Abstract
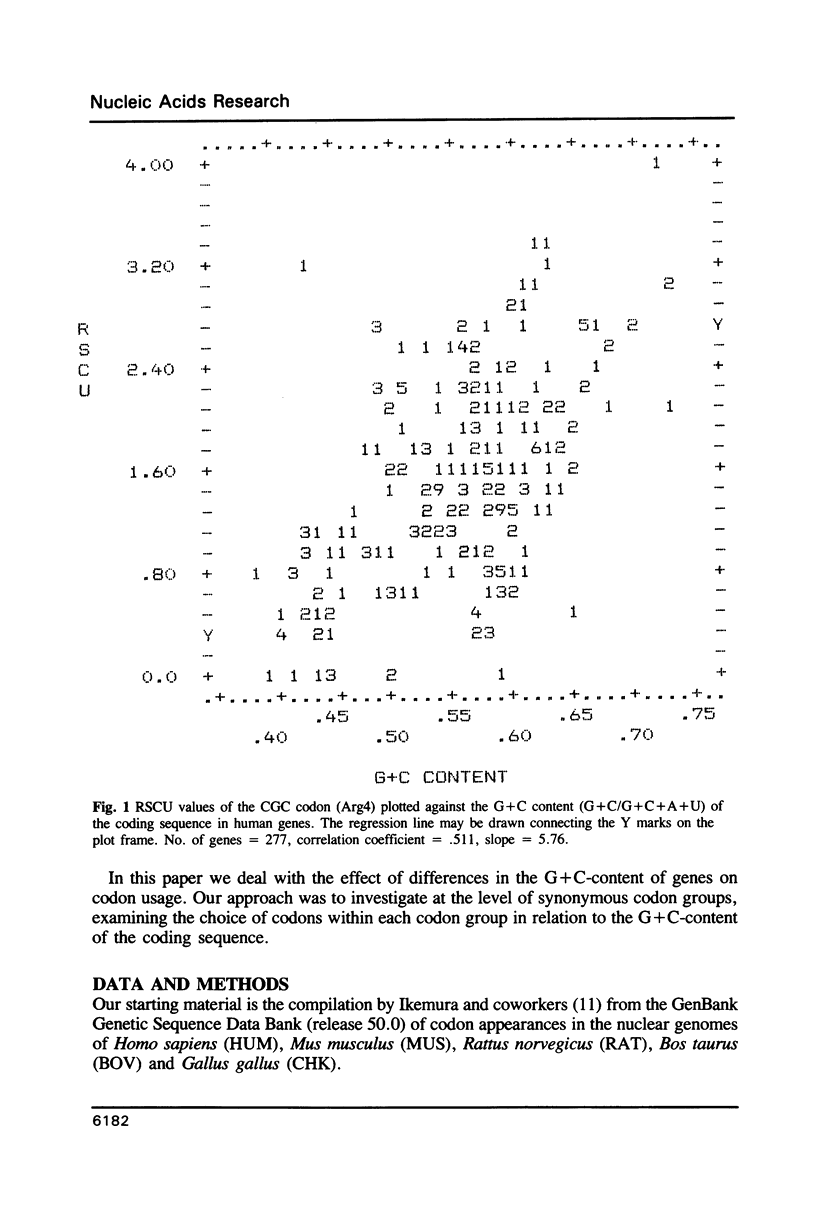
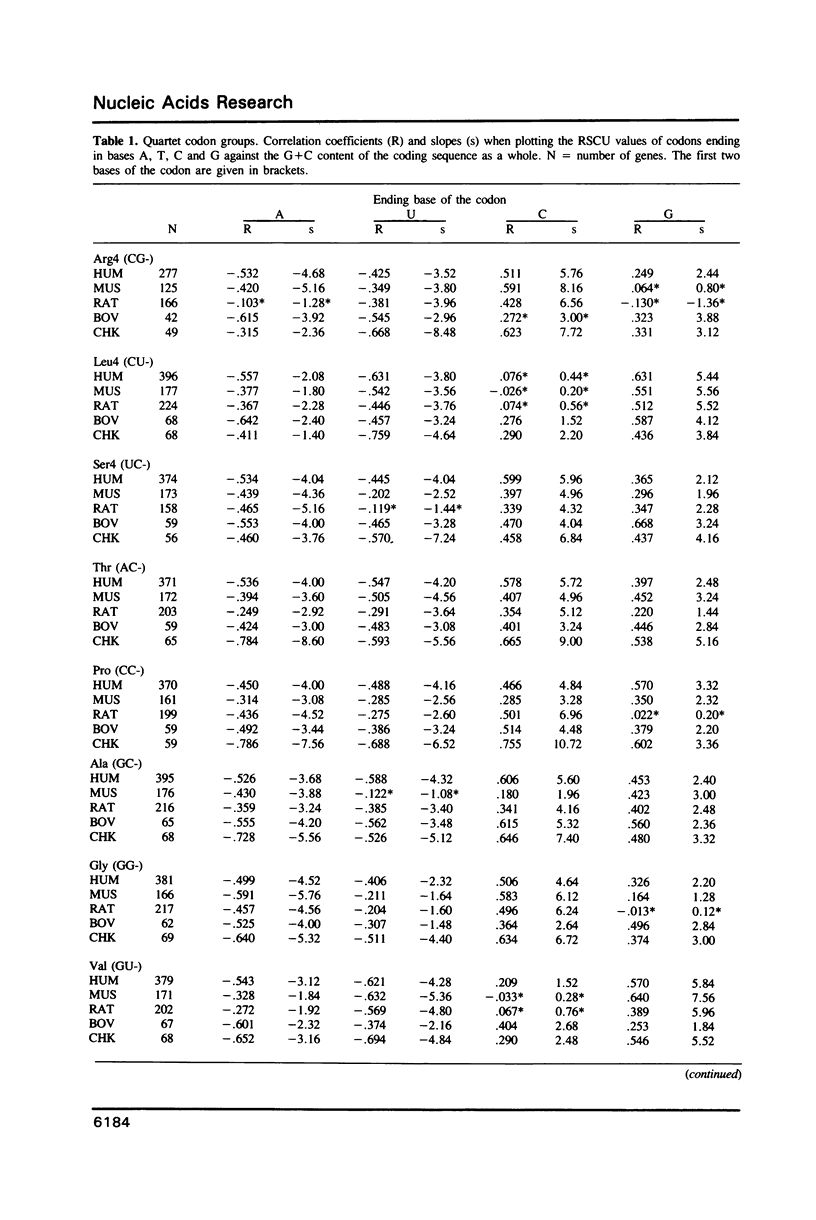
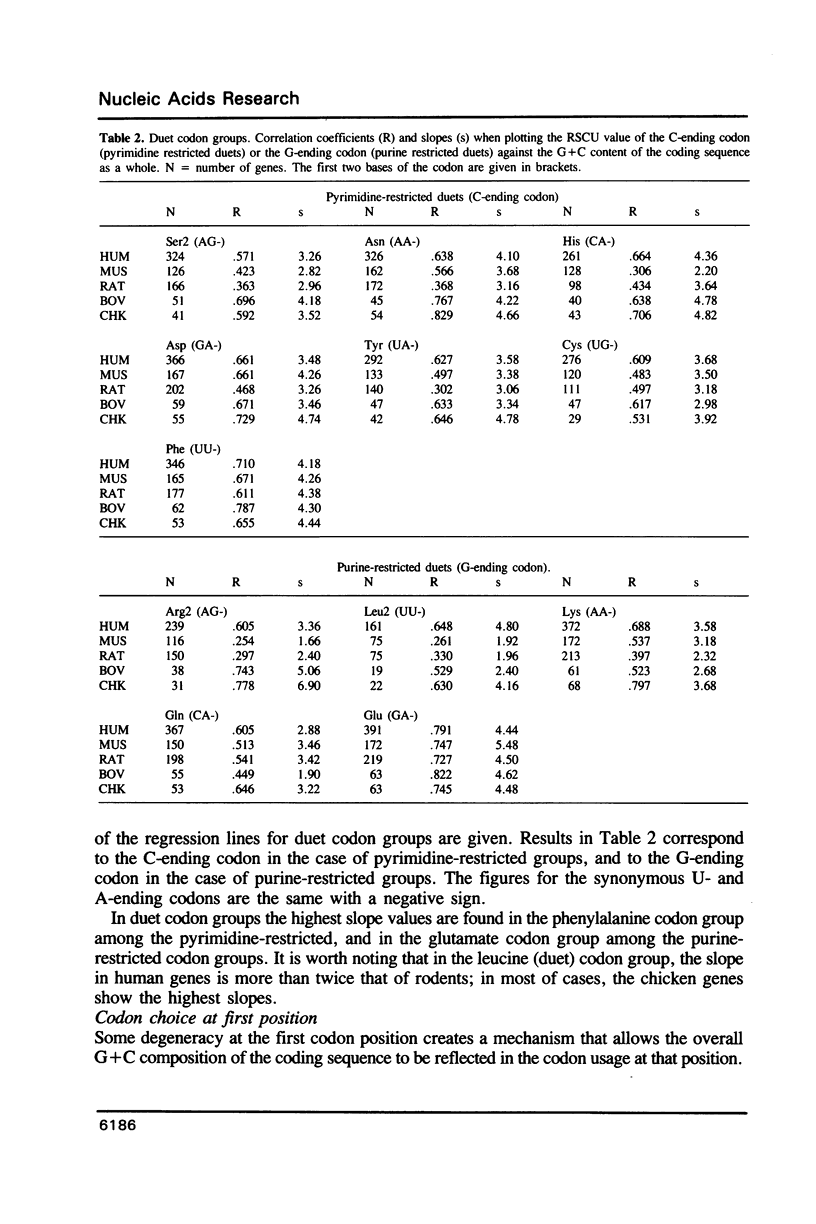
The relationship between G + C-content and codon usage in genes of human, mus, rat, bovine and chicken nuclear genomes was investigated. Correlation and lineal regression analyses were carried out on plots that related the frequency of each codon within each synonymous codon group to the G + C-content of the coding sequence as a whole. Under GC pressure, in most of the quartet codon groups there is a preferential choice of the C-ending codon, except in leucine and valine codon groups where the choice of the G-ending codon is preferred. Among ducts, the choice of codons specifying phenylalanine and glutamate shows the strongest dependence on G + C-content. The relationship found between G + C-content and codon usage in these genomes correlate with taxonomic distance.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aota S., Ikemura T. Diversity in G + C content at the third position of codons in vertebrate genes and its cause. Nucleic Acids Res. 1986 Aug 26;14(16):6345–6355. doi: 10.1093/nar/14.16.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Codon usage and genome composition. J Mol Evol. 1985;22(4):363–365. doi: 10.1007/BF02115693. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. Compositional constraints and genome evolution. J Mol Evol. 1986;24(1-2):1–11. doi: 10.1007/BF02099946. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Bernardi G. The human genome and its evolutionary context. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):479–487. doi: 10.1101/sqb.1986.051.01.059. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Brenner S. The molecular evolution of genes and proteins: a tale of two serines. Nature. 1988 Aug 11;334(6182):528–530. doi: 10.1038/334528a0. [DOI] [PubMed] [Google Scholar]
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987 Feb 19;325(6106):728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- Filipski J. Correlation between molecular clock ticking, codon usage fidelity of DNA repair, chromosome banding and chromatin compactness in germline cells. FEBS Lett. 1987 Jun 15;217(2):184–186. doi: 10.1016/0014-5793(87)80660-9. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. Is there selection against wobble in codon-anticodon pairing? Science. 1976 Dec 10;194(4270):1173–1174. doi: 10.1126/science.996548. [DOI] [PubMed] [Google Scholar]
- Gouy M., Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982 Nov 25;10(22):7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980 Jan 11;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Yasunaga T., Miyata T. Secondary structure of MS2 phage RNA and bias in code word usage. Nucleic Acids Res. 1979 Dec 11;7(7):2073–2079. doi: 10.1093/nar/7.7.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T., Aota S. Global variation in G+C content along vertebrate genome DNA. Possible correlation with chromosome band structures. J Mol Biol. 1988 Sep 5;203(1):1–13. doi: 10.1016/0022-2836(88)90086-1. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Ikemura T., Ozeki H. Codon usage and transfer RNA contents: organism-specific codon-choice patterns in reference to the isoacceptor contents. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1087–1097. doi: 10.1101/sqb.1983.047.01.123. [DOI] [PubMed] [Google Scholar]
- Li W. H. Models of nearly neutral mutations with particular implications for nonrandom usage of synonymous codons. J Mol Evol. 1987;24(4):337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Wilbur W. J. Contextual constraints on synonymous codon choice. J Mol Biol. 1983 Jan 25;163(3):363–376. doi: 10.1016/0022-2836(83)90063-3. [DOI] [PubMed] [Google Scholar]
- Nichols B. P., Blumenberg M., Yanofsky C. Comparison of the nucleoside sequence of trpA and sequences immediately beyond the trp operon of Klebsiella aerogenes. Salmonella typhimurium and Escherichia coli. Nucleic Acids Res. 1981 Apr 10;9(7):1743–1755. doi: 10.1093/nar/9.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R. Some rules in the ordering of nucleotides in the DNA. Nucleic Acids Res. 1980 Oct 10;8(19):4545–4562. doi: 10.1093/nar/8.19.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Codon preference is but an illusion created by the construction principle of coding sequences. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4378–4382. doi: 10.1073/pnas.85.12.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa S., Ohama T., Yamao F., Muto A., Jukes T. H., Ozeki H., Umesono K. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1124–1128. doi: 10.1073/pnas.85.4.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe R., Meselson M. THE RELATIVE HOMOGENEITY OF MICROBIAL DNA. Proc Natl Acad Sci U S A. 1959 Jul;45(7):1039–1043. doi: 10.1073/pnas.45.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUEOKA N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci U S A. 1962 Apr 15;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas L. K., Dix D. B., Thompson R. C. Codon choice and gene expression: synonymous codons differ in their ability to direct aminoacylated-transfer RNA binding to ribosomes in vitro. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4242–4246. doi: 10.1073/pnas.85.12.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. E., Iglewski B. H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988 Oct 11;16(19):9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Sharp P. M., Li W. H. Mutation rates differ among regions of the mammalian genome. Nature. 1989 Jan 19;337(6204):283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Zerial M., Salinas J., Filipski J., Bernardi G. Gene distribution and nucleotide sequence organization in the human genome. Eur J Biochem. 1986 Nov 3;160(3):479–485. doi: 10.1111/j.1432-1033.1986.tb10064.x. [DOI] [PubMed] [Google Scholar]


