Abstract
mRNA capping is a cotranscriptional event mediated by the association of capping enzyme with the phosphorylated carboxy-terminal domain (CTD) of RNA polymerase II. In the yeast Saccharomyces cerevisiae, capping enzyme is composed of two subunits, the mRNA 5′-triphosphatase (Cet1) and the mRNA guanylyltransferase (Ceg1). Here we map interactions between Ceg1, Cet1, and the CTD. Although the guanylyltransferase subunit can bind alone to the CTD, it cannot be guanylylated unless the triphosphatase subunit is also present. Therefore, the yeast mRNA guanylyltransferase is regulated by allosteric interactions with both the triphosphatase and CTD.
Keywords: Pol II, CTD, guanylyltransferase, triphosphatase
Eukaryotic pre-mRNAs are transcribed by RNA polymerase II (Pol II) and undergo several processing events before becoming mature mRNA. These events, including 5′ capping, splicing, and polyadenylation, are completed in the nucleus before the mRNA is transported to the cytoplasm and translated. Capping of the 5′ end of the mRNA is the first detectable mRNA processing event, occurring by the time the transcript is only 25–30 nucleotides long (Jove and Manley 1984; Rasmussen and Lis 1993). This cotranscriptional event is mediated by recruitment of the capping enzyme machinery to the phosphorylated carboxy-terminal domain (CTD) of the largest subunit of Pol II (Cho et al. 1997; McCracken et al. 1997; Yue et al. 1997; Ho et al. 1998).
Capping occurs by a series of three enzymatic reactions. The 5′ triphosphate end of the nascent RNA Pol II transcript is cleaved by 5′ RNA triphosphatase to produce a diphosphate terminus. RNA guanylyltransferase forms a covalent enzyme–GMP complex and subsequently caps the RNA substrate by adding the guanosine residue in a 5′-5′ triphosphate linkage. The cap is then methylated at the guanine N7 position by RNA (guanine-7) methyltransferase, completing the m7GpppN, or cap0, structure (for review, see Mizumoto and Kaziro 1987; Shuman 1995). In higher eukaryotes, a bifunctional monomeric polypeptide carries both RNA triphosphatase and guanylyltransferase activities (Mizumoto and Kaziro 1987). Recent characterizations of the capping enzymes from Caenorhabidits elegans, mouse, and human reveal an amino-terminal RNA triphosphatase domain and a carboxy-terminal guanylyltransferase domain (McCracken et al. 1997; Takagi et al. 1997; Wang et al. 1997; Yue et al. 1997; Tsukamoto et al. 1998; Yamada-Okabe et al. 1998). In contrast, the yeast Saccharomyces cerevisiae capping enzyme is a heterodimer. The CET1 gene encodes the 62-kD triphosphatase subunit (Tsukamoto et al. 1997), and the CEG1 gene encodes the 52-kD guanylyltransferase subunit (Itoh et al. 1987; Shibagaki et al. 1992). Both CET1 and CEG1 genes are essential for viability.
The guanylyltransferase mechanism is conserved among eukaryotes and virus, involving a covalent enzyme-guanylate intermediate in which GMP is linked to the ε-amino group of the active site lysine (for review, see Mizumoto and Kaziro 1987). The recently solved structures of the Chlorella virus PBCV-1 guanylyltransferase provides insight into the mechanism of guanylylation (Hakansson et al. 1997; Hakansson and Wigley 1998). The viral enzyme has two domains with the active site located in a cleft between them. The covalent linkage between enzyme and GMP may require a conformation change that transiently brings the two domains closer together. Although sequence similarity is mostly limited to regions around the active site, the viral guanylyltransferase serves as a useful model for the eukaryotic guanylyltransferases.
In contrast to the conservation of the guanylyltransferase, the RNA triphosphatase shows diversity over evolution. The triphosphatase domain of higher eukaryote capping enzyme is related to the protein tyrosine phosphatases (McCracken et al. 1997; Takagi et al. 1997; Wang et al. 1997; Yue et al. 1997; Tsukamoto et al. 1998; Yamada-Okabe et al. 1998), whereas the yeast Cet1 subunit does not resemble any known phosphatase (Tsukamoto et al. 1997).
Previously, we reported that the S. cerevisiae capping enzyme is recruited to the Pol II transcription complex by the phosphorylation of CTD (Cho et al. 1997). Here, we explore the nature of the Cet1–Ceg1 interaction and the mechanism of recruitment to the phosphorylated CTD. Using a combination of biochemical and genetic experiments, we map the subunit interaction to residues 205–265 of Cet1 and the carboxy-terminal region of Ceg1. Furthermore, we found that the phosphorylated CTD interacts directly with Ceg1, but inhibits its ability to be guanylylated. The interaction between Cet1 and Ceg1 reverses this inhibition. These results demonstrate that guanylyltransferase activity is regulated allosterically by interactions with the triphosphatase and CTD.
Results
Mutations in the carboxy-terminal region of CEG1 are suppressed by CET1 overexpression
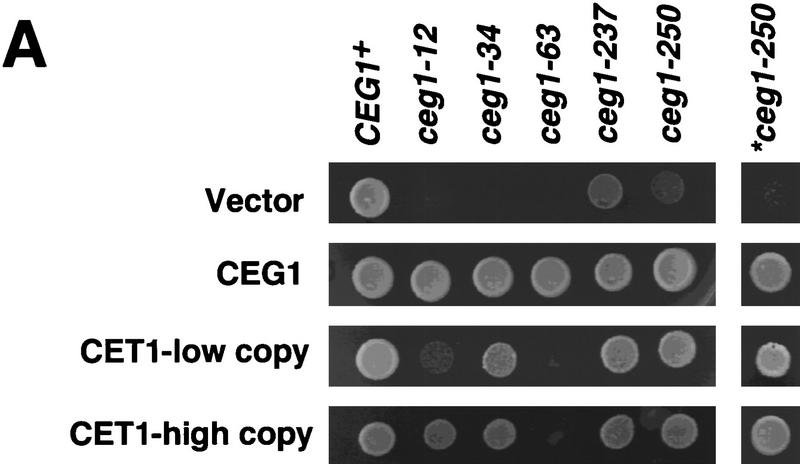
To explore the Ceg1–Cet1 heterodimer interaction, we used a combination of genetic analyses and in vitro protein interaction studies. Yeast strains carrying a variety of ceg1 temperature-sensitive alleles were generated previously (Fresco and Buratowski 1996). To determine whether any of these mutants might be defective in interactions with Cet1, the strains were transformed with an additional copy of CET1 on a low-copy centromeric plasmid. Overexpression of the triphosphatase subunit might compensate for weakened subunit interactions. Strains were also transformed with vector or with wild-type CEG1 and assayed for growth at the permissive or nonpermissive temperature by spotting.
As shown in Figure 1A, ceg1-34 (P346L) is suppressed by an additional centromeric copy of CET1 (CET1-low copy), whereas allele ceg1-63 (D152N) is not. Allele ceg1-12 (C354Y) showed partial suppression. Alleles ceg1-237 (G45A) and ceg1-250 (P288S) exhibit a leaky phenotype when present on a plasmid; the background growth in this assay was too high to visualize a different growth phenotype from vector alone. When ceg1-250 replaced CEG1 in the natural chromosomal locus, the temperature-sensitive phenotype was much more pronounced (*ceg1-250) and could be suppressed by a centromeric copy of CET1. To see whether higher levels of Cet1 overexpression would further suppress the ceg1 mutant phenotypes, a 2μ plasmid bearing CET1 (CET1-high copy) was transformed into the various ceg1 mutant strains. In this case, ceg1-12 as well as ceg1-34 and ceg1-250 were suppressed (Fig. 1A).
Figure 1.
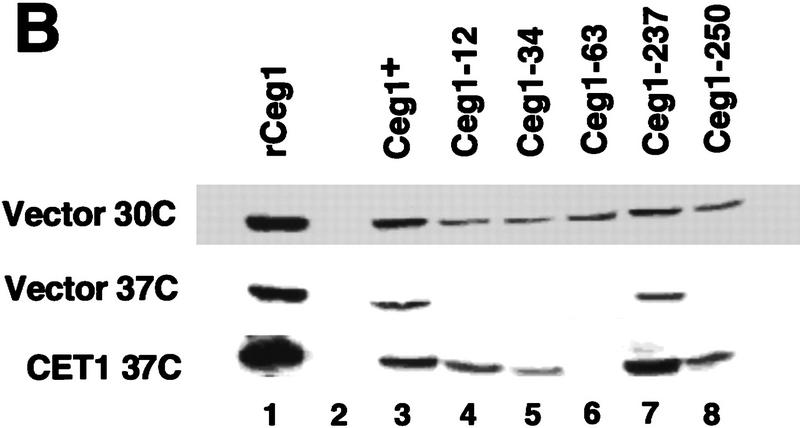
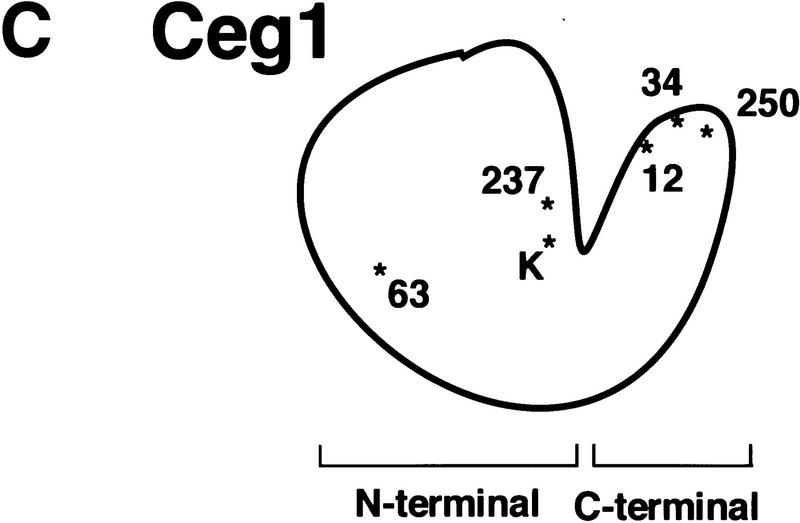
CET1 overexpression suppresses CEG1 temperature-sensitive mutants in an allele-specific manner. (A) Growth of wild-type and conditional ceg1 strains. Strains harboring the indicated plasmids and CEG1 alleles were assayed for growth at 30°C and 37°C. Low copy indicates a centromeric plasmid; high copy indicates a 2μ plasmid. All strains contain a plasmid-borne ceg1 allele covering a deletion of the chromosomal CEG1 locus, with the exception of *ceg1-250, in which the mutant allele is integrated at the chromosomal locus. (B) Whole-cell extracts were prepared from wild-type or ceg1 mutant strains grown for 4.5 hr at 30°C or 37°C. Extracts were assayed by immunoblotting with anti-Ceg1 antibody. (Lane 1) 100 ng Ceg1; (lane 2) no extract; (lanes 3–8) 40 μg of extract from the indicated strain. (C) Schematic representation of Ceg1 as modeled by the Chlorella virus PBCV-1 guanylyltransferase structure (Hakansson et al. 1997). Relative locations of mutant residues are denoted by * and the allele number; (K) active site lysine.
The temperature-sensitive phenotypes of alleles ceg1-12, ceg1-34, ceg1-63, and ceg1-250 are the result of Ceg1 protein instability at 37°C (Fresco and Buratowski 1996). Therefore, it seemed likely that the mechanism of suppression was Ceg1 protein stabilization. To test this hypothesis, extracts were prepared from the strains in Figure 1A after 4.5 hr of growth at 30°C or 37°C, and steady-state levels of Ceg1 protein were monitored by immunoblot analysis. As reported previously (Fresco and Buratowski 1996), Ceg1 is unstable in alleles ceg1-12, ceg1-34, ceg1-63, and ceg1-250, whereas allele ceg1-237 encodes a stable protein (Fig. 1B). In the extracts prepared from strains carrying a high copy CET1 plasmid, protein stabilization was observed with mutant proteins Ceg1-12, Ceg1-34, and Ceg1-250, but not for Ceg1-63 (Fig. 1B). This pattern matches the suppression pattern, confirming that the mechanism of high copy suppression is by protein stabilization of Ceg1 upon interaction with Cet1.
By use of the structure of Chlorella virus PBCV-1 guanylyltransferase as a model for Ceg1, the mutations were mapped to their approximate position in the enzyme (Fig. 1C). The suppressible ceg1-12, ceg1-34, and ceg1-250 mutations are in the predicted carboxyterminal region of Ceg1. The nonsuppressed ceg1-63 mutation is located within the larger amino-terminal domain. The allele-specific suppression pattern suggests that Cet1 might interact with the carboxy-terminal region of Ceg1 functionally and/or physically.
Allosteric effects of the CTD and triphosphatase subunit on guanylyltransferase activity
Coupling of capping with transcription is mediated by specific binding of capping enzyme to the Pol II transcription complex on phosphorylation of CTD of RNA Pol II. In higher eukaryotes, interaction with phosphorylated CTD is mediated by the guanylyltransferase domain (Yue et al. 1997; Ho et al. 1998). In yeast, it was shown that Ceg1 (assayed by immunoblotting) could interact directly with the phosphorylated CTD (McCracken et al. 1997). In apparent contradiction, we reported that yeast Ceg1 subunit alone was insufficient for recruitment of guanylyltransferase activity (assayed by enzyme-GMP formation, Cho et al. 1997). To resolve the matter, we examined the interaction of each subunit with CTD and the effect of Cet1 on CTD binding and guanylyltransferase activity (Fig. 2). Surprisingly, guanylyltransferase binds directly to the CTD but cannot be guanylylated unless the triphosphatase subunit is also present.
Figure 2.
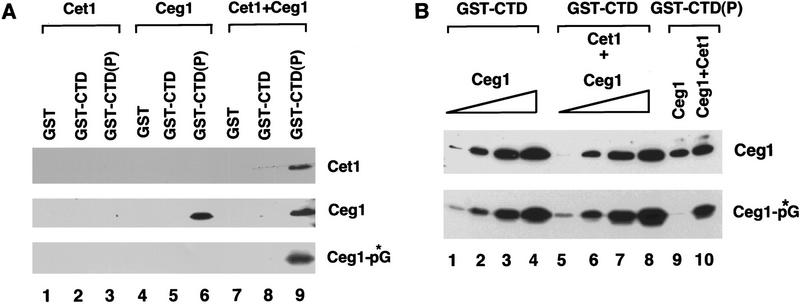
Ceg1 guanylylation is inhibited by binding to phosphorylated CTD and stimulated by Cet1. (A) Recombinant guanylyltransferase (Ceg1), RNA triphosphatase (Cet1) or both (Cet1 + Ceg1) were incubated with glutathione agarose beads carrying GST, GST–CTD, or phosphorylated GST–CTD protein [GST–CTD(P)]. Beads were pelleted, and bound proteins were detected by immunoblotting (top, middle). The ability of Ceg1 to be guanylylated was assayed by the addition of [α-32P]GTP (Ceg1-*pG, bottom). (B) 150 ng Ceg1 was incubated with GST–CTD(P) in the absence (lane 9) or presence (lane 10) of 200 ng of Cet1 and precipitated as in A. A standard curve was generated in the presence of GST–CTD with 1.4 ng (lanes 1,5), 7.0 ng (lanes 2,6), 14 ng (lanes 3,7), or 42 ng (lanes 4,8) of Ceg1. Lanes 5–8 also contained 100 ng of Cet1. Ceg1 used in this experiment was completely deguanylylated (see Materials and Methods). Ceg1 protein (top) and enzyme-GMP complex (bottom) were visualized as in A and quantitated by densitometry.
Recombinant histidine-tagged Cet1 (lanes 1–3), Ceg1 (lanes 4–6) or a mixture of both proteins (lanes 7–9) were incubated with GST (lanes 1,4,7) or a GST–CTD fusion protein that was either untreated (GST–CTD, lanes 2,5,8) or phosphorylated with the basal transcription factor TFIIH [GST–CTD(P), lanes 3,6,9]. Bound proteins were then precipitated with glutathione-agarose and the pellets were analyzed by immunoblot for Cet1 and Ceg1. Guanylyltransferase activity was assayed by formation of the enzyme-GMP covalent intermediate with [α-32P]GTP (Ceg1-pG).
In the absence of the guanylyltransferase subunit, Cet1 did not interact with phosphorylated CTD (lane 3). In contrast, Ceg1 subunit interacted either alone (lane 6) or complexed with the Cet1 subunit (lane 9), demonstrating that Cet1 recruitment to the CTD is mediated by Ceg1. In this experiment, the presence of Cet1 did not appear to dramatically affect the amount of Ceg1 bound to the CTD. However, in most experiments, the presence of Cet1 increased the amount of Ceg1 bound to the CTD somewhat (see Fig. 2B).
Surprisingly, when the ability to form the enzyme-GMP intermediate was assayed, CTD-bound Ceg1 could be guanylylated only when the triphosphatase subunit was present (Fig. 2A, cf. lanes 6 and 9), consistent with our earlier observations (Cho et al. 1997). To determine whether this result reflected inhibition of guanylylation by the CTD, stimulation by Cet1, or a combination of both, we compared the specific activity of guanylyltransferase bound to phosphorylated CTD in the presence or absence of triphosphatase with a standard curve generated in the presence of unphosphorylated CTD (Fig. 2B). Quantitation of Ceg1 protein (by immunoblotting) and guanylylation (by autoradiography) indicates that binding to phosphorylated CTD inhibits the ability of Ceg1 to be guanylylated and that Cet1 reverses this inhibition. In the absence of binding to the CTD, Cet1 only modestly (about twofold) increased guanylylation of Ceg1 (cf. lanes 1–4 with lanes 5–8).
The inability to guanylylate Ceg1 indicates that either the Ceg1 bound to the CTD is already guanylylated (a significant proportion of recombinant capping enzymes are guanylylated; see Ho et al. 1998) or that it is held in an inactive state. We favor the second explanation for two reasons. First, experiments performed with a Ceg1 mutant that is unable to be guanylylated (lysine 70 changed to threonine; see Fresco and Buratowski 1994) show that it is able to be recruited to the phosphorylated CTD in the presence or absence of Cet1 (data not shown). Therefore, guanylylation is not required for binding to the CTD. Second, we deguanylylated recombinant guanylyltransferase by incubation with 1 mm pyrophosphate (Hakansson and Wigley 1998). This preparation behaved similarly to untreated Ceg1 (Fig. 2B).
We conclude that both the CTD and the triphosphatase subunit affect the activity of the capping enzyme guanylyltransferase. Isolated Ceg1 bound to the CTD is inhibited from being guanylylated, and association with Cet1 reverses this inhibition.
A small region of Cet1 is required for interaction with and stimulation of Ceg1
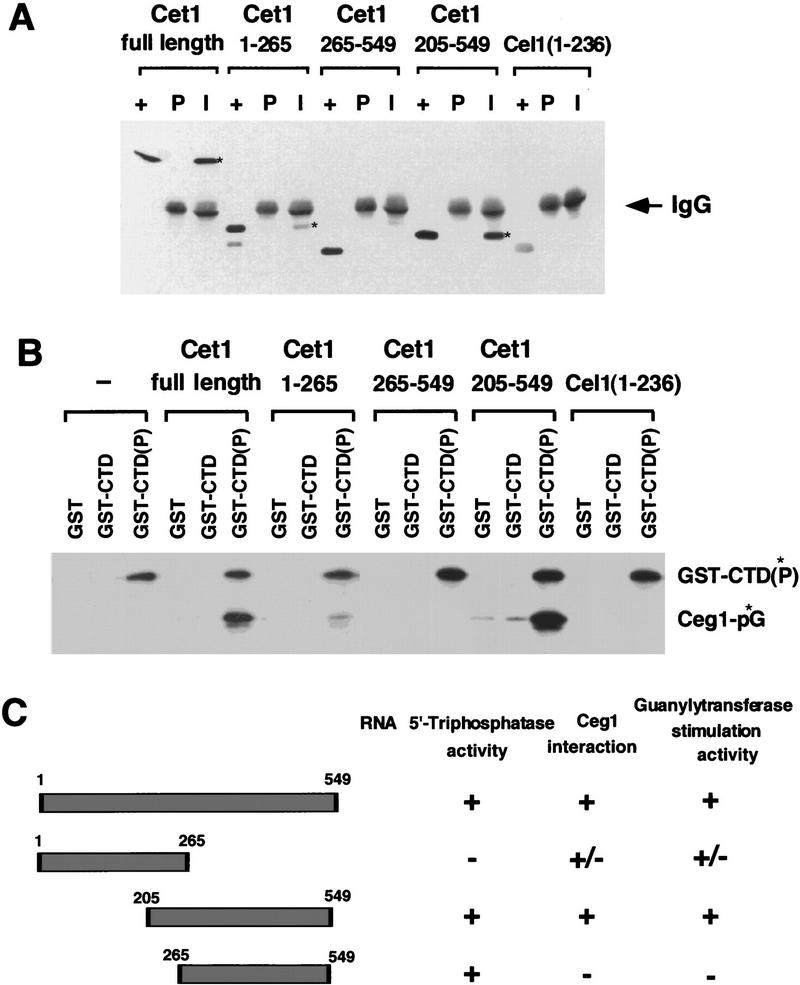
Amino acids 205–549 of Cet1 are sufficient for both RNA triphosphatase activity and the ability to bind to Ceg1 (Tsukamoto et al. 1997). We constructed several deletion mutations of Cet1 to further analyze their interaction with Ceg1 and their effect on guanylylation. Cet1(265–549) and Cet1(205–549) have RNA 5′triphosphatase activity, whereas Cet1(1–265) does not (data not shown). Coimmunoprecipitation with Ceg1 was used to identify the region of Cet1 required for subunit interaction (Fig. 3A). Reaction mixtures were precipitated with either preimmune (P) or anti-Ceg1 immune (I) serum, and the pellet was assayed for Cet1. Full-length protein and Cet1(205–549) were tightly bound to Ceg1. Cet1(1–265) was also precipitated with Ceg1, but with lower affinity. Cet1(265–549) could not bind to Ceg1. We also tested the RNA triphosphatase domain of C. elegans capping enzyme [Cel1(1–236)] (Takagi et al. 1997), which, as expected, did not show any binding.
Figure 3.
Amino acids 205–265 of Cet1 are required for interaction with and stimulation of Ceg1. (A) Coimmunoprecipitation of Ceg1 with various deletions of Cet1. Ceg1 was incubated with Cet1, Cet1(1–265), Cet1(265–549), Cet1(205–549), or Cel1(1–236) and the mixture precipitated with either preimmune serum (P) or anti-Ceg1 immune serum (I) as described in Materials and Methods. Proteins were visualized by anti-6× His monoclonal antibody. (+) A positive control lane containing the indicated triphosphatase derivative. The signal of Ceg1 protein was masked by the heavy chain of rabbit IgG which migrates with Ceg1. (*) The Cet1 derivatives coprecipitated with Ceg1. (B) Activation of guanylyltransferase by Cet1 derivatives. Ceg1 was incubated with Cet1, Cet1(1–265), Cet1(265–549), Cet1(205–549), or Cel1(1–236). The mixture was allowed to interact with GST, GST–CTD, or phosphorylated GST–CTD [GST–CTD(P) was radioactively labeled as described in Fig. 2]. Formation of the enzyme-GMP intermediate (Ceg1-p*G) was assayed on the precipitated beads. (C) Summary of Cet1 domains required for RNA 5′-triphosphatase activity, Ceg1 subunit interaction, and guanylyltransferase activity stimulation.
The Cet1 derivatives were next incubated with Ceg1 and the reactions precipitated with GST, GST–CTD, or GST–CTD(P) (Fig. 3B). Both full-length and Cet1(205–549) strongly stimulated Ceg1 guanylylation on the phosphorylated CTD. Cet1(1–265) also stimulated guanylylation but at a lower level correlating with its ability to bind Ceg1. Therefore, RNA triphosphatase activity is not required for Ceg1 stimulation. Cet1(265–549) did not stimulate Ceg1 activity.
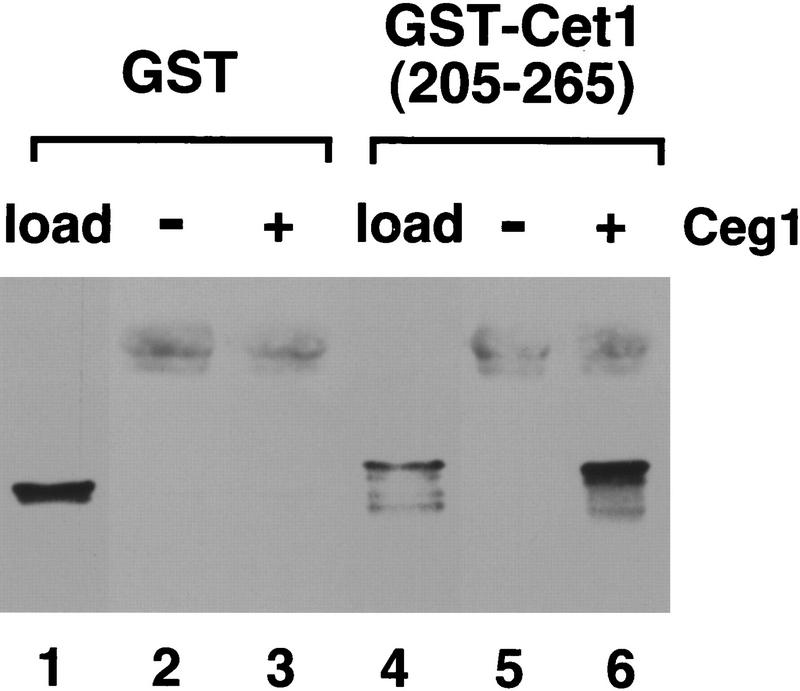
To summarize these results (Fig. 3C), we conclude that amino acids 205–265 of Cet1 are minimally required for both interaction with Ceg1 and stimulation of guanylyltransferase activity. Triphosphatase activity is not required for stimulation of Ceg1. To determine whether residues 205–265 were sufficient for interaction with Ceg1, they were fused to GST. Whereas GST showed no binding, GST–Cet1(205–265) was efficiently coimmunoprecipitated with Ceg1 (Fig. 4).
Figure 4.
Amino acids 205–265 of Cet1 are sufficient for interaction with Ceg1. GST (lane 1) or GST fused to amino acids 205–265 of Cet1 (lane 4) were incubated alone (lanes 2,4) or with Ceg1 (lanes 3,5). Reactions were precipitated with anti-Ceg1 antibody and pellets were washed and then immunoblotted with anti-GST monoclonal antibody.
Discussion
Capping is restricted to RNA Pol II transcripts in vivo even though capping enzyme can use a variety of RNA substrates in vitro. In vivo specificity is accomplished by the specific binding of capping enzyme to the phosphorylated CTD of Pol II. In the two subunit yeast capping enzyme, the guanylyltransferase subunit interacts with CTD (Fig. 2; McCracken et al. 1997), as does the homologous domain of the mammalian capping enzyme (Yue et al. 1997; Ho et al. 1998). Our results suggest that this interaction does not simply recruit capping enzyme to the transcription complex. Rather, guanylyltransferase activity is regulated allosterically by interactions with both the CTD and with the triphosphatase subunit.
Structural studies of the Chlorella virus enzyme guanylyltransferase suggest a possible mechanism for allosteric regulation. The viral guanylyltransferase (and presumably eukaryotic guanylyltransferases) consists of two domains. Two forms are observed in the crystal, suggesting that the enzyme undergoes a large conformational change during the guanyl transfer reaction (Hakansson et al. 1997). The active site lysine is located in the amino-terminal domain deep within the cleft between the two domains, and the enzyme adopts an open conformation when binding GTP. The enzyme then changes to a closed conformation, which brings the CTD of the enzyme in proximity of the β–γ phosphates while the covalent enzyme–GMP intermediate is formed. Subsequently, the enzyme reopens to allow release of pyrophosphate and binding of RNA. The guanosine residue is then transferred to the diphosphate terminus of the RNA.
Assuming Ceg1 is structurally similar to the viral guanylyltransferase, allosteric regulation could occur by affecting the conformation change required for enzyme activity. Binding of Ceg1 to the phosphorylated CTD might freeze the enzyme in either the open or closed conformation, thereby blocking guanylylation. Association of the Cet1 triphosphatase might facilitate the conformation change, thereby reversing the inhibition by CTD.
Tsukamoto et al. (1997) found that the carboxy-terminal (205–549) portion of Cet1 has RNA triphosphatase activity and is sufficient for interaction with Ceg1. Our analysis further showed that amino acids 265–549 retain triphosphatase activity, but Ceg1 interaction is completely lost. Furthermore, residues 205–265 of Cet1 are necessary and sufficient for binding to Ceg1. Interestingly, the ability to stimulate Ceg1 guanylyltransferase activity correlates exactly with binding. Therefore, the allostery does not require triphosphatase activity or any other regions of Cet1 outside of the interacting region.
The region of Ceg1 contacted by the triphosphatase has not been mapped conclusively. Attempts to coimmunoprecipitate the predicted amino-terminal or carboxy-terminal domain of Ceg1 with Cet1 were unsuccessful (data not shown). This suggests that both Ceg1 domains are required for this interaction or that the Ceg1 deletion proteins are not folded properly. However, our genetic results suggest that the triphosphatase subunit interacts with the carboxy-terminal region of Ceg1.
Overexpression of CET1 suppresses only certain ceg1 conditional alleles. Figure 1C shows a schematic of the ceg1 mutant allele positions mapped to its corresponding position in the Chlorella virus guanylyltransferase structure (Hakansson et al. 1997). Allele ceg1-63 is buried within the amino-terminal domain, and is not suppressed by additional CET1. In contrast, alleles ceg1-12, ceg1-34, and ceg1-250 are in the carboxy-terminal domain. Cysteine 354, mutated in allele ceg1-12, is part of a motif conserved throughout the viral, yeast, and mammalian capping enzymes. However, alleles ceg134 and ceg1-250 are located in a nonconserved region which corresponds to a surface accessible loop in the Chlorella guanylyltransferase structure. Interestingly, the sequence of this region (including the residues mutated in ceg1-34 and ceg1-250) is quite conserved between the S. cerevisiae, Schizosaccharomyces pombe, and Candida albicans guanylyltransferases, but significantly diverged from the higher eukaryotic and viral enzymes (see Wang et al. 1997 for alignment). The proteins mutated in this region are degraded at the nonpermissive temperature, but are stabilized by CET1 overexpression. We believe our findings are most consistent with a direct physical interaction between Cet1 and this part of Ceg1, although an indirect mechanism of suppression and stabilization (involving the conformational change in Ceg1, for example) cannot be ruled out.
Why regulate guanylyltransferase activity through interactions with the triphosphatase subunit and the Pol II CTD? The most likely reason is to coordinate the triphosphatase and guanylyltransferase activities and thereby prevent spurious enzyme activity. CTD inhibition of guanylyltransferase and reactivation on binding to triphosphatase helps ensure that newly synthesized mRNAs will be acted on by both enzymes. Furthermore, the stabilization of Ceg1 by overexpression of Cet1 suggests that excess guanylyltransferase may be degraded when not complexed with Cet1. These findings suggest that cells have mechanisms to carefully control the localization and specificity of mRNA capping.
Materials and methods
DNA cloning methods
The CET1 gene was amplified from S. cerevisiae genomic DNA by PCR using Vent DNA polymerase (New England BioLabs) and oligonucleotide primers derived from the published sequence (Tsukamoto et al. 1997). The sequences of PCR primers are as follows: CET1-A, 5′-GCGGATCCGGGTTTTATCTGCAAACAGACGCAAG-3′ (BamHI site boldface); CET1-B, 5′-GCGGATCCGTATTTGCATACAGAGGGTTGGATCT-3′ (BamHI site boldface); CET1-C, 5′-GCGGATCCATGGGTTACACTGACAACCCTCCTCAAAC-3′ (NcoI site underlined, BamHI site boldface).
A 2.5-kb PCR product (oligonucleotides CET1-A and CET1-C) containing the entire CET1 gene was subcloned into pCR-Script SK + (Stratagene) to generate pBS–CET1. A 2.5-kb BamHI fragment from this plasmid was then cloned into BamHI cut pRS316 or pRS426 to generate pRS316-CET1 (CEN/ARS, URA3) and pRS426-CET1 (2μ, URA3).
The CET1 ORF was amplified by primers CET1-B and CET1-C, creating an NcoI site at the initiation codon and BamHI sites at the both ends of the fragment. The resulting 2.0-kb fragment was cloned into pCR-Script SK+ to generate pBS-CET1orf. A 2.0-kb NcoI–BamHI fragment was then excised and cloned into the NcoI–BamHI sites of pET11-his7, a modified version of pET11d that contains a cassette coding for seven histidines at the amino terminus of the protein (pET-his7CET1). Subsequently, a 2.3-kb XbaI–EcoRI fragment of pET-his7CET1 containing T7 promoter and CET1 ORF replaced a 0.6-kb XbaI–EcoRI fragment of pSBETa vector (Schenk et al. 1995) to generate pSBET-his7CET1.
To express an amino-terminal fragment of Cet1, a 0.8-kb BglII–BglII fragment of pET-his7CET1 encoding amino acids 1–265 replaced a 0.17-kb BglII–BamHI fragment of pET-14b (Novagen) to generate pETb-his7CET1(1–265). A 0.6-kb BglII–EcoRI fragment of pSBETa was replaced with a 1.4-kb BglII–EcoRI fragment of pETb-his7CET1(1-265) to generate pSBET-his7CET1(1–265). This plasmid produces histidine-tagged Cet1(1–265) fused to 20 extra amino acid residues (PAANKARKEAELAAATAEQ) at the carboxyl terminus.
Two amino-terminal deletion mutants of the CET1 gene were constructed as follows. For CET1(205–549) , PCR was performed with pBS-CET1orf as a template and CET1(205) [5′-GGATCCATGGTAGATAATATTTTTGAAGA-3′ (NcoI site underlined, BamHI site boldface)] and CET1-B as primers. A 1.4-kb amplified fragment was subcloned into pCR-Script SK + [pBS-CET1(205–549)]. The NcoI–BamHI fragment was used to replace an NcoI–BamHI fragment of pSBET-his7CET1 to generate pSBET-his7CET1(205–549). To create CET1(265–549) mutant (amino acids 265–549), pSBET14b was constructed by replacing a 0.7-kb BglII–EcoRI fragment of pSBETa with a 0.6-kb BglII–EcoRI fragment of pET-14b. A 1.2-kb BglII–BamHI fragment of pET-his7CET1 was subcloned into BamHI site of pSBET14b to get pSBET14b-CET1(265–549) which encodes Cet1(265-549) fused with polyhistidine-tag and 13 extra residues (MGSSHHHHHHSSGLVPRGSHMLE) at the amino terminus. To express GST–Cet1(205–265), a BamHI–BglII fragment from pBS-CET1(205–549) was cloned into the BamHI site of pGEX-2T.
Recombinant protein production and purification
Full-length and truncated Cet1 proteins were expressed by a T7 promoter/polymerase system in Escherichia coli (Studier et al. 1990). Induction was performed as described (Higman et al. 1992). Lysates were prepared by sonication, and recombinant proteins were purified from soluble extract with Ni2+-NTA-agarose resin (Qiagen) (Takagi et al. 1997). Proteins were monitored by SDS-PAGE and Coomassie blue staining, immunoblot analysis with anti-6×His monoclonal antibody (Clontech), and RNA 5′-triphosphatase assay (Takagi et al. 1998). Polyhistidine tagged recombinant guanylyltransferase subunit of S. cerevisiae-capping enzyme (his7-Ceg1) and RNA triphosphatase domain of C. elegans-capping enzyme [his7-Cel-1(1-236)] were expressed and purified as described (Fresco and Buratowski 1994; Takagi et al. 1997). To generate nonguanylylated Ceg1, nickel column purified protein was dialyzed against buffer B [20 mm MOPS-KOH (pH 7.0), 20 mm potassium acetate, 1 mm MgCl2, 10% glycerol, 1 mm dithiothreitol, 1 mm EDTA, 1 mm PMSF, and 1 mm sodium pyrophosphate]. Deguanylylated Ceg1 was further purified by MonoS(HR5/5) FPLC-chromatography. Cet1 proteins were further purified by MonoQ(HR 5/5) FPLC-chromatography.
Protein–protein interaction
Coimmunoprecipitation with anti-Ceg1 antibody (Fresco and Buratowski 1996) was performed as described in Cho et al. (1997) with minor modifications. Ceg1 (150 ng) was incubated with 200 ng of the indicated Cet1 derivative in buffer A [20 mm HEPES-KOH at pH 7.6, 1 mm EDTA, 1 mm DTT, 15% (vol/vol) glycerol] containing 100 mm KOAc, 0.1% Triton X-100, 0.01% NP-40, and 750 μg/ml BSA. After preclearing the mixture with protein A-Sepharose CL-4B (Pharmacia), the reaction was incubated with 1 μl of either preimmune or anti-Ceg1 serum bound to protein A–Sepharose beads that were blocked with BSA in the same buffer. The precipitate was washed, and resuspended in sample loading buffer for SDS-PAGE. Proteins were detected by immunoblot analysis using anti-6xHis monoclonal antibody or anti-GST antibody (Sigma). GST pull-down experiments were performed as described in Cho et al. (1997) in the buffer described above. Formation of enzyme-GMP covalent intermediate was assayed as described (Fresco and Buratowski 1994).
Yeast strains
YSB243, YSB228, YSB229, YSB230, YSB231, and YSB232 containing wild-type or mutant CEG1 alleles have been described previously (Fresco and Buratowski 1994, 1996). YSB491 was generated in parallel with YSB517 as reported previously (Cho et al. 1997), except the mating type was not changed. For suppression studies, these strains were transformed with pRS316, pRS316-CEG1, pRS316-CET1, or pRS426-CET1. Plasmids were introduced into yeast using modified lithium acetate transformation protocol. To compare growth of strains at permissive and nonpermissive temperatures, cells were grown at 30°C in selective medium. Cultures were normalized to an OD600 = 0.3, and three serial 1:8 dilutions were prepared. Ten microliters of each dilution were then spotted on selective plates and incubated for 3 days at 30°C or 37°C. Medium preparation and other yeast manipulations were performed by standard methods.
Yeast whole-cell extract preparation and analysis of Ceg1 proteins
To characterize Ceg1 proteins in vivo, yeast cells were grown overnight at 30°C in selective medium and diluted to an OD600 = 0.5. An equal volume of medium at either 30°C or 46°C was added, and the cells were grown at 30°C or 37°C, respectively for 4.5 hr. Cells were harvested and extracts were prepared as described (Eisenmann et al. 1992). Immunoblot analysis was performed with rabbit anti-Ceg1 antibody as described (Fresco and Buratowski 1996).
Acknowledgments
This work was supported by a grant from the Tobacco Research Council USA. S.B. gratefully acknowledges support from the Pew Scholars program and a Junior Faculty Research Award from the American Cancer Society. T.T. is a Senior Postdoctoral Fellow of the American Cancer Society (Massachusetts Division).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steveb@warren.med.harvard.edu; FAX (617)738-0516.
References
- Cho E-J, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Fresco LD, Buratowski S. Active site of the mRNA capping enzyme guanylyltransferase from Saccharomyces cerevisiae: Similarity to the nucleotidyl attachment motif of DNA and RNA ligases. Proc Natl Acad Sci. 1994;91:6624–6628. doi: 10.1073/pnas.91.14.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Conditional mutants in the yeast mRNA capping enzyme show that the cap enhances, but is not required for, mRNA splicing. RNA. 1996;2:584–595. [PMC free article] [PubMed] [Google Scholar]
- Hakansson K, Wigley DB. Structure of a complex between a cap analogue and mRNA guanylyltransferase demonstrates the structural chemistry of RNA capping. Proc Natl Acad Sci. 1998;95:1505–1510. doi: 10.1073/pnas.95.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakansson K, Doherty AJ, Shuman S, Wigley DB. X-ray crystallography reveals a large conformational change during guanylyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- Higman HA, Bourgeois N, Niles EG. The vaccinia virus mRNA (guanine-N7-)-methyltransferase requires both subunits of the mRNA capping enzyme for activity. J Biol Chem. 1992;267:16430–16437. [PubMed] [Google Scholar]
- Ho CK, Sriskanda V, McCracken S, Bentley D, Schwer B, Shuman S. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- Itoh N, Yamada H, Kaziro Y, Mizumoto K. Messenger RNA guanylyltransferase from Saccharomyces cerevisae: Large scale purification, subunit functions, and subcellular localization. J Biol Chem. 1987;262:1989–1995. [PubMed] [Google Scholar]
- Jove R, Manley JL. In vitro transcription from adenovirus 2 major late promoter utilizing templates truncated at promoter-proximal siltes. J Biol Chem. 1984;259:8513–8521. [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL Amgen EST Program. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto K, Kaziro Y. Messenger RNA capping enzyme from eukaryotic cells. Prog Nucleic Acid Res Mol Biol. 1987;34:1–28. doi: 10.1016/s0079-6603(08)60491-2. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vitro transcriptional pausing and cap formation on three Drosophilla heat shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Baumann S, Mattes R, Steinbib H-H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare argtRNAs. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: Isolation and characterization of the gene encoding mRNA guanylyltransferase subunit from Saccharomyces cerevisiae. J Biol Chem. 1992;267:9521–9528. [PubMed] [Google Scholar]
- Shuman S. Capping enzyme in eukaryotic mRNA synthesis. Prog Nucleic Acid Res Mol Biol. 1995;50:101–129. doi: 10.1016/s0079-6603(08)60812-0. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Meth Enzymol. 1990;153:3–11. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Takagi T, Moore CR, Diehn F, Buratowski S. An RNA 5′-triphosphatase related to the protein tyrosine phosphatases. Cell. 1997;89:867–873. doi: 10.1016/s0092-8674(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Takagi T, Taylor GS, Kusakabe T, Charbonneau H, Buratowski S. A protein tyrosine phosphatase-like protein from baculovirus has RNA 5′-triphosphatase and diphosphatase activities. Proc Natl Acad Sci. 1998;95:9808–9812. doi: 10.1073/pnas.95.17.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Shibagaki Y, Imajoh-Ohmi S, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Isolation and characterization of the yeast mRNA capping enzyme β subunit encoding RNA 5′-triphosphatase, which is essential for cell viability. Biochem Biophys Res Commun. 1997;239:116–122. doi: 10.1006/bbrc.1997.7439. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Shibagaki Y, Murakoshi T, Suzuki M, Nakamura A, Gotoh H, Mizumoto K. Cloning and characterization of two human cDNAs encoding the mRNA capping enzyme. Biochem Biophys Res Commun. 1998;243:101–108. doi: 10.1006/bbrc.1997.8038. [DOI] [PubMed] [Google Scholar]
- Wang SP, Deng L, Ho CK, Shuman S. Phylogeny of mRNA capping enzymes. Proc Natl Acad Sci. 1997;94:9573–9578. doi: 10.1073/pnas.94.18.9573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada-Okabe T, Doi R, Shimmi O, Arisawa M, Yamada-Okabe H. Isolation and charaterization of human cDNA for mRNA 5′-capping enzyme. Nucleic Acids Res. 1998;26:1700–1706. doi: 10.1093/nar/26.7.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]