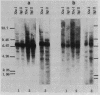
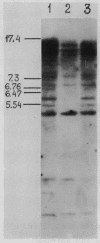
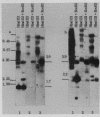
Abstract
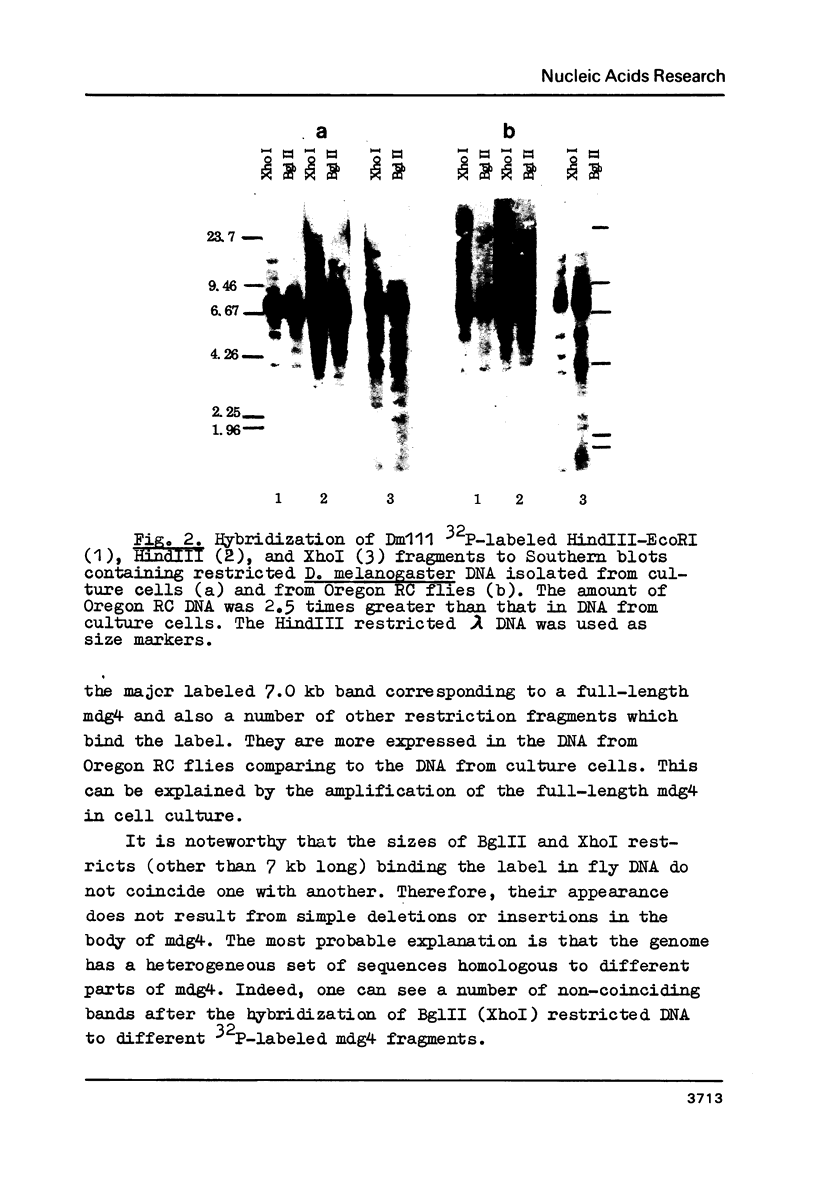
A mobile dispersed genetic element, mdg4 , approximately 7.5 kilobases (kb) long has been cloned from D. melanogaster genome. Chromosomal bands have only few sites of mdg4 , but it always hybridizes to the chromocenter. The location of mdg4 varies among D. melanogaster strains. Blot hybridization shows that, in contrast to other mdg elements, mdg4 sequences are rather heterogeneous. Only few copies are full-length. A strong amplification of mdg4 has occurred during the in vitro cultivation of cells involving only one mdg4 variant. Long terminal repeats (LTRs) and flanking sequences have been sequenced in two cloned copies of transposable element mdg4 . In both cloned copies of mdg4 , LTRs have an identical nucleotide sequence 479 bp long. The mdg4 is flanked by four-base-pair direct repeats, short mismatched palindromes being present at the ends of each LTR. The termini of the mdg4 body contain an oligopurine stretch and a region partially complementary to D. melanogaster tRNA-Lys. Thus, structural organization of mdg4 LTRs is similar to that of several other mdg elements and retroviral proviruses.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ananiev E. V., Gvozdev V. A., Ilyin Yu V., Tchurikov N. A., Georgiev G. P. Reiterated genes with varying location in intercalary heterochromatin regions of Drosophila melanogaster polytene chromosomes. Chromosoma. 1978 Dec 21;70(1):1–17. doi: 10.1007/BF00292211. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Brown A. J. Variation at the 87A heat shock locus in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5350–5354. doi: 10.1073/pnas.80.17.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower L. A., Huang A. L., Hager G. L. Regulatory and coding potential of the mouse mammary tumor virus long terminal redundancy. J Virol. 1981 Jan;37(1):226–238. doi: 10.1128/jvi.37.1.226-238.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Ish-Horowicz D. Extrachromosomal circular copies of the eukaryotic transposable element copia in cultured Drosophila cells. Nature. 1981 Aug 13;292(5824):591–595. doi: 10.1038/292591a0. [DOI] [PubMed] [Google Scholar]
- Georgiev G. P., Ilyin Y. V., Chmeliauskaite V. G., Ryskov A. P., Kramerov D. A., Skryabin K. G., Krayev A. S., Lukanidin E. M., Grigoryan M. S. Mobile dispersed genetic elements and other middle repetitive DNA sequences in the genomes of Drosophila and mouse: transcription and biological significance. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):641–654. doi: 10.1101/sqb.1981.045.01.082. [DOI] [PubMed] [Google Scholar]
- Ilyin Y. V., Chmeliauskaite V. G., Ananiev E. V., Lyubomirskaya N. V., Kulguskin V. V., Bayev A. A., Jr, Georgiev G. P. Mobile dispersed genetic element MDG1 of Drosophila melanogaster: structural organization. Nucleic Acids Res. 1980 Nov 25;8(22):5333–5346. doi: 10.1093/nar/8.22.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyin Y. V., Chmeliauskaite V. G., Georgiev G. P. Double-stranded sequences in RNA of Drosophila melanogaster: relation to mobile dispersed genes. Nucleic Acids Res. 1980 Aug 11;8(15):3439–3457. doi: 10.1093/nar/8.15.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyin Y. V., Tchurikov N. A., Ananiev E. V., Ryskov A. P., Yenikolopov G. N., Limborska S. A., Maleeva N. E., Gvozdev V. A., Georgiev G. P. Studies on the DNA fragments of mammals and Drosophila containing structural genes and adjacent sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):959–969. doi: 10.1101/sqb.1978.042.01.097. [DOI] [PubMed] [Google Scholar]
- Kugimiya W., Ikenaga H., Saigo K. Close relationship between the long terminal repeats of avian leukosis-sarcoma virus and copia-like movable genetic elements of Drosophila. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3193–3197. doi: 10.1073/pnas.80.11.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulguskin V. V., Ilyin Y. V., Georgiev G. P. Mobile dispersed genetic element MDG1 of Drosophila melanogaster: nucleotide sequence of long terminal repeats. Nucleic Acids Res. 1981 Jul 24;9(14):3451–3464. doi: 10.1093/nar/9.14.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. E., Varmus H. E. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981 Jan 22;289(5795):253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modolell J., Bender W., Meselson M. Drosophila melanogaster mutations suppressible by the suppressor of Hairy-wing are insertions of a 7.3-kilobase mobile element. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1678–1682. doi: 10.1073/pnas.80.6.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter S. S., Brorein W. J., Jr, Dunsmuir P., Rubin G. M. Transposition of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):415–427. doi: 10.1016/0092-8674(79)90168-5. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. Sequence at the 3' end of globin mRNA shows homology with immunoglobulin light chain mRNA. Nature. 1974 Nov 29;252(5482):359–362. doi: 10.1038/252359a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Scherer G., Tschudi C., Perera J., Delius H., Pirrotta V. B104, a new dispersed repeated gene family in Drosophila melanogaster and its analogies with retroviruses. J Mol Biol. 1982 May 25;157(3):435–451. doi: 10.1016/0022-2836(82)90470-3. [DOI] [PubMed] [Google Scholar]
- Shaw J. E., Huang E. S., Pagano J. S. Iodination of herpesvirus nucleic acids. J Virol. 1975 Jul;16(1):132–140. doi: 10.1128/jvi.16.1.132-140.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T., Saigo K. Retrovirus-like particles containing RNA homologous to the transposable element copia in Drosophila melanogaster. Nature. 1983 Mar 10;302(5904):119–124. doi: 10.1038/302119a0. [DOI] [PubMed] [Google Scholar]
- Shimotohno K., Mizutani S., Temin H. M. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980 Jun 19;285(5766):550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):429–439. doi: 10.1016/0092-8674(79)90169-7. [DOI] [PubMed] [Google Scholar]
- Swanstrom R., DeLorbe W. J., Bishop J. M., Varmus H. E. Nucleotide sequence of cloned unintegrated avian sarcoma virus DNA: viral DNA contains direct and inverted repeats similar to those in transposable elements. Proc Natl Acad Sci U S A. 1981 Jan;78(1):124–128. doi: 10.1073/pnas.78.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchurikov N. A., Ilyin Y. V., Skryabin K. G., Ananiev E. V., Bayev A. A., Jr, Krayev A. S., Zelentsova E. S., Kulguskin V. V., Lyubomirskaya N. V., Georgiev G. P. General properties of mobile dispersed genetic elements in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 2):655–665. doi: 10.1101/sqb.1981.045.01.083. [DOI] [PubMed] [Google Scholar]
- Thuring R. W., Sanders J. P., Borst P. A freeze-squeeze method for recovering long DNA from agarose gels. Anal Biochem. 1975 May 26;66(1):213–220. doi: 10.1016/0003-2697(75)90739-3. [DOI] [PubMed] [Google Scholar]
- Will B. M., Bayev A. A., Finnegan D. J. Nucleotide sequence of terminal repeats of 412 transposable elements of Drosophila melanogaster. A similarity to proviral long terminal repeats and its implications for the mechanism of transposition. J Mol Biol. 1981 Dec 25;153(4):897–915. doi: 10.1016/0022-2836(81)90458-7. [DOI] [PubMed] [Google Scholar]