Abstract
Background
It has been hypothesized that vitamin D deficiency (VDD) contributes to the development of food sensitization (FS) and then food allergy. However, the epidemiological evidence is conflicting. We aim to examine if cord blood VDD is associated with FS and if such association can be modified by genetic variants in a prospective birth cohort.
Methods
This study included 649 children who were enrolled at birth and followed from birth onward at the Boston Medical Center. We defined VDD as cord blood 25(OH)D < 11ng/ml, and FS as specific IgE ≥ 0.35kUA/L to any of eight common food allergens in early childhood. We genotyped potentially functional single nucleotide polymorphisms (SNPs) in 11 genes known to be involved in regulating IgE and 25(OH)D concentrations. Logistic regressions were used to test the effects of VDD on FS individually and jointly with SNPs.
Results
Among the 649 children, 44% had VDD and 37% had FS. When examined alone, VDD was not associated with FS. When examined jointly with SNPs, a significant interaction between IL4 gene polymorphism (rs2243250) and VDD (pinteraction=0.003, pFDR=0.10) was found: VDD increased the risk of FS among children carrying CC/CT genotypes (OR=1.79, 95%CI: 1.15–2.77). Similar but weaker interactions were observed for SNPs in MS4A2 (rs512555), FCER1G (rs2070901), and CYP24A1 (rs2762934). When all four SNPs were simultaneously considered, a strong gene-VDD interaction was evident (pinteraction=9×10−6).
Conclusions
Our data demonstrate that VDD may increase the risk of FS among individuals with certain genotypes, providing evidence of gene-vitamin D interaction on FS.
Keywords: cord blood plasma 25(OH)D, food sensitization, gene-vitamin D deficiency interaction, SNP
INTRODUCTION
Food allergy, a condition caused by an immunoglobulin (Ig) E-mediated hypersensitivity reaction to food, affects approximately 5% of children and 3–4% of adults(1) and is a growing clinical and public health problem in the U.S. and worldwide. The etiology underlying food allergy remains unclear, although several hypotheses have been suggested(2). The focus of the present report was to test the vitamin D hypothesis.
Serum 25 hydroxyvitamin D (25(OH)D) reflects the end result of both dietary intake and cutaneous synthesis after sun exposure, and serves as the objective measure of an individual’s vitamin D status(3). Approximately 1 billion people worldwide have low vitamin D status (25(OH)D <30 ng/ml)(4). In the U.S., mean serum 25(OH)D concentrations dropped from 30 to 24ng/ml during the last decade(5). Some studies conducted in the northern U.S. also have demonstrated that a high proportion of mothers and their infants have vitamin D deficiency (VDD) with the cutoff values of cord blood 25(OH)D concentration ranging from <11ng/ml to <15ng/ml(6–9). VDD or insufficiency has emerged as a rediscovered global public health problem.
Previous epidemiological studies have evaluated the associations between vitamin D status and allergic diseases and associated phenotypes(10–20), but the findings have been inconsistent. One reason for this discrepancy is the use of different methods of vitamin D assessment across studies. Some studies have used maternal/infant supplement intake, which is not an objective measure of serum 25(OH)D. With solar exposure serving as a major source of vitamin D(3), dietary/supplement intake alone cannot fully reflect an individual’s vitamin D status. Another reason may be differences in study design since the majority are cross-sectional studies that are unable to determine temporal and causal association. Furthermore, the effect of vitamin D status on allergic diseases may only exist among a subgroup of subjects with particular genotypes. However, genetic susceptibility has yet to be considered in these studies. Finally, only a few vitamin D studies have focused on serum IgE levels(11, 16, 18, 20), an important intermediate phenotype of allergic diseases including food allergy. A recent large German birth cohort study demonstrated that early-onset and persistent FS were independent risk factors for physician-diagnosed food allergy at age 6(21). Experimental evidence also strongly suggests that the hormonally-active form of vitamin D, 1, 25 dihydroxy-vitamin D (1,25(OH)2D), could influence IgE production because of its regulatory effects on the immune system(22). We speculate that a careful evaluation of vitamin D status along with genetic susceptibility in relation to the development of FS in a prospective birth cohort study will provide new insight into the role of VDD in the development of FS and subsequent food allergy.
Using a large, well-established U.S. prospective birth cohort, we evaluated the relationship of cord blood 25(OH)D concentration and the development of FS in early childhood, with simultaneous consideration of individual genetic variants in 11 genes known to be involved in IgE synthesis (IL4, IL13, IL4RA, IL13RA1), regulation of IgE function through its receptor complex (FCER1A, MS4A2, FCER1G)(23, 24), and modulation of vitamin D metabolism (CYP27B1,CYP24A1, VDR, GC) (22, 25). We were particularly interested in whether individual genetic variations could modify the VDD-FS association, that is, if there are gene-VDD interactions on FS.
MATERIALS AND METHODS
Study Population
The Boston Birth Cohort is an ongoing study that enrolls mother-infant pairs at birth and prospectively follows the infants at the Boston Medical Center (BMC). A detailed description of the initial recruitment(26) and follow-up(27) has been previously published.
We studied 649 children for whom we currently have available information on cord blood 25(OH)D concentration, specific IgE to common food allergens and genotyping data of 11 candidate genes. The Children's Memorial Hospital (CMH) Institutional Review Board (IRB) in Chicago and the BMC IRB approved the study protocol.
Definition of Food Sensitization
Plasma specific IgE (sIgE) for 8 food allergens (milk, egg white, peanut, soy, shrimp, walnut, cod fish, and wheat) were measured using PhadiaImmunoCAP performed at Quest Diagnostics Nichols Institute, Chantilly, VA (CLIA 49D0221801). FS cases were defined as children with sIgE ≥ 0.35kUA/L to any one of the above food allergens. Non-food sensitized controls were defined as children without detectable sIgE (<0.35kUA/L).
Cord Blood Plasma 25(OH)D Measurement
Cord blood plasma total 25(OH)D concentrations (sum of 25(OH)D2 and 25(OH)D3) were measured using HPLC-MS/MS assay in a well-established laboratory(28). Since there is no universal criterion for defining VDD in cord blood, we used a cutoff value of 11 ng/ml, as specified for infants, neonates, and young children by the Institute of Medicine (IOM)(29).
Candidate Genes and Single Nucleotide Polymorphisms (SNPs) Selection
The primary focus of this report was to test if well-established genetic variants can modify VDD-FS relationships. To this end, we chose to examine 11 genes that are critical to the synthesis and regulation of plasma IgE and 25(OH)D. Specifically, we selected genes encoding interleukin-4 and interleukin-13 (IL4 and IL13) and their receptors (IL4R and IL13RA1) given that IL-4 and IL-13 are the only cytokines known to induce the isotype class switching to IgE(23). We also selected genes encoding the IgE receptor complex (FCER1A, MS4A2, and FCER1G) because of recently significant findings for gene FCER1A from a genome-wide association study (GWAS) of total serum IgE (tIgE) levels(24) and a follow-up candidate gene study(30). Finally, we included the main genes encoding the molecules essential for 25(OH)D metabolism and regulation (CYP27B1, CYP24A1, VDR, GC) (22, 25).
To assure statistical power, we examined potentially functional SNPs with minor allele frequency (MAF) >0.05 in African Americans or Yoruba populations. We selected potentially functional SNPs that were predicted by two bioinformatics tools: PupaSuite (http://pupasuite.bioinfo.cipf.es/) and FuncPred (http://manticore.niehs.nih.gov/snpfunc.htm). For example, we included SNPs that result in non-synonymous amino acid changes, and SNPs located in predicted transcription factor binding sites, exonic splicing enhancers, exonic splicing silencers, microRNAs and their targets, and DNA triplexes. After excluding SNPs with low Illumina design score (<0.60), a total of 40 SNPs were selected for testing gene-VDD interaction on FS.
Genotyping
SNPs were genotyped with the Illumina GoldenGate custom panel at the Genome Center, Washington University at St. Louis as detailed in our previous publication(31). Thirty-nine SNPs (Table S1) had a call rate >98.0% and were examined herein. We also genotyped 150 ancestry informative markers (AIMs), which are highly informative among three ancestral populations (African, European, Asian). Individual ancestral proportion was estimated from 144 AIMs with a call rate >98.0%, and then included as covariates in subsequent analyses(31).
Statistical analyses
We conducted a χ2 goodness of fit test to assess deviation from Hardy-Weinberg equilibrium (HWE) for SNPs on the autosomal chromosomes among non-sensitized African American children using the program Haploview (http://www.broadinstitute.org/haploview/haploview). Similar tests were performed for SNPs on the X chromosome but among non-sensitized African American girls only. Linkage disequilibrium (LD) between the SNPs of each gene also was calculated with Haploview. Three SNPs were removed from further analyses due to deviation from HWE (p<0.01) or because they had an MAF less than 5% or were in high LD with other SNPs (Table S1).
We first evaluated the effect of VDD itself on the risk of FS to any food, and FS to the three most common food allergens (egg white, milk, and peanut: >10% of the studied samples). We used a logistic regression model with and without the adjustment of important covariates, which included the child’s age and gender, maternal smoking after birth, breastfeeding, and ancestral proportion estimates. We also evaluated the interactions between VDD and each SNP on any FS by adding main effects and cross-product terms to the logistic regression after adjustment of the same covariates listed above. Statistical significance was tested via the likelihood ratio test comparing full and reduced models (with and without the interaction term). With consideration for the small number of minor allele homozygotes for some SNPs under additive coding, we grouped together subjects carrying a minor allele were grouped together to assure statistical power and to obtain stable estimates. We corrected for multiple testing using the false discovery rate (FDR) method (PROC MULTTEST). All p-values were derived from two-sided tests, and all analyses were undertaken with SAS software (version9.2; SAS Institute, Inc., Cary, North Carolina).
RESULTS
Epidemiological Characteristics of Study Children
Among the 649 children, 240 developed FS (37%). Compared with non-sensitized children, those sensitized to any food allergen were slightly older (yrs: 2.08 vs. 1.77), were more likely to be male, have a mother who smoked, and were less likely to be non-black and exclusively bottle-fed (p<0.05) (Table 1). These significant covariates (excluding race) together with individual ancestral proportions were included in the subsequent regression analyses. FS was significantly associated with physician diagnosis of eczema and food allergy but not with wheezing. Cord blood plasma 25(OH)D concentrations (ng/ml) appeared to be lower for children with any FS than for non-sensitized controls (12.52 vs. 13.83, p=0.14 based on t-test of log-transformed 25(OH)D). When cord blood plasma 25(OH)D was dichotomized using the IOM cutoff of 11ng/ml, 286 children (44%) had VDD (48% vs. 42% for FS and control children, respectively; p=0.09).
Table 1.
Epidemiological characteristics of 649 BMC subjects by food sensitization.
| Variables # | Food Sensitization (N=240) |
Non-Food Sensitization (N=409) |
|---|---|---|
| N (%) | ||
| Maternal Race * | ||
| Black | 151 (63) | 221 (54) |
| White | 8 (3) | 31 (8) |
| Hispanic | 48 (20) | 100 (24) |
| Others | 33 (14) | 55 (13) |
| Maternal BMI (kg/m2) (pre-pregnancy) | ||
| <20 | 26 (11) | 35 (9) |
| 20–25 | 81 (34) | 154 (38) |
| 25–30 | 78 (33) | 129 (32) |
| >=30 | 55 (23) | 88 (22) |
| Gestational Diabetes | 14 (6) | 13 (3) |
| Household Income | ||
| <$30,000 | 106 (44) | 174 (43) |
| ≥$30,000 | 27 (11) | 64 (16) |
| unknown | 107 (45) | 171 (42) |
| Maternal Atopy | 89 (37) | 138 (34) |
| Paternal Atopy | 46 (20) | 66 (17) |
| Infant Sex (Male) * | 135 (56) | 194 (47) |
| Preterm (< 37 GWs) | 41 (17) | 75 (18) |
| Birth Season | ||
| Winter (Dec. to Mar.) | 81 (34) | 123 (30) |
| Spring (April to June) | 57 (24) | 109 (27) |
| Summer (July to Sep.) | 54 (23) | 103 (25) |
| Fall (Oct. to Nov.) | 48 (20) | 71 (17) |
| Maternal Smoking (post-natal) * | 31 (13) | 80 (20) |
| Breast Feeding * | ||
| Breast feeding only | 14 (6) | 27 (7) |
| Formula only | 45 (19) | 116 (29) |
| Both | 181 (75) | 264 (65) |
| Vitamin D Deficiency (25(OH)D<11ng/ml) | 116 (48) | 170 (42) |
| Wheezing | 34 (14) | 46 (11) |
| Eczema * | 121 (50) | 156 (38) |
| Food Allergy * | 13 (6) | 9 (2) |
| Mean ± SD | ||
| Age (yr) * | 2.08 ± 1.88 | 1.77 ± 1.65 |
| Cord Blood 25(OH)D (ng/ml) & | 12.52 ± 6.41 | 13.83 ± 8.18 |
Number of missing data for birth season and BMI is 3; Numbers of missing data for maternal smoking (post-natal), breast feeding, maternal and paternal atopy, and food allergy are 1, 2, 4, 22, and 22, respectively.
P-values of the associations between the variables and FS ≤0.05.
P=0.14 based on t-test of log-transformed 25(OH)D.
Association between Vitamin D Deficiency and Food Sensitization
There were no significant associations between VDD and FS to any food, or FS to egg white, milk, peanut (Table 2). VDD only slightly increased the risk of FS to any food with the ORs of 1.16 (95%CI: 0.83–1.63) and 1.32 (95%CI: 0.95–1.81), with and without the adjustment of the covariates, respectively.
Table 2.
Associations between cord blood 25(OH)D and food sensitization in 649 BMC subjects.
| Cord Blood 25(OH)D |
Cases /Controls |
Crude OR (95%CI) |
Crude p-values |
Adjusted OR * (95%CI) |
Adjusted* p-values |
|---|---|---|---|---|---|
| Any Food Sensitization | |||||
| ≥ 11ng/ml | 124/239 | Ref | Ref | ||
| < 11ng/ml | 116/170 | 1.32 (0.95 – 1.81) | 0.09 | 1.16 (0.83 – 1.63) | 0.38 |
| Egg Sensitization | |||||
| ≥ 11ng/ml | 86/239 | Ref | Ref | ||
| < 11ng/ml | 55/170 | 0.90 (0.61 – 1.33) | 0.59 | 0.84 (0.56 – 1.27) | 0.41 |
| Milk Sensitization | |||||
| ≥ 11ng/ml | 72/239 | Ref | Ref | ||
| < 11ng/ml | 68/170 | 1.33 (0.90 – 1.95) | 0.15 | 1.15 (0.76 – 1.73) | 0.52 |
| Peanut Sensitization | |||||
| ≥ 11ng/ml | 43/124 | Ref | Ref | ||
| < 11ng/ml | 38/170 | 1.24 (0.77 – 2.01) | 0.37 | 1.06 (0.64 – 1.75) | 0.84 |
All the OR estimates were adjusted for age, sex, maternal smoking after birth, breast feeding, and ancestry proportion.
Gene-Vitamin D Deficiency Interactions on Food Sensitization
To assure statistical power for the tests of gene-vitamin D interactions, we focused on FS to any food for the interaction tests. We presented the genetic associations of 36 SNPs with FS in Table S1. None of the associations were significant after correcting for multiple testing. Table 3 presents the significant gene-VDD interactions on FS based on a nominal significance level of 0.05. Of note, one SNP, rs2243250 (C-590T) in the gene IL4, remained marginally significant with an FDR correction (pinteraction=0.003, pFDR = 0.1). Specifically, VDD increased the risk of FS among those carrying the rs2243250 C allele (OR=1.79, 95%CI: 1.15–2.78); and VDD decreased the risk of FS among children with the rs2243250 TT genotype, but this was not statistically significant (OR=0.60, 95%CI: 0.34–1.06).
Table 3.
Gene-vitamin D deficiency interactions on any food sensitization in 649 BMC subjects.
| Interleukin-4 (IL4) | |||||||
| rs2243250=TT | rs2243250=CC/CT | ||||||
| CBP 25(OH)D | Case/Control | OR (95%CI) | p-value | Case/Control | OR (95%CI) | p-value | pinteraction |
| >=11 ng/ml | 63/69 | Ref | 61/168 | ref | |||
| <11 ng/ml | 39/61 | 0.60 (0.34 – 1.06) | 0.08 | 77/107 | 1.79 (1.15 – 2.77) | 0.01 | 0.003 * |
| membrane-spanning 4-domains, subfamily A, member 2 (MS4A2) | |||||||
| rs512555=AA/AG | rs512555=GG | ||||||
| CBP 25(OH)D | Case/Control | OR (95%CI) | p-value | Case/Control | OR (95%CI) | p-value | pinteraction |
| >=11 ng/ml | 30/29 | Ref | 94/210 | ref | |||
| <11 ng/ml | 18/32 | 0.40 (0.17 – 0.98) | 0.04 | 98/138 | 1.43 (0.99 – 2.08) | 0.06 | 0.009 |
| IgE Fc receptor subunit gamma (FCER1G) | |||||||
| rs2070901=GG | rs2070901=TT/TG | ||||||
| CBP 25(OH)D | Case/Control | OR (95%CI) | p-value | Case/Control | OR (95%CI) | p-value | pinteraction |
| >=11 ng/ml | 49/84 | Ref | 75/154 | ref | |||
| <11 ng/ml | 38/68 | 0.69 (0.38 – 1.25) | 0.23 | 78/102 | 1.52 (1.00 – 2.33) | 0.05 | 0.04 |
| cytochrome P450, family 24, subfamily A, polypeptide 1 | |||||||
| rs2762934=GG | rs2762934=AA/AG | ||||||
| CBP 25(OH)D | Case/Control | OR (95%CI) | p-value | Case/Control | OR (95%CI) | p-value | pinteraction |
| >=11 ng/ml | 86/157 | Ref | 38/82 | ref | |||
| <11 ng/ml | 75/131 | 0.88 (0.58 – 1.32) | 0.53 | 41/38 | 2.29 (1.22 – 4.32) | 0.01 | 0.02 |
FDR-corrected p=0.10. All the OR estimates were adjusted for age, sex, maternal smoking after birth, breast feeding, and ancestry proportion.
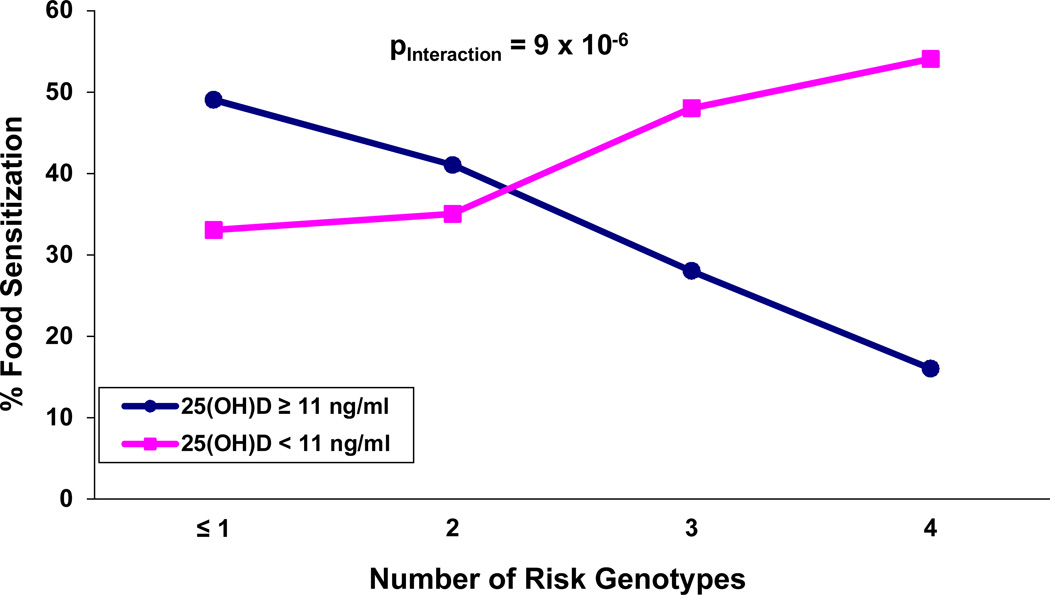
Similar interaction patterns were observed for the SNPs in MS4A2 (rs512555), FCER1G (rs2070901), and CYP24A1 (rs2762934) (Table 3). Given that qualitative interactions were found for all 4 SNPs in Table 3, we tested the interaction between VDD and the combined genotypes. We first defined risk genotypes as the groups in which VDD increased the risk of FS (right panel of Table 3) and then regrouped the subjects according to the number of such genotypes. The proportion of FS increased with an increasing number of risk genotypes among subjects having VDD. The opposite trend was observed for those having cord blood 25(OH)D ≥ 11ng/ml (Figure 1). The test for the combined SNPs-VDD interaction on FS was highly significant (pinteraction= 9x10−6).
Figure 1.
Interactive effect between the combined risk genotypesa of the genes IL4 (rs2243250), MS4A2 (rs512555), FCER1G (rs2070901), and CYP24A1 (rs2762934) with cord blood 25(OH)D on food sensitization in BMC birth cohort.
aRisk genotypes refer to the group for which vitamin D deficiency showed increased risk of food sensitization. They are CC/CT, GG, TT/TG and AA/AG for rs2243250, rs512555, rs2070901and, rs2762934 respectively. Pinteraction=9×10−6.
Further adjustment of maternal atopy or FS-associated phenotypes (having either eczema or food allergy) in the multivariate logistic models did not alter our findings (data not shown). In addition, we performed race-specific analyses (blacks vs. non-blacks) and found that gene-VDD interactions on FS appeared to be more pronounced in blacks than in non-blacks (Table S2) although the general pattern is similar.
DISCUSSION
This is the first study to evaluate the effect of low cord blood vitamin D and gene-VDD interactions on the development of FS during early childhood using a prospective birth cohort design. Consistent with previous studies, VDD was widespread and the incidence of FS was high in this urban U.S. birth cohort. We observed that VDD alone, as assessed in cord blood, was not associated with FS. However, when individual genetic susceptibility also was considered, VDD increased the risk of FS among children with certain high-risk genotypes. Our study may partially explain the conflicting results on VDD and allergic phenotypes in previous epidemiological studies. More importantly, it underscores the need to consider individual genetic susceptibility in assessing the effect of VDD in allergic diseases. Our findings, if confirmed, could ultimately help to assess individual risk in terms of both VDD and genotypes, and be used to design personalized health care that would maximize the benefits and minimize the risk.
Our findings on gene-VDD interactions may stimulate future laboratory investigations to better understand the molecular basis of VDD on FS. Previous studies have indicated that vitamin D could increase IgE production by shifting the Th1/Th2 balance toward Th2 dominance, or decrease IgE production through the inhibition of B cell proliferation/differentiation and induction of Treg cells and their suppressive capacity(22). The central role of IL-4 in IgE regulation also has been well recognized. The T allele of promoter polymorphism rs2243250 (C-590T) has been demonstrated to increase IL-4 promoter activity, and thus increase IgE production(32). Moreover, this SNP has been repeatedly reported to be associated with asthma and serum IgE in different populations(33). A negative association between rs2243250 and FS in 649 BMC subjects (predominantly blacks) was consistent with a previous report(33), although it was not statistically significant (Table S1). As such, mechanistic study underlying the observed IL4-VDD interaction on FS should be seriously considered in the near future.
A genetic variant, rs2427837 in gene FCER1A, which was a hit in a recent GWAS of total serum IgE levels (24), did not show a significant interaction with VDD for FS. Instead, we observed marginally significant interactions between VDD and SNPs in genes encoding the beta and gamma chains of the IgE high affinity receptor. The test for interaction between VDD and the combined genotypes of rs2243250 in IL4, rs512555 in MS4A2, rs2070901 in FCER1G, and rs2762934 in CYP24A1 appeared to be more substantial than an individual test, indicating that these four functional SNPs may jointly interact with VDD to influence the risk of FS, although the underlying biological mechanisms are unclear. This flip-flop pattern of association (Table 3 & Figure 1) is commonly observed in testing GxE interactions in asthma according to a recent review by Ober et al.(34). It underscores the need to examine VDD and multiple genetic polymorphisms simultaneously in order to elucidate the complex interplay of gene and VDD on FS. Our findings need to be replicated and underlying biological mechanisms need to be explored.
Our study is limited in several ways. The major limitations are the relatively modest sample size for testing the interaction effects on FS and the lack of replication. Our findings need to be confirmed in larger and independent studies. Also, this study only focused on 11 well-known candidate genes involved in IgE and 25(OH)D synthesis and regulation. Other genes that have been demonstrated to be associated with total serum IgE or IgE-mediated phenotypes or regulated by 1,25(OH)D, for example, HLA-DRB1, STAT6 and IL10 (25, 35), warrant further study. In addition, this study is the first attempt to exam the relationships of vitamin D, genes and FS with the adjustment for individual ancestral proportion. However, the confounding effect of population stratification could not be entirely excluded due to heterogeneous ancestral background in the Boston Birth Cohort. Finally, although cord serum 25(OH)D <11ng/ml has been suggested as a deficiency cutoff for neonates by the IOM, other cutoffs (15ng/ml(8) and 12ng/ml(9)) have been used before. Our findings remained similar when we used a cutoff of 12ng/ml (data not shown), but because close to half of our study subjects had cord blood 25(OH)D < 11ng/ml, the sample size was not enough to test the interactions at a 15ng/ml cutoff, and analyses among those with high vitamin D concentrations (25(OH)D>54 ng/ml(16)) could not be conducted. Interactions between log-transformed 25(OH)D concentrations and SNPs on FS were not explored because the assumption of linearity between 25(OH)D and FS in each genotype group may not be correct.
Our study has several strengths. This is a prospective birth cohort, which could overcome many drawbacks related to cross-sectional or retrospective studies. Objective and clinically relevant biomarkers for assessing vitamin D status in cord blood were used. Finally, this study expands previous studies by incorporating individual genotypes in the assessment of VDD-FS relationships.
In summary, we found that low cord blood vitamin D levels significantly increased the risk of FS among children carrying certain genotypes in a prospective urban U.S. birth cohort. Our study also raises the possibility that the simultaneous consideration of both VDD and individual genotypes may improve our ability to identify newborns who may be at high risk of developing FS and subsequent food allergy. If further confirmed in independent studies, our findings could lead to the design of targeted, cost-effective clinical and public health interventions with the goal of preventing or reducing the risk of FS and subsequent food allergy.
Supplementary Material
ACKNOWLEDGEMENTS
The parent study is supported in part by the March of Dimes PERI grants (PI: Wang, 20-FY02-56), NIEHS (PI: Wang, R21 ES011666), and NICHD (PI: Wang, R01 HD041702). The follow-up study is supported in part by the Food Allergy Initiative and the NIAID (PI Wang, R21AI079872, R21AI088609, U01AI090727) and the Department of Defense (PI Wang, W81XWH-10-1-0123). Drs. Liu and Arguelles are supported by the NIH/NCRR, through the Clinical and Translational Science Awards Program (CTSA), Northwestern University KL2RR025740. Dr. Liu also is supported by the NIAID (PI. Liu, R21AI087888). Dr. Kumar is supported by the NHLBI (PI: Kumar, K23HL093023).
Footnotes
Authors Contributions:
XL has the primary responsibility for this manuscript. XL, GW, XH, DW, HT, SZ, LA, RK, HW, RL, YZ, CP, KO, RS, PH, JP, HEP, CL, and XW all played a role in the conception, design, acquisition and analysis of data and interpretation of results. XL drafted the manuscript. HT, LA, RK, RS, PH, JP, CL, and XW have been involved in revision of the article. GW, XH, SZ, HW, RL, CP, KO, and HEP have been involved in lab and clinical data collection. All authors have read and approved the final manuscript.
None of the authors have a conflict of interest pertaining to this work.
REFERENCES
- 1.Sicherer SH, Sampson HA. Food allergy. J Allergy Clin Immunol. 2010;125(2) Suppl 2:S116–S125. doi: 10.1016/j.jaci.2009.08.028. [DOI] [PubMed] [Google Scholar]
- 2.Lack G. Epidemiologic risks for food allergy. J Allergy Clin Immunol. 2008;121(6):1331–1336. doi: 10.1016/j.jaci.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Holick MF. Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am J Clin Nutr. 2004;80(6) Suppl:1678S–1688S. doi: 10.1093/ajcn/80.6.1678S. [DOI] [PubMed] [Google Scholar]
- 4.Holick MF. Vitamin D deficiency. N Engl J Med. 2007;357(3):266–281. doi: 10.1056/NEJMra070553. [DOI] [PubMed] [Google Scholar]
- 5.Ginde AA, Liu MC, Camargo CA., Jr Demographic differences and trends of vitamin D insufficiency in the US population, 1988–2004. Arch Intern Med. 2009;169(6):626–632. doi: 10.1001/archinternmed.2008.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merewood A, Mehta SD, Grossman X, Chen TC, Mathieu JS, Holick MF, et al. Widespread vitamin D deficiency in urban Massachusetts newborns and their mothers. Pediatrics. 2010;125(4):640–647. doi: 10.1542/peds.2009-2158. [DOI] [PubMed] [Google Scholar]
- 7.Basile LA, Taylor SN, Wagner CL, Quinones L, Hollis BW. Neonatal vitamin D status at birth at latitude 32 degrees 72': evidence of deficiency. J Perinatol. 2007;27(9):568–571. doi: 10.1038/sj.jp.7211796. [DOI] [PubMed] [Google Scholar]
- 8.Bodnar LM, Simhan HN, Powers RW, Frank MP, Cooperstein E, Roberts JM. High prevalence of vitamin D insufficiency in black and white pregnant women residing in the northern United States and their neonates. J Nutr. 2007;137(2):447–452. doi: 10.1093/jn/137.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee JM, Smith JR, Philipp BL, Chen TC, Mathieu J, Holick MF. Vitamin D deficiency in a healthy group of mothers and newborn infants. Clin Pediatr (Phila) 2007;46(1):42–44. doi: 10.1177/0009922806289311. [DOI] [PubMed] [Google Scholar]
- 10.Back O, Blomquist HK, Hernell O, Stenberg B. Does vitamin D intake during infancy promote the development of atopic allergy? Acta Derm Venereol. 2009;89(1):28–32. doi: 10.2340/00015555-0541. [DOI] [PubMed] [Google Scholar]
- 11.Brehm JM, Celedon JC, Soto-Quiros ME, Avila L, Hunninghake GM, Forno E, et al. Serum vitamin D levels and markers of severity of childhood asthma in Costa Rica. Am J Respir Crit Care Med. 2009;179(9):765–771. doi: 10.1164/rccm.200808-1361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camargo CA, Jr, Rifas-Shiman SL, Litonjua AA, Rich-Edwards JW, Weiss ST, Gold DR, et al. Maternal intake of vitamin D during pregnancy and risk of recurrent wheeze in children at 3 y of age. Am J Clin Nutr. 2007;85(3):788–795. doi: 10.1093/ajcn/85.3.788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erkkola M, Kaila M, Nwaru BI, Kronberg-Kippila C, Ahonen S, Nevalainen J, et al. Maternal vitamin D intake during pregnancy is inversely associated with asthma and allergic rhinitis in 5-year-old children. Clin Exp Allergy. 2009;39(6):875–882. doi: 10.1111/j.1365-2222.2009.03234.x. [DOI] [PubMed] [Google Scholar]
- 14.Freishtat RJ, Iqbal SF, Pillai DK, Klein CJ, Ryan LM, Benton AS, et al. High prevalence of vitamin D deficiency among inner-city African American youth with asthma in Washington, DC. J Pediatr. 2010;156(6):948–952. doi: 10.1016/j.jpeds.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale CR, Robinson SM, Harvey NC, Javaid MK, Jiang B, Martyn CN, et al. Maternal vitamin D status during pregnancy and child outcomes. Eur J Clin Nutr. 2008;62(1):68–77. doi: 10.1038/sj.ejcn.1602680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hypponen E, Berry DJ, Wjst M, Power C. Serum 25-hydroxyvitamin D and IgE - a significant but nonlinear relationship. Allergy. 2009;64(4):613–620. doi: 10.1111/j.1398-9995.2008.01865.x. [DOI] [PubMed] [Google Scholar]
- 17.Miyake Y, Sasaki S, Tanaka K, Hirota Y. Dairy food, calcium and vitamin D intake in pregnancy, and wheeze and eczema in infants. Eur Respir J. 2010;35(6):1228–1234. doi: 10.1183/09031936.00100609. [DOI] [PubMed] [Google Scholar]
- 18.Nwaru BI, Ahonen S, Kaila M, Erkkola M, Haapala AM, Kronberg-Kippila C, et al. Maternal diet during pregnancy and allergic sensitization in the offspring by 5 yrs of age: a prospective cohort study. Pediatr Allergy Immunol. 2010;21(1 Pt 1):29–37. doi: 10.1111/j.1399-3038.2009.00949.x. [DOI] [PubMed] [Google Scholar]
- 19.Oren E, Banerji A, Camargo CA., Jr Vitamin D and atopic disorders in an obese population screened for vitamin D deficiency. J Allergy Clin Immunol. 2008;121(2):533–534. doi: 10.1016/j.jaci.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 20.Wjst M, Hypponen E. Vitamin D serum levels and allergic rhinitis. Allergy. 2007;62(9):1085–1086. doi: 10.1111/j.1398-9995.2007.01437.x. [DOI] [PubMed] [Google Scholar]
- 21.Schnabel E, Sausenthaler S, Schaaf B, Schafer T, Lehmann I, Behrendt H, et al. Prospective association between food sensitization and food allergy: results of the LISA birth cohort study. Clin Exp Allergy. 2010;40(3):450–457. doi: 10.1111/j.1365-2222.2009.03400.x. [DOI] [PubMed] [Google Scholar]
- 22.Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C. Vitamin D: modulator of the immune system. Curr Opin Pharmacol. 2010 doi: 10.1016/j.coph.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Jutel M, Akdis CA. T-cell subset regulation in atopy. Curr Allergy Asthma Rep. 2011;11(2):139–145. doi: 10.1007/s11882-011-0178-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4(8) doi: 10.1371/journal.pgen.1000166. e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosse Y, Lemire M, Poon AH, Daley D, He JQ, Sandford A, et al. Asthma and genes encoding components of the vitamin D pathway. Respir Res. 2009;10:98. doi: 10.1186/1465-9921-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Zuckerman B, Pearson C, Kaufman G, Chen C, Wang G, et al. Maternal cigarette smoking, metabolic gene polymorphism, and infant birth weight. Jama. 2002;287(2):195–202. doi: 10.1001/jama.287.2.195. [DOI] [PubMed] [Google Scholar]
- 27.Kumar R, Ouyang F, Story RE, Pongracic JA, Hong X, Wang G, et al. Gestational diabetes, atopic dermatitis, and allergen sensitization in early childhood. J Allergy Clin Immunol. 2009;124(5):1031–1038. doi: 10.1016/j.jaci.2009.06.052. e1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arguelles LM, Langman CB, Ariza AJ, Ali FN, Dilley K, Price H, et al. Heritability and environmental factors affecting vitamin D status in rural Chinese adolescent twins. J Clin Endocrinol Metab. 2009;94(9):3273–3281. doi: 10.1210/jc.2008-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietary reference intakes for calcium, phosphorus, magnesium, vitamin D, and fluoride. Washington: National Academy Press; 1997. Standing Committee on the Scientific Evaluation of Dietary Reference Intakes, Food and Nutrition Board, Institute of Medicine. [Google Scholar]
- 30.Chen CM, Weidinger S, Klopp N, Sausenthaler S, Bischof W, Herbarth O, et al. Common variants in FCER1A influence total serum IgE levels from cord blood up to six years of life. Allergy. 2009;64(9):1327–1332. doi: 10.1111/j.1398-9995.2009.02005.x. [DOI] [PubMed] [Google Scholar]
- 31.Hong X, Tsai HJ, Liu X, Arguelles L, Kumar R, Wang G, et al. Does genetic regulation of IgE begin in utero? Evidence from T(H)1/T(H)2 gene polymorphisms and cord blood total IgE. J Allergy Clin Immunol. 2010;126(5):1059–1067. doi: 10.1016/j.jaci.2010.08.029. 1067 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, et al. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy. 1995;25 Suppl 2:74–78. doi: 10.1111/j.1365-2222.1995.tb00428.x. discussion 95-6. [DOI] [PubMed] [Google Scholar]
- 33.Baye TM, Butsch Kovacic M, Biagini Myers JM, Martin LJ, Lindsey M, Patterson TL, et al. Differences in Candidate Gene Association between European Ancestry and African American Asthmatic Children. PLoS One. 2011;6(2) doi: 10.1371/journal.pone.0016522. e16522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ober C, Vercelli D. Gene-environment interactions in human disease: nuisance or opportunity? Trends Genet. 2011;27(3):107–115. doi: 10.1016/j.tig.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.