Abstract
Muscle fibres have different properties with respect to force, contraction speed, endurance, oxidative/glycolytic capacity etc. Although adult muscle fibres are normally post-mitotic with little turnover of cells, the physiological properties of the pre-existing fibres can be changed in the adult animal upon changes in usage such as after exercise. The signal to change is mainly conveyed by alterations in the patterns of nerve-evoked electrical activity, and is to a large extent due to switches in the expression of genes. Thus, an excitation-transcription coupling must exist. It is suggested that changes in nerve-evoked muscle activity lead to a variety of activity correlates such as increases in free intracellular Ca2+ levels caused by influx across the cell membrane and/or release from the sarcoplasmatic reticulum, concentrations of metabolites such as lipids and ADP, hypoxia and mechanical stress. Such correlates are detected by sensors such as protein kinase C (PKC), calmodulin, AMP-activated kinase (AMPK), peroxisome proliferator-activated receptor δ (PPARδ), and oxygen dependent prolyl hydroxylases that trigger intracellular signaling cascades. These complex cascades involve several transcription factors such as nuclear factor of activated T-cells (NFAT), myocyte enhancer factor 2 (MEF2), myogenic differentiation factor (myoD), myogenin, PPARδ, and sine oculis homeobox 1/eyes absent 1 (Six1/Eya1). These factors might act indirectly by inducing gene products that act back on the cascade, or as ultimate transcription factors binding to and transactivating/repressing genes for the fast and slow isoforms of various contractile proteins and of metabolic enzymes. The determination of size and force is even more complex as this involves not only intracellular signaling within the muscle fibres, but also muscle stem cells called satellite cells. Intercellular signaling substances such as myostatin and insulin-like growth factor 1 (IGF-1) seem to act in a paracrine fashion. Induction of hypertrophy is accompanied by the satellite cells fusing to myofibres and thereby increasing the capacity for protein synthesis. These extra nuclei seem to remain part of the fibre even during subsequent atrophy as a form of muscle memory facilitating retraining. In addition to changes in myonuclear number during hypertrophy, changes in muscle fibre size seem to be caused by alterations in transcription, translation (per nucleus) and protein degradation.
Keywords: skeletal muscle, exercise, plasticity, contraction speed, atrophy, hypertrophy, fiber type
Contents
-
Introduction…………565
Muscle fibre phenotypes…………565
Changes in muscle fibre phenotypes…………566
Determinants of muscle fibre phenotype…………567
The importance of cell lineage…………567
-
Cell external signals…………567
What are the signals from the nerve?…………567
-
The importance of nerve-evoked muscle activity…………568
The effects of normal and mismatching activity patterns on muscle contractile properties…………568
Dissecting the patterns…………569
Mechanical stress…………570
-
Intracellular signals…………571
-
Calcium…………571
The source of elevated [Ca2+]i during activity…………571
How does [Ca2+]i fluctuate with different activity patterns?…………572
-
Calcium sensors: decoding the calcium fluctuations…………572
Calmodulin and its targets CaMKII and calcineurin…………572
Protein kinase C…………574
-
Cascades downstream of [Ca2+]i…………575
Ras andMAPK…………575
Protein kinase D…………575
Histone deacetylase…………576
PGC-1α and—β…………576
-
Transcription factors downstream of [Ca2+]i…………577
NFAT…………577
Does NFAT bind to fast or slow promoters?…………578
MEF2…………579
Myogenin and MyoD: the yin and yang of muscle plasticity?…………579
MyoD in slow-to-fast transformations…………580
Myogenin as a glycolytic-to-oxidative transforming agent…………581
-
Metabolic activity correlates…………582
AMP-kinase…………582
PPARδ…………582
Oxygen…………583
Six 1 and eya 1…………584
MusTRD…………585
-
-
Plasticity of muscle force…………585
The cell biology of muscle fibre size…………585
-
Paracrine and autocrine mechanisms…………587
Myostatin…………587
Insulin-like growth factor I (IGF-1)…………587
-
Intracellular pathways of size regulation…………588
-
Regulation of protein production in muscle fibres…………588
Regulation of transcription…………588
Regulation of translation: the PI(3)K/Akt pathway…………588
Regulation of protein degradation…………588
Putative mechano-sensing mechanisms in muscle size regulation…………589
-
-
The excitation-transcription coupling: towards a synthesis?…………589
Plasticity of speed…………589
Plasticity offorce…………590
Conclusions…………590
Acknowledgements…………590
References…………590
I. INTRODUCTION
The ability of adult muscle fibres to change in response to external stimuli has been called muscle plasticity. Force, contraction speed, endurance and oxidative/glycolytic capacity are all examples of muscle properties that are plastic. Skeletal muscle is a permanent, post-mitotic tissue, and unless the muscle is damaged there is little turnover of cells (Bintliff & Walker, 1960; Moss & Leblond, 1970, 1971; Stockdale & Holtzer, 1961). Thus, it has been demonstrated that dramatic changes in gene expression, protein composition and physiological properties can occur in pre-existing fibres without de- or regeneration (Delp & Pette, 1994; Gorza et al., 1988; Mayne et al., 1993; Windisch et al., 1998). The plastic changes occur mainly by reprogramming the cell by turning on and off sets of relevant genes.
(1) Muscle fibre phenotypes
It has long been known that skeletal muscles differ in phenotype (Ranvier, 1874) and there is a tendency for sets of properties to be connected, or clustered which has led to the notion of distinct muscle fibre types. The notion of type was supported by the observation that different myosin heavy chain (MyHC) ATPases have different pH optima allowing histochemical procedures to display a checkerboard-like staining pattern with distinct black and white fibres observed in cross sections (Brooke & Kaiser, 1970). A more modern approach has been to categorize muscle fibre type based on immunohistochemical labeling of distinct MyHC isoforms (Schiaffino et al., 1989). The foundation for muscle fibre typing has been reviewed previously (Pette & Staron, 2000, 2001; Schiaffino & Reggiani, 1996), but a summary of fibre-type properties of limb muscles and the current fibre-type nomenclature is given in Fig. 1.
Fig. 1.
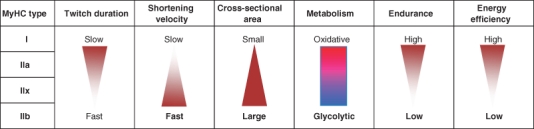
The properties of the four major fibre types found in mammalian muscle. There are exceptions to this general scheme. MyHC, myosin heavy chain.
Originally, fibres were classified into three types based on histochemistry: I, IIa and IIb, with an additional IIc as a hybrid form. In rodents a fourth form was subsequently identified based on monoclonal antibodies against MyHC (Schiaffino et al., 1989). This unknown “X” was dubbed IIx and is also called IId by some authors. In humans, fibres which had previously been called IIb express a MyHC gene that turned out to be homologous to the IIx gene in rodents (Ennion et al., 1995; Smerdu et al., 1994). Hence human fibre types are currently classified as I, IIa and IIx, where IIx is the former IIb described in older literature. The human genome also contains a gene homologous to the rodent IIb gene (Weiss, Schiaffino & Leinwand, 1999), but so far protein expression from this gene has not been demonstrated. In addition to MyHC, a large number of other contractile and structural proteins also come in distinct isoforms for which expression is more or less tightly connected to fibre type (Schiaffino & Reggiani, 1996). In addition to the three or four major MyHC genes expressed in adult limb muscles there are specialized forms expressed during development and in gill-arch-derived muscles (Hoh, 2002). In total 10 different MyHC genes have been connected to skeletal muscle (Desjardins et al., 2002).
Analysis of physiological properties such as strength, twitch speed and endurance of single motor units suggests that units can be categorized into distinct largely non-overlapping groups correlating with their histochemical properties (Burke, 1967; Burke et al., 1974; Burke, Levine & Zajac, 1971; Dum et al., 1982); but the physiological properties of units of the same histochemical type can differ among muscles (Burke, 1967). In addition, although fibres belonging to one motor unit are all of the same MyHC type, measurements at the motor unit level might underestimate heterogeneity at the single fibre level. For example it has been demonstrated that both the shortening velocity and the MyHC composition might vary along the length of the fibre (Edman, Reggiani & Kronnie, 1985). While histochemical or immunohistochemical staining might give the impression that the vast majority of fibres are positive only for one MyHC, single-fibre gel electrophoresis has revealed that 11–67% of the fibres from various limb muscles express more than one MyHC isoform even under steady-state activity conditions (Stephenson, 2001). It can be concluded that the concept of universal fibre types throughout the body is an oversimplification.
The molecular foundation for the variability in contractile properties is partly known. For example, the MyHC isoform determines shortening velocity (the sliding velocity between actin and myosin), but also the ratio between the myosin light chain 3f and 2f isoform has a major influence. This ratio varies widely among individual fibres of the same MyHC type giving rise to rather large variability in shortening velocity within the type (Bottinelli & Reggiani, 2000; Schiaffino & Reggiani, 1996). Twitch speed is dependent on shortening velocity, but also on Ca2+ sequestering systems such as parvalbumin and the sarcoplasmic reticulum Ca2+ ATPases (SERCAs) (Gundersen et al., 1988). Isoforms of SERCA are differentially expressed in different fibre types, thus SERCA2a is the predominant form in type I fibres while SERCA1a is expressed mainly in type II fibres (Periasamy & Kalyanasundaram, 2007). Although muscle endurance and fatigue seems to rely on many cellular factors (Allen, Lamb & Westerblad, 2008), endurance is correlated to high oxidative capacity with a high content of mitochondria and oxidative enzymes, and such properties are again linked to MyHC fibre type (Fig. 1).
(2) Changes in muscle fibre phenotypes
The physiological properties (shortening velocity, twitch duration, endurance, etc.) that are linked in a fibre type are related to highly different molecular families (MyHC, SERCA, metabolic enzymes, etc.). As discussed below, coupling regulation of different physiological properties may be beneficial from an energy conservation point of view, and/or it might reflect common signaling systems diverging to regulate several sets of diverse genes. To some extent however, different properties can be uncoupled and regulated independently during plastic changes. For example, some degree of uncoupling has been observed between twitch speed and shortening velocity (Eken & Gundersen, 1988). More importantly, endurance-exercise in man and in other animals can lead to pronounced changes in metabolic properties without MyHC fibre-type conversion (Henriksson & Hickner, 1994; Saltin & Gollnick, 1983; Fitts & Widrick, 1996), although exercise can also change MyHC type in particular within type II (e.g. from type IIb/IIx to IIa) (Abdelmalki et al., 1996; Andersen, Klitgaard & Saltin, 1994; Jansson et al., 1990; Kraemer et al., 1995; Staron et al., 1994; Wang et al., 1993), and under more severe experimental conditions fibre-type conversions are frequent.
When fibre-type conversions occur, it usually happens in a sequential order (Pette & Staron, 2000; Schiaffino & Reggiani, 1996): I ↔ IIa ↔ IIx ↔ IIb. During transitions hybrids between the “nearest neighbour” fibre type in this flow chart (e.g. I+IIa, IIa+IIx) are common, but aberrant hybrids such as I+IIb, I+IIa+IIb and I+IIx+IIb can also be seen under some experimental conditions (Caiozzo, Baker & Baldwin, 1998).
(3) Determinants of muscle fibre phenotype
This review will discuss the factors that determine the molecular make-up of already formed adult muscles, and how that make-up can change. At any point in time, a fibre's make-up appears to depend on previous: (1) cell history/lineage; (2) nerve-evoked electrical activity; (3) mechanical conditions; (4) para-/autocrine conditions; and (5) circulating hormones.
Muscle properties are strongly influenced by hormones such as testosterone and thyroid hormones, as reviewed previously (Kadi, 2008; Pette & Staron, 2001; Staron et al., 2000). This review will focus on the link between external factors related to activity and usage (points 2 and 3 above) and gene expression. The ability to change is, however, constrained by the muscle's cell lineage.
(4) The importance of cell lineage
Developmental studies suggest that initial fibre-type differentiation might be determined by myoblast cell lineage independent of external influences such as innervation or usage. Thus, in experiments with quail chick chimera, limb myoblast diversity arises prior to the migration of myoblasts into the limb (Van Swearingen & Lance-Jones, 1995), and fibre-type distribution resembling the normal pattern is displayed even if the nerve is absent during development (Butler, Cosmos & Brierley, 1982; Condon et al., 1990; Phillips & Bennett, 1984). When mammalian muscles are transplanted and made to regenerate in a different body location, some of the information determining fibre type is apparently derived from the muscle of origin rather than from the new position or innervation (Gutmann & Carlson, 1975; Hoh & Hughes, 1991; Pin et al., 2002; Pin & Merrifield, 1997).
In vitro avian myoblasts constitute clones that give rise to specific fibre types (DiMario, Fernyak & Stockdale, 1993; DiMario & Stockdale, 1997; Miller & Stockdale, 1986), and when satellite cells derived from single mouse fibres form new myotubes, these express a MyHC type reminiscent of the fibre from which the satellite cells were derived (Rosenblatt, Parry & Partridge, 1996; but see also Bonavaud et al., 2001). In adult rats, when different muscles are regenerating from myoblasts after myofibre destruction, the various regenerated muscles express different MyHC types reminiscent of the muscle of origin. This happens even if the muscles receive similar experimental patterns of activity. Thus, when regenerating soleus and extensor digitorum longus (EDL) were stimulated by the same slow pattern, the EDL failed to express the large amount of slow MyHC that was observed in the soleus under the same conditions (Kalhovde et al., 2005).
It can be concluded that muscle fibre pedigree matters, and that there is a cell line component to the resulting adult phenotype of a fibre. The relationship is, however, not simple, since experiments with genetically marked myoblasts suggest that single myoblast clones can contribute to both fast and slow fibres, clones are not restricted to contribute to subsets of fibre types, and clones show no detectable preference for fusion to a particular fibre type (Hughes & Blau, 1992).
II. CELL EXTERNAL SIGNALS
While it seems clear that cell lineage limits the adaptive range of muscle plasticity, it is equally clear that external signals can change muscle phenotype in the adult. In particular signals from the nerve appear to be important.
(1) What are the signals from the nerve?
The field of muscle plasticity began with the seminal paper of Buller, Eccles & Eccles (1960) who showed that when a nerve from a fast muscle is transplanted to a slow muscle, and vice versa, both the reinnervated muscles change phenotype according to the new nerve supply. Subsequently, several other studies have confirmed this principle (reviewed by Close, 1972).
There were originally two theories for how the nerve could influence the muscle. The preferred hypothesis at the time suggested that different trophic substances were released from nerves supplying fast and slow muscles. But it was also recognized that differences in the pattern of action potentials in fast and slow motor units somehow could contain a coded message for muscle fibre change (Buller et al., 1960).
While there is detailed information about the differences in the activity pattern of fast and slow motor units (Hennig & Lømo, 1985; Lømo, 2009), searches for relevant neurotrophic substances have failed. As discussed in detail in a previous review, there is little direct evidence for the existence of such factors regulating muscle contractile properties (Gundersen, 1998; but see Salviati, Biasia & Aloisi, 1986). In fact, stimulation of denervated muscles with patterns of activity resembling the activity in fast and slow motor units mimic virtually all the effects of cross innervation (Eken & Gundersen, 1988; Windisch et al., 1998).
The old concept of neurotrophic (from the Greek trophos = nourishing) substances was based on the observation that denervation led to severe atrophy, but stimulating denervated muscles with implanted electrodes largely restores strength even after long-term denervation in both rats (Hennig & Lømo, 1987), and humans (Boncompagni et al., 2007). Although the stimulated muscles never regain normal strength, activity is comparable to self-reinnervation in restoring maximal tetanic force production (Hennig & Lømo, 1987).
The converse experiment: to compare denervation to removal of action potential activity with an otherwise intact nerve supply, has led to more controversial results. Isolation of the spinal cord leads to less severe atrophy than denervation in the medial gastrocnemius and tibialis anterior muscles (but not in the soleus) (Hyatt et al., 2003, 2006). These findings have been claimed as evidence for an important non-activity source of neural control (Roy et al., 2008). It is however not unlikely that the difference is rather caused by small amounts of residual activity elicited in the isolated spinal cord. Stimulation of denervated muscles has shown that small amounts of electrical activity have a strong effect on muscle properties. For example a brief 0.6 s long high-frequency pulse train delivered every 100 min led to a sixfold increase in tetanic force of denervated muscles (Westgaard & Lømo, 1988). Moreover, in contrast to the experiments with spinal isolation, nerve impulse block with tetrodotoxin applied to the nerve, but with otherwise intact axons, induced muscle mass loss, fibre atrophy and reduction in force quantitatively indistinguishable from those induced by complete nerve transsection in rats (Buffelli, Pasino & Cangiano, 1997). It should be noted that extraordinary precautions were required to ensure complete absence of nerve-evoked activity even in this model.
In conclusion, there is currently no compelling evidence to suggest that there are any relevant sources of neural influence on the muscle other than activity, and in spite of intense searching for several decades, no neurotrophic substances have been identified that prevent atrophy or mimic other effects of normal innervation or cross-innervation outside the synaptic zone.
(2) The importance of nerve-evoked muscle activity
(a) The effects of normal and mismatching activity patterns on muscle contractile properties
There are distinct differences in the pattern of activity evoked in fast and slow muscles (Hennig & Lømo, 1985). Generally type I motor units seem to receive high amounts of impulses delivered in long low-frequency trains, while type II units seem to receive short bursts of high-frequency activity. The total amount of impulses delivered to type II units is lower, but the amount seems to vary among the type II subtypes (Table 1).
Table 1.
Firing characteristics of motor units in rats. Data are from Hennig & Lomo (1985) and represent the range of the means for 5–6 units followed over 24 h
| Assumed fibre type | Instantaneous frequency (Hz) | Number of impulses in 24 hrs. | Number of impulses per train | % time active | Longest train duration |
|---|---|---|---|---|---|
| I | 18–21 | 309 500–495 800 | 5–10 | 22–35 | 5–9 min |
| IIa or IIx | 41–71 | 89 500–243 100 | 3–39 | 1.6–5.0 | 1.5–2.4 min |
| IIb | 67–91 | 2 600–11 200 | 3–13 | 0.04–0.22 | 0.83–3.9 s |
The importance of electrical activity has been confirmed by a large number of studies subsequent to the pioneering work of G. Vrbová and T. Lømo. Vrbová and collaborators demonstrated the importance of activity by stimulating muscles via the nerve (Vrbová, 1963), while Lømo's group studied denervated muscles and demonstrated the importance of muscle activity per se in the absence of any nerve influence (Lømo, Westgaard & Dahl, 1974).
In nerve-stimulation studies large amounts of low-frequency activity have been superimposed on normal background activity, leading to a fast-to-slow transformation. D. Pette and collaborators have, in a series of papers, described these changes extensively at the physiological and molecular/biochemical level (for reviews, see Pette & Vrbová, 1999, 1992). Since during nerve stimulation, the exogenous activity is superimposed on activity from the central nervous system, this limits pattern control. In particular, since the external activity is always added, the effect of a reduced amount of activity cannot be studied in innervated muscles. In slow muscles the large amounts of background activity will dominate, and preclude a study of slow-to-fast transformation. In addition, nerve stimulation does not preclude the possibility that the activity effects are secondary to changes in the motor neurone, for example via the release of putative neurotrophic substances (Gundersen, 1998).
An ideal model is to block nerve impulse activity with tetrodotoxin, and then stimulate the inactive nerve below the block (Ekmark et al., 2007). The model is however technically demanding, and most of what we know about the importance of the pattern of activity is derived from stimulating denervated muscles directly. In such experiments pattern specificity has been demonstrated. If a pattern mimicking the native activity of a fibre type is used, more or less normal properties are maintained, while a mismatching pattern induces changes: a fast pattern leads to a slow-to-fast transformation in slow muscles; a slow pattern induces a fast-to-slow transformation in fast muscles. The physiological/molecular transformation encompasses metabolic properties/oxidative enzymes (Gundersen et al., 1988), twitch duration/Ca2+-ATPase activity (Lømo et al., 1974; Westgaard & Lømo, 1988), and shortening velocity/MyHC type (Ausoni et al., 1990; Eken & Gundersen, 1988; Gorza et al., 1988; Gundersen & Eken, 1992; Gundersen et al., 1988; Schiaffino et al., 1988, 1989; Windisch et al., 1998).
(b) Dissecting the patterns
Simply mimicking the complex differences in the firing characteristics of fast and slow motor units (Table 1) does not provide information about which aspect of the differences is important, and more detailed experiments have been designed to unravel the “Morse code”.
The most comprehensive study is by Westgaard & Lømo (1988). They studied the effects of different patterns of activity on isometric contraction. In particular, they showed a tight frequency dependence of twitch duration when the total number of impulses was kept constant. Thus, the twitch time-to-peak of the soleus fell from about 30 ms to 11 ms when the instantaneous frequency was increased from 1 Hz to 300 Hz (Fig. 2A). There is a good teleological explanation for the frequency dependence of the twitch: the twitch determines the fusion frequency of the tetanus, and in order to ensure efficient force regulation, the steep portion of the force-frequency curve should be matched to the firing frequency of the motorneurone. As can be seen in Fig. 2C, the steep portion of the curve moved towards the stimulation frequency. In addition to the frequency effect it was shown that when the frequency was kept constant at 10 or 100 Hz, respectively, higher amounts of stimuli led to slower twitches (Fig. 2B). This amount effect has led some authors to challenge the importance of frequency (Al-Amood & Lewis, 1987; Donselaar et al., 1987; Kernell, Donselaar & Eerbeek, 1989). A distinct effect of frequency was demonstrated however; when 100 Hz bursts of stimulation were superimposed on a 10 Hz pattern, the twitch speed was reduced compared to the effect of the low-frequency pattern alone (Westgaard & Lømo, 1988). Thus, the high-frequency bursts made the twitch faster in spite of adding to the total amount of activity.
Fig. 2.
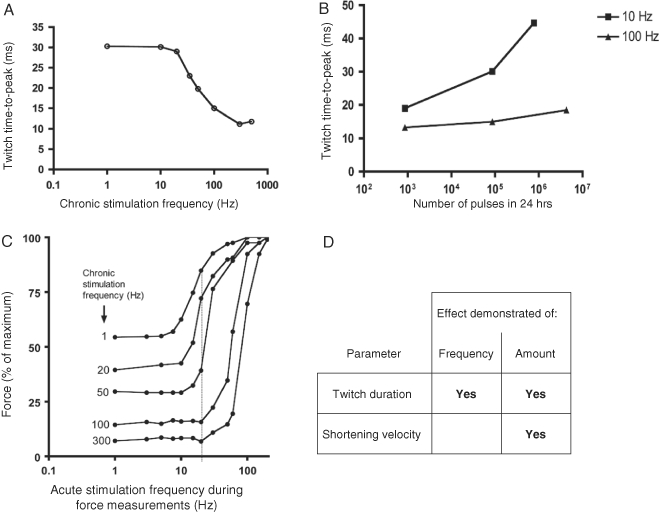
The effects of different chronic stimulation patterns on contractile properties of rat muscle. Twitch time-to peak seems to be regulated both by frequency (A), and amount (B). Teleologically, a frequency dependence of the twitch duration might ensure a match between the prevailing frequency received by the muscle and the frequency range that is efficient in regulating force by summation (C). In C note how the steep part of the force frequency curve moves towards the stimulation frequency. See for example how the force of an unfused tetanus with 20 Hz stimulation (broken vertical line) is on the steep portion of the curve for muscles receiving 20 Hz chronic stimulation. The data are replotted from Westgaard & Lømo (1988). (D) Summary of the dependence of different parameters on frequency and amount of chronic stimulation [for further details see text, Gundersen (1998) and Gundersen & Eken (1992)].
The twitch duration does not directly reflect sliding velocity between actin and myosin or MyHC type, as it is also heavily dependent on the kinetics of Ca2+ handling. There is much less information on the effects of electrical activity on isotonic shortening velocity or MyHC content. In a study where the train duration and the repetition rate of the trains (but not total number of impulses) was kept constant and the instantaneous frequency varied, low amounts of 20 Hz stimulation were as efficient in increasing shortening velocity of the soleus muscle as high-frequency patterns (Gundersen & Eken, 1992). Considering the effects of all the various activity patterns published there is no positive evidence that shortening velocity or MyHC composition is dependent on instantaneous frequency, and the amount of stimulation seems to be the important factor (Gundersen, 1998).
In conclusion, as summarized in Fig. 2D, the twitch duration seems to be controlled by both frequency and amount of activity, while there are no data demonstrating that frequency is important for shortening velocity or MyHC composition. Teleologically it would seem beneficial if muscles that are used frequently become more energy efficient. Both longer twitches and slower MyHC type contribute to higher energy efficiency for comparable amounts of external work (Gibbs & Gibson, 1972; Wendt & Gibbs, 1973).
Since the amount of activity seems to play a vital role, it seems important to determine what aspect of amount is of consequence. Amount has several attributes such as: (1) number of pulses per contraction; (2) duration of each contraction; (3) total number of pulses over long time periods (e.g. 24 h); (4) fraction of time a muscle remains contracted; and (5) duration of rest periods. Since these parameters do not change independently, it has been difficult to dissect the decisive factor, but for maintenance of normal fast contractile properties in the EDL, the number of impulses per train/contractile event (attribute 1) seems to be important for regulating shortening velocity and hence probably also MyHC type (Gundersen & Eken, 1992). This effect must somehow be decoded by a molecular impulse “counter” registering the number of impulses per activity episode, or if attribute 3 is important registering the number of impulses over longer periods of time. If attributes such as the fraction of time occupied by activity (4), the duration of rest periods (5) or the duration of each contractile event (2) are important, there must be a molecular “timer” rather than a “counter”. As discussed below, calcium-dependent kinases and phosphatases might serve as such “counters” or “timers” of nerve-evoked action potentials.
(3) Mechanical stress
Action potential activity leads not only to depolarization and triggering of intracellular signals downstream of the depolarization, but also leads to shortening and/or mechanical tension. It is widely assumed that contraction against a resistance leads to larger muscles than contraction against lower resistance, but this does not necessarily have a direct bearing on the importance of mechanical factors as such. Thus, both the recruitment and the activity pattern of each motor unit vary with the force output. One model that has been used in attempts to manipulate mechanical and electrical factors more independently is hind limb suspension. This is a procedure where rats are chronically lifted by the tail so as to unload the hind limbs leading to atrophy. Initially, there is a halt in electrical activity as judged by an integrated electromyogram (EMG). The integrated EMG however appears gradually to recover to normal levels within a few days whereas muscle atrophy continues to progress (Alford et al., 1987). This could mean that action potential activity and downstream events such as for example calcium release is of relatively little importance, and that a more important role should be postulated for force generation per se. This conclusion is however based on integrated EMGs only. Detailed information about firing properties of single motor units during hind limb suspension is not available, and, at least with intact feedback from proprioreceptors, it seems highly unlikely that the activity pattern in limbs that are not developing force should be identical to those of muscles exerting normal external force. Thus, hind limb suspension is not an optimal model for separating effects of mechanical stress and electrical activity.
The most compelling evidence for a mechano-dependent mechanism comes from experiments where limbs have been immobilized by a cast. This leads to atrophy, but studies over almost 100 years have shown that atrophy can be partly counteracted when muscles are immobilized in a lengthened position rather than a shortened position (Booth, 1977; Ferguson, Vaughan & Ward, 1957; Fournier et al., 1983; Froboese, 1922; Goldspink, 1977; Herbert & Balnave, 1993; Kurakami, 1966; Meyer, 1922; Ralston, Feinstein & Inman, 1952; Savolainen et al., 1988; Tabary et al., 1972; Tardieu et al., 1969; Thomsen & Luco, 1944; Yang et al., 1997).
There are also studies suggesting that muscle length influences contraction speed such that chronic stretch makes a muscle slower; immobilization of fast muscles in a lengthened position thus increases the fraction of slow fibres (Goldspink, 1999; Goldspink et al., 1992, 1991; Loughna et al., 1990; Pattullo et al., 1992). Similarly, overload elicited by ablation of synergists leads to pronounced changes in the slow direction (Gregory et al., 1990; Gregory, Low & Stirewalt, 1986; Ianuzzo et al., 1989; Ianuzzo, Gollnick & Armstrong, 1976; Kandarian, Schulte & Esser, 1992; Morgan & Loughna, 1989; Noble, Dabrowski & Ianuzzo, 1983; Periasamy et al., 1989; Roy et al., 1985; Tsika, Herrick & Baldwin, 1987). Again, the effect of length or load in these experiments could be secondary to the effect that these procedures have on the activity pattern. In fact, the properties of the motorneurones are dependent on muscle length, as it has been shown that the duration of the after-hyper-polarization, which is correlated to firing frequency, can be influenced by the length at which the muscles are immobilized (Gallego et al., 1979). Moreover, integrated EMG measurements indicated that the amount of activity is lower when the muscle is immobilized in the shortened position (Fournier et al., 1983; Hnik et al., 1985). Thus, the apparent effects of length and load could be largely secondary to activity changes and changes in motor neurons rather than to mechanical factors.
A few length studies have been performed on denervated muscles showing that immobilization in the stretched position (Cotlar, Thrasher & Harris, 1963; Summers & Hines, 1951; Loughna & Morgan, 1999; Savolainen et al., 1988) or tenotomy of synergistic muscles (Herbison, Jaweed & Ditunno, 1975; Schiaffino & Hanzlikova, 1970) counteracts atrophy in absence of the nerve. Moreover there is a minor counteracting effect of mechanical stretch on denervation atrophy, and similarly in tissue culture stretching myotubes increases protein synthesis (Vandenburgh & Kaufman, 1979) and decreases proteolysis (Vandenburgh & Kaufman, 1980).
Attempts have been made to train rat muscles by standardized electrical nerve stimulation, but letting the muscle contract under eccentric, isometric or shortening isotonic conditions. After eight weeks of training there were, however, no clear effects of the mechanical conditions on muscle mass (Adams et al., 2004), but levels of some signaling substances such as myostatin and insulin-like-growth factor 1 (IGF-1) varied (Heinemeier et al., 2007).
In summary, in most experimental conditions it is hard to separate electrical activity and mechanical stretch, but some experimental data point to the presence of an activity-independent mechanical mechanism influencing muscle size and perhaps contraction speed. The quantitative importance of mechanical mechanisms still seems somewhat elusive, and more studies where both activity and force are controlled separately should be performed.
III. INTRACELLULAR SIGNALS
There is a consensus that changes in muscle usage will transform muscle phenotype, but the precise biological signaling mechanisms responsible for such changes are less clear. A simplistic flow chart for how activity information could be processed by the muscle fibre is given in Fig. 3. As discussed above, muscle activity is important, and signaling pathways must somehow be triggered by an activity correlate. The relative importance of various activity correlates is not known, and in fact many factors may serve as “messengers”, such as: (1) free intracellular Ca2+ caused by cell membrane influx or release from the sarcoplasmatic reticulum; (2) metabolites (lipids, ADP, etc.); (3) hypoxia; and (4) tension (mechanosensation).
Fig. 3.
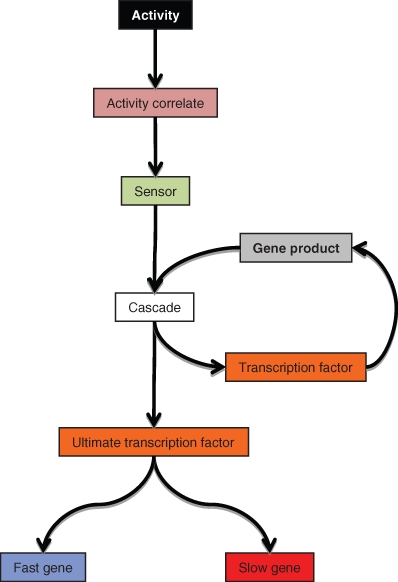
A simplified flow chart illustrating the excitation-transcription coupling, i.e. the flow of information from change in activity to change in fibre-type-specific gene expression.
Few attempts have been made to dissect the various activity correlates, and it is hard to manipulate one parameter without affecting the others, but evidence related to each of these activity correlates that might serve as messengers will be discussed below.
The activity correlate must somehow be registered via a sensor that can transduce the correlate into a signal in an intracellular signaling cascade pathway. A cascade allows amplification and complex interactions with other pathways. A cascade might involve transcription factors acting on genes taking part in the signaling, but ultimately a pathway would end in affecting one or more ultimate transcription factors binding to the promoters of fast and slow genes. It should in fact be emphasized that transcription factors can act indirectly by regulating genes for various factors and thus taking part in the cascade (Fig. 3). Therefore there has been a quest for the transcription factors working at the end of the signaling pathway by binding selectively to promoters for fast and slow genes, such factors will be called ‘ultimate transcription factors' in this review.
(1) Calcium
(a) The source of elevated [Ca2+]i during activity
Free intracellular Ca2+ (Ca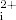 ) is the most abundant and potent of all second messengers, and action potential activity in muscle probably leads to the strongest Ca2+ release [mainly from the sarcoplasmic reticulum (SR)] that occurs in any cell type under physiological conditions. This has made fluctuations in cytosolic [Ca2+]i the prime candidate for mediating the effects of electrical activity on muscle phenotype. Although the release from the SR is massive, there is also an influx across the cell surface, and the relative role of these two sources is not clear. For instance, it has been shown in organ culture that electrical activity stabilizes the acetylcholine receptors at the neuromuscular junction by way of an influx of ions through dihydropyridine-sensitive Ca2+channels in the sarcolemma, while release from the SR is ineffective (Caroni et al., 1993; Rotzler, Schramek & Brenner, 1991). On the other hand, caffeine application leading to release of calcium from the SR elevated the level of several proteins regulated by activity in vivo such as glucose transporter type 4 (GLUT4) and oxidative enzymes (Ojuka, 2004; Ojuka et al., 2002). More research would be required to establish the relative importance of different sources of Ca2+, and the possible importance of compartmentalization of [Ca2+]i in regulating different plastic properties.
) is the most abundant and potent of all second messengers, and action potential activity in muscle probably leads to the strongest Ca2+ release [mainly from the sarcoplasmic reticulum (SR)] that occurs in any cell type under physiological conditions. This has made fluctuations in cytosolic [Ca2+]i the prime candidate for mediating the effects of electrical activity on muscle phenotype. Although the release from the SR is massive, there is also an influx across the cell surface, and the relative role of these two sources is not clear. For instance, it has been shown in organ culture that electrical activity stabilizes the acetylcholine receptors at the neuromuscular junction by way of an influx of ions through dihydropyridine-sensitive Ca2+channels in the sarcolemma, while release from the SR is ineffective (Caroni et al., 1993; Rotzler, Schramek & Brenner, 1991). On the other hand, caffeine application leading to release of calcium from the SR elevated the level of several proteins regulated by activity in vivo such as glucose transporter type 4 (GLUT4) and oxidative enzymes (Ojuka, 2004; Ojuka et al., 2002). More research would be required to establish the relative importance of different sources of Ca2+, and the possible importance of compartmentalization of [Ca2+]i in regulating different plastic properties.
(b) How does [Ca2+]i fluctuate with different activity patterns?
Early studies indicated that a slow stimulation pattern induced a sustained increased level of resting [Ca2+]i in muscle fibres (Sréter et al., 1987), More recently however, it was shown that short-term electrical stimulation (<24 h) of myotubes in culture did not induce a sustained increased [Ca2+]i, in spite of inducing expression of mRNA for slower MyHC types (Kubis et al., 2003). Thus, slow activity patterns seem to be able to induce changes before sustained increased levels of Ca2+ are detected, and more acute fluctuations in [Ca2+]i time-locked to the slow activity pattern are probably more important.
In resting mammalian muscle, various reports have indicated [Ca2+]i ranging from 26 to 145 nmol l−1 (Konishi, 1998). In isolated mouse flexor digitorum brevis fibres, which were most likely of type II, fura-2 measurements indicate resting levels of 30–50 nmol l−1 (Westerblad & Allen, 1991). With slow stimulation at 10 Hz, mean [Ca2+]i showed a modest increase to below 500 nmol l−1 in isolated fibres of mouse fast-twitch flexor digitorum brevis and slow-twitch soleus muscles (Aydin et al., 2008; Westerblad & Allen, 1993). With a fast pattern at 100 Hz mean tetanic [Ca2+]i increased to >1000 nmol l−1 (Allen et al., 2008). Thus, it seems clear that both amplitude and temporal differences in the [Ca2+]i signals downstream of different activity patterns might serve as a signal for changing gene transcription. There is currently however, no definitive evidence for such a role under physiological conditions in adult muscle in vivo. Future research should answer questions such as: what is the key source or relevant compartment for the [Ca2+]i fluctuations? Moreover, the transients that are connected to specific fast and slow patterns that are known to induce changes in the fast and slow direction, respectively, should be mapped, and the differences between them should be connected to downstream signaling cascades that might regulate fibre-type-specific genes.
(c) Calcium sensors: decoding the calcium fluctuations
As concluded above, the rise in [Ca2+]i connected to action potentials is a likely signal for muscle plasticity, but how are the signals decoded (for a previous review, see Buonanno & Fields, 1999)? The rather tight connection between that pattern of activity and variables such as the twitch time-to-peak suggest the existence of rather precise timing or counting mechanisms. Likely primary sensors for the signal are compounds such as protein kinase C and calmodulin.
(i) Calmodulin and its targets CaMKII and calcineurin
Calmodulin activates Ca2+/calmodulin-dependent protein kinase-II (CaMKII). CaMKII has been demonstrated to be activated during exercise (Rose & Hargreaves, 2003), but its function in synaptic plasticity in nerve cells is better characterized. In this intensely investigated field of neurobiology, CaMKII is thought to be involved in spike-timing-dependent plasticity. A central hypothesis has been that peak [Ca2+]i determines the plasticity outcome, such that a sufficiently high [Ca2+]i level leads to long-term potentiation (LTP) of the synapse, while moderately increased [Ca2+]i leads to long-term depression (LTD) (Bi & Rubin, 2005). By analogy, in muscle a fast activity pattern leads to short-lived but high peak [Ca2+]i, while slow activity leads to sustained but more moderate levels, and these might turn on a fast and slow gene expression program, respectively. In addition to the importance of the peak levels of [Ca2+]i that are discussed below, the rate of rise of [Ca2+]i during the onset of an activity event might play a role (Buonanno & Fields, 1999).
In the brain, CaMKII is activated by the Ca2+/calmodulin complex, such that half-maximal activation occurs at a [Ca2+]i of 500–1000 nmol l−1 (Rostas & Dunkley, 1992). In addition to the dependence on peak [Ca2+]i there is a time-dependent component. Thus pulse exposure of CaMKII to Ca2+in vitro at room temperature showed that the enzyme reacted differently when Ca2+ pulses were delivered at 1 or 4 Hz or when the Ca2+ pulse duration was varied between 80 and 1000 ms (De Koninck & Schulman, 1998). Thus CaMKII might decode not only [Ca2+]i levels, but also the temporal pattern of [Ca2+]i fluctuations at a timescale relevant for decoding fast and slow patterns of motor activity.
For LTD in the brain calcineurin also seems to be required (Mulkey et al., 1994), and as discussed below calcineurin has been implicated in the fast-to-slow shift in fibre type. Like CaMKII, calcineurin is activated by calmodulin, but at much lower [Ca2+]i levels because calmodulin has a much higher affinity for calcineurin (dissociation constant Kd = 0.1nmol l−1) than for CaMKII (Kd = 45nmol l−1) (Cohen & Klee, 1988). Hence, at moderate [Ca2+]i the phosphatase calcineurin would be active (Crabtree, 1999), while at higher [Ca2+]i (or at a higher frequency of the Ca2+spikes) the kinase CaMKII would also be active. The difference in the phosphorylation/dephosphorylation of substrates downstream of these two enzymes may trigger fast and slow gene transcription, respectively. In combination, calmodulin, CaMKII and calcineurin could decode both amplitude and temporal aspects of [Ca2+]i (blue triangle in Fig. 4).
Fig. 4.
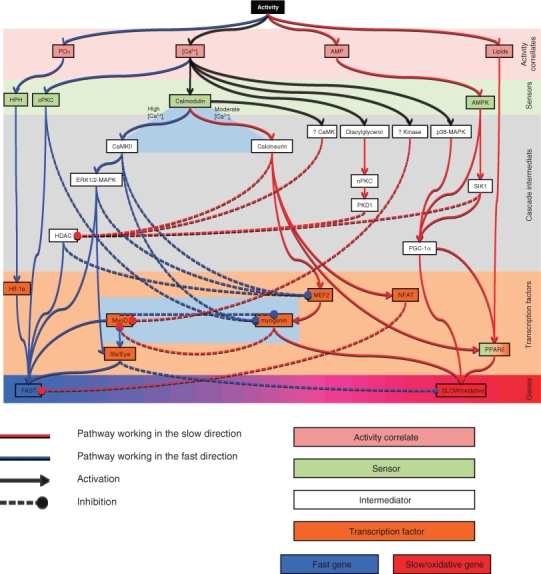
Diagram summarizing pathways currently believed to be involved in excitation-transcription coupling in skeletal muscle with respect to regulating contraction speed. The pathways have different degrees of scientific support, and their relative quantitative importance is still poorly understood. Abbreviations: adenosine monophosphate (AMP), AMP-activated kinase (AMPK), Ca2+/calmodulin-dependent protein kinase (CaMK), conventional protein kinase C (cPKC), extracellular signal-regulated kinase (ERK), free intracellular calcium concentration [Ca2+]i, HIF-1α prolyl hydroxsylases (HPH), histone deacetylase (HDAC), intracellular oxygen pressure (PO2i), mitogen-activated kinase MAPK, myocyte enhancer factor 2 (MEF2), myogenic differentiation factor (MyoD), novel protein kinase C (nPKC), nuclear factor of activated T-cells (NFAT), peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α), hypoxia inducible factor 1α (HIF-1α), peroxisome proliferator-activated receptor δ (PPARδ), protein kinase D1 (PKD1), salt inducible kinase 1 (SIK1), sine oculis homeobox 1/eyes absent 1 (Six1/Eya1).
Calcineurin knock-out mice show a reduction in the number of slow fibres (Parsons et al., 2003), and similar observations were made in mice where the knock-out was targeted to skeletal muscle only (Parsons et al., 2004) Similar results were obtained in mice that overexpressed the calcineurin-inhibiting protein Regulator of Calcineurin 1 (RCAN1) in muscle fibres from early in development (Oh et al., 2005). In tissue culture, the calcineurin-inhibitor cyclosporin A has been shown to block up-regulation of endogenous slow MyHC (Higginson et al., 2002), and overexpression of active calcineurin transactivates promoter-reporter constructs for slow genes (Chin et al., 1998). This finding has been criticized in another study where a larger variety of promoters were investigated, and it was found that calcineurin selectively activated several muscle-specific promoters, but with no selectivity for slow genes (Swoap et al., 2000). For the endogenous MyHC genes in culture however, calcineurin increases the expression of slow isoforms, without any effect on fast MyHC isoforms (Delling et al., 2000). Similar effects have been observed in transgenic mice overexpressing calcineurin driven by the muscle creatine kinase (MCK) promoter conferring early expression in myotubes (Naya et al., 2000; Wu et al., 2001). In regenerating muscle of wild-type mice the calcineurin inhibitors cyclosporin A, FK506 and Cain/cabin-1 prevent up-regulation of slow MyHC induced by slow patterned activity from motorneurones or electrodes (Serrano et al., 2001).
Experiments in culture, transgenic mice or regenerating muscle such as those described above may reflect developmental effects, rather than adult plasticity. In adult mice, systemic administration of cyclosporin A increased the proportion of slow fibres in one study (Chin et al., 1998), but failed to do so in another (Biring et al., 1998). Moreover, cyclosporine has systemic effects on the animals' activity level, so the effects may be secondary to activity changes.
Overexpression of the calcineurin inhibitor cain/cabin-1 in adult rat soleus fibres after somatic gene transfer by electroporation led to the appearance of transcripts for type IIb and IIx MyHC, which is normally rare in this muscle. The transfected fibres expressing these MyHCs did not express type I or IIa MyHC, which are normally the most common forms in this muscle, implying that calcineurin activates these genes (Serrano et al., 2001). These data support the idea that calcineurin is maintaining normal slow properties in adult slow muscles. Calcineurin may also be involved in fibre-type transformation in fast muscles since fast-to-slow transformation induced by overload was impaired when calcineurin blockers (FK506, cyclosporin A) were used (Dunn, Burns & Michel, 1999). Similar observations were made in calcineurin-deficient transgenic mice (Parsons et al., 2004).
As discussed below, nuclear factor of activated T-cells (NFAT) is an important substrate for calcineurin, but it should be emphasized that NFAT is by no means the only protein dephosphorylated by this phosphatase. Knocking out two different variants of the calcineurin catalytic subunit A (Aα and Aβ) both led to a dramatic reduction in slow fibre type and oxidative enzymes. However, only the Aα-null mouse displayed reduced NFAT activity. This suggests the existence of a relevant calcineurin-activated pathway that is not NFAT dependent (Parsons et al., 2003).
In conclusion, calcineurin may act in slow signaling both dependent on and independently of NFAT. Its effects might be both in activating slow genes, and in inhibiting fast genes (see Fig. 4).
(ii) Protein kinase C
The mammalian protein kinase C (PKC) family can be grouped into three classes: conventional (cPKC; α, γ, βI and βII), novel (nPKC; δ, ε, η/L, θ), and atypical (aPKC; ζ, ι/λ). PKC μ and ν have been considered to be a fourth class but are now classified as protein kinase D (see Section III.1dii). PKCs are activated by translocating to the plasma membrane where they can be activated by Ca2+ and lipids. cPKCs are in themselves Ca2+ sensors, but are also activated by diacylglycerol and phosphatidylserine. Other PKCs are not directly Ca2+ dependent, but nPKC is stimulated by diacylglycerol and phosphatidylserine, and aPKC by phosphatidylserine only (Newton, 2001).
In cultured mast cells, cPKCγ has been shown to translocate in response to Ca2+ spikes, and the kinetics suggest that it may serve as a decoding machine for patterned [Ca2+]i oscillation. In order to activate cPKC, [Ca2+]i should reach a threshold level of 400 nmol l−1 (Almholt et al., 1999; Mogami et al., 2003); and localization seems to be important since Ca2+ influx over the surface membrane seems to be much more efficient than release from internal stores (Mogami et al., 2003; Pinton et al., 2002). Muscle has however a massive Ca2+ release from its specialized endoplasmatic reticulum (SR), and the importance of this release has not been investigated. With respect to temporal aspects, in the timescale of seconds, high-frequency, but not low-frequency calcium spikes elicit high cPKCγ activity (Oancea & Meyer, 1998), suggesting that cPKC might be activated by fast, but not slow, patterns of activity (but see also Pedersen et al., 2009).
In rat skeletal muscle PKC has been demonstrated to be translocated and activated by electrical stimulation in vivo (Cleland et al., 1989; Huang, Tong & Schmidt, 1992), but cPKC did not seem to be affected by exercise in humans (Rose et al., 2004). In developing avian muscle and also tissue culture, PKC seemed to be negatively regulated by nerve-evoked activity, thus PKC activity was decreased by innervation and increased by blocking neuromuscular transmission in slow muscles. In the avian models PKC activity was also found to be higher in fast than in slow muscles, and elevation of cPKCα and nPKCθ reduced expression of a slow MyHC isoform in slow muscles (DiMario, 2001; DiMario & Funk, 1999; Jordan et al., 2004). In conclusion it could be speculated that various cPKCs might serve as a sensor for fast activity patterns mediating a suppression of slow MyHC in muscles receiving such activity (Fig. 4). However, although it has been demonstrated that a slow MyHC isoform can be regulated by PKCs, it remains to be demonstrated if they do so in adult muscle in vivo.
Among the novel PKCs, nPKCθ is the most abundant isoform in skeletal muscle (Chang et al., 1993; Osada et al., 1992), and the protein is 2.5-fold more abundant in fast than in slow muscles (Donnelly et al., 1994). Although nPKCs lack the Ca2+-binding domain, it has been shown in neuro-endocrine cells that Ca2+ influx produced diacylglycerol that in turn translocates and activates nPKCθ over a timescale of seconds (Mogami et al., 2003). nPKCs could hence act as pattern-specific signal decoders of nerve-evoked activity in muscle. Protein kinase D1 (PKD1) is an interesting downstream target for nPKCθ and other nPKCs such as nPKCε and nPKCη (Rykx et al., 2003; Yuan et al., 2002). As illustrated in Fig. 4 and discussed in Section III.1dii, PKD1 would act in the slow direction, and thus nPKCs might have the opposite role as cPKCs.
Little is known about aPKCs in muscle, but activity may regulate aPKC via AMP-activated kinase (AMPK) (Chen et al., 2002); aPKC isoforms are activated within minutes following the onset of a single bout of endurance exercise both in mice (Chen et al., 2002) and humans (Nielsen et al., 2003; Perrini et al., 2004; Rose et al., 2004). aPKC has not directly been implied in regulation of fast and slow genes, but interestingly, aPKC can activate mitogen-activated kinases (MAPKs) (Hirai & Chida, 2003), which might take part in such regulation. More research seems, however, required to clarify if aPKC has a role in muscle plasticity.
(d) Cascades downstream of [Ca2+]i
(i) Ras and MAPK
Ras is a subfamily of small GTP-binding proteins encoded by three different genes in mammals. Ras is localized to the inner face of the plasma membrane and functions as a molecular switch that transmits receptor signals, but it can also be regulated by pathways dependent on Ca2+ influx, including at least one calmodulin-dependent pathway. Downstream of Ras is a cascade involving mitogen-activated kinases (MAPKs) originally called extracellular signal-regulated kinases (ERKs). The MAPK family has many members such as eight ERKs (ERK1-8), p38 and Jun N-terminal kinase (JNK). The MAPKs are turned on by phosphorylation. Active MAPKs in turn have a multitude of downstream target genes (Bogoyevitch & Court, 2004; Bradley & Finkbeiner, 2002; Chang & Karin, 2001; Finkbeiner & Greenberg, 1996).
MAPKs are activated in a variety of endurance-type exercises and other activity models both in animals and in man (Kramer & Goodyear, 2007); in particular a slow, but not fast pattern increased ERK activity (Murgia et al., 2000). Blocking of ERK1/2 with U0126 decreased type I and increased type II MyHC mRNA expression in primary culture (Higginson et al., 2002). In regenerating slow muscles a constitutively active form of Ras and a MAPK both mimicked the effects of slow nerve activity, while a dominant negative Ras prevented nerve-evoked activity from inducing slow MyHC in regenerating muscle (Murgia et al., 2000). Thus Ras and MAPK might be involved in maintaining slow properties in this model; but since active Ras was unable to induce slow myosin in fast regenerating muscle (Murgia et al., 2000), Ras apparently does not take part in fast-to-slow transformation.
Findings in C2C12 tissue culture and in adult animals have, however, not supported the findings in regenerating muscle, as MAPKs rather seem to act in the fast direction. Thus, it was found that ERK1/2 activity was twofold higher in fast than in slow adult muscle (Shi et al., 2008, 2007). In addition, while the levels of ERK1 and 2 were similar in soleus, ERK2 was the predominant form in the fast EDL. No differences were found with respect to p38 and JNK (Shi et al., 2007), although these MAPKs are also activated by exercise (Kramer & Goodyear, 2007). In C2C12 cells in tissue culture when constitutively active ERK2 was overexpressed fast promoter-reporter genes such as MyHCIIb and SERCA1 were activated. The ERK1/2 inhibitor PD98059 had the opposite effect, as it was shown that the MyHCI promoter was stimulated while the MyHCIIb and SERCA1 promoters were inhibited. Similarly, endogenous expression of fast MyHC and SERCA1 was decreased while the levels of MyHCI and myoglobin increased (Shi et al., 2008).
In adults MAPK was inhibited by electroporation of an expression vector for MAPK phosphatase 1 (MKP1) which will dephosphorylate and inactivate ERK1/2. This procedure activated MyHCI promoter-reporter constructs, and reduced fast SERCA1 and MyHCIIb reporter levels; endogenous MyHC type I and IIa genes also were activated in fast IIx or IIb fibres. In slow soleus fibres, MyHC1 promoter-reporter genes were induced even more strongly than in fast muscles. When a constitutively active form of ERK2 was overexpressed in fast fibres it had no effect on fast or slow promoter-reporter constructs, but in the slow soleus MyHCIIb reporter level was increased, although MyHCI reporter was not decreased (Shi et al., 2008). Thus, ERK2 seems to activate fast genes in slow muscles, while its role in inhibiting slow genes remains more uncertain.
In conclusion, manipulating MAPKs has given somewhat conflicting results in different models. During regeneration MAPK seems to promote slow properties, while in adult muscle fibres there is evidence that MAPKs can be involved in slow-to-fast transformation of pre-existing fibres. How this role could be related to the findings that slow activity increases MAPK activity (Kramer & Goodyear, 2007; Murgia et al., 2000) remains unclear.
(ii) Protein kinase D
Protein kinase D1 (PKD1; previously also called PKCμ) is one of a three-member kinase family (Rykx et al., 2003). PKD is not directly regulated by calcium, but can be activated through phosphorylation by nPKC isoforms such as PKCε, PKCη and PKCθ (Rykx et al., 2003; Yuan et al., 2002). PKD1 is enriched in slow fibres. Forced expression of a constitutively active form of PKD1 from a MCK promoter, which confers expression from early muscle differentiation, developed transgenic mice with perturbed fibre-type distribution, smaller fibres and a somewhat reduced fibre number. The fraction of type I fibres was increased, and the level of IIa and IIx mRNA and protein was increased, while IIb and IIx MyHC and mRNA did not change significantly. Myoglobin levels also increased (Kim et al., 2008). On the other hand a muscle-specific knock-out of PKD1 had no effect on fibre types, but fatigability was increased (Kim et al., 2008). In these experiments positive effects might be attributed to effects during development, and the lack of effects to compensatory mechanisms. Nonetheless, the observed perturbation of fibre type composition opens up the possibility that PKD1 is involved in regulating fibre type in adult animals.
PKD1 influences many fundamental cell biological processes including membrane trafficking, cell survival, differentiation and migration. The Raf–MEK–ERK pathway (MEK is MAP/ERK kinase) and nuclear factor of κ light polypeptide gene enhancer in B-cells (NFκB) seem to be among the downstream targets it activates (Rykx et al., 2003; Van Lint et al., 2002). In cardiomyocytes PKD1 has been shown directly to phosphorylate histone deacetylase (HDAC) 5 and stimulate its nuclear export (Kim et al., 2008). A hypothetical pathway where calcium influx leads to elevation of diacylglycerol activating nPKCs, activating PKD1, inhibiting HDACs, and thus acting in the slow direction is illustrated in Fig. 4.
(iii) Histone deacetylase
HDACs are a family of proteins with the ability to deacetylate a variety of proteins, not only histones. Thus, HDACs might influence gene expression not only by affecting chromatin structure but also by interacting with other transcription factors. The HDAC subclass IIa has the highest expression in brain and muscle, and has been implicated in muscle plasticity. The IIa class consists of HDAC4, −5, −7 and 9. Class IIa HDACs have low deacetylase activity and might not be authentic deacetylases, they can however function as co-repressors of transcription in interaction with transcription factors. They might also interact with class I HDACs to regulate histone acetylation (Mejat et al., 2005). The class IIa HDACs are regulated not only transcriptionally but by several post-translational modifications including ubiquitination which leads to degradation, and phosphorylation which leads to nuclear export. Knock-out experiments suggest that the different class IIa HDACs can substitute for each other. HDAC biology was reviewed recently (Haberland, Montgomery & Olson, 2008; Walkinshaw et al., 2008).
In spite of lower RNA levels, HDAC IIa protein levels are higher in fast muscles compared to slow, apparently due to ubiquitin-proteasome-mediated degradation acting selectively in slow muscles (Potthoff et al., 2007). Knock-out mice lacking individual class IIa HDACs displayed no changes in muscle fibre type. However when double knock-outs of two HDAC IIa genes were created, the animals showed increased numbers of type I and IIa fibres, and elevated levels of mRNA for these slow MyHC types (Potthoff et al., 2007). These data suggest that HDACs might be involved in fibre-type differentiation during development, but not necessarily in muscle plasticity in adults.
The importance of HDACs was also investigated in adult mice by overexpressing HDAC5 from an inducible promoter. When such animals were subjected to treadmill running, the elevated levels of HDAC5 prevented an exercise-induced increase in type I and IIa fibres in the plantaris muscle (Potthoff et al., 2007). In isolated adult muscle fibres, it was shown that slow stimulation led to an export of HDAC4 (but not HDAC5) from the nucleus, and that this process was dependent on calmodulin kinases (Liu, Randall & Schneider, 2005).
These experiments suggest that HDACs somehow maintain fast properties, and that reduced HDAC signaling facilitates fast-to-slow transformation.
Signaling pathways coupling specific patterns of activity to HDAC activity are not well understood, but phosphorylation could provide a rapid activity-dependent response. Four putative inhibitory pathways acting on HDAC are illustrated in Fig. 4. (1) Experiments in C2C12 cells suggest that HDAC5 could be negatively regulated by AMPK by activating salt inducible kinase (SIK1) (Takemori et al., 2008). (2) Forced expression of constitutive active forms of calcineurin using the MCK promoter in transgenic mice leads to development of an increased number of slow fibres which is paralleled by a decrease in HDAC5 and -7 (Naya et al., 2000; Potthoff et al., 2007), and this mechanism might be relevant also in adult muscle. (3) Similar results to those found with calcineurin have been obtained with CaMKIV (Wu et al., 2002), but this kinase is not normally expressed in muscle (Bassel-Duby & Olson, 2006), so perhaps another CaMK is operating in this tissue? (4) Diacylglycerol might have an effect via PKD1.
(iv) PGC-1α and—β
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) and its homolog PGC-1β are co-activators of transcription factors, and may be among the best-studied examples of an increasingly recognized group of proteins that regulate transcription without themselves having independent DNA-binding capability. The PGC-1 molecules have been implicated mainly in regulating pathways related to mitochondrial oxidative metabolism, and to glucose, lipid and energy homeostasis. PGC-1α is a co-activator not only for PPARγ from which its name is derived, but also the other PPARs, including PPARδ (Lin, Handschin & Spiegelman, 2005) which has been implicated in muscle plasticity (discussed in Section III.2b). For PGC-1β, co-activation of PPARα, and—γ has been established but the ability to co-activate PPARδ has not been determined. In addition to the PPARs, PGC-1α and—β also have other downstream targets; and for PGC-1α it has been established that forkhead box O1 (FoxO1), myocyte enhancer factor 2 (MEF2), cAMP response element-binding (CREB) and sex determining region Y-box 9 (Sox9) are among them (Lin et al., 2005). As discussed below FoxO1 seems to be central for muscle size regulation, while MEF2 has been implicated in muscle plasticity, and another member of the Sox gene family (Sox6) has been implicated in the development of muscle fibre type (Hagiwara, Ma & Ly, 2005; Hagiwara, Yeh & Liu, 2007; Hofsten et al., 2008).
PGC-1α is preferentially expressed in slow muscles both at the mRNA and protein level (Lin et al., 2002b). In both humans and in rodents physical exercise increases PGC-1α levels (Atherton et al., 2005; Baar et al., 2002; Goto et al., 2000; Norrbom et al., 2004; Pilegaard, Saltin & Neufer, 2003; Terada et al., 2002, 2005; Wright et al., 2007). In tissue culture application of a Ca2+-ionophore increased PGC-1α levels. Both in culture and in vivo PGC-1α concentration is elevated by slow-patterned electrical stimulation (Irrcher et al., 2003). The latter probably indicates that activity per se influences PGC-1α levels; in addition catecholamine (Miura et al., 2007) and thyroid hormones (Irrcher et al., 2003) have been implicated in up-regulating PGC-1α levels in muscle. Activity seems to regulate not only the level of PGC-1α, but also the translocation of the protein to the nucleus (Wright et al., 2007).
A possible mediator for activity effects on PGC-1α is the MAPK p38, and interestingly, p38 activation seems to be related to mechanical stretch (Akimoto et al., 2005; Boppart et al., 2001; Goodyear et al., 1996; Long, Widegren & Zierath, 2004; Martineau & Gardiner, 2001; Wretman et al., 2001). In tissue culture, PGC-1α could also be regulated by SIK1 (Takemori et al., 2008) and AMPK (Irrcher, Ljubicic & Hood, 2009; Irrcher et al., 2008; Takemori et al., 2008) and in vivo (Suwa, Nakano & Kumagai, 2003).
When PGC-1α was overexpressed in muscle using the MCK promoter in transgenic mice, slower muscles were developed, with higher levels of mitochondrial enzymes, and appearance of 10–20% type I fibres in muscles that normally contain almost exclusively type IIb fibres (Lin et al., 2002b). Knock-out of PGC-1α had no clear effect on the development of MyHC fibre type, although oxidative capacity was reduced (Arany et al., 2005). The results are however difficult to interpret since the mice were hyperactive. When a muscle-specific knock-out was made these animals developed a slow-to-fast shift in MyHC fibre type and a reduced oxidative capacity, thus these animals showed approximately the reverse change compared to animals overexpressing PGC-1α. The animals were however spontaneously hypoactive (Handschin et al., 2007), and this might in itself lead to a shift in fibre type.
In conclusion there are indications that PGC-1α is involved in a slow phenotype. As hypothesized in Fig. 4, PGC-1α may be regulated by p38 and AMPK. PGC-1α might activate slow genes by acting as a PPARδ co-factor, but could also act on other downstream targets independently of PPARδ.
PGC-1β was cloned much later than PGC-1α (Lin et al., 2002a) and there is less accumulated information. Its role also seems to be more complex, not fitting into a simple fast-to-slow or slow-to-fast transformation model. PGC-1β is not clearly differentially expressed in fast and slow muscles, but at least in some muscles, PGC-1β seemed to be associated with type IIx fibres (Arany et al., 2007). Exercise has been reported either to have no effect on the level of PGC-1β (Meirhaeghe et al., 2003) or to induce a decrease (Mathai et al., 2008; Mortensen et al., 2007). When PGC-1α or PGC-1β were overexpressed in muscle tissue culture they had similar effects; oxidative enzyme activity was increased and mRNA for MyHCI was increased while IIb and IIx were reduced; PGC-1β also led to up-regulation of IIa (Mortensen et al., 2006).
PGC-1β was expressed in transgenic mice with the same MCK promoter as for PGC-1α, but with different results. In muscles that were developed in PGC-1β mice, there was no alteration in MyHCI, but IIa and IIb expression was suppressed. The IIx isoform was highly elevated in the fast/mixed muscles that were tested, and in situ hybridization suggested that IIx MyHC was highly expressed in nearly all fibres. PGC-1β also increased mitochondrial biogenesis and oxidative enzyme activity, and increased running distances to exhaustion for the mice (Arany et al., 2007).
In conclusion, although overexpression of PGC-1β changed fibre type to the intermediate IIx form, its relationship to muscle activity remains unclear. Moreover, both for PGC-1α and PGC-1β it remains to be investigated what the role of the cofactor is in regulating adult properties. The effects observed so far have all been in transgenic animals where PGC-1 is present during part of muscle development.
(e) Transcription factors downstream of [Ca2+]i
(i) NFAT
Nuclear factor of activated T-cells (NFAT) is a five-gene family of transcription factors with multiple splicing variants expressed from each gene. NFATc1-4 are similar in structure and have been investigated in muscle (Lopez-Rodriguez et al., 1999; Rao, Luo & Hogan, 1997).
In B-cells it was shown that low sustained levels of elevated [Ca2+]i activates NFAT (Dolmetsch et al., 1997), and a similar [Ca2+]i-calmodulin-calcineurin-NFAT pathway was subsequently suggested for the excitation-transcription coupling in muscle (Chin et al., 1998). It is believed that calcineurin acts by dephosphorylating NFAT, which again triggers a translocation of NFAT into the cell nuclei where it can act as a transcription factor (Crabtree & Olson, 2002; Rao et al., 1997). Re-phosphorylation of NFAT leads to export from the nucleus, and in muscle this seems to be mediated by glycogen synthase kinase 3β (GSK3β) or casein kinase 1 or 2 (CK1/2) (Shen et al., 2007).
In order to establish NFAT as a substrate for calcineurin in the context of muscle plasticity, several groups have used the peptide VIVIT, which inhibits the calcineurin-NFAT interaction without affecting general calcineurin phosphatase activity (Aramburu et al., 1999). In primary cultures, co-transfection with VIVIT blocked the positive effect of calcineurin on slow MyHC expression by 70%, but had no effect on fast MyHC. In adult slow muscle VIVIT was shown to block the expression of MyHCI, and was also shown to inhibit nerve-activity-induced expression of slow myosin in the regenerating soleus muscle (McCullagh et al., 2004). These experiments suggest that calcineurin-NFAT interaction could be important in maintaining slow properties in the slow soleus muscle. The critical experiment to demonstrate that calcineurin-NFAT interaction is crucial for activity-induced fast-to-slow fibre-type transformation, however, would be to investigate if VIVIT can block the effects of slow stimulation in an adult fast muscle, but such experiments have yet to be reported.
NFATc1-4 are all expressed in skeletal muscle, but only NFATc1 (also called NFATc or NFAT2a, but not to be confused with NFAT1c) seems to be preferably translocated to the nuclei in slow fibres. Although NFATc4 and to some extent NFATc2 and NFATc3 also show nuclear localization in muscle, it is not fibre-type specific (Calabria et al., 2009; Tothova et al., 2006). Translocation seems to be regulated by activity; when slow muscles are inactivated for 2 h NFATc1 is exported from the nucleus (Kubis et al., 2002; Liu et al., 2001; Tothova et al., 2006), while slow but not fast electrical stimulation translocates it to the nucleus (Calabria et al., 2009; Shen et al., 2006). NFATc2 and -3 are also to some extent translocated by activity but with no clear pattern specificity, NFATc4 is localized to the nuclei irrespective of activity (Calabria et al., 2009).
In order to investigate NFAT transactivating activity directly, experiments have been performed in vivo with artificial promoters where a high number of NFAT binding sites were coupled to luciferase as a reporter. It was shown that the transactivating activity of NFAT was higher in slow than in fast muscle, and it was reduced in slow muscle subjected to denervation or a fast stimulation pattern (McCullagh et al., 2004). This suggests that NFAT can take part in maintaining slow properties in slow muscles. It was not investigated if NFAT transactivating activity increases in fast muscles subjected to slow stimulation, hence NFATs role in fast-to-slow transformation was not directly addressed in these experiments.
A constitutively active form of NFATc1 has been overexpressed in various muscle models in vivo (McCullagh et al., 2004). Slow MyHC was increased in denervated, regenerating soleus and EDL muscles, which normally do not express this isoform. When co-electroporated with promoter-reporter constructs in the intact adult fast EDL muscle, the MyHCI promoter was induced sixfold while the IIb promoter was completely inhibited (but see also Swoap et al., 2000). For the endogenous genes in intact adult muscle, NFAT downregulated the IIb gene, but surprisingly no type I was induced (McCullagh et al., 2004). This discrepancy illustrates that neither regenerating muscle nor episomal promoter-reporter genes necessarily represent gene regulation of endogenous genes in intact adult muscle.
Even if overexpression of NFATc1 failed to activate MyHCI expression in EDL after seven days of treatment (McCullagh et al., 2004), a role in fast-to-slow transformation could not be excluded. Thus, during the first two weeks of slow stimulation IIb expression is inhibited, but with only a minor induction of MyHCI. Remarkably, massive MyHCI expression commenced only after prolonged stimulation (Ausoni et al., 1990; Windisch et al., 1998) suggesting different regulatory mechanisms in the long term. However, stimulation rapidly inhibited IIx expression and increased IIa expression in these experiments. Unfortunately the effects of overexpressing NFATc1 on these genes have not been reported.
Loss-of-function experiments with inhibitory RNA gainst NFATc1-4 have been performed in vivo (Calabria et al., 2009). With promoter-reporter constructs it was shown that activation of the MyHCI promoter was reduced by reducing any of the NFATs. Reducing NFATc1 had no effect on any of the type II MyHCs, while reducing NFATc4 reduced all of the type II MyHCs. NFATc2 and -3 reduced transactivation of IIa and IIx MyHC, but had no effect on IIb. The positive effect of NFATc1-4 on MyHCI expression was confirmed for the endogenous gene, but only in regenerating muscle.
Based on all these experiments it is tempting to suggest that NFATc1 is the factor activating a slow expression program, while NFATc4 is activating a fast program. NFATc4, however is not expressed in a fibre-type-specific fashion, nor is it regulated by activity. While NFATc1 seems to downregulate the endogenous MyHCIIb in intact adult muscle, it remains to be seen if it can turn on endogenous MyHCI under such conditions, which would be the hallmark of a fast-to-slow transition.
(ii) Does NFAT bind to fast or slow promoters?
Effects of various transcription factors on fast and slow promoters might reflect direct binding to gene regulatory sequences of such genes (regulation in cis as ultimate transcription factors), or they might regulate other genes that in turn regulate the genes determining fast and slow phenotype (Fig. 3). The original idea of the calcineurin-NFAT pathway was that calcineurin was activated by a slow stimulation pattern, the calcineurin then dephosphorylated NFAT that in turn transactivated slow genes by NFAT binding directly to slow promoters (Chin et al., 1998). Binding to relevant promoters, however, was not directly demonstrated in their study.
The promoters for the fast and the slow isoforms of troponin I are the best characterized of the fast and slow specific promoters and they have been used in the quest for the ultimate fast and slow transcription factors. Troponin I (Tn1) is the regulatory component of the troponin complex, and probably takes part in determination of twitch speed (Squire & Morris, 1998; Zot & Potter, 1987). In adults the fast and the slow gene are expressed in fast and slow muscle fibres, respectively (Hallauer & Hastings, 2002), and the expression can be modulated by electrical activity (Buonanno et al., 1998; Calvo et al., 1996; Rana, Gundersen & Buonanno, 2008; Rana et al., 2005). In transgenic mice, slow-specific expression is conferred by a 128 base pair (bp) rat sequence from the TnI slow gene dubbed the slow upstream regulatory element (SURE), and fast-specific expression by a 144 bp sequence from the fast TnI in quail called the fast intronic regulatory element (FIRE) (Banerjee-Basu & Buonanno, 1993; Hallauer & Hastings, 2002; Nakayama et al., 1996).
Putative NFAT binding sites have been identified in both FIRE and SURE. Constitutively active calcineurin was shown to activate SURE (but not FIRE) promoter-reporter constructs in vitro, and the response in SURE was attenuated by mutating the NFAT site (Wu et al., 2000). In vivo, the situation has been less clear. In one study on transgenic animals differential expression in fast and slow muscles from a SURE-reporter construct was abolished by mutating the NFAT site (Wu et al., 2000), while in a different study a SURE construct from which the NFAT site was removed continued to exhibit slow-specific expression (Calvo et al., 1999). Thus, the role of NFAT as an ultimate transcription factor directly activating slow genes still remains somewhat uncertain.
By contrast, in FIRE, a NFAT binding site has been documented by site-directed mutations electromobility shift assays and supershift assays. By utilizing novel time lapse in vivo imaging techniques (Rana et al., 2005) it was shown that the NFAT binding site was necessary for the suppression of the fast troponin I promoter by a slow stimulation pattern, while the destruction of the site had no effect on the positive regulation of this gene by fast activity (Rana et al., 2008).
In conclusion, current evidence suggests that NFAT signaling directly or indirectly connects slow activity to maintaining slow properties in slow muscles, and that NFAT is an ultimate transcription factor in inhibiting expression of the fast isoform of troponin I, and perhaps MyHC IIb during fast-to-slow transformation. The identification of NFAT as an inhibitory transcription factor for fast troponin I, might be one of the few documented examples of an ultimate transcription factor involved in differential expression of fibre-type-specific genes (see also Section III.3).
(iii) MEF2
The myocyte enhancer factor 2 (MEF2) transcription family of MADS box transcription factors are encoded by four different genes in vertebrates denoted as A, B, C, and D, each of which diplay various splicing variants. MEF2 is important during early muscle development (reviewed in Potthoff & Olson, 2007). The number of type I fibres was reduced in MEF2c and MEF2d, but not in MEF2a knock-outs. When a hyperactive MEF2c-VP16 fusion protein was overexpressed in muscle driven by the myogenin promoter, the number of type I fibres developed was increased, and the mice displayed improved running endurance (Potthoff et al., 2007). These experiments suggest that some of the MEF2 isoforms are implicated in establishing slow fibre identity during development.
In adult animals MEF2 protein is expressed at similar levels in fast and slow muscles (Potthoff et al., 2007), but MEF2 activity can be modified by mechanisms not related to its concentration. Thus, several kinases can inactivate MEF2 by phosphorylation including calmodulin-dependent kinases and MAPK, and it can be activated by dephosphorylation by calcineurin (Mao et al., 1999; Wu et al., 2000). MEF2 also seems to be regulated through HDAC class IIa proteins. When HDAC is present in the nucleus, it will bind to MEF2 and repress its transactivating ability (reviewed in Bassel-Duby & Olson, 2006; Walkinshaw et al., 2008).
In order to investigate MEF2 activity, a transgenic mouse was developed where the reporter lacZ is driven by triplicated MEF2 binding sites from the desmin promoter (Naya et al., 1999). In most mice no lacZ expression was detected in the adult, but lacZ-staining was visible in some soleus muscles, suggesting that MEF2 activity was higher in this slow muscle than in fast muscles. When the transgenic animals were subjected to treadmill running, the lacZ reporter became strongly increased in the soleus and in some, but not all fast muscles in the running lower leg. These findings suggest that exercise activity increased MEF2 transactivating activity. The effect was suggested to be calcineurin dependent, since injection of the calcineurin inhibitor cyclosporine A prevented a slow pattern of electrical stimulation from inducing lacZ. Moreover, when transgenic mice expressing the calcineurin inhibitor myocyte-enriched calcineurin-interacting protein 1 (MCIP1) driven by the MCK promoter were crossed with the MEF2-lacZ reporter mice, the mice developed muscles where treadmill running did not induce lacZ expression (Wu et al., 2001).
A cautionary remark should be made with respect to using lacZ as a reporter gene in muscle, as it has been shown that the level of lacZ protein is strongly influenced by muscle activity in adult muscle. Thus, in transgenic mice where lacZ is driven by a promoter for the acetylcholine receptor α subunit, denervation leads to a marked decrease in lacZ staining in spite of the lacZ mRNA being strongly up- regulated. It was suggested that inactivity strongly promotes lacZ protein degradation (Gundersen, Sanes & Merlie, 1993). In avian myotube cultures similar experiments to those described for lacZ, using luciferase as a reporter, revealed no difference between MEF2 activity in fast and slow muscle with or without innervation from a spinal cord explant (Jordan et al., 2004). The pattern of activity imposed by the explants was, however, not characterized.
The SURE and FAST elements from the slow and fast troponin I promoters respectively can also be used to illuminate the role of the MEF2 binding site, which are necessary for transcription of both enhancers in skeletal muscle (Calvo et al., 1996; Nakayama et al., 1996). In co-transfection experiments in tissue culture, various forms of MEF2 generally up-regulated expression from SURE more than from FIRE, and this effect is strongly potentiated by simultaneously expressing a constitutively active form of calcineurin. Overexpression of calcineurin alone also had a selective effect on SURE, and this effect was abolished by mutating the SURE MEF2 site into the homologous site in FIRE. This site provides weaker binding of MEF2 in electromobility shift assays (EMSA). These findings led to the suggestion that activation of calcineurin in slow muscles, or muscles subjected to slow stimulation, dephosphorylates and activates MEF2, which binds to and transactivates the slow but not the fast troponin I promoter (Wu et al., 2000). However, this hypothesis has not been supported by in vivo experiments (Calvo et al., 1999, 2001). In transgenic mice harbouring SURE elements, slow fibre-type-specific expression (but not muscle specificity) was lost when elements upstream of the MEF2 site were removed. When homologous elements from FIRE were ligated upstream of the part of SURE containing the MEF2 site, high expression was conferred in fast muscles, demonstrating that fibre-type-specific expression was not conferred by the MEF2 site, but rather by other promoter elements in SURE. In other words, when the SURE MEF2 site was added to the FIRE sequence, FIRE dominated (Calvo et al., 2001; Rana et al., 2008). Thus, the MEF2 site seems not to confer fibre-type specific expression of the slow troponin isoform.
Even if these experiments suggest that MEF2 is not a ultimate factor activating slow genes, MEF2 might be involved in slow-specific expression by activating other transcription factors, thus it is interesting that MEF2 has been shown to transactivate the myogenin promoter in tissue culture (Edmondson et al., 1992; Friday et al., 2003). As discussed below, myogenin may in turn induce oxidative properties, but apparently not slow MyHC fibre type (Ekmark et al., 2003).
(iv) Myogenin and MyoD: the yin and yang of muscle plasticity?
Myogenic differentiation factor (MyoD), myogenin, myogenic regulatory factor 4 (MRF4), and myogenic factor 5 (myf-5 ) are a group of proteins that are specific to skeletal muscle and belong to the basic-helix-loop-helix (bHLH) family of transcription factors. These four proteins are called the myogenic regulatory factors (MRFs), as they play a key role during early differentiation of muscle cells. Like other bHLH proteins they are thought to bind to DNA mainly as heterodimers with E-proteins. The dimers bind to E-boxes in the DNA with the canonical sequence CANNTG. Other partners for bHLH transcription factors are inhibition-of-DNA-binding-proteins (Id-proteins) that act negatively since the heterodimers do not bind to DNA. Thus, the activity of MRFs can be regulated by modifying the level of the MRF or one of its partners. In addition, the MRFs have phosphorylation sites that influence protein stability (Song et al., 1998), or prevent DNA binding (Bengal et al., 1994; Li et al., 1992b). There is evidence that these sites can be phosphorylated by PKC and CaMKII (Blagden, Fromm & Burden, 2004; Li et al., 1992b) but apparently not protein kinase A (PKA) (Li et al., 1992a; Winter, Braun & Arnold, 1993). It is unclear if different MRFs are selectively phosphorylated by different kinases.
MyoD acts early in myogenesis to determine myogenic fate, while myogenin acts later in the differentiation of myoblasts into myotubes (Berkes & Tapscott, 2005; Buckingham et al., 2003). MyoD is involved in chromatin remodeling when fibroblasts differentiate into myoblasts, and in this model MyoD seems to have little ability to transactivate genes independently. Myogenin on the other hand seems to act more as a conventional transcription factor (Bergstrom & Tapscott, 2001; Cao et al., 2006; Davie et al., 2007; Ohkawa, Marfella & Imbalzano, 2006). It should be kept in mind, however, that gene regulation even with the same transcription factors and genes, is different in tissue culture, during development, regeneration, and in the adult animal (e.g. see Davie et al., 2007; Zammit et al., 2008).
All the MRFs are expressed in adult muscle, although myf-5 only at very low levels. MRF4 is the factor with the highest concentration in adults. MRF4 seems to be predominantly expressed in slow/oxidative fibres in some, but not all muscles (Walters, Stickland & Loughna, 2000a). On the other hand, promoter elements conferring expression in fast muscle fibres have also been identified in the MRF4 promoter (Pin & Konieczny, 2002). Further investigation is required to establish if MRF4 might play a role in maintaining or regulating adult muscle fibre type.
In adults, MyoD and myogenin show reciprocal expression patterns in vertebrates as diverse as mammals and fishes. The MyoD level is high in fast muscles and myogenin level is high in slow muscles; in rodents MyoD cis regulatory regions seem to restrict expression of MyoD to IIx and IIb fibres (Delalande & Rescan, 1999; Ekmark et al., 2007; Hughes et al., 1993; Rescan, Gauvry & Paboeuf, 1995; Voytik et al., 1993). Contributing strongly to the difference in MyoD activity between fast and slow muscles are the differences in T115 phosphorylation. Thus the amount of phosphorylated, inactive MyoD was three times higher in soleus than in EDL, and since the total MyoD level was about half, the fraction of the MyoD that is inactive must be six times higher in soleus than in EDL (Ekmark et al., 2007).
In vitro experiments have suggested that the different MRFs regulate one other in a positive manner (Olson, 1990; Weintraub, 1993), but this might be different in vivo. Thus in transgenic mice starting to overexpress myogenin in differentiated muscle cells, MyoD expression is decreased (Gundersen et al., 1995). In mice with reduced MyoD expression, myf-5 expression is increased in a dose-dependent manner (Rudnicki et al., 1992). If MyoD and myogenin cross regulate each other negatively in vivo, such a coupling would tend to stabilize fibres as ‘MyoD’ or ‘myogenin’ fibres, respectively, and hence as distinct fibre types (blue rectangle in Fig. 4). Altered activity or forced expression of one of the factors would undermine the stability, and transform the phenotype.
In most exercise studies MyoD and myogenin mRNA or protein is found to be elevated both after resistance (Bickel et al., 2005; Drummond et al., 2008; Haddad & Adams, 2002; Hentzen et al., 2006; Hulmi et al., 2007; Ishido, Kami & Masuhara, 2004; Kosek et al., 2006; Kvorning et al., 2007; Liu et al., 2008; McKay et al., 2008; Peters et al., 2003; Psilander, Damsgaard & Pilegaard, 2003; Raue et al., 2006; Willoughby & Nelson, 2002; Yang et al., 2005), and endurance (Kadi et al., 2004; Siu et al., 2004; Vissing et al., 2008) training. A few studies that have investigated both factors find an elevation only in MyoD (Harber et al., 2008; Yang et al., 2005), or only in myogenin (Costa et al., 2007; Siu et al., 2004). Some studies report little change (Bamman et al., 2004; Coffey et al., 2006; Hulmi et al., 2008a; Jensky et al., 2007) or even a reduced level (Costa et al., 2007; Hulmi et al., 2008b).
Studies based on muscle homogenates are however hard to interpret. Resistance training in particular leads to focal muscle damage and repair, and myogenic factors are strongly elevated in regenerating muscle tissue (Fuchtbauer & Westphal, 1992; Mendler et al., 1998). The MyoD and myogenin relevant to muscle plasticity would have to be present in the myonuclei of pre-existing fibres. Importantly, an immunohistochemical analysis indicated that one bout of endurance training led to accumulation of myogenin in some myonuclei. This type of exercise had no effect on MyoD levels (Kadi et al., 2004). Overload exercise, on the other hand, elevated the level of both MyoD and myogenin in some myonuclei (Ishido et al., 2004).
(v) MyoD in slow-to-fast transformations
Thyroid hormone treatment results in activation of both MyoD and fast MyHC gene expression in the slow soleus in vivo (Hughes et al., 1993). The effect of fast stimulation on the MyoD protein content was investigated in the soleus muscle by blocking the endogenous slow impulse activity with tetrodotoxin, and then stimulating the nerve below the block with a fast pattern. After 14 days the average MyoD protein level was increased, but the change was not significant. When the fast rat EDL muscle with its high MyoD content was subjected to a slow pattern of electrical stimulation via the nerve, the level of MyoD was initially unaltered. Thus, although it was reduced after 14 days (Ekmark et al., 2007), regulation of the total MyoD protein level does not seem to be important for the initial phenotypic changes obtained by electrical stimulation. With an antibody specific for inactive MyoD phosphorylated at T115, it was found that levels of phosphorylated and inactive MyoD protein were significantly increased already after 10 h of slow stimulation, and after 14 days of stimulation levels more than doubled. At this time the level of total MyoD was reduced to half, so stimulation increased the fraction of inactive MyoD by four times. The analysis did not allow a quantification of the amount of MyoD that is active. However, it was suggested that in a normal EDL, the fraction of inactive MyoD could not be higher than 1/4 of the total. These data suggest that dephosphorylated MyoD could take part in maintaining fast properties in fast muscle, and that inactivation of MyoD could take part in fast-to-slow transformations. It is less clear if activation of MyoD could take part in slow-to-fast transformation. Thus, when slow rat soleus was stimulated with a fast pattern for two weeks, the level of phosphorylated MyoD was virtually unchanged (Ekmark et al., 2007).
MyoD-null mutant mice failed to display any major shift in MyHC expression, although subtle shifts in the fibre type of fast muscles towards a slower character were seen; in slow muscle a tendency towards a faster phenotype was observed (Hughes et al., 1997; Seward et al., 2001). The lack of change might however be attributed to compensatory mechanisms during development; in order to test the possibility that MyoD might act in adult muscle fibres of normal mice, MyoD was overexpressed in slow muscle fibres. Wild-type MyoD had rather mild effects on fibre type. This was attributed to the activity-dependent phosphorylation discussed above, as it was subsequently shown that after denervation, in the absence of slow activity, overexpression of wild-type MyoD increased the number of fibres displaying fast MyHC significantly in both mice and rats. In further experiments a T115A mutation was introduced, and when this constitutively active form of MyoD was overexpressed after electroporation, the number of MyHC type II positive soleus fibres increased from 50% to 85% in mice and from 13% to 62% in rats, in spite of the muscles receiving the slow endogenous activity pattern (Ekmark et al., 2007).
Thus, active MyoD is a very potent activator of fast transcription in adult muscle, and it could act in three different ways: (1) indirectly by activating genes for other signaling molecules; (2) directly by chromatin remodeling; or (3) as an ultimate transcription factor acting on type II MyHC promoters and other fibre-type-specific promoters.
There are few data to illuminate whether or not MyoD is acting to remodel chromatin in adult post-mitotic muscle fibres, but in the context of MyHC the structural organization of MyHC genes seems suitable for such regulation: in both mice and man the slow MyHCI gene is located on chromosome 14, while the fast IIa, IIx, and IIb forms are clustered on a 300–600 kb segment in the order IIa-IIx-IIb on human chromosome 17 and mouse chromosome 11. Other multi-gene families of contractile proteins are however dispersed throughout the genome, and may require different regulatory mechanisms (Vikstrom et al., 1997).
There is some evidence that MyoD could act as an ultimate transcription factor for MyHC genes. In mice, the IIa and IIb genes each have two E-boxes, while the IIx gene has seven (Allen et al., 2001); but after overexpression of MyoD in C2C12 cells only the IIb, and not the IIa and IIx, promoter is activated (Allen et al., 2001). This demonstrates that MyoD can work selectively on different MyHC promoters in myotubes in culture, and might provide a mechanism for MyoD maintaining IIb expression in normal EDL muscles. In the soleus, fibres overexpressing MyoD and MyHC IIa and IIx were induced, but no IIb was detected. Interestingly this mirrors the effect of fast-patterned electrical stimulation, which also turns on IIa and IIx myosin, while no MyHC IIb is detected in soleus even after more than two months of fast electrical stimulation (Ausoni et al., 1990). Even cross-innervation for more than a year failed to produce shortening velocities consistent with a high type IIb fibre content (Close, 1969; Eken & Gundersen, 1988). Thus, in vivo there seem to be similar limitations for cross-innervation, electrical stimulation, and MyoD overexpression.
A differential effect of MyoD and myogenin has been discussed also for genes other than MyHC relevant to fibre type, and it has been suggested that the E-boxes flanking sequences are involved in determining target sequence specificity for different MRFs. In chicken the fast myosin light chain 1 (MyLC1) has an upstream regulatory region where the distal part contains two E-boxes which are acting as an enhancer responsive to MyoD, while the proximal part contains a single E-box responsive to myogenin (Asakura et al., 1993).
In the fast troponin I gene from quail, the affinity of MyoD for various E-boxes was found to correlate with the ability to trans-activate, and substitution of E-boxes from other muscle-specific genes conferred less binding of MyoD, and less expression when introduced into the fast troponin I gene (Yutzey & Konieczny, 1992). On the other hand, for a chimeric construct combining the E-box parts of SURE with parts of FIRE not containing an E-box, the construct conferred fast expression in adult animals (Calvo et al., 1999, 2001; Rana et al., 2008). Thus, in these experiments the differences between the E-boxes or their flanking sequences were not important for the fibre-type-specific expression, suggesting that MyoD does not act as the ultimate fast transcription factor for TnI. There is more hope for MyHC, but further in vivo experiments are required to establish if MyoD is an ultimate transactivator for type II MyHC, or if it acts indirectly via other signaling systems or by chromatin remodeling.
(vi) Myogenin as a glycolytic-to-oxidative transforming agent
When fast rat EDL muscles are subjected to a slow pattern of electrical stimulation via the nerve, the level of myogenin protein and mRNA in the muscle is elevated after a few hours; conversely myogenin concentration is reduced in slow muscles upon fast stimulation (Ekmark et al., 2007), or fast cross innervation (Hughes et al., 1993). When fast muscles in hypothyroid rats are subjected to slow stimulation, myonuclei in fast-to-slow transforming fibres show accumulation of myogenin mRNA (Putman, Dusterhoft & Pette, 2000). Thus, changes in the myogenin protein level respond in a timely and pattern-specific fashion and could be responsible for changes in the slow/oxidative direction.
Myogenin knock-out mice die at birth (Hasty et al., 1993; Nabeshima et al., 1993), precluding their use in illuminating the role of myogenin in adult muscles. Mice overexpressing myogenin in muscle also had a high neonatal mortality, but for those that survived there were no changes in MyHC fibre type. The activity levels of oxidative mitochondrial enzymes in fast muscles were however increased two- to threefold, whereas levels of glycolytic enzymes were reduced to 30–60% of normal values. Histochemical analysis shows widespread increases in mitochondrial components and glycogen accumulation. The changes in enzyme content were accompanied by a reduction in fibre size, such that many fibres acquired a size typical of oxidative fibres (Hughes et al., 1999). In a subsequent study myogenin expression vectors were injected intracellularly into single adult muscle fibres, or introduced by electroporation into a limited number of fibres. These experiments essentially gave the same results as those observed in the transgenic animals (Ekmark et al., 2003), demonstrating that myogenin can alter the phenotype of pre-existing muscle fibres in adult mice.
Thus, myogenin seems to mimic the typical effects of endurance training since changes in metabolic properties without changes in MyHC type are observed after moderate endurance training in mammals, including humans (Fitts & Widrick, 1996; Henriksson & Hickner, 1994; Saltin & Gollnick, 1983), although changes in MyHC type at least within type II (e.g. from type IIb/IIx to IIa), can also occur (Abdelmalki et al., 1996; Kraemer et al., 1995; Wang et al., 1993).
Like MyoD, myogenin has an inhibitory phosphorylation site (T87) that was shown to be phosphorylated by a high-frequency stimulation pattern (Blagden et al., 2004), but more experiments would be required to establish pattern specificity. An interesting possibility would be if myogenin and MyoD were both phosphorylated by activity, but with a different pattern specificity.
Alterations in oxidative capacity are a question of regulating mitochondrial quantity and quality, and transcriptional regulation of mainly nuclear, but also mitochondrial genes is involved. In general, a large number of nuclear genes are regulated by a small number of transcription factors such as nuclear respiratory factor 1 (NRF-1) and NRF-2. These factors regulate nuclear genes for mitochondrial proteins, and expression of transcription factors regulating genes in the mitochondrial genome. Mitochondrial composition (Forner et al., 2006; Mootha et al., 2003) and relevant regulatory mechanisms (Scarpulla, 2008) are partially tissue specific. Thus, the muscle-specific protein myogenin could possibly work as an ultimate transcription factor binding directly to promoters of mitochondrial enzymes. Several of them contain the consensus sequence for the binding site of bHLH transcription factors (E-box), and in genes encoding muscle/heart-specific forms of the cytochrome c oxidase (COX) subunits VIII and VIa, it has been shown that an intact E-box is required for efficient tissue-specific transcription (Lenka et al., 1996; Wan & Moreadith, 1995). For the COX VIII subunit, negative effects of Id-1 were also demonstrated, strengthening the idea that bHLH proteins are important in regulating this gene.
Interestingly the protein zinc finger protein 106 (ZFP106) is directly regulated by both myogenin and NRF-1 binding to its promoter. The exact role of ZFP106 is not known, but it has been implicated in insulin receptor signaling (Grasberger et al., 2005).
Myogenin may also act indirectly by regulating other transcription factors such as the nuclear respiratory factors (NRFs). PGC-1α can act as a cofactor for NRF-1 and is thought to be crucial for the regulation of oxidative capacity (Lowell & Spiegelman, 2000; Scarpulla, 2008). PGC-1α in turn seems to be regulated by MRFs binding directly to the core PGC-1α promoter. The binding of myogenin to the relevant E-box is slightly stronger than that of MyoD, but in the in vitro assays tested so far myogenin did not have a stronger trans-activating ability than MyoD (Chang et al., 2006).
(2) Metabolic activity correlates
The availability of a variety of metabolites changes during activity. Two examples are ADP and lipids, and studies of signaling molecules that are sensors for ADP and lipids, respectively, suggest that these activity correlates can serve as messengers for muscle plasticity.
(a) AMP-kinase
Active muscles have elevated AMP/ATP ratios that may serve as an activity correlate sensed by AMP-activated kinase (AMPK). It is known that AMPK is activated by exercise; when the AMPK agonist aminoimidazole carboxamide ribonucleotide (AICAR) was administered to rats, metabolic genes were induced and running endurance was increased by 44% (Narkar et al., 2008).
Interestingly, the AMPKα subunit co-immuniprecipitates with peroxisome proliferator-activated receptor δ (PPARδ) (Narkar et al., 2008) suggesting that this kinase might also interact directly with this lipid sensor.
(b) PPARδ
PPARs are ligand-activated transcription factors of the nuclear receptor superfamily. Lipids such as free fatty acids (FFAs) serve as ligands for these receptors, and hence PPARs are lipid sensors. In the current context PPARs therefore combine the role of sensors and transcription factors.
FFAs are elevated in active muscles and might activate the PPARs during activity (Nawrocki & Gorski, 2004). Another interesting possibility would be if metabolites from food intake in itself may influence relevant signaling systems. Changes in fibre type and metabolic properties have been reported after feeding experiments (Nakazato & Song, 2008), but it can not be excluded that the effects are secondary to activity changes.
Three mammalian subtypes of PPARs, -α, -γ, and -δ (the latter also called -β), have been identified (Dreyer et al., 1992; Kliewer et al., 1994). The PPARs form dimers with retinoid X receptors (RXRs). PPARα and PPARγ are predominantly expressed in liver and adipose tissue, respectively, while PPARδ is the predominant isoform in heart and skeletal muscle (Braissant et al., 1996; Escher et al., 2001; Kliewer et al., 1994; Muoio et al., 2002).
In rats PPARδ mRNA is three times higher in slow than in fast muscles, and the protein appears to be accumulated in oxidative fibres (Lunde et al., 2007). Swimming exercise increased muscle PPARδ mRNA in rats on a high-fat diet, but had no effect in lean rats (Kannisto et al., 2006). In normally fed mice, PPARδ protein increased significantly after three weeks of moderate treadmill exercise, but short-term effects were not investigated in this study (Luquet et al., 2003). In a human microarray study, the PPARδ signal was found to be elevated during recovery after a single bout of cycling exercise to exhaustion (Mahoney et al., 2005). In rat, the effects of precisely defined activity patterns have been studied by implanting electrodes. When a slow activity pattern was imposed on a fast muscle via the nerve, the level of PPARδ mRNA trebled in 24 h. In order to study the effect of a fast pattern on slow muscles, the muscles had to be denervated to abolish slow activity. While denervation itself had little effect on the level of PPARδ mRNA, a fast stimulation pattern reduced the level to less than half within 24 h (Lunde et al., 2007). Since only a limited group of muscles below the knee in one leg were stimulated in otherwise normally active rats, and since the effects were compared to contralateral muscles, electrical activity apparently regulates PPARδ levels locally and not as a consequence of increased overall physical activity. A possible mediator of the activity effect is calcineurin, thus expression of PPARα, PPARδ and the PPAR cofactor PGC-1α were promoted by expression of a constitutively active form of calcineurin in both fast and slow muscles in transgenic mice (Long et al., 2007).
In transgenic mice, where the PPARδ gene was knocked out in muscle by α-actin driven Cre recombinase expression, decreased levels of oxidative enzymes, and a slow-to fast-shift in MyHC and troponin I isoenzymes were observed (Schuler et al., 2006). Transgenic mice overexpressing wild-type PPARδ selectively in skeletal muscle develop muscles with elevated oxidative capacity and some hyperplasia, but show no alterations in MyHC fibre type (Luquet et al., 2003). When a ligand-independent constitutively active VP16–PPARδ fusion protein was expressed, however, the transgenic animals also developed muscles with more type I fibres (Wang et al., 2004b). It is tempting to speculate that the inability of the wild-type PPARδ to induce MyHC fibre-type change is due to lack of ligand in muscles lacking slow activity. However, the transgenic technology also differed between the experiments; in the mice expressing the wild-type protein probably only some of the muscle nuclei were expressing the transgene. Nonetheless, the findings may indicate that a moderately elevated level of PPARδ-activity during late muscle development induces muscles to become more oxidative, while with higher activity, MyHC fibre type is also changed. This might be reminiscent of the effects of moderate and hard training in adults but based on the transgenic model developmental, rather than adult effects cannot be excluded.
In adult rats overexpression of PPARδ after electroporation showed that constitutively active VP16-PPARδ can induce both oxidative metabolism and changes in fibre type in adult muscle. In the fast EDL muscle the number of IIa fibres was almost doubled, mainly at the cost of IIb fibres. In all of the transfected fibres, irrespective of type, the level of the oxidative enzyme succinate dehydrogenase (SDH) was increased (Lunde et al., 2007).
These experiments suggest that active PPARδ can serve as a changing agent in adult muscle, but they do not illuminate the role of its ligands. Treatment with the PPAR agonist GW501516 up-regulates mRNA for slow fibre contractile proteins, promotes mitochondrial biogenesis (Wang et al., 2004a), and works synergistically with exercise so as to increase endurance (Narkar et al., 2008). Further experiments are needed to determine the role of lipid ligands in muscle plasticity.
In conclusion, the level of PPARδ is up-regulated by slow activity and down-regulated by fast activity. Activity can increase the PPARδ signal both by increasing the protein level and by activating ligands. An increased PPARδ protein level would increase the sensitivity of the muscle to lipids created by activity. In addition PPARδ might be regulated by its co-factors PGC-1α and—β as discussed above and in Fig. 4.
(c) Oxygen
Intracellular oxygen pressures (PO2i) have been investigated using 1H magnetic resonance spectroscopy of myoglobin. PO2i are reduced from >27 hPa at rest, to values lower than 5 hPa during exercise (Richardson, Newcomer & Noyszewski, 2001; Richardson et al., 1995). These numbers are averages, and the importance of variability in fibre type was not assessed (Hoppeler et al., 2003). In animals exposed to chronic hypoxia in general a slow-to-fast transformation is observed (Bigard et al., 2000; Faucher et al., 2005; Itoh et al., 1990), but this could be related to changes in activity, and the change can be partly counteracted by exercise. There are however well-characterized signaling pathways sensing oxygen levels.
Prolyl hydroxylases use molecular oxygen as a substrate and hence can act as oxygen sensors (Fig. 4). Specifically HIF-1α prolyl hydroxsylases (HPHs) affect the stability of hypoxia inducible factor 1α (HIF-1α) by hydroxylation of conserved prolines. Under normoxic conditions prolyl hydroxylation enables the interaction of HIF-1α and the von-Hippel-Lindau protein, subsequent ubiquitination and proteasomal degradation; under hypoxic conditions HIF-1α accumulates in the cells and acts as a transcription factor (Bracken, Whitelaw & Peet, 2003).
It is possible that HIF-1α may work on fibre-type-specific genes. The level of HIF-1α has been shown to be higher in fast/glycolytic than in slow/oxidative muscles, and the level was increased after hind limb suspension where the muscles become more glycolytic (Pisani & Dechesne, 2005). A HIF-1α knock-out mouse displayed a shift towards a more oxidative metabolism (Mason et al., 2004), but there were no shifts in fibre-type composition (Mason et al., 2007). This could however be due to compensatory mechanisms during development. We have observed that when HIF-1α was overexpressed in adult rat muscle a shift in the fast direction was observed both in the soleus and the EDL. In the soleus, which contains mainly type I fibres, the number of faster IIa fibres was trebled and in the fast EDL the number of IIb fibres was increased by about 50% at the expense of the slower IIa and IIx fibre types in this muscle (K. Gundersen and I.G. Lunde, unpublished data).
The finding that Hif-1α induced a slow-to-fast transformation in fibre type was not necessarily predicted, since most forms of exercise activity might reduce muscle PO2i. The PO2i experienced by individual muscle fibres would however be a complex function of oxygen supply and consumption: supply will vary between richly capillarized oxidative fibres and poorly vascularized glycolytic fibres. On the other hand, oxidative fibres use more oxygen than glycolytic fibres for the same amount of energy consumption. In addition, the pattern of activity varies between fibre types, and intramuscular pressure might restrict blood flow during high-intensity contraction. Adding to the complexity is the observation that the temporal aspects of hypoxia matter, thus intermittent hypoxia has been shown to be more efficient in accumulating HIF-1α than chronic hypoxia (Semenza, 2009).
Although precise information on the PO2i in different muscle fibre types during various activity conditions is not available (Hoppeler et al., 2003), there are indications that fast/glycolytic fibres experience a more severe hypoxia during contraction than do slow/oxidative fibres (McDonough et al., 2007). From a teleological point of view it might be argued that it is beneficial if muscles that experience hypoxia become more glycolytic and use less oxygen, or if severe hypoxia is related to high-intensity exercise, that fibres used in such become faster, stronger and more glycolytic to meet the demand. In sports physiology, the possibility of hypoxia acting as a changing agent for muscle also has interesting implications for altitude training or training with obstructed blood supply, and for understanding muscle changes correlated with respiratory disease.
(3) Six1 and eya1
The signaling molecules discussed above can easily be placed in a general signaling pathway such as outlined in Fig. 3. For sine oculis homeobox 1/eyes absent 1 (Six1/Eya1) and muscle TFII-I repeat-domain-containing protein 1 (MusTRD; see Section III.4) such placement is perhaps more difficult, and it is inspiration from developmental biology that has led to suggestions that these transcription factors might play a role in muscle plasticity.
The Six-family of transcription factors was established by structure homology to its first member the sine oculis gene (so) in Drosophila melanogaster. In mice the six-family has five member-genes (Six1–5). The factors have been implicated particularly in the development of eye and muscle. Six proteins seem to bind to ARE and MEF3 binding motifs of promoters (Kawakami et al., 2000). Eya is a metal-dependent protein tyrosine phosphatase that can act both as a transcription factor and an enzyme. In mammals there are four paralogs (Eya1-4). The role of Eya as a phosphatase is still unclear, but interestingly it has both itself and MAPK as substrates, and it is also itself a MAPK substrate (Hsiao et al., 2001; Jemc & Rebay, 2007). The activation of Eya by MAPK may provide a link to fast activity (Fig. 4).
Eya itself lacks a DNA binding motif and is considered a transcription co-factor. Eya interacts physically with Six, and although both Eya and Six proteins alone promote low levels of transcription, co-transfection studies have produced robust synergetic activation. Six seems to be responsible for accumulating Eya in cell nuclei where the complex can act as a transcription factor (Jemc & Rebay, 2007).
In zebrafish (Danio rerio) a Six1 homolog is essential for development of fast muscle fibres (Bessarab et al., 2008). Six1, −4 and −5 are expressed in vertebrate adult skeletal muscle (Spitz et al., 1998). Surprisingly Six1 mRNA was not accumulated in fast muscles, but western blotting of total protein fractions revealed protein accumulation. Similarly, Eya1, −2 and −4 were all expressed at the mRNA level in muscle, but judged by immunohistochemistry only Eya1 protein was accumulated in nuclei of fast fibres (Grifone et al., 2004).
When Six1 and Eya1 were overexpressed in adult soleus, products of endogenous fast genes such as MyHCIIb, MyHCIIx, MyLC3f, SERCA1 and the glycolytic enzyme β-enolase were induced. Moreover, MyHCI, MyHCIIa and the oxidative enzymes SDH and nicotine adenine dinucleotide tetrazolium reductase (NADH-TR) were supressed (Grifone et al., 2004). Thus, Six1/Eya1 seem to induce most of the phenomena connected to a slow-to-fast transformation such as induction of fast genes, inhibition of slow genes, and a shift from oxidative to glycolytic metabolism.
Interestingly blocking calcineurin with FK506 did not lead to accumulation of Six1 or Eya1 (Grifone et al., 2004). Since calcineurin is believed to mediate effects of slow activity, this suggests that Six1/Eya1 is not activated as a default in absence of slow activity, but as a specific fast mechanism. During development it has been demonstrated that Six1 might be a target gene for myogenic factors. In particular MyoD is strikingly efficient at activating Six1 (Ishibashi et al., 2005). If similar mechanisms operate in the adult, MyoD may be linking Six1 levels to fast activity (Fig. 4).
Six1/Eya1 might also work by regulating the MRFs, as during muscle development, Six1 seems to be implied in regulating all four of them (Giordani et al., 2007; Grifone et al., 2005; Spitz et al., 1998; Zhang & Stavnezer, 2008), but it remains to be seen if Six1/Eya1 regulates myogenic factors in the adult.
There is evidence that Six1/Eya1 could act directly by binding to relevant promoters. When Six1 and Eya1 were co-electroporated with reporter-promoter constructs for the glycolytic enzyme aldolase A, the promoter was activated. Reporter levels increased both when the transgene is episomal and when incorporated into the genome. Thus, in transgenic promoter-reporter mice, reporter was about 10-fold higher in fast than in slow muscle, and mutating the MRF3 site reduced expression in fast muscle, but had no effect on slow muscle (Grifone et al., 2004). Electromobility shift assays (EMSA) demonstrated that the Six1/Eya1 complex binds directly to a MEF3 binding site in the aldolase A promoter, and a trimerized MRF3 site from the promoter is transactivated by Six1/Eya1 in vivo. Thus, Six1/Eya1 might together with the inhibitory effect of NFAT on the fast troponin I promoter, be the best-documented example of an ultimate transcription factor involved in differential expression of fibre-type-specific genes.
(4) MusTRD
Muscle TFII-I repeat-domain-containing protein 1 (MusTRD1) is a protein with homology to the transcription factor TFII-1, and was cloned as a protein binding to an element within the SURE region of the troponin I promoter (O'Mahoney et al., 1998). MusTRD has been cloned independently by others and been given a variety of names such as GTF3, WBSCR11, GTF2IRD, CREAM and BEN. MusTRD is coded by a gene termed GTF21RD1 and is expressed in several splice forms with different DNA-binding abilities (Issa et al., 2006; Polly et al., 2003; Vullhorst & Buonanno, 2003). MusTRD mRNA was originally described as expressed predominantly in muscle (O'Mahoney et al., 1998), but others have found that it is ubiquitously expressed (Bayarsaihan & Ruddle, 2000; Calvo et al., 2001). MusTRD is found in high levels in C2C12 tissue culture and in developing or regenerating muscle, but the level in muscle decreases sharply between postnatal day 7 and 15. During development there is no difference between fast and slow muscle, although in the adult it is marginally higher (<1.4-fold) in the slow soleus compared to the fast EDL (Calvo et al., 2001). Taken together, 11 different splice forms have been found in C2C12 cells, and in developing and adult muscle. Although some of the splice variants were differentially expressed spatially in animals, a fibre-type-specific expression was not reported (Tay et al., 2003). A binding site for MusTRD has been demonstrated in the troponin I promoter by gel shift assays (Calvo et al., 2001; O'Mahoney et al., 1998), and when MusTRD was overexpressed after electroporation in adult muscle, expression from a SURE-luciferase promoter reporter construct was repressed (Calvo et al., 2001). This was essentially confirmed in COS cells by studying a trimerized human slow troponon I binding site connected to luciferase. It was suggested, however, that MusTRD could have a suppressive effect without binding to the SURE promoter since a MusTRD lacking the DNA binding domain also suppressed expression of the reporter. It was suggested that the indirect effect could be due to MusTRD physically interacting with nuclear receptor co-repressor (NoCR), or by interfering with MEF2 binding (Polly et al., 2003).
Recently, transgenic mice overexpressing MusTRD in muscle via the α-actin promoter were created. Although transgenic transcripts were detected already at 13.5 days post coitum the animals were born with muscles that had normal mass, fibre number and fibre type, however they displayed kyphoscoliosis. During the first few post-natal weeks the animals lost their type I fibres, in what appeared to be a type I to IIa transformation in the soleus. Most other slow isoenzymes were also lost, including slow troponin I but not myosin light chain 2 (MyLC2) and oxidative enzymes (Issa et al., 2006). The change in fibre type coincided with the time when distinct fast and slow adult nerve-evoked activity patterns are established (Buffelli et al., 2002; Personius & Balice-Gordon, 2001; Vrbova, Navarrete & Lowrie, 1985), so it is tempting to suggest that MusTRD is involved in activity regulation. In adults, however, an active role of MusTRD is hard to reconcile with its low protein levels, and the lack of differential expression in fast and slow muscle. However the reduced level of MusTRD may be crucial for maintaining slow fibre properties in the adult.
In conclusion, it is unclear if MusTRD plays any role in adult muscle plasticity. Its role might be purely developmental, in playing a restrictive role for development of fibre type prenatally; in addition its absence seems crucial for maintaining normal fibre-type distribution after birth. The complex effects of MusTRD underscore the notion that factors might have different roles at different developmental stages.
IV. PLASTICITY OF MUSCLE FORCE
(1) The cell biology of muscle fibre size
Regulation of force is mainly a question of regulating fibre size, and ultimately size is regulated by altering the balance between protein synthesis and degradation in each muscle fibre. As illustrated in the lower part of Fig. 5, change in fibre size can be achieved by regulating three major conditions: (1) the number of nuclei; (2) the rate of protein synthesis for each nucleus; and (3) the rate of protein degradation.
Fig. 5.
Diagram summarizing pathways currently believed to be involved in regulating muscle fibre size. The pathways have different degrees of scientific support, and their relative quantitative importance is still poorly understood. For key to line and box types, see key to Fig. 4. Abbreviations: forkhead box O (FoxO), glycogen synthase kinase 3 (GSK3), inhibition-of-DNA-binding-protein 1 (Id-1), insulin-like growth factor I (IGF-1), mammalian target of rapamycin (mTOR), myogenic regulatory factor (MRF), phosphatidylinositol 3-kinases/Akt (PI(3)K/Akt), serum response factor (SRF), κ light polypeptide gene enhancer in B-cells (NFκB).
With regard to the regulation of the number of nuclei, it has long been thought (Strassburger, 1893) that a nucleus can support a certain volume of cytoplasm, and that a constant myonuclear domain is maintained during changes in fibre size. Thus, it has generally been believed that hypertrophy was accompanied by addition of new nuclei by satellite cells fusing with the pre-existing muscle fibre syncytia (Lipton & Schultz, 1979; Moss & Leblond, 1970), and that atrophy was accompanied by loss of myonuclei by a selective apoptosis of some nuclei within the intact fibres (Adhihetty et al., 2007; Allen et al., 1997a,b; Alway et al., 2003a,b; Borisov & Carlson, 2000; Darr & Schultz, 1989; Dupont-Versteegden et al., 2000, 1999, 2006; Jin et al., 2001; Schmalbruch & Lewis, 2000; Siu & Alway, 2005; Siu, Pistilli & Alway, 2005a; Siu et al., 2005b; Tang et al., 2000; Tews, 2005; Tews et al., 1997; Viguie et al., 1997; Yoshimura & Harii, 1999).
As reviewed previously, recent research suggests that the nuclear domains may be less constant than previously thought (Gundersen & Bruusgaard, 2008). Moreover, although apoptosis is commonly observed in atrophying muscle tissue, myonuclei seem to be excluded from apoptosis under atrophy conditions. Time-lapse imaging indicates that the number of nuclei is not altered during atrophy, thus nuclei are not lost by a selective intracellular apoptosis as suggested previously (Bruusgaard & Gundersen, 2008; Gundersen & Bruusgaard, 2008).
The finding that nuclear domains may not be as tightly regulated as previously thought (Gundersen & Bruusgaard, 2008), and the lack of elimination of myonuclei during atrophy observed by in vivo imaging (Bruusgaard & Gundersen, 2008) also questions the idea that myonuclei are added to muscle fibres during hypertrophy. The issue of whether activation and fusion of satellite cells is obligatory for hypertrophy was recently debated in the Journal of Applied Physiology (Bodine, 2007; Booth, 2007; Hikida, 2007; Kadi, 2007; Lowe, 2007; Mantilla & Sieck, 2007; McCarthy & Esser, 2007; O'Connor & Pavlath, 2007; O'Connor et al., 2007; Rehfeldt, 2007). It was argued that molecular evidence suggests that there is an early increase in protein synthesis (per nucleus), and that addition of nuclei follow later if at all. Based on in vivo imaging we have observed a different temporal picture at the phenomenological level. When mice EDL muscle was overloaded by synergist ablation the addition of new nuclei occurred mainly after 6–8 days, while the increase in size occurred after 8–12 days (Bruusgaard et al., 2010). Thus addition of nuclei preceded the hypertrophy leading to a temporary decrease in myonuclear domain sizes. This sequence of events strongly suggests that the increased number of nuclei is a major cause of hypertrophy, but it does not preclude that at least some hypertrophy could occur in the absence of new nuclei under experimental conditions where satellite cells are eliminated.
Interestingly, newly added “hypertrophy nuclei” seem to be maintained during subsequent atrophy. Thus, the elevated number of nuclei was maintained even after three months of denervation resulting in severe atrophy (Bruusgaard et al., 2010). This might explain the phenomenon of “muscle memory”: that previously strong muscles seem to be more easily retrained. The number of nuclei might be limiting during the first hypertrophy, and the increased number of nuclei resulting from hypertrophy might facilitate increased synthesis upon retraining.
(2) Paracrine and autocrine mechanisms
While the regulation of protein synthesis per nucleus and protein degradation in principle could be obtained by an intracellular excitation-transcription coupling such as discussed for regulation of fibre type; involvement of satellite cells would require additional intercellular signaling systems. Interestingly, there seem to be at least two systems acting as local hormones in muscle, contributing to regulation of muscle fibre size in the adult: myostatin and IGF-1. The release of these factors is dependent on activity (upper part of Fig. 5).
(a) Myostatin
Myostatin (previously called GDF-8) is a member of the transforming growth factor β (TGF-β) superfamily and plays a major role during development where it acts as an inhibitor of muscle growth. Disruption of the myostatin gene leads to development of grossly enlarged muscles in mice (McPherron, Lawler & Lee, 1997), farm animals (McPherron & Lee, 1997), and man (Schuelke et al., 2004). The enlargement is caused both by an increase in the number of fibres (hyperplasia) and in fibre size (hypertrophy). Importantly, muscle enlargement obtained by myostatin deficiency is peculiar because it does not increase force in proportion to size, thus the amount of contractile proteins may not be properly regulated (Amthor et al., 2007; Bruusgaard et al., 2005). Thus, reducing myostatin alone might not mimic effects of strength training, although strength training in adults has been shown to be associated with reduced levels of myostatin in muscle and plasma (Heinemeier et al., 2007; Kim, Cross & Bamman, 2005; Roth et al., 2003; Walker et al., 2004). Conversely various disuse conditions have been associated with elevated levels (Gonzalez-Cadavid et al., 1998; Lalani et al., 2000; Reardon et al., 2001; Wehling, Cai & Tidball, 2000; Yarasheski et al., 2002). Interestingly, release of myostatin can be regulated by the titin-cap protein (Nicholas et al., 2002), opening up the possibility that stretch signals could regulate myostatin release.
In adult animals inhibition of myostatin with antibodies leads to hypertrophy without an increase in the number of fibres (Bogdanovich et al., 2002); conversely, overexpression of myostatin in muscle fibres after electroporation leads to muscle atrophy without loss of muscle fibres (Durieux et al., 2007). In the latter study it was suggested that myostatin acted by reducing muscle gene expression of myofibrillar proteins perhaps by reducing expression of MyoD and myogenin. In addition, myostatin might activate the ubiquitin-proteasome pathway for proteolysis (Gilson et al., 2007).
(b) Insulin-like growth factor I (IGF-1)
IGF-1 has been implicated as a factor promoting hypertrophy in the adult animal. The gene gives rise to several proteins by differential splicing with a rather confusing nomenclature (Barton, 2006a). Adding to the complexity is the observation that IGF-1 activity is regulated by IGF-binding proteins acting as carriers in blood, or locally binding IGF-1 to the extracellular matrix (Clemmons, 1998, 2007).
The liver supplies approximately 75% of the circulating IGF-1 (Schwander et al., 1983), and a selective abolishment of IGF-1 production in hepatocytes leads to a 75% reduction in circulating IGF-1 levels but without growth impairment (Sjogren et al., 1999). In humans increasing the circulating level of IGF-1 does not promote muscle protein synthesis (Yarasheski et al., 1992, 1993). IGF-1 is also expressed locally in several tissues including muscle where it is induced by stretch or high-resistance exercise (Adams & Haddad, 1996; Adams, Haddad & Baldwin, 1999; Bamman et al., 2001; DeVol et al., 1990; Petrella et al., 2006; Sakuma et al., 1998; Singh et al., 1999; Yan, Biggs & Booth, 1993). A specific splice variant of IGF-1 is particularly prone to be up-regulated under such conditions, and has been dubbed mechano growth factor (MGF) (Hameed et al., 2003; Yang et al., 1996). Muscle also produces IGF-1 during exercise (Brahm et al., 1997), although plasma IGF-1 levels are not significantly altered (Walker et al., 2004). Based on these findings, it is suggested that IGF-1 production is triggered by resistance exercise, and can act in a para- and autocrine fashion in muscle.
Transgenic mice over-expressing IGF-1 have increased muscle mass (Mathews et al., 1988) while mice deficient in IGF-1 have a severe muscular dystrophy (Powell-Braxton et al., 1993). Also transgenic mice overexpressing IGF-1 selectively in muscle cells display muscle hypertrophy (Coleman et al., 1995; Musaro et al., 2001). In adult muscles, localized infusion of IGF-1 protein increased muscle mass (Adams & McCue, 1998), and muscle transfected with expression vectors for IGF-1 by electroporation (Alzghoul et al., 2004) or viral infection (Barton, 2006b) displays hypertrophy. These findings suggest that IGF-1 plays a role in adult hypertrophy.
The selective effects of different splice forms have been investigated. In muscles of two-week-old animals where muscles were infected with IGF-1-producing adenoviruses, hypertrophy was observed with both the MGF (IGF-1b) and IGF-1a isoforms, but only the IGF-1a form had effects when introduced in mature mice (Barton, 2006b).
In muscle tissue culture, IGF-1 has been shown to increase myotube diameter, suppress proteolysis, stimulate protein synthesis, and induce a higher number of nuclei per length of myotube (Engert, Berglund & Rosenthal, 1996; Florini, Ewton & Coolican, 1996; Jacquemin et al., 2004; Rommel et al., 2001; Vandenburgh et al., 1991). IGF-1 seems to promote myoblast mitosis as well as myoblast differentiation (Rosenthal & Cheng, 1995). Thus IGF-1 might lead to production of new myoblasts, and promote them to fuse to existing myotubes. In addition, IGF-1 has a direct effect on protein synthesis mainly by stimulating translation (Philippou et al., 2007).
In vivo IGF-1 increases the DNA content in muscles (Adams & McCue, 1998), but it remains to be demonstrated if IGF-1 induces satellite cells to fuse to pre-existing fibres. Elevated IGF-1 immunoreactivity has been demonstrated in myoblasts and myotubes during muscle regeneration (Jennische & Hansson, 1987; Jennische, Skottner & Hansson, 1987), and a loss-of-function study with an IGF-1 antibody showed that the number and size of regenerating muscle fibres is reduced (Lefaucheur & Sebille, 1995).
In conclusion, IGF-1 seems to work as a local hormone that promotes hypertrophy in adult muscle. It might do so both by interfering with protein balance in muscle fibres and by activating satellite cells, but for the latter there is still little information in adult muscles.
(3) Intracellular pathways of size regulation
A discussion on signaling pathways for atrophy and hypertrophy can easily be rather complex since it involves signaling both inside and between at least two different cell types: satellite cells and myofibres. As discussed above, multiplication, differentiation and the ultimate fusion of satellite cells to pre-existing muscle fibres is thought to be important for hypertrophy, but satellite cell behaviour and regulatory pathways have been mostly studied during muscle formation and regeneration (Holterman & Rudnicki, 2005; Philippou et al., 2007; Wagers & Conboy, 2005; Zammit, 2008), and less so in intact adult muscles.
(a) Regulation of protein production in muscle fibres
(i) Regulation of transcription
During changes in muscle fibre size, protein production seems to be regulated both at the transcriptional and at the translational level (for review see Favier, Benoit & Freyssenet, 2008). It is tempting to suggest that transcription factors operating during development also act in the adult. For example the developmentally important MRFs have been implicated also in regulation of fibre size, and can be up-regulated by resistance exercise. Conversely Id proteins are up-regulated during inactivity (Alway et al., 2003b; Carlsen & Gundersen, 2000; Dupont-Versteegden et al., 1998; Gundersen & Merlie, 1994). When Id-1 was overexpressed, it induced atrophy in a dose-dependent manner (Gundersen & Merlie, 1994). However also the MRFs themselves (Buonanno et al., 1992; Duclert, Piette & Changeux, 1991; Eftimie, Brenner & Buonanno, 1991; Merlie et al., 1994; Neville, Schmidt & Schmidt, 1992; Walters, Stickland & Loughna, 2000b; Witzemann & Sakmann, 1991), and their positive partners; the E-proteins (Carlsen & Gundersen, 2000), are up-regulated during disuse. The putative role of the MRFs is illustrated in Fig. 5, but more research is required to investigate the role of MRFs, their phosphorylation state, and their partners in order to establish their importance for transcriptional regulation during atrophy/hypertrophy conditions in the adult.
The transcription factor serum response factor (SRF) is increased by stretch in avian muscle, and seems to bind to and transactivate the alpha actin gene (Carson & Booth, 1999; Carson, Schwartz & Booth, 1996; Fluck et al., 1999). If SRF is a major regulator of this important contractile protein, SRF might have a central role in regulating fibre size.
It has been suggested that the transcription factor NFκB contributes to muscle atrophy, activated by the cytokine tumor necrosis factor α (TNF-α) which is activated in disease states, leading to atrophy (Kandarian & Jackman, 2006; Tisdale, 2009). Disuse atrophy does not however, seem to trigger TNF-α production, but the concentration of NFκB is increased by unloading (Hunter et al., 2002). NFκB may downregulate MyoD and hence synthesis of muscle-specific proteins (Guttridge et al., 2000).
(ii) Regulation of translation: the PI(3)K/Akt pathway
An important regulated process is translation. During hypertrophy, IGF-I seems to activate the phosphatidylinositol 3-kinases/Akt/mammalian target of rapamycin (PI(3)K/Akt/mTOR) pathway, which seems to boost translation (Bodine et al., 2001b; Li, Li & Parkhouse, 2002; Rommel et al., 2001). This pathway is illustrated in Fig. 5, and has been reviewed extensively (Adams, 2002; Favier et al., 2008; Miyazaki & Esser, 2009; Sandri, 2008). mTOR seems to be crucial, since selective mTOR blockers inhibit hypertrophy in vivo. Interestingly such inhibition only prevented use-related hypertrophy, but did not induce atrophy (Bodine et al., 2001b).
In vitro evidence suggests, that there is also an alternative collateral mTOR-independent pathway where Akt activates glycogen synthase kinase 3 (GSK-3) (Fig. 5) (Holterman & Rudnicki, 2005; Rommel et al., 2001), but since the results might be confounded by effects on muscle differentiation in vitro, more research on this pathway should be performed in vivo in order to establish its importance for muscle plasticity.
The PI(3)K/Akt/mTOR pathway seems to operate in mice expressing a dominant negative IGF-1 receptor in muscle (Spangenburg et al., 2008), suggesting that overload hypertrophy is at least not solely dependent on activation of the IGF-1 receptor, and that the pathway can be activated also by other signals.
In addition to its effects on translation, the PI(3)K/Akt pathway inhibits FoxO, which is an activator of the ubiquitin-proteasome pathway, and thus protein degradation (Glass, 2005).
(b) Regulation of protein degradation
Atrophy is caused both by reduced protein production, and by increased proteolysis. The latter has received most of the attention recently, partly because the specific mechanisms are easier to study, but it has also been claimed that it is quantitatively the most important process during atrophy (Kandarian & Jackman, 2006). The large body of literature on the regulation of protein degradation in muscle during atrophy has been extensively reviewed (Favier et al., 2008; Glass, 2003, 2005; Kandarian & Jackman, 2006; Sandri, 2008), and the pathways will be only briefly outlined here.
The central pathway for proteolysis during atrophy is the ubiquitin-proteasome pathway. With this mechanism doomed proteins are “tagged” by multiple copies of a 76-amino-acid peptide, called ubiquitin, being ligated to a lysine residue. These ubiquitinated proteins are then degraded by a large protein complex called the 20 S proteasome (Jung, Catalgol & Grune, 2009). The ubiquitination is accomplished by a complex enzymatic reaction where members of a large group of ubiquitine-ligases participate, each one specific to a limited number of substrate proteins (Jung et al., 2009). Two such proteins have been shown to be up-regulated in at least 13 distinct atrophy models: muscle ring finger1 (MuRF1) and muscle atrophy F-box protein (MAFbx, also called atrogin) (Glass, 2005). Mice with null-mutations for one of these proteins appear normal, but atrophy is attenuated (Bodine et al., 2001a). MuRF1 has also been shown to inhibit nuclear localization and transcriptional activity of SRF (Lange et al., 2005), and may thus contribute to reduced transcription during atrophy.
(c) Putative mechano-sensing mechanisms in muscle size regulation
Interestingly, MuRF1 has been shown to bind specifically to titin (Centner et al., 2001; Lange et al., 2005; McElhinny et al., 2002; Mrosek et al., 2007), and it has been postulated that titin may serve as a mechanosensor relaying stretch signals to intracellular pathways. Titin appears to contain a kinase domain that binds Ca2+/calmodulin (Mayans et al., 1998). The domain is located in the M-line, a position that would be prone to stretch during active muscle contraction, and recent experiments combining atomic force microscopy, molecular dynamics simulations, and enzymatics suggest that the catalytic domain is exposed by stretch (Puchner et al., 2008). This makes titin a prime force-transducer candidate (Tskhovrebova & Trinick, 2008). In cardiomyocytes, an elaborate mechano-sensitive signaling system connected to the contractile apparatus and mediating stretch signals has been postulated (for review see Kruger & Linke, 2009), but its relevance to skeletal muscle is still unclear.
V. THE EXCITATION-TRANSCRIPTION COUPLING: TOWARDS A SYNTHESIS?
(1) Plasticity of speed
Nadal Ginard has termed the factor linking activity and adult muscle phenotype “the holy grail” of muscle biology. As evident from Fig. 4, there seems to be a large number of such grails, with few simplifying principles. It is a problem that we currently do not have an understanding of the relative importance of different pathways, but it is possible that more loss-of-function experiments could reveal which are the major pathways that are crucial for plasticity.
Several activity correlates might be sensed by various sensor molecules such as HPH, PKC, calmodulin, AMPK, and PPARδ. The activation of these sensors triggers complex cascades ending up with activation or inhibition of ultimate transcription factors binding to promoter regions of fast and slow genes. Most of the research has been on participants in cascades, and since they interact in complex ways and also might interfere with fundamental and general cell biological mechanisms; it is often difficult to assess their specific importance for muscle plasticity. It is possible that more emphasis should be put on unraveling signaling pathways from the distal end: which are the factors binding to the promoters for fast and slow isoforms?
A hard-earned lesson from the study of gene regulation is that the mechanisms seem to be different in different experimental systems. Thus, cultured, developing or regenerating muscles are different from adult muscles, and endogenous genes frequently behave differently from promoter-reporter constructs. Although inspiration might be drawn from other systems, ultimately any hypotheses would have to be tested on endogenous genes in adult muscle that have developed in the absence of genetic manipulation.
Based on experiments in developing or regenerating muscle, it has been suggested that there are pathways connecting slow activity to slow phenotype, but that fast phenotype develops as a default in the absence of slow activity (Butler-Browne et al., 1982; Esser, Gunning & Hardeman, 1993; Jerkovic et al., 1997). This idea of a default fast pathway is not supported in adult muscle. Thus, slow phenotype can develop in anural muscles (Condon et al., 1990; Phillips & Bennett, 1984), and in adult muscle, absence of activity does not lead to the same phenotype as stimulation with a fast pattern. Not only does a denervated or inactive muscle become extremely atrophic, but it also displays abnormal membrane properties such as supersensitivity for acetylcholine. This abnormality is not seen in denervated muscles given even very little activity (Lømo & Westgaard, 1975; Midrio, 2006). After denervation, the twitch at least initially becomes slower (Gundersen, 1985), and even if fast MyHC is eventually induced in a minority of fibres, fast stimulation will induce fast MyHC in all fibres (Windisch et al., 1998). Promoter-reporter studies in vivo showed that denervation inhibited promoters for both the fast and slow troponin I isoforms, while the fast promoter was selectively activated by a fast pattern and not a slow pattern (Rana et al., 2008, 2005). These findings suggest that there is a specific fast signaling pathway, at least for troponin I. The idea of a default fast pathway might be more relevant for MyHC regulation where a frequency effect has not been demonstrated. A slow activity pattern will phosphorylate and inactivate MyoD, and MyoD protein was shown to trigger expression of fast MyHC only in the absence of externally triggered activity in denervated slow muscles (Ekmark et al., 2007). It was argued that MyoD maintains fast properties in fast muscles where slow activity will not phosphorylate and inactivate it, and this could be an example of a fast signal acting only in the absence of a slow activity pattern.
(2) Plasticity of force
It is generally accepted that nerve-evoked activity plays a major role in determining muscle size in adults. During activity both electrical and mechanical factors are believed to play a major role in providing signaling (Fig. 5), but there is currently little precise information on the initial mechanisms (activity correlates) involved. Activity seems to act at least partly by altering paracrine conditions in the muscle tissue such as via IGF-1 and myostatin.
Our knowledge about pathways further downstream has increased over the last decade, in particular about the intracellular pathways responsible for increased proteolysis during atrophy, and increased translation during hypertrophy. Less is known about transcriptional regulation during such conditions.
The activation of satellite cells is still poorly understood, but it seems clear that fusion of satellite cells seems to play a role during hypertrophy, but contrary to previous ideas myonuclei do not seem to be lost during atrophy. This suggests that myonuclei recruited during strength training are permanent. Thus, effects of strength training might be more permanent than previously thought and the elevated number of nuclei may represent a form of muscle memory facilitating retraining (Bruusgaard et al., 2010).
VI. CONCLUSIONS
Muscle properties change in an adaptive way when the activity pattern delivered to the muscle is changed, and the changes are dependent on the specific fast or slow pattern of activity.
Several intracellular changes such as an increase in free intracellular Ca2+, alterations in metabolites, hypoxia, and mechanical stress are correlated to muscle activity and molecular sensors connected to intracellular signaling pathways sense various activity correlates and trigger intracellular signaling pathways.
The numerous putative signaling pathways reviewed herein as taking part in muscle plasticity rely on scientific support of variable strength. In addition, more quantitative approaches are desirable in order to establish the most important pathways, as opposed to those that have some effects under some experimental conditions. Thus, our current model seems to be in need of both pruning, and new data. To this end loss-of-function studies might be useful.
Signaling pathways might involve transcription factors and gene regulation as part of the cascade, but the pathways end up regulating “ultimate” transcription factors binding to the genes for fast and slow isoforms. Very few ultimate transcription factors have been unequivocally identified so far, and more knowledge about such factors might contribute to a more precise unraveling of signaling pathways from the distal end.
Mechanisms of adult plasticity should be confirmed by studies of endogenous genes in adult muscle in vivo. Insights from other more convenient models, although inspirational, have sometimes proven treacherous.
Regulating the balance between protein synthesis and proteolysis alters muscle fibre size. Myonuclei are recruited from satellite cells during hypertrophy, but not lost during atrophy. Thus, the idea of constant myonuclear domains seems oversimplified.
VII. ACKNOWLEDGEMENTS
I am grateful to the members of my group, and to Drs Andres Buonanno, Terje Lømo and Håkan Westerblad for comments on previous versions of this manuscript. They have improved it, but they have no responsibility for the end result.
VIII. REFERENCES
- Abdelmalki A, Fimbel S, Mayet-Sornay MH, Sempore B, Favier R. Aerobic capacity and skeletal muscle properties of normoxic and hypoxic rats in response to training. Pflügers Archiv. 1996;431:671–679. doi: 10.1007/BF02253829. [DOI] [PubMed] [Google Scholar]
- Adams GR. Invited review: autocrine/paracrine IGF-I and skeletal muscle adaptation. Journal of Applied Physiology. 2002;93:1159–67. doi: 10.1152/japplphysiol.01264.2001. [DOI] [PubMed] [Google Scholar]
- Adams GR, Cheng DC, Haddad F, Baldwin KM. Skeletal muscle hypertrophy in response to isometric, lengthening, and shortening training bouts of equivalent duration. Journal of Applied Physiology. 2004;96:1613–1618. doi: 10.1152/japplphysiol.01162.2003. [DOI] [PubMed] [Google Scholar]
- Adams GR, Haddad F. The relationships among IGF-1, DNA content, and protein accumulation during skeletal muscle hypertrophy. Journal of Applied Physiology. 1996;81:2509–2516. doi: 10.1152/jappl.1996.81.6.2509. [DOI] [PubMed] [Google Scholar]
- Adams GR, Haddad F, Baldwin KM. Time course of changes in markers of myogenesis in overloaded rat skeletal muscles. Journal of Applied Physiology. 1999;87:1705–1712. doi: 10.1152/jappl.1999.87.5.1705. [DOI] [PubMed] [Google Scholar]
- Adams GR, McCue SA. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. Journal of Applied Physiology. 1998;84:1716–1722. doi: 10.1152/jappl.1998.84.5.1716. [DOI] [PubMed] [Google Scholar]
- Adhihetty PJ, O'Leary MF, Chabi B, Wicks KL, Hood DA. Effect of denervation on mitochondrially mediated apoptosis in skeletal muscle. Journal of Applied Physiology. 2007;102:1143–1151. doi: 10.1152/japplphysiol.00768.2006. [DOI] [PubMed] [Google Scholar]
- Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates PGC-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. Journal of Biological Chemistry. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- Al-Amood WS, Lewis DM. The role of frequency in the effects of long-term intermittent stimulation of denervated slow-twitch muscle of the rat. Journal of Physiology. 1987;392:377–395. doi: 10.1113/jphysiol.1987.sp016786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford EK, Roy RR, Hodgson JA, Edgerton VR. Electromyography of rat soleus, medial gastrocnemius, and tibialis anterior during hind limb suspension. Experimental Neurology. 1987;96:635–649. doi: 10.1016/0014-4886(87)90225-1. [DOI] [PubMed] [Google Scholar]
- Allen DG, Lamb GD, Westerblad H. Skeletal muscle fatigue: cellular mechanisms. Physiological Reviews. 2008;88:287–332. doi: 10.1152/physrev.00015.2007. [DOI] [PubMed] [Google Scholar]
- Allen DL, Linderman JK, Roy RR, Bigbee AJ, Grindeland RE, Mukku V, Edgerton VR. Apoptosis: a mechanism contributing to remodeling of skeletal muscle in response to hindlimb unweighting. American Journal of Physiology. 1997a;273:C579–C587. doi: 10.1152/ajpcell.1997.273.2.C579. [DOI] [PubMed] [Google Scholar]
- Allen DL, Linderman JK, Roy RR, Grindeland RE, Mukku V, Edgerton VR. Growth hormone/IGF-I and/or resistive exercise maintains myonuclear number in hindlimb unweighted muscles. Journal of Applied Physiology. 1997b;83:1857–1861. doi: 10.1152/jappl.1997.83.6.1857. [DOI] [PubMed] [Google Scholar]
- Allen DL, Sartorius CA, Sycuro LK, Leinwand LA. Different pathways regulate expression of the skeletal myosin heavy chain genes. Journal of Biological Chemistry. 2001;276:43524–43533. doi: 10.1074/jbc.M108017200. [DOI] [PubMed] [Google Scholar]
- Almholt K, Arkhammar PO, Thastrup O, Tullin S. Simultaneous visualization of the translocation of protein kinase Calpha-green fluorescent protein hybrids and intracellular calcium concentrations. Biochemcal Journal. 1999;337:211–218. [PMC free article] [PubMed] [Google Scholar]
- Alway SE, Degens H, Krishnamurthy G, Chaudhrai A. Denervation stimulates apoptosis but not Id2 expression in hindlimb muscles of aged rats. Journals of Gerontology Series A: Biological Sciences and Medical Sciences. 2003a;58:687–697. doi: 10.1093/gerona/58.8.b687. [DOI] [PubMed] [Google Scholar]
- Alway SE, Martyn JK, Ouyang J, Chaudhrai A, Murlasits ZS. Id2 expression during apoptosis and satellite cell activation in unloaded and loaded quail skeletal muscles. American Journal of Physiology. 2003b;284:R540–R549. doi: 10.1152/ajpregu.00550.2002. [DOI] [PubMed] [Google Scholar]
- Alzghoul MB, Gerrard D, Watkins BA, Hannon K. Ectopic expression of IGF-I and Shh by skeletal muscle inhibits disuse-mediated skeletal muscle atrophy and bone osteopenia in vivo. FASEB Journal. 2004;18:221–223. doi: 10.1096/fj.03-0293fje. [DOI] [PubMed] [Google Scholar]
- Amthor H, Macharia R, Navarrete R, Schuelke M, Brown SC, Otto A, Voit T, Muntoni F, Vrbova G, Partridge T, Zammit P, Bunger L, Patel K. Lack of myostatin results in excessive muscle growth but impaired force generation. Proceedings of the National Academy of Sciences USA. 2007;104:1835–1840. doi: 10.1073/pnas.0604893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JL, Klitgaard H, Saltin B. Myosin heavy chain isoforms in single fibres from m. vastus lateralis of sprinters: Influence of training. Acta Physiologica Scandinavica. 1994;151:135–142. doi: 10.1111/j.1748-1716.1994.tb09730.x. [DOI] [PubMed] [Google Scholar]
- Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- Arany Z, He H, Lin J, Hoyer K, Handschin C, Toka O, Ahmad F, Matsui T, Chin S, Wu PH, Rybkin II, Shelton JM, Manieri M, Cinti S, Schoen FJ, Bassel-Duby R, Rosenzweig A, Ingwall JS, Spiegelman BM. Transcriptional coactivator PGC-1 alpha controls the energy state and contractile function of cardiac muscle. Cell Metabolism. 2005;1:259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Arany Z, Lebrasseur N, Morris C, Smith E, Yang W, Ma Y, Chin S, Spiegelman BM. The Transcriptional Coactivator PGC-1beta Drives the Formation of Oxidative Type IIX Fibers in Skeletal Muscle. Cell Metabolism. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Asakura A, Fujisawa SA, Komiya T, Nabeshima Y, Nabeshima YI. MyoD and myogenin act on the chicken myosin light-chain 1 gene as distinct transcriptional factors. Molecular and Cellular Biology. 1993;13:7153–7162. doi: 10.1128/mcb.13.11.7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB Journal. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- Ausoni S, Gorza L, Schiaffino S, Gundersen K, Lomo T. Expression of myosin heavy chain isoforms in stimulated fast and slow rat muscles. Journal of Neuroscience. 1990;10:153–160. doi: 10.1523/JNEUROSCI.10-01-00153.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin J, Shabalina IG, Place N, Reiken S, Zhang SJ, Bellinger AM, Nedergaard J, Cannon B, Marks AR, Bruton JD, Westerblad H. Nonshivering thermogenesis protects against defective calcium handling in muscle. FASEB Journal. 2008;22:3919–24. doi: 10.1096/fj.08-113712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB Journal. 2002;16:1879–86. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- Bamman MM, Ragan RC, Kim JS, Cross JM, Hill VJ, Tuggle SC, Allman RM. Myogenic protein expression before and after resistance loading in 26- and 64-yr-old men and women. Journal of Applied Physiology. 2004;97:1329–1337. doi: 10.1152/japplphysiol.01387.2003. [DOI] [PubMed] [Google Scholar]
- Bamman MM, Shipp JR, Jiang J, Gower BA, Hunter GR, Goodman A, McLafferty CL, Jr, Urban RJ. Mechanical load increases muscle IGF-I and androgen receptor mRNA concentrations in humans. American Journal of Physiology. 2001;280:E383–E390. doi: 10.1152/ajpendo.2001.280.3.E383. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu S, Buonanno A. cis-acting sequences of the rat troponin I slow gene confer tissue- and development-specific transcription in cultured muscle cells as well as fiber type specificity in transgenic mice. Molecular and Cellular Biology. 1993;13:7019–7028. doi: 10.1128/mcb.13.11.7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton ER. The ABCs of IGF-I isoforms: impact on muscle hypertrophy and implications for repair. Applied Physiology, Nutrition, and Metabolism. 2006a;31:791–797. doi: 10.1139/h06-054. [DOI] [PubMed] [Google Scholar]
- Barton ER. Viral expression of insulin-like growth factor-I isoforms promotes different responses in skeletal muscle. Journal of Applied Physiology. 2006b;100:1778–1784. doi: 10.1152/japplphysiol.01405.2005. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annual Review of Biochemistry. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D, Ruddle FH. Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains. Proceedings of the National Academy of Sciences USA. 2000;97:7342–7347. doi: 10.1073/pnas.97.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengal E, Flores O, Rangarajan PN, Chen A, Weintraub H, Verma IM. Positive control mutations in the MyoD basic region fail to show cooperative DNA binding and transcriptional activation in vitro. Proceedings of the National Academy of Sciences USA. 1994;91:6221–6225. doi: 10.1073/pnas.91.13.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom DA, Tapscott SJ. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Molecular and Cellular Biology. 2001;21:2404–2412. doi: 10.1128/MCB.21.7.2404-2412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Seminars in Cell and Developmental Biology. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Bessarab DA, Chong SW, Srinivas BP, Korzh V. Six1a is required for the onset of fast muscle differentiation in zebrafish. Developmental Biology. 2008;323:216–228. doi: 10.1016/j.ydbio.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Bi GQ, Rubin J. Timing in synaptic plasticity: from detection to integration. Trends in Neuroscience. 2005;28:222–228. doi: 10.1016/j.tins.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Bickel CS, Slade J, Mahoney E, Haddad F, Dudley GA, Adams GR. Time course of molecular responses of human skeletal muscle to acute bouts of resistance exercise. Journal of Applied Physiology. 2005;98:482–488. doi: 10.1152/japplphysiol.00895.2004. [DOI] [PubMed] [Google Scholar]
- Bigard AX, Sanchez H, Birot O, Serrurier B. Myosin heavy chain composition of skeletal muscles in young rats growing under hypobaric hypoxia conditions. Journal of Applied Physiology. 2000;88:479–486. doi: 10.1152/jappl.2000.88.2.479. [DOI] [PubMed] [Google Scholar]
- Bintliff S, Walker BE. Radioautographic study of skeletal muscle regeneration. American Journal of Anatomy. 1960;106:233–245. [Google Scholar]
- Biring MS, Fournier M, Ross DJ, Lewis MI. Cellular adaptations of skeletal muscles to cyclosporine. Journal of Applied Physiology. 1998;84:1967–1975. doi: 10.1152/jappl.1998.84.6.1967. [DOI] [PubMed] [Google Scholar]
- Blagden CS, Fromm L, Burden SJ. Accelerated response of the myogenin gene to denervation in mutant mice lacking phosphorylation of myogenin at threonine 87. Molecular and Cellular Biology. 2004;24:1983–1989. doi: 10.1128/MCB.24.5.1983-1989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1105–1106. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD, Glass DJ. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001a;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nature Cell biology. 2001b;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS. Functional improvement of dystrophic muscle by myostatin blockade. Nature. 2002;420:418–421. doi: 10.1038/nature01154. [DOI] [PubMed] [Google Scholar]
- Bogoyevitch MA, Court NW. Counting on mitogen-activated protein kinases–ERKs 3, 4, 5, 6, 7 and 8. Cell Signal. 2004;16:1345–1354. doi: 10.1016/j.cellsig.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bonavaud S, Agbulut O, Nizard R, D'Honneur G, Mouly V, Butler-Browne G. A discrepancy resolved: human satellite cells are not preprogrammed to fast and slow lineages. Neuromuscular Disorders. 2001;11:747–752. doi: 10.1016/s0960-8966(01)00222-x. [DOI] [PubMed] [Google Scholar]
- Boncompagni S, Kern H, Rossini K, Hofer C, Mayr W, Carraro U, Protasi F. Structural differentiation of skeletal muscle fibers in the absence of innervation in humans. Proceedings of the National Academy of Sciences USA. 2007;104:19339–19344. doi: 10.1073/pnas.0709061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth FW. Time course of muscular atrophy during immobilization of hindlimbs in rats. Journal of Applied Physiology. 1977;43:656–661. doi: 10.1152/jappl.1977.43.4.656. [DOI] [PubMed] [Google Scholar]
- Booth FW. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1105.. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Boppart MD, Hirshman MF, Sakamoto K, Fielding RA, Goodyear LJ. Static stretch increases c-Jun NH2-terminal kinase activity and p38 phosphorylation in rat skeletal muscle. American Journal of Physiology. 2001;280:C352–C358. doi: 10.1152/ajpcell.2001.280.2.C352. [DOI] [PubMed] [Google Scholar]
- Borisov AB, Carlson BM. Cell death in denervated skeletal muscle is distinct from classical apoptosis. Anatomical Records. 2000;258:305–318. doi: 10.1002/(SICI)1097-0185(20000301)258:3<305::AID-AR10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Bottinelli R, Reggiani C. Human skeletal muscle fibres: molecular and functional diversity. Progress in Biophysics and Molecular Biology. 2000;73:195–262. doi: 10.1016/s0079-6107(00)00006-7. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Whitelaw ML, Peet DJ. The hypoxia-inducible factors: key transcriptional regulators of hypoxic responses. Cellular and Molecular Life Sciences. 2003;60:1376–1393. doi: 10.1007/s00018-003-2370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley J, Finkbeiner S. An evaluation of specificity in activity-dependent gene expression in neurons. Progress in Neurobiology. 2002;67:469–477. doi: 10.1016/s0301-0082(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Brahm H, Piehl-Aulin K, Saltin B, Ljunghall S. Net fluxes over working thigh of hormones, growth factors and biomarkers of bone metabolism during short lasting dynamic exercise. Calcified Tissue International. 1997;60:175–180. doi: 10.1007/s002239900210. [DOI] [PubMed] [Google Scholar]
- Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- Brooke MH, Kaiser KK. Muscle fiber types: how many and what kind? Archives of Neurology. 1970;23 doi: 10.1001/archneur.1970.00480280083010. [DOI] [PubMed] [Google Scholar]
- Bruusgaard JC, Brack AS, Hughes SM, Gundersen K. Muscle hypertrophy induced by the Ski protein: cyto-architecture and ultrastructure. Acta Physiologica Scandinavica. 2005;185:141–149. doi: 10.1111/j.1365-201X.2005.01462.x. [DOI] [PubMed] [Google Scholar]
- Bruusgaard JC, Gundersen K. In vivo time-lapse microscopy reveals no loss of murine myonuclei during weeks of muscle atrophy. Journal of Clinical Investigation. 2008;118:1450–1457. doi: 10.1172/JCI34022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard JC, Johansen IB, Egner IM, Rana ZA, Gundersen K. Myonuclei acquired by overload exercise precede hypertrophy and are not lost upon detraining. Proceedings of the National Academy of Sciences USA. 2010;107:15111–15116. doi: 10.1073/pnas.0913935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. Journal of Anatomy. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffelli M, Busetto G, Cangiano L, Cangiano A. Perinatal switch from synchronous to asynchronous activity of motoneurons: link with synapse elimination. Proceedings of the National Academy of Sciences USA. 2002;99:13200–13205. doi: 10.1073/pnas.202471199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffelli M, Pasino E, Cangiano A. Paralysis of rat skeletal muscle equally affects contractile properties as does permanent denervation. Journal of Muscle Research and Cell Motility. 1997;18:683–695. doi: 10.1023/a:1018687923929. [DOI] [PubMed] [Google Scholar]
- Buller AJ, Eccles JC, Eccles RM. Interactions between motoneurones and muscles in respect of the characteristic speeds of their responses. Journal of Physiology. 1960;150:417–439. doi: 10.1113/jphysiol.1960.sp006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A, Apone L, Morasso MI, Beers R, Brenner HR, Eftimie R. The myoD family of myogenic factors is regulated by electrical activity: isolation and characterization of a mouse myf-5 cDNA. Nucleic Acids Research. 1992;20:539–544. doi: 10.1093/nar/20.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A, Cheng J, Venepally P, Weis J, Calvo S. Activity-dependent regulation of muscle genes: repressive and stimulatory effects of innervation. Acta Physiologica Scandinavica. 1998;163:S17–S26. doi: 10.1046/j.1365-201X.1998.1630s3S17.x. [DOI] [PubMed] [Google Scholar]
- Buonanno A, Fields RD. Gene regulation by patterned electrical activity during neural and skeletal muscle development. Current Opinion in Neurobiology. 1999;9:110–120. doi: 10.1016/s0959-4388(99)80014-2. [DOI] [PubMed] [Google Scholar]
- Burke RE. Motor unit types of cat triceps surae muscle. Journal of Physiology. 1967;193:141–160. doi: 10.1113/jphysiol.1967.sp008348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RE, Levine DN, Salcman M, Tsairis P. Motor units in cat soleus muscle: physiological, histochemical and morphological characteristics. Journal of Physiology. 1974;238:503–514. doi: 10.1113/jphysiol.1974.sp010540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke RE, Levine DN, Zajac FE., 3rd Mammalian motor units: physiological-histochemical correlation in three types in cat gastrocnemius. Science. 1971;174:709–712. doi: 10.1126/science.174.4010.709. [DOI] [PubMed] [Google Scholar]
- Butler J, Cosmos E, Brierley J. Differetiation of muscle fiber types in aneurogenic brachial muscle chick embryo. Journal of Experimental Zoology. 1982;224:65–80. doi: 10.1002/jez.1402240108. [DOI] [PubMed] [Google Scholar]
- Butler-Browne GS, Bugaisky LB, Cuenoud S, Schwartz K, Whalen RG. Denervation of newborn rat muscle does not block the appearance of adult fast myosin heavy chain. Nature. 1982;299:830–833. doi: 10.1038/299830a0. [DOI] [PubMed] [Google Scholar]
- Caiozzo VJ, Baker MJ, Baldwin KM. Novel transitions in MHC isoforms: separate and combined effects of thyroid hormone and mechanical unloading. Journal of Applied Physiology. 1998;85:2237–2248. doi: 10.1152/jappl.1998.85.6.2237. [DOI] [PubMed] [Google Scholar]
- Calabria E, Ciciliot S, Moretti I, Garcia M, Picard A, Dyar KA, Pallafacchina G, Tothova J, Schiaffino S, Murgia M. NFAT isoforms control activity-dependent muscle fiber type specification. Proceedings of the National Academy of Sciences USA. 2009;106:13335–13340. doi: 10.1073/pnas.0812911106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S, Stauffer J, Nakayama M, Buonanno A. Transcriptional control of muscle plasticity: differential regulation of troponin I genes by electrical activity. Developmental Genetics. 1996;19:169–181. doi: 10.1002/(SICI)1520-6408(1996)19:2<169::AID-DVG9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Calvo S, Venepally P, Cheng J, Buonanno A. Fiber-type-specific transcription of the troponin I slow gene is regulated by multiple elements. Molecular and Cellular Biology. 1999;19:515–525. doi: 10.1128/mcb.19.1.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S, Vullhorst D, Venepally P, Cheng J, Karavanova I, Buonanno A. Molecular dissection of DNA sequences and factors involved in slow muscle-specific transcription. Molecular and Cellular Biology. 2001;21:8490–84503. doi: 10.1128/MCB.21.24.8490-8503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Kumar RM, Penn BH, Berkes CA, Kooperberg C, Boyer LA, Young RA, Tapscott SJ. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO Journal. 2006;25:502–511. doi: 10.1038/sj.emboj.7600958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsen H, Gundersen K. Helix-loop-helix transcription factors in electrically active and inactive skeletal muscles. Muscle & Nerve. 2000;23:1374–1380. doi: 10.1002/1097-4598(200009)23:9<1374::aid-mus8>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Caroni P, Rotzler S, Britt JC, Brenner HR. Ca++ influx and protein phosphorylation mediate the metabolic stabilization of synaptic acetylcholine receptors in muscle. Journal of Neuroscience. 1993;13:1315–1325. doi: 10.1523/JNEUROSCI.13-03-01315.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JA, Booth FW. Serum response factor mRNA induction in the hypertrophying chicken patagialis muscle. Journal of Applied Physiology. 1999;86:377–82. doi: 10.1152/jappl.1999.86.1.377. [DOI] [PubMed] [Google Scholar]
- Carson JA, Schwartz RJ, Booth FW. SRF and TEF-1 control of chicken skeletal alpha-actin gene during slow-muscle hypertrophy. American Journal of Physiology. 1996;270:C1624–C1633. doi: 10.1152/ajpcell.1996.270.6.C1624. [DOI] [PubMed] [Google Scholar]
- Centner T, Yano J, Kimura E, McElhinny AS, Pelin K, Witt CC, Bang ML, Trombitas K, Granzier H, Gregorio CC, Sorimachi H, Labeit S. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. Journal of Molecular Biology. 2001;306:717–726. doi: 10.1006/jmbi.2001.4448. [DOI] [PubMed] [Google Scholar]
- Chang JD, Xu Y, Raychowdhury MK, Ware JA. Molecular cloning and expression of a cDNA encoding a novel isoenzyme of protein kinase C (nPKC). A new member of the nPKC family expressed in skeletal muscle, megakaryoblastic cells, and platelets. Journal of Biological Chemistry. 1993;268:14208–14214. [PubMed] [Google Scholar]
- Chang JH, Lin KH, Shih CH, Chang YJ, Chi HC, Chen SL. Myogenic basic helix-loop-helix proteins regulate the expression of peroxisomal proliferator activated receptor-gamma coactivator-1alpha. Endocrinology. 2006;147:3093–3106. doi: 10.1210/en.2005-1317. [DOI] [PubMed] [Google Scholar]
- Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Chen HC, Bandyopadhyay G, Sajan MP, Kanoh Y, Standaert M, Farese RV, Jr, Farese RV. Activation of the ERK pathway and atypical protein kinase C isoforms in exercise- and aminoimidazole-4-carboxamide-1-beta-D-riboside (AICAR)-stimulated glucose transport. Journal of Biological Chemistry. 2002;277:23554–23562. doi: 10.1074/jbc.M201152200. [DOI] [PubMed] [Google Scholar]
- Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes & Development. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland PJ, Appleby GJ, Rattigan S, Clark MG. Exercise-induced translocation of protein kinase C and production of diacylglycerol and phosphatidic acid in rat skeletal muscle in vivo. Relationship to changes in glucose transport. Journal of Biological Chemistry. 1989;264:17704–17711. [PubMed] [Google Scholar]
- Clemmons DR. Role of insulin-like growth factor binding proteins in controlling IGF actions. Molecular and Cellular Endocrinology. 1998;140:19–24. doi: 10.1016/s0303-7207(98)00024-0. [DOI] [PubMed] [Google Scholar]
- Clemmons DR. Modifying IGF1 activity: an approach to treat endocrine disorders, atherosclerosis and cancer. Nature Reviews Drug Discovery. 2007;6:821–833. doi: 10.1038/nrd2359. [DOI] [PubMed] [Google Scholar]
- Close RI. Dynamic properties of mammalian skeletal muscles of the rat after nerve corss-union. Journal of Physiology. 1969;204:331–346. doi: 10.1113/jphysiol.1969.sp008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close RI. Dynamic properties of mammalian skeletal muscles. Physiological Reviews. 1972;52:129–197. doi: 10.1152/physrev.1972.52.1.129. [DOI] [PubMed] [Google Scholar]
- Coffey VG, Shield A, Canny BJ, Carey KA, Cameron-Smith D, Hawley JA. Interaction of contractile activity and training history on mRNA abundance in skeletal muscle from trained athletes. American Journal of Physiology. 2006;290:E849–E855. doi: 10.1152/ajpendo.00299.2005. [DOI] [PubMed] [Google Scholar]
- Cohen P, Klee CB. Calmodulin. Amsterdam: Elsevier; 1988. [Google Scholar]
- Coleman ME, DeMayo F, Yin KC, Lee HM, Geske R, Montgomery C, Schwartz RJ. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. Journal of Biological Chemistry. 1995;270:12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]
- Condon K, Silberstein L, Blau HM, Thompson WJ. Differentiation of fiber types in aneural musculature of the prenatal rat hindlimb. Developmental Biology. 1990;138:275–295. doi: 10.1016/0012-1606(90)90197-q. [DOI] [PubMed] [Google Scholar]
- Costa A, Dalloul H, Hegyesi H, Apor P, Csende Z, Racz L, Vaczi M, Tihanyi J. Impact of repeated bouts of eccentric exercise on myogenic gene expression. European Journal of Applied Physiology. 2007;101:427–436. doi: 10.1007/s00421-007-0510-z. [DOI] [PubMed] [Google Scholar]
- Cotlar AM, Thrasher JP, Harris AS. Effect of Muscle Length on Denervation Atrophy. Journal of Bone and Joint Surgery. 1963;45:1234–1240. [PubMed] [Google Scholar]
- Crabtree GR. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- Crabtree GR, Olson EN. NFAT signaling: choreographing the social lives of cells. Cell. 2002;109(Suppl):S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- Darr KC, Schultz E. Hindlimb suspension suppresses muscle growth and satellite cell proliferation. Journal of Applied Physiology. 1989;67:1827–1834. doi: 10.1152/jappl.1989.67.5.1827. [DOI] [PubMed] [Google Scholar]
- Davie JK, Cho JH, Meadows E, Flynn JM, Knapp JR, Klein WH. Target gene selectivity of the myogenic basic helix-loop-helix transcription factor myogenin in embryonic muscle. Developmental Biology. 2007;311:650–664. doi: 10.1016/j.ydbio.2007.08.014. [DOI] [PubMed] [Google Scholar]
- De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- Delalande JM, Rescan PY. Differential expression of two nonallelic MyoD genes in developing and adult myotomal musculature of the trout (Oncorhynchus mykiss) Development Genes and Evolution. 1999;209:432–437. doi: 10.1007/s004270050274. [DOI] [PubMed] [Google Scholar]
- Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Molecular and Cellular Biology. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delp MD, Pette D. Morphological changes during fiber type transitions in low-frequency-stimulated rat fast-twitch muscle. Cell and Tissue Research. 1994;277:363–371. doi: 10.1007/BF00327784. [DOI] [PubMed] [Google Scholar]
- Desjardins PR, Burkman JM, Shrager JB, Allmond LA, Stedman HH. Evolutionary implications of three novel members of the human sarcomeric myosin heavy chain gene family. Molecular Biology and Evolution. 2002;19:375–393. doi: 10.1093/oxfordjournals.molbev.a004093. [DOI] [PubMed] [Google Scholar]
- DeVol DL, Rotwein P, Sadow JL, Novakofski J, Bechtel PJ. Activation of insulin-like growth factor gene expression during work-induced skeletal muscle growth. American Journal of Physiology. 1990;259:E89–E95. doi: 10.1152/ajpendo.1990.259.1.E89. [DOI] [PubMed] [Google Scholar]
- DiMario JX. Protein kinase C signaling controls skeletal muscle fiber types. Experimental Cell Research. 2001;263:23–32. doi: 10.1006/excr.2000.5094. [DOI] [PubMed] [Google Scholar]
- DiMario JX, Fernyak SE, Stockdale FE. Myoblasts transferred to the limbs of embryos are committed to specific fibre fates. Nature. 1993;362:165–167. doi: 10.1038/362165a0. [DOI] [PubMed] [Google Scholar]
- DiMario JX, Funk PE. Protein kinase C activity regulates slow myosin heavy chain 2 gene expression in slow lineage skeletal muscle fibers. Developmental Dynamics. 1999;216:177–189. doi: 10.1002/(SICI)1097-0177(199910)216:2<177::AID-DVDY8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- DiMario JX, Stockdale FE. Both myoblast lineage and innervation determine fiber type and are required for expression of the slow myosin heavy chain 2 gene. Developmental Biology. 1997;188:167–180. doi: 10.1006/dbio.1997.8619. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Donnelly R, Reed MJ, Azhar S, Reaven GM. Expression of the major isoenzyme of protein kinase-C in skeletal muscle, nPKC theta, varies with muscle type and in response to fructose-induced insulin resistance. Endocrinology. 1994;135:2369–2374. doi: 10.1210/endo.135.6.7988419. [DOI] [PubMed] [Google Scholar]
- Donselaar Y, Eerbeek O, Kernell D, Verhey BA. Fibre sizes and histochemical staining characteristics in normal and chronically stimulated fast muscle of cat. Journal of Physiology. 1987;382:237–254. doi: 10.1113/jphysiol.1987.sp016365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- Drummond MJ, Fujita S, Abe T, Dreyer HC, Volpi E, Rasmussen BB. Human muscle gene expression following resistance exercise and blood flow restriction. Medicine & Science in Sports & Exercise. 2008;40:691–698. doi: 10.1249/MSS.0b013e318160ff84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclert A, Piette J, Changeux JP. Influence of innervation on myogenic factors and acetylcholine receptor a-subunit mRNAs. NeuroReport. 1991;2:25–28. doi: 10.1097/00001756-199101000-00006. [DOI] [PubMed] [Google Scholar]
- Dum RP, Burke RE, O'Donovan MJ, Toop J, Hodgson JA. Motor-unit organization in flexor digitorum longus muscle of the cat. Journal of Neurophysiology. 1982;47:1108–1125. doi: 10.1152/jn.1982.47.6.1108. [DOI] [PubMed] [Google Scholar]
- Dunn SE, Burns JL, Michel RN. Calcineurin is required for skeletal muscle hypertrophy. Journal of Biological Chemistry. 1999;274:21908–21912. doi: 10.1074/jbc.274.31.21908. [DOI] [PubMed] [Google Scholar]
- Dupont-Versteegden EE, Houle JD, Gurley CM, Peterson CA. Early changes in muscle fiber size and gene expression in response to spinal cord transection and exercise. American Journal of Physiology. 1998;275:C1124–C1133. doi: 10.1152/ajpcell.1998.275.4.C1124. [DOI] [PubMed] [Google Scholar]
- Dupont-Versteegden EE, Murphy RJ, Houle JD, Gurley CM, Peterson CA. Mechanisms leading to restoration of muscle size with exercise and transplantation after spinal cord injury. American Journal of Physiology. 2000;279:C1677–C1684. doi: 10.1152/ajpcell.2000.279.6.C1677. [DOI] [PubMed] [Google Scholar]
- Dupont-Versteegden EE, Murphy RJL, Houlè JD, Gurley CM, Peterson CA. Activated satellite cells fail to restore myonuclear number in spinal cord transected and exercised rat. American Journal of Physiology. 1999;277:C589–C597. doi: 10.1152/ajpcell.1999.277.3.C589. [DOI] [PubMed] [Google Scholar]
- Dupont-Versteegden EE, Strotman BA, Gurley CM, Gaddy D, Knox M, Fluckey JD, Peterson CA. Nuclear translocation of EndoG at the initiation of disuse muscle atrophy and apoptosis is specific to myonuclei. American Journal of Physiology. 2006;291:R1730–R1740. doi: 10.1152/ajpregu.00176.2006. [DOI] [PubMed] [Google Scholar]
- Durieux AC, Amirouche A, Banzet S, Koulmann N, Bonnefoy R, Pasdeloup M, Mouret C, Bigard X, Peinnequin A, Freyssenet D. Ectopic expression of myostatin induces atrophy of adult skeletal muscle by decreasing muscle gene expression. Endocrinology. 2007;148:3140–3147. doi: 10.1210/en.2006-1500. [DOI] [PubMed] [Google Scholar]
- Edman K, Reggiani C, Kronnie G. Differences in maximum velocity of shortening along single muscle fibres of the frog. Journal of Physiology. 1985;365:147–163. doi: 10.1113/jphysiol.1985.sp015764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson DG, Cheng TC, Cserjesi P, Chakraborty T, Olson EN. Analysis of the myogenin promoter reveals an indirect pathway for positive autoregulation mediated by the muscle-specific enhancer factor MEF-2. Molecular and Cellular Biology. 1992;12:3665–3677. doi: 10.1128/mcb.12.9.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eftimie R, Brenner HR, Buonanno A. Myogenin and myoD join a family of skeletal muscle genes regulated by electrical activity. Proceedings of the National Academy of Sciences USA. 1991;88:1349–1353. doi: 10.1073/pnas.88.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eken T, Gundersen K. Electrical stimulation resembling normal motor-unit activity: effects on denervated fast and slow rat muscles. Journal of Physiology. 1988;402:651–669. doi: 10.1113/jphysiol.1988.sp017227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmark M, Gronevik E, Schjerling P, Gundersen K. Myogenin induces higher oxidative capacity in pre-existing mouse muscle fibres after somatic DNA transfer. Journal of Physiology. 2003;548:259–269. doi: 10.1113/jphysiol.2002.036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekmark M, Rana ZA, Stewart G, Hardie DG, Gundersen K. De-phosphorylation of MyoD is linking nerve-evoked activity to fast myosin heavy chain expression in rodent adult skeletal muscle. Journal of Physiology. 2007;584:637–650. doi: 10.1113/jphysiol.2007.141457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert JC, Berglund EB, Rosenthal N. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. Journal of Cell Biology. 1996;135:431–440. doi: 10.1083/jcb.135.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennion S, Sant'ana Pereira J, Sargeant AJ, Young A, Goldspink G. Characterization of human skeletal muscle fibres according to the myosin heavy chains they express. Journal of Muscle Research and Cell Motility. 1995;16:35–43. doi: 10.1007/BF00125308. [DOI] [PubMed] [Google Scholar]
- Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142:4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- Esser K, Gunning P, Hardeman E. Nerve-dependent and -independent patterns of mRNA expression in regenerating skeletal muscle. Developmental Biology. 1993;159:173–183. doi: 10.1006/dbio.1993.1231. [DOI] [PubMed] [Google Scholar]
- Faucher M, Guillot C, Marqueste T, Kipson N, Mayet-Sornay MH, Desplanches D, Jammes Y, Badier M. Matched adaptations of electrophysiological, physiological, and histological properties of skeletal muscles in response to chronic hypoxia. Pflügers Archiv. 2005;450:45–52. doi: 10.1007/s00424-004-1370-6. [DOI] [PubMed] [Google Scholar]
- Favier FB, Benoit H, Freyssenet D. Cellular and molecular events controlling skeletal muscle mass in response to altered use. Pflügers Archiv. 2008;456:587–600. doi: 10.1007/s00424-007-0423-z. [DOI] [PubMed] [Google Scholar]
- Ferguson AB, Jr, Vaughan L, Ward L. A study of disuse atrophy of skeletal muscle in the rabbit. Journal of Bone and Joint Surgery. 1957;39-A:583–596. [PubMed] [Google Scholar]
- Finkbeiner S, Greenberg ME. Ca(2+)-dependent routes to Ras: mechanisms for neuronal survival, differentiation, and plasticity? Neuron. 1996;16:233–236. doi: 10.1016/s0896-6273(00)80040-9. [DOI] [PubMed] [Google Scholar]
- Fitts RH, Widrick JJ. Muscle mechanics: adaptions with exercise-training. In: Holloszy JO, editor. Exercise and Sport Sciences Reviews. Vol. 24. Williams & Wilkins; 1996. pp. 427–473. [PubMed] [Google Scholar]
- Florini JR, Ewton DZ, Coolican SA. Growth hormone and the insulin-like growth factor system in myogenesis. Endocrine Reviews. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- Fluck M, Carson JA, Schwartz RJ, Booth FW. SRF protein is upregulated during stretch-induced hypertrophy of rooster ALD muscle. Journal of Applied Physiology. 1999;86:1793–1799. doi: 10.1152/jappl.1999.86.6.1793. [DOI] [PubMed] [Google Scholar]
- Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Molecular & Cellular Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- Fournier M, Roy RR, Perham H, Simard CP, Edgerton VR. Is limb immobilization a model of muscle disuse? Experimental Neurology. 1983;80:147–156. doi: 10.1016/0014-4886(83)90011-0. [DOI] [PubMed] [Google Scholar]
- Friday BB, Mitchell PO, Kegley KM, Pavlath GK. Calcineurin initiates skeletal muscle differentiation by activating MEF2 and MyoD. Differentiation. 2003;71:217–227. doi: 10.1046/j.1432-0436.2003.710303.x. [DOI] [PubMed] [Google Scholar]
- Froboese C. Histologische befunde zur theorie der muskelatrophie. Mitteilungen aus den Grenzgebieten der Medizin und Chirurgie. 1922;35:683–690. [Google Scholar]
- Fuchtbauer EM, Westphal H. MyoD and myogenin are coexpressed in regenerating skeletal muscle of the mouse. Developmental Dynamics. 1992;193:34–39. doi: 10.1002/aja.1001930106. [DOI] [PubMed] [Google Scholar]
- Gallego R, Kuno M, Nunez R, Snider WD. Dependence of motoneurone properties on the length of immobilized muscle. Journal of Physiology. 1979;291:179–189. doi: 10.1113/jphysiol.1979.sp012806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs CL, Gibson WR. Energy production of rat soleus muscle. American Journal of Physiology. 1972;223:864–870. doi: 10.1152/ajplegacy.1972.223.4.864. [DOI] [PubMed] [Google Scholar]
- Gilson H, Schakman O, Combaret L, Lause P, Grobet L, Attaix D, Ketelslegers JM, Thissen JP. Myostatin gene deletion prevents glucocorticoid-induced muscle atrophy. Endocrinology. 2007;148:452–460. doi: 10.1210/en.2006-0539. [DOI] [PubMed] [Google Scholar]
- Giordani J, Bajard L, Demignon J, Daubas P, Buckingham M, Maire P. Six proteins regulate the activation of Myf5 expression in embryonic mouse limbs. Proceedings of the National Academy of Sciences USA. 2007;104:11310–11315. doi: 10.1073/pnas.0611299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nature Cell Biology. 2003;5:87–90. doi: 10.1038/ncb0203-87. [DOI] [PubMed] [Google Scholar]
- Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. International Journal of Biochemistry and Cell Biology. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Goldspink DF. The influence of immobilization and stretch on protein turnover of rat skeletal muscle. Journal of Physiology. 1977;264:267–282. doi: 10.1113/jphysiol.1977.sp011667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink G. Changes in muscle mass and phenotype and the expression of autocrine and systemic growth factors by muscle in response to stretch and overload. Journal of Anatomy. 1999;194:323–334. doi: 10.1046/j.1469-7580.1999.19430323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldspink G, Scutt A, Loughna PT, Wells DJ, Jaenicke T, Gerlach GF. Gene expression in skeletal muscle in response to stretch and force generation. American Journal of Physiology. 1992;262:R356–R363. doi: 10.1152/ajpregu.1992.262.3.R356. [DOI] [PubMed] [Google Scholar]
- Goldspink G, Scutt A, Martindale J, Jaenicke T, Turay L, Gerlach GF. Stretch and force generation induce rapid hypertrophy and myosin isoform gene switching in adult skeletal muscle. Biochemical Society Transactions. 1991;19:368–373. doi: 10.1042/bst0190368. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cadavid NF, Taylor WE, Yarasheski K, Sinha-Hikim I, Ma K, Ezzat S, Shen R, Lalani R, Asa S, Mamita M, Nair G, Arver S, Bhasin S. Organization of the human myostatin gene and expression in healthy men and HIV-infected men with muscle wasting. Proceedings of the National Academy of Sciences USA. 1998;95:14938–43. doi: 10.1073/pnas.95.25.14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodyear LJ, Chang PY, Sherwood DJ, Dufresne SD, Moller DE. Effects of exercise and insulin on mitogen-activated protein kinase signaling pathways in rat skeletal muscle. American Journal of Physiology. 1996;271:E403–E408. doi: 10.1152/ajpendo.1996.271.2.E403. [DOI] [PubMed] [Google Scholar]
- Gorza L, Gundersen K, Lømo T, Schiaffino S, Westgaard RH. Slow-to-fast transformation of denervated soleus muscles by chronic high-frequency stimulation in the rat. Journal of Physiology. 1988;402:627–649. doi: 10.1113/jphysiol.1988.sp017226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Terada S, Kato M, Katoh M, Yokozeki T, Tabata I, Shimokawa T. cDNA Cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochemical and Biophysical Research Communications. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- Grasberger H, Ye H, Mashima H, Bell GI. Dual promoter structure of ZFP106: regulation by myogenin and nuclear respiratory factor-1. Gene. 2005;344:143–159. doi: 10.1016/j.gene.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Gregory P, Gagnon J, Essig DA, Reid SK, Prior G, Zak R. Differential regulation of actin and myosin isoenzyme synthesis in functionally overloaded skeletal muscle. Biochemcal Journal. 1990;265:525–532. doi: 10.1042/bj2650525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory P, Low RB, Stirewalt WS. Changes in skeletal-muscle myosin isoenzymes with hypertrophy and exercise. Biochemcal Journal. 1986;238:55–63. doi: 10.1042/bj2380055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone R, Demignon J, Houbron C, Souil E, Niro C, Seller MJ, Hamard G, Maire P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Development. 2005;132:2235–2249. doi: 10.1242/dev.01773. [DOI] [PubMed] [Google Scholar]
- Grifone R, Laclef C, Spitz F, Lopez S, Demignon J, Guidotti JE, Kawakami K, Xu PX, Kelly R, Petrof BJ, Daegelen D, Concordet JP, Maire P. Six1 and Eya1 expression can reprogram adult muscle from the slow-twitch phenotype into the fast-twitch phenotype. Molecular and Cellular Biology. 2004;24:6253–6267. doi: 10.1128/MCB.24.14.6253-6267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K. Early effects of denervation on isometric and isotonic contractile properties of rat skeletal muscles. Acta Physiologica Scandinavica. 1985;124:549–555. doi: 10.1111/j.1748-1716.1985.tb00047.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K. Determination of muscle contractile properties: The importance of the nerve. Acta Physiologica Scandinavica. 1998;162:333–341. doi: 10.1046/j.1365-201X.1998.0336e.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K, Bruusgaard JC. Nuclear domains during muscle atrophy: nuclei lost or paradigm lost? Journal of Physiology. 2008;586:2675–2681. doi: 10.1113/jphysiol.2008.154369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Eken T. The importance of frequency and amount of electrical stimulation for contractile properties of denervated rat muscles. Acta Physiologica Scandinavica. 1992;145:49–57. doi: 10.1111/j.1748-1716.1992.tb09335.x. [DOI] [PubMed] [Google Scholar]
- Gundersen K, Leberer E, Lomo T, Pette D, Staron RS. Fibre types, calcium-sequestering proteins and metabolic enzymes in denervated and chronically stimulated muscles of the rat. Journal of Physiology. 1988;398:177–189. doi: 10.1113/jphysiol.1988.sp017037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Merlie JP. Id-1 as a possible transcriptional mediator of muscle disuse atrophy. Proceedings of the National Academy of Sciences USA. 1994;91:3647–3651. doi: 10.1073/pnas.91.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Rabben I, Klocke BJ, Merlie JP. Overexpression of myogenin in muscles of transgenic mice: interaction with Id-1, negative crossregulation of myogenic factors, and induction of extrasynaptic acetylcholine receptor expression. Molecular and Cellular Biology. 1995;15:7127–7134. doi: 10.1128/mcb.15.12.7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen K, Sanes JR, Merlie JP. Neural regulation of muscle acetylcholine receptor epsilon- and alpha-subunit gene promoters in transgenic mice. Journal of Cell Biology. 1993;123:1535–1544. doi: 10.1083/jcb.123.6.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann E, Carlson BM. Contractile and histochemical properties of regenerating cross-transplanted fast and slow muscles in the rat. Pflügers Archiv. 1975;353:227–239. doi: 10.1007/BF00584286. [DOI] [PubMed] [Google Scholar]
- Guttridge DC, Mayo MW, Madrid LV, Wang CY, Baldwin AS., Jr NF-kappaB-induced loss of MyoD messenger RNA: possible role in muscle decay and cachexia. Science. 2000;289:2363–2366. doi: 10.1126/science.289.5488.2363. [DOI] [PubMed] [Google Scholar]
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature Reviews Genetics. 2008;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad F, Adams GR. Selected contribution: acute cellular and molecular responses to resistance exercise. Journal of Applied Physiology. 2002;93:394–403. doi: 10.1152/japplphysiol.01153.2001. [DOI] [PubMed] [Google Scholar]
- Hagiwara N, Ma B, Ly A. Slow and fast fiber isoform gene expression is systematically altered in skeletal muscle of the Sox6 mutant, p100H. Developmental Dynamics. 2005;234:301–311. doi: 10.1002/dvdy.20535. [DOI] [PubMed] [Google Scholar]
- Hagiwara N, Yeh M, Liu A. Sox6 is required for normal fiber type differentiation of fetal skeletal muscle in mice. Developmental Dynamics. 2007;236:2062–2076. doi: 10.1002/dvdy.21223. [DOI] [PubMed] [Google Scholar]
- Hallauer PL, Hastings KE. TnIfast IRE enhancer: multistep developmental regulation during skeletal muscle fiber type differentiation. Developmental Dynamics. 2002;224:422–431. doi: 10.1002/dvdy.10122. [DOI] [PubMed] [Google Scholar]
- Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. Journal of Physiology. 2003;547:247–254. doi: 10.1113/jphysiol.2002.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. Journal of Biological Chemistry. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- Harber MP, Crane JD, Dickinson JM, Jemiolo B, Raue U, Trappe TA, Trappe SW. Protein Synthesis and the Expression of Growth Related Genes are Altered by Running in Human Vastus Lateralis and Soleus Muscles. American Journal of Physiology. 2008;296:R708–R714. doi: 10.1152/ajpregu.90906.2008. [DOI] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Heinemeier KM, Olesen JL, Schjerling P, Haddad F, Langberg H, Baldwin KM, Kjaer M. Short-term strength training and the expression of myostatin and IGF-I isoforms in rat muscle and tendon: differential effects of specific contraction types. Journal of Applied Physiology. 2007;102:573–581. doi: 10.1152/japplphysiol.00866.2006. [DOI] [PubMed] [Google Scholar]
- Hennig R, Lømo T. Firing patterns of motor units in normal rats. Nature. 1985;314:164–166. doi: 10.1038/314164a0. [DOI] [PubMed] [Google Scholar]
- Hennig R, Lømo T. Effects of chronic stimulation on the size and speed of long-term denervated and innervated rat fast and slow skeletal muscles. Acta Physiologica Scandinavica. 1987;130:115–131. doi: 10.1111/j.1748-1716.1987.tb08118.x. [DOI] [PubMed] [Google Scholar]
- Henriksson J, Hickner J. Training-induced adaptions in skeletal muscle. In: Harries M, editor. Oxford textbook of sports medicine. Oxford: Oxford University Press; 1994. [Google Scholar]
- Hentzen ER, Lahey M, Peters D, Mathew L, Barash IA, Friden J, Lieber RL. Stress-dependent and -independent expression of the myogenic regulatory factors and the MARP genes after eccentric contractions in rats. Journal of Physiology. 2006;570:157–167. doi: 10.1113/jphysiol.2005.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert RD, Balnave RJ. The effect of position of immobilisation on resting length, resting stiffness, and weight of the soleus muscle of the rabbit. Journal of Orthopaedic Research. 1993;11:358–366. doi: 10.1002/jor.1100110307. [DOI] [PubMed] [Google Scholar]
- Herbison GJ, Jaweed MM, Ditunno JF. Synergistic tenotomy: effect on chronically denervated slow and fast muscles of rat. Archives of Physical Medicine and Rehabilitation. 1975;56:483–487. [PubMed] [Google Scholar]
- Higginson J, Wackerhage H, Woods N, Schjerling P, Ratkevicius A, Grunnet N, Quistorff B. Blockades of mitogen-activated protein kinase and calcineurin both change fibre-type markers in skeletal muscle culture. Pflügers Archiv. 2002;445:437–443. doi: 10.1007/s00424-002-0939-1. [DOI] [PubMed] [Google Scholar]
- Hikida RS. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1104–1105. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Hirai T, Chida K. Protein kinase Czeta (PKCzeta): activation mechanisms and cellular functions. Journal of Biochemistry. 2003;133:1–7. doi: 10.1093/jb/mvg017. [DOI] [PubMed] [Google Scholar]
- Hnik P, Vejsada R, Goldspink DF, Kasicki S, Krekule I. Quantitative evaluation of electromyogram activity in rat extensor and flexor muscles immobilized at different lengths. Experimental Neurology. 1985;88:515–528. doi: 10.1016/0014-4886(85)90067-6. [DOI] [PubMed] [Google Scholar]
- Hofsten J, Elworthy S, Gilchrist MJ, Smith JC, Wardle FC, Ingham PW. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Report. 2008;9:683–689. doi: 10.1038/embor.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh JF. ‘Superfast’ or masticatory myosin and the evolution of jaw-closing muscles of vertebrates. Journal of Experimental Biology. 2002;205:2203–2210. doi: 10.1242/jeb.205.15.2203. [DOI] [PubMed] [Google Scholar]
- Hoh JF, Hughes S. Basal lamina and superfast myosin expression in regenerating cat jaw muscle. Muscle & Nerve. 1991;14:398–406. doi: 10.1002/mus.880140503. [DOI] [PubMed] [Google Scholar]
- Holterman CE, Rudnicki MA. Molecular regulation of satellite cell function. Seminars in Cell and Developmental Biology. 2005;16:575–584. doi: 10.1016/j.semcdb.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Hoppeler H, Vogt M, Weibel ER, Fluck M. Response of skeletal muscle mitochondria to hypoxia. Experimental Physiology. 2003;88:109–119. doi: 10.1113/eph8802513. [DOI] [PubMed] [Google Scholar]
- Hsiao FC, Williams A, Davies EL, Rebay I. Eyes absent mediates cross-talk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Developmental Cell. 2001;1:51–61. doi: 10.1016/s1534-5807(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Huang CF, Tong J, Schmidt J. Protein kinase C couples membrane excitation to acetylcholine receptor gene inactivation in chick skeletal muscle. Neuron. 1992;9:671–678. doi: 10.1016/0896-6273(92)90030-h. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Blau HM. Muscle fiber pattern is independent of cell lineage in postnatal rodent development. Cell. 1992;68:659–671. doi: 10.1016/0092-8674(92)90142-y. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Chi MM, Lowry OH, Gundersen K. Myogenin induces a shift of enzyme activity from glycolytic to oxidative metabolism in muscles of transgenic mice. Journal of Cell Biology. 1999;145:633–642. doi: 10.1083/jcb.145.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SM, Koishi K, Rudnicki M, Maggs AM. MyoD protein is differentially accumulated in fast and slow skeletal muscle fibres and required for normal fibre type balance in rodents. Mechanisms of Development. 1997;61:151–163. doi: 10.1016/s0925-4773(96)00631-4. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Taylor JM, Tapscott SJ, Gurley CM, Carter WJ, Peterson CA. Selective accumulation of myoD and myogenin mRNAs in fast and slow adult skeletal muscle is controlled by innervation and hormones. Development. 1993;118:1137–1147. doi: 10.1242/dev.118.4.1137. [DOI] [PubMed] [Google Scholar]
- Hulmi JJ, Ahtiainen JP, Kaasalainen T, Pollanen E, Hakkinen K, Alen M, Selanne H, Kovanen V, Mero AA. Postexercise myostatin and activin IIb mRNA levels: effects of strength training. Medicine & Science in Sports & Exercise. 2007;39:289–297. doi: 10.1249/01.mss.0000241650.15006.6e. [DOI] [PubMed] [Google Scholar]
- Hulmi JJ, Kovanen V, Lisko I, Selanne H, Mero AA. The effects of whey protein on myostatin and cell cycle-related gene expression responses to a single heavy resistance exercise bout in trained older men. European Journal of Applied Physiology. 2008a;102:205–213. doi: 10.1007/s00421-007-0579-4. [DOI] [PubMed] [Google Scholar]
- Hulmi JJ, Kovanen V, Selanne H, Kraemer WJ, Hakkinen K, Mero AA. Acute and long-term effects of resistance exercise with or without protein ingestion on muscle hypertrophy and gene expression. Amino Acids. 2008b;37:297–308. doi: 10.1007/s00726-008-0150-6. [DOI] [PubMed] [Google Scholar]
- Hunter RB, Stevenson E, Koncarevic A, Mitchell-Felton H, Essig DA, Kandarian SC. Activation of an alternative NF-kappaB pathway in skeletal muscle during disuse atrophy. FASEB Journal. 2002;16:529–538. doi: 10.1096/fj.01-0866com. [DOI] [PubMed] [Google Scholar]
- Hyatt JP, Roy RR, Baldwin KM, Edgerton VR. Nerve activity-independent regulation of skeletal muscle atrophy: role of MyoD and myogenin in satellite cells and myonuclei. American Journal of Physiology. 2003;285:C1161–C1173. doi: 10.1152/ajpcell.00128.2003. [DOI] [PubMed] [Google Scholar]
- Hyatt JP, Roy RR, Baldwin KM, Wernig A, Edgerton VR. Activity-unrelated neural control of myogenic factors in a slow muscle. Muscle & Nerve. 2006;33:49–60. doi: 10.1002/mus.20433. [DOI] [PubMed] [Google Scholar]
- Ianuzzo CD, Blank S, Crassweller A, Spalding J, Hamilton N, Dabrowski B, Rooks N. Effect of hindlimb immobilization and recovery on compensatory hypertrophied rat plantaris muscle. Molecular and Cellular Biochemistry. 1989;90:57–68. doi: 10.1007/BF00225221. [DOI] [PubMed] [Google Scholar]
- Ianuzzo CD, Gollnick PD, Armstrong RB. Compensatory adaptations of skeletal muscle fiber types to a long-term functional overload. Life Sciences. 1976;19:1517–1523. doi: 10.1016/0024-3205(76)90096-5. [DOI] [PubMed] [Google Scholar]
- Irrcher I, Adhihetty PJ, Sheehan T, Joseph AM, Hood DA. PPARgamma coactivator-1alpha expression during thyroid hormone- and contractile activity-induced mitochondrial adaptations. American Journal of Physiology. 2003;284:C1669–C1677. doi: 10.1152/ajpcell.00409.2002. [DOI] [PubMed] [Google Scholar]
- Irrcher I, Ljubicic V, Hood DA. Interactions between ROS and AMP kinase activity in the regulation of PGC-1{alpha} transcription in skeletal muscle cells. American Journal of Physiology. 2009;296:C116–C123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- Irrcher I, Ljubicic V, Kirwan AF, Hood DA. AMP-activated protein kinase-regulated activation of the PGC-1alpha promoter in skeletal muscle cells. PLoS ONE. 2008;3:e3614.. doi: 10.1371/journal.pone.0003614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J, Perry RL, Asakura A, Rudnicki MA. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. Journal of Cell Biology. 2005;171:471–482. doi: 10.1083/jcb.200502101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishido M, Kami K, Masuhara M. Localization of MyoD, myogenin and cell cycle regulatory factors in hypertrophying rat skeletal muscles. Acta Physiologica Scandinavica. 2004;180:281–289. doi: 10.1046/j.0001-6772.2003.01238.x. [DOI] [PubMed] [Google Scholar]
- Issa LL, Palmer SJ, Guven KL, Santucci N, Hodgson VR, Popovic K, Joya JE, Hardeman EC. MusTRD can regulate postnatal fiber-specific expression. Developmental Biology. 2006;293:104–115. doi: 10.1016/j.ydbio.2006.01.019. [DOI] [PubMed] [Google Scholar]
- Itoh K, Moritani T, Ishida K, Hirofuji C, Taguchi S, Itoh M. Hypoxia-induced fibre type transformation in rat hindlimb muscles. Histochemical and electro-mechanical changes. European Journal of Applied Physiology and Occupational Physiology. 1990;60:331–336. doi: 10.1007/BF00713495. [DOI] [PubMed] [Google Scholar]
- Jacquemin V, Furling D, Bigot A, Butler-Browne GS, Mouly V. IGF-1 induces human myotube hypertrophy by increasing cell recruitment. Experimental Cell Research. 2004;299:148–158. doi: 10.1016/j.yexcr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Jansson E, Esbjornsson M, Holm I, Jacobs I. Increase in the proportion of fast-twitch muscle fibres by sprint training in males. Acta Physiologica Scandinavica. 1990;140:359–363. doi: 10.1111/j.1748-1716.1990.tb09010.x. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annual Review of Biochemistry. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- Jennische E, Hansson HA. Regenerating skeletal muscle cells express insulin-like growth factor I. Acta Physiologica Scandinavica. 1987;130:327–332. doi: 10.1111/j.1748-1716.1987.tb08144.x. [DOI] [PubMed] [Google Scholar]
- Jennische E, Skottner A, Hansson HA. Satellite cells express the trophic factor IGF-I in regenerating skeletal muscle. Acta Physiologica Scandinavica. 1987;129:9–15. doi: 10.1111/j.1748-1716.1987.tb08034.x. [DOI] [PubMed] [Google Scholar]
- Jensky NE, Sims JK, Rice JC, Dreyer HC, Schroeder ET. The influence of eccentric exercise on mRNA expression of skeletal muscle regulators. European Journal of Applied Physiology. 2007;101:473–480. doi: 10.1007/s00421-007-0521-9. [DOI] [PubMed] [Google Scholar]
- Jerkovic R, Argentini C, Serrano-Sanchez A, Cordonnier C, Schiaffino S. Early myosin switching induced by nerve activity in regenerating slow skeletal muscle. Cell Structure and Function. 1997;22:147–153. doi: 10.1247/csf.22.147. [DOI] [PubMed] [Google Scholar]
- Jin H, Wu Z, Tian T, Gu Y. Apoptosis in atrophic skeletal muscle induced by brachial plexus injury in rats. Journal of Trauma. 2001;50:31–35. doi: 10.1097/00005373-200101000-00005. [DOI] [PubMed] [Google Scholar]
- Jordan T, Jiang H, Li H, DiMario JX. Inhibition of ryanodine receptor 1 in fast skeletal muscle fibers induces a fast-to-slow muscle fiber type transition. Journal of Cell Science. 2004;117:6175–6183. doi: 10.1242/jcs.01543. [DOI] [PubMed] [Google Scholar]
- Jung T, Catalgol B, Grune T. The Proteasomal System. Molecular Aspects Medicine. 2009;30:191–296. doi: 10.1016/j.mam.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Kadi F. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1105.. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Kadi F. Cellular and molecular mechanisms responsible for the action of testosterone on human skeletal muscle. A basis for illegal performance enhancement. British Journal of Pharmacology. 2008;154:522–528. doi: 10.1038/bjp.2008.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadi F, Johansson F, Johansson R, Sjostrom M, Henriksson J. Effects of one bout of endurance exercise on the expression of myogenin in human quadriceps muscle. Histochemistry and Cell Biology. 2004;121:329–334. doi: 10.1007/s00418-004-0630-z. [DOI] [PubMed] [Google Scholar]
- Kalhovde JM, Jerkovic R, Sefland I, Cordonnier C, Calabria E, Schiaffino S, Lomo T. “Fast” and “slow” muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. Journal of Physiology. 2005;562:847–857. doi: 10.1113/jphysiol.2004.073684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandarian SC, Jackman RW. Intracellular signaling during skeletal muscle atrophy. Muscle & Nerve. 2006;33:155–165. doi: 10.1002/mus.20442. [DOI] [PubMed] [Google Scholar]
- Kandarian SC, Schulte LM, Esser KA. Age effects on myosin subunit and biochemical alterations with skeletal muscle hypertrophy. Journal of Applied Physiology. 1992;72:1934–1939. doi: 10.1152/jappl.1992.72.5.1934. [DOI] [PubMed] [Google Scholar]
- Kannisto K, Chibalin A, Glinghammar B, Zierath JR, Hamsten A, Ehrenborg E. Differential expression of peroxisomal proliferator activated receptors alpha and delta in skeletal muscle in response to changes in diet and exercise. International Journal of Molecular Medicine. 2006;17:45–52. [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kernell D, Donselaar Y, Eerbeek O. Effects of physiological amounts of high- and low-rate chronic stimulation on fast-twitch muscle of the cat hindlimb. II. Endurance-related properties. Journal of Neurophysiology. 1987;58:614–627. doi: 10.1152/jn.1987.58.3.614. [DOI] [PubMed] [Google Scholar]
- Kim JS, Cross JM, Bamman MM. Impact of resistance loading on myostatin expression and cell cycle regulation in young and older men and women. American Journal of Physiology. 2005;288:E1110–E1119. doi: 10.1152/ajpendo.00464.2004. [DOI] [PubMed] [Google Scholar]
- Kim MS, Fielitz J, McAnally J, Shelton JM, Lemon DD, McKinsey TA, Richardson JA, Bassel-Duby R, Olson EN. Protein kinase D1 stimulates MEF2 activity in skeletal muscle and enhances muscle performance. Molecular and Cellular Biology. 2008;28:3600–3609. doi: 10.1128/MCB.00189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proceedings of the National Academy of Sciences USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M. Cytoplasmic free concentrations of Ca2+ and Mg2+ in skeletal muscle fibers at rest and during contraction. Japanese Journal of Physiology. 1998;48:421–438. doi: 10.2170/jjphysiol.48.421. [DOI] [PubMed] [Google Scholar]
- Kosek DJ, Kim JS, Petrella JK, Cross JM, Bamman MM. Efficacy of 3 days/wk resistance training on myofiber hypertrophy and myogenic mechanisms in young vs. older adults. Journal of Applied Physiology. 2006;101:531–544. doi: 10.1152/japplphysiol.01474.2005. [DOI] [PubMed] [Google Scholar]
- Kraemer WJ, Patton JF, Gordon SE, Harman EA, Deschenes MR, Reynolds K, Newton RU, Triplett NT, Dziados JE. Compatibility of high-intesity strength and endurance training on hormonal and skeletal muscle adaptions. Journal of Applied Physiology. 1995;78:976–989. doi: 10.1152/jappl.1995.78.3.976. [DOI] [PubMed] [Google Scholar]
- Kramer HF, Goodyear LJ. Exercise, MAPK, and NF-kappaB signaling in skeletal muscle. Journal of Applied Physiology. 2007;103:388–395. doi: 10.1152/japplphysiol.00085.2007. [DOI] [PubMed] [Google Scholar]
- Kruger M, Linke WA. Titin-based mechanical signalling in normal and failing myocardium. Journal of Molecular and Cellular Cardiology. 2009;46:490–498. doi: 10.1016/j.yjmcc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Kubis HP, Hanke N, Scheibe RJ, Meissner JD, Gros G. Ca2+ transients activate calcineurin/NFATc1 and initiate fast-to-slow transformation in a primary skeletal muscle culture. American Journal of Physiology. 2003;285:C56–C63. doi: 10.1152/ajpcell.00377.2002. [DOI] [PubMed] [Google Scholar]
- Kubis HP, Scheibe RJ, Meissner JD, Hornung G, Gros G. Fast-to-slow transformation and nuclear import/export kinetics of the transcription factor NFATc1 during electrostimulation of rabbit muscle cells in culture. Journal of Physiology. 2002;541:835–847. doi: 10.1113/jphysiol.2002.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakami K. Studies on changes of rabbit skeletal muscle components induced by immobilization with plaster cast. Nagoya Medical Journal. 1966;12:165–184. [PubMed] [Google Scholar]
- Kvorning T, Andersen M, Brixen K, Schjerling P, Suetta C, Madsen K. Suppression of testosterone does not blunt mRNA expression of myoD, myogenin, IGF, myostatin or androgen receptor post strength training in humans. Journal of Physiology. 2007;578:579–593. doi: 10.1113/jphysiol.2006.122671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani R, Bhasin S, Byhower F, Tarnuzzer R, Grant M, Shen R, Asa S, Ezzat S, Gonzalez-Cadavid NF. Myostatin and insulin-like growth factor-I and -II expression in the muscle of rats exposed to the microgravity environment of the NeuroLab space shuttle flight. Journal of Endocrinology. 2000;167:417–428. doi: 10.1677/joe.0.1670417. [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edstrom L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Lefaucheur JP, Sebille A. Muscle regeneration following injury can be modified in vivo by immune neutralization of basic fibroblast growth factor, transforming growth factor beta 1 or insulin-like growth factor I. Journal of Neuroimmunology. 1995;57:85–91. doi: 10.1016/0165-5728(94)00166-l. [DOI] [PubMed] [Google Scholar]
- Lenka N, Basu A, Mullick J, Avadhani NG. The role of an E box binding basic helix loop helix protein in the cardiac muscle-specific expression of the rat cytochrome oxidase subunit VIII gene. Journal of Biological Chemistry. 1996;271:30281–30289. doi: 10.1074/jbc.271.47.30281. [DOI] [PubMed] [Google Scholar]
- Li L, Heller-Harrison R, Czech M, Olson EN. Cyclic AMP-dependent protein kinase inhibits the activity of myogenic helix-loop-helix proteins. Molecular and Cellular Biology. 1992a;12:4478–4485. doi: 10.1128/mcb.12.10.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou J, James G, Heller-Harrison R, Czech MP, Olson EN. FGF inactivates myogenic helix-loop-helix proteins through phosphorylation of a conserved protein kinase C site in their DNA-binding domains. Cell. 1992b;71:1181–1194. doi: 10.1016/s0092-8674(05)80066-2. [DOI] [PubMed] [Google Scholar]
- Li M, Li C, Parkhouse WS. Differential effects of des IGF-1 on Erks, AKT-1 and P70 S6K activation in mouse skeletal and cardiac muscle. Molecular and Cellular Biochemistry. 2002;236:115–122. doi: 10.1023/a:1016164601887. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta), a novel PGC-1-related transcription coactivator associated with host cell factor. Journal of Biological Chemistry. 2002a;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002b;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lipton BH, Schultz E. Developmental fate of skeletal muscle satellite cells. Science. 1979;205:1292–1294. doi: 10.1126/science.472747. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cseresnyes Z, Randall WR, Schneider MF. Activity-dependent nuclear translocation and intranuclear distribution of NFATc in adult skeletal muscle fibers. Journal of Cell Biology. 2001;155:27–39. doi: 10.1083/jcb.200103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Heinichen M, Wirth K, Schmidtbleicher D, Steinacker JM. Response of growth and myogenic factors in human skeletal muscle to strength training. British Journal of Sports Medicine. 2008;42:989–993. doi: 10.1136/bjsm.2007.045518. [DOI] [PubMed] [Google Scholar]
- Liu Y, Randall WR, Schneider MF. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. Journal of Cell Biology. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lømo T. Neuromuscular Junction (NMJ): Activity-Dependent Muscle Fiber Modulation. In: Squire LR, editor. Encyclopedia of Neuroscience. Vol. 6. Oxford: Academic press; 2009. pp. 559–568. [Google Scholar]
- Lømo T, Westgaard RH. Further studies on the control of ACh sensitivity by muscle activity in the rat. Journal of Physiology. 1975;252:603–626. doi: 10.1113/jphysiol.1975.sp011161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lømo T, Westgaard RH, Dahl HA. Contractile properties of muscle: control by pattern of muscle activity in the rat. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1974;187:99–103. doi: 10.1098/rspb.1974.0064. [DOI] [PubMed] [Google Scholar]
- Long YC, Glund S, Garcia-Roves PM, Zierath JR. Calcineurin regulates skeletal muscle metabolism via coordinated changes in gene expression. Journal of Biological Chemistry. 2007;282:1607–1614. doi: 10.1074/jbc.M609208200. [DOI] [PubMed] [Google Scholar]
- Long YC, Widegren U, Zierath JR. Exercise-induced mitogen-activated protein kinase signalling in skeletal muscle. Proc Nutr Soc. 2004;63:227–232. doi: 10.1079/PNS2004346. [DOI] [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Aramburu J, Rakeman AS, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proceedings of the National Academy of Sciences USA. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughna PT, Izumo S, Goldspink G, Nadal-Ginard B. Disuse and passive stretch cause rapid alterations in expression of developmental and adult contractile protein genes in skeletal muscle. Development. 1990;109:217–223. doi: 10.1242/dev.109.1.217. [DOI] [PubMed] [Google Scholar]
- Loughna PT, Morgan MJ. Passive stretch modulates denervation induced alterations in skeletal muscle myosin heavy chain mRNA levels. Pflügers Archiv. 1999;439:52–55. doi: 10.1007/s004249900130. [DOI] [PubMed] [Google Scholar]
- Lowe DA. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1106.. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Lowell BB, Spiegelman BM. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404:652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- Lunde IG, Ekmark M, Rana ZA, Buonanno A, Gundersen K. PPAR{delta} expression is influenced by muscle activity and induces slow muscle properties in adult rat muscles after somatic gene transfer. Journal of Physiology. 2007;582:1277–1287. doi: 10.1113/jphysiol.2007.133025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquet S, Lopez-Soriano J, Holst D, Gaudel C, Jehl-Pietri C, Fredenrich A, Melki J, Rassoulzadegan M, Grimaldi PA. Peroxisome proliferator-activated receptor delta controls muscle development and oxidative capability. FASEB Journal. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- Mahoney DJ, Parise G, Melov S, Safdar A, Tarnapolsky MA. Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB Journal. 2005;19:1498–1500. doi: 10.1096/fj.04-3149fje. [DOI] [PubMed] [Google Scholar]
- Mantilla CB, Sieck GC. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1104.. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Martineau LC, Gardiner PF. Insight into skeletal muscle mechanotransduction: MAPK activation is quantitatively related to tension. Journal of Applied Physiology. 2001;91:693–702. doi: 10.1152/jappl.2001.91.2.693. [DOI] [PubMed] [Google Scholar]
- Mason SD, Howlett RA, Kim MJ, Olfert IM, Hogan MC, McNulty W, Hickey RP, Wagner PD, Kahn CR, Giordano FJ, Johnson RS. Loss of skeletal muscle HIF-1alpha results in altered exercise endurance. PLoS Biology. 2004;2:e288.. doi: 10.1371/journal.pbio.0020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason SD, Rundqvist H, Papandreou I, Duh R, McNulty WJ, Howlett RA, Olfert IM, Sundberg CJ, Denko NC, Poellinger L, Johnson RS. HIF-1alpha in endurance training: suppression of oxidative metabolism. American Journal of Physiology. 2007;293:R2059–R2069. doi: 10.1152/ajpregu.00335.2007. [DOI] [PubMed] [Google Scholar]
- Mathai AS, Bonen A, Benton CR, Robinson DL, Graham TE. Rapid exercise-induced changes in PGC-1alpha mRNA and protein in human skeletal muscle. Journal of Applied Physiology. 2008;105:1098–1105. doi: 10.1152/japplphysiol.00847.2007. [DOI] [PubMed] [Google Scholar]
- Mathews LS, Hammer RE, Behringer RR, D'Ercole AJ, Bell GI, Brinster RL, Palmiter RD. Growth enhancement of transgenic mice expressing human insulin-like growth factor I. Endocrinology. 1988;123:2827–2833. doi: 10.1210/endo-123-6-2827. [DOI] [PubMed] [Google Scholar]
- Mayans O, van der Ven PF, Wilm M, Mues A, Young P, Furst DO, Wilmanns M, Gautel M. Structural basis for activation of the titin kinase domain during myofibrillogenesis. Nature. 1998;395:863–869. doi: 10.1038/27603. [DOI] [PubMed] [Google Scholar]
- Mayne CN, Mokrusch T, Jarvis JC, Gilroy SJ, Salmons S. Stimulation-unduced expression of slow muscle myoein in a fast muscle of the rat. Evidence of an un. FEBS. 1993;327:297–300. doi: 10.1016/0014-5793(93)81008-n. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA. Counterpoint: Satellite cell addition is not obligatory for skeletal muscle hypertrophy. Journal of Applied Physiology. 2007;103:1100–1102. doi: 10.1152/japplphysiol.00101.2007a. [DOI] [PubMed] [Google Scholar]
- McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lømo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proceedings of the National Academy of Sciences USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough P, Behnke BJ, Padilla DJ, Musch TI, Poole DC. Control of microvascular oxygen pressures during recovery in rat fast-twitch muscle of differing oxidative capacity. Experimental Physiology. 2007;92:731–738. doi: 10.1113/expphysiol.2007.037721. [DOI] [PubMed] [Google Scholar]
- McElhinny AS, Kakinuma K, Sorimachi H, Labeit S, Gregorio CC. Muscle-specific RING finger-1 interacts with titin to regulate sarcomeric M-line and thick filament structure and may have nuclear functions via its interaction with glucocorticoid modulatory element binding protein-1. Journal of Cell Biology. 2002;157:125–136. doi: 10.1083/jcb.200108089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay BR, O'Reilly CE, Phillips SM, Tarnopolsky MA, Parise G. Co-expression of IGF-1 family members with myogenic regulatory factors following acute damaging muscle-lengthening contractions in humans. Journal of Physiology. 2008;586:5549–5560. doi: 10.1113/jphysiol.2008.160176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proceedings of the National Academy of Sciences USA. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirhaeghe A, Crowley V, Lenaghan C, Lelliott C, Green K, Stewart A, Hart K, Schinner S, Sethi JK, Yeo G, Brand MD, Cortright RN, O'Rahilly S, Montague C, Vidal-Puig AJ. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochemcal Journal. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejat A, Ramond F, Bassel-Duby R, Khochbin S, Olson EN, Schaeffer L. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nature Neuroscience. 2005;8:313–321. doi: 10.1038/nn1408. [DOI] [PubMed] [Google Scholar]
- Mendler L, Zador E, Dux L, Wuytack F. mRNA levels of myogenic regulatory factors in rat slow and fast muscles regenerating from notexin-induced necrosis. Neuromuscular Disorders. 1998;8:533–541. doi: 10.1016/s0960-8966(98)00070-4. [DOI] [PubMed] [Google Scholar]
- Merlie JP, Mudd J, Cheng TC, Olson EN. Myogenin and acetylcholine receptor alpha gene promoters mediate transcriptional regulation in response to motor innervation. Journal of Biological Chemistry. 1994;269:2461–2467. [PubMed] [Google Scholar]
- Meyer AW. Theorie der muskelatrophie. (Nach experimentellen untersuchungen) Mitteilungen aus den Grenzgebieten der Medizin und Chirurgie. 1922;35:651–682. [Google Scholar]
- Midrio M. The denervated muscle: facts and hypotheses. A historical review. European Journal of Applied Physiology. 2006;98:1–21. doi: 10.1007/s00421-006-0256-z. [DOI] [PubMed] [Google Scholar]
- Miller JB, Stockdale FE. Developmental origins of skeletal muscle fibers: Clonal analysis of myogenic cell lineages based on expression of fast and slow myosin heavy chains. Proceedings of the National Academy of Sciences USA. 1986;83:3860–3864. doi: 10.1073/pnas.83.11.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Kawanaka K, Kai Y, Tamura M, Goto M, Shiuchi T, Minokoshi Y, Ezaki O. An increase in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to exercise is mediated by beta-adrenergic receptor activation. Endocrinology. 2007;148:3441–3448. doi: 10.1210/en.2006-1646. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Esser KA. Cellular mechanisms regulating protein synthesis and skeletal muscle hypertrophy in animals. Journal of Applied Physiology. 2009;106:1367–1373. doi: 10.1152/japplphysiol.91355.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogami H, Zhang H, Suzuki Y, Urano T, Saito N, Kojima I, Petersen OH. Decoding of short-lived Ca2+ influx signals into long term substrate phosphorylation through activation of two distinct classes of protein kinase C. Journal of Biological Chemistry. 2003;278:9896–9904. doi: 10.1074/jbc.M210653200. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Loughna PT. Work overload induced changes in fast and slow skeletal muscle myosin heavy chain gene expression. FEBS Lett. 1989;255:427–430. doi: 10.1016/0014-5793(89)81138-x. [DOI] [PubMed] [Google Scholar]
- Mortensen OH, Frandsen L, Schjerling P, Nishimura E, Grunnet N. PGC-1alpha and PGC-1beta have both similar and distinct effects on myofiber switching toward an oxidative phenotype. American Journal of Physiology. 2006;291:E807–E816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- Mortensen OH, Plomgaard P, Fischer CP, Hansen AK, Pilegaard H, Pedersen BK. PGC-1beta is downregulated by training in human skeletal muscle: no effect of training twice every second day vs. once daily on expression of the PGC-1 family. Journal of Applied Physiology. 2007;103:1536–1542. doi: 10.1152/japplphysiol.00575.2007. [DOI] [PubMed] [Google Scholar]
- Moss FP, Leblond CP. Nature of dividing nuclei in skeletal muscle of growing rats. Journal of Cell Biology. 1970;44:459–462. doi: 10.1083/jcb.44.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss FP, Leblond CP. Satellite cells as the source of nuclei in muscles in growing rats. Anatomical Records. 1971;170:421–436. doi: 10.1002/ar.1091700405. [DOI] [PubMed] [Google Scholar]
- Mrosek M, Labeit D, Witt S, Heerklotz H, von Castelmur E, Labeit S, Mayans O. Molecular determinants for the recruitment of the ubiquitin-ligase MuRF-1 onto M-line titin. FASEB Journal. 2007;21:1383–1392. doi: 10.1096/fj.06-7644com. [DOI] [PubMed] [Google Scholar]
- Mulkey RM, Endo S, Shenolikar S, Malenka RC. Involvement of a calcineurin/inhibitor-1 phosphatase cascade in hippocampal long-term depression. Nature. 1994;369:486–488. doi: 10.1038/369486a0. [DOI] [PubMed] [Google Scholar]
- Muoio DM, MacLean PS, Lang DB, Li S, Houmard JA, Way JM, Winegar DA, Corton JC, Dohm GL, Kraus WE. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor alpha knock-out mice. Journal of Biological Chemistry. 2002;277:26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- Murgia M, Serrano L, Calabria E, Pallafacchina G, Lømo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nature Cell Biology. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- Musaro A, McCullagh K, Paul A, Houghton L, Dobrowolny G, Molinaro M, Barton ER, Sweeney HL, Rosenthal N. Localized Igf-1 transgene expression sustains hypertrophy and regeneration in senescent skeletal muscle. Nature Genetics. 2001;27:195–200. doi: 10.1038/84839. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Stauffer J, Cheng J, Banerjee-Basu S, Wawrousek E, Buonanno A. Common core sequences are found in skeletal muscle slow- and fast-fiber- type-specific regulatory elements. Molecular and Cellular Biology. 1996;16:2408–2417. doi: 10.1128/mcb.16.5.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazato K, Song H. Increased oxidative properties of gastrocnemius in rats fed on a high-protein diet. Journal of Nutritional Biochemistry. 2008;19:26–32. doi: 10.1016/j.jnutbio.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, Evans RM. AMPK and PPARdelta agonists are exercise mimetics. Cell. 2008;134:405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki A, Gorski J. Effect of plasma free fatty acid concentration on the content and composition of the free fatty acid fraction in rat skeletal muscles. Horm Metab Res. 2004;36:601–606. doi: 10.1055/s-2004-825922. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. Journal of Biological Chemistry. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Wu C, Richardson JA, Overbeek P, Olson EN. Transcriptional activity of MEF2 during mouse embryogenesis monitored with a MEF2-dependent transgene. Development. 1999;126:2045–2052. doi: 10.1242/dev.126.10.2045. [DOI] [PubMed] [Google Scholar]
- Neville CM, Schmidt M, Schmidt J. Response of myogenic determination factors to cessation and resumption of electrical activity in skeletal muscle: a possible role for myogenin in denervation supersensitivity. Cellular and Molecular Neurobiology. 1992;12:511–527. doi: 10.1007/BF00711232. [DOI] [PubMed] [Google Scholar]
- Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chemical Reviews. 2001;101:2353–2364. doi: 10.1021/cr0002801. [DOI] [PubMed] [Google Scholar]
- Nicholas G, Thomas M, Langley B, Somers W, Patel K, Kemp CF, Sharma M, Kambadur R. Titin-cap associates with, and regulates secretion of, Myostatin. Journal of Cell Physiology. 2002;193:120–131. doi: 10.1002/jcp.10158. [DOI] [PubMed] [Google Scholar]
- Nielsen JN, Frosig C, Sajan MP, Miura A, Standaert ML, Graham DA, Wojtaszewski JF, Farese RV, Richter EA. Increased atypical PKC activity in endurance-trained human skeletal muscle. Biochemical and Biophysical Research Communications. 2003;312:1147–1153. doi: 10.1016/j.bbrc.2003.11.041. [DOI] [PubMed] [Google Scholar]
- Noble EG, Dabrowski BL, Ianuzzo CD. Myosin transformation in hypertrophied rat muscle. Pflügers Archiv. 1983;396:260–262. doi: 10.1007/BF00587864. [DOI] [PubMed] [Google Scholar]
- Norrbom J, Sundberg CJ, Ameln H, Kraus WE, Jansson E, Gustafsson T. PGC-1alpha mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. Journal of Applied Physiology. 2004;96:189–194. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- O'Connor RS, Pavlath GK. Point:Counterpoint: Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy. Journal of Applied Physiology. 2007;103:1099–1100. doi: 10.1152/japplphysiol.00101.2007. [DOI] [PubMed] [Google Scholar]
- O'Connor RS, Pavlath GK, McCarthy JJ, Esser KA. Last Word on Point:Counterpoint: Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy. Journal of Applied Physiology. 2007;103:1107.. doi: 10.1152/japplphysiol.00502.2007. [DOI] [PubMed] [Google Scholar]
- O'Mahoney JV, Guven KL, Lin J, Joya JE, Robinson CS, Wade RP, Hardeman EC. Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1. Molecular and Cellular Biology. 1998;18:6641–6652. doi: 10.1128/mcb.18.11.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95:307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- Oh M, Rybkin II, Copeland V, Czubryt MP, Shelton JM, van Rooij E, Richardson JA, Hill JA, De Windt LJ, Bassel-Duby R, Olson EN, Rothermel BA. Calcineurin is necessary for the maintainance but not embryonic development of slow musle fibers. Molecular and Cellular Biology. 2005;25:6629–6638. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkawa Y, Marfella CG, Imbalzano AN. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO Journal. 2006;25:490–501. doi: 10.1038/sj.emboj.7600943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojuka EO. Role of calcium and AMP kinase in the regulation of mitochondrial biogenesis and GLUT4 levels in muscle. Proceedings of the Nutrition Society. 2004;63:275–278. doi: 10.1079/PNS2004339. [DOI] [PubMed] [Google Scholar]
- Ojuka EO, Jones TE, Nolte LA, Chen M, Wamhoff BR, Sturek M, Holloszy JO. Regulation of GLUT4 biogenesis in muscle: evidence for involvement of AMPK and Ca(2+) American Journal of Physiology. 2002;282:E1008–E1013. doi: 10.1152/ajpendo.00512.2001. [DOI] [PubMed] [Google Scholar]
- Olson EN. MyoD family: a paradigm for development? Genes & Development. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- Osada S-S, Mizuno K, Saido TC, Suzuki K, Kuroki T, Ohno S. A new member of the protin kinase C family nPKCq, predominantly expressed in skeletal muscle. Molecular and Cellular Biology. 1992;12:3930–3938. doi: 10.1128/mcb.12.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilson JR, Liberatore CM, Yutzey KE, Crabtree GR, Tsika RW, Molkentin JD. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. Journal of Biological Chemistry. 2004;279:26192–26200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Altered skeletal muscle phenotypes in calcineurin Aalpha and Abeta gene-targeted mice. Molecular and Cellular Biology. 2003;23:4331–4343. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattullo MC, Cotter MA, Cameron NE, Barry JA. Effects of lenghtened immobilization on functional and histochemical properties of rabbit tibialis anterior muscle. Experimental Physiology. 1992;77:433–442. doi: 10.1113/expphysiol.1992.sp003604. [DOI] [PubMed] [Google Scholar]
- Pedersen TH, Macdonald WA, de Paoli FV, Gurung IS, Nielsen OB. Comparison of regulated passive membrane conductance in action potential-firing fast- and slow-twitch muscle. Journal of General Physiology. 2009;134:323–337. doi: 10.1085/jgp.200910291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy M, Gregory P, Martin BJ, Stirewalt WS. Regulation of myosin heavy-chain gene expression during skeletal-muscle hypertrophy. Biochemcal Journal. 1989;257:691–698. doi: 10.1042/bj2570691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy M, Kalyanasundaram A. SERCA pump isoforms: their role in calcium transport and disease. Muscle & Nerve. 2007;35:430–442. doi: 10.1002/mus.20745. [DOI] [PubMed] [Google Scholar]
- Perrini S, Henriksson J, Zierath JR, Widegren U. Exercise-induced protein kinase C isoform-specific activation in human skeletal muscle. Diabetes. 2004;53:21–24. doi: 10.2337/diabetes.53.1.21. [DOI] [PubMed] [Google Scholar]
- Personius KE, Balice-Gordon RJ. Loss of correlated motor neuron activity during synaptic competition at developing neuromuscular synapses. Neuron. 2001;31:395–408. doi: 10.1016/s0896-6273(01)00369-5. [DOI] [PubMed] [Google Scholar]
- Peters D, Barash IA, Burdi M, Yuan PS, Mathew L, Friden J, Lieber RL. Asynchronous functional, cellular and transcriptional changes after a bout of eccentric exercise in the rat. Journal of Physiology. 2003;553:947–957. doi: 10.1113/jphysiol.2003.048462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella JK, Kim JS, Cross JM, Kosek DJ, Bamman MM. Efficacy of myonuclear addition may explain differential myofiber growth among resistance-trained young and older men and women. American Journal of Physiology. 2006;291:E937–E946. doi: 10.1152/ajpendo.00190.2006. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Myosin isoforms, muscle fiber types, and transitions. Microscopy Research and Technique. 2000;50:500–509. doi: 10.1002/1097-0029(20000915)50:6<500::AID-JEMT7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Pette D, Staron RS. Transitions of muscle fiber phenotypic profiles. Histochemistry and Cell Biology. 2001;115:359–372. doi: 10.1007/s004180100268. [DOI] [PubMed] [Google Scholar]
- Pette D, Vrbova G. What does chronic electrical stimulation teach us about muscle plasticity? Muscle & Nerve. 1999;22:666–677. doi: 10.1002/(sici)1097-4598(199906)22:6<666::aid-mus3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Pette D, Vrbová G. Adaption of mammalian skeletal muscle fibers to chronic electrical stimulation. Reviews of Physiology, Biochemistry and Pharmacology. 1992;120:116–201. doi: 10.1007/BFb0036123. [DOI] [PubMed] [Google Scholar]
- Philippou A, Halapas A, Maridaki M, Koutsilieris M. Type I insulin-like growth factor receptor signaling in skeletal muscle regeneration and hypertrophy. Journal of Musculoskeletal & Neuronal Interactions. 2007;7:208–218. [PubMed] [Google Scholar]
- Phillips WD, Bennett MR. Differentiation of fiber types in wing muscles during embryonic development: effect of neural tube removal. Developmental Biology. 1984;106:457–468. doi: 10.1016/0012-1606(84)90245-8. [DOI] [PubMed] [Google Scholar]
- Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. Journal of Physiology. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin CL, Hrycyshyn AW, Rogers KA, Rushlow WJ, Merrifield PA. Embryonic and fetal rat myoblasts form different muscle fiber types in an ectopic in vivo environment. Developmental Dynamics. 2002;224:253–266. doi: 10.1002/dvdy.10106. [DOI] [PubMed] [Google Scholar]
- Pin CL, Konieczny SF. A fast fiber enhancer exists in the muscle regulatory factor 4 gene promoter. Biochemical and Biophysical Research Communications. 2002;299:7–13. doi: 10.1016/s0006-291x(02)02571-8. [DOI] [PubMed] [Google Scholar]
- Pin CL, Merrifield PA. Developmental potential of rat L6 myoblasts in vivo following injection into regenerating muscles. Developmental Biology. 1997;188:147–166. doi: 10.1006/dbio.1997.8624. [DOI] [PubMed] [Google Scholar]
- Pinton P, Tsuboi T, Ainscow EK, Pozzan T, Rizzuto R, Rutter GA. Dynamics of glucose-induced membrane recruitment of protein kinase C beta II in living pancreatic islet beta-cells. Journal of Biological Chemistry. 2002;277:37702–37710. doi: 10.1074/jbc.M204478200. [DOI] [PubMed] [Google Scholar]
- Pisani DF, Dechesne CA. Skeletal muscle HIF-1alpha expression is dependent on muscle fiber type. Journal of General Physiology. 2005;126:173–178. doi: 10.1085/jgp.200509265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polly P, Haddadi LM, Issa LL, Subramaniam N, Palmer SJ, Tay ES, Hardeman EC. hMusTRD1alpha1 represses MEF2 activation of the troponin I slow enhancer. Journal of Biological Chemistry. 2003;278:36603–36610. doi: 10.1074/jbc.M212814200. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Wu H, Arnold MA, Shelton JM, Backs J, McAnally J, Richardson JA, Bassel-Duby R, Olson EN. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. Journal of Clinical Investigation. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell-Braxton L, Hollingshead P, Warburton C, Dowd M, Pitts-Meek S, Dalton D, Gillett N, Stewart TA. IGF-I is required for normal embryonic growth in mice. Genes & Development. 1993;7:2609–2617. doi: 10.1101/gad.7.12b.2609. [DOI] [PubMed] [Google Scholar]
- Psilander N, Damsgaard R, Pilegaard H. Resistance exercise alters MRF and IGF-I mRNA content in human skeletal muscle. Journal of Applied Physiology. 2003;95:1038–1044. doi: 10.1152/japplphysiol.00903.2002. [DOI] [PubMed] [Google Scholar]
- Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schafer LV, Brandmeier B, Grater F, Grubmuller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proceedings of the National Academy of Sciences USA. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putman CT, Dusterhoft S, Pette D. Satellite cell proliferation in low frequency-stimulated fast muscle of hypothyroid rat. American Journal of Physiology. 2000;279:C682–C690. doi: 10.1152/ajpcell.2000.279.3.C682. [DOI] [PubMed] [Google Scholar]
- Ralston HJ, Feinstein B, Inman VT. Rate of atrophy in muscles immobilized at different lengths. Fed. Proc. 1952;11:127.. [Google Scholar]
- Rana ZA, Gundersen K, Buonanno A. Activity-dependent repression of muscle genes by NFAT. Proceedings of the National Academy of Sciences USA. 2008;105:5921–5926. doi: 10.1073/pnas.0801330105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana ZA, Gundersen K, Buonanno A, Vullhorst D. Imaging transcription in vivo: distinct regulatory effects of fast and slow activity patterns on promoter elements from vertebrate troponin I isoform genes. Journal of Physiology. 2005;562.3:815–828. doi: 10.1113/jphysiol.2004.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranvier L. De quelques faits relatifs a l'histologie et a la physiologie des muscles stries. Archives de Physiologie normale et Pathologique. 1874;6:1–15. [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Raue U, Slivka D, Jemiolo B, Hollon C, Trappe S. Myogenic gene expression at rest and after a bout of resistance exercise in young (18–30 yr) and old (80–89 yr) women. Journal of Applied Physiology. 2006;101:53–59. doi: 10.1152/japplphysiol.01616.2005. [DOI] [PubMed] [Google Scholar]
- Reardon KA, Davis J, Kapsa RM, Choong P, Byrne E. Myostatin, insulin-like growth factor-1, and leukemia inhibitory factor mRNAs are upregulated in chronic human disuse muscle atrophy. Muscle & Nerve. 2001;24:893–899. doi: 10.1002/mus.1086. [DOI] [PubMed] [Google Scholar]
- Rehfeldt C. In response to Point:Counterpoint: “Satellite cell addition is/is not obligatory for skeletal muscle hypertrophy”. Journal of Applied Physiology. 2007;103:1104.. doi: 10.1152/japplphysiol.00466.2007. [DOI] [PubMed] [Google Scholar]
- Rescan PY, Gauvry L, Paboeuf G. A gene with homology to myogenin is expressed in developing myotomal musculature of the rainbow trout and in vitro during the conversion of myosatellite cells to myotubes. FEBS Letters. 1995;362:89–92. doi: 10.1016/0014-5793(95)00215-u. [DOI] [PubMed] [Google Scholar]
- Richardson RS, Newcomer SC, Noyszewski EA. Skeletal muscle intracellular PO(2) assessed by myoglobin desaturation: response to graded exercise. Journal of Applied Physiology. 2001;91:2679–2685. doi: 10.1152/jappl.2001.91.6.2679. [DOI] [PubMed] [Google Scholar]
- Richardson RS, Noyszewski EA, Kendrick KF, Leigh JS, Wagner PD. Myoglobin O2 desaturation during exercise. Evidence of limited O2 transport. Journal of Clinical Investigation. 1995;96:1916–1926. doi: 10.1172/JCI118237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nature Cell Biology. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- Rose AJ, Hargreaves M. Exercise increases Ca2+-calmodulin-dependent protein kinase II activity in human skeletal muscle. Journal of Physiology. 2003;553:303–309. doi: 10.1113/jphysiol.2003.054171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Michell BJ, Kemp BE, Hargreaves M. Effect of exercise on protein kinase C activity and localization in human skeletal muscle. Journal of Physiology. 2004;561:861–870. doi: 10.1113/jphysiol.2004.075549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt JD, Parry DJ, Partridge TA. Phenotype of adult mouse muscle myoblasts reflects their fibertype of origin. Differentiation. 1996;60:39–45. doi: 10.1046/j.1432-0436.1996.6010039.x. [DOI] [PubMed] [Google Scholar]
- Rosenthal SM, Cheng ZQ. Opposing early and late effects of insulin-like growth factor I on differentiation and the cell cycle regulatory retinoblastoma protein in skeletal myoblasts. Proceedings of the National Academy of Sciences USA. 1995;92:10307–10311. doi: 10.1073/pnas.92.22.10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostas JA, Dunkley PR. Multiple forms and distribution of calcium/calmodulin-stimulated protein kinase II in brain. Journal of Neurochemistry. 1992;59:1191–202. doi: 10.1111/j.1471-4159.1992.tb08428.x. [DOI] [PubMed] [Google Scholar]
- Roth SM, Martel GF, Ferrell RE, Metter EJ, Hurley BF, Rogers MA. Myostatin gene expression is reduced in humans with heavy-resistance strength training: a brief communication. Exp Biol Med (Maywood) 2003;228:706–709. doi: 10.1177/153537020322800609. [DOI] [PubMed] [Google Scholar]
- Rotzler S, Schramek H, Brenner HR. Metabolic stabilization of endplate acetylcholine receptors regulated by Ca2+ influx associated with muscle activity. Nature. 1991;349:337–339. doi: 10.1038/349337a0. [DOI] [PubMed] [Google Scholar]
- Roy RR, Baldwin KM, Martin TP, Chimarusti SP, Edgerton VR. Biochemical and physiological changes in overloaded rat fast- and slow-twitch ankle extensors. Journal of Applied Physiology. 1985;59:639–646. doi: 10.1152/jappl.1985.59.2.639. [DOI] [PubMed] [Google Scholar]
- Roy RR, Pierotti DJ, Garfinkel A, Zhong H, Baldwin KM, Edgerton VR. Persistence of motor unit and muscle fiber types in the presence of inactivity. Journal of Experimental Biology. 2008;211:1041–1049. doi: 10.1242/jeb.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of myoD in mice leads to up-regulation of myogenic HLH gene myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- Rykx A, De Kimpe L, Mikhalap S, Vantus T, Seufferlein T, Vandenheede JR, Van Lint J. Protein kinase D: a family affair. FEBS Letters. 2003;546:81–86. doi: 10.1016/s0014-5793(03)00487-3. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Watanabe K, Totsuka T, Uramoto I, Sano M, Sakamoto K. Differential adaptations of insulin-like growth factor-I, basic fibroblast growth factor, and leukemia inhibitory factor in the plantaris muscle of rats by mechanical overloading: an immunohistochemical study. Acta Neuropathol. 1998;95:123–130. doi: 10.1007/s004010050775. [DOI] [PubMed] [Google Scholar]
- Saltin B, Gollnick PD. Skeletal muscle adaptability: significance for metabolism and performance. In: Peachey LD, Adrian RH, Geiger SR, editors. Handbook of Physiology. Vol. 10. Bethesda: American Physiological Society; 1983. pp. 555–631. [Google Scholar]
- Salviati C, Biasia E, Aloisi M. Synthesis of fast myosin induced by fast ectopic innervation of rat soleus muscle is restricted to ectopic endplate region. Nature. 1986;322:637–639. doi: 10.1038/322637a0. [DOI] [PubMed] [Google Scholar]
- Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- Savolainen J, Vaananen K, Puranen J, Takala TE, Komulainen J, Vihko V. Collagen synthesis and proteolytic activities in rat skeletal muscles: effect of cast-immobilization in the lengthened and shortened positions. Archives of Physical Medicine and Rehabilitation. 1988;69:964–969. [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiological Reviews. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Ausoni S, Gorza L, Saggin L, Gundersen K, Lømo T. Myosin heavy chain isoforms and velocity of shortening of type 2 skeletal muscle fibres. Acta Physiologica Scandinavica. 1988;134:575–576. doi: 10.1111/j.1748-1716.1998.tb08539.x. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Gorza L, Sartore S, Saggin L, Ausoni S, Vianello M, Gundersen K, Lomo T. Three myosin heavy chain isoforms in type 2 skeletal muscle fibres. Journal of Muscle Research and Cell Motility. 1989;10:197–205. doi: 10.1007/BF01739810. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Hanzlikova V. On the mechanism of compensatory hypertrophy in skeletal muscles. Experientia. 1970;26:152–153. doi: 10.1007/BF01895548. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiological Reviews. 1996;76:371–423. doi: 10.1152/physrev.1996.76.2.371. [DOI] [PubMed] [Google Scholar]
- Schmalbruch H, Lewis DM. Dynamics of nuclei of muscle fibers and connective tissue cells in normal and denervated rat muscles. Muscle & Nerve. 2000;23:617–626. doi: 10.1002/(sici)1097-4598(200004)23:4<617::aid-mus22>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. New England Journal of Medicine. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- Schuler M, Ali F, Chambon C, Duteil D, Bornert JM, Tardivel A, Desvergne B, Wahli W, Chambon P, Metzger D. PGC1alpha expression is controlled in skeletal muscles by PPARbeta, whose ablation results in fiber-type switching, obesity, and type 2 diabetes. Cell Metabolism. 2006;4:407–414. doi: 10.1016/j.cmet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Schwander JC, Hauri C, Zapf J, Froesch ER. Synthesis and secretion of insulin-like growth factor and its binding protein by the perfused rat liver: dependence on growth hormone status. Endocrinology. 1983;113:297–305. doi: 10.1210/endo-113-1-297. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 2009;24:97–106. doi: 10.1152/physiol.00045.2008. [DOI] [PubMed] [Google Scholar]
- Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proceedings of the National Academy of Sciences USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seward DJ, Haney JC, Rudnicki MA, Swoap SJ. bHLH transcription factor MyoD affects myosin heavy chain expression pattern in a muscle-specific fashion. American Journal of Physiology. 2001;280:C408–C413. doi: 10.1152/ajpcell.2001.280.2.C408. [DOI] [PubMed] [Google Scholar]
- Shen T, Cseresnyes Z, Liu Y, Randall WR, Schneider MF. Regulation of the nuclear export of the transcription factor NFATc1 by protein kinases after slow fibre type electrical stimulation of adult mouse skeletal muscle fibres. Journal of Physiology. 2007;579:535–551. doi: 10.1113/jphysiol.2006.120048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen T, Liu Y, Cseresnyes Z, Hawkins A, Randall WR, Schneider MF. Activity- and calcineurin-independent nuclear shuttling of NFATc1, but not NFATc3, in adult skeletal muscle fibers. Mol Biol Cell. 2006;17:1570–1582. doi: 10.1091/mbc.E05-08-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Scheffler JM, Pleitner JM, Zeng C, Park S, Hannon KM, Grant AL, Gerrard DE. Modulation of skeletal muscle fiber type by mitogen-activated protein kinase signaling. FASEB Journal. 2008;22:2990–3000. doi: 10.1096/fj.07-097600. [DOI] [PubMed] [Google Scholar]
- Shi H, Zeng C, Ricome A, Hannon KM, Grant AL, Gerrard DE. Extracellular signal-regulated kinase pathway is differentially involved in beta-agonist-induced hypertrophy in slow and fast muscles. American Journal of Physiology. 2007;292:C1681–C1689. doi: 10.1152/ajpcell.00466.2006. [DOI] [PubMed] [Google Scholar]
- Singh MA, Ding W, Manfredi TJ, Solares GS, O'Neill EF, Clements KM, Ryan ND, Kehayias JJ, Fielding RA, Evans WJ. Insulin-like growth factor I in skeletal muscle after weight-lifting exercise in frail elders. American Journal of Physiology. 1999;277:E135–E143. doi: 10.1152/ajpendo.1999.277.1.E135. [DOI] [PubMed] [Google Scholar]
- Siu PM, Alway SE. Mitochondria-associated apoptotic signalling in denervated rat skeletal muscle. Journal of Physiology. 2005;565:309–323. doi: 10.1113/jphysiol.2004.081083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu PM, Donley DA, Bryner RW, Alway SE. Myogenin and oxidative enzyme gene expression levels are elevated in rat soleus muscles after endurance training. Journal of Applied Physiology. 2004;97:277–285. doi: 10.1152/japplphysiol.00534.2004. [DOI] [PubMed] [Google Scholar]
- Siu PM, Pistilli EE, Alway SE. Apoptotic responses to hindlimb suspension in gastrocnemius muscles from young adult and aged rats. American Journal of Physiology. 2005a;289:R1015–R1026. doi: 10.1152/ajpregu.00198.2005. [DOI] [PubMed] [Google Scholar]
- Siu PM, Pistilli EE, Butler DC, Alway SE. Aging influences cellular and molecular responses of apoptosis to skeletal muscle unloading. American Journal of Physiology. 2005b;288:C338–C349. doi: 10.1152/ajpcell.00239.2004. [DOI] [PubMed] [Google Scholar]
- Sjogren K, Liu JL, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Tornell J, Isaksson OG, Jansson JO, Ohlsson C. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proceedings of the National Academy of Sciences USA. 1999;96:7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdu V, Karsch-Mizrachi I, Campione M, Leinwand L, Schiaffino S. Type IIx myosin heavy chain transcripts are expressed in type IIb fibers of human skeletal muscle. American Journal of Physiology. 1994;267:C1723–C1728. doi: 10.1152/ajpcell.1994.267.6.C1723. [DOI] [PubMed] [Google Scholar]
- Song A, Wang Q, Goebl MG, Harrington MA. Phosphorylation of nuclear MyoD is required for its rapid degradation. Molecular and Cellular Biology. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenburg EE, Le Roith D, Ward CW, Bodine SC. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. Journal of Physiology. 2008;586:283–291. doi: 10.1113/jphysiol.2007.141507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Demignon J, Porteu A, Kahn A, Concordet JP, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proceedings of the National Academy of Sciences USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squire JM, Morris EP. A new look at thin filament regulation in vertebrate skeletal muscle. FASEB Journal. 1998;12:761–771. doi: 10.1096/fasebj.12.10.761. [DOI] [PubMed] [Google Scholar]
- Sréter FA, Lopez JR, Alamo L, Mabuchi K, Gergely J. Changes in intracellular ionized Ca concentration associated with muscle fiber type transformation. American Journal of Physiology. 1987;253:C296–C300. doi: 10.1152/ajpcell.1987.253.2.C296. [DOI] [PubMed] [Google Scholar]
- Staron RS, Hagerman FC, Hikida RS, Murray TF, Hostler DP, Crill MT, Ragg KE, Toma K. Fiber type composition of the vastus lateralis muscle of young men and women. Journal of Histochemistry and Cytochemistry. 2000;48:623–629. doi: 10.1177/002215540004800506. [DOI] [PubMed] [Google Scholar]
- Staron RS, Karapondo DL, Kraemer WJ, Fry AC, Gordon SE, Falkel JE, Hagerman FC, Hikida RS. Skeletal muscle adaptations during early phase of heavy-resistance training in men and women. Journal of Applied Physiology. 1994;76:1247–1255. doi: 10.1152/jappl.1994.76.3.1247. [DOI] [PubMed] [Google Scholar]
- Stephenson GM. Hybrid skeletal muscle fibres: a rare or common phenomenon? Clinical and Experimental Pharmacology & Physiology. 2001;28:692–702. doi: 10.1046/j.1440-1681.2001.03505.x. [DOI] [PubMed] [Google Scholar]
- Stockdale FE, Holtzer H. DNA synthesis and myogenesis. Experimental Cell Research. 1961;24:508–520. doi: 10.1016/0014-4827(61)90450-5. [DOI] [PubMed] [Google Scholar]
- Strassburger E. Ûber die Wirkungssphäre der kerne und die zellgrösse. Histologische Beiträge. 1893;5:97–124. [Google Scholar]
- Summers TB, Hines HM. Effect of immobilization in various positions upon the weight and strength of skeletal muscle. Archives of Physical Medicine and Rehabilitation. 1951;32:142–145. [PubMed] [Google Scholar]
- Suwa M, Nakano H, Kumagai S. Effects of chronic AICAR treatment on fiber composition, enzyme activity, UCP3, and PGC-1 in rat muscles. Journal of Applied Physiology. 2003;95:960–968. doi: 10.1152/japplphysiol.00349.2003. [DOI] [PubMed] [Google Scholar]
- Swoap SJ, Hunter RB, Stevenson EJ, Felton HM, Kansagra NV, Lang JM, Esser KA, Kandarian SC. The calcineurin-NFAT pathway and muscle fiber-type gene expression. American Journal of Physiology. 2000;279:C915–C924. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- Tabary JC, Tabary C, Tardieu C, Tardieu G, Goldspink G. Physiological and structural changes in the cat's soleus muscle due to immobilization at different lengths by plaster casts. Journal of Physiology. 1972;224:231–244. doi: 10.1113/jphysiol.1972.sp009891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemori H, Katoh Hashimoto Y, Nakae J, Olson EN, Okamoto M. Inactivation of HDAC5 by SIK1 in AICAR-treated C2C12 Myoblasts. Endocrine Journal. 2008;56:121–130. doi: 10.1507/endocrj.k08e-173. [DOI] [PubMed] [Google Scholar]
- Tang H, Cheung WM, Ip FC, Ip NY. Identification and characterization of differentially expressed genes in denervated muscle. Molecular and Cellular Neurosciences. 2000;16:127–140. doi: 10.1006/mcne.2000.0864. [DOI] [PubMed] [Google Scholar]
- Tardieu JC, Tarddieu G, Gagnard L, Tabary C. Les rétractions musculaires. Etude expérimentale. Conséquences thérapautiques. Revue Pratn. 1969;19:1535–1543. [Google Scholar]
- Tay ES, Guven KL, Subramaniam N, Polly P, Issa LL, Gunning PW, Hardeman EC. Regulation of alternative splicing of Gtf2ird1 and its impact on slow muscle promoter activity. Biochemcal Journal. 2003;374:359–367. doi: 10.1042/BJ20030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada S, Goto M, Kato M, Kawanaka K, Shimokawa T, Tabata I. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochemical and Biophysical Research Communications. 2002;296:350–354. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- Terada S, Kawanaka K, Goto M, Shimokawa T, Tabata I. Effects of high-intensity intermittent swimming on PGC-1alpha protein expression in rat skeletal muscle. Acta Physiologica Scandinavica. 2005;184:59–65. doi: 10.1111/j.1365-201X.2005.01423.x. [DOI] [PubMed] [Google Scholar]
- Tews DS. Muscle-fiber apoptosis in neuromuscular diseases. Muscle & Nerve. 2005;32:443–458. doi: 10.1002/mus.20348. [DOI] [PubMed] [Google Scholar]
- Tews DS, Goebel HH, Schneider I, Gunkel A, Stennert E, Neiss WF. DNA-fragmentation and expression of apoptosis-related proteins in experimentally denervated and reinnervated rat facial muscle. Neuropathology and applied neurobiology. 1997;23:141–149. [PubMed] [Google Scholar]
- Thomsen P, Luco JV. Changes of weight and neuromuscular transmission in muscles of immobilised joints. Journal of Neurophysiology. 1944;7:245–251. [Google Scholar]
- Tisdale MJ. Mechanisms of cancer cachexia. Physiological Reviews. 2009;89:381–410. doi: 10.1152/physrev.00016.2008. [DOI] [PubMed] [Google Scholar]
- Tothova J, Blaauw B, Pallafacchina G, Rudolf R, Argentini C, Reggiani C, Schiaffino S. NFATc1 nucleocytoplasmic shuttling is controlled by nerve acticity in skeletal muscle. Journal of Cell Science. 2006;119:1604–1611. doi: 10.1242/jcs.02875. [DOI] [PubMed] [Google Scholar]
- Tsika RW, Herrick RE, Baldwin KM. Interaction of compensatory overload and hindlimb suspension on myosin isoform expression. Journal of Applied Physiology. 1987;62:2180–2186. doi: 10.1152/jappl.1987.62.6.2180. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L, Trinick J. Giant proteins: sensing tension with titin kinase. Current Biology. 2008;18:R1141–R1142. doi: 10.1016/j.cub.2008.10.035. [DOI] [PubMed] [Google Scholar]
- Van Lint J, Rykx A, Maeda Y, Vantus T, Sturany S, Malhotra V, Vandenheede JR, Seufferlein T. Protein kinase D: an intracellular traffic regulator on the move. Trends in Cell Biology. 2002;12:193–200. doi: 10.1016/s0962-8924(02)02262-6. [DOI] [PubMed] [Google Scholar]
- Van Swearingen J, Lance-Jones C. Slow and fast muscle fibers are preferentially derived from myoblasts migrating into the chick limb bud at different developmental times. Developmental Biology. 1995;170:321–337. doi: 10.1006/dbio.1995.1218. [DOI] [PubMed] [Google Scholar]
- Vandenburgh H, Kaufman S. In vitro model for stretch-induced hypertrophy of skeletal muscle. Science. 1979;203:265–268. doi: 10.1126/science.569901. [DOI] [PubMed] [Google Scholar]
- Vandenburgh H, Kaufman S. Protein degradation in embryonic skeletal muscle. Effect of medium, cell type, inhibitors, and passive stretch. Journal of Biological Chemistry. 1980;255:5826–5833. [PubMed] [Google Scholar]
- Vandenburgh HH, Karlisch P, Shansky J, Feldstein R. Insulin and IGF-I induce pronounced hypertrophy of skeletal myofibers in tissue culture. American Journal of Physiology. 1991;260:C475–C484. doi: 10.1152/ajpcell.1991.260.3.C475. [DOI] [PubMed] [Google Scholar]
- Viguie CA, Lu DX, Huang SK, Rengen H, Carlson BM. Quantitative study of the effects of long-term denervation on the extensor digitorum longus muscle of the rat. Anatomical Records. 1997;248:346–354. doi: 10.1002/(SICI)1097-0185(199707)248:3<346::AID-AR7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Vikstrom KL, Seiler SH, Sohn RL, Strauss M, Weiss A, Welikson RE, Leinwand LA. The vertebrate myosin heavy chain: genetics and assembly properties. Cell Structure and Function. 1997;22:123–129. doi: 10.1247/csf.22.123. [DOI] [PubMed] [Google Scholar]
- Vissing K, McGee SL, Roepstorff C, Schjerling P, Hargreaves M, Kiens B. Effect of sex differences on human MEF2 regulation during endurance exercise. American Journal of Physiology. 2008;294:E408–E415. doi: 10.1152/ajpendo.00403.2007. [DOI] [PubMed] [Google Scholar]
- Voytik SL, Przyborsky M, Badylak SF, Koenieczny SF. Differential expression of muscle regulatory factor genes in normal and denervated adult rat hindlimb muscles. Developmental Dynamics. 1993;198:214–224. doi: 10.1002/aja.1001980307. [DOI] [PubMed] [Google Scholar]
- Vrbová G. The effect of motoneurone activity on the speed of contraction of striated muscle. Journal of Physiology. 1963;169:513–526. doi: 10.1113/jphysiol.1963.sp007276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrbova G, Navarrete R, Lowrie M. Matching of muscle properties and motoneurone firing patterns during early stages of development. Journal of Experimental Biology. 1985;115:113–123. doi: 10.1242/jeb.115.1.113. [DOI] [PubMed] [Google Scholar]
- Vullhorst D, Buonanno A. Characterization of general transcription factor 3, a transcription factor involved in slow muscle-specific gene expression. Journal of Biological Chemistry. 2003;278:8370–8379. doi: 10.1074/jbc.M209361200. [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Conboy IM. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell. 2005;122:659–667. doi: 10.1016/j.cell.2005.08.021. [DOI] [PubMed] [Google Scholar]
- Walker KS, Kambadur R, Sharma M, Smith HK. Resistance training alters plasma myostatin but not IGF-1 in healthy men. Medicine & Science in Sports & Exercise. 2004;36:787–793. doi: 10.1249/01.mss.0000126384.04778.29. [DOI] [PubMed] [Google Scholar]
- Walkinshaw DR, Tahmasebi S, Bertos NR, Yang XJ. Histone deacetylases as transducers and targets of nuclear signaling. Journal of Cellular Biochemistry. 2008;104:1541–1552. doi: 10.1002/jcb.21746. [DOI] [PubMed] [Google Scholar]
- Walters EH, Stickland NC, Loughna PT. MRF-4 exhibits fiber type- and muscle-specific pattern of expression in postnatal rat muscle. American Journal of Physiology. 2000a;278:R1381–R1384. doi: 10.1152/ajpregu.2000.278.5.R1381. [DOI] [PubMed] [Google Scholar]
- Walters EH, Stickland NC, Loughna PT. The expression of the myogenic regulatory factors in denervated and normal muscles of different phenotypes. Journal of Muscle Research and Cell Motility. 2000b;21:647–653. doi: 10.1023/a:1005683825960. [DOI] [PubMed] [Google Scholar]
- Wan B, Moreadith RW. Structural characterization and regulatory element analysis of the heart isoform of cytocrome c oxidase VIa. Journal of Biological Chemistry. 1995;270:26433–26440. doi: 10.1074/jbc.270.44.26433. [DOI] [PubMed] [Google Scholar]
- Wang N, Hikida RS, Staron RS, Simoneau JA. Muscle fiber types of women after resistance training - quantitative ultrastructure and enzyme activity. Pflügers Archiv. 1993;424:494–502. doi: 10.1007/BF00374913. [DOI] [PubMed] [Google Scholar]
- Wang Y-X, Zhang CL, Tou RT, Cho HK, Nelson MC, Bayagua-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPAR-d. PLoS Biology. 2004a;2:1–8. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YX, Zhang C-L, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPAR delta. PLoS Biology. 2004b;2:e294.. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehling M, Cai B, Tidball JG. Modulation of myostatin expression during modified muscle use. FASEB Journal. 2000;14:103–110. doi: 10.1096/fasebj.14.1.103. [DOI] [PubMed] [Google Scholar]
- Weintraub H. The myoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Weiss A, Schiaffino S, Leinwand LA. Comparative sequence analysis of the complete human sarcomeric myosin heavy chain family: Implications for functional diversity. Journal of Molecular Biology. 1999;290:61–75. doi: 10.1006/jmbi.1999.2865. [DOI] [PubMed] [Google Scholar]
- Wendt IR, Gibbs CL. Energy production of rat extensor digitorum longus muscle. American Journal of Physiology. 1973;224:1081–1086. doi: 10.1152/ajplegacy.1973.224.5.1081. [DOI] [PubMed] [Google Scholar]
- Westerblad H, Allen DG. Changes of myoplasmic calcium concentration during fatigue in single mouse muscle fibers. Journal of General Physiology. 1991;98:615–635. doi: 10.1085/jgp.98.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerblad H, Allen DG. The contribution of [Ca2+]i to the slowing of relaxation in fatigued single fibres from mouse skeletal muscle. Journal of Physiology. 1993;468:729–740. doi: 10.1113/jphysiol.1993.sp019797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgaard RH, Lømo T. Control of contractile properties within adaptive ranges by patterns of impulse activity in the rat. Journal of Neuroscience. 1988;8:4415–4426. doi: 10.1523/JNEUROSCI.08-12-04415.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby DS, Nelson MJ. Myosin heavy-chain mRNA expression after a single session of heavy-resistance exercise. Medicine & Science in Sports & Exercise. 2002;34:1262–1269. doi: 10.1097/00005768-200208000-00006. [DOI] [PubMed] [Google Scholar]
- Windisch A, Gundersen K, Szabolcs MJ, Gruber H, Lomo T. Fast to slow transformation of denervated and electrically stimulated rat muscle. Journal of Physiology. 1998;510:623–632. doi: 10.1111/j.1469-7793.1998.623bk.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter B, Braun T, Arnold HH. cAMP-dependent protein kinase represses myogenic differentiation and the activity of the muscle-specific helix-loop-helix transcription factors myf-5 and myoD. Journal of Biological Chemistry. 1993;268:9869–9878. [PubMed] [Google Scholar]
- Witzemann V, Sakmann B. Differential regulation of myoD and myogenin mRNA by nerve induced muscle activity. FEBS Letters. 1991;282:259–264. doi: 10.1016/0014-5793(91)80490-t. [DOI] [PubMed] [Google Scholar]
- Wretman C, Lionikas A, Widegren U, Lannergren J, Westerblad H, Henriksson J. Effects of concentric and eccentric contractions on phosphorylation of MAPK(erk1/2) and MAPK(p38) in isolated rat skeletal muscle. Journal of Physiology. 2001;535:155–164. doi: 10.1111/j.1469-7793.2001.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DC, Han DH, Garcia-Roves PM, Geiger PC, Jones TE, Holloszy JO. Exercise-induced mitochondrial biogenesis begins before the increase in muscle PGC-1alpha expression. Journal of Biological Chemistry. 2007;282:194–199. doi: 10.1074/jbc.M606116200. [DOI] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- Wu H, Naya FJ, McKinsey TA, Mercer B, Shelton JM, Chin ER, Simard AR, Michel RN, Bassel-Duby R, Olson EN, Williams RS. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO Journal. 2000;19:1963–1973. doi: 10.1093/emboj/19.9.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Rothermel B, Kanatous S, Rosenberg P, Naya FJ, Shelton JM, Hutcheson KA, DiMaio JM, Olson EN, Bassel-Duby R, Williams RS. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO Journal. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Biggs RB, Booth FW. Insulin-like growth factor immunoreactivity increases in muscle after acute eccentric contractions. Journal of Applied Physiology. 1993;74:410–414. doi: 10.1152/jappl.1993.74.1.410. [DOI] [PubMed] [Google Scholar]
- Yang H, Alnaqeeb M, Simpson H, Goldspink G. Changes in muscle fibre type, muscle mass and IGF-I gene expression in rabbit skeletal muscle subjected to stretch. Journal of Anatomy. 1997;190:613–622. doi: 10.1046/j.1469-7580.1997.19040613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Alnaqeeb M, Simpson H, Goldspink G. Cloning and characterization of an IGF-1 isoform expressed in skeletal muscle subjected to stretch. Journal of Muscle Research and Cell Motility. 1996;17:487–495. doi: 10.1007/BF00123364. [DOI] [PubMed] [Google Scholar]
- Yang Y, Creer A, Jemiolo B, Trappe S. Time course of myogenic and metabolic gene expression in response to acute exercise in human skeletal muscle. Journal of Applied Physiology. 2005;98:1745–1752. doi: 10.1152/japplphysiol.01185.2004. [DOI] [PubMed] [Google Scholar]
- Yarasheski KE, Bhasin S, Sinha-Hikim I, Pak-Loduca J, Gonzalez-Cadavid NF. Serum myostatin-immunoreactive protein is increased in 60–92 year old women and men with muscle wasting. Journal of Nutrition, Health & Aging. 2002;6:343–348. [PubMed] [Google Scholar]
- Yarasheski KE, Campbell JA, Smith K, Rennie MJ, Holloszy JO, Bier DM. Effect of growth hormone and resistance exercise on muscle growth in young men. American Journal of Physiology. 1992;262:E261–E267. doi: 10.1152/ajpendo.1992.262.3.E261. [DOI] [PubMed] [Google Scholar]
- Yarasheski KE, Zachweija JJ, Angelopoulos TJ, Bier DM. Short-term growth hormone treatment does not increase muscle protein synthesis in experienced weight lifters. Journal of Applied Physiology. 1993;74:3073–3076. doi: 10.1152/jappl.1993.74.6.3073. [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Harii K. A regenerative change during muscle adaptation to denervation in rats. Journal of Surgical Research. 1999;81:139–146. doi: 10.1006/jsre.1998.5504. [DOI] [PubMed] [Google Scholar]
- Yuan J, Bae D, Cantrell D, Nel AE, Rozengurt E. Protein kinase D is a downstream target of protein kinase Ctheta. Biochemical and Biophysical Research Communications. 2002;291:444–452. doi: 10.1006/bbrc.2002.6469. [DOI] [PubMed] [Google Scholar]
- Yutzey KE, Konieczny SF. Different E-box regulatory sequences are functionally distinct when placed within the context of the troponin I enhancer. Nucleic Acids Res. 1992;20:5105–5113. doi: 10.1093/nar/20.19.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit PS. All muscle satellite cells are equal, but are some more equal than others? Journal of Cell Science. 2008;121:2975–2982. doi: 10.1242/jcs.019661. [DOI] [PubMed] [Google Scholar]
- Zammit PS, Cohen A, Buckingham ME, Kelly RG. Integration of embryonic and fetal skeletal myogenic programs at the myosin light chain 1f/3f locus. Developmental Biology. 2008;313:420–433. doi: 10.1016/j.ydbio.2007.10.044. [DOI] [PubMed] [Google Scholar]
- Zhang H, Stavnezer E. Ski regulates muscle terminal differentiation by transcriptional activation of Myog in a complex with Six1 and Eya3. Journal of Biological Chemistry. 2008;284:2867–2879. doi: 10.1074/jbc.M807526200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zot AS, Potter JD. Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annual Review of Biophysics and Biophysical Chemistry. 1987;16:535–559. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]


