Abstract
Purpose
Tumor cells expressing elevated aldehyde dehydrogenase (ALDH) activity attributed to ALDH1/3 isoforms have been identified as ALDHbright cells and have the properties attributed to cancer initiating cells (CIC). CIC represent the subpopulation of tumor cells that are resistant to conventional cancer treatments and highly tumorigenic in immunodeficient mice. They are considered to be responsible for tumor recurrence and metastasis. The ALDH1A1 isoform was previously identified as a tumor antigen recognized by CD8+ T cells. This study examines the ability of ALDH1A1-specific CD8+ T cells to eliminate ALDHbright cells and control tumor growth and metastases.
Experimental Design
ALDHbright cells were isolated by flow cytometry from HLA-A2+ human head and neck, breast and pancreas carcinoma cell lines using ALDEFLUOR® and tested for their tumorigenicity in immunodeficient mice. ALDH1A1-specific CD8+ T cells were generated in vitro and tested for their ability to eliminate CIC in vitro and in vivo by adoptive transfer to immunodeficient mice bearing human tumor xenografts.
Results
ALDHbright cells isolated by flow cytometry from HLA-A2+ breast, head and neck and pancreas carcinoma cell lines at low numbers (500 cells) were tumorigenic in immunodeficient mice. ALDHbright cells present in these cell lines, xenografts or surgically removed lesions were recognized by ALDH1A1-specific CD8+ T cells in vitro. Adoptive therapy with ALDH1A1-specific CD8+ T cells eliminated ALDHbright cells, inhibited tumor growth, metastases or prolonged survival of xenograft-bearing immunodeficient mice.
Conclusions
The results of this translational study strongly support the potential of ALDH1A1-based immunotherapy to selectively target CIC in human cancer.
Introduction
Cancer initiating cells (CIC) are characterized as a subpopulation of tumor cells in tumors which exhibit “stem-cell like” properties, such as self-renewal, chemo- and radio-resistance, and high tumorigenicity at low cell numbers in immunodeficient mice (1–5). Therefore, they are considered responsible for tumor recurrence and metastasis. In several types of tumors, cell populations enriched for cancer initiating activity are being readily identified and isolated by flow cytometry analysis based on their high level of aldehyde dehydrogenase (ALDH) activity using the ALDEFLUOR® reagent and can be referred to as ALDHbright cells (6–12). The ALDH activity detected by this reagent is primarily attributed to members of the ALDH1 and ALDH3 family of ALDH isoforms. The ability to readily identify and isolate ALDHbright cells by flow cytometry is facilitating the efforts to develop therapeutic approaches that would target CIC and elicit long term and effective responses in subjects with cancer (13). ALDH1-targeted immunotherapy represents such an approach, since in a previous study we have shown that the ALDH1A1 isoform can mediate the recognition and lysis of ALDH1A1+ squamous cell carcinoma of the head and neck (SCCHN) cell lines by cognate CD8+ cytotoxic T cells (CTL). Relevant to the potential clinical use of ALDH1A1-specific CTL-based immunotherapy, ALDH1A1-specific CTL recognize neither normal differentiated cells, such as fibroblasts, unless they are transfected to express high levels of ALDH1A1 nor normal CD34+ hematopoietic stem cells. The latter express ALDH1A1 at a level which is higher than that found in most normal differentiated cells and tissues, but lower than that in CIC (14).
In the present study we have investigated the ability of in vitro-generated ALDH1A1-specific CTL to eliminate ALDHbright cells present in HLA-A2+ human carcinoma cell lines, xenografts and surgically removed lesions in vitro and the anti-tumor activity of adoptive immunotherapy with ALDH1A1-specific CTL in vivo. In addition we have analyzed the expression of ALDH1A1 and HLA class I Ag expression in normal liver, since normal hepatocytes have been reported to express a high level of this ALDH1 isoform (15) and, therefore, represent a potential concern in implementing ALDH1A1-based immunotherapy.
Materials and Methods
Human cell lines, tumor specimens and blood
The human SCCHN cell lines used in these studies have been previously described (14). The MDA-MB-231 breast and MIA PaCa-2 pancreatic carcinoma cell lines were obtained from American Type Culture Collection (ATCC). The luciferase-transfected MDA-MB-231-Luc cell line was obtained from Xenogen. KT-64 feeder cells were generously supplied by Dr. Bruce Levine (Univ. Pennsylvania, Philadelphia, PA) (16–18). Cell lines were maintained in RPMI-1640 medium supplemented with 10% (v/v) FBS, 2 mM L-glutamine, 50µg/ml streptomycin and 50 IU/ml penicillin (Life Technologies, Inc.). Tumor and blood specimens were obtained from consented subjects with SCCHN and pancreatic cancer under the auspices of the University of Pittsburgh Tissue Bank with Institutional Review Board (IRB) approval #991206 and Massachusetts General Hospital IRB approval #08-265, respectively. Blood was obtained from HLA-A2+ normal donors with IRB approval #980633.
Antibodies
ALDH1A1-specific rabbit monoclonal antibody (mAb) (cat. no. ab52492) was purchased from Abcam. The HLA-A,B,C,E,F,G Ag-specific mAb W6/32 (IgG2a), HLA-A2,-A28 Ag-specific mAb KS1(IgG1), HLA-DR Ag-specific mAb L243 (IgG2a), HLA-A heavy chain -specific mAb HC-2A (IgG1), and HLA-B,C heavy chain -specific mAb HC-10 (IgG2a) have been previously described (19–23). APC anti-CD5, FITC anti-CD8, ECD anti-CD45RA, PC7 anti-CCR7 mAb were purchased from BD Biosciences. Rabbit anti-histoneH3 phosphoserine10 mAb was purchased from Cell Signaling Technology, Inc. The ApopTag® Plus Peroxidase In Situ Apoptosis Detection Kit was purchased from Millipore Corp.
Flow cytometry analysis of cell surface stained cells
Tumor cell lines were harvested using 1 mM EDTA (Sigma), and xenografts and lesions disaggregated using Collagenase Type IV (Worthington Biochemical). Duplicate aliquots of tumor cell samples were incubated with ALDEFLUOR® (Stem Cell Technologies), with or without the ALDH inhibitor, diethylaminobenzaldehyde (DEAB) (control) according to the manufacturer’s instructions (14). To identify ALDH+ and ALDHbright cells, the control aliquot of the sample was analyzed by flow cytometry and set for detection of ≤0.2% ALDH+ and 0% ALDHbright cells in the aliquot. Using this cutoff, the test aliquot was analyzed to identify its ALDH+/ALDHbright cell content. The results for human tumor cell lines or lesion samples can vary depending on in vitro propagation of cell lines, lesion disaggregation conditions and/or reagent lot. Cells were sorted using a DakoCytomation MoFlo (Dako North America) at 1.5 ×103 events/second.
Cells were surface stained for HLA class I Ag-specific mAb using standard procedures. Flow cytometry was performed using an FC500 cytometer (Beckman Coulter), which was calibrated daily with fluorescent beads; all samples were run using identical settings to collect a minimum of 10,000 gated events, when possible. Analyses were performed using EXPO32 ADC software (Beckman Coulter) or Summit V4.3 (Dako).
Real time RT/PCR (qRT-PCR) analysis of ALDH1 mRNA
Expression of ALDH1 isoform mRNA relative to that of β-glucuronidase (GUS, an endogenous control or housekeeping gene) mRNA were determined using commercially available and custom designed ALDH1 isoform primer and probe sets and the Applied Biosystems 7700 Sequence Detection Instrument as previously described (14). The following primers/probe sets were used to measure ALDH1A1 mRNA, Forward 5’-cg caagacaggcttttcag-3’, Reverse 5’-tgtataatagtcgccccctctc-3’, Probe: 5’-FAM-attggatccccgtggcgtactatggat-3’; and ALDH1A2 mRNA, Forward 5’-agctttgtgctgtggcaata-3’, Reverse 5’-gatgagggctcccatgtaga-3’, Probe 5’-FAM-taagccagcagagcaaacaccactcag-3’. The Applied Biosystems TaqMan® Gene Expression Assay systems Hs00167476_m1 and Mm03003537_s1 were used to measure ALDH1A3 mRNA and GUS mRNA, respectively.
Tumorigenicity of ALDHbright cells in immunodeficient mice
ALDHbright and ALDHneg cells sorted from tumor cell lines were collected in 2 ml RPMI-1640 medium with 20% FBS and irradiated (300 Gy) bulk parental tumor cells, centrifuged and the supernatant saved for later use. The pellets were suspended in a pre-defined volume of the saved supernatant and equal volume of Matrigel (BD Biosciences), so that a 100µl aliquot contained 500 sorted ALDHbright or ALDHneg tumor cells and 1 × 104 irradiated carrier/feeder cells. These aliquots were injected s.c. in the right and left flanks or ip, respectively, in groups of NOD.CB17-Prckscid/J (NODscid) (The Jackson Laboratories) female (6–8 weeks of age) mice each. The tumorigenicity of MDA-MB-231-Luc cells was monitored by bioluminescence imaging (BLI) using Xenogen IVIS 50 instrument (Xenogen) according to the manufacturer’s recommended protocol at the UPCI In vivo Imaging Facility.
HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells
HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells were induced/expanded by in vitro stimulation (IVS) of CD8+ T cells isolated from peripheral blood obtained from normal HLA-A2+ donors with either ALDH1A188–96 peptide-pulsed autologous dendritic cells (DC) and OKT-3 mAb-activated KT64 feeder cells (the ratio of CD8+ T cells: dendritic cells: KT64 cells being 2:1:2) or ALDH1A188–96 peptide-pulsed artificial antigen presenting cells (aAPC) (16–18). The yields of effector cells using aAPC as stimulators was 3-fold greater than that using peptide-pulsed DC and feeder cells and more than 10-fold greater than the use of peptide-pulsed DC only (data not shown). CD8+ T cells obtained from HLA-A2+ normal donors and IVS with the HLA-A2-restricted, HIVgag362–370 peptide were used as controls in adoptive therapy experiments. Peripheral blood of HLA-A2+ normal donors and patients with SCCHN as well as IVS cultures was analyzed for HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells by flow cytometry using PE-conjugated HLA-A2/ ALDH1A188–96 peptide tetramer complexes obtained from the NIH Tetramer Facility as previously described (24).
Enzyme linked immunospot (ELISPOT) assays
ELISPOT INFγ assays were performed as previously described (14) using the ELISPOT 4.14.3 analyzer (Zeiss). Values were considered significantly different from control values based on the double permutation test. Assay performance and reproducibility were monitored using aliquots of cryopreserved PBMC obtained from a single donor stimulated with PMA (10ng/ml) and ionomycin (250ng/ml) (Sigma). The coefficient of variation (CV) for the assay was 15% (n=50). For mAb blocking experiments, target cells were pre-incubated with either the blocking mAb or an isotype matched mAb (10µg/ml) for 30 min at room temperature.
Flow cytometry-based cell mediated cytotoxicity (CMC) assay
Tumor cell lines, disaggregated xenografts, and lesions (5 × 105 cells) and HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells (2.5:1 E/T cell ratio) were incubated for 4h at 37°C, centrifuged, trypsinized, washed, incubated with ALDEFLUOR®, and analyzed for ALDH+ and ALDHbright cells by flow cytometry. For mAb blocking experiments, target cells were pre-incubated with mAb (10µg/ml) for 30 min at room temperature.
Adoptive therapy with HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells
Three distinct types of adoptive therapy experiments involving xenograft-bearing immunodeficient CB17scid female mice (Taconic Farms) and HLA-A2-restricted, ALDH1A1-specific CD8+ T cells were performed in this study. In a fixed end point experiment, CB17scid mice (N=15) were challenged with surgically implanted 5 mm pieces of a serial passage PCI-13 - derived xenograft. Seven days later, the mice were randomized into 3 groups of 5 mice each with equivalent tumor burden and adoptive therapy initiated biweekly by iv injection with HLA-A2 restricted, ALDH1A1-specific CD8+ T cells, irrelevant HIVgag362–370-specific CD8+ T cells (2×106/ mouse) or left untreated. Tumor volumes (mm3) were calculated using the formula: (a × b2)/2, where b is the smaller of the two diameter measurements (25). The experiment was terminated ~21 days later, the mice euthanized and xenografts removed for analyses.
In the second model, experimental lung metastases of MDA-MB-231 cells were established in CB17scid mice (N=9) following iv injection of 1×106 cells to each mouse. On day 3, mice were randomly divided into 3 groups of 3 mice each. Group 1 was injected i.v. with HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells (2×106/ mouse) twice per week for 5 weeks, then once per week for 4 weeks; Group 2 was injected i.v. with irrelevant HIVgag362–370-specific CD8+ T cells (2×106/mouse) using the same schedule; and Group 3 received no CD8+ T cells. All three groups received PEG-IL2 (equivalent of 6.6×104 I.U. /mouse) by ip injection twice on the day CD8+ T cells were administered (26). All the mice were euthanized on day 56 and their lungs harvested and fixed in 10% formalin for further analysis.
The third model employed was a post surgery and metastases survival model with a survival end point. Immunodeficient mice (N=27) were challenged in the mammary fat pad with 1 × 106 MDA-MB-231 cells and 30 days later, when the tumors were, on average, 0.8cm in diameter, the xenografts were surgically removed. The mice were randomized into 3 groups of 9 mice each, two of which received weekly i.v. injections of either HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells or irrelevant HIVgag362–370 peptide-specific CD8+ T cells (2×106/ mouse). All three groups of mice received PEG-IL2 (equivalent of 6.6×104 I.U. /mouse) by ip injection twice on the day CD8+ T cells were administered.
Immunohistochemistry (IHC) analyses
A liver tissue microarray (TMA cat. no. BN03011, Biomax USA) was stained and analyzed for HLA class I Ag and ALDH1A1 expression in normal liver hepatocytes. The HC-10 and HC-A2 mAb were used to stain for HLA class I Ag and ALDH1A1 was detected with the rabbit anti-ALDH1A1 mAb using standard procedures.
Tumor areas in experimentally induced pulmonary metastases were analyzed using 4 µm thick formalin fixed paraffin embedded sections (FFPS) of lung tissues stained with 0.5% alcoholic solution of Hematoxylin (Sigma-Aldrich, Inc.). Photos were taken using a Nikon Eclipse E800Microscope and the areas of tumor nodules in five randomly selected fields per section (magnification ×200) were measured and calculated by the SPOT Advanced Imaging software (Diagnostic Instruments, Inc.). For analysis of proliferative and apoptotic cells in untreated and treated xenografts, FFPS of xenografts were stained for Histone3 phosphoserine10+ and TUNEL+ cells and analyzed using standard procedures. A minimum of five sections from each xenograft was stained and five microscopic fields per section were counted manually in a double-blinded fashion by board certified pathologists.
Statistical Methods
The two-tailed Student’s t test was performed to interpret the differences between experimental groups. Kaplan-Meier analysis was used to calculate significance of median survival in the adoptive therapy in the post surgery and metastases xenograft experiment.
Results
ALDHbright cells present in established human carcinoma cell lines
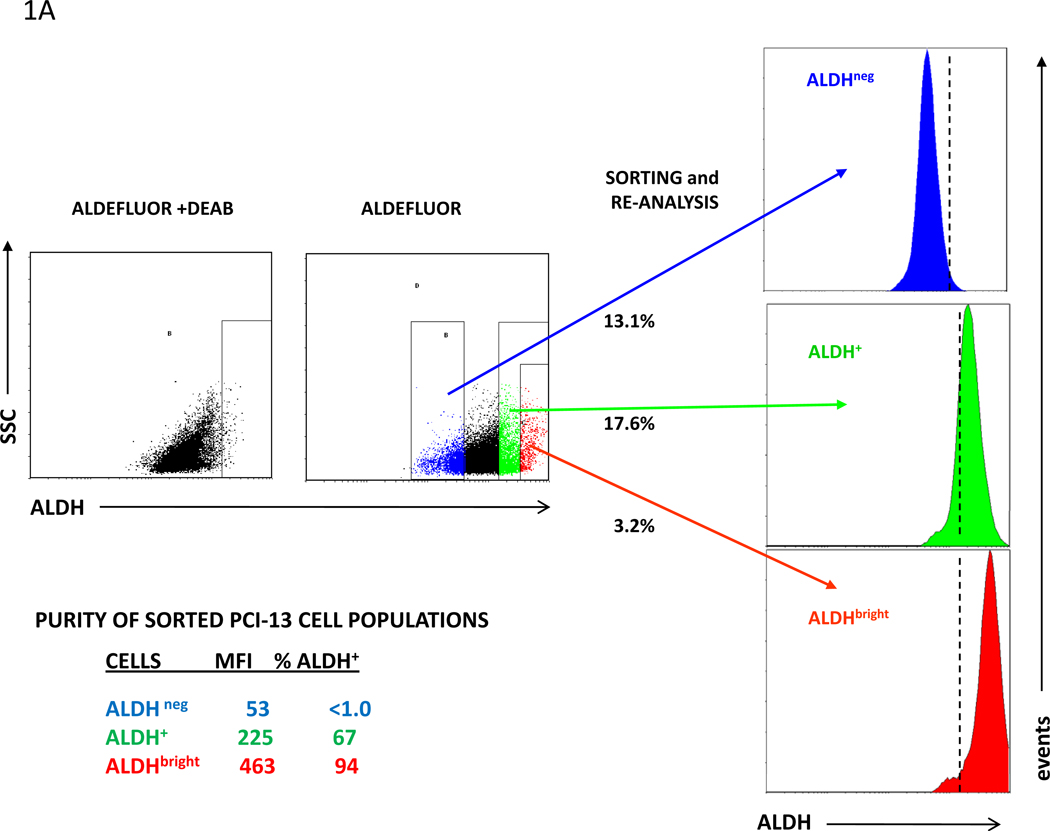
Using flow cytometry and the ALDEFLUOR® reagent, established human tumor cell lines and digests of tumor xenografts or surgically removed lesions were analyzed for their ALDH+ cell content. Sorting by flow cytometry was used to isolate the ALDHbright cell population from the cell lines. The ALDHbright cells identified in these samples had an ALDH MFI twice that of the bulk ALDH+ cell population. A representative flow cytometry analysis of the human squamous cell carcinoma of the head and neck (SCCHN) cell line, PCI-13, to identify ALDH+ and ALDHbright cells in the cell line and set the parameters for sorting ALDHneg, ALDH+ and ALDHbright cells together with the reanalysis of the sorted populations for ALDH+ cells and ALDH MFI are shown in Fig. 1A. Whereas sorted ALDH+ PCI-13 cells were found to contain only 67% ALDH+ cells, the sorted ALDHbright cell population had a purity of 94%. The sorted ALDHneg cells contained <1% ALDH+ cells and no ALDHbright cells.
Figure 1. ALDH+ and ALDHbright cells present in human carcinoma cell lines, cell line-derived xenografts and a surgically removed SCCHN lesion.
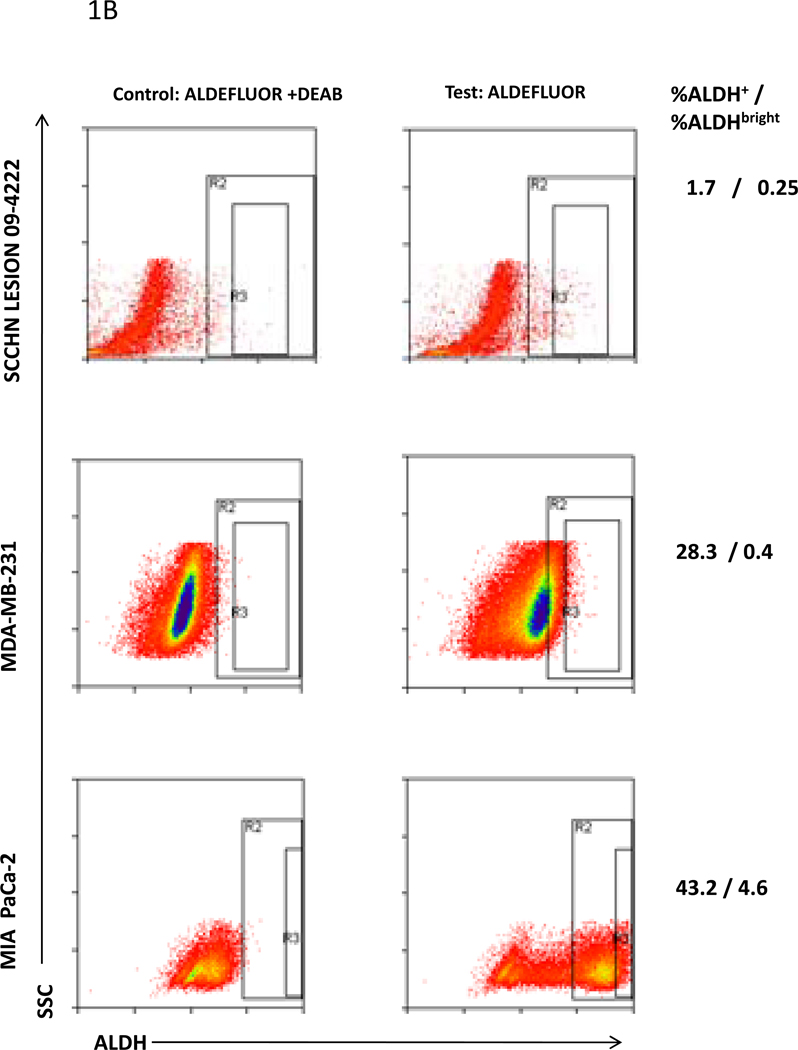
ALDH+ and ALDHbright were identified by flow cytometry analysis following incubation of cells with ALDEFLUOR® in the presence or absence of DEAB inhibitor. A, Purity of sorted ALDH+ and ALDHbright cells PCI-13 cells for ALDH+ cells: Percentage ALDH+ cells and ALDH MFI of each re-analzyed sorted population are indicated. The dotted lines in the re-analysis of the sorted populations indicate the gates set for identifying ALDH+ cells in the sample using ALDEFUOR + DEAB. B, ALDH+ and ALDHbright cells in the SCCHN PCI-13 cell line, a digest of a surgically removed SCCHN lesion, and MDA-MB-231 breast carcinoma and MIA PaCa-2 pancreatic carcinoma cell lines. The percentages of ALDH+ and ALDHbright cells are listed.
A panel of established human breast, pancreatic and SCCHN cell lines and digests of disaggregated surgically removed pancreatic and SCCHN lesions were then analyzed by flow cytometry to identify their ALDH+ and ALDHbright cell content. Representative analyses are shown in Fig. 1B, and the results are listed in S Table 1. In summary, the data indicate that the percentages of ALDH+ and ALDHbright cells varied with each sample regardless of its tumor type or source. High percentages of ALDH+ cells in a sample did not automatically correlate with high percentages of ALDHbright cells but, on average, the ALDHbright cell content was about 10% of the ALDH+ cell content. The ALDHbright cell content ranged from a low of 0.02% in the SCCHN PCI-30 cell line to a high of 4.6% in the MIA PaCa-2 pancreatic carcinoma cell line-derived xenograft. The percentages of ALDHbright cells in the MDA-MB-231 and MIA PaCa-2-derived xenografts were higher than in the parental cell lines, whereas the ALDHbright cell content of the PCI-13-derived xenograft was lower than that of the parental cell line. Additional passages of the xenografts did not result in significant changes in their ALDHbright cell content (data not shown).
Tumorigenicity of ALDHbright cells sorted from human carcinoma cell lines
To confirm that the ALDHbright cell population was highly tumorigenic, a critical characteristic of CIC, ALDHbright cells sorted from the PCI-13, MIA PaCa-2 and MDA-MB-231-Luc cell lines were tested for their tumorigenicity by challenging groups of 3 or 5 immunodeficient mice each at a dosage of 500 cells. Xenografts were established in 3/3 mice challenged with ALDHbright PCI-13 cells, 2/3 mice challenged with ALDHbright MIA PaCa-2 cells and 4/5 mice challenged with ALDHbright MDA-MB-231-Luc cells within 6 months of challenge (S Fig. 1). The tumorigenicity of the ALDHbright MDA-MB-231-Luc cells was monitored by BLI. None of the ALDHbright cell-derived xenografts can be attributed to the irradiated tumor feeder cells used in the inoculums, since ALDHneg challenges, which also included the same number of irradiated tumor feeder cells, failed to yield xenografts in the same mice.
ALDH1A1 mRNA expression levels in ALDHbright cells
According to its manufacturer, ALDH activity detected by the ALDEFLUOR reagent can be attributed to ALDH1 and ALDH 3 isoforms, with the emphasis on ALDH1 isoforms. Four ALDH1/3 isoforms have been identified, ALDH1A1,-1A2 and -1A3 and ALDH3A1. A qRT/PCR analysis of the levels of expression of these four isoform mRNA in bulk PCI-13 cells indicated predominate expression of ALDH1A1 mRNA compared to the other three isoform mRNA. Little to no ALDH1A2 mRNA was expressed and the level of ALDH1A1 mRNA was ~50X greater than that of ALDH1A3 and ALDH3A1 mRNA, a finding consistent with ALDH1A1 contributing to the ALDH activity detected by ALDEFLUOR. Furthermore, the analysis indicated that ALDHbright cells expressed ~8X fold higher level of ALDH1A1 RNA than bulk population of tumor cells (48.2±5.6 vs 6.2±0.3). This result correlates well with the nearly 10X higher ALDH MFI of ALDHbright PCI-13 cells compared to that of the ALDHneg PCI-13 cells. In addition, qRT/PCR analysis of the sorted ALDHbright populations indicated that ALDHbright PCI-13, MDA-MB-231 and MIA PaCa-2 cells uniformly express higher levels of ALDH1A1 mRNA than ALDH1A3 mRNA (S Table 2). Essentially no ALDH1A2 mRNA was detected in these cells (data not shown).
Detection of HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells in the peripheral circulation of subjects with SCCHN
We previously identified ALDH1A1 as a tumor associated antigen defined by HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells based on the ability of the ALDH1A188–96 peptide to stimulate the in vitro induction/generation of these effector cells from PBMC obtained from most normal donors, as well as a subject with SCCHN (14). In vivo, the immunogenicity of ALDH1A188–96 epitope was confirmed in a limited HLA-A2/ALDH1A188–96 peptide tetramer-based flow cytometry analysis of peripheral blood mononuclear cells (PBMC) obtained from HLA-A2+ subjects with SCCHN and normal donors. Representative results of this analysis are shown in S Fig. 2. Based on a cutoff frequency of 1/8,000 determined with PBMC obtained from HLA-A2neg normal donors, the frequency of tetramer+ cells detected in PBMC of HLA-A2+ normal donors was comparable to that of negative controls. In contrast, relatively high frequencies of tetramer+ cells in the range of 1/500 to 1/2,000 were detected in the peripheral circulation of subjects with SCCHN. The CCR7/CD45RA phenotypes of the tetramer+ CD8+ T cells varied with each subject; they had a predominately memory and terminally differentiated phenotype in subject #1, a predominately naïve phenotype in subject #2, and a mixture of naïve and terminally differentiated phenotypes in subject #3.
In vitro recognition of ALDHbright cells by HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells
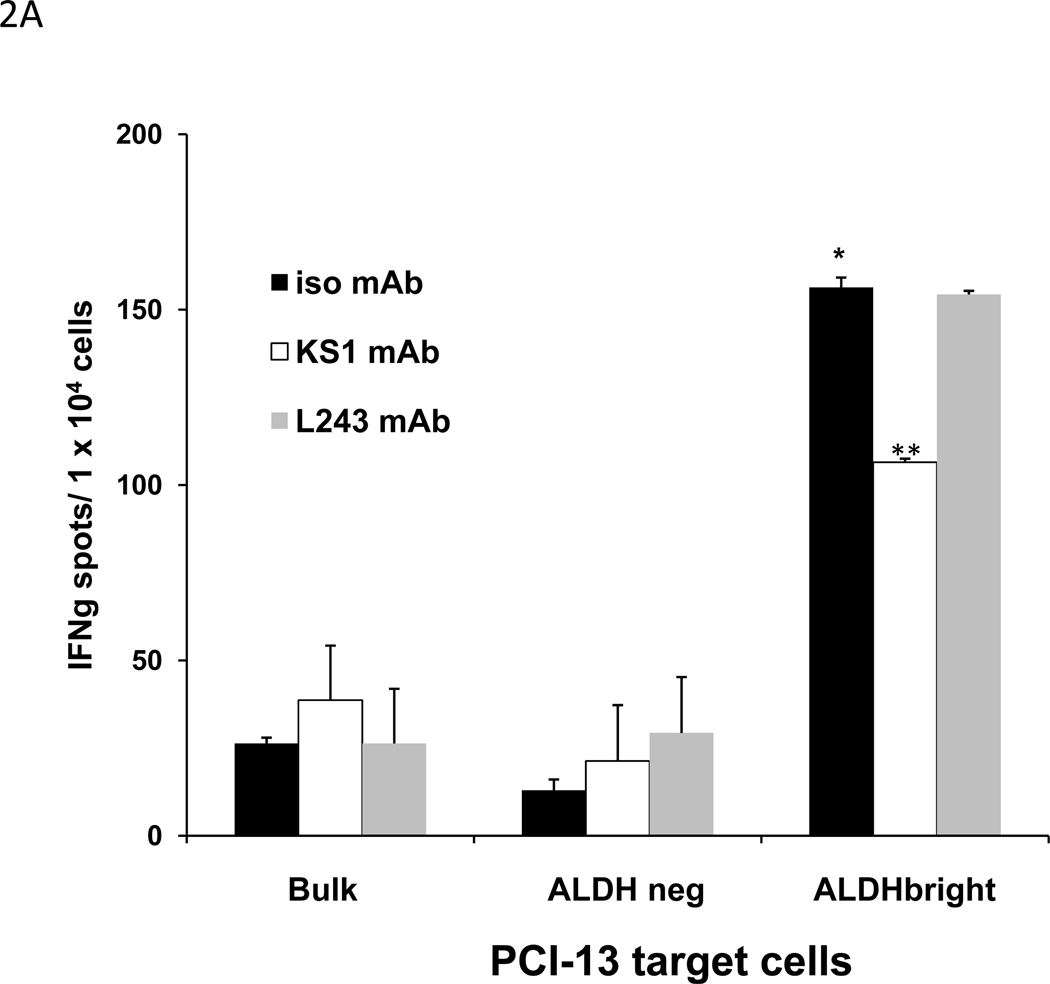
The specificity of the HLA-A2 restricted, ALDH1A1-specific CD8+ T cells used in this study for the ALDH1A188–96 peptide in ELISPOT IFNγ assays is shown in S Fig 3A. These effectors recognize ALDHbright target cells but not bulk cell population or ALDHneg target cells sorted from the HLA-A2+ PCI-13 SCCHN cell line (Fig. 2A). Recognition of the ALDHbright cells by the ALDH1A188–96 peptide-specific CD8+ T cells was blocked by the HLA-A2,-28 specific mAb KS1, but was not affected by the HLA-DR-specific mAb L243. In an HLA-A2-restricted manner, these effector T cells also recognize ALDHbright target cells but neither bulk cell population nor ALDHneg target cells sorted from the HLA-A2+ basal breast carcinoma MDA-MB-231 and pancreatic carcinoma MIA PaCa-2 cell lines, as well. (S Fig. 3B, C).
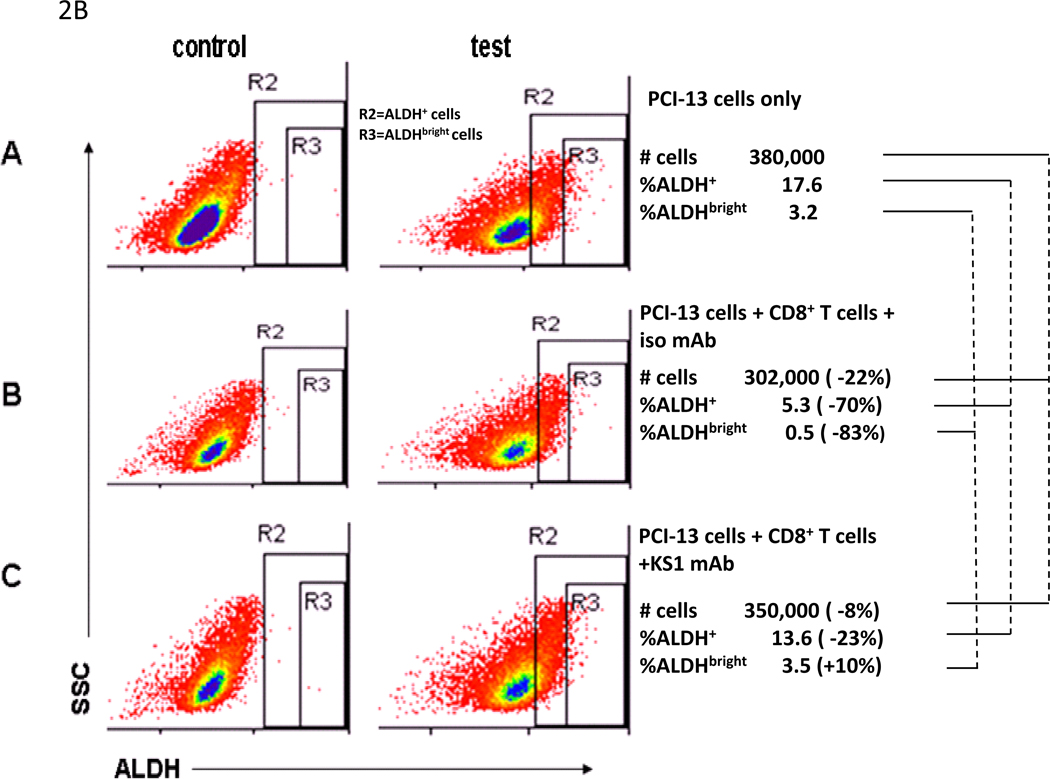
Figure 2. In vitro recognition of ALDHbright cells sorted from HLA-A2+ PCI-13 human carcinoma cell line by HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells in ELISPOT IFNγ and flow cytometry-based CMC assays.
A, recognition of sorted ALDHbright PCI-13 cells but not sorted ALDHneg or bulk PCI-13 cells by HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells in ELISPOT IFNγ assays. The bulk populations of tumor cells used as targets were cells that had been incubated with ALDEFLUOR and collected without gating. Recognition of ALDHbright cells blocked by HLA-A2, A28-specific KS1 mAb, but not affected by HLA-DR specific L243 mAb. Assays performed at E/T ratio of 2:1. Asterisk (*) indicates significant recognition (p<0.05) of ALDHbright cells relative to bulk or ALDHneg cells. Asterisks (**) indicates significant inhibition of recognition (p<0.05) of ALDHbright cells in the presence of KS1 mAb. B, flow cytometry analyses of ALDH+ and ALDHbright cells present in SCCHN PCI-13 (5 × 105 cells) following incubation with HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells at an E/T cell ratio of 2.5:1. Lysis was blocked by KS1 mAb. The decreases in the identified number of cells, and percentages ALDH+ and ALDHbright PCI-13 cells incubated alone or with HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells in the presence of isotype mAb or KS1 mAb are indicated.
In vitro recognition by HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells of ALDH+/ALDHbright cells present in the established human carcinoma cell lines was also measured in a CMC assay using flow cytometry. As indicated in Fig. 2B, 83% and 70% decreases in the percentages of ALDHbright and ALDH+ PCI-13 cells were observed following incubation of the tumor cells with the effectors, which can be attributed to the differential levels of ALDH1A1 expression in these cells. Recognition of ALDHbright/ALDH+ cells was blocked by the HLA-A2, -28 Ag-specific mAb KS1. Importantly, comparable results also were obtained using cells derived from in vivo-propagated tumor cells, namely, a PCI-13-derived xenograft and a HLA-A2+ surgically removed SCCHN lesion (Table 1). These results further confirm that HLA-A2+ ALDHbright tumor cells are recognized by HLA-A2 restricted, ALDH1A188–96 peptide-specific CD8+ T cells.
Table 1.
In vitro recognition of ALDH+ and ALDHbright cells present in human carcinoma cell lines, xenograft and surgically removed lesion by HLA-A2-restricted, ALDH1A188–96 peptide- CD8+ T cells in flow cytometry-based CMC assaysa.
| Cells | Cells only | Cells + ALDH1A1-specific CD8+ T cell + isotype mAb |
Cells + ALDH1A1-specific CD8+ T cells + KS1 mAb |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell number |
% ALDH+ |
% ALDHbright |
Cell number |
% ALDH+ |
% ALDHbright |
Cell number |
% ALDH+ |
% ALDHbright |
|
| PCI-13 | 380,000 | 17.6 | 3.2 | 302,000 (−22%) |
5.3 (−70%) |
0.5 (−83%) |
350,000 (−8%) |
13.6 (−23%) |
3.5 (+10%) |
| MDA-MB-231 | 204,000 | 28.3 | 0.4 | 188,000 (−8%) |
14.2 (−50%) |
0.3 (−75%) |
200,000 (−2%) |
31.1 (+11%) |
0.3 (−25%) |
| MIA PaCa-2 | 365,000 | 43.2 | 4.6 | 194,000 (−47%) |
37.2 (−14%) |
1.8 (−60%) |
299,000 (−18%) |
40 (−7%) |
3.8 (−18%) |
| PCI-13 xenograft | 350,000 | 13.1 | 2.1 | 213,000 (−29%) |
3.5 (−73%) |
0.6 (−72%) |
298,000 (−15%) |
13 (−5%) |
7-Jan (−22%) |
| SCCHN lesion 084124 | 150,000 | 3.4 | 0.9 | 100,000 (−33%) |
0.9 (−74%) |
0.1 (−89%) |
NDb | ND | ND |
Flow cytometry analyses of ALDH+ and ALDHbright cells present in SCCHN PCI-13, breast carcinoma MDA-MB-231 and pancreatic carcinoma MIA PaCa-2 cells, and digests of a PCI-13-derived xenograft and a SCCHN lesion following incubation with HLA-A2-restricted, ALDH1A188–96 peptide--specific CD8+ T cells at an E/T cell ratio of 2.5:1, followed by flow cytometry analysis with ALDEFLUOR±DEAB. 5×10 target cells were used with the exception of the analysis of the SCCHN lesion. Lysis was blocked by HLA-A2, A28-specific KS1 mAb. The percentages of ALDH+ and ALDHbright cells in each sample following incubation with HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells in the presence of isotype mAb or KS1 mAb are indicated. The decreases in these values compared to the “Cells only” control are indicated in parentheses
ND indicates not done due to insufficient cells
Adoptive immunotherapy of tumor-bearing immunodeficient mice with ALDH1A1 peptide-specific CD8+ T cells
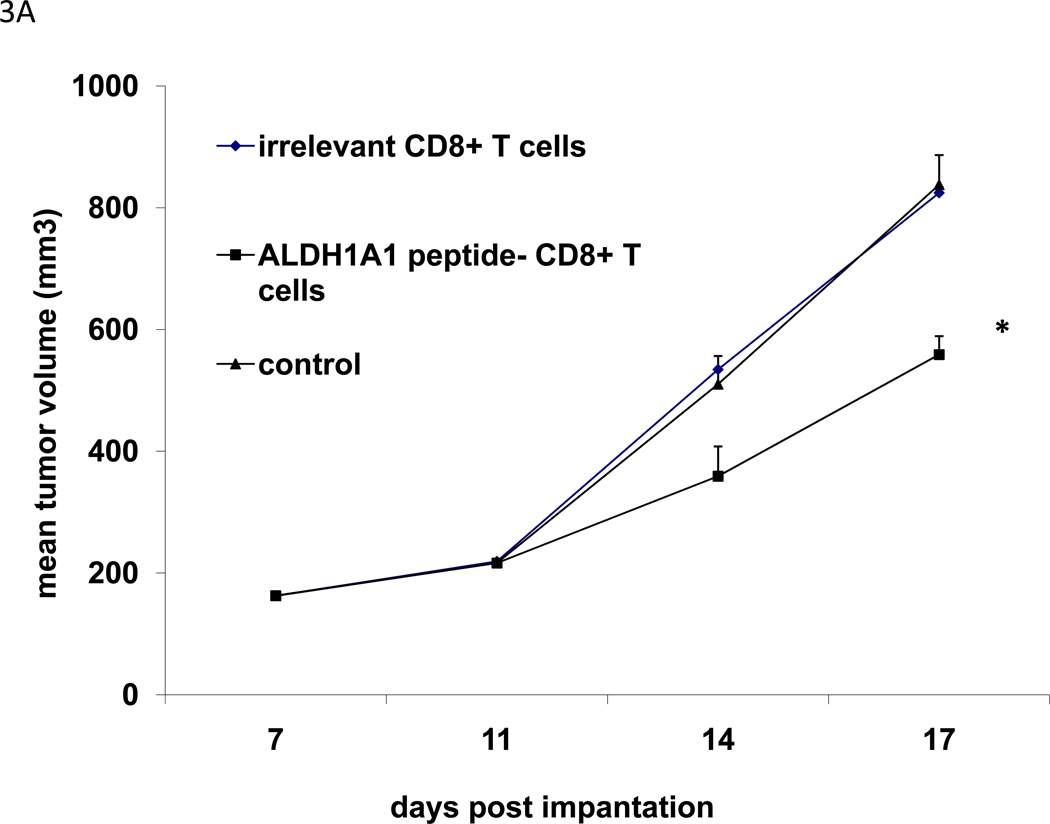
The efficacy of adoptive therapy with ALDH1A188–96 peptide-specific CD8+ T cells was evaluated in immunodeficient mice bearing either subcutaneous xenografts derived from SCCHN PCI-13 cells or experimental or spontaneous pulmonary metastases derived from basal breast carcinoma MDA-MB-231 cells. In a fixed time-point experiment involving immunodeficient mice bearing subcutaneous PCI-13-derived xenografts, adoptive therapy with ALDH1A1-specific CTL was administered i.v. The experiment was terminated on or about day 20 post-implantation in order to obtain sufficient residual xenograft specimens for subsequent analyses of their ALDHbright cell content as well as proliferation and apoptotic indices, measured by staining for histoneH3 phosphoserine10+ cells (27, 28) and TUNEL+ cells, respectively. The results indicate that treatment of the xenografts with ALDH1A188–96 peptide-specific CD8+ T cells, but not with irrelevant CD8+ T cells significantly inhibited their growth (Fig. 3A). This inhibition was concordant with significant decreases in their ALDHbright cell content and proliferative index, but a significant increase in the apoptotic index compared to xenografts obtained from the control groups of mice (Table 2 and S Fig. 4).
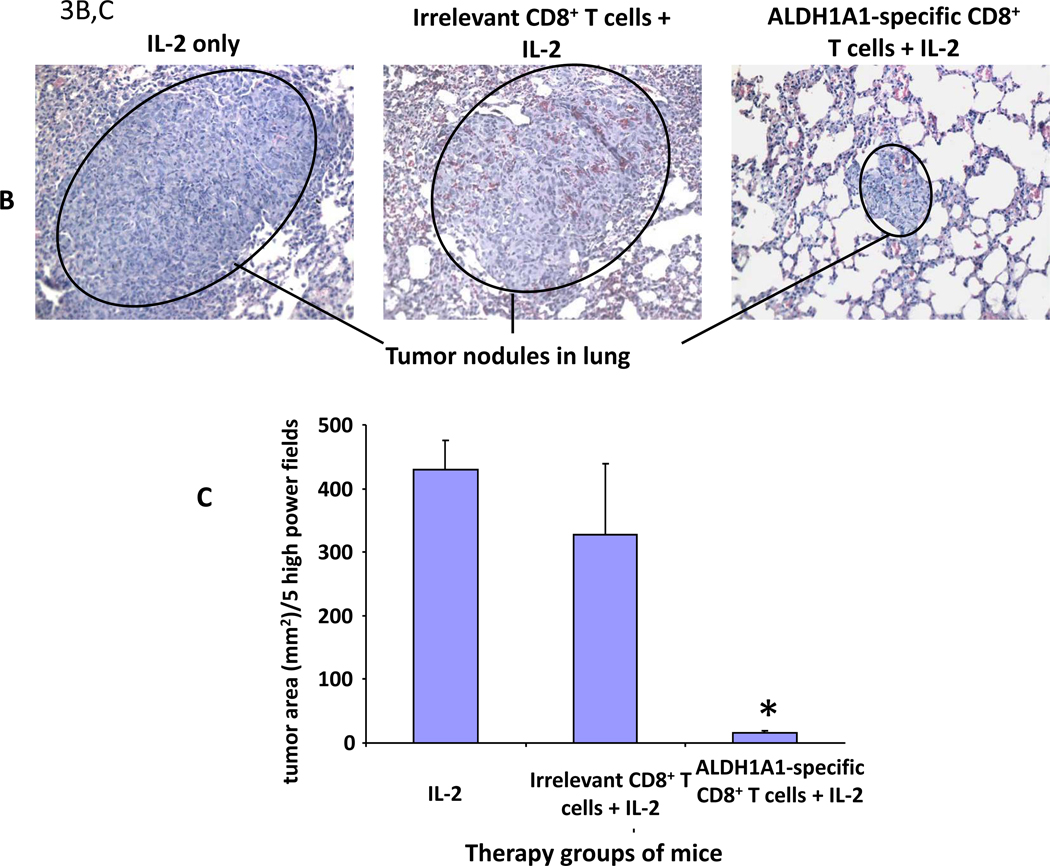
Figure 3. Control of xenograft growth in immunodeficient mice by adoptive therapy with ALDH1A1-specific CD8+ T cells.
Adoptive therapy of CB17scid mice bearing xenografts derived from PCI-13 or MDA-MB-231 cells by i.v. injection of HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells. A, adoptive therapy of mice bearing 7-day subcutaneously growing PCI-13-derived xenografts by i.v. injection of ALDH1A1-specific CD8 + T cells. Mean tumor volume (mm3 ± SD) of PCI-13-derived xenografts in each group of mice of 3 mice each on the indicated days is shown. Asterisk (*) indicates p=0.0001 relative to untreated control. B,C: adoptive therapy of mice bearing experimentally induced pulmonary metastases derived from the breast carcinoma MDA-MB-231 cell line: B, tumor lesions in representative FFPS of lung tissue of indicated groups of treated mice bearing experimentally induced pulmonary metastases derived from the breast carcinoma MDA-MB-231 cell line. Note differences in lesion sizes. C, total tumor area (mm2) ± SD of lesions in lungs of the indicated groups of mice. Asterisk (*) indicates p<0.001 relative to mice treated with IL-2 only.
Table 2.
Effects of adoptive transfer of ALDH1A188–96 peptide-specific CD8+ T cells on PCI-13-derived xenografts in immunodeficient micea.
| Group | Δ MTV±SDb | % ALDHbright cellsc |
Proliferation Indexd |
Apoptotic Indexe |
|---|---|---|---|---|
| Control | 676±45 | 1.3±0.8 | 172±18 | 22±7 |
| Irrelevant CD8+ T cells | 662±11 | 1.8±0.5 | 176±33 | 25±7 |
| ALDH1A188–96 peptide-specific CD8+ T cells | 398±33 p = 8×10−7 |
0.4±0.3 p = 0.009 |
90±25 p = 4×10−5 |
41±9 p = 0.01 |
See Material and Methods sections for protocol used. Two-tailed student’s t test based on values of untreated control groups of mice was used to determine significance.
The difference (Δ) in mean volume (mm3) of each tumor in a mice on days 7 and 20 was determined and the values expressed as mean tumor volume (MTV) ±SD for each group.
Percentages based on gated events.
Mean ± SD of histoneH3 phosphoserine10+ cells per tumor as analyzed by IHC as detailed in Materials and Methods section.
Mean ± SD of TUNEL+ cells per tumor as analyzed by IHC as detailed in Materials and Methods section.
MDA-MB-231 cells readily form pulmonary metastases following iv injection or spontaneously following surgical removal of primary orthotopic xenografts. Adoptive therapy of mice bearing experimentally induced MDA-MB-231 pulmonary metastases by systemic administration of ALDH1A1-specific CD8+ T cells resulted in fewer and smaller tumor nodules with a significantly reduced total tumor area in the lungs of mice compared to that of the control groups of mice (irrelevant CTL + IL-2) and (IL-2) (Fig. 3B). The total tumor area in the lungs of mice was quantified to determine the efficacy of the treatment in this experiment, because the metastatic lesions in the lungs of the control groups of mice had grown so extensively, they fused to form large tumor masses and were not individually discernible (Fig. 3C) (29).
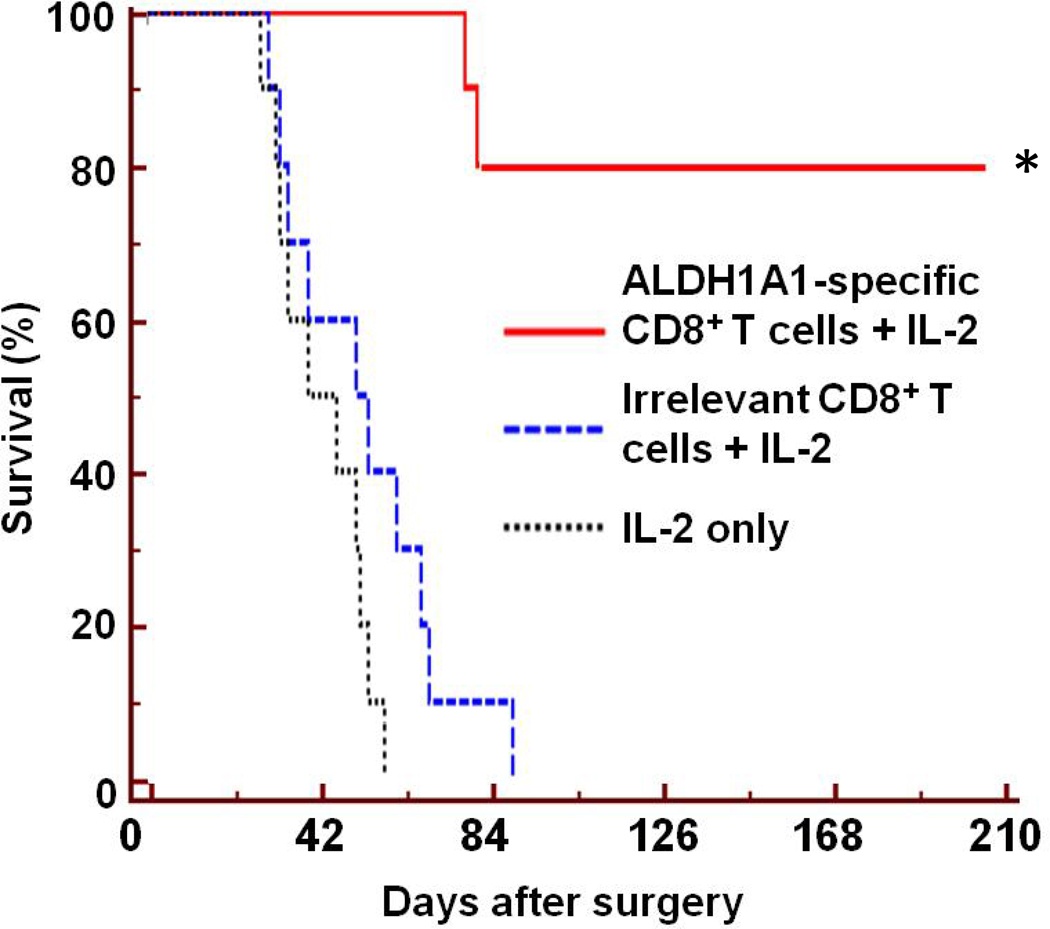
In the clinically relevant, post-surgery and metastasis survival model, mice succumb primarily to lung metastases following surgical removal of the primary MDA-MB-231-derived orthotopic xenograft. Groups of immunodeficient mice were treated with ALDH1A1-specific CD8+ T cells + IL-2, irrelevant CD8+ T cells + IL-2 or IL-2 only following their surgery. Only adoptive therapy with ALDH1A1-specific CTL significantly prolonged their survival (p<0.001) compared to the control groups of mice, as shown in Fig. 4. Whereas all mice in the two control groups died from lung metastases by day 87 post surgery, 80% of the ALDH1A1-specific CD8+ T cells -treated mice exhibited no signs of disease at day 210 post-surgery. The results of these in vivo human tumor xenograft experiments demonstrate the efficacy of adoptive therapy with ALDH1A188–96 peptide-specific CD8+ T cells to effectively target ALDHbright cells and control tumor growth and metastases.
Figure 4. Adoptive therapy with ALDH1A1-specific CD8+ T cells of immunodeficient mice following surgical removal of primary MDA-MB-231 orthotopic xenograft.
HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells were administered iv. to groups of 9 mice each following surgical removal of their primary MDA-MB-231 orthotopic xenografts. Survival in each group of mice relative to time of surgery is shown. Asterisk (*) indicates significant survival (p<0.0001) of group of mice treated with ALDH1A1-specific CD8+ T cells and IL-2 relative to groups of mice treated with irrelevant CD8+ T cells and IL-2 or IL-2 only.
Immunohistochemical analysis of ALDH1A1 and HLA class I Ag expression in normal liver hepatocytes
In view of potential clinical application of these results, we sought to address the question of whether ALDH1A1-based immunotherapy could target normal liver hepatocytes, which are reported to express a high level of ALDH1A1, and cause deleterious side effects (15). Therefore, we tested for the expression of HLA class I Ag and ALDH1A1 at the protein level in normal liver tissue by immunohistochemical staining with mAb. A liver TMA comprised of 63 cores derived from 3 non-diseased and 16 diseased livers (e.g. cirrhosis, fatty degeneration), 3 hepatocelluar carcinomas and an abnormal spleen was analyzed: 23 cores were considered to be normal liver tissue, 9 of which came from 3 non-diseased livers. Only 1 of these 9 cores stained for HLA class I Ag; it showed weakly patchy staining. The remaining 14 “normal liver tissue” cores came from non-diseased regions of diseased livers; 2 showed strong cytoplasmic HLA class I Ag but weak ALDH1A1 expression, while 2 others showed HLA class I Ag membrane staining but no ALDH1A1 expression (see S Table 3 and S Fig. 5). In contrast, HLA class I Ag expression was prevalent in multiple cores of diseased tissue taken from diseased livers.
Discussion
The results of this translational pre-clinical study demonstrate that a subset of tumor cells in human carcinomas identified as ALDHbright cells are recognized and eliminated in vitro and in vivo by HLA-A2-restricted, ALDH1A188–96 peptide-specific CD8+ T cells. In human tumor xenograft models, we have demonstrated that adoptive transfer of ALDH1A1-specific CD8+ T cells inhibited growth of subcutaneously growing xenografts and experimental-induced lung metastases. In addition, following surgery to remove a primary tumor, this therapy inhibited spontaneous metastases and prolonged survival of mice.
It was recently reported that the ALDH activity in breast cancer stem cells detected by ALDEFLUOR is primarily due to ALDH1A3 expression rather than ALDH1A1 (30). In our study, however, we have shown that sorted ALDHbright cells express higher levels of ALDH1A1 mRNA than ALDH1A3 mRNA. Nonetheless, as previously reported (14), the ALDH1A-specific CD8+ T cells used in this study recognize the ALDH1A188–96 peptide (LLYKLADLI), but not the highly related peptides derived from the ALDH1A2 (LLDKLADLV) and ALDH1A3 (LLHQLADLV) isoforms. Therefore, regardless of which ALDH1 isoform is prevalently expressed, the recognition of ALDHbright cells by the ALDH1A 88–96 peptide-specific CD8+ T cells used in this study is independent of ALDH1A3 expression.
Even though ALDH1A1 is expressed by many cell types, it is highly unlikely that ALDH1A1-based immunotherapy would induce toxicity. Normal stem cells, such as hematopoietic stem cells, which express ALDH1A1 but at a lower level than detected in tumors, have been shown not to be recognized by ALDH1A1-specific CD8+ T cells (14). Furthermore, although ALDH1A1 is expressed by normal hepatocytes, in agreement with the information in the literature, we have shown that these cells express little to no HLA class I antigen (Ag) on their cell surface; as a result, normal hepatocytes are highly unlikely to be recognized by HLA-class I restricted, ALDH1A1-specific CD8+ T cells (31–33).
Presently, there is little information about the recognition of CIC by HLA class I restricted, CD8+ T cell effectors. To the best of our knowledge, only three studies have investigated this subject; two involve gliobastoma multiforme (GBM) stem cells isolated using selective culture conditions and the third involves sorted colon cancer stem cells identified as a side-staining population. Utilizing a non-tumor related, cytomegalovirus (CMV) antigen as a model TA, Brown et al. (34) demonstrated recognition of CMV-transfected GBM stem cells by CMV pp65 peptide-specific CTL. Recognition of the targets, however, required targets pulsed with exogenous CMV pp65 peptide. This finding suggests that GBM stem cells expressed HLA class I Ag, but required the exogenous peptide to form a sufficient level of HLA class I Ag-peptide complexes for recognition by the cognate CTL. DiTommaso et al (35) detected defects in HLA class I Ag and APM component expression in the cultured population of GBM stem cells. As a result, recognition of these target cells by autologous anti-tumor CTL required pretreatment with IFNγ to upregulate HLA class I Ag expression and, presumably, HLA class I Ag/TA peptide complexes, a common situation observed in targeting tumor cells with HLA-class I restricted, TA peptide-specific T cell effectors (36,37). In the third study, Inoda et al. (38) showed that colon carcinoma stem cells are sensitive in vitro and in vivo to HLA class I-restricted CTL recognizing an epitope derived from the tumor associated centrosomal protein 55kDa protein, CEP55, which is expressed by the tumor initiating cells as well as the bulk population of cells in the colon carcinoma cell lines studied. Since ALDH1A1 is expressed by CIC present in colon carcinomas and gliomas (10, 39), targeting CIC populations in these tumors with ALDH1A1-specific CD8+ T cells is also possible and should be more selective.
Our results strongly support further development of strategies that would incorporate ALDH1A1-based immunotherapy to target CIC. The constraints of a practical evaluation of a T cell-based immunotherapy using human xenograft mouse models required adoptive transfer of the immune effector cells. Using recombinant DNA or optimized traditional protocols, sufficient numbers of TA-specific T cells can be generated in vitro for adoptive T-cell based immunotherapy; this strategy has been shown in recent years to yield beneficial clinical responses in subjects with cancer (40, 41). Nonetheless, the development and implantation of ALDH1A1-based immunotherapy need not preclude a vaccine-based approach.
In accordance with the cancer stem cell theory, the elimination CIC should be the critical criteria used to define the efficacy of a therapy, rather than only reduction in tumor volume. Our research demonstrates for the first time the potential ability of an immunotherapy to achieve the objective of targeting CIC in tumors. However, while we are aware that therapeutic protocols can promote tumor escape, our findings highlight the benefit that T cell-based immunotherapy offers, which combined other independent therapeutic modalities, such as tumor antigen- specific mAb and/or inhibitors of aberrantly regulated stem cell signaling pathways (13), would minimize the potential of tumor escape.
Translational Relevance.
Tumor cells expressing high levels of aldehyde dehydrogenase (ALDH) have been identified by flow cytometry as ALDHbright cells and shown to have the properties attributed to cancer initiating cells (CIC). CIC are resistant to conventional cancer treatments and considered responsible for recurrence and metastasis. Pertinent to developing immunotherapy for targeting CIC, these cells express ALDH1A1, a tumor associated antigen recognized by HLA class I restricted, CD8+ T cells, which can be induced/generated in vitro and are present in human subjects with cancer.
This study demonstrates that ALDHbright cells are sensitive to cytolysis by ALDH1A1-specific CTL in vitro. In preclinical models of human tumor xenografts growing in immunodeficient mice, adoptive therapy with ALDH1A1-specific CD8+ T cells was shown to target ALDHbright cells and inhibit xenograft growth, metastases or prolong survival. Our results demonstrate the usefulness of ALDH1A1 as a target of T cell-based immunotherapy to eliminate CIC in tumors.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by National Cancer Institute at the National Institutes of Health Grants DE12321 and CA109688 (C. Visus, A.B. DeLeo, T.L. Whiteside), P50 CA097190 (C. Visus, A.B. DeLeo, R.L. Ferris) and CA138188 (Y. Wang, S. Ferrone), the Hillman Foundation (A. B. DeLeo), Hirshberg Foundation for Pancreatic Cancer Research (C. Visus), DOD Concept Award BC085485 (C. Visus, X. Wang, A. B. DeLeo), the Elsa U. Pardee Foundation (X. Wang), RO3 CA141086 (C. R. Ferrone, X. Wang) and the Pennsylvania Department of Health, (A.B. DeLeo, S. Ferrone) which specifically disclaims responsibility for any analyses, interpretations or conclusions detailed in this report. M.J.S. is on leave from the Departments of Clinical Immunology and Otolaryngology, Poznan University of Medical Sciences, Pozan, Poland.
References
- 1.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 2.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 3.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Visvader JE. Cells of origin in cancer. Nature. 2011;469:314–322. doi: 10.1038/nature09781. [DOI] [PubMed] [Google Scholar]
- 6.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charafe-Jauffret E, Ginestier C, Iovino F, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69:1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasheed ZA, Yang J, Wang Q, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102:340–351. doi: 10.1093/jnci/djp535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boonyaratanakornkit JB, Yue L, Strachan LR, et al. Selection of Tumorigenic Melanoma Cells Using ALDH. J Invest Dermatol. 2010;130:2799–2808. doi: 10.1038/jid.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009 Apr 15;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen YC, Chen YW, Hsu HS, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385:307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- 12.Clay MR, Tabor M, Owen JH, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32:1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–4012. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visus C, Ito D, Amoscato A, et al. Identification of human aldehyde dehydrogenase 1 family member A1 as a novel CD8+ T-cell-defined tumor antigen in squamous cell carcinoma of the head and neck. Cancer Res. 2007;67:10538–10545. doi: 10.1158/0008-5472.CAN-07-1346. [DOI] [PubMed] [Google Scholar]
- 15.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008 Jun;4(6):697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Snyder KM, Suhoski MM, et al. 4-1BB is superior to CD28 costimulation for generating CD8+ cytotoxic lymphocytes for adoptive immunotherapy. J Immunol. 2007;179:4910–4918. doi: 10.4049/jimmunol.179.7.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suhoski MM, Golovina TN, Aqui NA, et al. Engineering artificial antigen-presenting cells to express a diverse array of co-stimulatory molecules. Mol Ther. 2007;15:981–988. doi: 10.1038/mt.sj.6300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sluijter BJ, van den Hout MF, Stam AG, et al. 4-1BB-mediated expansion affords superior detection of in vivo primed effector memory CD8(+) T cells from melanoma sentinel lymph nodes. Clin Immunol. 2010;137:221–233. doi: 10.1016/j.clim.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 19.Barnstable CJ, Bodmer WF, Brown G, et al. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 20.Lampson LA, Levy R. Two populations of Ia-like molecules on a human B cell line. J Immunol. 1980;125:293–299. [PubMed] [Google Scholar]
- 21.Tsujisaki M, Sakaguchi K, Igarashi M, Richiardi P, Perosa F, Ferrone S. Fine specificity and idiotype diversity of the murine anti-HLA-A2, A28 monoclonal antibodies CR11-351 and KS1. Transplantation. 1988;45:632–639. doi: 10.1097/00007890-198803000-00026. [DOI] [PubMed] [Google Scholar]
- 22.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
- 23.Stam NJ, Vroom TM, Peters PJ, Pastoors EB, Ploegh HL. HLA-A- and HLA-B-specific monoclonal antibodies reactive with free heavy chains in western blots, in formalin-fixed, paraffin-embedded tissue sections and in cryo-immuno-electron microscopy. Int Immunol. 1990;2:113–125. doi: 10.1093/intimm/2.2.113. [DOI] [PubMed] [Google Scholar]
- 24.Hoffmann TK, Donnenberg AD, Finkelstein S, Donnenberg VS, Friebe-Hoffmann U, Meyer EM, et al. Frequencies of tetramer+ T cells specific for the wild-type sequence p53264-272 peptide in the circulation of patients with head and neck cancer. Cancer Res. 2002;62:3521–3529. [PubMed] [Google Scholar]
- 25.Carlsson G, Fullberg B, Haftstrom L. Estimation of liver tumor volume using different formulas-an experimental study in rats. J. Cancer Res Clin Oncol. 1983;105:20–23. doi: 10.1007/BF00391826. [DOI] [PubMed] [Google Scholar]
- 26.Katre NV, Knauf MJ, Laird WJ. Chemical modification of recombinant interleukin 2 by polyethylene glycol increases its potency in the murine Meth A sarcoma model. Proc Natl Acad Sci U S A. 1987;84:1487–1491. doi: 10.1073/pnas.84.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendzel MJ, Wei Y, Mancini MA, et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- 28.Choi HS, Choi BY, Cho YY, et al. Phosphorylation of histone H3 at serine 10 is indispensable for neoplastic cell transformation. Cancer Res. 2005;65:5818–5827. doi: 10.1158/0008-5472.CAN-05-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, McCarty J, Brufsky A, Chivukula M, Khoury T, Hsu D, Lyerly H, Clay T, Ferrone S. CSPG4 as new target for antibody-based immunotherapy of triple negative breast cancer. J. Natl. Cancer Inst. 2010;102:1496–1512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32–45. doi: 10.1002/stem.563. [DOI] [PubMed] [Google Scholar]
- 31.Natali PG, Bigotti A, Nicotra MR, Viora M, Manfredi D, Ferrone S. Distribution of human Class I (HLA-A,B,C) histocompatibility antigens in normal and malignant tissues of nonlymphoid origin. Cancer Res. 1984;44:4679–4687. [PubMed] [Google Scholar]
- 32.Sung CH, Hu CP, Hsu HC, Ng AK, Chou CK, Ting LP, et al. Expression of class I and class II major histocompatibility antigens on human hepatocellular carcinoma. J Clin Invest. 1989;83:421–429. doi: 10.1172/JCI113900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kurokohchi K, Carrington M, Mann DL, Simonis TB, Alexander-Miller MA, Feinstone SM, et al. Expression of HLA class I molecules and the transporter associated with antigen processing in hepatocellular carcinoma. Hepatology. 1996;23:1181–1188. doi: 10.1002/hep.510230537. [DOI] [PubMed] [Google Scholar]
- 34.Brown CE, Starr R, Martinez C, et al. Recognition and killing of brain tumor stem-like initiating cells by CD8+ cytolytic T cells. Cancer Res. 2009;69:8886–8893. doi: 10.1158/0008-5472.CAN-09-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Tomaso T, Mazzoleni S, Wang E, et al. Immunobiological characterization of cancer stem cells isolated from glioblastoma patients. Clin Cancer Res. 2010;16:800–813. doi: 10.1158/1078-0432.CCR-09-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marincola FM, Jaffee EM, Hicklin DJ, Ferrone S. Escape of human solid tumors from T-cell recognition: molecular mechanisms and functional significance. Adv Immunol. 2000;74:181–273. doi: 10.1016/s0065-2776(08)60911-6. [DOI] [PubMed] [Google Scholar]
- 37.Lopez-Albaitero A, Nayak JV, Ogino T, Machandia A, Gooding W, DeLeo AB, et al. Role of antigen-processing machinery in the in vitro resistance of squamous cell carcinoma of the head and neck cells to recognition by CTL. J Immunol. 2006;176:3402–3409. doi: 10.4049/jimmunol.176.6.3402. [DOI] [PubMed] [Google Scholar]
- 38.Inoda S, Hirohashi Y, Torigoe T, Morita R, Takahashi A, Asanuma H, et al. Cytotoxic T lymphocytes efficiently recognize human colon cancer stem-like cells. Am J Pathol. 2011;178:1805–1813. doi: 10.1016/j.ajpath.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasper M, Schäfer A, Piontek G, Teufel J, Brockhoff G, Ringel F, et al. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro Oncol. 2010;12:1024–1033. doi: 10.1093/neuonc/noq070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.June CH. Adoptive T cell therapy for cancer in the clinic. J Clin Invest. 2007;117:1466–1476. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg SA, Dudley ME. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr Opin Immunol. 2009;21:233–240. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.