Abstract
Bazedoxifene (BZA) is a third-generation selective estrogen receptor modulator (SERM) that has been approved for the prevention and treatment of postmenopausal osteoporosis. It has antitumor activity; however, its mechanism of action remains unclear. In the present study, we characterized the effects of BZA and several other SERMs on the proliferation of hormone-dependent MCF-7 and T47D breast cancer cells and hormone-independent MCF-7:5C and MCF-7:2A cells and examined its mechanism of action in these cells. We found that all of the SERMs inhibited the growth of MCF-7, T47D, and MCF-7:2A cells; however, only BZA and fulvestrant (FUL) inhibited the growth of hormone-independent MCF-7:5C cells. Cell cycle analysis revealed that BZA and FUL induced G1 blockade in MCF-7:5C cells; however, BZA down-regulated cyclin D1, which was constitutively overexpressed in these cells, whereas FUL suppressed cyclin A. Further analysis revealed that small interfering RNA knockdown of cyclin D1 reduced the basal growth of MCF-7:5C cells, and it blocked the ability of BZA to induce G1 arrest in these cells. BZA also down-regulated estrogen receptor-α (ERα) protein by increasing its degradation and suppressing cyclin D1 promoter activity in MCF-7:5C cells. Finally, molecular modeling studies demonstrated that BZA bound to ERα in an orientation similar to raloxifene; however, a number of residues adopted different conformations in the induced-fit docking poses compared with the experimental structure of ERα-raloxifene. Together, these findings indicate that BZA is distinct from other SERMs in its ability to inhibit hormone-independent breast cancer cell growth and to regulate ERα and cyclin D1 expression in resistant cells.
Introduction
Bazedoxifene acetate (BZA) is a new third-generation selective estrogen receptor modulator (SERM) (Silverman et al., 2008) that is approved in Europe and is under regulatory review in the United States for the prevention and treatment of postmenopausal osteoporosis. In phase III clinical trials (Miller et al., 2008; Archer et al., 2009; Pinkerton et al., 2009) BZA (20 or 40 mg/daily) has been shown to prevent bone loss and to reduce bone turnover in postmenopausal women at risk for osteoporosis, with a favorable endometrial, ovarian, and breast safety profile. BZA also significantly reduces the risk of new vertebral fractures in postmenopausal women with osteoporosis compared with placebo (Silverman et al., 2008). In addition, recent studies indicate that BZA combined with conjugated estrogens relieves hot flashes and improves vulvovaginal atrophy and its symptoms (Kagan et al., 2010).
BZA is an indole-based ER ligand with unique structural characteristics with respect to tamoxifen (TAM) and raloxifene (RAL). It was assembled by using RAL as a template and substituting an indole ring for the benzothiophene core (Miller et al., 2001; Komm et al., 2005). BZA binds to both ERα and ERβ, with a slightly higher affinity for ERα; however, it is less ERα-selective than RAL, with an affinity for ERα that is approximately 10-fold lower than 17β-estradiol (E2) (Miller et al., 2001). ERα is a well studied member of the steroid/nuclear receptor family of transcription regulators. ERα acts in the nucleus to regulate gene expression by binding to estrogen response elements (EREs) and related DNA sequences and through association with transcription factors bound at SP1 and AP-1 DNA binding sites. In response to high-affinity estrogen binding, ERα dimerizes, binds to ERE DNAs, and undergoes a conformational change in the ligand binding domain that facilitates the recruitment of coactivators. In contrast, antagonist-occupied ERα recruits corepressors. Although previous studies have reported that BZA antagonizes E2-dependent MCF-7 breast cancer cell proliferation in vitro (Komm et al., 2005), little is known about the actions of BZA on ERα expression and functionality. In addition, not known is whether BZA has antitumor activity in breast cancer cells that have acquired resistance to endocrine therapies.
We have reported previously the development of two ERα-positive human breast cancer cell lines, MCF-7:5C (Jiang et al., 1992; Lewis et al., 2005a) and MCF-7:2A (Pink et al., 1995; Lewis-Wambi et al., 2008b), that were clonally selected from hormone-dependent MCF-7 breast cancer cells after long-term (>1 year) estrogen deprivation. An interesting phenotype of MCF-7:5C and MCF-7:2A cells is that, unlike MCF-7 cells, which require estrogen to grow and are inhibited by antiestrogens, they do not require estrogen to grow and they undergo apoptosis when exposed to physiological levels of E2 (Lewis et al., 2005a; Jordan, 2008; Lewis-Wambi et al., 2008b). However, the effects of SERMs on MCF-7:5C and MCF-7:2A cells have not been fully examined. In this study, we investigated the effects of BZA, 4-hydroxytamoxifen (4OHT), endoxifen (ENDOX), RAL, and the pure antiestrogen fulvestrant (ICI 182,780) on the growth of MCF-7:5C and MCF-7:2A breast cancer cells and determined the mechanism of action of BZA in these cells. We found that all of the SERMs inhibited E2-stimulated MCF-7 and T47D breast cancer cell growth; however, only BZA and FUL significantly inhibited the hormone-independent growth of MCF-7:5C cells. The inhibitory effect of BZA was associated with cell cycle arrest and cyclin D1 and ERα down-regulation, which was reversed by small interfering RNA (siRNA) knockdown of cyclin D1 and ERα. It is noteworthy that we found that FUL also inhibited MCF-7:5C cell growth; however, this compound partially down-regulated cyclin D1. Together, these data show that BZA is distinct from the other members of the SERM family in its ability to inhibit the growth of breast cancer cells that are resistant to long-term estrogen deprivation.
Materials and Methods
Reagents and Cell Culture.
E2, 4OHT (the active metabolite of TAM), and N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal (MG132) were purchased from Sigma-Aldrich (St. Louis, MO). Fulvestrant (ICI 182,780, Faslodex) was a generous gift from Dr. A. E. Wakeling (Zeneca Pharmaceuticals, Macclesfield, UK). ENDOX was a kind gift from Dr. James Ingle of the Mayo Clinic (Rochester, MN). RAL was a generous gift from Lilly Research Laboratories (Indianapolis, IN). BZA was synthesized by authors R.G. and M.A.S. using a protocol described previously (Miller et al., 2001). All of the compounds were dissolved in 100% ethanol except for MG132, which was dissolved in dimethyl sulfoxide. The compounds were added to the medium such that the total solvent concentration was never higher than 0.1%. An untreated group served as a control. The chemical structures of the compounds used in this study have been cited before (Komm et al., 2005; Jordan, 2007, 2009) and are shown in Supplemental Fig. 1.
MCF-7:WS8 and T47D:A18 human mammary carcinoma cells, clonally selected from their parental counterparts for sensitivity to growth stimulation by E2 (Pink and Jordan, 1996), were used in all experiments indicating MCF-7 and T47D cells. Cells were maintained in estrogenized medium (phenol red RPMI 1640 plus 10% fetal bovine serum), but 3 days before all experiments, they were cultured in steroid-free media as described previously (Pink and Jordan, 1996; Lewis et al., 2005a,b). MCF-7:5C (Jiang et al., 1992; Lewis et al., 2005a,b), and MCF-7:2A cells (Pink and Jordan, 1996; Lewis-Wambi et al., 2008b) were derived from the MCF-7 line by growth in estrogen-free media and two rounds of limiting dilution cloning and were maintained in phenol red-free RPMI 1640 medium containing 10% 3× dextran-coated charcoal-treated fetal bovine serum. MC2 cells were derived by stably transfecting ER-negative MDA-MB-231 breast cancer cells with the wild-type ERα (Jiang and Jordan, 1992), and these cells were grown in phenol red-free minimal essential medium supplemented with 5% 3× dextran-coated charcoal-treated calf serum, 0.5 mg/ml G-418. All cell culture reagents were from Invitrogen (Carlsbad, CA).
Cell Proliferation Assay.
These procedures have been reported previously (Lewis et al., 2005; Lewis-Wambi et al., 2008). In brief, MCF-7 and T47D cells were grown in fully estrogenized medium, whereas MCF-7:5C and MCF-7:2A cells were grown in nonestrogenized media. Cells were seeded in 24-well plates (30,000/well), and after overnight incubation, cells were treated with various concentrations of the tested compounds for 7 days. Media were changed on days 3 and 5, the experiment was ended on day 7, and the DNA content of the cells was determined as described previously (Labarca and Paigen, 1980) using a Fluorescent DNA Quantitation kit (Bio-Rad Laboratories, Hercules, CA). Cell proliferation was also determined by cell counting using a hemocytometer.
Western Blot Analyses.
Immunoblotting was performed using 30 μg of protein per well as described previously (Lewis et al., 2005a). Membranes were probed with primary antibodies against ERα, progesterone receptor, cyclin A, cyclin B1, or cyclin D1 (Santa Cruz Biotechnology, Santa Cruz, CA) with β-actin (AC-15; Sigma-Aldrich) used to standardize loading. The appropriate secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology) was used to visualize the stained bands with an enhanced chemiluminescence visualization kit (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). Bands were quantitated by densitometry using ImageQuant (GE Healthcare), and densitometric values were corrected for loading control.
Cell Cycle Analyses.
MCF-7 and MCF-7:5C cells were treated with E2 or BZA for 24 and 48 h and then fixed using ice-cold 70% ethanol. Cell cycle distribution was determined by propidium iodide staining using a fluorescence-activated cell sorter (BD Biosciences, San Jose, CA) as described previously (Ariazi et al., 2010). Data were analyzed using FlowJo 7.2.5 for Windows (Tree Star, Ashland, OR).
Knockdown of ERα and Cyclin D1 by siRNA.
MCF-7:5C cells were seeded at 105 cells/well in a 24-well plate overnight and then transfected with 100 nM nonspecific, ERα, or cyclin D1 siRNA (Dharmacon RNA Technologies, Lafayette, CO) using Lipofectamine 2000 (Invitrogen), as described previously (Lewis et al., 2005a). Transfected cells were either harvested for Western blot analysis or reseeded for cell growth or cell cycle analysis.
Quantitative Real-Time PCR.
The detail procedures have been reported previously (Lewis et al., 2005). MCF-7 and MCF-7:5C cells were treated with either E2 (10−9 M) or BZA (10−7 M) for 48 h, and total RNA was isolated and then reverse-transcribed to cDNA using the SuperScript II RNase H reverse transcriptase system (Invitrogen). Aliquots of the cDNA were combined with the SYBR green kit (Superarray) and primers and assayed in triplicate by quantitative PCR over 40 cycles using a GeneAmp 5700 Sequence detection system (Applied Biosystems, Foster City, CA) as described previously (Lewis et al., 2005a). Quantitation was done using the comparative CT method with 18S rRNA as the normalization gene, as described previously (Lewis-Wambi et al., 2008a). PCR primer sequences used were as follow: ERα forward, 5′-GGAGGGCAGGGGTGAA-3′; ERα reverse, 5′-GGCCAGGCTGTTCTTC TTAGA-3′; cyclin D1 forward, 5′-TCCTGTGCTGCGA AGTGGAAAC-3′; cyclin D1 reverse, 5′-AAATCGTGCGGGGTCATTGC-3; pS2 forward, 5′-GAGGCCCAGACAGAGACGTG-3′; and pS2 reverse, 5′-CCCTGCAGAAGTGTCTAAAATTCA-3′.
Transient Transfections and Luciferase Assays.
Cells were cultured in estrogen-free RPMI 1640 media for 48 h before transfection. On the day of the experiment, cells were seeded in estrogen-free media at a density of 1.5 × 105 cells per well in 24-well plates. After 24 h, cells were transfected with the firefly luciferase reporter plasmid pERE(5×)TA-ffLuc (containing five copies of a consensus ERE and a TATA-box driving firefly luciferase) and the pTA-srLuc Renilla reniformis luciferase plasmid (containing a TATA-box element driving R. reniformis luciferase) (Promega, Madison, WI) using LT1 (Mirus) transfection reagent, according to the manufacturer's protocol. After 24 h, transfection reagents were removed, and fresh media were added. Cells were then treated with ethanol (vehicle), 10−9 M E2, 10−8 M BZA, or E2 + BZA combined for 24 h. At the indicated time point, cells were washed, lysed, and ERE luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's recommendations. Samples were then read on a Mithras MB540 luminometer (Berthold Technologies, Oak Ridge, TN).
For the cyclin D1 promoter assay, MCF-7:5C cells were transiently transfected with the full-length cyclin D1 promoter plasmid (−1745CD1-LUC) as described previously (Lewis et al., 2005c,d). The full-length cyclin D1 plasmid (−1745CD1-LUC) (Albanese et al., 1995) was a gift from Dr. Richard Pestell (Thomas Jefferson Kimmel Cancer Center, Philadelphia, PA).
Molecular Modeling.
The molecular modeling performed in this study has been described previously (Maximov et al., 2010). In brief, the coordinates for the agonist and antagonist conformations of human ERα ligand binding domain cocrystallized with E2, RAL, and 4OHT were extracted from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB) (Berman et al., 2000). Entries 1gwr for E2 (Wärnmark et al., 2002), 1err for RAL (Brzozowski et al., 1997), and 3ert for 4OHT (Shiau et al., 1998) were selected for further modeling, and these structures were prepared for docking using the Protein Preparation Workflow (Friesner et al., 2004; Guallar et al., 2004) implemented in Schrödinger suite and accessible from within the Maestro 8.5 program (Schrödinger, Cambridge, MA). To study the molecular basis of interaction of bazedoxifene in the antagonist conformation of ERα, the ligands were docked into the binding site of the receptor cocrystallized with RAL (PDB code 1err). For comparison reasons, RAL was also docked in its native protein structure.
The input geometries of the ligands were generated with CORINA (online demo, http://www.molecular-networks.com/online_demos/corina_demo) and were further prepared for docking using the LigPrep2.2 utility (Friesner et al., 2004; Guallar et al., 2004). The prepared structure of ERα cocrystallized with RAL was used to generate the scoring grid for docking simulations. A grid box of 26 × 26 × 26 Å3 centered on the ligand was created, using the default parameters and without constraints.
Flexible ligand docking simulations were carried out with Glide 5.0 (Friesner et al., 2004; Guallar et al., 2004) using the default settings, and the best 10 poses for each ligand were evaluated using Glide (Schrödinger) in Standard-Precision (GlideSP) and Extra-Precision (GlideXP) mode. The results obtained from the docking runs were compared, and GlideXP docking poses were selected for analysis.
Statistical Analysis.
All quantitative experiments were performed in triplicate and/or repeated three times. Data were expressed as mean ± S.D. Statistical significances between vehicle treatment versus drug treatment were determined by one-way analysis of variance and the Student's t test. A value of p < 0.05 was considered statistically significant.
Results
BZA Inhibits the Growth of Hormone-Independent MCF-7:5C and MCF-7:2A Breast Cancer Cells.
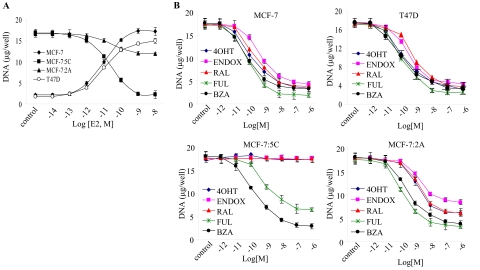
We first compared the growth characteristics of hormone-dependent MCF-7 and T47D breast cancer cells with those of long-term estrogen deprived MCF-7:5C and MCF-7:2A cells in the presence of E2. Cells were grown in estrogen-free media and then treated with 10−14 M to 10−8 M E2 for 7 days, and cellular DNA was measure as an index of growth. In parallel, cells were also treated with 10−9 M E2 for 2 to 12 days and then harvested and counted using a hemocytometer. Figure 1A shows that E2 treatment stimulated the growth of MCF-7 and T47D cells in a concentration-dependent manner with maximum stimulation at 10−9 M, whereas in MCF-7:5C and MCF-7:2A cells, E2 treatment had the opposite effect causing either complete growth inhibition in MCF-7:5C cells or partial growth inhibition in MCF-7:2A cells. This finding is consistent with our previous work (Lewis et al., 2005a; Lewis-Wambi et al., 2008b), which showed that physiological concentrations of E2 induced programmed cell death (apoptosis) in MCF-7:5C and MCF-7:2A cells through activation of the mitochondrial death pathway and suppression of glutathione synthesis, respectively. Specifically, we found that E2 induced apoptosis in MCF-7:5C cells by activating proapoptotic proteins Bax, Bak, Bim, and p53 and by suppressing antiapoptotic proteins. E2 also down-regulated survival proteins such as nuclear factor-κB, phospho-Akt, and Her2/neu, which were overexpressed in MCF-7:5C cells. In contrast, we found that MCF-7:2A cells underwent apoptosis after 10 to 12 days of E2 treatment and that these cells expressed elevated levels of the antioxidant glutathione as a result of overexpression of glutathione synthetase and glutathione peroxidase 2, the two main enzymes involved in glutathione synthesis. By selectively blocking the glutathione pathway in MCF-7:2A cells, we were able to sensitize these cells to E2-induced apoptosis, which was mediated by activation of the c-Jun NH2-terminal kinase signaling pathway.
Fig. 1.
Effects of E2 and SERMs on the growth of hormone-dependent MCF-7 and T47D cells versus hormone-independent MCF-7:5C and MCF-7:2A cells. A, MCF-7 and T47D cells were grown in phenol red-free RPMI medium supplemented with 10% charcoal-stripped fetal bovine serum for 3 days before the start of the experiment. On the day of the experiment, all cell lines were seeded in phenol red-free RPMI medium supplemented with 10% charcoal-stripped fetal bovine serum at 30,000 per well in 24-well dishes and after 24 h were treated with 10−14 to 10−8 M E2 for 7 days, with retreatment every other day. At the conclusion of the experiment, cells were harvested, and proliferation was assessed as cellular DNA mass (in micrograms per well) using a DNA quantitation kit. B, the effects of antihormones on the growth of hormone-dependent MCF-7 and T47D cells and hormone-independent MCF-7:5C and MCF-7:2A cells. Cells were seeded as described above, except MCF-7 and T47D cells were grown in fully estrogenized media and then treated with 10−12 to 10−6 M FUL, BZA, RAL, 4OHT, or ENDOX for 7 days with retreatment on alternate days. Proliferation was assessed as cellular DNA mass (in micrograms per well) as described under Materials and Methods. Each point represents the mean of three determinations ± S.E.M.
Next, we determined the inhibitory effects of BZA and other SERMs (see Supplemental Fig. 1 for chemical structures) on MCF-7, T47D, MCF-7:5C, and MCF-7:2A cells. For experiments, MCF-7 and T47D cells were grown in fully estrogenized media, and MCF-7:5C and MCF-7:2A cells were grown in estrogen-free media and then treated with 10−12 to 10−6 M BZA, RAL, FUL, 4OHT, or ENDOX for 7 days, and cellular DNA was measured as an index of growth. Figure 1B shows that all of the tested SERMs along with the pure antiestrogen FUL inhibited E2-stimulated growth in MCF-7 and T47D cells and hormone-independent growth in MCF-7:2A cells in a concentration-dependent manner; however, in MCF-7:5C cells, only BZA and FUL inhibited the growth of these cells with no effects observed with RAL, 4OHT, or ENDOX. BZA reduced the growth of MCF-7:5C cells in a concentration-dependent manner, causing an 80% reduction at 10−8 M, whereas FUL reduced the growth by 55% at a similar concentration.
BZA Down-Regulates ERα Protein in MCF-7:5C and MCF-7:2A Cells.
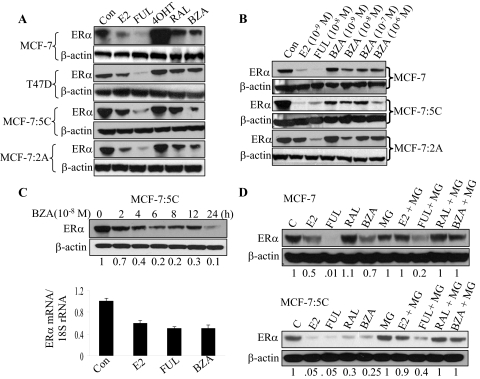
Because BZA dramatically reduced the growth of MCF-7:5C cells, we next determined whether BZA had actions similar to that of 4OHT or FUL at the level of ERα stability/degradation. We treated MCF-7:5C, MCF-7:2A, MCF-7, and T47D cells with 10−9 M E2 or 10−7 M FUL, 4OHT, RAL, or BZA for 24 h and monitored ERα protein level. As shown in Fig. 2A, ERα protein was highly expressed in MCF-7:5C and MCF-7:2A cells compared with MCF-7 and T47D cells and treatment with BZA markedly down-regulated ERα protein in MCF-7:5C and MCF-7:2A cells; however, it did not significantly reduce ERα levels in MCF-7 and T47D cells. The ability of BZA to down-regulate ERα in MCF-7:5C and MCF-7:2A cells was greater than that of RAL and almost comparable with that of the pure antiestrogen FUL, which strongly down-regulated ERα in all of the cell lines. E2 treatment also markedly down-regulated ERα protein in all of the cell lines including MCF-7:5C (Fig. 2A); however, 4OHT stabilized ERα against degradation in MCF-7 and T47D cells, as reported previously (Pink and Jordan, 1996), with marginal stabilization observed in MCF-7:5C and MCF-7:2A cells (Fig. 2A). We also examined the effect of the tamoxifen metabolite ENDOX on ERα expression in the different cell lines and found that endoxifen did not down-regulate ERα in any of the tested cell lines (Supplemental Fig. 2). Our finding differs from that of Wu et al. (2009), who reported that endoxifen degrades ERα in breast cancer cells.
Fig. 2.
Effects of SERMs on ERα expression and stability in hormone-dependent MCF-7 and T47D cells and hormone-independent MCF-7:5C and MCF-7:2A cells. A, Western blot analysis of ERα protein levels in MCF-7, T47D, MCF-7:5C, and MCF-7:2A cells in response to 24-h treatment with 10−9 M E2 or 10−7 M FUL, 4OHT, RAL, or BZA. β-actin was used as a loading control. B, Western blot analysis of ERα protein levels in MCF-7, MCF-7:5C, and MCF-7:2A cells after treatment with 10−9 to 10−6 M BZA for 24 h. For comparison, cells were also treated with 10−9 M E2 or 10−8 M FUL. C, Western blot analysis of ERα protein levels in MCF-7:5C cells in response to 10−8 M BZA treatment over a 24-h time period. Quantitated protein levels were normalized to β-actin. Densitometric quantitation relative to the control is shown on the bottom of the immunoreactive bands. Also shown is ERα mRNA levels in MCF-7:5C cells after treatment with E2 (10−9 M), FUL (10−8 M), or BZA (10−8 M) for 24 h. The amount of ERα mRNA was determined by real-time RT-PCR and normalized to the internal control 18S rRNA. Each data point represents the average of four biological replicates from three independent experiments. D, Western blot analysis of ERα protein levels in MCF-7 and MCF-7:5C cells pretreated with the proteosome inhibitor MG132 (4 μM) for 4 h and then treated as indicated for 8 h. β-Actin levels are shown as protein loading controls. Each point represents the mean of three determinations ± S.E.M.
We also performed dose-response studies in MCF-7, MCF-7:5C, and MCF-7:2A cells to determine the optimal concentration at which BZA down-regulated ERα protein. Figure 2B showed that BZA reduced ERα protein level in MCF-7:5C cells in a concentration-dependent manner with maximum inhibition at 10−6 M, whereas in MCF-7 and MCF-7:2A cells, BZA only marginally reduced ERα protein in these cells. It is noteworthy that the inhibitory effect of BZA on ERα protein was less pronounced than that observed with E2 or FUL, which almost completely reduced ERα protein level in MCF-7:5C cells. Time course studies revealed that BZA down-regulated ERα protein as early as 2 h after treatment with maximum suppression at 24 h (Fig. 2C, top). BZA also down-regulated ERα mRNA in MCF-7:5C cells to a level similar to that observed with E2 and FUL (Fig. 2C, bottom). To show that the decreased ERα protein by BZA was due to protein degradation, we used MG132 to inhibit the proteosome in MCF-7:5C and MCF-7 cells. We found that inhibition of proteosome activity completely blocked ERα degradation by BZA and E2 with partial reversal with fulvestrant (Fig. 2D). We further determined whether BZA might affect ERα protein expression by inhibiting its synthesis. We treated MCF-7:5C cells with 0.5 to 5 μM cycloheximide for 4 h to address this question. The impact of cycloheximide on ERα protein expression was much less dramatic than that of BZA (data not shown), which suggest that BZA-induced down-regulation of ERα protein is not likely to involve protein synthesis inhibition. Together, these data show that BZA differs from the other SERMs in its ability to regulate cell growth and ERα protein expression in MCF-7:5C cells.
BZA Inhibits ERα Transcriptional Activity in MCF-7:5C Cells.
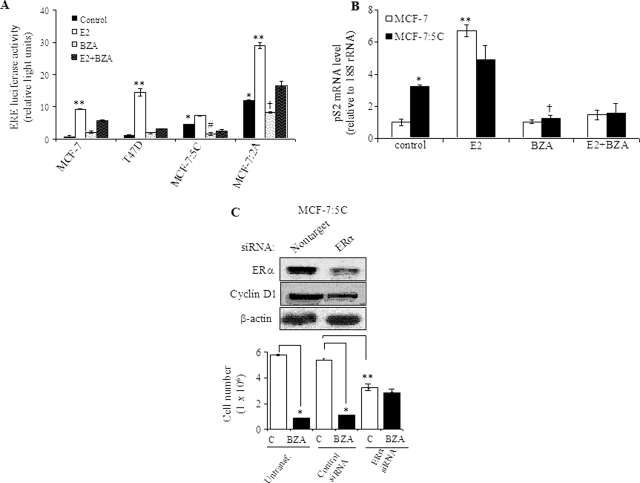
To determine whether BZA blocks ERα function, we next examined the transcriptional activation of an ERE in MCF-7, T47D, MCF-7:5C, and MCF-7:2A cells. Cells were transiently transfected with a 5× ERE-luciferase reporter plasmid and treated with 10−10 M E2, 10−8 M BZA, or E2 + BZA for 24 h. The results of these studies showed that basal ERE activity was elevated 5-fold in MCF-7:5C and 10-fold in MCF-7:2A cells compared with MCF-7 cells and treatment with BZA significantly reduced the basal ERE activity in these cells (Fig. 3A). E2 treatment further increased ERE activity in MCF-7:5C and MCF-7:2A cells by 1.5- and 2.5-fold, respectively; however, in MCF-7 and T47D cells, the response was markedly more robust with a 12- and 20-fold increase, respectively (Fig. 3A).
Fig. 3.
BZA inhibits constitutive ERα transcriptional activity in hormone-independent and hormone-dependent breast cancer cells. A, ERE luciferase activity in hormone-dependent MCF-7 and T47D cells and hormone-independent MCF-7:5C and MCF-7:2A cells. For experiment, cells were transiently transfected with a 5× ERE-luciferase reporter construct and treated with 10−9 M E2, 10−7 M BZA, E2 + BZA, or nothing (control) for 24 h. Luciferase values for the treatment groups are reported as relative luciferase units. *, p < 0.001 compared with MCF-7 and T47D cells (control); **, p < 0.0001 compared with control for each cell line; #, p < 0.01 compared with untreated MCF-7:5C cells (control); †, p < 0.05 compared with untreated MCF-7:2A cells. B, real-time RT-PCR analysis of pS2 mRNA gene expression in MCF-7 and MCF-7:5C cells after treatments with E2 (10−9 M), BZA (10−7 M), or E2 + BZA for 24 h. Each data point represents the average of three biological replicates. *, p < 0.01 compared with untreated MCF-7 cells (control); **, p < 0.001 compared with untreated MCF-7 cells (control); †, p < 0.001 compared with untreated MCF-7:5C cells (control). C, MCF-7:5C cells were transfected with 100 nM nonspecific control or ERα siRNA for 48 h. Transfected cells were then harvested for Western blot analysis to detect ERα and cyclin D1 protein (top) or treated with 10−7 M BZA for an additional 4 days followed by cell counting using a hemocytometer (bottom). Data shown are representative of three independent experiments. *, p < 0.001 compared with untransfected control and nonspecific transfected cells; **, p < 0.01 compared with nonspecific transfected cells.
To further test whether BZA is able to block ERα-regulated genes, we analyzed the expression level of pS2 mRNA in MCF-7:5C cells using quantitative RT-PCR. The pS2 gene is often used as a prognostic marker in breast cancer cells and is frequently used in studies of ER action. Furthermore, it is suggested that estrogen regulates the expression of pS2 through an imperfect ERE in the pS2 promoter (Berry et al., 1989). Our results showed that basal pS2 mRNA level was ∼3.5-fold higher in MCF-7:5C cells compared with wild-type MCF-7 cells, and E2 treatment increased pS2 mRNA level by ∼5.5-fold in MCF-7 cells and MCF-7:5C cells, which was completely blocked by BZA (Fig. 3B). It is noteworthy that we also found that siRNA knockdown of ERα (Fig. 3C) significantly reduced the basal growth of MCF-7:5C cells and markedly reduced the inhibitory effect of BZA in these cells (Fig. 3C, bottom). In addition, suppression of ERα significantly reduced cyclin D1 protein in MCF-7:5C cells. Overall, these data indicate that in the absence of estrogen, the unliganded ERα drives the proliferation of hormone-independent breast cancer cells; however, in the presence of BZA, the ability to inhibit cell proliferation is dependent on receptor degradation.
BZA Blocks Cell Cycle Progression in MCF-7:5C Cells and Down-Regulates Cyclin D1.
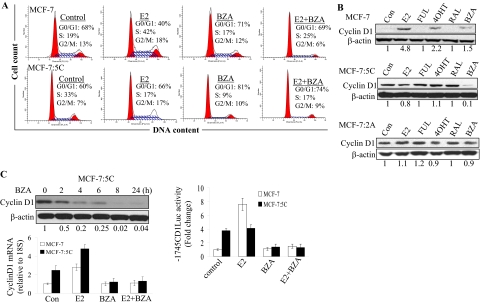
Because BZA significantly reduced the growth of MCF-7:5C cells, we next examined its effect on cell cycle progression. For experiment, MCF-7 and MCF-7:5C cells were treated with 10−9 M E2, 10−8 M BZA, or E2 plus BZA for 48 h followed by propidium iodide staining and flow cytometric analysis. The results showed that in MCF-7:5C cells, E2 treatment significantly reduced the percentage of cells in S phase from 33 to 17% and marginally increased the percentage of cells in G1 phase from 60 (control) to 66%, whereas BZA treatment increased the proportion of cells in the G1 phase from 60 to 81%, and it reduced the proportion of S phase cells from 33 to 9% at 48 h. In MCF-7 cells, treatment with E2 increased the proportion of S phase cells from 19 to 42% at 48 h with no effect observed with BZA alone (Fig. 4A). It is noteworthy that the inhibitory effect of BZA on cell cycle in MCF-7:5C cells was somewhat comparable with the pure antiestrogen fulvestrant; however, none of the other tested SERMs had any effect on cell cycle (data not shown).
Fig. 4.
Effects of BZA on cell cycle progression and cyclin D1 regulation in MCF-7 and MCF-7:5C cells. A, cell cycle distribution was determined by propidium iodide staining of DNA content and flow cytometry. Cells were treated with 10−9 M E2, 10−7 M BZA, or E2 plus BZA for 24 and 48 h. Thirty thousand cells per sample and three replicates per group were collected. Representative histograms are shown. B, Western blot analysis of cyclin D1 expression level in MCF-7 and MCF-7:5C cells after treatment with BZA and other SERMs. Before the experiment, MCF-7 cells were switched from fully estrogenized media to estrogen-free media for 3 days and then treated with ethanol vehicle (control), 10−9 M E2 alone, or 10−9 M E2 plus FUL (10−7 M), RAL (10−7 M), 4OHT (10−7 M), or BZA (10−7 M) for 24 h. MCF-7:5C cells, however, did not require a media switch because they are hormone-independent and are routinely grown in estrogen-free media. MCF-7:5C cells were treated as described above for MCF-7 cells. Quantitated protein levels normalized to β-actin are indicated. C, BZA regulation of cyclin D expression and promoter activity in MCF-7:5C cells. Cells were treated with 10−7 M BZA for the indicated time points. Cyclin D1 protein and mRNA levels were determined by Western blot and quantitative RT-PCR, respectively, with β-actin and 18S rRNA as internal controls. For cyclin D1 promoter activity experiment, MCF-7 and MCF-7:5C cells were cotransfected with a full-length cyclin D1 promoter plasmid (−1745CDLUC) and Renilla reniformis luciferase control plasmid overnight and then treated with 10−9 M E2, 10−8 M BZA, or E2 + BZA for 24 h. Luciferase activity was measured as described under Materials and Methods. Each point represents the mean of three determinations ± S.E.M.
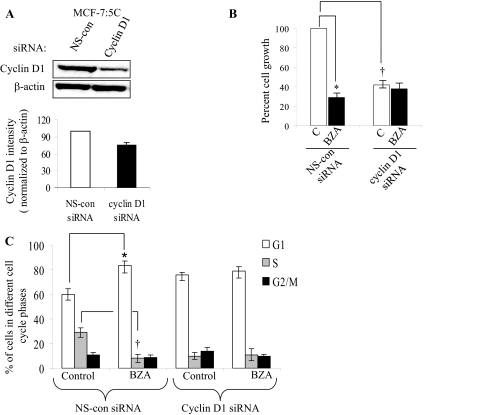
Because BZA induced G1-phase cell cycle block in MCF-7:5C cells, we further investigated the G1-specific protein cyclin D1 in these cells. MCF-7 and MCF-7:5C cells were treated with BZA, E2, RAL, 4OHT, or FUL for 24 h, and lysates were prepared and analyzed by immunoblotting. Figure 4B shows that cyclin D1 was undetectable in untreated MCF-7 cells; however, treatment with E2 and, to a lesser extent, with 4OHT markedly increased cyclin D1 protein in these cells. In contrast, we found that cyclin D1 protein was constitutively overexpressed in MCF-7:5C and MCF-7:2A cells, and treatment with BZA completely reduced cyclin D1 protein in MCF-7:5C cells but not MCF-7:2A cells (Fig. 4B). It is noteworthy that none of the other SERMs inhibited cyclin D1 in MCF-7:5C cells; however, FUL significantly reduced cyclin D1 protein level at 96 h, and it markedly reduced cyclin A protein in these cells (Supplemental Fig. 3). Time course experiments revealed that BZA inhibited basal cyclin D1 protein in a time-dependent manner with measurable effects observed as early as 2 h after treatment and maximum reduction at 24 h (Fig. 4C, top). BZA also reduced cyclin D1 mRNA (Fig. 4C, bottom) and cyclin D1 promoter activity (Fig. 4C, top right) in MCF-7:5C cells. Finally, we found that siRNA knockdown of cyclin D1 (Fig. 5A) significantly reduced the hormone-independent growth of MCF-7:5C cells (Fig. 5B), and it significantly reduced the ability of BZA to induce G1 blockade in these cells (Fig. 5C), thus confirming the importance of cyclin D1 in the inhibitory action of BZA in these cells.
Fig. 5.
Effect of cyclin D1 knockdown on proliferation and cell cycle in MCF-7:5C cells. A, Western blot analysis of cyclin D1 protein expression in MCF-7:5C cells transfected with 100 nM cyclin D1 siRNA or the nonspecific (NS)-control siRNA, as determined 72 h after transfection. B, cell growth of transfected cells treated with 100 nM BZA or vehicle (control). Transfected cells (30,000/well) were seeded in 24-well dishes overnight and then treated with BZA for 5 days. After treatment, cells were collected and counted using a hemocytometer. Data is presented as percentage and is based on the mean from three independent experiments with duplicate (*, p < 0.01 versus nontarget transfected cells). C, cell cycle analysis of cyclin D1 siRNA-transfected and control siRNA transfected MCF-7:5C cells after treatment with BZA for 48 h. Data are based on the mean from three independent experiments with duplicate. *, p < 0.01; **, p < 0.001.
Molecular Modeling and Docking of BZA into the Ligand Binding Site of ERα.
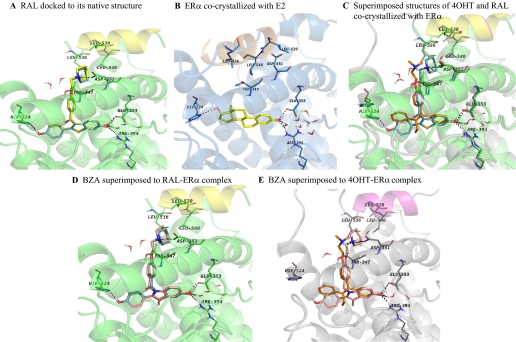
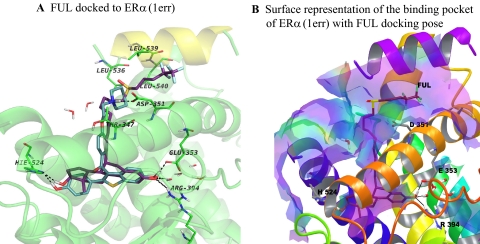
Molecular modeling and docking studies were carried out in an attempt to predict the bioactive conformation of BZA and to understand the molecular basis of interaction of this ligand with ERα. Using the available X-ray crystallographic data, the flexible docking of BZA into the ligand binding domain of ERα cocrystallized with RAL was performed, and for comparison reasons, FUL and RAL were also docked in their native protein structure. The superimposition of the docked solution and experimental structure of RAL shows that the docking model recapitulates the orientation of the native ligand in the active site, and the same interactions with the key amino acids of the binding cavity are formed with a ligand root mean square deviation of 0.362 compared with the crystal structure (Fig. 6A). The experimental structure of ERα cocrystallized with E2 (PDB code 1gwr), the agonist conformation of the receptor, is displayed in Fig. 6B, whereas the experimental antagonist conformation of ERα bound to 4OHT and RAL are superimposed and presented in Fig. 6C. The docking results analysis reveals that BZA binds to ERα in an antagonist orientation similar with RAL (Fig. 6D) and has the tendency to form the same hydrophobic contacts with the amino acids lining the binding cavity. In addition, the same complex H-bond network is formed with Asp351, Glu353, Arg394, His524, and a highly ordered water molecule, located in the vicinity of residues Glu353 and Arg394 (Fig. 6D). However, we should note that a number of residues adopt different conformations in the Induced Fit Docking (IFD) poses compared with the experimental structure of ERα, PDB code 1err (Supplemental Fig. 4). The most significant difference has been observed for Leu539 of helix 12. The larger ring of BZA causes the side chain of Leu539 to be pushed away from its original position by approximately 1 Å. In all top-ranked IFD structures (four poses having the composite score of 0.5 kcal/mol), Leu529 side chain is moved up from its original orientation toward the ring of BZA to optimize the hydrophobic contacts between the ligand and residue side chain (Supplemental Fig. 4). We also compared the docked structure of BZA with the binding mode of 4OHT to ERα (Fig. 6C) and superimposed it in the binding site of 4OHT-ERα complex (Fig. 6E). The 4OHT bound receptor shows that the H-bond between BZA and H524 is missing (Fig. 6E) because of the different orientation of this amino acid in the binding site compared with the RAL-ERα complex (Fig. 6C). When FUL was docked to RAL-ERα complex (Fig. 7A), the H-bond network was recapitulated with one exception: the interaction with Asp351 is missing, whereas the flexible side chain of FUL fills the groove between helix 3 and helix 12 (Fig. 7B).
Fig. 6.
Molecular modeling of ERα binding site with various ligands. A, comparison between the experimental (yellow sticks) and top ranked docking pose (cyan sticks) of RAL to ERα binding site. The docking pose recapitulates very well the alignment of the cocrystallized ligand in the receptor binding site having a ligand root mean square deviation of 0.36 Å. B, agonist conformation of ERα cocrystallized with E2; helix 12 is depicted in orange and lays over the binding site sealing the ligand inside it. The antagonist conformations of the receptor are shown in C, D, and E. X-ray structures of ERα cocrystallized with 4OHT (C), raloxifene (D), and bazedoxifene (E) docked into the ERα-raloxifene crystal structure. Helix 12 is depicted in magenta for 4OHT bound conformation and yellow for raloxifene and bazedoxifene. In addition, the key amino acids lining the binding site are displayed and the network of hydrogen bonds in which they are involved with the ligands is shown in black dashed lines. Carbon atoms are colored in yellow for E2, orange for 4OHT, cyan for raloxifene, and pink for bazedoxifene. These images show the differences between the agonist (B) and antagonist conformation (C, D, and E) of ERα and present the alignment of bazedoxifene in the binding site of ERα, which is similar to raloxifene's orientation, and the same interactions with the key amino acids of the binding cavity are encountered.
Fig. 7.
Simplified representations of the ERα binding site with fulvestrant. A, representation of the ERα binding site with the best docking pose for fulvestrant (FUL, purple sticks). B, surface representation of ERα binding site accommodating FUL. Hydrophobic areas are mapped in purple, whereas the hydrophilic parts are colored in light yellow-green. The binding site accommodates very well the ligand, which forms the H-bond contacts with the same amino acids like E2 or RAL, whereas the aliphatic side chain protrudes from the binding site and lies in the groove between helix 3 (orange cartoon) and helix 12 (purple cartoon). Only the key amino acids underlying the binding site are shown.
Overall, these findings indicate that the alignment of BZA in the binding pocket of ERα predicted by the IFD is similar with that predicted via the rigid docking method (Glide) and with the alignment of RAL in the experimental structure, PDB code 1err. However, there are a few differences in the orientation of some residues in the binding site when the docking of BZA is performed with IFD protocol, and these differences might help to explain the different biological effects of BZA versus RAL in our cell model.
Discussion
In the present study, we report for the first time that BZA inhibits the growth of breast cancer cells that have acquired resistance to long-term estrogen deprivation (i.e., hormone-independent/aromatase inhibitor resistant). Specifically, we found that BZA at 10−8 M inhibited the growth of hormone-independent MCF-7:5C and MCF-7:2A breast cancer cells by 80 and 55%, respectively. The inhibitory effect of BZA in MCF-7:5C cells was associated with G1 arrest and cyclin D1 and ERα down-regulation, whereas in MCF-7:2A cells, BZA suppressed cyclin A with marginal effects on cyclin D1. The pure antiestrogen FUL also inhibited the growth of MCF-7:5C cells by inducing G1 arrest; however, it did not down-regulate cyclin D1 until 96 h, which was 48 h after its effect on cell cycle. Strikingly, RAL, 4OHT, and ENDOX failed to inhibit cyclin D1 expression in MCF-7:5C cells, and these compounds did not have any growth-inhibitory effect in MCF-7:5C cells. Although it is not entirely clear why BZA was more potent than fulvestrant at inhibiting the growth of MCF-7:5C cells, one possibility might be due to the fact that BZA down-regulated both ERα and cyclin D1, whereas FUL down-regulated ERα and had marginal effects on cyclin D1, which was observed at 96 h. Molecular modeling studies indicated that BZA bound the ligand binding domain of ERα in an antagonist orientation similar to RAL (Fig. 6D) but distinct from 4OHT (Fig. 6E) and fulvestrant (Fig. 7). However, a few differences were noticed in the orientation of some residues in the binding site when the docking of BZA was performed with the IFD protocol. The most significant difference was observed for the Leu539 of helix 12. The larger ring of BZA caused the side chain of Leu539 to be pushed away from its original position by approximately 1 Å. This alteration in the orientation of Leu539 side chain could trigger a conformational change of helix 12, which in turn could lead to the recruitment of other proteins by the BZA-ERα compared with the RAL-ERα complex. Indeed, these findings help to further distinguish BZA from the other SERMs such as TAM and RAL, and they support the concept that subtle but moderate structural differentiation can dramatically affect the ability of a ligand to regulate cell proliferation.
Previous research has indicated that deregulation of ERα expression is a driving force in the initiation and progression of estrogen-sensitive breast tumors (Garcia-Closas and Chanock, 2008; Garcia-Closas et al., 2008). It has been suggested that alterations in pathways leading to ERα synthesis and/or degradation underlie the deregulation of ERα and its consequent manifestations, including enhanced proliferation in breast tumors (Sommer and Fuqua, 2001). ERα is the predominant receptor isoform expressed in breast cancer cells, and increased numbers of ERα-expressing cells can be observed at the earliest stages of breast tumorigenesis. We have shown previously that ERα mRNA and protein levels are significantly elevated in breast cancer cells that have been adapted to grow in an estrogen-depleted environment (Murphy et al., 1990; Pink et al., 1996; Lewis et al., 2005a). This particular type of regulation in which ERα levels are increased after estrogen deprivation has been described as a model I response (Pink and Jordan, 1996). A model I response is characterized by an ERα that is expressed at high levels in the absence of estrogen and is subsequently down-regulated after estrogen binding, primarily through repression of the steady-state level of the mRNA. In the present study, we found that basal ERα protein levels were up-regulated greater than 3-fold in hormone-independent MCF-7:5C and MCF-7:2A breast cancer cells compared with MCF-7 and T47D cells, and treatment with BZA (10−8 M) induced proteasome-mediated degradation of ERα in these cells, which was reversed by the proteasome inhibitor MG132. The ability of BZA to degrade ERα in MCF-7:5C cells was rapid and robust, occurring as early as 4 h after treatment with maximum degradation at 24 h. It is noteworthy that BZA and fulvestrant were the only compounds that markedly reduced the growth of both MCF-7:5C and MCF-7:2A breast cancer cells, and blocking BZA-induced ERα degradation with MG132 dramatically reduced its growth inhibitory effects on these cells (data not shown). The importance of ERα in mediating the antagonist effects of BZA in hormone-independent MCF-7:5C cells was further confirmed by siRNA knockdown experiments, which showed a 60% reduction in the ability of BZA to inhibit the growth of these cells. Suppression of ERα also significantly reduced the basal growth of MCF-7:5C cells and E2-induced growth in wild-type MCF-7 cells, which is consistent with recent findings by Ariazi et al. (2010). It should be noted, however, that degradation or suppression of ERα is not the only mechanism by which an antagonist can inhibit cell proliferation. For example, TAM has been shown to stabilize ERα protein against degradation in breast cancer cells (Murphy et al., 1990; Pink et al., 1995, 1996; Pink and Jordan, 1996); however, it is a potent antagonist in the breast with the ability to block E2-stimulated proliferation and E2-induced ERE activity in these cells.
Apart from ERα, BZA also significantly reduced cyclin D1 expression in hormone-independent MCF-7:5C breast cancer cells. Cyclin D1 is a breast cancer oncogene whose overexpression has been linked to poor prognosis in ERα and progesterone receptor-positive breast cancers (Lammie and Peters, 1991). It is a multifunctional G1-phase cyclin whose regulatory effects are particularly important in breast development and cancer (Sutherland and Musgrove, 2004). Cyclin D1 is highly induced by estrogen (Said et al., 1997), and it contributes to poor treatment response of ER-positive tumors by acting downstream to promote hormone agonist- and antagonist-independent proliferation (Wilcken et al., 1997). We found that cyclin D1 protein was constitutively elevated by 3-to 5-fold in hormone-independent MCF-7:5C and MCF-7:2A cells compared with wild-type MCF-7 and T47D cells, and treatment with BZA reduced it to an undetectable level in MCF-7:5C cells but not MCF-7:2A cells. In addition, we found that suppression of cyclin D1 in MCF-7:5C cells reduced the hormone-independent growth of these cells, and it significantly reduced the ability of BZA to inhibit cell growth and induce cell cycle arrest in these cells. Suppression of cyclin D1 also significantly reduced ERα protein levels in MCF-7:5C cells with similar effects observed after ERα suppression, thus suggesting a link between cyclin D1 and ERα in these cells. Indeed, a connection between ER and cyclin D1 was demonstrated previously when cyclin D1 was shown to interact directly with the ligand-binding domain of ER and stimulate ER transactivation in a ligand-independent fashion (Zwijsen et al., 1997). More recently, cyclin D1 was shown to interact with coactivators of the SRC-1 family through a motif that resembles the leucine-rich coactivator binding motif of nuclear receptors. By acting as a bridging factor between ER and SRCs, it is believed that cyclin D1 can recruit SRC family coactivators to ER in the absence of ligand. It is worth noting that hormone-independent MCF-7:5C cells express elevated levels of SRC-1 protein compared with hormone-dependent MCF-7 cells, and BZA treatment significantly reduces basal SRC-1 levels in these cells (data not shown).
Although cyclin D1 gene transcription is directly induced by estrogen, there is no estrogen response element in it. Instead, the cyclin D1 promoter contains multiple regulatory elements, including binding sites for activator protein-1, signal transducer and activator of transcription 5, nuclear factor-κB, cAMP response element, SP1, and E2F. A fragment between −994 and −136 of the cyclin D1 promoter was shown previously to be estrogen-responsive, and this region has binding sites for AP-1 and SP-1 (Altucci et al., 1996). We have reported that estrogen-induced cyclin D1 transactivation in MCF-7 breast cancer cells was mediated by the CRE region, which is known to bind activating transcription factor 2 (Lewis et al., 2005c,d). A notable finding of our study was that basal cyclin D1 promoter activity was significantly elevated in hormone-independent MCF-7:5C cells compared with hormone-dependent MCF-7 cells and treatment with BZA completely reduced the promoter activity in these cells to the level seen in the untreated MCF-7 cells. In contrast, E2 did not induce cyclin D1 expression or promoter activity in hormone-independent MCF-7:5C cells, whereas in hormone-dependent MCF-7 cells, it increased cyclin D1 protein level by 3-fold and its promoter activity by 4-fold, which is consistent with its function as a proapoptotic agent in MCF-7:5C cells versus an agonist in MCF-7 cells.
In conclusion, it is clear from clinical data that BZA in combination with conjugated estrogens represents a new form of therapeutic agents for the treatment of postmenopausal symptoms and prevention of postmenopausal osteoporosis. The fact that it does not stimulate the breast or endometrium and is very effective at inhibiting the proliferation of endocrine-resistant breast cancer cells highlights its widespread therapeutic potential and demonstrates that not all SERMs are alike. Our data also suggest that the overexpression of ERα and cyclin D1 in MCF-7:5C cells might be driving the hormone-independent growth of these cells and that the ability of BZA to down-regulate ERα and cyclin D1 is critical to treat and possibly reverse antihormone resistance in breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. James Ingle for the endoxifen compound and Drs. Richard Pestell and Chris Albanese for the full-length cyclin D1 promoter plasmid.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by the National Institutes of Health National Cancer Institute [Grants K01-CA12005101A2, P30-CA006927]; the American Cancer Society [Grant IRG-9202714]; the Department of Defense [Grant BC050277]; and the National University Research Council of Romania-CNCSIS-UEFISCU [Grant PN-II-PCE-ID1268].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.111.072249.
- BZA
- bazedoxifene acetate
- ER
- estrogen receptor
- SERM
- selective estrogen receptor modulator
- TAM
- tamoxifen
- RAL
- raloxifene
- E2
- 17β-estradiol
- FUL
- fulvestrant
- siRNA
- small interfering RNA
- 4OHT
- 4-hydroxytamoxifen
- ENDOX
- endoxifen
- Luc
- luciferase
- ERE
- estrogen response element
- PCR
- polymerase chain reaction
- RT-PCR
- reverse transcriptase-polymerase chain reaction
- IFD
- Induced Fit Docking
- SRC
- steroid receptor coactivator
- ICI 182,780
- fulvestrant
- MG132
- N-benzoyloxycarbonyl (Z)-Leu-Leu-leucinal
- PDB
- Protein Data Bank.
Authorship Contributions
Participated in research design: Lewis-Wambi and Jordan.
Conducted experiments: Lewis-Wambi and Kim.
Contributed new reagents or analytic tools: Curpan, Grigg, and Sarker.
Performed data analysis: Lewis-Wambi.
Wrote or contributed to the writing of the manuscript: Lewis-Wambi and Jordan.
References
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. (1995) Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem 270:23589–23597 [DOI] [PubMed] [Google Scholar]
- Altucci L, Addeo R, Cicatiello L, Dauvois S, Parker MG, Truss M, Beato M, Sica V, Bresciani F, Weisz A. (1996) 17beta-Estradiol induces cyclin D1 gene transcription, p36D1–p34cdk4 complex activation and p105Rb phosphorylation during mitogenic stimulation of G(1)-arrested human breast cancer cells. Oncogene 12:2315–2324 [PubMed] [Google Scholar]
- Archer DF, Pinkerton JV, Utian WH, Menegoci JC, de Villiers TJ, Yuen CK, Levine AB, Chines AA, Constantine GD. (2009) Bazedoxifene, a selective estrogen receptor modulator: effects on the endometrium, ovaries, and breast from a randomized controlled trial in osteoporotic postmenopausal women. Menopause 16:1109–1115 [DOI] [PubMed] [Google Scholar]
- Ariazi EA, Brailoiu E, Yerrum S, Shupp HA, Slifker MJ, Cunliffe HE, Black MA, Donato AL, Arterburn JB, Oprea TI, et al. (2010) The G protein-coupled receptor GPR30 inhibits proliferation of estrogen receptor-positive breast cancer cells. Cancer Res 70:1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M, Nunez AM, Chambon P. (1989) Estrogen-responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc Natl Acad Sci USA 86:1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engström O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 389:753–758 [DOI] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749 [DOI] [PubMed] [Google Scholar]
- Garcia-Closas M, Chanock S. (2008) Genetic susceptibility loci for breast cancer by estrogen receptor status. Clin Cancer Res 14:8000–8009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG, Axelsson CK, Arias JI, et al. (2008) Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet 4:e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guallar V, Jacobson M, McDermott A, Friesner RA. (2004) Computational modeling of the catalytic reaction in triosephosphate isomerase. J Mol Biol 337:227–239 [DOI] [PubMed] [Google Scholar]
- Jiang SY, Jordan VC. (1992) Growth regulation of estrogen receptor-negative breast cancer cells transfected with complementary DNAs for estrogen receptor. J Natl Cancer Inst 84:580–591 [DOI] [PubMed] [Google Scholar]
- Jiang SY, Wolf DM, Yingling JM, Chang C, Jordan VC. (1992) An estrogen receptor positive MCF-7 clone that is resistant to antiestrogens and estradiol. Mol Cell Endocrinol 90:77–86 [DOI] [PubMed] [Google Scholar]
- Jordan VC. (2007) New insights into the metabolism of tamoxifen and its role in the treatment and prevention of breast cancer. Steroids 72:829–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VC. (2008) The 38th David A. Karnofsky lecture: the paradoxical actions of estrogen in breast cancer–survival or death? J Clin Oncol 26:3073–3082 [DOI] [PubMed] [Google Scholar]
- Jordan VC. (2009) A century of deciphering the control mechanisms of sex steroid action in breast and prostate cancer: the origins of targeted therapy and chemoprevention. Cancer Res 69:1243–1254 [DOI] [PubMed] [Google Scholar]
- Kagan R, Williams RS, Pan K, Mirkin S, Pickar JH. (2010) A randomized, placebo- and active-controlled trial of bazedoxifene/conjugated estrogens for treatment of moderate to severe vulvar/vaginal atrophy in postmenopausal women. Menopause 17:281–289 [DOI] [PubMed] [Google Scholar]
- Komm BS, Kharode YP, Bodine PV, Harris HA, Miller CP, Lyttle CR. (2005) Bazedoxifene acetate: a selective estrogen receptor modulator with improved selectivity. Endocrinology 146:3999–4008 [DOI] [PubMed] [Google Scholar]
- Labarca C, Paigen K. (1980) A simple, rapid, and sensitive DNA assay procedure. Anal Biochem 102:344–352 [DOI] [PubMed] [Google Scholar]
- Lammie GA, Peters G. (1991) Chromosome 11q13 abnormalities in human cancer. Cancer Cells 3:413–420 [PubMed] [Google Scholar]
- Lewis-Wambi JS, Cunliffe HE, Kim HR, Willis AL, Jordan VC. (2008a) Overexpression of CEACAM6 promotes migration and invasion of oestrogen-deprived breast cancer cells. Eur J Cancer 44:1770–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis-Wambi JS, Kim HR, Wambi C, Patel R, Pyle JR, Klein-Szanto AJ, Jordan VC. (2008b) Buthionine sulfoximine sensitizes antihormone-resistant human breast cancer cells to estrogen-induced apoptosis. Breast Cancer Res 10:R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JS, Meeke K, Osipo C, Ross EA, Kidawi N, Li T, Bell E, Chandel NS, Jordan VC. (2005a) Intrinsic mechanism of estradiol-induced apoptosis in breast cancer cells resistant to estrogen deprivation. J Natl Cancer Inst 97:1746–1759 [DOI] [PubMed] [Google Scholar]
- Lewis JS, Osipo C, Meeke K, Jordan VC. (2005b) Estrogen-induced apoptosis in a breast cancer model resistant to long-term estrogen withdrawal. J Steroid Biochem Mol Biol 94:131–141 [DOI] [PubMed] [Google Scholar]
- Lewis JS, Thomas TJ, Pestell RG, Albanese C, Gallo MA, Thomas T. (2005c) Differential effects of 16alpha-hydroxyestrone and 2-methoxyestradiol on cyclin D1 involving the transcription factor ATF-2 in MCF-7 breast cancer cells. J Mol Endocrinol 34:91–105 [DOI] [PubMed] [Google Scholar]
- Lewis JS, Vijayanathan V, Thomas TJ, Pestell RG, Albanese C, Gallo MA, Thomas T. (2005d) Activation of cyclin D1 by estradiol and spermine in MCF-7 breast cancer cells: a mechanism involving the p38 MAP kinase and phosphorylation of ATF-2. Oncol Res 15:113–128 [DOI] [PubMed] [Google Scholar]
- Maximov PY, Myers CB, Curpan RF, Lewis-Wambi JS, Jordan VC. (2010) Structure-function relationships of estrogenic triphenylethylenes related to endoxifen and 4-hydroxytamoxifen. J Med Chem 53:3273–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CP, Collini MD, Tran BD, Harris HA, Kharode YP, Marzolf JT, Moran RA, Henderson RA, Bender RH, Unwalla RJ, et al. (2001) Design, synthesis, and preclinical characterization of novel, highly selective indole estrogens. J Med Chem 44:1654–1657 [DOI] [PubMed] [Google Scholar]
- Miller PD, Chines AA, Christiansen C, Hoeck HC, Kendler DL, Lewiecki EM, Woodson G, Levine AB, Constantine G, Delmas PD. (2008) Effects of bazedoxifene on BMD and bone turnover in postmenopausal women: 2-yr results of a randomized, double-blind, placebo-, and active-controlled study. J Bone Miner Res 23:525–535 [DOI] [PubMed] [Google Scholar]
- Murphy CS, Pink JJ, Jordan VC. (1990) Characterization of a receptor-negative, hormone-nonresponsive clone derived from a T47D human breast cancer cell line kept under estrogen-free conditions. Cancer Res 50:7285–7292 [PubMed] [Google Scholar]
- Pink JJ, Bilimoria MM, Assikis J, Jordan VC. (1996) Irreversible loss of the oestrogen receptor in T47D breast cancer cells following prolonged oestrogen deprivation. Br J Cancer 74:1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pink JJ, Jiang SY, Fritsch M, Jordan VC. (1995) An estrogen-independent MCF-7 breast cancer cell line which contains a novel 80-kilodalton estrogen receptor-related protein. Cancer Res 55:2583–2590 [PubMed] [Google Scholar]
- Pink JJ, Jordan VC. (1996) Models of estrogen receptor regulation by estrogens and antiestrogens in breast cancer cell lines. Cancer Res 56:2321–2330 [PubMed] [Google Scholar]
- Pinkerton JV, Archer DF, Utian WH, Menegoci JC, Levine AB, Chines AA, Constantine GD. (2009) Bazedoxifene effects on the reproductive tract in postmenopausal women at risk for osteoporosis. Menopause 16:1102–1108 [DOI] [PubMed] [Google Scholar]
- Said TK, Conneely OM, Medina D, O'Malley BW, Lydon JP. (1997) Progesterone, in addition to estrogen, induces cyclin D1 expression in the murine mammary epithelial cell, in vivo. Endocrinology 138:3933–3939 [DOI] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927–937 [DOI] [PubMed] [Google Scholar]
- Silverman SL, Christiansen C, Genant HK, Vukicevic S, Zanchetta JR, de Villiers TJ, Constantine GD, Chines AA. (2008) Efficacy of bazedoxifene in reducing new vertebral fracture risk in postmenopausal women with osteoporosis: results from a 3-year, randomized, placebo-, and active-controlled clinical trial. J Bone Miner Res 23:1923–1934 [DOI] [PubMed] [Google Scholar]
- Sommer S, Fuqua SA. (2001) Estrogen receptor and breast cancer. Semin Cancer Biol 11:339–352 [DOI] [PubMed] [Google Scholar]
- Sutherland RL, Musgrove EA. (2004) Cyclins and breast cancer. J Mammary Gland Biol Neoplasia 9:95–104 [DOI] [PubMed] [Google Scholar]
- Wärnmark A, Treuter E, Gustafsson JA, Hubbard RE, Brzozowski AM, Pike AC. (2002) Interaction of transcriptional intermediary factor 2 nuclear receptor box peptides with the coactivator binding site of estrogen receptor alpha. J Biol Chem 277:21862–21868 [DOI] [PubMed] [Google Scholar]
- Wilcken NR, Prall OW, Musgrove EA, Sutherland RL. (1997) Inducible overexpression of cyclin D1 in breast cancer cells reverses the growth-inhibitory effects of antiestrogens. Clin Cancer Res 3:849–854 [PubMed] [Google Scholar]
- Wu X, Hawse JR, Subramaniam M, Goetz MP, Ingle JN, Spelsberg TC. (2009) The tamoxifen metabolite, endoxifen, is a potent antiestrogen that targets estrogen receptor alpha for degradation in breast cancer cells. Cancer Res 69:1722–1727 [DOI] [PubMed] [Google Scholar]
- Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. (1997) CDK-independent activation of estrogen receptor by cyclin D1. Cell 88:405–415 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.