Abstract
Gamma secretase inhibitors (GSIs) comprise a growing class of compounds that interfere with the membrane-bound Notch signaling protein and its downstream intra-nuclear transcriptional targets. As GSI-I (Z-LLNle-CHO) is also a derivative of a widely used proteosome inhibitor MG-132, we hypothesized that this compound might be active in precursor-B acute lymphoblastic leukemia (ALL) cell lines and patient samples. We found that GSI-I treatment of precursor-B ALL blasts induced apoptotic cell death within 18–24 h. With confirmation using RNA and protein analyses, GSI-I blocked nuclear accumulation of cleaved Notch1 and Notch2, and inhibited Notch targets Hey2 and Myc. Microarray analyses of 207 children with high-risk precursor-B ALL demonstrate that Notch pathway expression is a common feature of these neoplasms. However, microarray studies also implicated additional transcriptional targets in GSI-I-dependent cell death, including genes in the unfolded protein response, nuclear factor-κB and p53 pathways. Z-LLNle-CHO blocks both γ-secretase and proteosome activity, inducing more robust cell death in precursor-B ALL cells than either proteosome-selective or γ-secretase-selective inhibitors alone. Using Z-LLNle-CHO in a nonobese diabetes/severe combined immunodeficiency (NOD/SCID) precursor-B ALL xenograft model, we found that GSI-I alone delayed or prevented engraftment of B-lymphoblasts in 50% of the animals comprising the experimental group, suggesting that this compound is worthy of additional testing.
Keywords: precursor-B acute lymphoblastic leukemia, gamma secretase inhibitor, Notch, proteosome
Introduction
Precursor-B acute lymphoblastic leukemia (precursor-B ALL) is the most common neoplasm in children and young adults. Although most patients appear to be cured, survivors commonly develop morbidities that result from dose-intensified treatment with cytotoxic drugs. And, despite the use of aggressive salvage treatments, those who relapse have a dismal prognosis, underscoring the need for improved treatment options. Among a number of novel therapeutic options, γ-secretase and proteosome inhibitors have recently shown biologic activity in T-lineage ALL (T-ALL),1 but remain to be fully defined for precursor-B ALL.2 When administered in animal models, GSIs have a range of biological activities, including an ability to reduce the accumulation of amyloid peptides associated with Alzheimer's disease, and can induce apoptosis in a variety of tumors, included breast cancer cells.3 In this report, we focus on gamma secretase inhibitor-I (GSI-I; Z-LLNle-CHO), which is structurally similar to the widely used proteosome inhibitor MG-132, and describe its biological activity in precursor-B ALL.
Recent studies in multiple myeloma and in lymphoma-derived B-cell lines suggest that GSI sensitivity may be a general property of mature B-cell neoplasms.4, 5, 6 Herein, we establish that GSI-I induces apoptosis in cultured precursor-B ALL cell lines and freshly isolated blasts. GSI-I also showed antitumor activity in a xenograft of human precursor-B ALL established in immuno-deficient nonobese diabetes/severe combined immunodeficiency (NOD/SCID) mice. We confirm that Notch receptors are at least one important downstream target impacted by GSI-I, consistent with their established role as γ-secretase substrates that control multiple aspects of lymphoid development and function. This work raises important questions regarding the complexity of Notch signaling in early B-cell development,7 because precursor-B blast cells are shown to express three of the four Notch receptors (Notch 1–4) and five ligands (Delta-like-1, Delta-like-3, Delta-like-4, Jagged-1 and Jagged-2) (see ref. 8 for a review of these proteins). We evaluate specific Notch target genes, including members of the HES, HEY and Myc.8, 9, 10, 11, 12 We confirm the observations of Han et al.3 that GSI-I is also a potent proteosome inhibitor, broadening the range of targets in leukemia blasts. We show that GSI-I treatment disrupts Akt-dependent pro-survival pathways13 and elevates reactive oxygen species (ROS) levels,14 consistent with previous reports that γ-secretase and proteosome targets regulate apoptotic cell death pathways.14, 15 We extend these observations to show that GSI-I has biological activity in precursor-B ALL cells and may be a suitable candidate for further testing in phase 1 clinical trials.
Materials and methods
Cell lines and institutional patients
B-linage cell lines (Nalm-6, 697, MHH-Call3, RS4;11; Tanoue) were from DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). Cells were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum. Bone marrow samples were acquired at diagnosis from pediatric patients with written, informed consent per local institutional human research guidelines. Mononuclear cells were enriched by Ficoll–Paque centrifugation; for DNA/RNA preparation, B-lineage cells were isolated at 4 °C using anti-CD19 magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Patients received risk-adjusted therapy according to protocols sponsored by the Children's Oncology Group. Peripheral blood B cells were isolated from the buffy coats of normal blood donors (United Blood Services, Albuquerque, NM, USA).
Reagents and antibodies
GSI-I (Z-LLNle-CHO), L-685,458, GSI-IX (DAPT) and GSI-XII (X-IL-CHO) were from Calbiochem (San Diego, CA, USA). Compound E was from Axxora LLC (San Diego, CA, USA). Cell Signaling Technology (Danvers, MA, USA) was the source of the PathScan Multiplex western cocktail I, the Phospho-Akt Pathway Sampler Kit, and antibodies to caspase 9 (Asp330), caspase 3 (8G10), Notch1 (6A5) and phospho-FoxO1/FoxO3. Notch 2 antibodies (ab8927) were from Abcam (Cambridge, MA, USA); Notch 3 (active) antibodies were from Epitomics (Burlingame, CA, USA).
Western blotting
Cells (5–10 × 106) were lysed in ice-cold buffer (50 m Tris-HCl pH 7.2, 150 m NaCl, 1 m NaVO3, protease inhibitor cocktail from Boehringer-Mannheim (Indianapolis, IN, USA), 1% NP-40). Lysates were clarified by centrifugation (15 000 g for 5 min) and protein levels measured by the Pierce bovine serum albumin method to normalize loading for sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Where indicated, nuclear/cytosol fractions were prepared (NE-PER reagents, Pierce, Rockford, IL, USA). Blocked membranes were sequentially incubated with 1° antibodies and horseradish peroxidase-conjugated 2° antibodies (Jackson, West Grove, PA, USA) and imaged using the chemiluminescence protocol.
Apoptosis and ROS production
Cell viability was assessed by Trypan blue exclusion and WST-1 assay (Roche Diagnostics Corporation, Indianapolis, IN, USA). To measure apoptosis, cells were labeled with annexin V-PE detection kit I (Pharmingen, San Diego, CA, USA) and data acquired on a FACS Calibur (Becton Dickinson, Franklin Lakes, NJ, USA) Screening for 35 apoptosis-related proteins was accomplished using the human apoptosis array from R&D Systems (Minneapolis, MN, USA). ROS production was measured by flow cytometry, after loading cells with 5-(and 6-)chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA; Invitrogen), a dye that increases fluorescence with oxidation.
PCR analysis
Qiagen RNAeasy Mini and OneStep RT-PCR kits (Qiagen, Valencia, CA, USA) were used for RNA isolation and reverse transcriptase-polymerase chain reactions (RT-PCR). Primer sets were designed such that amplifications crossed intron–exon boundaries to exclude genomic DNA. Quantitative RT-PCR used the QuantiTect Reverse Transcription and SYBR Green PCR kits, with specific primer sets.
Microarray analyses
RNA from 697 precursor-B ALL cell line and cryo-preserved bone marrow or peripheral blood patient samples was extracted using Trizol (Invitrogen) followed by amplification, and hybridization to Affymetrix HG-U133Plus2.0 oligonucleotide microarrays (https://www.affymetrix.com). The gene expression data set was derived from a cohort of high-risk B precursor ALL patients enrolled on COG ALL biology and treatment trials 9900 and 9906, respectively. Analysis of patient samples was performed using Affymetrix GCOS (GeneChip Operating Software) v1.4 (Affymetrix, Santa Clara, CA, USA). The Microarray Suite 5.0 statistical algorithm was applied and signal intensities and present/marginal/absent calls obtained. Control data were obtained from bone marrow CD19+ cells of six healthy subjects, analyzed as a separate cohort. The entire gene expression data set may be accessed via the National Cancer Institute caArray portal (https://array.nci.nih.gov/caarray) or at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession no. GSE11877).
Microarray data of drug-treated 697 cell line samples were subjected to quantile normalization using the PLIER-sketch algorithm in Affymetrix Power Tools 1.8, with the pm-mm background correction flag turned on. Normalized data were analyzed using R (https://www.r-project.org) and Bioconductor. Briefly, data were filtered to find expressed genes by requiring that at least two samples gave a signal greater than 256 normalized Affymetrix units. The LIMMA16 package was used to identify 383 probesets that were significantly (P-value 0.02) changed at least threefold between the two experimental conditions, using a Benjamini–Hochberg false discovery rate correction. Independently, a set of 342 probesets was identified using the SAMR algorithrm17 and a two class paired significance test with a delta of 4.0. The final list of genes with significant changes in expression was based on intersection of these two lists.
Murine xenograft model
Female SCID/NOD mice (6–8-week-old; pathogen free; Harlan Sprague Dawley Inc., Indianapolis, IN, USA) were purchased and housed in a specific pathogen free, AAALAC accredited facility in autoclaved static microisolation caging and paper bedding with ad libitum irradiated rodent diet (Harlan Teklad Tek-Fresh Bedding and 2918 Teklad Global rodent diet, Madison WI, USA) and acidified filtered drinking water (HydroPac water pouches, Lab Products Inc., Seaford, DE, USA). In addition to pathogen-free vendor health reports, the animal resource facility conducts periodic quality assurance health testing and assures that these mouse populations have remained free of pathogenic and opportunistic infectious agents. The macroenvironmental conditions within the room were maintained at 20–24.4 °C (68–76 °F) and 28–35% relative humidity, 10–15 air changes hourly and 12:12 h light:dark cycle. All procedures with animals complied with our Public Health Services Policy on humane care and use of laboratory animals and according to the animal protocol approved by the Institutional Animal Care and Use Committee. Within a set of 24 SCID/NOD mice, one was used to harvest control normal histologies, 11 were assigned to the control group (precursor-B ALL without GSI-I) and 12 animals were assigned to the treatment group (precursor-B ALL with GSI-I). After 28 days, mice were subcutaneously injected with phosphate-buffered saline-vehicle or GSI-I (5 mg/kg) in 250 μl of normal saline daily over a 12-day period (5 days on, 2 days off, 5 days on). Surviving mice were killed on day 72 after engraftment. In cases where the animals lost >10% bodyweight, exhibited organomegally or showed signs of decreased movement, following euthanasia, necropsies were performed using standard IACUC-approved techniques.
Histology and immunohistochemistry
Blood and bone marrow samples from control or engrafted mice were slide mounted and Wright stained. For each sample, percent blasts were scored in triplicate for groups of 100 peripheral mononuclear blood cells. Liver, spleen and bone marrow sections were fixed in 10% neutral-buffered formalin (Sigma-Aldrich, St Louis, MO, USA), dehydrated and embedded in paraffin, sectioned and stained with hematoxylin and eosin. For immunohistochemistry, bone marrow sections were rehydrated through a xylene followed by the graded alcohol series. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide and antigen retrieval was performed by pressurized heating in a decloaking chamber (Biocare Medical, Concord, CA, USA) in 0.01 citrate buffer (pH 6.0). Slide-mounted samples were blocked in phosphate-buffered saline containing 5% goat serum/0.5% Tween 20 for 30 min then sequentially incubated with rabbit anti-CD10 antibodies (Abcam, 1:50) at 4 °C and goat anti-rabbit immunoglobulin G–horseradish peroxidase secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:500) with washes between steps. Slides were stained with 3′3-diaminobenzidine (BD Pharmingen, Sparks, MD, USA), counterstained with hematoxylin (Sigma-Aldrich) and dehydrated before coverslips were mounted. Analysis was performed using a Zeiss Axioscope upright microscope (Thornwood, NY, USA) equipped with a digital camera.
Results
GSI-I induces death in cultured precursor-B ALL cells and freshly isolated lymphoblasts
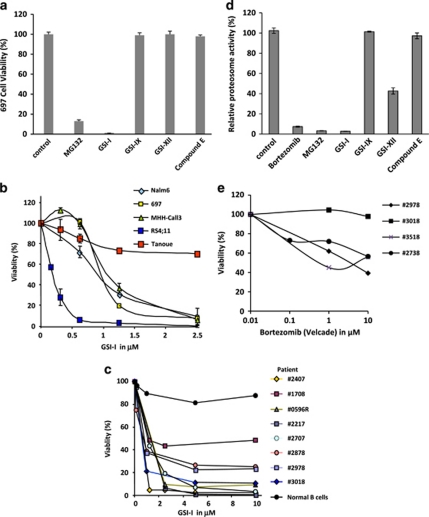
Mutations in the Notch signaling complex have been identified in approximately 50% of patients with T-ALL, leading to the understanding that Notch1 activation may be an important aspect of T-ALL leukemogenesis.18 As GSIs are important inhibitors of Notch, select lead candidates are being investigated in T-ALL phase 1 clinical trials.1 Equally important, Notch signaling has been shown to modulate early B-cell developmental events,4, 5, 6 leading us to hypothesize that GSIs may also have biological importance in precursor-B ALL. To evaluate the biological activity of GSI compounds in precursor-B ALL blasts, we tested a panel of GSI compounds including GSI-I (Z-LLNle-CHO), GSI-IX (DAPT), GSI-XII (X-IL-CHO) and compound E for their biological activity in the 697 precursor-B cell line. Among the compounds tested, GSI-I and the proteosome inhibitor MG132 caused a >80% reduction in cell viability (Figure 1a), while GSI-IX (DAPT), GSI-XII and compound E had no significant effects. On the basis of these results, we selected GSI-I as our lead candidate for further testing in four precursor-B ALL cell lines (Nalm6, 697, MHH-Call3 and RS4:11), which represent important precursor-B ALL cytogenetic and phenotypic subclasses (Supplementary Table 1). For comparison, we included the B-ALL Tanoue B-cell line, which is derived from a Burkitt's lymphoma and expresses mature BCR. Data in Figure 1b compare the dose response for GSI-I treatment of these cell lines, as measured by WST-1 cell viability assay. Nalm6, 697 and MHH-Call3 cells had similar responses, with ∼50% cell death after 24 h treatment with 1.25 μ GSI-I and >90% cell death for 2.5 μ GSI-I. The RS4;11 pre-B-cell line was very sensitive to the drug, with cell death occurring at only 0.5 μ GSI-I. In contrast, 2.5 μ GSI-I caused only about 20% cell death in the more mature Tanoue cell line. The next set of experiments (Figure 1c) show that GSI-I (2.5 μ, 18 h) induced cell death in CD19+ precursor-B ALL blasts isolated from eight patients, including two relapse cases (#0596R and #2707R). In contrast, peripheral blood B cells, isolated from the buffy coat of a healthy control were >90% viable after the 18-h treatment period.
Figure 1.
GSI-I treatment induces apoptosis in pre-B-cell lines and primary patient blasts. (a) Precursor-B ALL cell viability for 697 cells was abolished after 18 h treatment with GSI-I (Z-LLNle-CHO), reduced by 80% after treatment with MG132 and unaffected by treatment with three other GSI compounds. (b) GSI-1 induces cell death in precursor-B ALL cell lines with different cytogenetic characteristics. Tanoue B cells were relatively resistant to GSI-I. (c) Overnight treatment of blast cells from children and young adult patients led to 50–99% loss in cell viability, compared with only 10% loss of viability for normal peripheral blood B cells. (d) GSI-I inhibits proteosome activity at levels comparable to MG132 and bortezomib. (e) Bortezomib-induced cell death in patient samples is comparably less than that induced by GSI-I, as demonstrated in (d).
As the precursor-B ALL cell lines showed a response to the MG132 proteosome inhibitor, we next tested whether the same panel of GSIs had comparable effects to proteosomal inhibitorial effects of bortezomib. In Figure 1d, we compare the relative inhibition of proteosome activity for bortezomib and MG132 (two known proteosome inhibitors) with four γ-secretase inhibitors. GSI-I essentially abolished proteosome activity while GSI-XII showed ∼50% inhibition. Neither GSI-IX nor compound E had significant effects on proteosome activity. Results in Figure 1e show that blasts from individual patients can be relatively resistant to the proteosome inhibitor, bortezomib. Of the four samples tested, blasts from patient 3018 remained 100% viable after 24-h incubation with 1–10 μ bortezomib. This contrasts with only 10% viability for blasts from this patient after 24-h incubation with GSI-I (Figure 1c). Blasts from three other patients showed only 40–60% loss of viability at the highest bortezomib concentration. Together, these data suggested to us that the effectiveness of GSI-I might be explained by disruption of other pathways that synergize with Notch pathway inhibition to induce cell death. These results also allowed us to conclude that GSI-I has biological activity in precursor-B ALL cell lines and, equally important, in a diverse array of patient samples having intermediate and high-risk characteristics.
Evidence that GSI-I causes cell death through both Notch-dependent and Notch-independent pathways
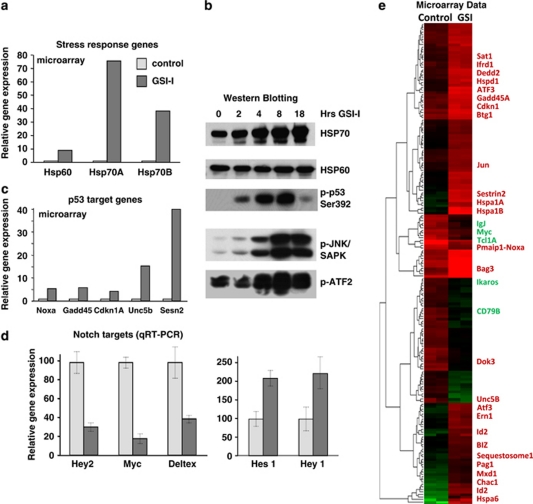
To address our hypothesis that GSI-I induces cell death in pathways that interact synergistically with Notch, we evaluated global gene expression changes in GSI-I-treated 697 precursor-B ALL cells (6 h) using Affymetrix microarrays. Results were dramatic, with >2-fold changes in expression of over 1300 genes within 6 h of GSI-I treatment. Figures 2a–d illustrate results for three classes of transcriptional targets that stood out in the data set. First, we noted that many of the genes upregulated after GSI-I treatment were signatures of stress response. The most remarkable examples are mRNA transcripts encoding HSP70 family members: HSPA6 and HSPA1A, whose expression increased 500- and 75-fold, respectively (Figure 2a). Levels of DNAJB1 mRNA, which encodes an HSP40 protein, also rose at 12–14-fold. In addition, there are transcriptional changes typical for unfolded protein response. GSI-I treatment of precursor-B ALL cells induced remarkable upregulation of CHAC1 (up 56-fold) and ATF3 (up 15-fold), two genes whose expression was recently associated with chemically induced unfolded protein stress response.19 For probeset IDs, see Supplementary Tables 3A and B. Importantly, western blots in Figure 2b confirm protein signatures consistent with stress response (elevated levels of HSP70 as well as phosphorylated forms of JNK and ATF2).
Figure 2.
Changes in levels of gene transcripts and proteins after GSI-I treatment in 697 cells. Graphs show relative changes in gene expression for heat-shock proteins (a), p53 target genes (c) or Notch targets (d) after GSI treatment. Western blots in (b) show compare levels of heat shock proteins and show increases in phoshorylation of JNK and ATF2. (e) Heatmap of signal intensity (in Affymetrix units) for probesets that were more than threefold increased or decreased after 6 h treatment of 697 cells with 2.5 μ GSI-I. Select genes are listed on the right and labeled either in red (increased) or green (decreased). Data are representative of two independent experiments.
A second important category of transcriptional alterations were signatures of p53 induction (Figure 2c), including marked upregulation of SESN2 and UNC5B expression. GSI-treatment induced smaller, but significant, induction of p53 target genes NOXA, GADD45 and CDKN1A. Consistent with these results, western blots in Figure 2b show an increase in p53 phosphorylation after GSI-I treatment.
The third important category reflects changes in known Notch target genes, an expected outcome of GSI-I treatment. Figure 2d reports these data, as measured by microarray methods and confirmed by quantitative PCR. Expression of Hey2, Myc and Deltex were reduced 2–4-fold after 6 h of GSI-I treatment. In contrast, expression of Hes1 and Hey1 increased by 4–20-fold in cells treated with GSI-I.
Expression profiling demonstrate GSI-I-induced transcriptional changes in B cells
In addition to the three main categories detailed in Figures 2a–d, the heatmap representation in Figure 2e illustrates the top 190 genes displaying the most dramatic and consistent changes in the precursor-B ALL transcription after GSI-I treatment. Genes of particular interest with respect to B-cell function or development are indicated to the right of the heatmap. A more extensive listing of gene expression differences after GSI-I treatment is found in Supplementary Tables 3A, B. Notable downregulated genes include CD79B, which encodes the immunoglobulin-β subunit common to both the mature and pre-BCR. There was an 85% drop in levels of CXXC5, implicated in nuclear factor (NF)κB activation.20 In contrast, there was a 12-fold induction of ID2, whose overexpression is associated with a block of B-cell development at the pre–pro B-cell state and with inhibition of the B-cell commitment factor, E2A.21 These results show that multiple pathways are affected by GSI-I, including signaling pathways that regulate NFκB activity and B-cell differentiation.
GSI-I treatment blocks Akt-mediated pro-survival pathways and induces caspase- and ROS-dependent cell death
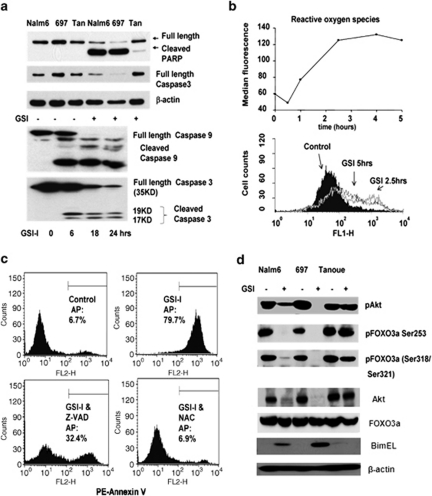
Cell death in GSI-treated cells exhibits typical hallmarks of apoptosis, including increases in annexin V-PE and 7-AAD staining, as well as fragmentation of nuclei (Supplementary Figure 1). To evaluate the mechanisms leading to apoptosis, we next evaluated the effect of GSI-I on potential regulators of apoptosis. The proapoptotic caspases become active following proteolytic cleavage of their proenzyme forms. GSI-I caused a reduction in full-length caspase 3 in Nalm6 and 697 cells (Figure 3a), with a complementary increase in cleaved PARP, a caspase 3 substrate. As shown in Figure 3a, caspase 3 cleavage began as early as 6 h into GSI-I treatment and was mostly complete by 18 h, and the cleaved form of caspase 9 was also detected in GSI-treated 697 cells. GSI-I treatment also resulted in the accumulation of ROS, as demonstrated in 697 cells loaded with the dye, CM-H2DCFDA (Figure 3b). These results suggest that GSI-I-mediated cell death in precursor-B ALL is importantly dependent on both caspase activation and ROS production. Supporting this, data in Figure 3c show that the general caspase inhibitor Z-VAD significantly prevented cell death in 697 precursor-B ALL cells treated with GSI-I. The oxidative scavenger, NAC, completely protected the viability of GSI-I-treated 697 cells, indicating a critical role for ROS in this process. By comparison, NAC failed to protect 697 cells from cell death induced by bortezomib (data not shown).
Figure 3.
Induction of proapoptotic pathways after GSI-I treatment. (a) Top: GSI-I treatment leads to PARP and caspase 3 cleavage/activation in Nalm6 and 697 cells but not Tanoue cells. Bottom: time course of caspase 9 and caspase 3 activation in GSI-I-treated 697 cells. (b) Time course of ROS production in 697 cells after addition of 2.5 μ GSI-I. (c) Protection of GSI-induced apoptosis in 697 cells by the caspase inhibitor, Z-VAD or the ROS scavenger, N-acetylcysteine (NAC). (d) Western blotting results show GSI-induced changes in the total AKT, phospho-AKT, phospho-FoxO3a and level of BimEL in lysates from 697, Nalm6 and Tanoue cells.
Results in Figure 3d show that GSI-I treatment led to a drop in total AKT protein levels, with no detectable phospho-AKT in 697 and >70% drop in phospho-Akt in Nalm6 cells (top). GSI-I treatment of both 697 and Nalm6 cells also led to detection of the BH3 domain-only protein, Bim. We hypothesized that the loss of Akt and the production of Bim might be linked, through regulation of the transcription factor FoxO3a (Forkheadbox, class O) that contains multiple Akt consensus sequences (RXRSSS/T).22 FoxO3 phosphorylation prevents nuclear transport of FoxO3a where it targets the FHER (FoxO-binding site) in the Bim promoter.23 We utilized phospho-specific FoxO3a antibodies to probe membranes bearing sodium dodecyl sulfate-polyacrylamide gel electrophoresis separated proteins from untreated and GSI-I-treated cells. Levels of total FoxO3 were unperturbed by GSI-I treatment, but FoxO3 serine/threonine phosphorylation was essentially ablated. Increase in FoxO3 transcriptional activity, associated with its dephosphorylation after GSI-I treatment, was confirmed by a DNA-pull down assay (Supplementary Figure 2).
Notch receptors, ligands and target genes are all expressed in freshly isolated pre-B lymphoblasts
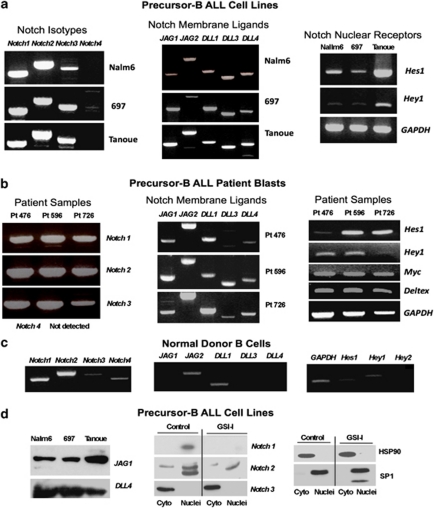
Notch signaling events upregulate the expression of several downstream targets, including HES1, HEY2 and MYC. On the basis of the findings of others,4, 5, 6 Notch receptors and ligands appear to be important targets of γ-secretase activity in cells of B-cell derivation. To evaluate Notch gene expression in our precursor-B ALL cell lines and patient samples, we isolated RNA from the Nalm6, 697 and Tanoue cell lines, as well as from CD19+ cells recovered from the bone marrow of pediatric precursor-B ALL patients. RT-PCR analysis showed that all cell lines had strong signals for Notch1, Notch2 and Notch3, but not for Notch4 (Figure 4a). All cell lines also had strong expression Notch targets JAG1, JAG2, DLL1, DLL3 and DLL4. Similar results in the GSI-I sensitive pre-B ALL cell lines, MHH-Call3 and RS4;11 (not shown). We note that the more mature Tanoue B-cell line has similar profile of Notch receptors and ligands, but is resistant to GSI-I treatment (Figure 1b). This is consistent with the concept that Notch-independent targets of GSI-I are important contributors to GSI-I toxicity. We next extended our observations for Notch signaling into a cohort of patients with precursor-B ALL. This group of 11 patients harbored a range of cytogenetic abnormalities (Supplementary Table 2). RT-PCR analysis showed that all the patients had strong signals for Notch1, Notch2 and Notch3; with one exception (patient #476), all also expressed all five Notch ligands. Representative results are shown in Figure 4b. To rule out a role for activating mutations in Notch1, the heterodimerization and PEST domains were amplified by RT-PCR and sequenced; no mutations were found. Representative results for Hes1 expression in this patient group are also shown in Figure 4a; again only patient #476 had barely detectable message. In contrast to the findings shown for precursor-B ALL cell lines and patient blasts, Notch signaling is weak in mature, non-malignant donor B cells (Figure 4c). For all experiments, RNA quality was confirmed by RT-PCR analysis for the housekeeping gene, GAPDH. These data show that Notch signaling constitutively occurs in precursor-B ALL lines and most pediatric B-ALL primary patient samples.
Figure 4.
Leukemic pre-B cells express Notch receptors, ligands and target genes. RT-PCR results for Notch receptors, ligands and target genes in blasts from (a) three precursor-B ALL patients and normal peripheral blood B cells, (b) B lineage cell lines or (c) normal peripheral blood B cells. (d) Western blots results at left detect JAG1 and DLL4 in 697 cell lysates. Results at right report relative distribution of Notch1, Notch2 and Notch3 ICNs between cytoplasmic and nuclear fractions prepared from 697 cells, ±GSI treatment. SP1 and Hsp90 serve as controls for nuclear and cytosol fractions. Note that a truncated form of SP1 is found in the nucleus of GSI-I-treated cells, consistent with induction of apoptosis.
Protein levels of ligands and cleaved Notch receptors
Western blotting was used to confirm the presence of Notch ligand and receptor expression in precursor-B ALL cells. Figure 4d documents presence of Jagged 1 and DLL4 protein expression in precursor-B ALL cell lines. This figure also illustrates detection of the cleaved intracellular Notch (ICN) forms of Notch 1 and Notch 2 in nuclear fractions prepared from the 697 cell line. Two bands were detected for ICN2, potentially representing different posttranslational modifications. In contrast, the cleaved form of Notch 3 (MW 97 kD) was detected only in the cytosol fraction. HSP90 and the nuclear factor, SP1, served as markers for cytosol and nuclear fractions. Importantly, GSI-I treatment led to a loss of both cleaved Notch 1 (ICN1) and cleaved Notch 2 (ICN2) in the nuclear fractions of 697 cells. We note that these results were obtained using pan-reactive antibodies to Notch 1, not the commonly used NCT-Val 1744 mAb that is specific to ICN1.24 The reasons for our inability to detect ICN1 using this reagent is unknown; possible explanations include poor sensitivity because of low levels (ICN1 is rapidly degraded24), or differential proteolysis by the presenilin-1 form of γ-secretase. GSI-I failed to reduce levels of activated Notch3 in pre-B cells. Together with the lack of Notch3 ICN in the nucleus, we assume that Notch3 has negligible roles in the events that follow GSI-I treatment.
Genomic analysis of Notch-related genes in a high-risk pediatric cohort
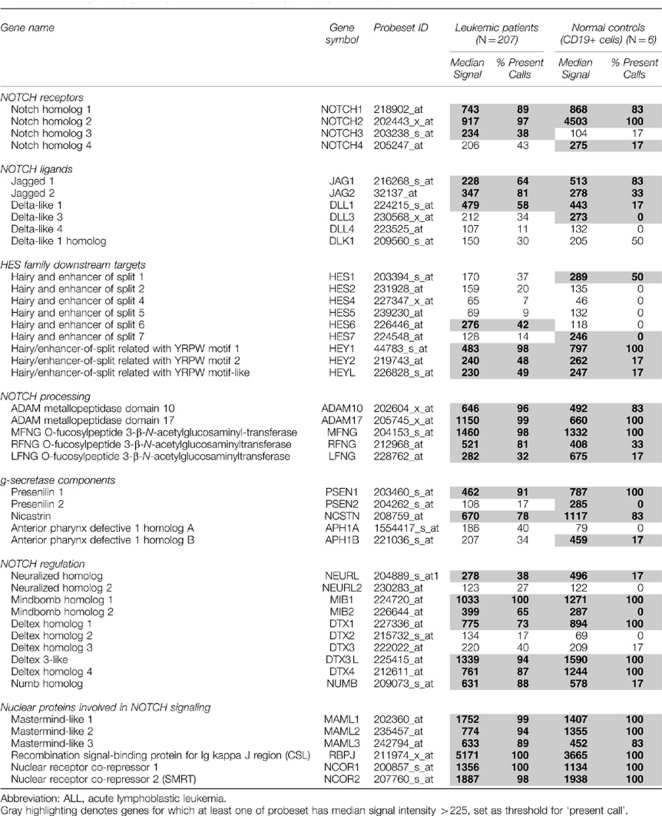
Gene profiling provides a platform to screen a large number of patients for alterations in RNA expression levels. We chose to extend our observations regarding Notch signaling in precursor-B ALL cells for three reasons. First, our findings in 11 patients may have been unique to a limited subset of patients, and not representative of a larger sample size. Second, while Notch-activating mutations have been well described in patients with T-ALL, the link between Notch signaling and precursor-B development is poorly understood. Third, we wished to investigate whether Notch expression patterns are restricted to specific, genetically defined patient subsets. Table 1 reports analysis of an extensive Affymetrix data set recently completed for 207 children and young adults enrolled on COG 9906 for high-risk precursor-B ALL. CD19+ cells from healthy bone marrow donors served as a normal control. Expression patterns of 45 genes are reported here, representing all Notch receptor and ligands as well as proteins critical to Notch processing, signaling and regulation. Gray shading in the table highlights genes where at least one probe set had signal intensity above a threshold value of 225 normalized Affymetrix units. The table reports the median signal for all patient samples across the gene probe set, as well as the number of patients with values above the threshold for expression of the gene. Approximately 97% of the high-risk precursor-B ALL patients expressed significant amounts of NOTCH2, comparable to normal CD19+ controls. NOTCH1 expression was significant in 89% of patients, while NOTCH3 was present in <40% of the samples. The signal for NOTCH4 was typically below the threshold, as in our patient sub-set (Figure 4b). Ligand expression was more variable, with only 60–80% expressing JAG1, JAG2 and DLL1. This finding is expected in the high-risk group, at least for JAG1, whose expression was previously correlated with good outcome.25 Importantly, the majority of Notch pathway-related genes were significantly expressed in the high-risk ALL cohort.
Table 1. Notch pathway gene expression in 207 high-risk pediatric pre-B ALL patients.
Interesting exceptions were noted. For example, 91% of patient samples and 100% of normal CD19+ controls had significant expression of the gene (PSEN1) encoding the γ-secretase component, presenilin 1. In contrast, expression of the gene (PSEN2) encoding presenilin 2 was virtually absent from patient samples and very low in normal CD19+ controls. We observed strong correlation between expression of pairs of genes in this analysis set (Supplementary Table 4). Thus, genomic analyses reveal consistent expression of >30 additional Notch genes whose products are necessary for Notch trafficking and processing or that act to amplify or repress Notch transcriptional events. Finally, by including data from two study groups, we demonstrate that Notch gene expression is not linked specifically to any of the genetic abnormalities that define precursor-B ALL subsets. The 11 patient institutional group included t(4;11) and t(1;19) translocations, as well as both hyperdiploid and hypodiploid cases. The high-risk cohort used for genomic analysis includes E2A/PBX (t(1;19)), BCR/ABL (t(9;22)) and mixed lineage leukemia rearrangements, as well as many others.
GSI-I shows therapeutic potential in a murine xenograft model of human precursor-B ALL
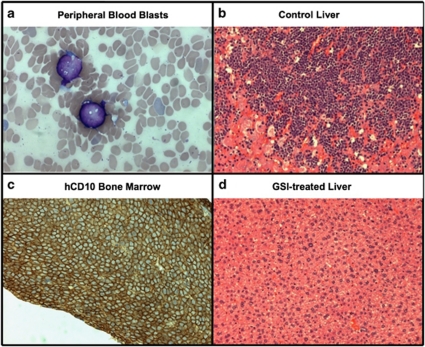
Given our findings that GSI-I is effective in eliciting cell death in precursor-B ALL cell lines and primary patients samples, it is reasonable to ask if the compound might be an effective therapeutic agent. We explored this possibility using NOD/SCID mice engrafted with human pre-B ALL blasts. On day 1 of the study, 23 mice were injected subcutaneously with 5 × 106 viable 697 cells; an additional mouse was used as a negative histology control. To avoid potential drug–drug interactions, GSI-I was used as a single agent in the experimental group (n=12). After a 1-month incubation period, GSI treatment was initiated in the experimental group with dosing schedule consisting of 5 days on, 2 days off and 5 days on. The 11 mice in the engrafted control group were given normal saline injections under the same schedule. Within 50–64 days, two mice in the control group and one mouse in the treated group were killed because of signs of progressive disease, as evidenced by weight loss, loss of appetite and decreased movement. At necropsy, affected mice showed significant disease burden in the liver and bone marrow (Figures 5a–c). We performed post-euthanasia necropsies on all remaining mice in both groups at day 72 and found that all eight surviving mice in the control group had profound histologic evidence of disease.
Figure 5.
Detection of precursor-B ALL blasts in blood (a), bone marrow (b) and liver (c) after engraftment of nonobese diabetes/severe combined immunodeficiency (NOD/SCID) mice with 697 cells. The image in (d) shows a liver free of blasts in a GSI-treated animal. (a), (c) and (d) are hematoxylin and eosin (H&E)-stained sections; the bone marrow sample in (b) shows immunohistochemical staining for human CD10, to distinguish blast cells from any potential leukocyte progenitors in this immunocompromised mouse.
Notably, only half of the 12 GSI-I-treated group showed signs of disease (Table 2). The effect on GSI-I on disease progression in the NOD/SCID xenograft model of precursor-B ALL was statistically significant (P=0.0091, log-rank test). This is a remarkable finding, because it was used as a single agent for only one course of therapy (Figure 5d).
Table 2. Summary of results in human precursor-B ALL xenograft model.
| Mouse number | % Blasts in blood smear | % Blasts in bone marrow aspirate | Liver infiltration | hCD10 in bone marrow |
|---|---|---|---|---|
| Untreated | ||||
| 1 | 0 | 8–11 | + | +++ |
| 2 | 0 | 3–6 | ++ | + |
| 3 | 4–6 | 39–44 | − | ++ |
| 4 | ND | ∼100 | +++ | +++ |
| 5 | 0–1 | 2 | + | + |
| 6 | 0 | 2 | − | + |
| 7 | 51–56 | ∼100 | +++ | +++ |
| 8 | 41–43 | ∼100 | +++ | +++ |
| 9 | 8–10 | 41–46 | ++ | +++ |
| 10 | 12–15 | 39–43 | ++ | ++ |
| 11 | 0 | 0–1 | + | + |
| GSI-I treated | ||||
| 1a | 0 | 0 | − | − |
| 2 | 41–45 | ∼100 | +++ | +++ |
| 3a | 0 | 0 | − | − |
| 4 | 1–3 | 6–11 | ++ | +++ |
| 5a | 0 | 0 | − | − |
| 6 | 3–6 | ND | ++ | + |
| 7 | 8–11 | ND | ++ | ++ |
| 8 | 19–22 | 56–58 | +++ | +++ |
| 9a | 0 | 0 | − | − |
| 10a | 0 | 0 | − | − |
| 11 | 8–12 | 44–46 | +++ | +++ |
| 12a | 0 | 0 | − | − |
Abbreviations: ALL, acute lymphoblastic leukemia; GSI, gamma secretase inhibitor; ND, not done.
In all, 6 of 12 treated mice free of disease after GSI-I treatment.
+, few nodules (1–5).
++, multiple nodules (>5).
+++, large areas infiltrated.
Discussion
The therapeutic potential of γ-secretase inhibitors in the human acute leukemias has focused on T-ALL,1 where up to 50% of cases harbor activating Notch 1 mutations.26 The involvement of Notch in early B-cell malignancies was largely dismissed, based on Notch1 association with the T-cell lineage commitment and evidence that overexpression of ICNs or Hes1 induces apoptosis in pre-B cell lines.27 Nevertheless, there are previous reports of JAG1 expression in precursor-B ALL cells25 and Notch receptors, ligands and targets have been implicated at various stages of B-cell development.28 Notch dysregulation has been documented in mature B-cell neoplasms, including multiple forms of lymphoma, B-cell chronic lymphoid leukemia and multiple myeloma.5, 9, 29, 30, 31 In mice, retroviral-mediated gene disruption implicated Notch-related genes (Notch 2, presenilin 1, Dtx2, Numb and ADAM) in pro-B and pre-B leukemogenesis,32 while long-term GSI treatment led to altered maturation of peripheral B cells.33 Here, we definitively show that expression of both Notch ligands and receptors are common features of leukemic B-cell precursors. This conclusion is based on analysis of 11 institutional patients and 207 high-risk patients enrolled in COG P9906. We further show that GSI-I induces cell death in precursor-B ALL cells ex vivo and in vivo using cell survival assays and a NOD/SCID xenograft model.
There are clear differences between Notch gene expression patterns and mutational status in the two lymphoid forms of acute leukemia, precursor-B ALL and T-ALL. First, comparison of gene expression profiles reported of T-ALL cells treated with GSIs34 shows only a small percentage of overlap between the gene sets in these studies, most notably the Notch target Myc. Massive induction of HSP70 and other stress response genes was not seen in T-ALL cells treated with DAPT or compound E,11, 34 despite previous connections between presenilin function and the unfolded protein response.35 The unexpected induction of Hes1 by GSI treatment (Figure 4) was also reported by others for B-cell leukemia and acute myeloid leukemia cell lines.4 Hes1 expression is also linked to activation of the JNK stress response pathway, which is elevated after GSI-I treatment (Figure 2b).36 Another explanation may be cross-talk between the Notch and NFκB pathways,37 because IκBα can bind the Hes1 promoter and suppress Hes1 gene expression.38 Active Notch may compete with IκBα to bind with NFκB, thereby enabling increased Hes1 expression. By 6 h treatment with GSI, several genes associated with the NFκB pathway have significantly altered expression levels in precursor-B ALL cells (Supplementary Tables 3A and B).
We found that transcription of most Notch target genes in precursor-B ALL is modest compared with that reported for T cells. As a result of low expression levels, confirmation by quantitative PCR methods is an important part of our study. We suspect that Notch signaling at this stage of B-cell development is held in check by inhibitory cis-interactions between receptors and ligands expressed in the same cell, as recently demonstrated in elegant studies by Sprinzak et al.39 We note also that γ-secretase may have other significant targets in precursor-B ALL leukemia cells. CD44 (ref. 8) is one likely candidate that merits further study.
Others have raised the possibility that GSIs might inhibit other cellular proteases.3 Our study confirms that GSI-I also inhibits the proteosome, an important cancer therapeutic target.14, 15, 39 Importantly, both protease pathways are mechanistically linked to apoptosis. Caspase activation and oxidative stress was found to be synergistic in chronic lymphoid leukemia cells treated with the proteasome inhibitor NPI-0052.40 ROS production distinguishes the killing activity of GSI-I from bortezomib (Figure 3), consistent with disruption of Notch-mediated anti-apoptotic pathways that protect mitochondrial function and prevent induction of pro-apoptotic Bcl2 family members like Bim41 and Bax.42
The significant enhancement in ALL blast cell killing because of GSI-I's dual inhibitory properties follows the recent trend of combined, targeted therapeutic regimens. In T-ALL, recent studies in mice have suggested that the combination of dexamethasone with compound E could enhance toxicity while protecting against GSI-induced gut toxicity.40 Other promising studies in mice combined a 3-day on/4-day off GSI dosing schedule with an mammalian target of rapamycin inhibitor.43 Our animal studies also showed a significant protective effect on an intermittent schedule, supporting the concept that short-term or periodic treatment with inhibitors (like GSI-I) that target multiple proteases might sensitize precursor-B ALL leukemia cells to chemotherapeutic reagents with less severe toxicity to normal organs. With these findings, we wish to further explore the effects of GSI-I in additional pre-clinical studies.
Acknowledgments
We thank the COG Cell Bank for the provision of patient samples and annotated demographic data, the UNM Pediatric Hematology/Oncology division for participation in acquiring consented samples, and Dr Kevin O'Hair for his administrative and technical support of the animal research studies. We acknowledge the Flow Cytometry, Microscopy and Keck-UNM Genomics Resource Core Facilities at UNM Cancer Center. We also thank Anna Holmes for technical assistance. This work was funded by the Leukemia Lymphoma Society SCOR program (LLS 7388–06), NIH (NCI SPECS 114762) and COG Grants CA98543 and CA114766.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15:50–58. doi: 10.1038/nm.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton TM, Pati D, Plon SE, Thompson PA, Bomgaars LR, Adamson PC, et al. A phase 1 study of the proteasome inhibitor bortezomib in pediatric patients with refractory leukemia: a Children's Oncology Group study. Clin Cancer Res. 2007;13:1516–1522. doi: 10.1158/1078-0432.CCR-06-2173. [DOI] [PubMed] [Google Scholar]
- Han J, Ma I, Hendzel MJ, Allalunis-Turner J. The cytotoxicity of gamma-secretase inhibitor I to breast cancer cells is mediated by proteasome inhibition, not by gamma-secretase inhibition. Breast Cancer Res. 2009;11:R57. doi: 10.1186/bcr2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoshi H, Sato T, Koyama T, Nara N, Tohda S. Gamma-secretase inhibitors suppress the growth of leukemia and lymphoma cells. Oncol Rep. 2007;18:77–80. [PubMed] [Google Scholar]
- Nefedova Y, Sullivan DM, Bolick SC, Dalton WS, Gabrilovich DI. Inhibition of Notch signaling induces apoptosis of myeloma cells and enhances sensitivity to chemotherapy. Blood. 2008;111:2220–2229. doi: 10.1182/blood-2007-07-102632. [DOI] [PubMed] [Google Scholar]
- Rosati E, Sabatini R, Rampino G, Tabilio A, Di Ianni M, Fettucciari K, et al. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood. 2009;113:856–865. doi: 10.1182/blood-2008-02-139725. [DOI] [PubMed] [Google Scholar]
- Masuda K, Germeraad WT, Satoh R, Itoi M, Ikawa T, Minato N, et al. Notch activation in thymic epithelial cells induces development of thymic microenvironments. Mol Immunol. 2009;46:1756–1767. doi: 10.1016/j.molimm.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Miele L, Miao H, Nickoloff BJ. NOTCH signaling as a novel cancer therapeutic target. Curr Cancer Drug Targets. 2006;6:313–323. doi: 10.2174/156800906777441771. [DOI] [PubMed] [Google Scholar]
- Ehebauer M, Hayward P, Arias AM. Notch, a universal arbiter of cell fate decisions. Science. 2006;314:1414–1415. doi: 10.1126/science.1134042. [DOI] [PubMed] [Google Scholar]
- Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci USA. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungamuri SK, Yang X, Thor AD, Somasundaram K. Survival signaling by Notch1: mammalian target of rapamycin (mTOR)-dependent inhibition of p53. Cancer Res. 2006;66:4715–4724. doi: 10.1158/0008-5472.CAN-05-3830. [DOI] [PubMed] [Google Scholar]
- Bheeshmachar G, Purushotaman D, Sade H, Gunasekharan V, Rangarajan A, Sarin A. Evidence for a role for notch signaling in the cytokine-dependent survival of activated T cells. J Immunol. 2006;177:5041–5050. doi: 10.4049/jimmunol.177.8.5041. [DOI] [PubMed] [Google Scholar]
- Bianchi G, Oliva L, Cascio P, Pengo N, Fontana F, Cerruti F, et al. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood. 2009;113:3040–3049. doi: 10.1182/blood-2008-08-172734. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui CH. T cell acute lymphoblastic leukemia: NOTCHing the way toward a better treatment outcome. Cancer Cell. 2009;15:85–87. doi: 10.1016/j.ccr.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendino F, Nguyen E, Jonassen I, Dysvik B, Azouz A, Lanotte M, et al. Functional involvement of RINF, retinoid-inducible nuclear factor (CXXC5), in normal and tumoral human myelopoiesis. Blood. 2009;113:3172–3181. doi: 10.1182/blood-2008-07-170035. [DOI] [PubMed] [Google Scholar]
- Johnson NA, Boyle M, Bashashati A, Leach S, Brooks-Wilson A, Sehn LH, et al. Diffuse large B-cell lymphoma: reduced CD20 expression is associated with an inferior survival. Blood. 2009;113:3773–3780. doi: 10.1182/blood-2008-09-177469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenkamp KU, Coffer PJ. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem Soc Trans. 2003;31 (Part 1:292–297. doi: 10.1042/bst0310292. [DOI] [PubMed] [Google Scholar]
- Essafi A, Fernandez de Mattos S, Hassen YA, Soeiro I, Mufti GJ, Thomas NS, et al. Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene. 2005;24:2317–2329. doi: 10.1038/sj.onc.1208421. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario G, Stanulla M, Fine BM, Teuffel O, Neuhoff NV, Schrauder A, et al. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105:821–826. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- Gridley T. Kick it up a Notch: NOTCH1 activation in T-ALL. Cancer Cell. 2004;6:431–432. doi: 10.1016/j.ccr.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Zweidler-McKay PA, He Y, Xu L, Rodriguez CG, Karnell FG, Carpenter AC, et al. Notch signaling is a potent inducer of growth arrest and apoptosis in a wide range of B-cell malignancies. Blood. 2005;106:3898–3906. doi: 10.1182/blood-2005-01-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Pear WS. Notch signalling in B cells. Semin Cell Dev Biol. 2003;14:135–142. doi: 10.1016/s1084-9521(02)00182-9. [DOI] [PubMed] [Google Scholar]
- Tohda S, Sato T, Kogoshi H, Fu L, Sakano S, Nara N. Establishment of a novel B-cell lymphoma cell line with suppressed growth by gamma-secretase inhibitors. Leuk Res. 2006;30:1385–1390. doi: 10.1016/j.leukres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Houde C, Li Y, Song L, Barton K, Zhang Q, Godwin J, et al. Overexpression of the NOTCH ligand JAG2 in malignant plasma cells from multiple myeloma patients and cell lines. Blood. 2004;104:3697–3704. doi: 10.1182/blood-2003-12-4114. [DOI] [PubMed] [Google Scholar]
- Shin DM, Shaffer DJ, Wang H, Roopenian DC, Morse HC., III NOTCH is part of the transcriptional network regulating cell growth and survival in mouse plasmacytomas. Cancer Res. 2008;68:9202–9211. doi: 10.1158/0008-5472.CAN-07-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser KC, Liu B, Hansen GM, Skapura D, Hentges KE, Yarlagadda S, et al. Retroviral insertions in the VISION database identify molecular pathways in mouse lymphoid leukemia and lymphoma. Mamm Genome. 2007;18:709–722. doi: 10.1007/s00335-007-9060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson MB, Thakkar S, Hix JK, Bhandarkar ND, Wong A, Schreiber MJ. Pathophysiology, clinical consequences, and treatment of tumor lysis syndrome. Am J Med. 2004;116:546–554. doi: 10.1016/j.amjmed.2003.09.045. [DOI] [PubMed] [Google Scholar]
- Dohda T, Maljukova A, Liu L, Heyman M, Grander D, Brodin D, et al. Notch signaling induces SKP2 expression and promotes reduction of p27Kip1 in T-cell acute lymphoblastic leukemia cell lines. Exp Cell Res. 2007;313:3141–3152. doi: 10.1016/j.yexcr.2007.04.027. [DOI] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Sato N, Miyoshi K, Kudo T, Hitomi J, et al. Presenilin-1 mutations downregulate the signalling pathway of the unfolded-protein response. Nat Cell Biol. 1999;1:479–485. doi: 10.1038/70265. [DOI] [PubMed] [Google Scholar]
- Curry CL, Reed LL, Nickoloff BJ, Miele L, Foreman KE. Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab Invest. 2006;86:842–852. doi: 10.1038/labinvest.3700442. [DOI] [PubMed] [Google Scholar]
- Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- Aguilera C, Hoya-Arias R, Haegeman G, Espinosa L, Bigas A. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc Natl Acad Sci USA. 2004;101:16537–16542. doi: 10.1073/pnas.0404429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak D, Lakhanpal A, Lebon L, Santat LA, Fontes ME, Anderson GA, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465:86–90. doi: 10.1038/nature08959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CP, Rudra S, Keating MJ, Wierda WG, Palladino M, Chandra J. Caspase-8 dependent histone acetylation by a novel proteasome inhibitor, NPI-0052: a mechanism for synergy in leukemia cells. Blood. 2009;113:4289–4299. doi: 10.1182/blood-2008-08-174797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz JB, Bremen D, Reed JC, Lommatzsch J, Takayama S, Wullner U, et al. Cooperative interception of neuronal apoptosis by BCL-2 and BAG-1 expression: prevention of caspase activation and reduced production of reactive oxygen species. J Neurochem. 1997;69:2075–2086. doi: 10.1046/j.1471-4159.1997.69052075.x. [DOI] [PubMed] [Google Scholar]
- Perumalsamy LR, Nagala M, Sarin A. Notch-activated signaling cascade interacts with mitochondrial remodeling proteins to regulate cell survival. Proc Natl Acad Sci USA. 2010;107:6882–6887. doi: 10.1073/pnas.0910060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113:6172–6181. doi: 10.1182/blood-2008-02-136762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.