Abstract
p19ARF suppresses the growth of cells lacking p53 through an unknown mechanism. p19ARF was found to complex with transcription factors E2F1, -2, and -3. Levels of endogenous or ectopically expressed E2F1, -2, and -3, but not E2F6, were reduced after synthesis of p19ARF, through a mechanism involving increased turnover. p19ARF-induced degradation of E2F1 depended on a functional proteasome, and E2F1 was relocalized to nucleoli when coexpressed with p19ARF. Consistent with reduced levels of E2F1 and E2F3, the proliferation of cells defective for p53 function was suppressed by p19ARF, and the effect was partially reversed by ectopic overexpression of E2F1. These results suggest a broader role for p19ARF as a tumor suppressor, in which targeting of certain E2F species may cooperate with stimulation of the p53 pathway to counteract oncogenic growth signals.
The alternative reading frame (ARF) protein leads to growth arrest or apoptosis of cells exposed to inappropriate mitogenic stimuli (for review, see refs. 1 and 2). ARF-expressing cells undergo p53 pathway activation, followed by cell cycle arrest or apoptosis, depending on the cell context (for review, see refs. 1 and 2). In multiple settings, the biological effects of ARF, such as growth arrest or suppression of transformation, appeared to depend largely on the maintenance of intact p53 signal transduction (3–5).
Recent reports demonstrate that ARF can also inhibit cell growth in the absence of p53. In one case, growth inhibition depended on the simultaneous presence of p16INK4A and MDM2 proteins (6). In another, it depended on the absence of MDM2 (7). Thus, how ARF engenders p53-independent growth suppression seems, at the very least, to depend on cell context.
The mechanism underlying ARF-dependent growth inhibition of p53 null cells remains obscure, although ectopic overexpression of E2F1 overcame this effect in certain cell species (6). These findings have led to speculation that, in addition to p53, ARF targets E2F1 and/or other E2F family members leading to a decrease in function.
Because ARF-mediated growth suppression is MDM2-dependent in at least one p53-null cell line (6), the mechanism of ARF action in p53-containing cells may be relevant to its mechanism in p53- null cells. ARF stably interacts with MDM2, and the two colocalize in the nucleolus (3, 4, 8–10). ARF inhibits MDM2 nuclear export, rendering MDM2 unable to export p53 to the cytoplasm for degradation(8, 9, 11). p53 ubiquitination, mediated by MDM2, is also impaired by ARF (3, 12, 13). Therefore, it is possible that other targets of MDM2 and/or related E3 proteins are modulated similarly by ARF, resulting in growth arrest of p53-null cells.
E2F1 and -3 would make logical ARF targets, given their roles in promoting cell cycle progression (14–16). Both are highly regulated at the transcriptional and posttranscriptional levels, and some elements of this complex set of regulatory events occur in a cell cycle-dependent manner (for review, see ref. 17). Herein we describe results suggesting a potential mechanism by which ARF could suppress proliferation of p53-null cells. The data reveal specific interactions between ARF and several E2F species paralleled by enhanced degradation of these proteins.
Materials and Methods
Cell Lines, Transfections, and Plasmids.
U2OS, 293T, and MDA-MB231 cells were cultured in DMEM containing 10% FBS at 37°C in an atmosphere of 10% CO2. Late-passage immortalized cultures of p19ARF−/− (5) and p53−/− (18) mouse embryo fibroblasts were similarly maintained. All transfections were performed with Fugene reagent by the manufacturer's instructions (Roche) with cells plated on 10- or 15-cm dishes. pRclCMVHA-E2F1, -2, and -3 and pCDNA3-HAE2F6 plasmids have been described (19, 20). pBabe-p19 contains a full-length p19ARF cDNA coding unit inserted into pBabe (21). pCD-p19 contains the same p19ARF encoding sequence cloned into pCDNA3.
293T Growth Assay.
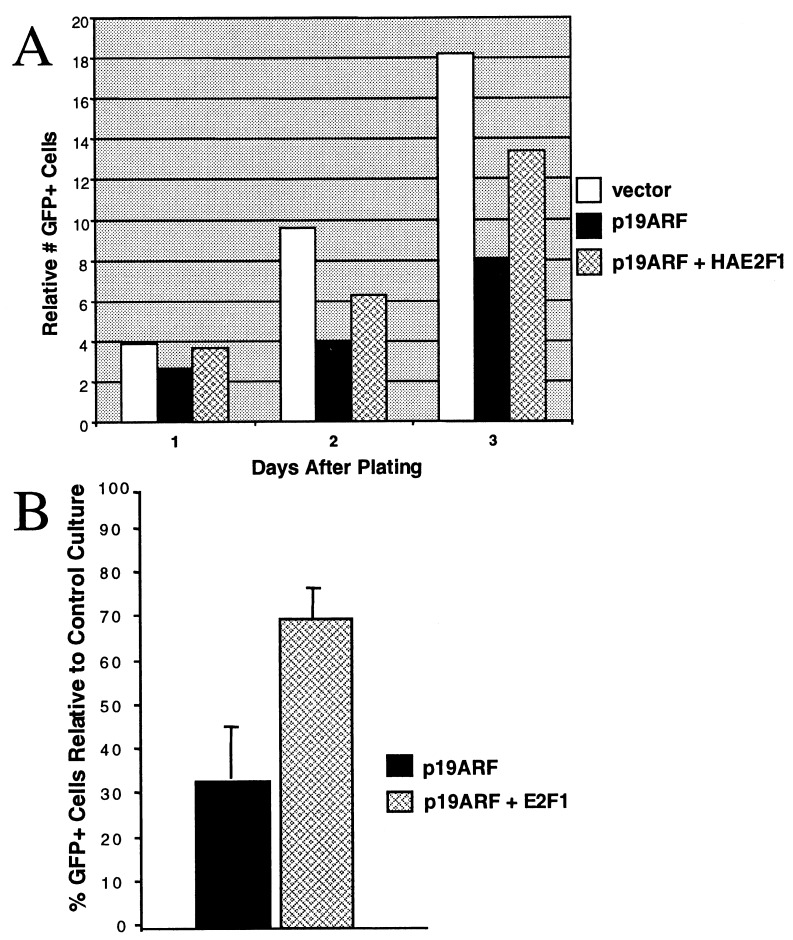
293T cells in 15-cm dishes were transfected with green fluorescent protein (GFP) expression vector and pCDNA3, p19ARF, and pRC/CMV-HA-E2F1, as indicated. Fifteen hours after transfection, transfected cells were split and seeded in two six-well dishes per transfection at 10%–20% confluence. The number of GFP-positive cells in five microscope fields of each well were counted, and the total number of cells counted per six-well plate summed to give the number of GFP-positive cells. GFP-positive cells were counted on the day of posttransfection plating and at 24, 48, and 72 h thereafter. Medium in each well was removed, and cells were given fresh medium every day. For a given transfection condition, a total of four plates from two transfections were analyzed. Within each transfection, numbers of GFP-positive cells were averaged between two replicate six-well dishes. The ratio of GFP-positive cells in ARF-transfected vs. vector-transfected plates then was calculated. These normalized ratios were then averaged for each time to yield the values depicted in Fig. 5B.
Figure 5.
Rescue of ARF growth suppression of p53-defective cells by ectopic E2F1. (A) Representative growth curves of 293T cells transfected with vector (open bars), ARF (solid bars), or ARF and E2F1 (hatched bars). A representative sample of GFP- positive cells from each transfected culture was counted each day for 3 days after plating. (B) The average percentage of GFP-positive cells in cultures transfected with ARF (solid bar) or ARF and E2F1 (hatched bar) relative to control-transfected cultures 48 h after plating. The data are the average from two experiments; error bars show ± 1 SD.
Retroviral Infection.
p19ARF- and GFP-encoding retroviruses and control backbone retroviruses were prepared by transfecting 293GPG cells with pBabe, pBabe-GFP, and pBabe-p19, followed by supernatant harvesting (22). Amphotropic retrovirus encoding a dominant negative p53 protein (143A) was constructed by transfer of a p53 m143A cDNA (23) into pMMP and cotransfection of pMMP-p53 m143A with pMD.G and pMD.MLV plasmids into 293T cells followed by harvesting of amphotropic viral supernatants (22). The resultant amphotropic retroviral supernatants were used to infect U2OS or MDA-MB231 cells by two serial 3-h applications followed by a third overnight application, all in Polybrene (8 μg/ml). The next day, cells were rinsed and incubated in complete medium for 40 h, followed by selection with puromycin (2 μg/ml).
Pulse–Chase Analysis.
Pulse–chase analysis of E2F1 protein was performed as described (24). 35S-labeled protein-containing lysates were immunoprecipitated with C-20 anti-E2F1 antibody (Santa Cruz Biotechnology), and the specific E2F1 autoradiographic signal was quantitated with a STORM PhosphorImager (Molecular Dynamics).
Northern Blot Analysis.
Total RNA was prepared from transfected cells with Rneasy (Qiagen, Chatsworth, CA). Identical amounts of RNA from each culture were loaded onto a formaldehyde-agarose gel. After electrophoresis, the gel was blotted onto a nylon membrane and hybridized as described (25) with specific E2F and 36B4 (loading control) probes as indicated (20, 26). Northern blots were analyzed and quantitated with a STORM PhosphorImager. For Western blotting experiments performed to determine relative protein stability in a set of cultures (Northern/Western analysis), the quantities of protein-containing lysate subjected to analysis were inversely proportional to the quantity of specific E2F RNA detected by Northern blotting of RNA-containing lysates of the same cultures.
Antibodies, Binding Assays, and Western Blotting.
For E2F protein analysis, cells were lysed in NETT300 or NETT240 (20 mM Tris⋅HCl, pH 8.0/0.1 mM EDTA/300 or 240 mM NaCl, respectively/0.5% Triton X-100), and the protein concentration of each lysate was determined by a Bradford assay. p19ARF was detected with AEC40 polyclonal antibody prepared against a C-terminal p19ARF peptide. E2F–p19ARF interactions were detected by lysing the relevant cells in NETT250 (20 mM Tris⋅HCl, pH 8.0/0.1 mM EDTA/250 mM NaCl/0.5% Triton X-100), followed by addition of the indicated antibody to the lysate for 1 h at 4°C. Protein A-Sepharose was then added, reaction mixtures were rocked at 4°C for 45 min, and the protein A-beads were washed four times in NETT250. Immunoprecipitated proteins were analyzed by SDS/PAGE followed by Western blotting for E2F1, -2, or -3 or p19ARF.
Immunofluorescence.
U2OS cells were transfected on coverslips, fixed with 3% paraformaldehyde/2% sucrose, and stained with the indicated antibodies as described (27).
Results
p19ARF Targets E2F Proteins for Degradation.
E2F1 overexpression can override ARF-induced growth arrest in certain p53-null cells (6). Hence, the possibility that p19ARF can modulate the abundance of E2F1 was explored. E2F1 cDNA was cotransfected with a control or a p19ARF-encoding expression vector into U2OS cells, which do not express INK4A and ARF proteins and are p53+/+ (10). Lysates were analyzed for E2F1 abundance. Synthesis of p19ARF resulted in a sharp decrease in the abundance of ectopically expressed E2F1, and a shift toward more rapidly migrating forms of E2F1 was also noted (Fig. 1A, compare lane 1 and 2). The multiple electrophoretic forms of E2F1 are products of differential phosphorylation (28).
Figure 1.
p19ARF promotes E2F1 degradation. (A) p19ARF affects E2F1 abundance and electrophoretic mobility. pRclCMV-HA-E2F1 (1 μg), pCD-p19ARF (5 μg), or vector alone (5 μg) were cotransfected into U2OS cells. Cell extracts were prepared and examined by SDS/PAGE on 12% gels and Western blotting using antibody specific for E2F1 (C20, Santa Cruz Biotechnology). Endogenous E2F1 is not detected in this experiment. (B) Pulse–chase analysis of E2F1 in the absence (◊) or presence (□) of p19ARF. pRclCMV-HA-E2F1 (1 μg) and pCD-p19ARF (5 μg) or vector alone (5 μg) were cotransfected into U2OS cells. Cells were pulsed-labeled with [35S]methionine for 30 min and chased for 1 or 3 h, followed by immunoprecipitation with anti-E2F1 antibodies (C20). Under these conditions, endogenous E2F1 was not detected (data not shown). The amount of transfected E2F1 in specific bands (as detected with a PhosphorImager) was plotted as a function of time. (The E2F1 signal detected at time 0 was set at 100% and used to normalize the signal at the subsequent times.) (C and D) E2F2 and E2F3 are targeted for degradation by p19ARF. pRclCMV-HAE2F2 or -3 expression plasmids (2 μg) were transfected into 293T cells with an empty vector (5 μg) or pCD-p19ARF (5 μg), and cell lysates were analyzed for ectopic E2F2 or -3 mRNA and protein levels. Lysates were analyzed by Western blotting for HA (F-7, Santa Cruz Biotechnology) and for p19ARF (AEC40), and gel loading of each lysate was normalized for E2F2 or -3 mRNA levels. The upper band in both lanes of C and D is nonspecific. (E) E2F6 is not targeted for degradation by ARF. pCDNA3-HAE2F6 (2 μg) or pRclCMV-HAE2F1 (2 μg,-a positive control for ARF-induced degradation) were cotransfected with empty vector (5 μg) or pCD-p19ARF(5 μg) into 293T cells, and lysates analyzed for ectopic E2F6 and E2F1 protein and mRNA levels. Lysates were analyzed by Western blotting for HA (F-7) and p19ARF (R562, Abcam, Cambridge, U.K.), and gel loading of each lysate was normalized for E2F6 or E2F1 mRNA levels, respectively. (F) LLnL suppresses p19ARF-mediated destabilization of E2F1. pRclCMV-HAE2F1 (2 μg) was cotransfected with control vector (5 μg) or pCD-p19ARF (5 μg) into U2OS (Upper) or 293T cells (Lower). Twenty-four hours after transfection, plates were incubated in 50 μM LLnL/0.1% DMSO or 0.1% DMSO for 12 h, and ectopically expressed E2F1 mRNA levels were measured in the transfected cells by Northern blotting. Cell lysates from the same cultures were loaded onto an SDS gel with loading normalized for the E2F1 mRNA levels present in the cells of origin. HA-E2F1 and p19ARF levels were determined by Western blotting with 12CA5 (Roche) anti-HA and AEC40 antibodies, respectively.
The possibility that the p19ARF-induced decrease in E2F1 abundance was caused by increased E2F1 protein turnover was directly investigated by pulse–chase analysis of 35S-labeled, ectopically overexpressed E2F1 (Fig. 1B). Consistent with the decreased abundance of E2F1 observed with coexpression of p19ARF (Fig. 1A), the half-life of ectopically expressed E2F1 was decreased from 2.5 h to ≈1.5 h when p19ARF was coexpressed (Fig. 1B). Thus, increased turnover accompanied the decrease in abundance of E2F1 triggered by p19ARF coexpression.
Furthermore, ectopically expressed E2F2 and E2F3 were targeted for degradation by coexpressed p19ARF. As shown in Fig. 1 C and D, E2F2 and E2F3 levels were drastically reduced in cells expressing the indicated ectopic E2F protein and p19ARF compared with E2F2 or -3 levels in cells expressing only the ectopic E2F protein. As for E2F1, the effect was most likely posttranscriptional, because the quantity of lysate protein loaded on the gels in Fig. 1 C and D was normalized for the amount of ectopically expressed E2F2 and 3 mRNA in the relevant transfected culture. Importantly, E2F6 was not targeted for degradation by coexpression of p19ARF (Fig. 1E Left), whereas in a parallel transfection E2F1 was targeted by ARF for degradation, as expected (Fig. 1E Right). Thus the effects of p19ARF on E2F1, -2, and -3 are relatively specific and presumably related to sequences not present in the related E2F family member, E2F6. Indeed, E2F6 lacks certain sequences common to E2F1 through -5 (19, 29, 30).
p19ARF-Mediated Destabilization of E2F1 Requires a Functional Proteasome.
E2F1 degradation is normally controlled by the ubiquitin/proteasome system, and the protein is stabilized by the exposure of cells to specific proteasome inhibitors (24, 31–33). To determine whether the acceleration of E2F1 turnover by p19ARF depended on proteasome function, U2OS (wild-type p53) and 293T (p53 disabled by E1B and large T antigen) cells, expressing a transfected allele of E2F1 alone or with a cotransfected allele of p19ARF, were exposed to the proteasome inhibitor LLnL. The relative abundance of E2F1 was analyzed by Western blotting, with gel loading of lysate protein normalized for the abundance of E2F1 mRNA. As shown in Fig. 1F, LLnL, but not DMSO, interfered significantly with the ability of p19ARF to destabilize E2F1 in U2OS and 293T cells. These results suggest that proteasome action contributes to p19ARF-mediated destabilization of E2F1.
The Stability of Endogenous E2F1 and E2F3 Is Reduced in the Presence of p19ARF.
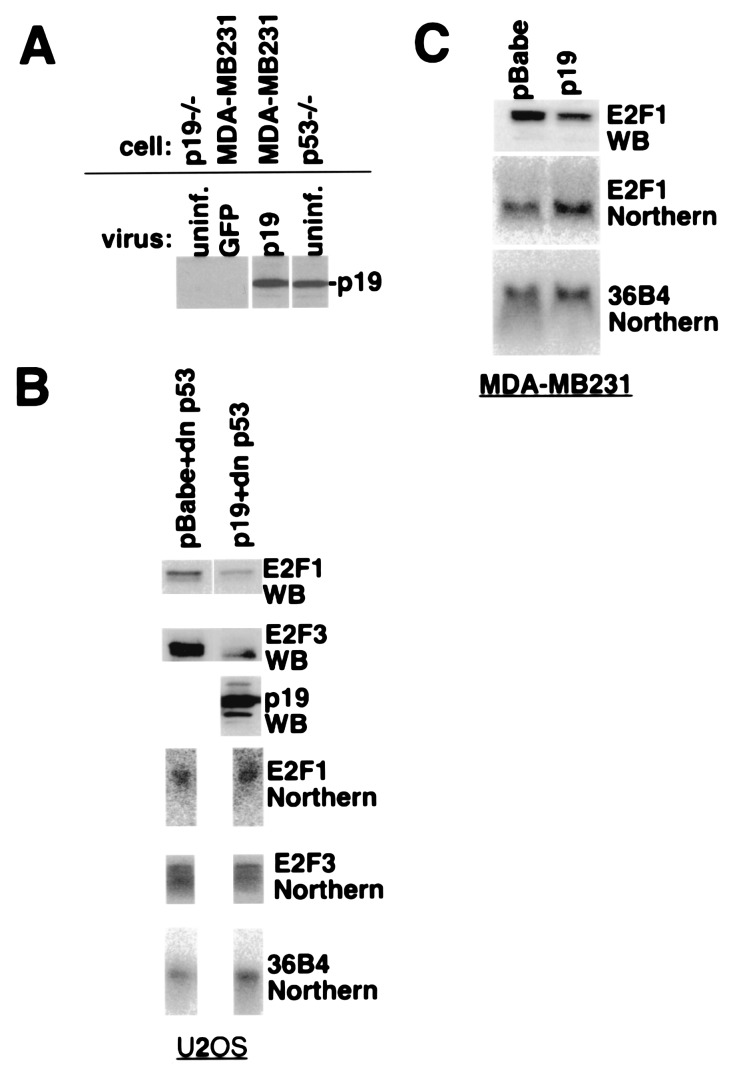
To gauge whether the effect of ARF on ectopically expressed E2F1 is physiologically relevant, U2OS (wild-type p53) or MDA/MB231 (p53 mutated) cells, neither of which synthesizes ARF protein (10), were infected with control or p19ARF-encoding retrovirus. The level of p19ARF expression achieved in infected MDA-MB231 cells paralleled that normally present in mouse embryo fibroblasts (MEFs) nullizygous for p53 (Fig. 2A). Cultures of both infected lines were analyzed for E2F1 protein and mRNA abundance by Western and Northern blotting, respectively. To avoid confounding effects on E2F1 abundance resulting from ARF/p53-mediated cell cycle arrest (5), the p53 pathway was disabled in the U2OS cells by coinfection with a retrovirus encoding a dominant negative p53 allele. As with the ectopically expressed protein, endogenous E2F1 levels in both cell types decreased relative to control virus-infected cells (Fig. 2 B and C). The source of this decrease was likely posttranscriptional, because there was no significant difference in E2F1 mRNA between control and p19ARF-transduced cells.
Figure 2.
p19ARF degrades endogenous E2F1 and E2F3. (A) Expression level of p19ARF in infected MDA-MB231 cells and MEFs. Lysates of control GFP-expressing virus- and p19ARF virus-infected MDA-MB231 cells and uninfected p19−/− MEFs and p53−/− (p19 +/+) MEFs were analyzed for p19ARF levels by Western blotting with AEC40. (B) Effect of p19ARF on endogenous E2F1 and E2F3. U2OS cells were infected with an amphotropic retrovirus encoding a dominant negative p53 allele, in combination with a control backbone retrovirus or a virus encoding p19ARF. Puromycin-resistant cells were harvested and analyzed by Western and Northern blotting for E2F1 and E2F3 protein (antibodies C-20/E2F1 and C-20/E2F3, Santa Cruz Biotechnology) and mRNA levels. Northern and Western blots were, respectively, loaded with equivalent amounts of total RNA or protein per lane. 36B4 mRNA served as an mRNA loading control. (C) Effect of p19ARF on endogenous E2F1 in p53 mutant cells. MDA-MB231 breast cancer cells (null for INK4A with mutant p53) were infected in parallel with control backbone or ARF-encoding retrovirus (in a separate experiment from that depicted in A). Puromycin-resistant cells were analyzed for E2F1 protein and mRNA levels as in B.
Analysis of retrovirus-infected U2OS cells for levels of E2F3, an E2F family member involved in G1/S-phase progression (15, 16), demonstrated that its abundance also decreased with p19ARF expression (Fig. 2B). The mechanism underlying the decrease in E2F3 concentration is also likely to be increased protein turnover, because no significant change in E2F3 mRNA levels was noted in the p19ARF-expressing cells (Fig. 2B).
E2F1 and p19ARF Interact in Vivo.
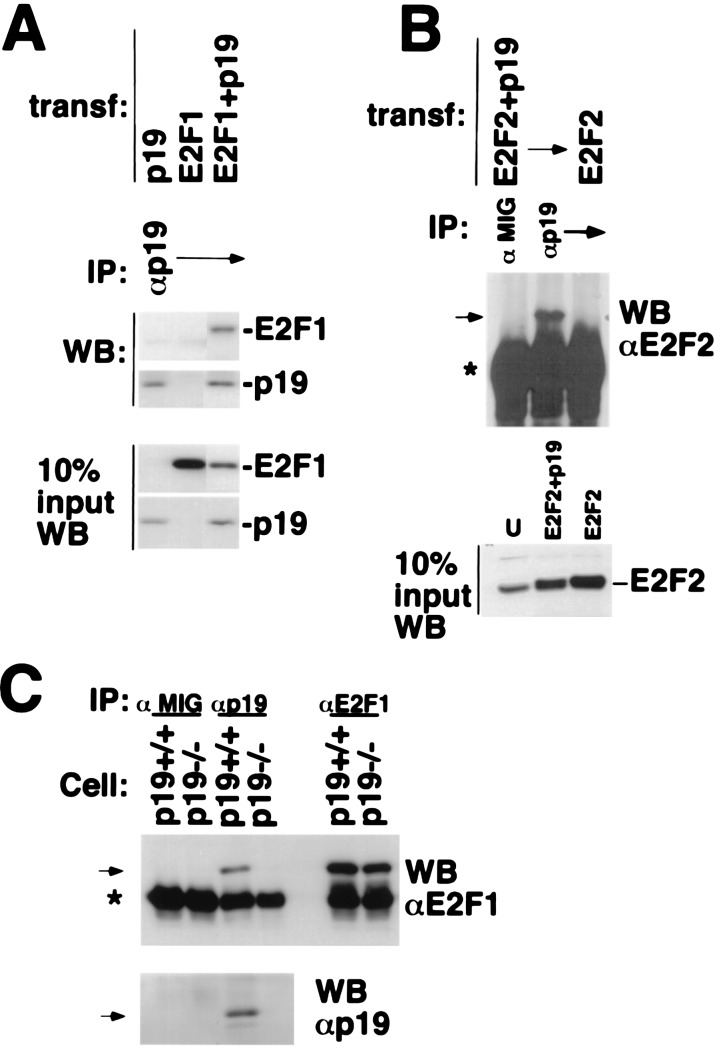
The effect of p19ARF on the turnover of E2F could, in principle, be associated with the formation of complex(es) containing p19ARF and E2F. U2OS cells were transfected with expression vectors encoding the indicated E2F proteins, p19ARF, or both; and transfected cell lysates were immunoprecipitated with p19ARF antibody (Fig. 3). E2F1 was readily detected in p19ARF immunoprecipitates from cells expressing both proteins (Fig. 3A) but not from cells expressing either E2F1 or p19ARF alone (Fig. 3A). Similarly, E2F2 and E2F3 proteins were detected in the respective anti-ARF immunoprecipitates of lysates from cells cotransfected with ARF and E2F2 or E2F3 expression vectors (Fig. 3B and data not shown). Neither E2F2 nor E2F3 appeared in anti-ARF immunoprecipitates of lysates from cells expressing either E2F protein in the absence of cotransfected ARF, nor was E2F2 or E2F3 immunoprecipitated with an irrelevant antibody in extracts of ARF/E2F cotransfectants (Fig. 3B and data not shown). p19ARF was also immunoprecipitated with an anti-E2F1 antibody in a lysate from cells cotransfected with p19ARF and E2F1 (data not shown).
Figure 3.
p19/E2F1 and E2F2 interactions. (A and B) p19ARF interacts with E2F1 and E2F2. U2OS cells were transfected with plasmids encoding p19ARF, HA-E2F1, or HA-E2F2, as indicated. Cell extracts were prepared and then immunoprecipitated with antibody to p19ARF (AEC40). HA-E2F1, HA-E2F2, and p19ARF, present in the p19ARF immunoprecipitates, were analyzed by Western blotting for E2F1 (C20/E2F1), E2F2 (C20/E2F2, Santa Cruz Biotechnology), and p19ARF (AEC40), respectively. In A and B, 10% of each cell extract used for the immunoprecipitations was directly analyzed for relevant protein abundance by Western blotting, with the antibodies above for E2F1, E2F2, or p19ARF. (C) Endogenous p19ARF binds to E2F1. Cell extracts derived from asynchronously growing, immortalized murine p19ARF+/+ (also p53 null) or p19ARF−/− MEFs were immunoprecipitated with antibody to p19ARF (AEC40) or E2F1 (C20) or with mouse IgG (Cappel, negative control). E2F1 and p19ARF present in the p19ARF immunoprecipitates were analyzed by Western blotting for E2F1 (C20) and p19ARF (AEC40). Specific bands are indicated by arrows. The asterisk indicates the Ig heavy chain.
To confirm the physiologic relevance of the p19ARF–E2F complexes containing ectopic E2F proteins, lysates of p53-null MEFs (p19ARF+/+; Fig. 3C) were analyzed for endogenous p19ARF–E2F1 complexes. E2F1 and p19ARF were present in an anti-p19ARF immunoprecipitate, but not in that of an irrelevant control immunoprecipitate (Fig. 3C) of p53-null MEF lysate, suggesting an endogenous ARF–E2F1 interaction. To rule out nonspecific or cross-reactive interactions of E2F1 with the ARF antibody, lysate from p19ARF-null MEFs (5) was subjected to anti-ARF and anti-E2F1 immunoprecipitation (Fig. 3C). Although E2F1 was readily detected when anti-E2F1 was used with the p19-null MEF lysate, it was absent when anti-p19ARF was used (Fig. 3C), demonstrating that ARF protein is required for detection of endogenous E2F1 in an anti-ARF immunoprecipitate.
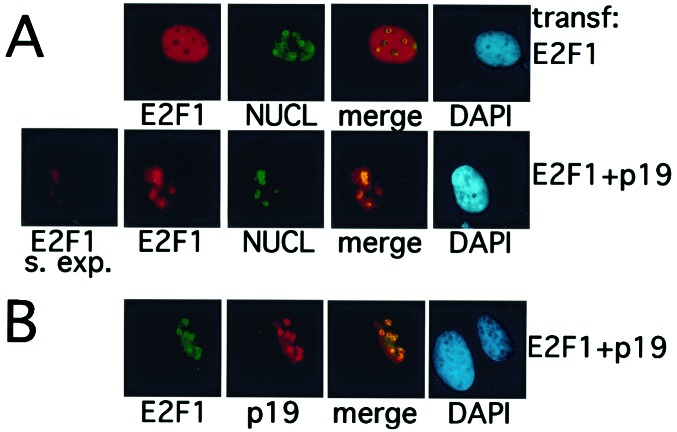
p19ARF Relocalizes E2F1 to the Nucleolus.
p19ARF normally resides in the nucleolus (3, 10) and can stably retain MDM2 in this organelle (8, 9). Given the evidence that p19ARF interacts with E2F1, the intranuclear localization of E2F1 was analyzed in p19ARF-expressing cells. U2OS cells expressing ectopic E2F1 with or without ectopic p19ARF were stained, in parallel, with E2F1 and nucleolin antibodies (Fig. 4A). When ectopically expressed, E2F1 is normally nucleoplasmic and excluded from nucleoli (34) (Fig. 4A Upper). By contrast, there was clear and quantitative relocalization of E2F1 to nucleoli (defined by nucleolin costaining) when it was coexpressed with p19ARF (Fig. 4A Lower).
Figure 4.
p19ARF and E2F1 colocalize in nucleoli. (A) E2F1 colocalizes with nucleolin in ARF-expressing cells. U2OS cells were transfected with plasmids encoding HA-E2F1, with or without a p19ARF expression vector, as indicated on the right. Immunostaining of transfected cells was performed with the following antibodies: α-E2F1(C20, red), α-nucleolin (C23, Santa Cruz Biotechnology, green). In the merged image, yellow indicates merger of the red and green signals. In S. exp., the exposure time for the E2F1 image (in cells containing coexpressed ARF) was the same as that used to generate the E2F1 image in the row above (in cells in which there was no coexpressed ARF). (B) p19ARF and E2F1 colocalize in nucleoli. U2OS cells were cotransfected with plasmids encoding HA-E2F1 and p19ARF. Immunostaining was performed with the following antibodies: α-E2F1(KH95, Santa Cruz Biotechnology, green) and α-p19 (AEC40, red). Colocalization of E2F1 with p19ARF in nucleoli is reflected by the merged yellow signal.
The absolute level of E2F1 was also greatly reduced in the presence of p19ARF, as was apparent when the exposure times for E2F1 immunofluorescent imagings of control and p19ARF-expressing cells were equalized (Fig. 4A, compare Upper and Lower). When cells coexpressing E2F1 and p19ARF were simultaneously stained for the two proteins, they were colocalized in nucleoli (Fig. 4B). Thus, the E2F1–p19ARF interaction results not only in the destabilization of E2F1 protein but also in subnuclear relocalization of this protein into nucleoli with p19ARF.
p19ARF Suppresses the Proliferation of Cells Defective for p53 Function.
Excessive degradation of E2F1 and E2F3 might be expected to alter cell cycle progression in p53-null cells expressing p19ARF (14–16). p19ARF-expressing 293T cells, rendered p53-defective by the expression of viral transforming proteins (35, 36), grew more slowly than control-transfected cells (Fig. 5). In a representative experiment depicted in Fig. 5A, ARF-expressing cells underwent only three cell doublings by 72 h after plating, whereas control cells underwent more than four doublings in this period. Moreover, E2F1 and E2F3 levels were lower in the ARF-transfected 293T cells than in vector-transfected cells (data not shown). Consistent with earlier results (6) and with a role for E2F degradation in the mechanism of ARF growth suppression in p53-defective cells, coexpression of ectopic E2F1 partially, albeit significantly, ameliorated ARF-induced growth suppression of these cells (Fig. 5, hatched bars).
Discussion
Herein we demonstrate an effect of p19ARF on the abundance of multiple members of the E2F transcription factor family, at least two of which (E2F1 and -3) play defined roles in promoting cell cycle progression (14–16). Until recently, ARF proteins were known to signal through the p53 pathway, providing a sensor for oncogenic signals (3, 5, 37, 38). However, recent evidence suggests that p19ARF can support a tumor suppressor function in the absence of an active p53 pathway (6, 7). Thus, additional ARF targets exist; given these recent results, we wondered whether certain E2F species were ARF targets.
Results herein suggest a role for p19ARF in posttranslational regulation of E2F stability. Biochemically, the effect is the opposite of the ARF effect on p53 stability, which increases largely because of ARF-mediated inhibition of MDM2 E3 ubiquitin ligase and nuclear export functions (8, 9, 12, 13). Whether MDM2 is involved in the ARF–E2F effect remains to be determined, although it must be emphasized that others have suggested a necessary role for MDM2 in E2F1 degradation (39) and in ARF-mediated proliferation arrest of p53−/− cells (6).
Although loss of ARF lessens the selective pressure for loss of p53 during immortalization and tumorigenesis in mice (5, 40–43), a number of human tumors have lost both p53 and ARF functions (2, 10, 44). In these tumors, there has likely been a selection for loss of both loci, although ARF expression is always lost in combination with p16INK4A, making it difficult to understand the individual contributions of the loss of p16INK4A and of ARF to the promotion of tumorigenesis.
Another potential role for p19ARF-mediated destabilization of E2F might be as part of a negative feedback loop. Activation of the p53 tumor suppressor by DNA damage is eventually switched off by the p53-induced MDM2 protein, which promotes the ubiquitin-mediated degradation of p53 (45). An analogous mechanism might be relevant to the interrelationships of ARF and E2F1. E2F1 can specifically activate the ARF promoter, leading to p53-dependent apoptosis (46–49). Attenuation of a major stimulus to ARF synthesis should, in theory, be an outcome of ARF-driven E2F1 depletion, which, in turn, would have cell survival value.
Ideally, future analyses will help to define the biochemical mechanisms by which p19ARF targets E2F proteins for degradation. It appears that multiple, independent domains within E2F1 serve as targets for p19ARF-mediated degradation (S.G., F.M., and D.M.L., unpublished observations). Whether the ARF–E2F interaction is direct or not has also not been established. Given the LLnL results, the biochemical mechanism of ARF-mediated E2F degradation may be connected with E2F ubiquitination (24, 32, 33, 50), where ARF might serve as an adaptor that brings E2F and the ubiquitination apparatus together and/or to augment the delivery of properly modified E2F protein to the proteasome, perhaps through the nuclear export pathway.
Acknowledgments
We thank Dr. C. Sherr for ARF-null cells, Drs. J.-S. Lee, R. C. Mulligan, and J. You for pMMP, pMD.G, and pMD.MLV, Drs. J. Pomerantz and L. Chin for AEC40 antibody, Dr. J. DeCaprio for sharing unpublished data, Drs. S. Gaubatz and M. Ewen for critically reading the manuscript, and members of the Livingston lab for helpful discussions. This work was supported by Grant PO1 CA50661–12 from the National Cancer Institute and by a fellowship from the Leukemia and Lymphoma Society to F.M.
Abbreviations
- ARF
alternative reading frame
- GFP
green fluorescent protein
- MEF
mouse embryo fibroblast
References
- 1.Sherr C J. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 2.Sharpless N E, DePinho R A. Curr Opin Genet Dev. 1999;9:22–30. doi: 10.1016/s0959-437x(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz J, Schreiber-Agus N, Liegeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H W, et al. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 5.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 6.Carnero A, Hudson J D, Price C M, Beach D H. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- 7.Weber J D, Jeffers J R, Rehg J E, Randle D H, Lozano G, Roussel M F, Sherr C J, Zambetti G P. Genes Dev. 2000;14:2358–2365. doi: 10.1101/gad.827300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber J, Taylor L, Roussel M, Sherr C, Bar-Sagi D. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 9.Tao W, Levine A J. Proc Natl Acad Sci USA. 1999;96:6937–6941. doi: 10.1073/pnas.96.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, et al. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Xiong Y. Mol Cell. 1999;3:579–591. doi: 10.1016/s1097-2765(00)80351-2. [DOI] [PubMed] [Google Scholar]
- 12.Honda R, Yasuda H. EMBO J. 1999;18:22–27. doi: 10.1093/emboj/18.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Midgley C A, Desterro J M, Saville M K, Howard S, Sparks A, Hay R T, Lane D P. Oncogene. 2000;19:2312–2323. doi: 10.1038/sj.onc.1203593. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z M, Yang H, Livingston D M. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humbert P O, Verona R, Trimarchi J M, Rogers C, Dandapani S, Lees J A. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- 17.Muller H, Helin K. Biochim Biophys Acta. 2000;1470:M1–M12. doi: 10.1016/s0304-419x(99)00030-x. [DOI] [PubMed] [Google Scholar]
- 18.Montes de Oca Luna R, Wagner D S, Lozano G. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 19.Gaubatz S, Wood J G, Livingston D M. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krek W, Ewen M E, Shirodkar S, Arany Z, Kaelin W G, Jr, Livingston D M. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 21.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ory D S, Neugeboren B A, Mulligan R C. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinds P W, Finlay C A, Quartin R S, Baker S J, Fearon E R, Vogelstein B, Levine A J. Cell Growth Differ. 1990;1:571–580. [PubMed] [Google Scholar]
- 24.Hofmann F, Martelli F, Livingston D M, Wang Z. Genes Dev. 1996;10:2949–2959. doi: 10.1101/gad.10.23.2949. [DOI] [PubMed] [Google Scholar]
- 25.Martelli F, Cenciarelli C, Santarelli G, Polikar B, Felsani A, Caruso M. Oncogene. 1994;9:3579–3590. [PubMed] [Google Scholar]
- 26.Masiakowski P, Breathnach R, Bloch J, Gannon F, Krust A, Chambon P. Nucleic Acids Res. 1982;10:7895–7903. doi: 10.1093/nar/10.24.7895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lill N L, Grossman S R, Ginsberg D, DeCaprio J A, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 28.Peeper D S, Keblusek P, Helin K, Toebes M, van der Eb A J, Zantema A. Oncogene. 1995;10:39–48. [PubMed] [Google Scholar]
- 29.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartwright P, Muller H, Wagener C, Holm K, Helin K. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 31.Martelli F, Livingston D M. Proc Natl Acad Sci USA. 1999;96:2858–2863. doi: 10.1073/pnas.96.6.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hateboer G, Kerkhoven R M, Shvarts A, Bernards R, Beijersbergen R L. Genes Dev. 1996;10:2960–2970. doi: 10.1101/gad.10.23.2960. [DOI] [PubMed] [Google Scholar]
- 33.Campanero M R, Flemington E K. Proc Natl Acad Sci USA. 1997;94:2221–2226. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magae J, Wu C L, Illenye S, Harlow E, Heintz N H. J Cell Sci. 1996;109:1717–1726. doi: 10.1242/jcs.109.7.1717. [DOI] [PubMed] [Google Scholar]
- 35.Grand R J, Lecane P S, Owen D, Grant M L, Roberts S, Levine A J, Gallimore P H. Virology. 1995;210:323–334. doi: 10.1006/viro.1995.1349. [DOI] [PubMed] [Google Scholar]
- 36.Steegenga W T, Van Laar T, Shvarts A, Terleth C, Van der Eb A J, Jochemsen A G. Virology. 1995;212:543–554. doi: 10.1006/viro.1995.1512. [DOI] [PubMed] [Google Scholar]
- 37.de Stanchina E, McCurrach M E, Zindy F, Shieh S Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blattner C, Sparks A, Lane D. Mol Cell Biol. 1999;19:3704–3713. doi: 10.1128/mcb.19.5.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chin L, Pomerantz J, Polsky D, Jacobson M, Cohen C, Cordon-Cardo C, Horner J W, 2nd, DePinho R A. Genes Dev. 1997;11:2822–2834. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs J J, Scheijen B, Voncken J W, Kieboom K, Berns A, van Lohuizen M. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt C A, McCurrach M E, de Stanchina E, Wallace-Brodeur R R, Lowe S W. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez-Cespedes M, Reed A L, Buta M, Wu L, Westra W H, Herman J G, Yang S C, Jen J, Sidransky D. Oncogene. 1999;18:5843–5849. doi: 10.1038/sj.onc.1203003. [DOI] [PubMed] [Google Scholar]
- 45.Prives C, Hall P A. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 46.Dimri G P, Itahana K, Acosta M, Campisi J. Mol Cell Biol. 2000;20:273–285. doi: 10.1128/mcb.20.1.273-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bates S, Phillips A C, Clark P A, Stott F, Peters G, Ludwig R L, Vousden K H. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 48.Tsai K Y, Hu Y, Macleod K F, Crowley D, Yamasaki L, Jacks T. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 49.Kowalik T F, DeGregori J, Leone G, Jakoi L, Nevins J R. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 50.Marti A, Wirbelauer C, Scheffner M, Krek W. Nat Cell Biol. 1999;1:14–19. doi: 10.1038/8984. [DOI] [PubMed] [Google Scholar]