Abstract
Diploid budding yeast exhibits two developmental programs in response to nitrogen starvation, pseudohyphal growth, and sporulation. Here we show that both programs are repressed by activation of the unfolded protein response (UPR), a stress-signal transduction pathway responsible for induction of endoplasmic reticulum (ER)-resident chaperones when protein folding in the ER is impaired. Pseudohyphal growth was derepressed in ire1Δ/ire1Δ and hac1Δ/hac1Δ strains. Activation of the UPR or overexpression of the transcription factor Hac1ip, the product of an unconventional splicing reaction regulated by the UPR, was sufficient for repression of pseudohyphal growth and meiosis. HAC1 splicing occurred in a nitrogen-rich environment but ceased rapidly on nitrogen starvation. Further, addition of ammonium salts to nitrogen-starved cells was sufficient to rapidly reactivate HAC1 splicing. We propose that high translation rates in a nitrogen-rich environment are coupled to limited protein unfolding in the ER, thereby activating the UPR. An activated UPR then represses pseudohyphal growth and meiosis. Nitrogen starvation slows translation rates, allowing for more efficient folding of nascent polypeptide chains, down-regulation of the UPR, and subsequent derepression of pseudohyphal growth and meiosis. These findings significantly broaden the range of physiological functions of the UPR and define a role for the UPR in nitrogen sensing.
Keywords: unfolded protein response, pseudohyphal growth, meiosis, sporulation, nitrogen sensing, translation
Nitrogen starvation triggers one of two developmental responses in diploid cells of the yeast Saccharomyces cerevisiae. Nitrogen starvation in the presence of a fermentable carbon source, such as glucose, results in a morphological change from the yeast to a filamentous or pseudohyphal growth form. The pseudohyphal growth form is characterized by an elongated cell shape, a change to a unipolar budding pattern, adhesion of the cells to each other after cell division has been completed, invasion of the agar, and synchronization of cell division between mother and daughter cells (Kron et al. 1994). Macroscopically, these changes result in colonies with multiple projections radiating away from the center of the colony.
Two signaling cascades are required for induction of pseudohyphal growth. They partially overlap with the mitogen-activated protein kinase (MAPK) pathway that signals the mating pheromone response (Liu et al. 1993) and the cAMP-dependent protein kinase A (PKA) pathway (Gimeno et al. 1992). How both pathways are activated by nitrogen starvation is not very well understood. The high-affinity ammonium permease Mep2p is one of the most upstream elements of the PKA pathway (Lorenz and Heitman 1997) and, thus, may link activation of the PKA pathway to nitrogen starvation (Lorenz and Heitman 1998). However, how Mep2p senses nitrogen and transduces the signal is unknown. In addition to these two pathways, additional genes are involved in regulation of pseudohyphal growth. These include genes that induce pseudohyphal growth when overexpressed (PHD1, PHD3, PHD4, PHD6, PHD7, and MSN1; Gimeno and Fink 1994) and genes with recessive mutations that cause constitutive pseudohyphal growth (ELM1-3, ELM5-8, ELM12, ELM14, and GRR1; Blacketer et al. 1995).
Nitrogen starvation on nonfermentable carbon sources as acetate or ethanol induces meiosis and differentiation into asci in diploid a/α cells (Herskowitz 1988; Kupiec et al. 1997). Meiosis can be divided into at least three distinctive phases: early, middle, and late phases of gene expression. A transcriptional cascade governs entry into meiosis. The first known event is induction of IME1 mRNA by relieving nutritional repression by fermentable carbon sources, that is, glucose, and nitrogen. Once synthesized, Ime1p binds to the transcriptional repressor Ume6p and converts it to an activator, resulting in transcriptional induction of early meiotic genes such as IME2, HOP1, and SPO13 (Rubin-Bejerano et al. 1996; Malathi et al. 1997). Once early genes, including IME2, are induced the protein kinase, Ime2p activates a subset of early genes independent of IME1. Only after completion of the early phase are middle genes expressed.
The nature of the carbon source controls the decision whether filaments or asci are formed when nitrogen becomes limiting. The presence of a fermentable carbon source such as glucose results in formation of filaments, whereas the presence of a nonfermentable carbon source results in sporulation (Gimeno et al. 1992; Donzeau and Bandlow 1999). How this decision is made on a molecular level is not understood.
The unfolded protein response (UPR; for reviews see Chapman et al. 1998; Kaufman 1999) is an unconventional signal transduction pathway that transduces the stress signal for unfolded proteins in the lumen of the endoplasmic reticulum (ER) to the nucleus. Drugs that interfere with N-linked glycosylation, for example, tunicamycin and 2-deoxyglucose, or reduce disulfide bonds, for example, β-mercaptoethanol and dithiothreitol, induce protein unfolding in the ER. Unfolded proteins in the ER sequester Kar2p/BiP/GRP78 from the type I transmembrane kinase/endoribonuclease Ire1p, resulting in its dimerization and autophosphorylation (Bertolotti et al. 2000). In yeast, activated Ire1p cleaves the mRNA for the transcription factor Hac1p. Unspliced HAC1 mRNA (HAC1u) consists of two exons that are separated by a 252-bp intron harboring a translational attenuator. After cleavage of both exon–intron junctions by Ire1p, the exons are joined by tRNA ligase, thereby bypassing the spliceosome. Consequently, the translational block is relieved and spliced HAC1 mRNA (HAC1i) is translated. Hac1ip activates transcription of genes encoding ER-resident chaperones such as KAR2, PDI1, and LHS1 through binding to the UPR-element (UPRE, CAGCGTG) as a homodimer. Globally, the UPR regulates transcription of 381 open reading frames (ORFs) in response to ER stress. Of these, ∼50% of the ORFs with a known function play a role in the secretory pathway (Travers et al. 2000). However, ∼100 ORFs with a known function are induced by the UPR on ER stress and have no known connection to the secretory pathway, indicating that aspects of the secretory pathway are involved in the regulation of more cellular processes than currently appreciated.
Here, we provide evidence that the UPR represses both nitrogen-starvation induced developmental programs, pseudohyphal growth, and meiosis and, thus, contributes to nitrogen sensing in budding yeast. We demonstrate that HAC1 splicing is regulated by extracellular nitrogen sources. HAC1 splicing was stimulated by high extracellular nitrogen levels and ceased rapidly on nitrogen starvation. To explain the dependence of HAC1 splicing on high environmental levels of nitrogen sources, we propose the following model. In a nitrogen-rich environment, protein unfolding is a byproduct of rapid translation, resulting in low-level activation of the UPR and subsequent repression of pseudohyphal growth and meiosis. When nitrogen becomes limiting, translation slows, the amount of unfolded protein in the ER drops below a threshold level that is no longer sufficient to activate the UPR, and repression of pseudohyphal growth and meiosis is relieved. Taken together, these findings demonstrate a novel function for the UPR and the secretory pathway in controlling the global physiology of eukaryotic cells in response to nutrient availability.
Results
Diploid ire1Δ and hac1Δ strains grow constitutively as pseudohyphae
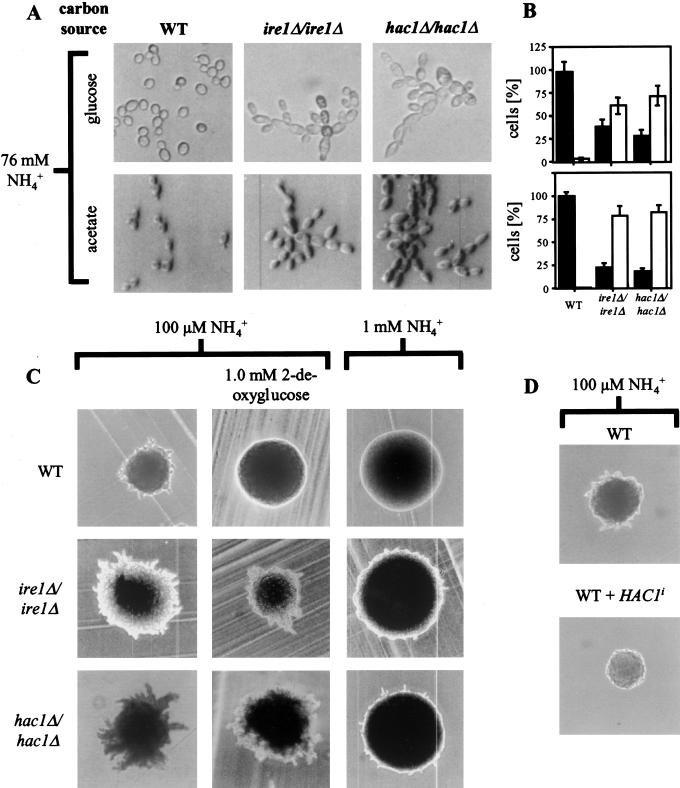
To our surprise, microscopic examination of diploid ire1Δ/ire1Δ and hac1Δ/hac1Δ cells in mid-log phase revealed a striking morphological change (Fig. 1A). Wild-type (WT) diploid cells displayed the familiar yeast morphology, a round or oval cell shape, and mother cells with buds of every different size. In contrast, ire1Δ/ire1Δ and hac1Δ/hac1Δ cells were elongated beyond a length to width ratio of two and formed chains of cells sticking together even after cell division was completed. This morphology of the ire1Δ/ire1Δ and hac1Δ/hac1Δ strains is identical to yeast growing as filaments or pseudohyphae (Gimeno et al. 1992). The same observation was made in the W303 genetic background. ire1Δ/ire1Δ and hac1Δ/hac1Δ strains did not only form pseudohyphae when grown on glucose but also when grown on acetate (Fig. 1A), a condition that represses pseudohyphae formation in WT strains (Donzeau and Bandlow 1999). To quantify this observation, the cells were classed into two groups after their morphological appearance: yeast-form (YF) cells with a length to width ratio less than two, and pseudohyphal (PH) cells with a length to width ratio of greater than or equal to two (Mösch et al. 1996). More than 50% of the ire1Δ/ire1Δ and hac1Δ/hac1Δ cells belonged to the PH group. Surprisingly, the percentage of PH cells was more pronounced on a nonfermentable carbon source like acetate (Fig. 1B).
Figure 1.
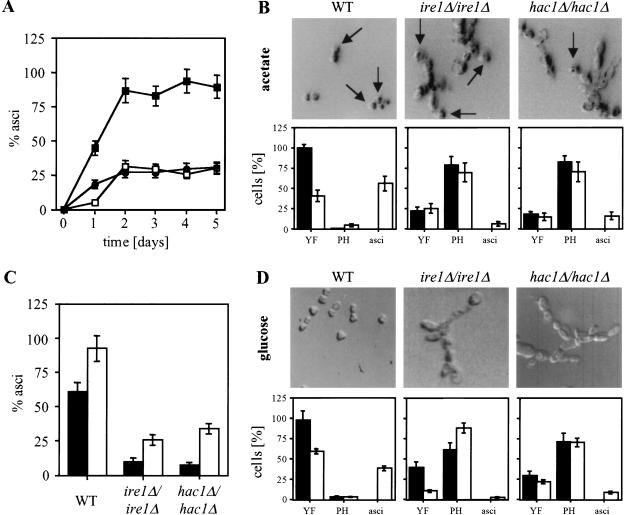
Pseudohyphal growth of diploid ire1Δ/ire1Δ and hac1Δ/hac1Δ strains. (A) Morphology of WT (AMP1618 × AMP1619), ire1Δ/ire1Δ (MSY17-46 × MSY18-59) and hac1Δ/hac1Δ (MSY19-29 × MSY20-19) cells in mid-log phase in liquid culture grown on glucose (YPD) or acetate (YPAc) as carbon source. (B) Quantitation of yeast form (length to width ratio less than two; black bars) and pseudohyphal (length to width ratio greater than or equal to two; open bars) from the experiment shown in part A. For each strain, >100 cells were counted. (C) Pseudohyphal growth is derepressed in homozygous diploid ire1Δ and hac1Δ strains and insensitive to activation of the UPR by 2-deoxyglucose or repression by ammonium sulfate. The strains are the same as in A and were grown on SLAD-plates for 7 d with the indicated concentrations of ammonium sulfate and 2-deoxyglucose. (D) Overexpression of Hac1ip represses pseudohyphal growth in nitrogen-starved diploid WT cells (AMP1618 × AMP1619). The strains were grown on SLAD-plates as described in B. The same result was obtained with three independent transformants.
Cell separation in pseudohyphae of S. cerevisiae is complete, and pseudohyphae can easily be disrupted in shaking liquid cultures. Further, the strong tendency of some strain backgrounds for flocculation can complicate the identification of pseudohyphae in liquid culture. We therefore tested whether our ire1Δ/ire1Δ and hac1Δ/hac1Δ strains also displayed another macroscopic change characteristic for pseudohyphal growth. Colonies growing as pseudohyphae are characterized by multiple projections radiating away from the center of the colony, whereas vegetatively growing colonies form round and smooth colonies (Gimeno et al. 1992). Pseudohyphae formation on plates can be induced with low–ammonium sulfate media (50 μM, SLAD-medium; Lorenz and Heitman 1997). Colonies of ire1Δ/ire1Δ and hac1Δ/hac1Δ cells displayed more vigorous pseudohyphae formation than WT cells (Fig. 1C, left panel). Invasive growth of haploid cells was not altered in ire1Δ and hac1Δ strains (data not shown).
Spliced Hac1p represses pseudohyphal development
As both IRE1 and HAC1 were required to repress pseudohyphal growth, we speculated that repression of pseudohyphal growth requires HAC1 splicing. To test this hypothesis, we overexpressed spliced Hac1p (Hac1ip) from a single-copy plasmid carrying the HAC1i gene under control of its own promoter (pRS316-HAC1i) in a WT strain. Cells carrying this plasmid constitutively expressed Hac1ip (e.g., see Fig. 4B, below). When grown on SLAD plates, only WT cells formed pseudohyphae, whereas pseudohyphae formation was strongly repressed in cells expressing Hac1ip (Fig. 1D). Thus, Hac1ip is sufficient to repress pseudohyphal growth.
Figure 4.
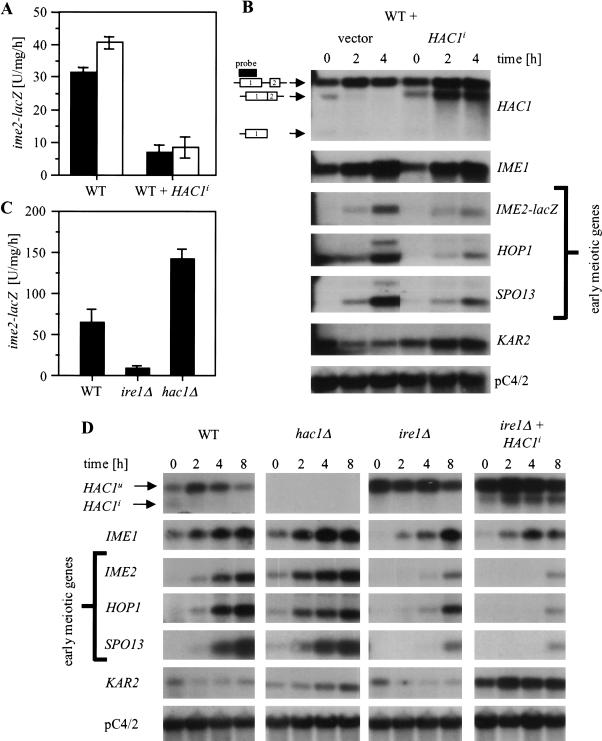
HAC1 is a negative regulator of early meiotic gene expression. (A,C) ime2–lacZ expression after induction of meiosis with C-SPO medium. The average and standard error of three independent clones (A) or experiments (C) are shown. (A) WT cells (AMP1618 and AMP1619) were transformed with an empty plasmid (pRS316) or with a plasmid that carries the HAC1i gene under control of its own promoter (pRS316-HAC1i). Uracil was omitted from acetate (PSP2) and C–SPO medium. The black and open bars represent two independent experiments. (C) WT (AMP1619), ire1Δ (MSY18-59), and hac1Δ cells (MSY20-19) were grown on YPAc and shifted to C–SPO medium. (B,D) Northern analysis to the experiments described in A and C, respectively. All lanes were taken from the same blot with the same exposure time. Two mRNAs for HOP1 and SPO13 were detected, as reported by Smith et al. (1990). Similar results were obtained in two independent experiments.
Unfolded proteins in the ER repress pseudohyphal growth through activation of the UPR
So far, the role of components of the UPR in repression of pseudohyphal growth resembles their role in the UPR. Both IRE1 and HAC1 were required, and overexpression of Hac1ip was sufficient to repress pseudohyphal growth. Therefore, we speculated that the whole UPR, starting with activation of Ire1p by unfolded proteins and resulting in splicing of HAC1 mRNA might be a repressing pathway for pseudohyphal growth. We therefore tested directly whether induction of unfolded proteins in the ER by disrupting N-linked glycosylation with tunicamycin or 2-deoxyglucose was sufficient to repress pseudohyphal growth in WT cells at low nitrogen concentrations. Homozygous diploid WT, ire1Δ, and hac1Δ strains were grown on SLAD-plates in the presence of low concentrations of tunicamycin or 2-deoxyglucose. Sublethal concentrations of tunicamycin (0.2 μg/mL; data not shown) and 2-deoxyglucose (1 mM; Fig. 1C center panel) inhibited pseudohyphae formation in WT cells. Both drugs did not interfere with pseudohyphae formation by homozygous diploid ire1Δ and hac1Δ strains. This demonstrated that accumulation of unfolded proteins in the ER is sufficient to repress pseudohyphal development through activation of Ire1p and HAC1 splicing.
Repression of pseudohyphal growth by nitrogen is defective in cells with a compromised UPR
Pseudohyphae formation in ire1Δ/ire1Δ and hac1Δ/hac1Δ strains was observed in nitrogen-rich media independent of carbon source (Fig. 1A). As pseudohyphal growth is repressed by high extracellular nitrogen concentrations (Gimeno et al. 1992), we speculated that homozygous diploid ire1Δ and hac1Δ cells are defective in nitrogen repression of pseudohyphal growth. To test this idea more directly, we grew WT, ire1Δ/ire1Δ, and hac1Δ/hac1Δ strains at different ammonium sulfate concentrations. Whereas pseudohyphae formation in the WT strain was completely repressed by 1 mM ammonium sulfate, the strains defective in the UPR still formed hyphae at this ammonium sulfate concentration (Fig. 1C, right panel). This suggested that the UPR is, indeed, transmitting a nitrogen signal to repress pseudohyphal growth. However, hyphae formation at 1 mM ammonium sulfate was not as vigorous as at 100 μM in ire1Δ/ire1Δ and hac1Δ/hac1Δ strains, indicating the existence of additional pathways that repress pseudohyphal growth in response to ammonium.
Basal activity of the UPR during vegetative growth
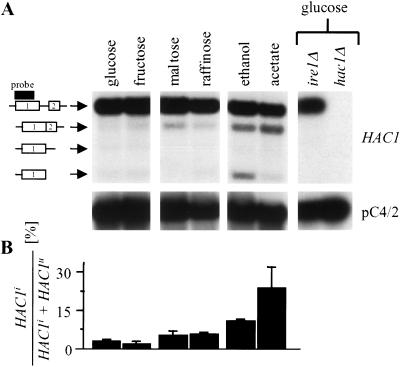
The data presented up to this point suggest that the UPR contributes to repression of pseudohyphal growth in a nitrogen-rich environment. If true, synthesis of the readout of the UPR, spliced Hac1ip, in unstressed, vegetatively growing cells must occur. To test this hypothesis, a WT strain was grown to mid-log phase on different carbon sources, RNA extracted, and analyzed by Northern blotting and PhosphorImaging. Synthesis of Hac1ip is controlled by splicing of HAC1 precursor mRNA (Chapman et al. 1998; Kaufman 1999). Thus, HAC1i mRNA is a hallmark of activation of Ire1p and the UPR. On all carbon sources, HAC1 splicing was observed (Fig. 2A). On glucose, ∼1%–3% of HAC1 mRNA was spliced (Fig. 2B). ire1Δ and hac1Δ cells showed no signals that corresponded to HAC1i mRNA or cleavage intermediates (Fig. 2A), thus demonstrating the specificity of these signals for HAC1 mRNA species in WT cells. The amount of HAC1i mRNA increased five- to 10-fold during growth on a nonfermentable carbon source such as ethanol or acetate (Fig. 2). Growth on glucose-repressible carbon sources like maltose or raffinose resulted in a twofold increase in HAC1 splicing (Fig. 2B). HAC1 splicing on nonfermentable carbon sources peaked in the exponential growth phase and was lower in the stationary growth phase (data not shown). HAC1 splicing in unstressed, vegetatively growing yeast was observed in both haploid and diploid cells (data not shown), demonstrating that basal low-level UPR activity is not dependent on cell type. Similar results were obtained with different strain backgrounds.
Figure 2.
Basal UPR activity during vegetative growth. (A) HAC1 splicing in a WT strain (AMP1619) grown to mid-log phase on different carbon sources (2%) on rich medium. All lanes are from one Northern blot and were exposed for the same time. Symbols to the left of the HAC1 blot, starting at the top: unspliced, spliced HAC1 mRNA, products of the endonucleolytic cleavage of HAC1 mRNA by Ire1p at the 3′- and 5′-splice site. The two exons of HAC1 mRNA are labeled 1 and 2. A black bar indicates the region recognized by the HAC1 probe. (B) Quantitative analysis of HAC1 splicing on different carbon sources by PhosphorImaging. The average and standard error from two independent experiments are shown.
The small percentage of HAC1 splicing detected in WT cells grown on glucose (Fig. 2A) must be sufficient for repression of pseudohyphal growth, as hac1Δ/hac1Δ cells do not synthesize any Hac1p and were constitutively growing as pseudohyphae. Further, as ire1Δ/ire1Δ cells also grew constitutively as pseudohyphae, it can be concluded that the splicing reaction is necessary to produce the inhibitor; in other words, unspliced HAC1 mRNA does not contribute to the repression. Therefore, either Hac1p levels are too low in ire1Δ/ire1Δ cells for efficient repression or the change in the C terminus of Hac1p introduced by the splicing reaction (Mori et al. 2000) is required for repression. Unspliced HAC1 mRNA levels in ire1Δ cells are the same as in WT cells (Fig. 2A, glucose as carbon source) or elevated (see below, Fig. 4D, acetate as carbon source) showing that a loss in HAC1 transcription is not responsible for constitutive activation of pseudohyphal growth in diploid ire1Δ strains. On the basis of these different lines of evidence, we conclude that the UPR represses pseudohyphal growth in a nitrogen-rich environment in vegetatively growing cells.
HAC1 splicing is regulated by extracellular nitrogen
Repression of pseudohyphal growth by the UPR must be relieved when nitrogen becomes limiting to allow for hyphae formation. On the basis of the data discussed before (Fig. 1D), the repressing activity resides in Hac1ip. Therefore, a mechanism has to exist that inactivates Hac1ip during nitrogen starvation. That the activity of Hac1ip is modulated can be ruled out, as overexpression of Hac1ip was sufficient to repress pseudohyphal growth at low nitrogen concentrations (Fig. 1D). Regulation of Hac1p synthesis is sufficient to control Hac1p levels, as both forms of Hac1p have a half-life of ∼2 min (Kawahara et al. 1997). Several mechanisms for how synthesis of Hac1ip can be regulated by nitrogen sources can be envisioned, such as transcription of HAC1 or the splicing reaction. Nitrogen starvation did not result in a dramatic decrease in HAC1 mRNA levels (Fig. 3; Fig. 4B,D), showing that rapid transcriptional regulation of Hac1ip synthesis by nitrogen sources does not occur.
Figure 3.
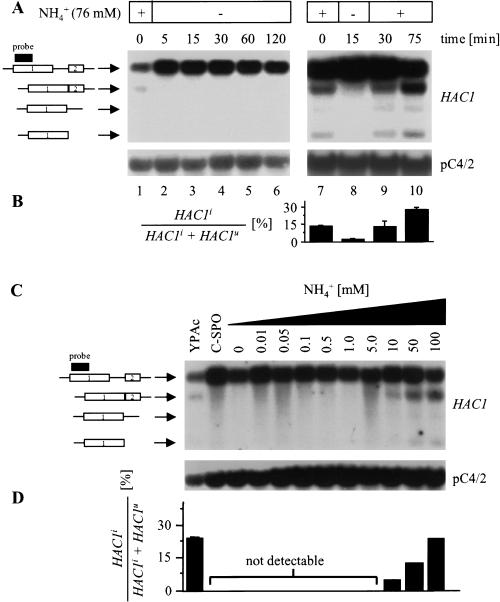
HAC1 splicing is regulated by extracellular nitrogen. (A) Northern analysis of a WT strain (AMP1619) grown to mid-log phase on acetate (lanes 1,7) after nitrogen starvation with C-SPO medium (lanes 2–6,8), and 15 and 60 min after addition of ammonium sulfate (5 g/L) to cells starved for nitrogen for 15 min (lanes 9,10). Similar results were obtained in two independent experiments. (B) PhosphorImaging analysis of the experiment in part A. The average and standard error from two independent experiments are shown. (C) Titration of the ammonium concentration necessary to stimulate HAC1 splicing in nitrogen-starved WT cells. Northern analysis of the same WT strain as in A grown to mid-log phase on YPAc, starved for nitrogen with C-SPO medium for 15 min and stimulated with different concentrations of ammonium sulfate for 30 min. (D) PhosphorImaging analysis of the Northern shown in C. The standard error for each data point is <2%. Similar results were obtained in two independent experiments.
To test whether the splicing reaction is regulated by nitrogen, we looked at the effect of nitrogen starvation on HAC1 splicing in cells grown on acetate, as this carbon source yielded levels of HAC1 splicing that allowed us to detect changes in the level of HAC1 splicing by Northern blotting (Fig. 2). RNA extracted from WT cells grown to mid-log phase on acetate (YPAc) or after nitrogen starvation with C–SPO medium was subjected to Northern analysis. As early as 5 min after establishing nitrogen starvation, virtually no HAC1 splicing was seen (Fig. 3A, lanes 1–6). HAC1 splicing was not restored under these conditions over a period of 2 h. Next, we wanted to demonstrate that the level of nitrogen sources actually regulates HAC1 splicing and that regulation of HAC1 splicing is not caused by changes in metabolite levels other than nitrogen sources. We therefore tried to reactivate HAC1 splicing in cells starved for nitrogen with C–SPO medium for 15 min by adding ammonium sulfate to the concentration in vegetative growth media (5 g/L or 37.9 mM). Indeed, HAC1 splicing was seen 15 min after nitrogen stimulation and persisted for at least 1 h under these conditions (Fig. 3A, lanes 7–10). Moreover, the level of HAC1 splicing 15 min after reactivation with ammonium sulfate was identical to the level seen in exponentially growing cells (Fig. 3B, cf. lane 9 with lane 7). This demonstrated that the only environmental change to which HAC1 splicing responds is the change in the nitrogen source ammonium sulfate. Other ammonium salts such as ammonium chloride and ammonium acetate stimulated HAC1 splicing in a similar manner (data not shown); showing that ammonium is the regulatory component and not sulfate. The smallest concentration of ammonium able to reactivate HAC1 splicing was 10 mM (Fig. 3C). Thus, the amount of ammonium present in SLAD-medium (100 μM) used to induce pseudohyphal growth is not sufficient to activate HAC1 splicing. The same response of the splicing reaction to environmental nitrogen levels was observed in haploid and diploid cells and in different genetic backgrounds, showing that it is independent of cell type.
Two steps in the splicing reaction may be regulated by nitrogen: either the endoribonuclease activity of Ire1p or tRNA-ligase activity. In the first case, not only does HAC1i mRNA decrease on nitrogen starvation but the mRNAs for the cleavage intermediates at the 5′- and 3′-splice junction do also. In the latter case, only HAC1i mRNA decreases on nitrogen starvation, and mRNAs for the intermediates should be unaffected. Nitrogen starvation resulted in a loss of HAC1i mRNA and the mRNAs for the cleavage intermediates (Fig. 3A), demonstrating that the endoribonuclease activity of Ire1p is regulated by extracellular nitrogen sources.
On the basis of these results, we conclude that the UPR represses pseudohyphal growth in response to nitrogen. If nitrogen starvation is encountered, the UPR and HAC1 splicing are significantly down-regulated to permit formation of pseudohyphae. Induction of unfolded proteins in the ER by drug treatment was sufficient to repress pseudohyphal growth in an IRE1- and HAC1-dependent manner. In addition, high extracellular nitrogen levels activated the endoribonuclease activity of Ire1p. Therefore, the most attractive hypothesis for how nitrogen activates Ire1p is to assume that it acts through increased synthesis of unfolded proteins, which may be a byproduct of high translation rates in a nitrogen-rich environment.
Spliced Hac1p represses entry into meiosis
In addition to pseudohyphal growth, nitrogen starvation can trigger other physiological events, such as derepression of arginine catabolism (Strich et al. 1994) or induction of meiosis (Kupiec et al. 1997). We therefore asked whether the repressing role of the UPR in response to a nitrogen-rich environment is limited to pseudohyphal growth or whether it extends to other adaptational responses to nitrogen starvation. Meiosis, besides pseudohyphal growth, is the second and only other known developmental response of S. cerevisiae to nitrogen starvation (Kupiec et al. 1997). Meiosis is triggered by the presence of poor carbon sources (e.g., acetate), whereas pseudohyphal growth is believed to require the presence of a fermentable carbon source (Gimeno et al. 1992). A role for HAC1 in meiosis was proposed earlier on more circumstantial data. First, a GGCGG-element, reminiscent of the URS1 site, was noted in the HAC1 promoter (Nojima et al. 1994). Second, HAC1 mRNA levels were reported to oscillate during meiosis but not during vegetative growth (Nojima et al. 1994). Most interesting, tunicamycin was reported to inhibit ascus formation if added during the early phase of meiosis but to not do so during the later phases (Weinstock and Ballou 1987). Differences in tunicamycin uptake during the three stages of meiosis did not account for this observation. It seemed reasonable to assume that this inhibitory effect was caused by activation of HAC1 splicing and repression of early genes by Hac1ip. Last, nitrogen starvation resulted in rapid loss of HAC1 splicing (Fig. 3A), further supporting the idea that HAC1 may play the role of a negative regulator of early meiotic genes.
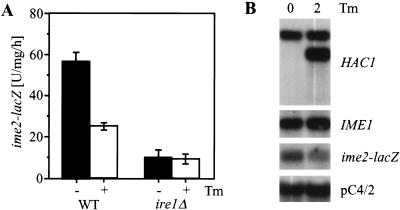
To test whether Hac1ip inhibits early meiotic gene expression, we studied the effect of Hac1ip overexpression on activation of the promoter for the early meiotic gene IME2. To follow IME2 induction, an integrated reporter was used in which nucleotides −983 to +114 of IME2 were fused to Escherichia coli at codon 39 of IME2 (Su and Mitchell 1993). We found that pseudohyphal growth persisted in homozygous diploid ire1Δ and hac1Δ strains after induction of meiosis (e.g., see Fig. 6B, below) and interfered with efficient sporulation. We therefore decided to use haploid cells in which the cell type specific repressor of meiosis RME1 was deleted to study the role of IRE1 and HAC1 in regulation of early meiotic genes. The kinetics and extent of early meiotic gene induction in rme1Δ haploid cells is comparable to WT diploid cells (Su and Mitchell 1993). Haploid cells do not induce filamentation in response to nitrogen starvation (Roberts and Fink 1994), and haploid invasive growth was not altered in ire1Δ and hac1Δ cells (see above). Overexpression of Hac1ip in these cells did repress expression of the ime2–lacZ reporter (Fig. 4A), proving that Hac1ip is a negative regulator of IME2.
Figure 6.
Pseudohyphal growth persists during meiosis in ire1Δ/ire1Δ and hac1Δ/hac1Δ strains. (A) Time course of ascus formation by homozygous diploid WT (filled squares), ire1Δ (open squares), and hac1Δ (filled circles) strains. (B,D) Meiosis was induced in homozygous diploid WT, ire1Δ, and hac1Δ strains grown to mid-log phase on glucose (D) or acetate (B) by shifting to C–SPO medium. Cells were photographed and counted before (filled bars) and 30 (D) or 20 h (B; open bars) after induction of meiosis. Representative pictures of the majority of the population after induction of meiosis are shown. Abbreviations: YF, yeast form (length to width [l/w] ratio less than 2), PH, pseudohyphal cells (l/w greater than or equal to two). More than 100 cells were counted for each strain. (C) Sporulation efficiency after 5 d. Growth to mid-log phase before induction of meiosis was on glucose (filled bars) or acetate (open bars). The average and standard error of two experiments are shown.
Induction of early meiotic genes such as IME2, HOP1, and SPO13 is mainly controlled at the transcriptional level and requires the conversion of the transcriptional repressor Ume6p to an activator by binding it to Ime1p (Rubin-Bejerano et al. 1996; Malathi et al. 1997). Ime1p activity is controlled by its abundance in the cell. Glucose and nitrogen repress its transcription (Kupiec et al. 1997). Thus, glucose and nitrogen starvation induce IME1 and activate transcription of early meiotic genes by the Ume6p–Ime1p complex. Therefore, repression of IME2 by Hac1ip may be indirect, that is, a consequence of repression of IME1 by Hac1ip. To address this question, we performed Northern analysis on WT and on WT cells carrying the extra HAC1i gene. Overexpression of Hac1ip had no or only little effect on transcription of IME1, whereas the early meiotic genes HOP1, IME2, and SPO13 were repressed four- to fivefold (Fig. 4B). To ensure that the HAC1i gene was expressed, we looked at HAC1 and KAR2 mRNA levels. HAC1i mRNA was detected in cells carrying the HAC1i gene even in the presence of nitrogen starvation (Fig. 4B). Further, transcription of the chaperone gene KAR2, characterized as one target gene induced by Hac1ip (Kaufman 1999), was elevated compared to WT cells (Fig. 4B), showing that Hac1ip levels were elevated and functional in these cells. Taken together, these data demonstrate that Hac1ip is a negative regulator of early meiotic genes but not of their positive regulator IME1.
Different roles for IRE1 and HAC1 in regulation of entry into meiosis
If Hac1ip is a negative regulator of early meiotic genes, cells defective in HAC1 splicing should show enhanced induction of early meiotic genes. Therefore, we analyzed induction of early meiotic genes in ire1Δ and hac1Δ strains. First, expression of the ime2–lacZ reporter during nitrogen starvation on acetate was monitored. Indeed, hac1Δ cells showed enhanced expression of the reporter (Fig. 4C), and ime2–lacZ expression was also derepressed during vegetative growth (data not shown). Surprisingly, ire1Δ cells were found to be defective in induction of IME2 (Fig. 4C). To confirm this result, we performed Northern analysis on these cells. Consistent with the previous results, deletion of HAC1 had no major effect on induction of IME1 (Fig. 4D). Further, the early meiotic genes IME2, HOP1, and SPO13 were induced faster than in the WT strain, and transcript levels of these genes were elevated during vegetative growth (Fig. 4D). This shows that HAC1 is a negative regulator for these genes but not of their activator IME1. In contrast, ire1Δ cells displayed a slower induction of IME1 than the WT strain (Fig. 4D). As transcription of early meiotic genes is primarily regulated by the abundance of Ime1p in the cell, this defect in IME1 induction in the ire1Δ strain is sufficient to explain the delay in induction of IME2 (Fig. 4C) and other early genes (Fig. 4D).
In the classical UPR, Ire1p activates transcription of chaperone genes through up-regulation of synthesis of Hac1ip (Chapman et al. 1998). However, HAC1 was not required for induction of IME1, raising the possibility that activation of the IME1 promoter by IRE1 is independent of HAC1 function. To confirm this observation, we asked whether overexpression of Hac1ip from pRS316-HAC1i in ire1Δ cells could rescue the defect in activation of IME1 transcription. Indeed, IME1 mRNA levels in cells overexpressing Hac1ip were similar to the levels in an ire1Δ strain (Fig. 4D), showing that HAC1i cannot rescue the defect in IME1 transcription. Hac1ip was synthesized in these cells as HAC1i mRNA was detected and KAR2 mRNA levels were elevated throughout the experiment (Fig. 4D). Further, overexpression of Hac1ip in WT cells did not affect IME1 mRNA levels (Fig. 4B). These results demonstrate that IRE1, but not HAC1, is a positive regulator of IME1.
Unfolded proteins in the ER repress entry into meiosis
Repression of early meiotic genes by Hac1ip resembled repression of pseudohyphal growth by the UPR. Therefore, we were interested in whether the most upstream element of the UPR is also part of the pathway repressing early meiotic genes. Meiosis was induced in WT cells by nitrogen starvation for 4 h in the presence or absence of 2 μg/mL tunicamycin and induction of IME1 and IME2 was compared. We observed a twofold reduction of ime2–lacZ mRNA levels (Fig. 5B) and expression of the ime2–lacZ reporter (Fig. 5A) in tunicamycin-treated WT cells but no change in IME1 mRNA levels (Fig. 5B). In ire1Δ cells, tunicamycin did not further decrease ime2–lacZ expression (Fig. 5A), showing that repression of IME2 by unfolded proteins in the ER requires activation of Ire1p and most likely HAC1 splicing. Other commonly used activators of the UPR, 2-deoxyglucose and β-mercaptoethanol (Chapman et al. 1998), severely interfered with cell viability on nonfermentable carbon sources (data not shown). Thus, unfolded proteins are the upstream element of the pathway that represses early meiotic genes such as IME2 during vegetative growth through activation of HAC1 splicing. Taken together, these results establish the UPR pathway as a negative regulator for early meiotic genes that are targets of the Ume6p–Ime1p transcriptional activator complex. In addition, IRE1 acts independent of HAC1 as a positive regulator for IME1.
Figure 5.
Induction of unfolded proteins in the ER represses IME2. (A) Induction of the ime2–lacZ reporter in WT (AMP1619) and ire1Δ cells (MSY18-59) treated with 2 μg/mL tunicamycin (Tm) during meiosis. Similar results were obtained in two independent experiments. (B) Analysis of IME1 and ime2–lacZ mRNA levels by Northern blotting in a WT strain under the conditions described in A.
Derepression of pseudohyphal growth interferes with efficient sporulation
To this point, our data establish the UPR, that is, activation of HAC1 splicing through protein unfolding in the ER, as a nitrogen-dependent repressor of both pseudohyphal growth and meiosis. On nitrogen starvation, HAC1 splicing ceases, Hac1ip levels drop dramatically, and repression of both developmental responses is relieved. Mechanisms must exist that ensure that only the correct adaptational response to certain conditions is chosen. Carbon source influences this choice. On glucose, the predominant response to nitrogen starvation is pseudohyphal growth, whereas in its absence meiosis prevails (Gimeno et al. 1992; Donzeau and Bandlow 1999). Homozygous diploid ire1Δ and hac1Δ strains grew as pseudohyphae under conditions that normally favor vegetative growth (Fig. 2A). We now wanted to know how these cells respond to conditions that induce meiosis in a WT strain, that is, nitrogen starvation in the presence of a nonfermentable carbon source. ire1Δ/ire1Δ and hac1Δ/hac1Δ strains displayed a very similar phenotype when sporulated, indicating that abrogation of HAC1 splicing in these cells is the main cause for their behavior. At the onset of meiosis, ascus formation was slower in the ire1Δ/ire1Δ strain than in the hac1Δ/hac1Δ strain (Fig. 6A), consistent with the observation that only IRE1 was required for efficient induction of IME1 (Fig. 4D). However, induction of IME1 mRNA in the ire1Δ strain was not completely abolished (Fig. 4D), and IME1 mRNA accumulated after prolonged times to levels comparable to WT cells (Fig. 4D). Sporulation efficiency was decreased to one-third of the WT in both strains (Fig. 6A,C). Pseudohyphal growth persisted for at least the first 20 h of meiosis (Fig. 6B), and chains of pseudohyphal cells were also observed 2 d after induction of meiosis. Homozygous diploid ire1Δ and hac1Δ strains were able to form asci that originated from pseudohyphal cells (Fig. 6B). This indicates that pseudohyphal cells are capable of inducing and completing the meiotic program. However, asci formed by ire1Δ/ire1Δ and hac1Δ/hac1Δ cells were greatly enlarged compared with asci formed by WT cells (Fig. 6B). We propose that derepression of pseudohyphal growth under vegetative conditions may interfere with efficient induction of meiosis in homozygous diploid ire1Δ and hac1Δ strains.
We also tried to induce meiosis in cells grown to mid-log phase on glucose, which resulted in an ∼70% reduction of sporulation in the UPR-defective strains, compared to only a 30% reduction for the WT (Fig. 6C). Under these conditions, the ire1Δ/ire1Δ strain induced pseudohyphal growth as an increase in the pseudohyphal population was observed (Fig. 6D). In contrast, WT and even hac1Δ/hac1Δ cells responded by forming asci. This suggests that ire1Δ/ire1Δ cells are defective in a second pathway that represses pseudohyphal growth during meiosis. Interestingly, overexpression of the IME1 target IME2 repressed pseudohyphal growth (Donzeau and Bandlow 1999). We therefore propose that the physiological significance of HAC1 independent activation of the IME1 promoter by IRE1 is repression of pseudohyphal growth during meiosis through subsequent induction of IME2.
Discussion
Regulation of vegetative growth, filamentation, and meiosis by the UPR
We report here that an activated UPR represses nitrogen starvation–induced developmental responses of budding yeast, pseudohyphal growth, and meiosis. The UPR is responsible for transcriptional induction of 381 ORFs in response to ER stress, including genes for ER-resident chaperones and ER-associated protein-degrading machineries (Travers et al. 2000). Our study significantly broadens the range of physiological functions of the UPR and defines a role for the UPR in nitrogen sensing.
The readout of the UPR, the spliced version of the transcription factor Hac1p, or activation of the UPR by induction of unfolded proteins in the ER were all sufficient to mediate this repression. Thus, all the elements of the UPR—unfolded proteins, IRE1, and HAC1—appear to act in the same order as they do in the classical UPR. Deletion of IRE1 and HAC1 proved that, indeed, very low levels of HAC1 splicing are sufficient to repress pseudohyphal growth. Therefore, activation of the UPR at low levels during vegetative growth represses pseudohyphal growth and meiosis (Fig. 7A). On nonfermentable carbon sources HAC1 splicing was increased, possibly because of less efficient synthesis of ATP or altered core oligosaccharide synthesis under these conditions. This increase in HAC1 splicing may also reflect the need for more Hac1ip to repress meiosis in addition to pseudohyphal growth.
Figure 7.
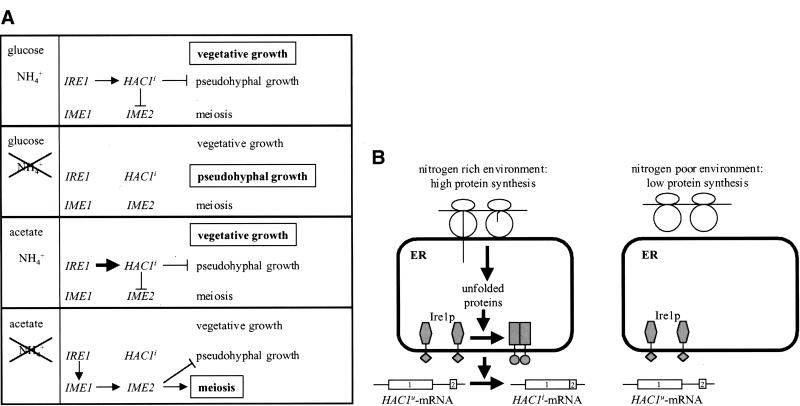
(A) Regulation of developmental responses to nitrogen starvation by the UPR. The carbon source and the presence or absence of a nitrogen source are shown in the left column. The right column shows how the UPR regulates the two developmental programs of diploid cells under each condition. The outcome is shown in bold letters in a box. Arrows stand for activating pathways, blunt-ended lines for inhibitory pathways. For discussion see text. (B) Model of a nitrogen-sensing pathway including translation and protein unfolding in the ER.
Once nitrogen becomes limiting, the UPR is turned off and a developmental decision is made that is dependent on the carbon source (Gimeno et al. 1992; Donzeau and Bandlow 1999). In accordance with their different roles in the regulation of early meiotic genes, homozygous diploid ire1Δ and hac1Δ strains displayed different kinetics of ascus formation but a similar decrease in sporulation efficiency after prolonged times (Fig. 6A). At the same time, pseudohyphal growth persisted in these strains during conditions that are favorable for meiosis in WT cells. Pseudohyphal growth in S. cerevisiae is thought to be a form of movement that allows the cells to forage for nutrients during starvation (Gimeno et al. 1992). Cells in which pseudohyphal growth is derepressed continue to grow, whereas vegetative cells arrest growth in G1 on encountering starvation conditions that are severe enough to trigger meiosis. A growth arrest in G1 triggered by starvation is considered a very early event necessary to induce the meiotic program (Herskowitz 1988). The behavior of diploid strains that are constitutively growing as pseudohyphae because of loss of the UPR after induction of meiosis demonstrated that this growth arrest is important for efficient induction of meiosis.
In addition, ire1Δ/ire1Δ, but not WT or hac1Δ/hac1Δ, cells responded by induction of pseudohyphal growth when sporulated after growth on glucose (Fig. 6D). Thus, IRE1 seems to play a role in transducing a signal in response to carbon source that commits the cell to meiosis when nitrogen starvation is encountered on a nonfermentable carbon source. As hac1Δ/hac1Δ cells responded correctly to nitrogen starvation under these conditions (Fig. 6D), it seems reasonable to assume that this IRE1 function is identical to its stimulatory effect on the IME1 promoter and subsequent induction of IME2, a repressor of pseudohyphal growth (Donzeau and Bandlow 1999). This model is supported by the observation that HAC1 was not required for activation of the IME1 promoter by IRE1.
The change from the yeast to a filamentous growth form is common to many fungi and is in many cases associated with a pathogenic phenotype. The best-studied examples are the human pathogens Candida albicans and Cryptococcus neoformans and the corn pathogen Ustilago maydis (Madhani and Fink 1998). Interestingly, tunicamycin inhibited germ tube formation in C. albicans (Chaffin 1985), indicating that the UPR plays a similar role in this organism. Therefore, drugs that impair protein folding in the ER or otherwise activate the UPR may serve as new lead structures in the search for new antifungal agents.
Protein unfolding and UPR activation as a sensor for translation rate and a nitrogen-rich environment
The physiological event common to pseudohyphal growth and meiosis is induction by nitrogen starvation (Gimeno et al. 1992; Kupiec et al. 1997). Both programs are repressed by the UPR, and we found pseudohyphal growth to be resistant to nitrogen repression in UPR-defective cells. This led us to the idea that the UPR represses both programs in a nitrogen-rich environment; in other words, it transduces a signal for nitrogen. But how does nitrogen regulate the UPR? We have seen that HAC1 splicing in unstressed cells depends on the presence of ammonium as an extracellular nitrogen source. As long as nitrogen was present, HAC1 mRNA was spliced. However, HAC1 splicing was dramatically reduced during nitrogen starvation. Furthermore, activation of the UPR by induction of unfolded proteins in the ER with tunicamycin or 2-deoxyglucose was sufficient to repress pseudohyphal growth (Fig. 1B) and meiosis (Fig. 5; Weinstock and Ballou 1987) under nitrogen-limiting conditions. This suggests that protein unfolding in the ER can substitute for high extracellular nitrogen levels to repress both developmental programs. Thus, nitrogen may act through protein unfolding in the ER to activate the UPR. Activation of HAC1 splicing by ammonium salts required protein synthesis and could be completely blocked with cycloheximide (M. Schröder and R.J. Kaufman, unpubl.), supporting this idea.
On the basis of these observations, we propose the following model for how the UPR senses nitrogen levels (Fig. 7B). Unfolded proteins in the ER are a by-product of rapid translation in a nitrogen-rich environment and, ultimately, activate the UPR to enhance their chances to fold correctly and to also repress developmental responses to nitrogen starvation. Indirect proof that unfolded proteins exist in vegetatively growing, unstressed cells comes from the observation that abrogation of both known pathways that deal with unfolded proteins in the ER (UPR and ERAD) is synthetic lethal (Travers et al. 2000). It was also estimated that as much as one-third of all proteins are sensed as unfolded during their synthesis (Ellgaard et al. 1999). This should be sufficient for constitutive low-level activation of the UPR that we see during vegetative growth in a nitrogen-rich environment (Fig. 2). Under nitrogen-limiting conditions, translation is slower, thus providing for more efficient folding of nascent chains (Fig. 7B). In this view, the efficiency of protein folding in the ER and subsequent activation of the UPR is a sensor for translation rate and, at least downstream, elements of a nitrogen sensing pathway. Translation is inhibited by amino acid starvation, a condition that closely resembles nitrogen starvation, and results in activation of the general amino acid control system (Hinnebusch 1992). We therefore think that transient inhibition of translation, triggered by nitrogen starvation, is a very early event in initiating pseudohyphal development and meiosis and may precede the G1 arrest and induction of IME1 in meiosis. Inhibition of translation before meiosis has not been observed to date (Miller 1989), but on the basis of the published data, an inhibition for only a limited time early in meiosis cannot be ruled out. Future experiments will address both this question and whether activation of Ire1p by nitrogen is strictly dependent on protein unfolding in the ER or if it occurs by another mechanism that is also dependent on protein synthesis and that bypasses the ER.
Molecular mechanisms of regulation of early meiotic gene expression and pseudohyphal growth by the UPR
Hac1ip-mediated repression of both meiosis and pseudohyphal growth. Hac1ip is a bZIP transcription factor and activates transcription of genes containing a UPR element (Kaufman 1999). In addition, it was suggested that Hac1p activated transcription of INO1 by titrating out the transcriptional repressor Opi1p by formation of a heterodimeric complex between these two leucine zipper proteins (Chapman et al. 1998). However, a genome-wide two-hybrid interaction screen failed to detect any interaction of Hac1ip with any protein other than itself (Uetz et al. 2000). Therefore, the most attractive scenario is that Hac1ip represses pseudohyphal growth and meiosis through transcriptional activation of a repressor. The global scope of the UPR has been characterized (Travers et al. 2000) and should be a very valuable tool for identifying the transcriptional targets of Hac1ip that mediate its repression of pseudohyphal growth and meiosis. Interestingly, SIN3, which is part of the Sin3p–Rpd3p histone deacetylase complex required for transcriptional repression of early meiotic genes by Ume6p (Kadosh and Struhl 1997), is induced approximately twofold on ER-stress in an IRE1- and HAC1-dependent manner (Travers et al. 2000). It also possesses a well-conserved UPR-like element at position −406 (GGgCAGCGcGT, nucleotides identical to the UPRE-consensus sequence in capitals), making it a likely target to mediate repression of early meiotic genes by Hac1ip.
The UPR also regulates transcription of GRR1 in response to ER stress (Travers et al. 2000). Grr1p is the F-box protein in the SCFGrr1 (Skp1/Cdc53/Grr1) ubiquitin–ligase complex (Loeb et al. 1999) and is required for repression of pseudohyphal growth (Blacketer et al. 1995). SCFGrr1 is responsible for degradation of the G1 cyclins Cln1p and Cln2p. Both CLN1 and CLN2 are required for pseudohyphal growth (Loeb et al. 1999), and grr1 mutants exhibit derepression of pseudohyphal growth. Activation of GRR1 by Hac1ip or the UPR should result in increased destruction of Cln1p and Cln2p and in a phenotype similar to cln1/cln1 and cln2/cln2 strains. It is interesting to note here that the UPR is also responsible for disposal of unfolded proteins through an ubiquitinylation-dependent degradation pathway (Casagrande et al. 2000). This may indicate a more global control of protein-degrading machineries by the UPR than appreciated previously.
In response to ER stress, both mammalian homologs of Ire1p activate the MAPK Jnk1p (Urano et al. 2000) by phosphorylation. In addition, the endoribonuclease activity of Ire1αp is necessary for activation of the BiP-promoter (W. Tirasophon and R.J. Kaufman, unpubl.). In yeast, the only known signaling event downstream of Ire1p in relieving unfolded protein stress in the ER is the splicing of HAC1 mRNA. In contrast to yeast, the mammalian UPR uses at least two mechanisms to transduce the unfolded protein signal. Our finding that activation of the IME1 promoter by Ire1p does not require HAC1 suggests that signal transduction pathways other than HAC1 splicing are regulated by Ire1p in yeast. This is the first report that divergence of pathways downstream of Ire1p also exists in yeast. Whether this divergence involves a second endonucleolytic cleavage event or solely requires the kinase function of yeast Ire1p is under investigation.
The findings reported here help to understand the regulation of developmental responses of yeast by nitrogen starvation. Intriguingly, deletion of IRE1 in mice had no effect on induction of the UPR but resulted in a lethal developmental defect (Urano et al. 2000; W. Tirasophon and R.J. Kaufman, unpubl.). In light of our findings, it seems clear that the role of the UPR in development is conserved from yeast to mammals. The UPR may have evolved from the need of archaic unicellular organisms to cope with long times of starvation by differentiation into a starvation resistant cell type. Deletion of IRE1 in other model organisms such as Caenorhabditis elegans or Drosophila melanogaster will help to study the role of IRE1 in development. Finally, our data show that protein folding is not solely the last event in gene expression but is regulated by and reports the nutritional status of the cell. Protein folding is also involved in deciding the fate of the cell, be it adaptation or apoptosis.
Materials and methods
Yeast strains and plasmids
Yeast strains AMP1618 (MATα arg6 IME2-20-lacZ::LEU2 rme1Δ5::LEU2 ura3 leu2 trp1 lys2 ho::LYS2; Vidan and Mitchell 1997), AMP1619 (MATa met4 IME2-20-lacZ::LEU2 rme1Δ5::LEU2 ura3 leu2 trp1 lys2 ho::LYS2, kindly provided by A.P. Mitchell) were described previously. IRE1 and HAC1 were deleted as described previously (Welihinda et al. 2000) to yield strains MSY17-46 and MSY18-59 (both ire1Δ::kanMX2) and MSY19-29 and MSY20-19 (both hac1Δ::URA3) derived from AMP1618 and AMP1619, respectively. Both deletions were confirmed by PCR and Southern blotting. ire1Δ and hac1Δ strains in the W303 background were derived from W303 1b (MATα ade2-1 can1-100 leu2-3,112 his3-11,15 trp1-1 ura3-52; Welihinda et al. 1998) and W303 1a (as W303 1b but MATa) as described above. Plasmid pRS316-HAC1i was described previously (Welihinda et al. 2000).
Yeast media and growth conditions
For vegetative growth, rich medium containing 1% bacto-yeast extract, 2% bacto-peptone, and 2% of the indicated carbon source (YPD, 2% glucose; YPAc, 2% potassium acetate; Vidan and Mitchell 1997), synthetic dextrose medium (SD; Vidan and Mitchell 1997), and minimal acetate vegetative medium (PSP2; Kassir et al. 1988) were used. Pseudohyphal growth was induced with synthetic low ammonium dextrose medium (SLAD; Lorenz and Heitman 1997). The UPR was induced with 2 μg/mL tunicamycin. Sporulation was induced by washing cells grown to mid-log phase with water and resuspending in complete sporulation medium (C-SPO; Vidan and Mitchell 1997).
ime2–lacZ reporter assays and sporulation efficiency
To monitor expression of the ime2–lacZ reporter during sporulation, samples were taken before and 4 and 8 h after induction of sporulation, and β-galactosidase activity was assayed and standardized to the protein concentration of the samples as described previously (Welihinda et al. 2000). The later time points were corrected for the 0-h-value and divided by the length of the induction period to express β-galactosidase induction per milligram intracellular protein and hour. Sporulation efficiency was scored 5 d after induction of sporulation by counting >200 cells and expressed as percentage asci per total cells counted.
Pseudohyphal growth assays
Pseudohyphal growth in liquid culture was observed in mid-log phase cultures. Photographs were taken with an Edge R400 microscope (Edge Scientific Instruments) at 400× magnification after fixing the cells with 3% formaldehyde. The length to width (l/w) ratio of at least 100 cells was determined from representative photographs and cells grouped into yeast form (YF, l/w < 2) and pseudohyphal cells (PH, l/w ≥ 2). Pseudohyphal growth on SLAD plates was scored after growth for 7 d, and pictures from representative colonies were taken at 100× magnification with an inverted microscope (Nikon TMS, Nikon). Agar invasion by haploid strains was assayed as described by Roberts and Fink (1994).
Northern blots
Isolation of RNA, the Northern blotting protocol, and probes for HAC1 and KAR2 were described previously (Welihinda et al. 1997, 2000). For lacZ, the 2.8-kb PvuII fragment of Z691 (Mori et al. 1993) was used. The probes for IME1; the early meiotic genes HOP1, IME2, and SPO13; and the loading control pC4/2, which hybridizes to an RNA unaffected by starvation (Smith et al. 1990) were described elsewhere (Vidan and Mitchell 1997). All mRNAs were quantified by PhosphorImager scanning (Molecular Dynamics) and standardized to the level of the loading control pC4/2.
Acknowledgments
We are very grateful to A.P. Mitchell (Columbia University) for generously providing strains and plasmids, his advice, and helpful discussions. We thank B. Athey (University of Michigan, Ann Arbor) for use of the Edge 400 microscope. We also thank D. Thiele for critically reading the manuscript.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kaufmanr@umich.edu; FAX (734) 763-9323.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.852300.
References
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- Blacketer MJ, Madaule P, Myers AM. Mutational analysis of morphologic differentiation in Saccharomyces cerevisiae. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zúñiga M, Brown PO, Ploegh H. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/s1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- Chaffin WL. Effect of tunicamycin on germ tube and yeast bud formation in Candida albicans. J Gen Microbiol. 1985;131:1853–1861. doi: 10.1099/00221287-131-8-1853. [DOI] [PubMed] [Google Scholar]
- Chapman R, Sidrauski C, Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Dev Biol. 1998;14:459–485. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- Donzeau M, Bandlow W. The yeast trimeric guanine nucleotide-binding protein α subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol Cell Biol. 1999;19:6110–6119. doi: 10.1128/mcb.19.9.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: Quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: Regulation by starvation and RAS. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/mr.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae. In: Jones EW, et al., editors. The molecular and cellular biology of the yeast Saccharomyces, Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 319–414. [Google Scholar]
- Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52:853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes & Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–1862. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M, Byers B, Esposito RE, Mitchell AP. Meiosis and sporulation in Saccharomyces cerevisiae. In: In: Pringle JR, et al., editors. The molecular and cellular biology of the yeast Saccharomyces, Vol. 3. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 889–1036. [Google Scholar]
- Liu H, Styles CA, Fink GR. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Loeb JDJ, Kerentseva TA, Pan T, Sepulveda-Becerra M, Liu H. Saccharomyces cerevisiae G1 cyclins are differentially involved in invasie and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- Malathi K, Xiao Y, Mitchell AP. Interaction of yeast repressor-activator protein Ume6p with glycogen synthase kinase 3 homolog Rim11p. Mol Cell Biol. 1997;17:7230–7236. doi: 10.1128/mcb.17.12.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JJ. Sporulation in Saccharomyces cerevisiae. In: Rose AH, Harrison JS, editors. The yeasts. 2nd ed. Vol. 3. London: Academic Press; 1989. pp. 489–550. [Google Scholar]
- Mori K, Ma W, Gething M-J, Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci. 2000;97:4660–4665. doi: 10.1073/pnas.050010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H-U, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Leem S-H, Araki H, Sakai A, Nakashima N, Kanaoka Y, Ono Y. Hac1: A novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5279–5288. doi: 10.1093/nar/22.24.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: Mating and invasive growth. Genes & Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional repressor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith HE, Su SSY, Neigeborn L, Driscoll SE, Mitchell AP. Role of IME1 expression in regulation of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:6103–6113. doi: 10.1128/mcb.10.12.6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strich R, Surosky RT, Steber C, Dubois E, Messenguy F, Esposito RE. UME6 is a key regulator of nitrogen repression and meiotic development. Genes & Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- Su SSY, Mitchell AP. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics. 1993;133:67–77. doi: 10.1093/genetics/133.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Chung YP, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Vidan S, Mitchell AP. Stimulation of yeast meiotic gene expression by the glucose-repressible protein kinase Rim15p. Mol Cell Biol. 1997;17:2688–2697. doi: 10.1128/mcb.17.5.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock KG, Ballou CE. Tunicamycin inhibition of epispore formation in Saccharomyces cerevisiae. J Bacteriol. 1987;169:4384–4387. doi: 10.1128/jb.169.9.4384-4387.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Green SR, Kaufman RJ. Gene induction in response to unfolded protein in the endoplasmic reticulum is mediated through Ire1p kinase interaction with a transcriptional coactivator complex containing Ada5p. Proc Natl Acad Sci. 1997;94:4289–4294. doi: 10.1073/pnas.94.9.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol Cell Biol. 1998;18:1967–1977. doi: 10.1128/mcb.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welihinda AA, Tirasophon W, Kaufman RJ. The transcriptional co-activator ADA5 is required for HAC1 mRNA processing in vivo. J Biol Chem. 2000;275:3377–3381. doi: 10.1074/jbc.275.5.3377. [DOI] [PubMed] [Google Scholar]