Abstract
In the yeast Saccharomyces cerevisiae, sequence-specific DNA binding by the origin recognition complex (ORC) is responsible for selecting origins of DNA replication. In metazoans, origin selection is poorly understood and it is unknown whether specific DNA binding by metazoan ORC controls replication. To address this problem, we used in vivo and in vitro approaches to demonstrate that Drosophila ORC (DmORC) binds to replication elements that direct repeated initiation of replication to amplify the Drosophila chorion gene loci in the follicle cells of egg chambers. Using immunolocalization, we observe that ACE3, a 440-bp chorion element that contains information sufficient to drive amplification, directs DmORC localization in follicle cells. Similarly, in vivo cross-linking and chromatin immunoprecipitation assays demonstrate association of DmORC with both ACE3 and two other amplification control elements, AER-d and ACE1. To demonstrate that the in vivo localization of DmORC is related to its DNA-binding properties, we find that purified DmORC binds to ACE3 and AER-d in vitro, and like its S. cerevisiae counterpart, this binding is dependent on ATP. Our findings suggest that sequence-specific DNA binding by ORC regulates initiation of metazoan DNA replication. Furthermore, adaptation of this experimental approach will allow for the identification of additional metazoan ORC DNA-binding sites and potentially origins of replication.
Keywords: ORC, origin of replication, DNA replication, amplification
The origin recognition complex (ORC) is a critical factor required for initiation of eukaryotic DNA replication. Originally identified as a six-protein complex that specifically bound Saccharomyces cerevisiae origins of DNA replication (Bell and Stillman 1992), ORC plays an essential role in replication by binding to the origin DNA and recruiting additional replication factors (Aparicio et al. 1997; Donovan et al. 1997; Liang and Stillman 1997; Tanaka et al. 1997; for review, see Leatherwood 1998). Sequence analogs of ORC subunits have been identified in metazoans including Drosophila, Xenopus, and humans (Gossen et al. 1995; Carpenter et al. 1996; Pak et al. 1997; Quintana et al. 1997, 1998; Carpenter and Dunphy 1998; Tugal et al. 1998; Chesnokov et al. 1999; Pinto et al. 1999). In vivo studies in Drosophila and in vitro studies using Xenopus egg or Drosophila embryo extracts have shown that ORC is required for DNA replication in metazoans (Carpenter et al. 1996; Rowles et al. 1996; Landis et al. 1997; Chesnokov et al. 1999). Furthermore, in vitro studies in Xenopus egg extracts indicate that, as in yeast, ORC acts by recruiting essential replication factors to the DNA (Coleman et al. 1996; Romanowski et al. 1996). S. cerevisiae ORC requires ATP for sequence-specific DNA binding in vitro. The amino acid sequences required for S. cerevisiae ORC to bind and hydrolyze ATP (Klemm et al. 1997) are conserved in the metazoan ORC analogs (Gavin et al. 1995; Rowles et al. 1996; Pak et al. 1997), suggesting that ATP will also regulate ORC–DNA interactions in metazoans.
Studies of DNA replication in S. cerevisiae are facilitated greatly by the availability of well-defined replication origins. These origins are typically less than 0.2 kb in size and have a modular composition. Each contains an 11-bp AT-rich sequence, known as the ARS consensus sequence, that is recognized by ORC. Two additional elements, termed B1 and B2, are functionally interchangeable between origins, although they lack clear consensus sequences (Marahrens and Stillman 1992; Rao et al. 1994; Theis and Newlon 1994; Huang and Kowalski 1996). The B1 element is bound by ORC (Bell and Stillman 1992; Rao and Stillman 1995; Rowley et al. 1995), although this element may have other important functions because there are mutations in the B1 sequence that affect replication but do not affect ORC DNA binding (Rao and Stillman 1995). The function of the B2 element is not known, but it has been suggested to be involved in DNA unwinding (Lin and Kowalski 1997). The yeast Schizosaccharomyces pombe has more complex origins that are about 0.5 to 1 kb in size. Within these origins, multiple 50-bp elements contribute to origin function, but unlike S. cerevisiae there are no small conserved elements that are essential for origin function (Clyne and Kelly 1995; Dubey et al. 1996; Kim and Huberman 1998).
In contrast to the small, well-defined S. cerevisiae origins, metazoan organisms have complex replication origins with no clearly defined elements. Using a variety of genetic and molecular techniques thus far ∼20 DNA loci have been found to contain metazoan replication origins (for review, see DePamphilis 1996, 1999). The initiation sites within these origins have been mapped to varying resolutions (0.5–11 kb), and the cis-acting sequences that allow these elements to act as origins of replication remain poorly defined. For example, deletion analysis of an 8-kb region of the human β-globin locus indicates that, like in S. pombe, multiple elements within this locus contribute to origin activity (Aladjem et al. 1998). Consistent with their complex structure thse origins are frequently found to initiate replication from one of a number of different sites within their boundaries. Because so little is known about metazoan origins, it has not yet been possible to identify the elements bound by metazoan ORC.
Metazoan origins are subject to developmental regulation (Callan 1972). In early Drosophila and Xenopus embryo development, DNA replication initiates approximately every 8–12 kb within the genome (Blumenthal et al. 1974; Hyrien and Mechali 1993; for review, see Spradling and Orr-Weaver 1987) arguing that there is limited sequence specificity for origin selection at this time in development. This close origin spacing allows for very short S phases and rapid proliferation of zygotic nuclei. Later in development, the length of S phase increases and corresponds to the onset of zygotic transcription, changes in chromatin structure, and an increase in the nuclear-to-cytoplasmic ratio (Newport and Kirschner 1982; Walter and Newport 1997; for review, see Carminati and Orr-Weaver 1996). In Drosophila, length of S phase has been correlated with an increase in the inter-origin spacing. Transitions of increased specificity in origin selection during development have been observed at the rDNA repeats in Xenopus and within the Drosophila DNApolα–dE2F genomic region (Hyrien et al. 1995; Sasaki et al. 1999).
DNA replication in the follicle cells of Drosophila egg chambers is highly regulated and provides a powerful system to study the regulation of metazoan DNA replication (for review, see Royzman and Orr-Weaver 1998). In stage 10 egg chambers (late in oogenesis), genomic DNA replication in the somatic follicle cells ceases with the exception of four DNA loci that continue to initiate replication (Calvi et al. 1998). Two of these loci encompass the chorion genes, and amplification of the chorion genes is necessary for eggshell formation and female fertility. DNA elements controlling amplification of the third chromosome chorion locus have been characterized (for review, see Orr-Weaver 1991). One element, termed ACE3, is a 440-bp DNA sequence that is necessary for amplification within the context of a 7.7-kb third chromosome chorion fragment (Orr-Weaver et al. 1989). A second control element, termed AER-d (oriβ), is located ∼1.5 kb away from ACE3, and two-dimensional gel analysis of replication intermediates during chorion amplification indicate that the majority of initiation events occurs within or near AER-d (Delidakis and Kafatos 1989; Heck and Spradling 1990). Two lines of evidence indicate that Drosophila ORC (DmORC) is involved in chorion amplification. First, flies with mutations in the second largest subunit of DmORC exhibit reduced amplification (Landis et al. 1997). Second, at stage 10A of egg chamber development just before the onset of amplification, DmORC is localized as two large discrete foci within the follicle cell nuclei that correspond to the two chorion gene clusters (Asano and Wharton 1999; Royzman et al. 1999). Mutations that affect this specific DmORC localization show reduced levels of chorion amplification (Royzman et al. 1999).
Although it is clear that ORC is required for metazoan replication, the role of ORC in selecting metazoan origins is not understood, in part because identified metazoan origins are limited in number and imprecisely characterized. Previously, we observed that introduction of a 7.7-kb fragment of chorion DNA through P-element mediated transformation resulted in a third major focus of DmORC localization (Royzman et al. 1999). This result strongly suggested that this 7.7-kb region contained a DmORC localization signal. In particular, the ACE3 and AER-d elements represented likely candidates for DmORC DNA-binding sites. Here we use both in vivo and in vitro approaches to demonstrate that ACE3 and AER-d are DNA-binding sites for DmORC. These findings suggest that DmORC DNA-binding specificity is likely to control origin selection and that identifying DmORC-binding sites in vivo will be a powerful tool to identify other Drosophila origins.
Results
ACE3 is sufficient to localize DmORC
To examine the role of ACE3 in DmORC localization, we compared the ability of two P-element constructs (Fig. 1A) containing or lacking ACE3 to localize DmORC using an anti-DmORC serum. This serum recognizes a single protein that comigrates with purified DmORC2 on Western blots of crude extracts (Fig. 1E). These constructs were A48O28, which contains a 7.7-kb region surrounding ACE3 and previously was observed to amplify in follicle cells, and A54O18, which is identical to A48O28 except for a 319-bp deletion within ACE3 and never amplifies (Orr-Weaver et al. 1989). As expected, we observed that flies transformed with the A48O28 construct consistently resulted in a third strong focus of DmORC localization in follicle cells (Fig. 1B). In contrast, integration of the A54O18 construct generally resulted in no additional focus of DmORC localization in ∼90% of the nuclei (Fig. 1C). The remaining 10% (10 of 102 counted) displayed only a weak third focus of DmORC localization (data not shown). Because P-element amplification constructs are known to be influenced by the chromatin surrounding the insertion site (de Cicco and Spradling 1984), we verified that the general inability of the A54O18 construct to localize DmORC was reproducible in 10 independent transformant lines of A54O18 (data not shown). From these data, we conclude that ACE3 is important, and in most nuclei necessary, for localizing DmORC in the follicle cells. To determine whether ACE3 was sufficient to localize DmORC, we examined the fly line M9-2 (Carminati et al. 1992). This line is transformed with a P-element containing nine repeats of ACE3 (Fig. 1A). We consistently observed a third major focus of DmORC localization in the M9-2 line (59 of 65 counted; Fig. 1D). Thus, ACE3 is sufficient to localize DmORC in follicle cells.
Figure 1.
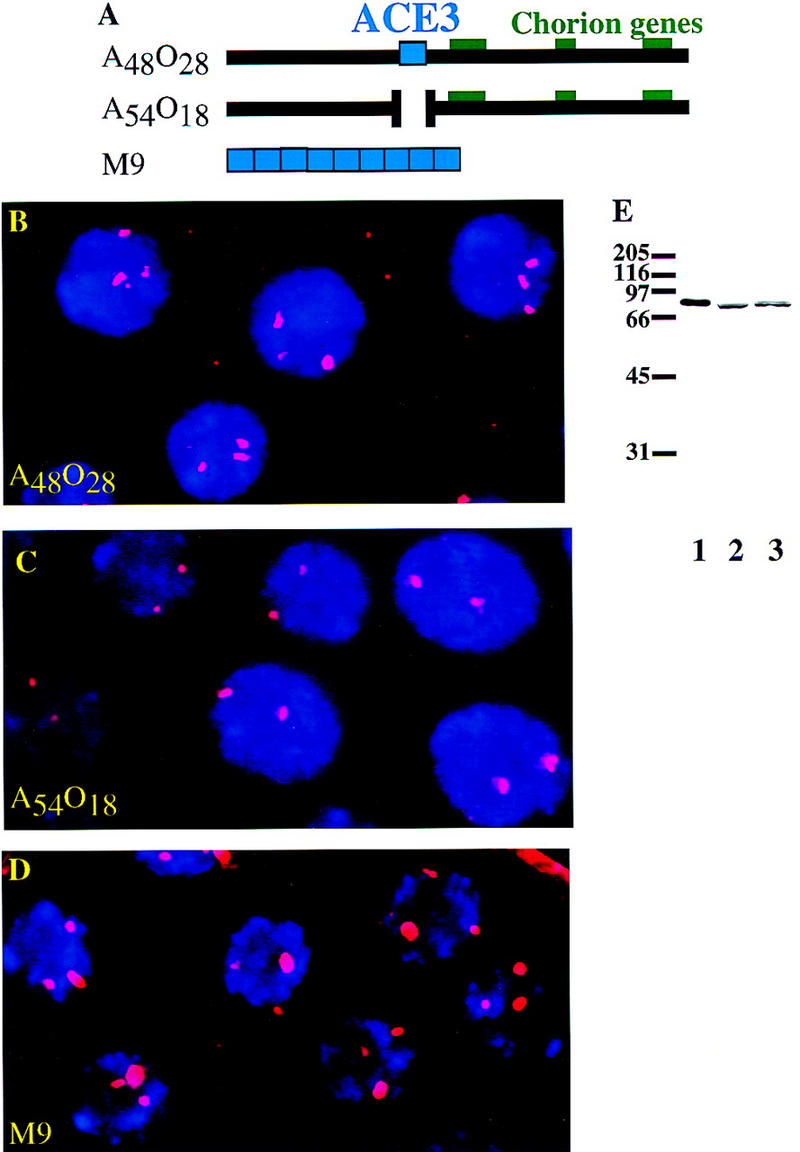
A construct containing nine repeats of ACE3 localizes DmORC. (A) Diagram of P-element constructs (Orr-Weaver et al. 1989; Carminati et al. 1992). Construct A48O28 has 7.7 kb of chorion DNA. Construct A54O18 is the same as A48O28 except that 319 bp of ACE3 is deleted. Construct M9 has nine repeats of the 440-bp ACE3 fragment. (B–D) DmORC protein was visualized by immunofluorescence microscopy in the follicle cells of stage 10A egg chambers of fly lines transformed with P-element constructs containing third chromosome chorion DNA. DNA was visualized by staining with DAPI. The merged red (DmORC) and blue (DNA) channels are shown. (B) Localization of DmORC in flies transformed with the A48O28 construct. Three foci were observed per nucleus. (C) Localization of DmORC in flies transformed with construct A54O18. Two foci were observed per nucleus. (D) Localization of DmORC in flies transformed with the M9 construct. Three foci of were observed per nucleus. (E) Western blot performed using anti-DmORC2 serum. Samples on the blot are 80 ng of recombinant DmORC2 protein (lane 1), extract from 0- to 12-hr embryos (lane 2), and nuclear extract from Schneider line 2 cells (lane 3).
High-resolution microscopy revealed an additional intriguing aspect of the DmORC immunolocalization (Fig. 2). In the small, early, stage 10B egg chambers, the foci of DmORC localization display a doughnut-like (toroidal) appearance. Because these doughnut-foci are seen after the chorion loci have undergone several rounds of amplification (Calvi et al. 1998; Royzman et al. 1999), we speculate that they reflect the onion skin DNA structures generated by the amplification process and that DmORC may only be associated with the external portion of the onion skin (Osheim and Miller 1983). Consistent with this hypothesis, these doughnut-like structures were not readily observed in the DmORC foci of stage 10A egg chambers (data not shown), which have undergone minimal amplification (Calvi et al. 1998).
Figure 2.
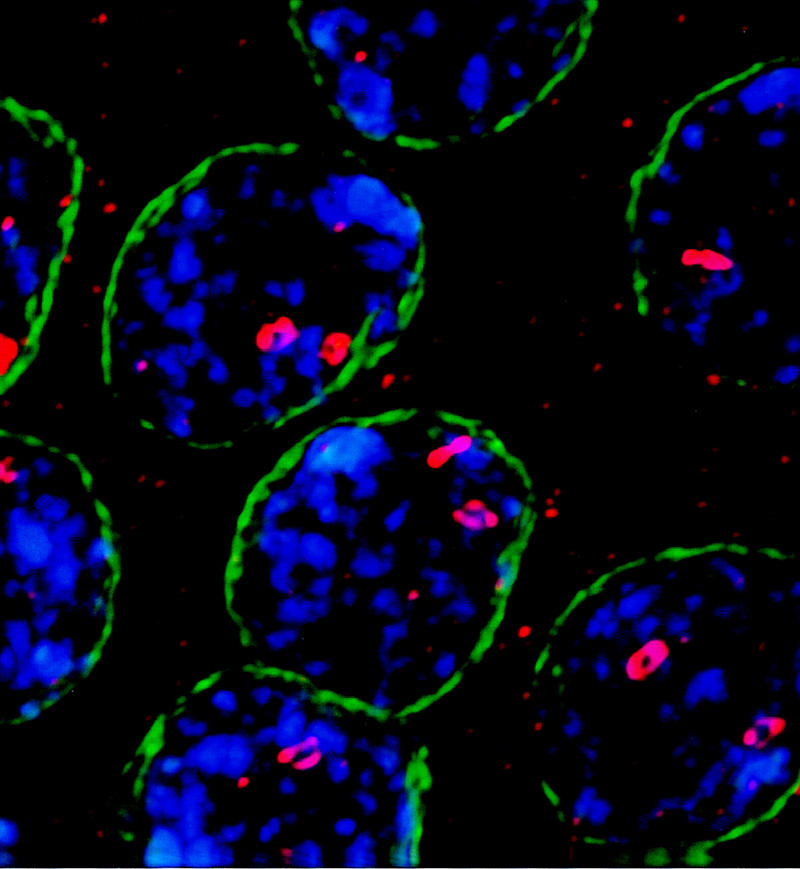
High-resolution microscopy of DmORC foci in follicle cells. Immunofluorescence microscopy was used to visualize DmORC (red), DNA (stained with DAPI, in blue), and lamin (green) in a small, early stage 10B egg chamber (this staging is defined in Royzman et al. 1999). The doughnut-like structures are consistently observed in early 10B egg chambers, and are not readily visible in stage 10A egg chambers (data not shown).
In vivo cross-linking of DmORC to chorion DNA elements
To demonstrate that DmORC is associated with chorion DNA in the follicle cells of stage 10 egg chambers, we used a chromatin immunoprecipitation assay (ChIP) (Strahl-Bolsinger et al. 1997). In these experiments, stage 10 egg chambers were dissected from ovaries and fixed with formaldehyde to cross-link proteins to DNA. The fixed samples were sonicated to disrupt the egg chambers and to shear the DNA. To purify DNA cross-linked to DmORC, the resulting material was immunoprecipitated using anti-DmORC2 serum. After reversing the protein–DNA cross-links, PCR reactions were performed on the immunoprecipitates to determine whether chorion sequences were present at enriched levels compared to control DNAs.
We first analyzed egg chambers isolated from wild-type flies to determine whether DmORC associates with ACE3. Primer sets specific for ACE3 and actin, a control DNA, yielded similar amounts of product in PCR reactions containing input (preimmunoprecipitation) DNA (Fig. 3A, lanes 1,2), indicating that the egg chambers tested in this experiment had undergone only limited chorion amplification, consistent with previous reports (Calvi et al. 1998; Royzman et al. 1999). In contrast, PCR reactions performed with DNA that coprecipitated with DmORC yield significantly more ACE3 product compared to actin (Fig. 3A, lane 3), demonstrating the anti-DmORC immunoprecipitate was enriched with ACE3 DNA. Control reactions containing DNA obtained with preimmune serum yielded neither ACE3 nor actin products (Fig. 3A, lane 4). Similar data are obtained when primers for Hsp70 or DmORC1 are used as a negative controls (data not shown). Therefore, we conclude that DmORC associates with ACE3 in vivo and that this association occurs before significant amplification.
Figure 3.
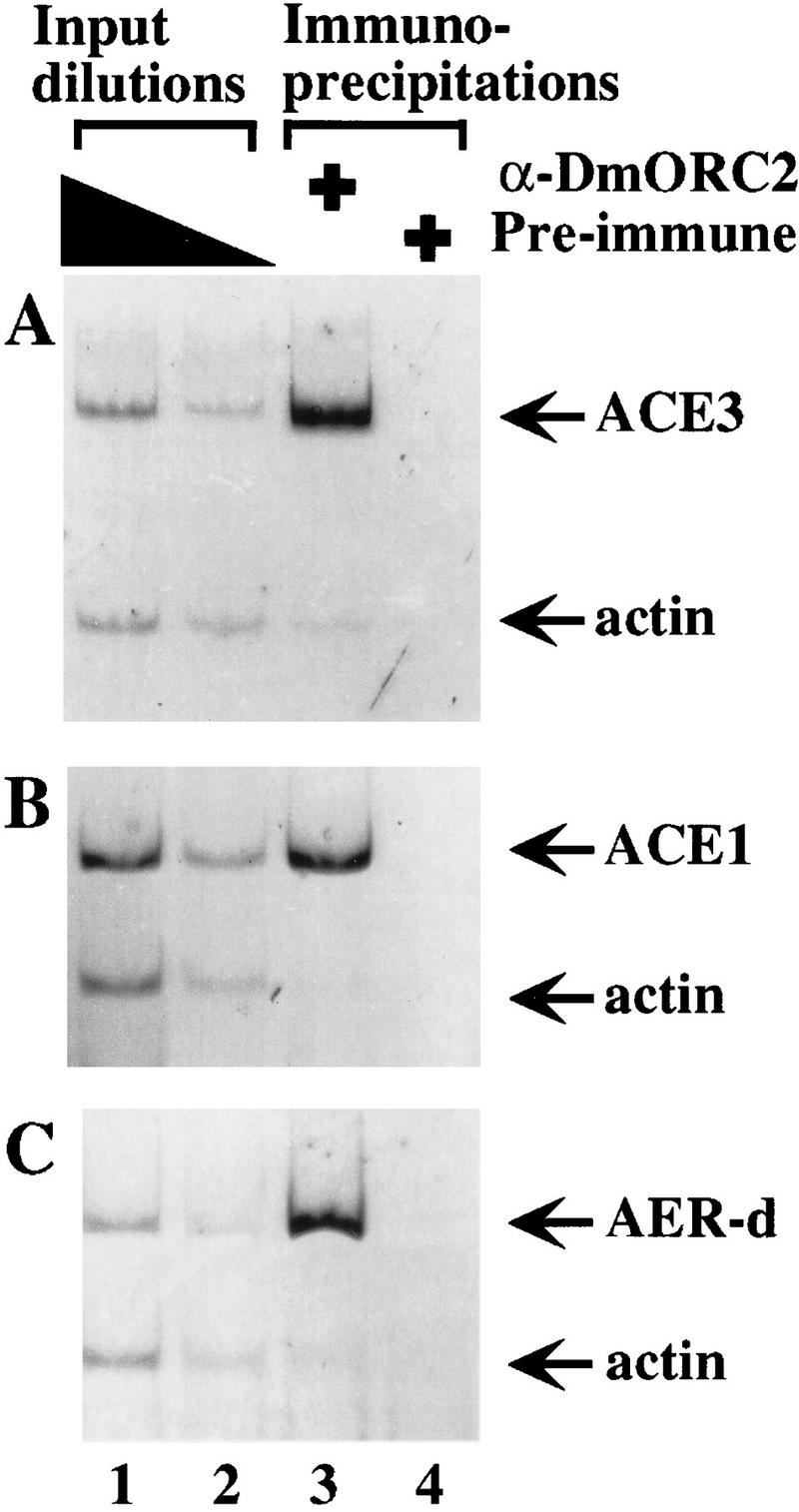
Association of DmORC with chorion loci elements in vivo. Chromatin-containing extracts were prepared from formaldehyde-treated stage 10 egg chambers and were immunoprecipitated with either anti-DmORC2 serum (lane 3) or preimmune serum (lane 4). DNA was amplified using PCR primers specific to ACE3 (A), ACE1 (B), or AER-d (C), and primers specific to actin (A–C). Quantitation of the PCR products indicates the following enrichments of ACE elements compared to actin DNA in the precipitation: 28-fold enrichment for ACE3 compared to actin; 10-fold enrichment of ACE1 compared to actin; and 14-fold enrichment of AER-d compared to actin. These primers were also used to amplify dilutions of DNA isolated from the extracts before immunoprecipitation (Input DNA, lanes 1,2). The amount of input DNA used in these reactions was the equivalent of 0.5% (lane 1) and 0.125% (lane 2) of the total DNA present before immunoprecipitation.
We performed additional experiments to test the association of DmORC with other chorion loci DNA elements. Using primers specific to the amplification control region on the X-chromosome, ACE1 (Spradling et al. 1987), or to AER-d, we observed that both of these elements are enriched in the anti-DmORC2 immunoprecipitate compared to actin (Fig. 3B,C). Because ACE3 and AER-d are only separated by a distance of ∼1.5 kb and the average size of the cross-linked DNA fragments in this experiment is 1.6 kb, this experiment does not definitively show that DmORC recongized both sequences. Additional experiments with more extensive sonication (average DNA size of 450 bp) resulted in similar enrichment of both AER-d and ACE-3 relative to actin, strongly suggesting that DmORC recognizes both sequences (data not shown).
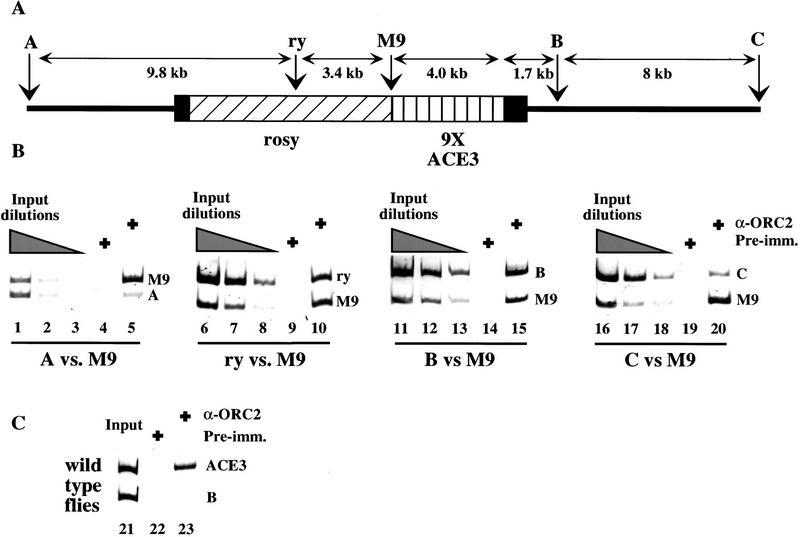
To show that DmORC binds to ACE3 independent of other chorion DNA sequences, the ChIP assay was performed on stage 10 egg chambers isolated from the M9-2 transformant line. For this experiment, we designed primer sets specific to the M9 repeat as well as primer sets recognizing sequences distal from (13.2 and 9.7 kb) and proximal to (3.4 and 1.7 kb) the M9 repeat (respectively, primer sets A, C, ry, and B; see diagram, Fig. 4). PCR analyses indicate the anti-DmORC immunoprecipitate is enriched with the M9 DNA compared to the sequences recognized by the distal primer sets (Fig. 4; cf. lane 5 to lanes 1 and 4, and cf. lane 20 to lanes 16 and 19). The proximal primer sets yield a similar amount of product to the M9-specific primer set in reactions containing anti-DmORC immunoprecipitate DNA (Fig. 4, lanes 10,15). Given that the DNA in the immunoprecipitation was sonicated to an average size of 450 bp, the signal from the ry and B primer sets indicates that DmORC is associated with DNA that proximally flanks the M9 repeat. To demonstrate that these proximal sequences are not associated with DmORC in the absence of ACE3, we performed a PCR analysis of DNA immunoprecipitated from wild-type flies (lacking the M9 construct) using primer set B (Fig. 4C, lanes 21–23). The results of this analysis indicate that sequence recognized by primer set B is not enriched after anti-DmORC precipitation in wild-type flies. Thus, DmORC only localizes to this region of DNA when it is adjacent to the M9 repeat. These findings demonstrate that ACE3 is sufficient to localize DmORC to ACE3 as well as adjacent linked sequences (see Discussion).
Figure 4.
Association of DmORC with the M9 P-element construct in vivo. Chromatin immunoprecipitation analysis of stage 10 egg chambers isolated from the M9-2 transformant line. (A) Diagram of the P-element insertion site of the transformant line M9-2. The diagram shows the location of the PCR primers used for this experiment, primers sets A, B, C, ry, and M9. (B) PCR analysis was performed on DNA isolated from immunoprecipitations with preimmune serum (lanes 4,9,14,19) or anti-DmORC2 serum (lanes 5,10,15,20). PCR analysis was also performed on input DNA equivalent to 2% (lanes 1,6,11,16), 0.5% (lanes 2,7,12,17), and 0.125% (lanes 3,8,13,18) of the total DNA present before immunoprecipitation. (C) PCR analysis was performed on DNA isolated from the wild-type flies after immunoprecipitations with preimune serum (lane 22) or anti-DmORC2 serum (lane 23). The wild-type flies lack the M9 construct. In the M9-2 transformant line, the M9 construct is present at one copy per genome equivalent whereas the A, B, and C. primer sets recognize loci that are present in two copies per genome equivalent. The rosy sequence recognized by the ry primer set is present at three copies per genome equivalent in the M9-transformant line. Consequently, PCR reactions performed with input DNA from the M9-2 transformant line always yield less product for the M9 DNA compared to the other sequences.
ATP-dependent binding of DmORC to ACE3 in vitro
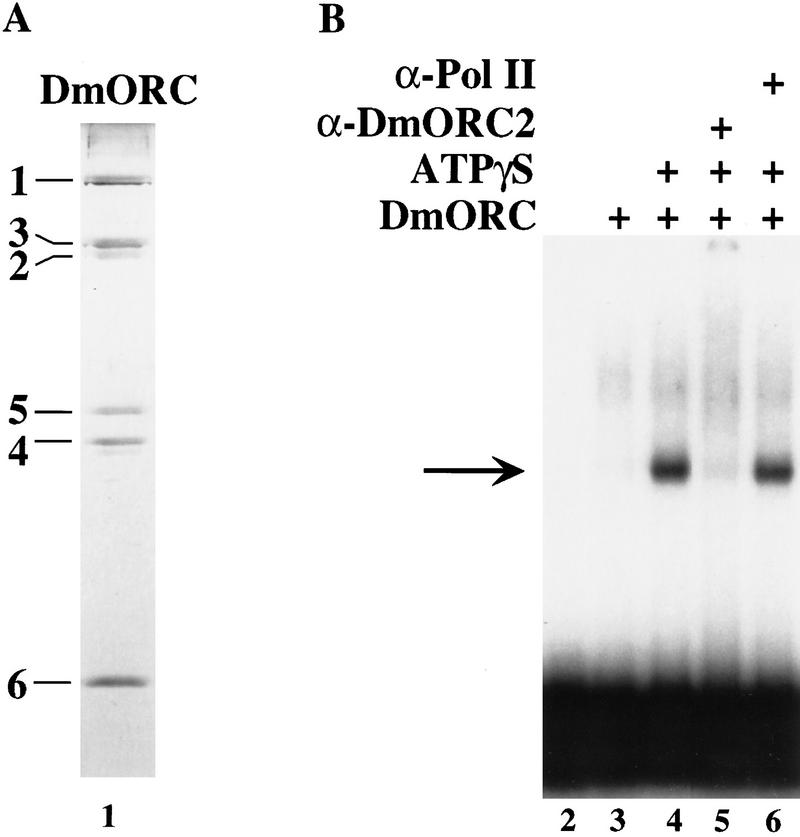
To determine whether DmORC binds to ACE3 in the absence of other cellular proteins, gel mobility-shift assays were performed using DmORC protein purified from embryos (Fig. 5A; Gossen et al. 1995) and radiolabeled ACE3 DNA. Binding reactions containing DmORC yield a discrete shifted species in the presence but not absence of ATPγS (Fig. 5B, lanes 2–4). ATPγS was used in these experiments to ensure that any potential ATPase activities present in our DmORC fraction did not affect the results of our binding assays; however, similar results are obtained when we use ATP instead of ATPγS (data not shown). This ATP-dependent binding activity coelutes with DmORC protein in a glycerol gradient (data not shown). Addition of affinity-purified anti-DmORC2 antibody reduces the amount of this shifted species and results in a corresponding increase in material migrating near the top of the gel or retained in the wells (Fig. 5B, lane 5). In contrast, an anti-RNA polymerase II antibody has no effect the mobility of the ATP-dependent DNA-binding activity (Fig. 5B, lane 6), nor does the anti-DmORC2 antibody affect the mobility of a S. cerevisiae ORC–DNA complex (data not shown). In the gel mobility-shift assay we do observe smeared slow mobility species in the presence of DmORC that are independent of ATP. This binding activity does not coelute with DmORC protein on a glycerol gradient (data not shown) and addition of anti-DmORC2 antibody does not affect the mobility of these species. We conclude DmORC binds ACE3 and, like S. cerevisiae ORC, requires ATP to interact with target DNA.
Figure 5.
ATP-dependent binding of DmORC to ACE3 in vitro. (A) DmORC was purified from embryo extracts. The peak fraction (2 μl) from the final purification step was separated on a 10% polyacrylamide gel and subsequently silver stained (lane 1). The mobility of each DmORC subunit is located to the left of lane 1. (B) Electrophoretic mobility-shift assays were performed with radiolabeled ACE3 DNA. Binding reactions contained, as indicated, DmORC protein (lanes 3–6), ATPγS (lane 4–6), 50 ng affinity purified anti-DmORC2 antibody (lane 5), 50 ng anti-Drosophila RNA polymearse II (PolII) antibody (lane 6). The ATP-dependent species is indicated by the arrow.
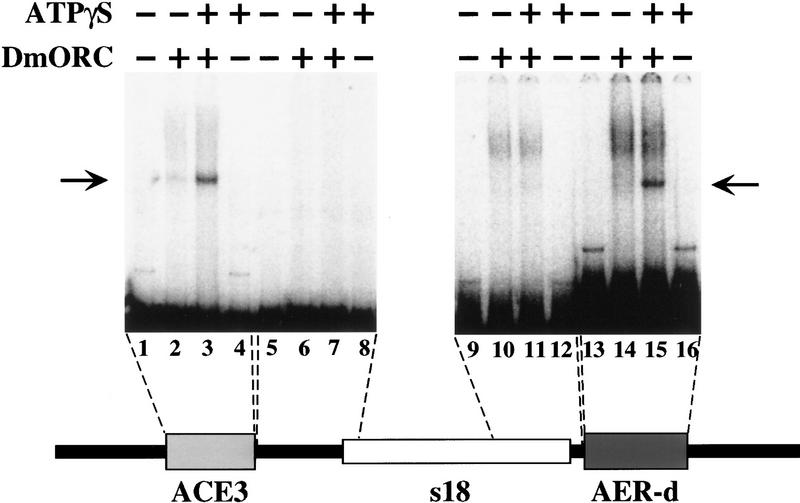
To address the sequence specificity of the ATP-dependent DmORC DNA-binding activity, we compared binding of DmORC to ACE3, AER-d, and DNA sequences that flank these fragments in the Drosophila genome (Fig. 6). We detect ATP-dependent binding to ACE3, as expected, and to AER-d. Little if any ATP-dependent binding to the flanking fragments was detected; however, some ATP-independent binding is observed. Thus, the ATP-dependent binding of DmORC to ACE3 and AER-d is sequence specific. To determine which sequences within ACE3 were recognized by DmORC, we divided ACE3 into three fragments. Individually these ACE3 fragments were not bound by DmORC (data not shown), which could indicate that DmORC does not interact with small (100 bp or less) DNA fragments. To circumvent this potential problem, these fragments were subcloned into the ACE3 flanking fragment (RF, Fig. 7) previously shown not to bind DmORC (Fig. 6, lane 7) to determine whether introduction of these ACE3 sequences was sufficient to allow DmORC recognition. Addition of the middle third of ACE3, which contains an 80-bp AT-rich noncoding sequence highly conserved between four Drosophila species, resulted in strong DmORC-binding activity (Fig. 7, lane 10; cf. lane 2) that is equivalent to that observed with ACE3 (data not shown). Inclusion of either remaining ACE3 fragments (Fig. 7, lanes 6,14) modestly stimulated binding to the RF fragment. Therefore, the sequences in the center of ACE3 are most critical for DmORC DNA binding; however, adjacent sequences also stimulate DmORC-binding, suggesting the presence of several potential DmORC binding sites within ACE3. Consistent with this idea, previous amplification studies found that no single internal deletion within ACE3 completely eliminated amplification (Orr-Weaver et al. 1989).
Figure 6.
DmORC binds specifically to ACE3 and AER-d in vitro. DmORC electrophoretic mobility-shift assays were performed with labeled DNA fragments. The DNA fragments, as diagramed at the bottom of the figure, were ACE3 DNA (lanes 1–4), DNA that flanks ACE3 (lanes 5–8), AER-d DNA (lanes 13–16), or DNA that flanks AER-d (lanes 9–12). Binding reactions contained DmORC (lanes 2,3,6,7,10,11,14,15) and ATPγS (lanes 3,4,7,8,11,12,15,16). The ATP-dependent species are indicated by the arrow.
Figure 7.
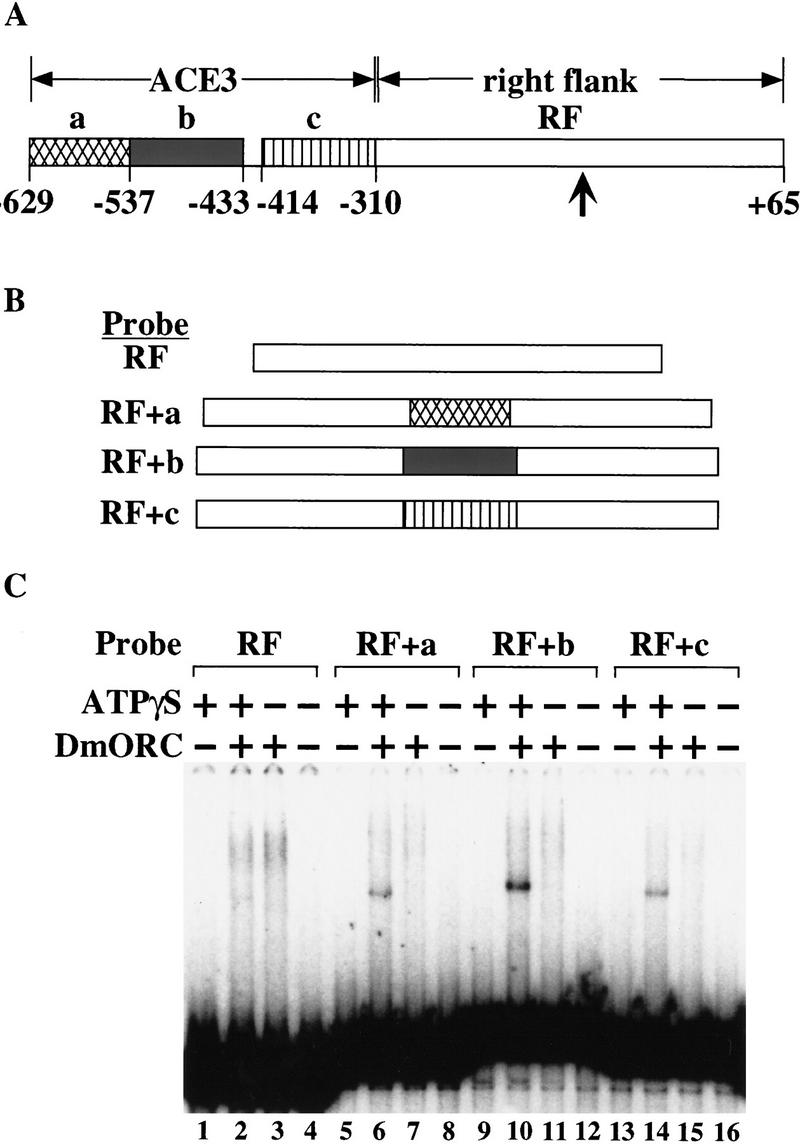
DmORC interacts most strongly with the middle-third of ACE-3. (A) Diagram of ACE3 and flanking DNA (RF fragment). Different parts of the ACE3 (fragments a,b, and c) were subcloned into the StuI site (indicated by a verticle arrow) located in the center of the RF fragment. These subclones were used to make the DNA probes (B), for an electrophoretic mobility-shift assay (C). Binding reactions contained probe RF (lanes 1–4) probe RF + a (lanes 5–8), probe RF + b (lanes 9–12), or probe RF + c (lanes 13–16). Binding reactions also contained DmORC (lanes 2,3,6,7,10,11,14,15) or ATPγS (lanes 1,2,5,6,9,10,13,14).
Discussion
We have used in vivo and in vitro methods to demonstrate that ACE3 can serve as a DNA-binding site for Drosophila ORC. Immunofluoresence and in vivo cross-linking experiments indicate that ACE3 element is sufficient to localize DmORC in vivo. In vitro DNA-binding studies demonstrate that purified DmORC protein specifically binds to ACE3 in an ATP-dependent manner. We also provide data that suggest DmORC binds to another third chromosome chorion DNA element, AER-d, and associates with the X-chromosome chorion element ACE1. Identification of these DNA elements as DmORC-binding sites is an important step toward understanding of how DmORC DNA binding regulates DNA replication and indicates that metazoan ORC can exhibit sequence-specific DNA binding. The results and methods described here also suggest new methods to identify additional ORC-binding sites and therefore, potential metazoan origins of replication.
Several lines of evidence suggest that DmORC will use similar mechanisms to S. cerevisiae ORC (ScORC) to direct initiation of DNA replication. An important role of ScORC in DNA replication is sequence-specific binding to yeast origins of replication, and we observed specific binding of DmORC to amplification control elements. Sequence-specific DNA binding by ScORC is dependent on ATP (Bell and Stillman 1992). In addition, ScORC binds and hydrolyzes ATP, and this hydrolysis activity is inhibited by the presence of origin DNA, suggesting that ATP hydrolysis controls ScORC activity (Klemm et al. 1997). Consistent with a similar mechanism in Drosophila, we observe ATP-dependent binding of DmORC to ACE3 and AER-d. In addition, the amino acid residues in ScORC that are important for ATP binding are conserved in DmORC (Pak et al. 1997). In S. cerevisiae, an essential role for ORC in DNA replication is the recruitment of additional replication control proteins to yeast origins of replication, including Cdc6p, the MCM proteins, and Cdc45p (Aparicio et al. 1997; Tanaka et al. 1997). Because sequence analogs for most of these replication control proteins have been identified in Drosophila (Feger et al. 1995; Su et al. 1996, 1997; Ohno et al. 1998; Feger 1999; Shaikh et al. 1999), it is likely that DmORC will direct replication by recruiting these proteins to Drosophila origins of DNA replication.
Although the sequence-specific DNA binding by DmORC is ATP dependent, we also observed DNA binding by DmORC that is nonspecific and ATP independent (R. Austin, unpubl.). Specifically, DmORC DNA mobility-shift assays performed in the absence of competitor DNA result in the DNA probe being retained in the wells of the gel, suggesting that multiple DmORC molecules are binding to sequences throughout the entire probe, and this binding is independent of ATP. Because the ATP-dependent activity only binds a small amount of probe in the mobility-shift assay, we suggest that only a subset of our purified DmORC protein possesses ATP-dependent DNA-binding activity. The DmORC protein used in these experiments was isolated from 0- to 12-hr embryos, and given that DmORC is present at higher levels in early embryos versus late embryos (Gossen et al. 1995), the majority of the protein should be derived from early embryos. Consistent with reported observations (Chesnokov et al. 1999), we speculate that the nonspecific ATP-independent DNA activity represents DmORC protein from younger embryos in which inter-origin spacing is relatively small (Spradling and Orr-Weaver 1987). We propose that the ATP-dependent sequence-specific DNA-binding activity is due to the subset of DmORC protein derived from older embryos where origin usage is more restricted. This raises the possibility that the developmental change in inter-origin spacing could correlate with a shift from ATP-independent to ATP-dependent DmORC function. Future studies of DmORC purified from more restricted embryo populations will allow a direct test of this hypothesis.
We have shown that DmORC specifically binds ACE3, but studies of DmORC localization in follicle cells suggests mechanisms must also exist to prevent DmORC from binding to nonamplifying loci. These mechanisms could include cofactors that possess chromatin remodeling activity or that physically interact with DmORC to restrict binding to the amplified regions. Interestingly, recent data indicate that Drosophila E2F controls binding of DmORC to the chorion loci. E2F mutants predicted to have reduced DNA-binding activity fail to form discrete foci of DmORC localization (Royzman et al. 1999). One interpretation of these data is that E2F affects expression of the putative cofactors mentioned above. Alternatively, E2F may regulate DmORC complex function by controlling expression of one or more of the DmORC subunits. Indeed, E2F-binding sites present in the DmORC1 promoter are important for promoter activity, and overexpression of DmORC1 in the follicle cells leads to induction of inappropriate genomic replication (Asano and Wharton 1999). The DNA-binding domain of E2F itself may be sufficient to restrict DmORC–DNA binding because a dE2F mutation predicted to lack transcriptional activity but with an intact DNA-binding domain exhibits normal DmORC localization in follicle cells (Royzman et al. 1999). Although these models are based on our analysis of DmORC localization in follicle cells, similar mechanisms could be used to regulate DmORC–DNA binding and origin usage in development.
Our data support previously suggested hypotheses that metazoan origins have multiple ORC DNA-binding sites. These hypotheses were based on two types of observations. First, replication has been observed to initiate at multiple sites within metazoan origins (for review, see DePamphilis 1999), including the Drosophila third chromosome chorion locus (Delidakis and Kafatos 1989; Heck and Spradling 1990). Second, metazoan origins of replication appear to have multiple elements that contribute their function. This modular organization has been observed at the chorion locus, where multiple sequences contribute to the overall levels of amplification (for review, see Orr-Weaver 1991), and at the human β-globin locus, where multiple elements are required for origin function (Aladjem et al. 1998). These observations were believed to be the consequences of ORC binding multiple sites within an origin.
Our in vivo studies are consistent with multiple DmORC molecules associating with ACE3-linked DNA. Insertion of the ACE3 element at a new locus in the genome results in DmORC association with sequences at least 3.4 kb but less than 9 kb from ACE3. Our data are consistent with a model in which ACE3 nucleates DmORC–DNA binding, and in turn this would lead to DmORC interacting with adjacent binding sites in the chorion locus. Although DmORC appears to associate with sequences outside of ACE3 in the chromatin immunoprecipitation experiments (Fig. 4B), these associations are still absolutely dependent on the presence of the M9 construct (Fig. 4C). It is possible that these associations are the result of the formation of a higher order complex at the origin that contains multiple DmORC molecules. This situation would be analogous to the ability of four DNA-binding sites to direct the assembly of 20–40 molecules of the dnaA initiator protein at the Eschrichia coli chromosomal origin of replication (oriC; for review, see Skarstad and Boye 1994). Similarly, S. pombe ORC contains a domain that binds to AT-rich DNA sequences, and it has been suggested that S. pombe ORC may interact with the multiple AT-rich elements that contribute to S. pombe origin function (Chuang and Kelly 1999). Currently we do not know the mechanism that leads to what appears to be multiple DmORC molecules associating with ACE3-linked DNA. It is possible that this association is related to the two populations of DmORC that we observe in our in vitro DNA-binding studies (see above), with some DmORC molecules making specific contact with ACE3 and others making nonspecific contacts with adjacent sequences. Alternatively, this type of extensive association may be the result of a specialized function of the ACE3 element that assists in the amplification process. Characterization of DmORC association with other nonamplifying origins of replication and further studies of the DNA-binding specificity of the different popluations of DmORC will be required to distinguish between these possibilities.
The in vivo methods described here for studying DmORC–DNA interactions in Drosophila follicle cells can be adapted to identify additional DmORC DNA-binding sites and potential replication origins in other tissues. Identification of these sequences will be important for understanding how ORC regulates metazoan replication and how origin usage is developmentally regulated. Using these sequences in combination with in vitro replication systems (Crevel and Cotterill 1991; Walter et al. 1998; Chesnokov et al. 1999) and the future purification of ORC from different tissues will allow for the design of more precise experiments to understand how metazoan DNA replication is regulated.
Materials and methods
Antibodies
The anti-Drosophila ORC2 serum has been described previously (Pinto et al. 1999; Royzman et al. 1999). For affinity purification of the anti-DmORC2 antibody, purified DmORC2 protein (Royzman et al. 1999) was coupled to cyanogen bromide-activated Sepharose (Pharmacia) and the antibody was purified from sera using standard methods (Harlow and Lane 1988). For the Western blot, samples were applied to a 10% SDS-PAGE and subsequently transferred to nitrocellulose (Schleicher and Schuell BA85). After blocking with 5% nonfat dry milk, the blot was probed with using a 1:2500 dilution of the anti-DmORC serum, and bound primary antibody was detected using an HRP-conjugated anti-rabbit antibody (Amersham) and a chemiluminescent substrate (Pierce SuperSignal).
Immunofluorescence of egg chambers
Egg chambers were stained as described previously (Royzman et al. 1999) except that the anti-DmORC2 serum was diluted in lysis buffer (Strahl-Bolsinger et al. 1997). The washes after incubating with the primary antibody were also performed with lysis buffer. The egg chambers shown in Figure 1 were mounted in clearing solution (2:1 benzyl benzoate:benzyl alcohol; Theurkauf and Hawley 1992) containing 50 mg/ml n-propyl gallate, and the egg chambers shown in Figure 2 were mounted in VectaShield (Vector Laboratories). Nuclear lamin Dm0 staining was performed using Ab101, an antibody provided by K. McCall and H. Steller (Smith et al. 1987). The antibody was diluted 1:1.5 and detected using a FITC-conjugated anti-mouse secondary antibody (Jackson Immuno Research Laboratories). Images were collected with a 100× oil objective using a CCD camera and were processed with the CELLscan 2.1 system (Scanalytics) for the images in Figure 1 or were processed with the DeltaVision system for the image in Figure 2.
Drosophila transformant lines
Anti-DmORC staining was performed on the previously characterized transformant lines: M9-2 is described in Carminati et al. (1992); A48O28-2 (shown in Fig. 1), A48O28-7, A54O18-1 (shown in Fig. 1), A54O18-2, A54O18-4, A54O18-6, A54O18-7, A54O18-8, A54O18-9, A54O18-11, A54O18-12, and A54O18-13 are described in Orr-Weaver et al. (1989).
Chromatin immunoprecipitation assay
Stage 10 egg chambers were dissected from ovaries of fattened flies in nonsupplemented Grace's medium (GIBCO-BRL) and were stored on ice for up to 2 hr. The nonsupplemented Grace's medium was replaced with room temperature supplemented Grace's medium (GIBCO-BRL). Formaldehyde (Mallinckrodt) was added to a final concentration of 2% and cross-linking was allowed to proceed for 15 min at room temperature on a rotator. The cross-linking reaction was stopped by addition of glycine at a final concentration of 0.125 mm and incubating 5 min. The cross-linked egg chambers were washed twice with 1 ml of TBS [20 mm Tris (pH 7.6), 140 mm NaCl], then twice with 1 ml of lysis buffer (Strahl-Bolsinger et al. 1997). The egg chambers were suspended in 500 μl of lysis buffer, frozen in liquid nitrogen, and stored at −70°C. When sufficient egg chambers had been collected and processed (300 egg chambers per immunoprecipitation reaction), the fixed egg chambers were thawed and disrupted by sonication. Sonication consisted of three treatments of 12 sec each using a Branson 250 sonicator at power setting 1.5 and 100% duty cycle. To obtain an average sonication size of 450 bp, an additional three rounds of sonication were performed. Between pulses the egg chambers were incubated on ice for 2 min or more. All postsonication procedures were performed as described previously (Strahl-Bolsinger et al. 1997) with the following exceptions: the immunoprecipitation reactions were performed with either 2 μl of preimmune serum or 2 μl of anti-DmORC2 serum; the immunoprecipitated DNA was dissolved in a final volume of 20 μl; RNase treatment was omitted; and PCR reactions were subjected to 32 rounds of cycling. The PCR products were analyzed and quantitated using a Molecular Dynamics FluorImager and ImagQuant software.
The ACE3, ACE1, actin, ry (rosy) primer sets were described previously (Royzman et al. 1999). These primers yield PCR products that, respectively, are 323, 252, 165, and 207 bp in size. The sequences of the other primers used here are: AAAGCTAAAACTAAATTAATTTGTGGGG and GGTTCCAGCCGGTTTTTCTGATAAAACC for AER-d which produce a 276-bp, product; GGCGTAAATGTTCTCGAATTCCCG and GACGCCAACACGAACGCGTCGGTC for DmORC1, which produce a 277-bp product; CCGATTTCGGCGCGACTGCTACCCG and ATGCGGTCGGAATCTTACGTATGGG for the M9specific primer set, which produce a 160-bp product; CTGCTGACCTCTTTGGTCAACTTCAC and GGCACCCAGTGTGCCTGTTGACCC for primer set A and produce a 139-bp product; GGCAATATGCCGTGGGTTGGGTAGG and GCTGACCTGACAATATCATTAAGGG for primer set C, which produce a 204-bp product; and GTGTTTCTTTGTGTGTGCGAAAGCGCTC and AGAACCGAATTTTTGCTAGATCTTCCTC for primer set B, which produce a 219-bp product. To design the A, B, and C primer sets, portions of genomic clones surrounding the M9-2 P-element insertion site (Carminati et al. 1992) were sequenced.
Plasmids
The constructs used in Figure 7 were made by inserting the Asp-718/MfeI, MfeI/ApoI, or ApoI/BamHI blunted fragments from ACE3 into the StuI site of plasmid A31O4 (Orr-Weaver et al. 1989) (the StuI site is located at −120 relative to the s18 transcription start site).
Purification of Drosophila ORC
Drosophila ORC was purified from 0- to 12-hr embryo extracts as outlined previously (Gossen et al. 1995) using anti-DmORC2 antiserum to follow DmORC activity. Details on extract preparation, heparin–agarose chromatography, Sephacryl S-300 chromatography, and Mono Q chromatography are described elsewhere (Austin and Biggin 1996). For the Mono S chromatography, the Mono Q fractions with the peak DmORC2 activity from two chromatography runs were pooled together and loaded onto a Mono S HR5/5 (Pharmacia) column and eluted with a 10-column volume 0.1 m KCl to a 0.6 m KCl salt gradient. The peak Mono S fractions were pooled, diluted with buffer HEMG [25 mm HEPES (K+) (pH 7.6), 12.5 mm MgCl2, 0.1 mm EDTA, 10% glycerol, 1 mm dithiothreitol, 0.01% NP-40] to a final salt concentration of 0.1 m KCl, bound to a 50-μl SP-Sepharose (Pharmacia) column, and eluted with 0.6 m KCl/HEMG. The SP-Sepharose eluate (50 μl) was fractionated on a 15%–35% glycerol gradient (Bell and Stillman 1992) containing 0.1 m KCl and buffer HEMG.
Gel mobility-shift assay
Binding reactions (5.5 μl) contained 0.5 mm ATPγS, 90 μg/ml poly[d(G-C)][d(G-C)], 9 μg/ml MspI-digested phage lambda DNA, 0.14 mg/ml BSA, ∼1.5 fmole of end-labeled probe, and 4.35 μl total volume of DmORC protein (typically 1–2 μl), with or without the addition of 0.1 m KCl/HEMG buffer. Binding reactions were set up on ice, incubated for 10 min at room temperature, loaded onto a native polyacrylamide gel (Rao and Stillman 1995), and electrophoresed for 150 min at 300 V. The resulting polyacrylamide gel was dried and exposed to X-ray film or a PhosphorImager screen (Molecular Dynamics). Probes used for these assays were a 319-bp Asp-718/BamHI fragment for ACE3; a 374-bp BamHI/AflII fragment for the ACE3 flanking probe; a 364-bp BglII/SpeI fragment for AER-d, and a 312-bp fragment for the AER-d flanking probe. Relative to the transcription start site for the s18 chorion gene, these probes are located at −629 to −310, −310 to +65, +878 to +1242, and +566 to +878, respectively. To achieve equivalent labeling activity between the ACE3 and flanking fragment probes, the plasmid A31O4 was cut and labeled on the unique BamHI site by fill-in reaction, and the probes were released by secondary digestion with the appropriate restriction enzyme. Similarly, AER-d and its flanking fragment were labeled using the unique BglII site and releasing with the appropriate enzyme. The probes for Figure 7 were all labeled on the BamHI site and released with AflII.
Acknowledgments
We thank Frank Gertler for microscopy assistance, Arno Greenleaf for the RNA polymerase II antibody, Mark Biggin for storage of extract fractions, and Giovanni Bosco and members of the Bell and Orr-Weaver laboratories for discussion and technical assistance. Jacqueline Lees and Anindya Dutta provided constructive comments on this manuscript. These studies were supported by NIH grants GM39341 and GM57960 to T.O.-W., GM52339 to S.P.B., and GM18170 to R.J.A. and awards from the Searle/Chicago Community Trust and The Rita Allen Foundation to S.P.B.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL spbell@mit.edu; FAX (617) 253-8699
References
- Aladjem MI, Rodewald LW, Kolman JL, Wahl GM. Genetic dissection of a mammalian replicator in the human beta-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Asano M, Wharton RP. E2F mediates developmental and cell cycle regulation of ORC1 in Drosophila. EMBO J. 1999;18:2435–2448. doi: 10.1093/emboj/18.9.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RJ, Biggin MD. Purification of the Drosophila RNA polymerase II general transcription factors. Proc Natl Acad Sci. 1996;93:5788–5792. doi: 10.1073/pnas.93.12.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Callan HG. Replication of DNA in the chromosomes of eukaryotes. Proc R Soc Lond B Biol Sci. 1972;181:19–41. doi: 10.1098/rspb.1972.0039. [DOI] [PubMed] [Google Scholar]
- Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes & Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati JL, Orr-Weaver TL. Changes in DNA replication during animal development. In: DeDamphilis ML, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 409–434. [Google Scholar]
- Carminati JL, Johnston CG, Orr-Weaver TL. The Drosophila ACE3 chorion element autonomously induces amplification. Mol Cell Biol. 1992;12:2444–2453. doi: 10.1128/mcb.12.5.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter PB, Dunphy WG. Identification of a novel 81-kDa component of the Xenopus origin recognition complex. J Biol Chem. 1998;273:24891–24897. doi: 10.1074/jbc.273.38.24891. [DOI] [PubMed] [Google Scholar]
- Carpenter PB, Mueller PR, Dunphy WG. Role for a Xenopus Orc2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- Chesnokov I, Gossen M, Remus D, Botchan M. Assembly of functionally active Drosophila origin recognition complex from recombinant proteins. Genes & Dev. 1999;13:1289–1296. doi: 10.1101/gad.13.10.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang RY, Kelly TJ. The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK, Kelly TJ. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Crevel G, Cotterill S. DNA replication in cell-free extracts from Drosophila melanogaster. EMBO J. 1991;10:4361–4369. doi: 10.1002/j.1460-2075.1991.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cicco DV, Spradling AC. Localization of a cis-acting element responsible for the developmentally regulated amplification of Drosophila chorion genes. Cell. 1984;38:45–54. doi: 10.1016/0092-8674(84)90525-7. [DOI] [PubMed] [Google Scholar]
- Delidakis C, Kafatos FC. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989;8:891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML. Origins of DNA replication. In: DePamphilis ML, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 45–86. [Google Scholar]
- ————— Replication origins in metazoan chromosomes: Fact or fiction? BioEssays. 1999;21:5–16. doi: 10.1002/(SICI)1521-1878(199901)21:1<5::AID-BIES2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JF. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey DD, Kim SM, Todorov IT, Huberman JA. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- Feger G. Identification and complete cDNA sequence of the missing Drosophila MCMs: DmMCM3, DmMCM6 and DmMCM7. Gene. 1999;227:149–155. doi: 10.1016/s0378-1119(98)00596-4. [DOI] [PubMed] [Google Scholar]
- Feger G, Vaessin H, Su TT, Wolff E, Jan LY, Jan YN. dpa, a member of the MCM family, is required for mitotic DNA replication but not endoreplication in Drosophila. EMBO J. 1995;14:5387–5398. doi: 10.1002/j.1460-2075.1995.tb00223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin KA, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1995;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- Gossen M, Pak DT, Hansen SK, Acharya JK, Botchan MR. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Heck MM, Spradling AC. Multiple replication origins are used during Drosophila chorion gene amplification. J Cell Biol. 1990;110:903–914. doi: 10.1083/jcb.110.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RY, Kowalski D. Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res. 1996;24:816–823. doi: 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Mechali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- Kim SM, Huberman JA. Multiple orientation-dependent, synergistically interacting, similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol. 1998;18:7294–7303. doi: 10.1128/mcb.18.12.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- Landis G, Kelley R, Spradling AC, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leatherwood J. Emerging mechanisms of eukaryotic DNA replication initiation. Curr Opin Cell Biol. 1998;10:742–748. doi: 10.1016/s0955-0674(98)80117-8. [DOI] [PubMed] [Google Scholar]
- Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes & Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Kowalski D. Functional equivalency and diversity of cis-acting elements among yeast replication origins. Mol Cell Biol. 1997;17:5473–5484. doi: 10.1128/mcb.17.9.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- Ohno K, Hirose F, Inoue YH, Takisawa H, Mimura S, Hashimoto Y, Kiyono T, Nishida Y, Matsukage A. cDNA cloning and expression during development of Drosophila melanogaster MCM3, MCM6 and MCM7. Gene. 1998;217:177–185. doi: 10.1016/s0378-1119(98)00358-8. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL. Drosophila chorion genes: Cracking the eggshell's secrets. BioEssays. 1991;13:97–105. doi: 10.1002/bies.950130302. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Johnston CG, Spradling AC. The role of ACE3 in Drosophila chorion gene amplification. EMBO J. 1989;8:4153–4162. doi: 10.1002/j.1460-2075.1989.tb08600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osheim YN, Miller OL., Jr Novel amplification and transcriptional activity of chorion genes in Drosophila melanogaster follicle cells. Cell. 1983;33:543–553. doi: 10.1016/0092-8674(83)90435-x. [DOI] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell. 1997;91:311–323. doi: 10.1016/s0092-8674(00)80415-8. [DOI] [PubMed] [Google Scholar]
- Pinto S, Quintana DG, Smith P, Mihalek RM, Hou Z-H, Boynton S, Jones CJ, Hendricks M, Velinzon K, Wohlschlegel JA, Austin RJ, Lane WS, Tully T, Dutta A. latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron. 1999;23:45–54. doi: 10.1016/s0896-6273(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Quintana DG, Hou Z, Thome KC, Hendricks M, Saha P, Dutta A. Identification of HsORC4, a member of the human origin of replication recognition complex. J Biol Chem. 1997;272:28247–28251. doi: 10.1074/jbc.272.45.28247. [DOI] [PubMed] [Google Scholar]
- Quintana DG, Thome KC, Hou ZH, Ligon AH, Morton CC, Dutta A. ORC5L, a new member of the human origin recognition complex, is deleted in uterine leiomyomas and malignant myeloid diseases. J Biol Chem. 1998;273:27137–27145. doi: 10.1074/jbc.273.42.27137. [DOI] [PubMed] [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Marahrens Y, Stillman B. Functional conservation of multiple elements in yeast chromosomal replicators. Mol Cell Biol. 1994;14:7643–7651. doi: 10.1128/mcb.14.11.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanowski P, Madine MA, Rowles A, Blow JJ, Laskey RA. The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Curr Biol. 1996;6:1416–1425. doi: 10.1016/s0960-9822(96)00746-4. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royzman I, Orr-Weaver TL. S phase and differential DNA replication during Drosophila oogenesis. Genes Cells. 1998;3:767–776. doi: 10.1046/j.1365-2443.1998.00232.x. [DOI] [PubMed] [Google Scholar]
- Royzman I, Austin RJ, Bosco G, Bell SP, Orr-Weaver TL. ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Genes & Dev. 1999;13:827–840. doi: 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Sawado T, Yamaguchi M, Shinomiya T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha–dE2F locus of Drosophila melanogaster. Mol Cell Biol. 1999;19:547–555. doi: 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TH, Gottlieb S, Sellinger B, Chen F, Roe BA, Oakey RJ, Emanuel BS, Budarf ML. Characterization of CDC45L: A gene in the 22q11.2 deletion region expressed during murine and human development. Mamm Genome. 1999;10:322–326. doi: 10.1007/s003359900996. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Boye E. The initiator protein DnaA: Evolution, properties and function. Biochem Biophys Acta. 1994;1217:111–130. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- Smith DE, Gruenbaum Y, Berrios M, Fisher PA. Biosynthesis and interconversion of Drosophila nuclear lamin isoforms during normal growth and in response to heat shock. J Cell Biol. 1987;105:771–790. doi: 10.1083/jcb.105.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A, Orr-Weaver T. Regulation of DNA replication during Drosophila development. Annu Rev Genet. 1987;21:373–403. doi: 10.1146/annurev.ge.21.120187.002105. [DOI] [PubMed] [Google Scholar]
- Spradling AC, de Cicco DV, Wakimoto BT, Levine JF, Kalfayan LJ, Cooley L. Amplification of the X-linked Drosophila chorion gene cluster requires a region upstream from the s38 chorion gene. EMBO J. 1987;6:1045–1053. doi: 10.1002/j.1460-2075.1987.tb04857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Su TT, Feger G, O'Farrell PH. Drosophila MCM protein complexes. Mol Biol Cell. 1996;7:319–329. doi: 10.1091/mbc.7.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su TT, Yakubovich N, O'Farrell PH. Cloning of Drosophila MCM homologs and analysis of their requirement during embryogenesis. Gene. 1997;192:283–289. doi: 10.1016/s0378-1119(97)00107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Theis JF, Newlon CS. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol Cell Biol. 1994;14:7652–7659. doi: 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugal T, Zou-Yang XH, Gavin K, Pappin D, Canas B, Kobayashi R, Hunt T, Stillman B. The Orc4p and Orc5p subunits of the Xenopus and human origin recognition complex are related to Orc1p and Cdc6p. J Biol Chem. 1998;273:32421–32429. doi: 10.1074/jbc.273.49.32421. [DOI] [PubMed] [Google Scholar]
- Walter J, Newport JW. Regulation of replicon size in Xenopus egg extracts. Science. 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]