genes were unaffected and phosphoserine-5 pol II was not observed upstream of the IL-8 promoter region (Fig. 8A). These results imply that a phosphorylation event essential for transcription initiation, possibly mediated by TFIIH-associated cdk7, occurs even under repressing conditions (Fig. 7B). Formally, however, because maintenance of phosphoserine-5 on just one of the 52 heptapeptide repeats might be sufficient for a positive signal in this assay, a potentially substantial effect of repression might go undetected.
Figure 8.
Effect of TNFα and glucocorticoids on carboxy-terminal domain phosphorylation of the promoter bound pol II large subunit. Chromatin immunoprecipitation assays were performed on A549-k9 cells by using either (A) a phosphoserine-5 monoclonal antibody (H14, BAbCO) or (B) a phosphoserine-2 monoclonal antibody (H5, BAbCo). Left panel (INPUT) shows that the starting chromatin extracts had equal amounts of the various chromatin regions; right panel (A, P-serine 5; B, P-serine 2) shows enrichment of the interleukin-8 and ICAM-1 experimental regions; fold enrichment values for the experimental regions were determined by normalizing to the internal control U6 snRNA intensity. (C) Chromatin immunoprecipitation assays against total pol II (sc-899, Santa Cruz Biotechnology), phosphoserine-5 pol II (H14, BabCO), and phosphoserine-2 pol II (H5, BabCO) were analyzed for the presence of IκBα promoter sequences as indicated; fold enrichment values were determined by normalizing to the internal control U6 snRNA intensity.
Phosphorylation of CTD serine-2 is reduced by GR-mediated repression
Phosphorylation of the serine-2 residue of the CTD heptapeptide repeat also accompanies transcription initiation; this modification is essential for viability and has been demonstrated to be required for transcription of various genes in Saccharomyces cerevisiae (West and Corden 1995; Trigon et al. 1998; Patturajan et al. 1999). To determine whether glucocorticoids affect phosphorylation of the CTD serine-2 position, we used a monoclonal antibody specific for the phosphoserine-2 heptapeptide repeat in the chromatin immunoprecipitation assay (Fig. 8B). Treatment with TNFα induced approximately 14-fold recruitment of phosphoserine-2 pol II to the IL-8 gene, whereas recruitment was reduced to sixfold under repressing conditions. As noted earlier, this effect corresponds to a very extensive reduction in CTD phosphorylation at the serine-2 position. Parallel results were obtained in the ICAM-1 promoter region (Fig. 8B). The treatments did not affect the control HSP70 and U6 snRNA promoter regions, and phosphoserine-2 pol II binding was specific to the IL-8 promoter region relative to the upstream control segment (Fig. 8B). In addition, little phosphorylated pol II was detected at the HSP70 gene (Fig. 8A,B), consistent with its occupancy by a stalled and unphosphorylated pol II complex (O'Brien et al. 1994; cf. Fig. 7A). We conclude that GR represses NFκB activation by selectively reducing the level of phosphoserine-2 pol II complexes.
Phosphorylation of CTD serine-2 is not blocked at the IκBα promoter
NFκB activates transcription of the IκBα gene through multiple NFκB response elements (Le Bail et al. 1993; Sun et al. 1993). However, IκBα transcription is not repressed by glucocorticoids (Fig. 1A). If in fact GR represses IL-8 and ICAM-1 by reducing phosphoserine-2 pol II levels at those promoters, we would predict that phosphoserine-2 pol II levels at the IκBα promoter would be unaffected by glucocorticoids. We therefore analyzed the chromatin immunoprecipitates in Figures 7A, 8A,B for the presence of IκBα promoter sequences (Fig. 8C). Indeed, TNFα treatment induced IκBα promoter occupancy by phosphoserine-containing pol II complexes, and in contrast to the IL-8 and ICAM-1 promoter results (Fig. 8A,B), dexamethasone did not significantly affect the relative levels of the pol II isoforms at the IκBα promoter. Thus, GR represses NFκB activity in a context-specific manner, acting at some but not all NFκB regulated promoters by interfering selectively with formation of phosphoserine-2-containing pol II complexes.
Discussion
Transcriptional activation by NFκB
RelA interacts with the general coactivators CBP and p300, which act in part as histone acetyltransferases (HATs) (Zhong et al. 1998), increasing factor accessibility to chromatin packaged templates (Grunstein 1997). RelA also interacts with the ARC/DRIP coactivator complex (Naar et al. 1999), which appears to stimulate PIC assembly by forming a bridge between activation domains and pol II (Chiba et al. 2000). In addition, the RelA activation domain itself interacts functionally and physically with several GTFs, including TFIIB, TBP, and TAFII105 (Schmitz et al. 1995; Yamit-Hezi and Dikstein 1998). Collectively, these findings imply that NFκB stimulates PIC assembly by multiple mechanisms. At the IL-8 and ICAM-1 promoters, we showed that RelA indeed enhances PIC assembly, producing a complete and phosphorylated promoter-bound pol II complex (Figs. 7,8). Thus, at these response elements, we conclude that RelA achieves transcriptional activation by using a multifunctional activation domain that stimulates several facets of PIC assembly and, either directly or indirectly, CTD phosphorylation.
Proinflammatory gene NFκB sites are tethering GREs
The fact that GR regulates NFκB response elements without direct binding to the DNA suggested to us that these sequences are tethering GREs. Consistent with this view, we showed in chromatin immunoprecipitation assays that RelA DNA binding was unaffected by repressing conditions (Fig. 3B). Efforts to detect the tethered GR at tethering response elements have been only sporadically successful (R.M. Nissen, unpubl.); however, several lines of evidence support the conclusion that GR is indeed associated. First, GR mutant K461A further enhanced RelA-driven transcription (Fig. 1B). Second, the GR–ZBR interacted directly with the RelA-RDD in a reaction that was inhibited by a GRE oligonucleotide (Fig. 4C). Finally, the MAD–ZBR fusion protein was selectively and functionally recruited to RelA in vivo (Fig. 5B,C). We conclude that proinflammatory gene NFκB response elements serve as tethering GREs.
These findings indicate that regulators that carry DNA binding domains can be recruited into certain regulatory complexes through protein–protein rather than through protein–DNA interactions. How general this mode of regulation might be is unknown, but it is clearly not limited to IRs such as GR. For example, the yeast Ste12 protein regulates certain yeast genes by directly binding simple Ste12 response elements; in contrast, at other genes, Ste12 tethers to the α1 protein, which, together with MCM1, binds to response element sequences (Yuan et al. 1993). Therefore, the α1/MCM1 composite response elements are tethering response elements for Ste12. Tethering demonstrates strikingly that DNA binding domains in fact can carry multiple functional surfaces whose use is contextually determined.
Repression affects a step after activation domain function
Unlike GAL80, GR can repress transcription mediated by more than one activation domain (i.e., NFκB and AP-1). Thus, it was unsurprising that in the RHD–VP16 fusion test, the GR–LBD inhibitory function was activation-domain independent (Fig. 6). Moreover, GR bypassed the multiple distinct mechanisms by which NFκB appears to stimulate PIC assembly, instead repressing initiation itself by interfering with pol II CTD phosphorylation (Figs. 7,8). These findings demonstrate a global repression mechanism, and underscore the notion that regulation of another regulator is not limited merely to enhancing or suppressing the effects of the targeted regulator.
In vivo detection of a hemi-phosphorylated pol II species?
The lack of intermediate species migrating on SDS–polyacrylamide gels between the unphosphorylated IIa and the maximally phosphorylated IIo forms suggests that CTD-kinases and CTD-phosphatases are highly processive (Lehman and Dahmus 2000). However, it has not been proved that a single heptapeptide repeat can be phosphorylated at multiple positions, nor has it been determined whether phosphorylation by one enzyme influences phosphorylation by another (e.g., might cdk7-mediated conversion of YSPTSPS to YSPTS*PS be further processed by another kinase to YS*PTS*PS, or vice versa?). Different phosphorylation sites can be functionally distinct and regulators might read a putative “CTD code” as a series of binding sites or allosteric effectors. For example, mammalian mRNA capping activity is stimulated by CTD peptides phosphorylated at serine-5 but not at serine-2 (Ho and Shuman 1999). Thus, differential recruitment of CTD kinases might generate functionally distinct CTD phosphorylation patterns. Our findings suggest that glucocorticoids repress NFκB activity by generating a transcriptionally inactive DNA-bound pol II species phosphorylated at serine-5 but not at serine-2. Notably, Bonnet et al. (1999) detected a similar pol II species, denoting it pol IIm. We speculate that pol IIm and the pol II species we detect are identical and represent one of several hemi-phosphorylated pol II isoforms. The specificity of the glucocorticoid effects reveals that differential posttranslational modifications of pol II are an important facet of gene regulation.
Recruitment of a TSA resistant corepressor?
Acetylation of nucleosomes appears to facilitate gene activation by increasing factor access to genomic binding sites (Grunstein 1997; Blackwood and Kadonaga 1998). Some (Mizzen et al. 1996; Chen et al. 1997; Grant et al. 1997; Blanco et al. 1998; Kraus and Kadonaga 1998) but not all (Naar et al. 1999; Orphanides et al. 1999; Rachez et al. 1999; Ryu et al. 1999; Rachez et al. 2000) coactivator complexes harbor HAT activity. Conversely, the IR corepressors identified to date carry HDAC activity; both coactivators and corepressors can interact with the LBD (Heery et al. 1997; Nagy et al. 1997; Darimont et al. 1998; Voegel et al. 1998; Perissi et al. 1999).
Repression of AP-1 mediated activation by the thyroid hormone receptor (TR) is blocked by trichostatin A (TSA) (M. Cronin and K.R. Yamamoto, unpubl.), a general inhibitor of most HDACs (Taunton et al. 1996). In contrast, GR regulation of AP-1 activity, at the same response element used in the TR experiments, is resistant even to high TSA concentrations (M. Cronin and K.R. Yamamoto, unpubl.). Similarly, we found that GR repression of NFκB activity was TSA resistant in A549 cells (data not shown). Together with our findings that neither PIC assembly (Fig. 7A) nor PIC incorporation of TFIIH (Fig. 7B) is affected by glucocorticoid treatment, we suggest that promoter occlusion or HDAC recruitment are unlikely to be the mechanisms by which GR represses NFκB action.
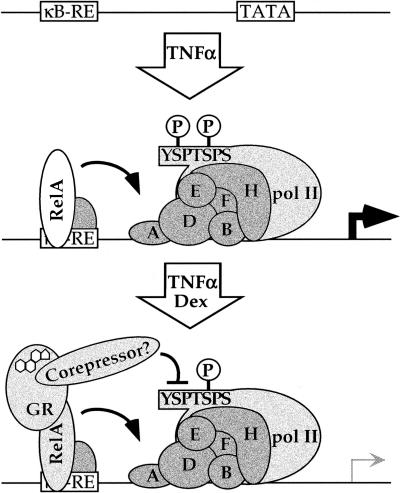
In principle, the GR–LBD might have an intrinsic activity for blocking CTD serine-2 phosphorylation, but the modularity of regulatory complexes leads us to postulate the existence of a novel corepressor, recruited by the GR–LBD to tethering GREs in a manner analogous to previously established cofactors (Fig. 9). According to this scheme, the resultant regulatory complex would yield complete PICs deficient in serine-2 phosphorylation, with a consequent decline in initiation or promoter clearance.
Figure 9.
A model for repression of NFκB-mediated activation by glucocorticoid receptor (GR). TNFα treatment induces NFκB response element binding, preinitiation complex assembly, and transcription. Dexamethasone treatment induces tethering of GR to RelA-occupied NFκB response elements. The GR-ligand binding domain, perhaps via recruitment of an unidentified corepressor, bypasses RelA activation domain functions, interfering with phosphorylation of the pol II CTD at serine-2.
The putative corepressor might be a serine-2 phosphatase or a serine-2 kinase inhibitor. Some potential candidates are intriguing to consider. The human CTD phosphatase FCP1 fully dephosphorylates the CTD before assembly into PICs (Cho et al. 1999); hence, although FCP1 itself may not be the putative corepressor, there are four identified human FCP1 homologs (Archambault et al. 1997), one of which might encode a specific CTD serine-2 phosphatase.
The mammalian CTD-kinase cdk9, a subunit of the positive transcript elongation factor (P-TEFb; Price 2000) is a potential target for a GR-recruited inhibitor. Yeast strains lacking CTK1, the homolog of cdk9, display reduced phosphorylation of the pol II CTD at serine-2 (Patturajan et al. 1999), slow growth and cold sensitivity, and reduced transcription of various genes (Lee and Greenleaf 1991; Patturajan et al. 1999). Because CTK1 is not the only yeast serine-2 kinase (Patturajan et al. 1999), higher eukaryotes are also likely to carry multiple kinases that affect this residue of the CTD repeat. Thus, GR might target, either directly or through recruitment of a kinase inhibitor, a cdk9-like factor.
Several regulators that function as kinase inhibitors have been described. For example, the Caenorhabditis elegans repressor protein PIE-1 appears to abrogate pol II transcription in germ-line blastomeres by preventing phosphorylation of the CTD at serine-2 (Seydoux and Dunn 1997; Tenenhaus et al. 1998). Similarly, the cdc2/cyclinB kinase complex mediates global mitotic repression of transcription by phosphorylating and inactivating the CTD-kinase of TFIIH (Akoulitchev and Reinberg 1998; Long et al. 1998). Likewise, the NAT complex, a global negative regulator for pol II that contains the cyclin dependent kinase cdk8/Srb10 (Hengartner et al. 1998; Sun et al. 1998), uses that kinase activity to phosphorylate TFIIH and downregulate the CTD-kinase activity of cdk7 (D. Reinberg, pers. comm.).
Mechanisms for transcriptional repression
Several members of the IR gene family, including mineralocorticoid receptor (Liden et al. 1997), estrogen receptor (ER; Ray et al. 1994; Stein and Yang 1995), progesterone receptor (PR; Caldenhoven et al. 1995; Kalkhoven et al. 1996), androgen receptor (AR; Palvimo et al. 1996), thyroid hormone receptor (R.M. Nissen, unpubl.), and retinoic acid receptor (R.M. Nissen, unpubl.) can regulate NFκB-mediated activation. Consistent with results obtained from studies on PR, ER, and AR (Stein and Yang 1995; Kalkhoven et al. 1996; Palvimo et al. 1996), we found that the GR–LBD was essential for repression of NFκB (Fig. 5A). By extension, it seems likely that these and perhaps other IRs might similarly regulate NFκB activity by affecting the pol II CTD.
Our finding that the IκBα gene is induced by TNFα but not repressed by dexamethasone (Fig. 1A), together with the failure of dexamethasone to alter the level of phosphoserine-2 pol II at IκBα (Fig. 8C) suggests that unidentified contextual features of response elements and promoters impart selectivity on GR action. A detailed comparative analysis of the IL-8 and IκBα promoter regions would likely provide insights into the determinants for context-dependent glucocorticoid regulation of transcriptional activation activity.
The distinct modes used by different transcriptional regulators can provide insights about the contexts in which those regulators might function. Repressors that interfere with intrinsic activator functions (e.g., DNA binding or activation domain accessibility) are more likely to be activator-specific (Baeuerle and Baltimore 1988b; Small et al. 1991; Ansari et al. 1998); affected promoters could still be activated by other activators. In contrast, repressors that act on activator targets or initiation events downstream from those targets would function more globally on the affected promoters; repressors that affect late steps of initiation would down-regulate targeted promoters regardless of the range or mechanisms of associated activators. As we have shown here, for example, GR bypasses RelA stabilization of PIC assembly by affecting CTD phosphorylation at serine-2.
Multiple signaling networks regulate natural promoters, requiring that pol II integrate multiple inputs from positive and negative regulators. This combinatorial approach to gene expression enables fine-tuning of transcriptional activity at discrete chromosomal loci. Transcriptional regulators such as GR are likely to exploit a diversity of mechanisms across different cellular and promoter contexts.
Materials and methods
Plasmid DNAs
The plasmids pSG5 (Promega), pSG5–rGR (Darimont et al. 1998), p6R–N525 (Iniguez-Lluhi et al. 1997), TAT3–DLO (Iniguez-Lluhi et al. 1997), pSG5–MAD–GR (gift of M. Cronin, UCSF), pGEX4T-1 (Pharmacia), 5xGAL4–e1b–Luc (gift of R. Uht, UCSF), pRS423–N577–Δ108–317 (gift of B. Darimont, UCSF), CMV–βgal (Spaete and Mocarski 1985), pSG424–MAD–GAL4, and PGK–Neo were described previously. Subcloning the BspEI–BbsI fragment of GR containing the K461A mutation from p6R–K461A (Starr et al. 1996) into pSG5–rGR generated plasmid pSG5–GR–K461A. PCR amplification of nucleotide sequences encoding amino acids 407–795 of rat GR from p6R–rGR (Starr et al. 1996) by using forward primer 7 5′-AAAAGGATCCATAATGTCAGTGTTTTCTAATGGG-3′ and reverse primer 8 5′-AAAAGGATCCTCATTTTTGATGAAACAGAAGC-3′ generated plasmid pSG5–GR–407C by digesting the PCR fragment with BamHI followed by ligation into pSG5 digested with BamHI. The BamHI–ScaI fragment of pRS423–N577–Δ(108–317) ligated into pSG5–rGR digested with EagI (blunt) and BamHI generated plasmid pSG5–N577–Δ(108–317). PCR amplification of the mouse RelA open reading frame from plasmid J134 (Blank et al. 1991) by using forward primer 1 5′-GGCGCGAATTCATGGACGATGTGTTTCCCC-3′ and reverse primer 1 5′-GGCGCGAATTCTTAGGAGCTGATCTGACTCAAA-3′ followed by digestion with EcoRI and ligation into the EcoRI site of pSG5 generated plasmid pSG5–RelA. PCR amplification of two tandem copies of the VP16 amino acids 413–454 from plasmid pGAL4–VP16 (gift of M. Carey, UCLA) by using forward primer 2 5′-CCCCCGAATTCCAGCCCGGGCGATCCGCC-3′ and reverse primer 2 5′-CCCCCGGATCCTTATCTAGAGGATCTCGG-3′ followed by digestion with EcoRI and BamHI, and ligation into pSG5 generated intermediate plasmid pSG5–VP16. PCR amplification of RelA nucleotide sequences encoding amino acids 1–312 by using forward primer 1 and reverse primer 3 5′-GGCGCGAATTCGATACTCTTGAAGGTCTCATAGGT-3′ followed by digestion with EcoRI and ligation into EcoRI-cleaved pSG5–VP16 yielded plasmid pSG5–RHD–VP16.
Inserting three copies of the annealed and kinased IL-2Rα NFκB response element oligonucleotides 5′-TCGACGGAGAGGGAGATTCCCCTGCCGTC-3′ and 5′-TCGAGACGGCAGGGGAATCTCCCTCTCCG-3′ into the SalI site of plasmid pΔODLO (Iniguez-Lluhi et al. 1997) generated the reporter plasmid κB3–DLO. Subcloning the double GAL4 binding site oligonucleotide 5′-AGCTCGGAGGACTGTCCTCCGTTCTCGAGAACGGAGGACAGTCCTCCG-3′ into the HindIII site of pΔODLO generated reporter plasmid 2xGAL4–DLO, which has a higher basal activity than 5xGAL4–e1b–Luc, allowing for analysis of Mad–GAL4 repression activity.
The Escherichia coli expression vector pGEX–RelA for the GST–RelA fusion protein was described previously (Stein and Yang 1995). PCR amplification of mouse RelA nucleotide sequences encoding amino acids 1–312 by using forward primer 1 and reverse primer 9 5′-GGCGCGAATTCGATACTCTTGAAGGTCTCATAGGT-3′ followed by digestion with EcoRI and ligation into the EcoRI site of pGEX4T-1 generated plasmid pGEX–RelA–RHD. PCR amplification of mouse RelA sequences encoding amino acids 1–196 by using forward primer 1 and reverse primer 10 5′-GACTGATCGCGGCCGCTCAGATCTTGAGCTCGGCAGT-3′, followed by EcoRI–NotI digestion and ligation into the corresponding sites of pGEX4T-1 yielded plasmid pGEX–RelA–N196. PCR amplification of mouse RelA sequences encoding amino acids 192–312 by using forward primer 8 5′-GGCGCGAATTCACTGCCGAGCTCAAGATC-3′ and reverse primer 9, followed by EcoRI digestion and ligation into the EcoRI site of pGEX4T-1 generated plasmid pGEX–RelA-RDD. PCR amplification of mouse p50 sequences encoding amino acids 245–372 from plasmid J130 (Blank et al. 1991) by using forward primer 9 5′-GAAGAGGATCCATGGCATCCAACCTGAAAATCGT-3′ and reverse primer 11 5′-GAAGAGAATTCTTAGAAGCTGTCCGAGAAGTTC-3′, followed by BamHI–EcoRI digestion and ligation into the corresponding sites of pGEX4T-1 yielded plasmid pGEX–p50-RDD. PCR amplification of mouse RelA sequences encoding amino acids 304–550 by using forward primer 6 and reverse primer 1 followed by EcoRI digestion and ligation into the corresponding site of pGEX4T-1 generated plasmid pGEX–RelA-304–550.
Subcloning the PCR amplified human IL-8 coding sequence into pBLUESCRIPT KS+ (Stratagene) at the XhoI to SmaI sites generated the anti-sense IL-8 plasmid pBS–IL8–AS. Subcloning the XhoI–XmnI fragment of the human IκBα coding sequence into pBLUESCRIPT KS+ at the XhoI and EcoRV sites generated the anti-sense IκBα plasmid pBS–IκBα–AS. Digestion of the anti-sense plasmids with XhoI and in vitro transcription from the T7 promoter yielded the anti-sense probes for RNase protection.
Cell culture and transfections
CV-1 cells and A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal calf serum and split every third day.
A549-k9 cells carrying stably integrated NFκB-responsive luciferase reporter genes were derived by linearizing both the kB3DLO luciferase reporter and the PGK–Neo plasmid with XmnI followed by electroporation and selection in 1.5 mg/ml G418. Selected colonies were then screened for TNFα-inducible luciferase activity; subsequent culturing of A549-k9 was performed in the absence of G418.
For transient transfections, CV-1 cells were plated in 24 well plates (5 × 104 cells per well in 500 μl of DMEM containing 5% charcoal-stripped [Freeman et al. 2000] fetal calf serum) approximately 16 hr before addition of lipid–DNA complexes. Cationic lipid stock was prepared as described (Hong et al. 1997). A typical transfection contained 100 ng luciferase reporter, 100 ng β-galactosidase control plasmid, regulatory factor expression plasmids where indicated, and empty pSG5 plasmid to 450 ng total DNA in 40 μl of OPTI-MEM I (GIBCO BRL). The 5-mM cationic lipid stock was diluted to 0.5 μl lipid per 40 μl in OPTI-MEM I. Then, 40 μl of the diluted lipid was added to the 40 μl of DNA, mixed, and incubated at room temperature for 15 min. After incubation, the lipid–DNA complexes were further diluted with 300 μl of OPTI-MEM I and the entire 380 μl was added to a single aspirated well of the 24-well plate. Lipid–DNA complexes were typically prepared as a stock of 6.5 identical reactions and aliquoted onto cells for replicates. Approximately 6 hr later, 400 μl of DMEM containing 5% stripped FCS and 2× concentrations of hormone or ethanol vehicle was added. Approximately 16 hr posthormone addition, cells were harvested and assayed for β-galactosidase and luciferase activity as described (Iniguez-Lluhi et al. 1997).
Protein purification
Proteins fused to GST were expressed in E. coli strain BL21 at 18°C with 1 mM IPTG induction at approximately 0.3 OD600 followed by an additional 12–14 hr of growth before harvest. All steps were performed at 4°C unless noted otherwise, and all buffers for GST fusions contained 0.5 mM PMSF and 1 μg/ml each of leupeptin, pepstatin, and aprotinin. Cell pellets were resuspended in one volume of phosphate buffered saline containing 1 mM EDTA and 14 mM β-mercaptoethanol. Extracts were prepared by lysozyme treatment followed by sonication to reduce turbidity. The extracts were then centrifuged in a Beckman 70.1Ti rotor at 45,000 rpm for 2 hr. The supernatant was batch bound onto 1 ml of 50% slurry glutathione–agarose beads (Sigma) in the same buffer adjusted to 0.1% NP-40 for 30 min. The beads were then loaded into a disposable column and washed with 10 ml of Wash Buffer (10 mM Tris at pH 8, 1 mM EDTA, 2 M NaCl, 0.1% NP-40, 14 mM β-mercaptoethanol) followed by 10 ml DnaK Buffer (50 mM Tris at pH 8, 10 mM MgSO4, 2 mM ATP). A final 5-ml wash in Binding Buffer (10 mM Tris at pH 8, 1 mM EDTA, 150 mM NaCl, 14 mM β-mercaptoethanol) was then performed and the purified proteins were stored with the beads at −80°C.
GST-fusion protein interaction assays
Equal amounts, ∼1 μg, of each fusion protein were used as judged by Coomassie gel loading titrations. GR and derivatives in the pSG5 vector were in vitro translated by using the Promega TNT coupled transcription/translation kit in the presence of 35S-methionine. Binding reactions were performed in a total volume of 100 μl binding buffer containing 12.5 μl packed glutathione beads and ∼5 nM receptor (estimated by immunoblotting comparisons to purified GR standards). Reactions were conducted for 45 min at room temperature with mild agitation to keep the beads in suspension. Samples were then pelleted and beads were washed four times with 500 μl of binding buffer containing 500 mM NaCl. The washed pellets were resuspended in 10 μl of SDS–polyacrylamide gel loading buffer and the entire reaction was resolved over an appropriate percentage (12% or 15%) SDS–polyacrylamide gel. Dried gels were exposed to a Phosphor screen overnight, and Molecular Dynamics software was used to generate and quantify gel images.
Western blot and ribonuclease protection assays
Immunoblotting was performed on A549-k9 whole cell extracts with a RelA antibody (sc-372, Santa Cruz Biotechnology) according to manufacturer's recommendations. The ribonuclease protection assays were performed on 20 μg total RNA according to manufacturer's recommendations (Ambion).
Electrophoretic gel-mobility shift assays
Nuclear extracts were prepared from A549-k9 by the Dignam method (Dignam et al. 1983). Gel shifts were performed with 2.5 μg nuclear extract in a 10 μl volume of final concentrations 10 mM HEPES (pH 8), 80 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 5% glycerol, 0.05% NP-40, 0.1 mg/ml poly-dG/dC (Pharmacia Biotech). After 5 min at room temperature, antibodies were added where indicated (anti-p50, sc-114, Santa Cruz Biotechnology). After an additional 5 min, 32P end-labeled NFκB IL-8 site probe 5′-CAAATCGTGGAAATTTCCTCTGAC-3′ was added to a final concentration of 2 nM followed immediately by cold oligonucleotide or not. The nonspecific probe was the collagenase AP-1 site 5′-AGTCATGAGTCAGACACCTCTGGC-3′.
Chromatin immunoprecipitation assays
A modified protocol was developed on the basis of several previous reports (Braunstein et al. 1996; Orlando et al. 1997; Parekh and Maniatis 1999). Antibodies were used at levels that maximized selective immunoprecipitation of signal. Identical conditions were used for the RelA antibody (Santa Cruz, sc-109) and the total RNA polymerase II antibody (Santa Cruz, sc-899). Following the 2-hr treatments with various hormone combinations, approximately 5 × 108 adherent A549-k9 cells were cross-linked for 40 min at 4°C by the addition of 11× formaldehyde Stock (50 mM Hepes-KOH at pH 8; 1 mM EDTA; 0.5 mM EGTA; 100 mM NaCl; 11% formaldehyde) to a final of 1× for 40 min. Cross-linking was stopped by the addition of glycine to a final concentration of 125 mM for 5 min.
The cell monolayers were rinsed with ice cold phosphate buffered saline, scraped into 50-ml conical tubes, and centrifuged at 600g for 5 min at 4°C. Pellets were aspirated and resuspended in 10 ml Chro-IP Lysis Buffer (50 mM Hepes-KOH at pH 8; 1 mM EDTA; 0.5 mM EGTA; 140 mM NaCl; 10% glycerol; 0.5% NP-40; 0.25% Triton X-100; 1 mM PMSF; 5 μg/ml each of leupeptin, pepstatin A, and aprotinin) and nutated for 10 min at 4°C. The crude nuclei were collected by centrifugation (600g for 5 min at 4°C) and resuspended in 10 ml Wash Buffer (10 mM Tris-HCl at pH 8; 1 mM EDTA; 0.5 mM EGTA; 200 mM NaCl; 1 mM PMSF; 5 μg/ml each of leupeptin, pepstatin A, and aprotinin) and nutated again. Washed nuclei were centrifuged as described earlier and resuspended in 2 ml of 1× RIPA Buffer (10 mM Tris-HCl at pH 8; 1 mM EDTA; 0.5 mM EGTA; 140 mM NaCl; 1% Triton X-100; 0.1% Na-deoxycholate; 0.1% SDS; 1 mM PMSF; 5 μg/ml each of leupeptin, pepstatin A, and aprotinin).
Samples were sonicated (power setting 5) with a Branson Sonifier 250 with a microtip in 20-sec bursts followed by 1 min of cooling on ice for a total sonication time of 3 min per sample. This procedure resulted in DNA fragment sizes of 0.3–1.5 kb. Samples were then centrifuged in an Eppendorf 5415C at 16,000g for 10 min at 4°C. Either 2 μg of sc-899 antibody or 2 μg of sc-109 antibody were added to 600-μl aliquots of cleared chromatin extract and incubated with rotation at 4°C for 6 hr. Samples were then centrifuged again and supernatants were transferred to fresh tubes containing 20 μl of precleared 50% slurry Protein A/G beads (ICN Pharmaceuticals) in 1× RIPA Buffer containing 100 μg/ml sonicated salmon sperm DNA. After 3 hr with the beads, samples were centrifuged at 600g and the pellets washed twice with 1× RIPA buffer, once with 1× RIPA buffer containing 100 μg/ml salmon sperm DNA for 5 min with rotation, five times with 1× RIPA buffer containing 500 mM NaCl final plus 100 μg/ml salmon sperm DNA for 5 min with rotation, and once with 1× RIPA buffer. Then, 100 μl of digestion buffer was added (50 mM Tris at pH 8; 1 mm EDTA; 100 mM NaCl; 0.5% SDS; 100 μg/ml proteinase K) and placed at 55°C for 3 hr, followed by 6 hr at 65°C to reverse cross-links. DNA was phenol–CHCl3 extracted once, CHCl3 extracted once, and ethanol precipitated in the presence of 20 μg glycogen. Pellets were resuspended in 20 μl TE.
PCR reactions, 50 μl, were programmed for 30 cycles with 4 μl of DNA sample and 50 nM each of appropriate 32P end-labeled (500,000 cpm/reaction) primer oligonucleotides (GIBCO BRL buffers, nucleotides, and Taq enzyme). Titrations were performed to ensure a linear range of amplification. One-fifth of each PCR reaction was electrophoresed on a 6% 0.5× TBE gel (19:1 acrylamide:bisacrylamide; Bio-Rad), dried, and exposed on a Phosphor cassette for quantification and image generation. The IL-8 or ICAM-1 promoter intensities were first normalized by dividing them by the internal control intensity of the U6 snRNA gene. Fold inductions are defined as the ratio of the normalized intensities for the treated lanes to the untreated lane.
The TFIIH XPB/ERCC3 subunit antibody (sc-283, Santa Cruz) was treated similarly except for the following modifications. Eight micrograms antibody was used per aliquot of chromatin extract and incubated for 16–24 hr with rotation at 4°C. Immunoprecipitates were washed twice with 1× RIPA buffer, once with 1× RIPA containing 100 μg/ml salmon sperm DNA for 5 min with rotation, once with 1× RIPA containing 500 mM NaCl final plus 100 μg/ml salmon sperm DNA for 5 min with rotation, and once with LiCl buffer (10 mM Tris-HCl at pH 8; 1 mM EDTA; 0.5 mM EGTA; 250 mM LiCl; 1% Triton X-100; 1% Na-deoxycholate; 1 mM PMSF; 5 μg/ml each of leupeptin, pepstatin A, and aprotinin).
The CTD phosphoserine-2 specific monoclonal antibody H5 (BAbCO) and the CTD phosphoserine-5 specific monoclonal antibody H14 (BAbCO) were treated similarly except for the following modifications. Extracts were prepared with all buffers including the general phosphatase inhibitor 10 mM Na-pyrophosphate (pH 8). Five microliters of H5 ascites fluid or 10 μl of H14 ascites fluid were used per aliquot of chromatin extract and incubated with rotation 16–24 hr at 4°C. Protein A/G beads were preincubated with goat IgG anti-mouse IgM (2 μg/μl packed beads) overnight in 1× RIPA and washed three times with 1× RIPA before use. Immunoprecipitates were washed twice with 1× RIPA buffer, once with 1× RIPA containing 100 μg/ml salmon sperm DNA for 5 min with rotation, once with 1× RIPA containing 300 mM NaCl final plus 100 μg/ml salmon sperm DNA for 5 min with rotation, and once with LiCl buffer.
PCR primer sets for the chromatin immunoprecipitation assay
The human IL-8 promoter region −121/ +61 was amplified with the primer pairs 5′-GGGCCATCAGTTGCAAATC-3′ and 5′-TTCCTTCCGGTGGTTTCTTC-3′. The human IL-8 upstream region −1042/−826 was amplified with the PCR primer pairs 5′-AACAGTGGCTGAACCAGAG-3′ and 5′-AGGAGGGCT TCAATAGAGG-3′. The human U6 snRNA promoter region −245/+85 was amplified with the PCR primer pairs 5′-GGC CTATTTCCCATGATTCC-3′ and 5′-ATTTGCGTGTCATCCT TGC-3′. The human ICAM-1 promoter region −305/−91 was amplified with the PCR primer pairs 5′-ACCTTAGCGCGGTGT AGACC-3′ and 5′-CTCCGGAACAAATGCTGC-3′. The human HSP70 promoter region +153/+423 was amplified with the PCR primer pairs 5′-GGATCCAGTGTTCCGTTTCC-3′ and 5′-GTCA AACACGGTGTTCTGCG-3′. The human IκBα promoter region –168/+21 was amplified with the PCR primer pairs 5′-CTCATC GCAGGGAGTTTCT-3′ and 5′-ACTGCTGTGGGCTCTGCA-3′.
Acknowledgments
We are grateful to M. Cronin, B. Darimont, and R. Uht for plasmids, and M. Dahmus, D. Reinberg, and members of the Yamamoto lab for useful discussions. We also thank C. Bargmann, A. Frankel, N. Freedman, B. Freeman, H. Luecke, and I. Rogatsky for critiques of the manuscript. Research support was from the National Institutes of Health.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL yamamoto@cgl.ucsf.edu; FAX (415) 476-6129.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.827900.
References
- Adcock IM, Nasuhara Y, Stevens DA, Barnes PJ. Ligand-induced differentiation of glucocorticoid receptor (GR) trans-repression and transactivation: Preferential targetting of NF-κB and lack of I-κB involvement. Br J Pharmacol. 1999;127:1003–1011. doi: 10.1038/sj.bjp.0702613. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Akoulitchev S, Reinberg D. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes & Dev. 1998;12:3541–3550. doi: 10.1101/gad.12.22.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Ansari AZ, Reece RJ, Ptashne M. A transcriptional activating region with two contrasting modes of protein interaction. Proc Natl Acad Sci. 1998;95:13543–13548. doi: 10.1073/pnas.95.23.13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault J, Chambers RS, Kobor MS, Ho Y, Cartier M, Bolotin D, Andrews B, Kane CM, Greenblatt J. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:14300–14305. doi: 10.1073/pnas.94.26.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: Inhibition of NF-κ B activity through induction of I κ B synthesis. Science. 1995;270:286–290. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- Ayer DE, Laherty CD, Lawrence QA, Armstrong AP, Eisenman RN. Mad proteins contain a dominant transcription repression domain. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κ B transcription factor. Cell. 1988a;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- ————— I κ B: A specific inhibitor of the NF-κ B transcription factor. Science. 1988b;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Baldwin AS., Jr The NF-κ B and I κ B proteins: New discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Halden NF, Cullen CR, Corden JL. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnstiel ML. Structure and function of major and minor small nuclear ribonucleoprotein particles. New York, NY: Springer Verlag; 1988. [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: A current view of enhancer action. Science. 1998;281:61–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes & Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V, Kourilsky P, Israel A. Cytoplasmic retention, DNA binding and processing of the NF-κ B p50 precursor are controlled by a small region in its C-terminus. EMBO J. 1991;10:4159–4167. doi: 10.1002/j.1460-2075.1991.tb04994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet F, Vigneron M, Bensaude O, Dubois MF. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2) Nucleic Acids Res. 1999;27:4399–4404. doi: 10.1093/nar/27.22.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brostjan C, Anrather J, Csizmadia V, Stroka D, Soares M, Bach FH, Winkler H. Glucocorticoid-mediated repression of NFκB activity in endothelial cells does not involve induction of IκBα synthesis. J Biol Chem. 1996;271:19612–19616. doi: 10.1074/jbc.271.32.19612. [DOI] [PubMed] [Google Scholar]
- Caldenhoven E, Liden J, Wissink S, Van de Stolpe A, Raaijmakers J, Koenderman L, Okret S, Gustafsson JA, Van der Saag PT. Negative cross-talk between RelA and the glucocorticoid receptor: A possible mechanism for the antiinflammatory action of glucocorticoids. Mol Endocrinol. 1995;9:401–412. doi: 10.1210/mend.9.4.7659084. [DOI] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF- κB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets I κ B α to the ubiquitin-proteasome pathway. Genes & Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- Chiba N, Suldan Z, Freedman LP, Parvin JD. Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J Biol Chem. 2000;275:10719–10722. doi: 10.1074/jbc.275.15.10719. [DOI] [PubMed] [Google Scholar]
- Cho H, Kim TK, Mancebo H, Lane WS, Flores O, Reinberg D. A protein phosphatase functions to recycle RNA polymerase II. Genes & Dev. 1999;13:1540–1552. doi: 10.1101/gad.13.12.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismowski MJ, Laff GM, Solomon MJ, Reed SI. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva JA. Sex hormones and glucocorticoids: Interactions with the immune system. Ann N Y Acad Sci. 1999;876:102–117. doi: 10.1111/j.1749-6632.1999.tb07628.x. ; discussion 117–108. [DOI] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes & Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bosscher K, Schmitz ML, Vanden Berghe W, Plaisance S, Fiers W, Haegeman G. Glucocorticoid-mediated repression of nuclear factor-κB-dependent transcription involves direct interference with transactivation. Proc Natl Acad Sci. 1997;94:13504–13509. doi: 10.1073/pnas.94.25.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenkov IJ, Webster EL, Torpy DJ, Chrousos GP. Stress, corticotropin-releasing hormone, glucocorticoids, and the immune/inflammatory response: Acute and chronic effects. Ann N Y Acad Sci. 1999;876:1–11. doi: 10.1111/j.1749-6632.1999.tb07618.x. ; discussion 11–13. [DOI] [PubMed] [Google Scholar]
- Freeman BC, Felts SJ, Toft DO, Yamamoto KR. The p23 molecular chaperones act at a late step in intracellular receptor action to differentially affect ligand efficacies. Genes & Dev. 2000;14:422–434. [PMC free article] [PubMed] [Google Scholar]
- Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-κ B p50 homodimer bound to a κ B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-κ B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Godowski PJ, Rusconi S, Miesfeld R, Yamamoto KR. Glucocorticoid receptor mutants that are constitutive activators of transcriptional enhancement. Nature. 1987;325:365–368. doi: 10.1038/325365a0. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Guzman E, Lis JT. Transcription factor TFIIH is required for promoter melting in vivo. Mol Cell Biol. 1999;19:5652–5658. doi: 10.1128/mcb.19.8.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada A, Sekido N, Akahoshi T, Wada T, Mukaida N, Matsushima K. Essential involvement of interleukin-8 (IL-8) in acute inflammation. J Leukoc Biol. 1994;56:559–564. [PubMed] [Google Scholar]
- Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Heck S, Bender K, Kullmann M, Gottlicher M, Herrlich P, Cato AC. I κB α-independent downregulation of NF-κB activity by glucocorticoid receptor. EMBO J. 1997;16:4698–4707. doi: 10.1093/emboj/16.15.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hengartner CJ, Myer VE, Liao SM, Wilson CJ, Koh SS, Young RA. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- Hong K, Zheng W, Baker A, Papahadjopoulos D. Stabilization of cationic liposome-plasmid DNA complexes by polyamines and poly(ethylene glycol)-phospholipid conjugates for efficient in vivo gene delivery. FEBS Lett. 1997;400:233–237. doi: 10.1016/s0014-5793(96)01397-x. [DOI] [PubMed] [Google Scholar]
- Howard KJ, Holley SJ, Yamamoto KR, Distelhorst CW. Mapping the HSP90 binding region of the glucocorticoid receptor. J Biol Chem. 1990;265:11928–11935. [PubMed] [Google Scholar]
- Iniguez-Lluhi JA, Lou DY, Yamamoto KR. Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem. 1997;272:4149–4156. doi: 10.1074/jbc.272.7.4149. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E, Wissink S, van der Saag PT, van der Burg B. Negative interaction between the RelA(p65) subunit of NF-κB and the progesterone receptor. J Biol Chem. 1996;271:6217–6224. doi: 10.1074/jbc.271.11.6217. [DOI] [PubMed] [Google Scholar]
- Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Konig H, Ponta H, Rahmsdorf HJ, Herrlich P. Interference between pathway-specific transcription factors: Glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. EMBO J. 1992;11:2241–2246. doi: 10.1002/j.1460-2075.1992.tb05283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes & Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunsch C, Lang RK, Rosen CA, Shannon MF. Synergistic transcriptional activation of the IL-8 gene by NF-κ B p65 (RelA) and NF-IL-6. J Immunol. 1994;153:153–164. [PubMed] [Google Scholar]
- Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Le Bail O, Schmidt-Ullrich R, Israel A. Promoter analysis of the gene encoding the I κ B-α/MAD3 inhibitor of NF-κ B: Positive regulation by members of the rel/NF-κ B family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Lis JT. Transcriptional activation independent of TFIIH kinase and the RNA polymerase II mediator in vivo. Nature. 1998;393:389–392. doi: 10.1038/30770. [DOI] [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Thomas JR, Yamamoto KR. Influence of a steroid receptor DNA-binding domain on transcriptional regulatory functions. Genes & Dev. 1994;8:2842–2856. doi: 10.1101/gad.8.23.2842. [DOI] [PubMed] [Google Scholar]
- Lehman AL, Dahmus ME. The sensitivity of RNA polymerase II in elongation complexes to C-terminal domain phosphatase. J Biol Chem. 2000;275:14923–14932. doi: 10.1074/jbc.275.20.14923. [DOI] [PubMed] [Google Scholar]
- Liden J, Delaunay F, Rafter I, Gustafsson J, Okret S. A new function for the C-terminal zinc finger of the glucocorticoid receptor. Repression of RelA transactivation. J Biol Chem. 1997;272:21467–21472. doi: 10.1074/jbc.272.34.21467. [DOI] [PubMed] [Google Scholar]
- Long JJ, Leresche A, Kriwacki RW, Gottesfeld JM. Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol Cell Biol. 1998;18:1467–1476. doi: 10.1128/mcb.18.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue NF, Chasman DI, Buchman AR, Kornberg RD. Interaction of GAL4 and GAL80 gene regulatory proteins in vitro. Mol Cell Biol. 1987;7:3446–3451. doi: 10.1128/mcb.7.10.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi BF, Xu BF, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Melcher K, Johnston SA. GAL4 interacts with TATA-binding protein and coactivators. Mol Cell Biol. 1995;15:2839–2848. doi: 10.1128/mcb.15.5.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, et al. IKK-1 and IKK-2: Cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Miner JN, Yamamoto KR. Regulatory crosstalk at composite response elements. Trends Biochem Sci. 1991;16:423–426. doi: 10.1016/0968-0004(91)90168-u. [DOI] [PubMed] [Google Scholar]
- Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- Moreland RJ, Tirode F, Yan Q, Conaway JW, Egly JM, Conaway RC. A role for the TFIIH XPB DNA helicase in promoter escape by RNA polymerase II. J Biol Chem. 1999;274:22127–22130. doi: 10.1074/jbc.274.32.22127. [DOI] [PubMed] [Google Scholar]
- Mukaida N, Gussella GL, Kasahara T, Ko Y, Zachariae CO, Kawai T, Matsushima K. Molecular analysis of the inhibition of interleukin-8 production by dexamethasone in a human fibrosarcoma cell line. Immunology. 1992;75:674–679. [PMC free article] [PubMed] [Google Scholar]
- Mukaida N, Morita M, Ishikawa Y, Rice N, Okamoto S, Kasahara T, Matsushima K. Novel mechanism of glucocorticoid-mediated gene repression. Nuclear factor-κ B is target for glucocorticoid-mediated interleukin 8 gene repression. J Biol Chem. 1994;269:13289–13295. [PubMed] [Google Scholar]
- Naar AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Newton R, Hart LA, Stevens DA, Bergmann M, Donnelly LE, Adcock IM, Barnes PJ. Effect of dexamethasone on interleukin-1β-(IL-1β)-induced nuclear factor-κB (NF-κB) and κB-dependent transcription in epithelial cells. Eur J Biochem. 1998;254:81–89. doi: 10.1046/j.1432-1327.1998.2540081.x. [DOI] [PubMed] [Google Scholar]
- Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Oro AE, McKeown M, Evans RM. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature. 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Palvimo JJ, Reinikainen P, Ikonen T, Kallio PJ, Moilanen A, Janne OA. Mutual transcriptional interference between RelA and androgen receptor. J Biol Chem. 1996;271:24151–24156. doi: 10.1074/jbc.271.39.24151. [DOI] [PubMed] [Google Scholar]
- Parada CA, Roeder RG. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature. 1996;384:375–378. doi: 10.1038/384375a0. [DOI] [PubMed] [Google Scholar]
- Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Conrad NK, Bregman DB, Corden JL. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J Biol Chem. 1999;274:27823–27828. doi: 10.1074/jbc.274.39.27823. [DOI] [PubMed] [Google Scholar]
- Perissi V, Staszewski LM, McInerney EM, Kurokawa R, Krones A, Rose DW, Lambert MH, Milburn MV, Glass CK, Rosenfeld MG. Molecular determinants of nuclear receptor-corepressor interaction. Genes & Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6:3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D, Salser SJ, Yamamoto KR. A movable and regulable inactivation function within the steroid binding domain of the glucocorticoid receptor. Cell. 1988;54:1073–1080. doi: 10.1016/0092-8674(88)90122-5. [DOI] [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20:2629–2634. doi: 10.1128/mcb.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C, Gamble M, Chang CP, Atkins GB, Lazar MA, Freedman LP. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Mol Cell Biol. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Naar AM, Erdjument-Bromage H, Tempst P, Freedman LP. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Ray A, Prefontaine KE. Physical association and functional antagonism between the p65 subunit of transcription factor NF-κ B and the glucocorticoid receptor. Proc Natl Acad Sci. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A, Prefontaine KE, Ray P. Down-modulation of interleukin-6 gene expression by 17 β-estradiol in the absence of high affinity DNA binding by the estrogen receptor. J Biol Chem. 1994;269:12940–12946. [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Rusconi S, Yamamoto KR. Functional dissection of the hormone and DNA binding activities of the glucocorticoid receptor. EMBO J. 1987;6:1309–1315. doi: 10.1002/j.1460-2075.1987.tb02369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu S, Zhou S, Ladurner AG, Tjian R. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature. 1999;397:446–450. doi: 10.1038/17141. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS., Jr Role of transcriptional activation of I κ B α in mediation of immunosuppression by glucocorticoids. Science. 1995a;270:283–286. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin AS., Jr Characterization of mechanisms involved in transrepression of NF-κ B by activated glucocorticoid receptors. Mol Cell Biol. 1995b;15:943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Freedman LP, Yamamoto KR. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes & Dev. 1989;3:1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- Scherer DC, Brockman JA, Chen Z, Maniatis T, Ballard DW. Signal-induced degradation of I κ B α requires site-specific ubiquitination. Proc Natl Acad Sci. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz ML, Stelzer G, Altmann H, Meisterernst M, Baeuerle PA. Interaction of the COOH-terminal transactivation domain of p65 NF-κ B with TATA-binding protein, transcription factor IIB, and coactivators. J Biol Chem. 1995;270:7219–7226. doi: 10.1074/jbc.270.13.7219. [DOI] [PubMed] [Google Scholar]
- Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- Sil AK, Alam S, Xin P, Ma L, Morgan M, Lebo CM, Woods MP, Hopper JE. The Gal3p-Gal80p-Gal4p transcription switch of yeast: Gal3p destabilizes the Gal80p-Gal4p complex in response to galactose and ATP. Mol Cell Biol. 1999;19:7828–7840. doi: 10.1128/mcb.19.11.7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Kraut R, Hoey T, Warrior R, Levine M. Transcriptional regulation of a pair-rule stripe in Drosophila. Genes & Dev. 1991;5:827–839. doi: 10.1101/gad.5.5.827. [DOI] [PubMed] [Google Scholar]
- Spaete RR, Mocarski ES. Regulation of cytomegalovirus gene expression: Alpha and beta promoters are trans activated by viral functions in permissive human fibroblasts. J Virol. 1985;56:135–143. doi: 10.1128/jvi.56.1.135-143.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DB, Matsui W, Thomas JR, Yamamoto KR. Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes & Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]
- Stein B, Yang MX. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-κ B and C/EBP β. Mol Cell Biol. 1995;15:4971–4979. doi: 10.1128/mcb.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun SC, Ganchi PA, Ballard DW, Greene WC. NF-κ B controls expression of inhibitor I κ B α: Evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- Sun X, Zhang Y, Cho H, Rickert P, Lees E, Lane W, Reinberg D. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol Cell. 1998;2:213–222. doi: 10.1016/s1097-2765(00)80131-8. [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ, Vichi P, Egly JM. The multiple roles of transcription/repair factor TFIIH. Trends Biochem Sci. 1996;21:346–350. [PubMed] [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Tenenhaus C, Schubert C, Seydoux G. Genetic requirements for PIE-1 localization and inhibition of gene expression in the embryonic germ lineage of Caenorhabditis elegans. Dev Biol. 1998;200:212–224. doi: 10.1006/dbio.1998.8940. [DOI] [PubMed] [Google Scholar]
- Triezenberg SJ, Kingsbury RC, McKnight SL. Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes & Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- Tsai MJ, O'Malley BW. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu Rev Biochem. 1994;63:451–486. doi: 10.1146/annurev.bi.63.070194.002315. [DOI] [PubMed] [Google Scholar]
- van de Stolpe A, Caldenhoven E, Raaijmakers JA, van der Saag PT, Koenderman L. Glucocorticoid-mediated repression of intercellular adhesion molecule-1 expression in human monocytic and bronchial epithelial cell lines. Am J Respir Cell Mol Biol. 1993;8:340–347. doi: 10.1165/ajrcmb/8.3.340. [DOI] [PubMed] [Google Scholar]
- van de Stolpe A, Caldenhoven E, Stade BG, Koenderman L, Raaijmakers JA, Johnson JP, van der Saag PT. 12-O-tetradecanoylphorbol-13-acetate- and tumor necrosis factor α-mediated induction of intercellular adhesion molecule-1 is inhibited by dexamethasone. Functional analysis of the human intercellular adhesion molecular-1 promoter. J Biol Chem. 1994;269:6185–6192. [PubMed] [Google Scholar]
- Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissink S, van Heerde EC, Schmitz ML, Kalkhoven E, van der Burg B, Baeuerle PA, van der Saag PT. Distinct domains of the RelA NF-κB subunit are required for negative cross-talk and direct interaction with the glucocorticoid receptor. J Biol Chem. 1997;272:22278–22284. doi: 10.1074/jbc.272.35.22278. [DOI] [PubMed] [Google Scholar]
- Wissink S, van Heerde EC, vand der Burg B, van der Saag PT. A dual mechanism mediates repression of NF-κB activity by glucocorticoids. Mol Endocrinol. 1998;12:355–363. doi: 10.1210/mend.12.3.0081. [DOI] [PubMed] [Google Scholar]
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- Yamamoto KR. Multilayered control of intracellular receptor function. Harvey Lect. 1995;91:1–19. [PubMed] [Google Scholar]
- Yamamoto KR, Darimont BD, Wagner RL, Iniguez-Lluhi JA. Building transcriptional regulatory complexes: Signals and surfaces. Cold Spring Harb Symp Quant Biol. 1998;63:587–598. doi: 10.1101/sqb.1998.63.587. [DOI] [PubMed] [Google Scholar]
- Yamit-Hezi A, Dikstein R. TAFII105 mediates activation of anti-apoptotic genes by NF-κB. EMBO J. 1998;17:5161–5169. doi: 10.1093/emboj/17.17.5161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Fukasawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YO, Stroke IL, Fields S. Coupling of cell identity to signal response in yeast: Interaction between the α 1 and STE12 proteins. Genes & Dev. 1993;7:1584–1597. doi: 10.1101/gad.7.8.1584. [DOI] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-κ B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]