Summary
Chromosomal rearrangements, including translocations, require formation and joining of DNA double strand breaks (DSBs). These events disrupt the integrity of the genome and are frequently involved in producing leukemias, lymphomas and sarcomas. Despite the importance of these events, current understanding of their genesis is limited. To examine the origins of chromosomal rearrangements we developed Translocation Capture Sequencing (TC-Seq), a method to document chromosomal rearrangements genome-wide, in primary cells. We examined over 180,000 rearrangements obtained from 400 million B lymphocytes, revealing that proximity between DSBs, transcriptional activity and chromosome territories are key determinants of genome rearrangement. Specifically, rearrangements tend to occur in cis and to transcribed genes. Finally, we find that activation-induced cytidine deaminase (AID) induces the rearrangement of many genes found as translocation partners in mature B cell lymphoma.
Introduction
Lymphomas, leukemias, and solid tumors frequently carry gross genomic rearrangements, including chromosomal translocations (Kuppers, 2005; Nussenzweig and Nussenzweig, 2010; Tsai and Lieber, 2010; Tsai et al., 2008; Zhang et al., 2010). Recurrent chromosomal translocations are key pathogenic events in hematopoietic tumors and sarcomas; they may juxtapose proto-oncogenes to constitutively active promoters, delete tumor suppressors, or produce chimeric oncogenes (Rabbitts, 2009). For example, the c-myc/IgH translocation, a hallmark of human Burkitt’s lymphoma and mouse plasmacytomas, deregulates the expression of c-myc by bringing it under the control of Immunoglobulin (Ig) gene transcriptional regulatory elements (Gostissa et al., 2009; Kuppers, 2005; Potter, 2003). Alternatively, in chronic myeloid leukemia, the Bcr/Abl translocation fuses two disparate coding sequences to produce a novel, constitutively active tyrosine kinase (Goldman and Melo, 2003; Wong and Witte, 2004).
Chromosome translocation requires formation and joining of paired DNA double strand breaks (DSBs), a process that may be limited in part by the proximity of two breaks in the nucleus (Nussenzweig and Nussenzweig, 2010; Zhang et al., 2010). B lymphocytes are particularly prone to translocation-induced malignancy, and mature B cell lymphomas are the most common lymphoid cancer (Kuppers, 2005). This enhanced susceptibility appears to be the direct consequence of activation-induced cytidine deaminase (AID) expression in activated B cells (Nussenzweig and Nussenzweig, 2010). AID normally diversifies antibody genes by initiating Ig class switch recombination (CSR) and somatic hypermutation (SHM) (Muramatsu et al., 2000; Revy et al., 2000). It does so by deaminating cytosine residues in single-stranded DNA (ssDNA) exposed by stalled RNA polymerase II during transcription (Chaudhuri and Alt, 2004; Pavri et al., 2010; Storb et al., 2007). The resulting U:G mismatches are then processed by one of several repair pathways to yield mutations or DSBs, which are obligate intermediates in CSR, but may also serve as substrates for translocation (Di Noia and Neuberger, 2007; Honjo, 2002; Peled et al., 2008; Stavnezer et al., 2008). Although AID has a strong preference for targeting Ig genes, it also mutates a large number of non-Ig loci, including Bcl6, Pax5, miR142, Pim1, and c-myc (Gordon et al., 2003; Liu et al., 2008; Pasqualucci et al., 2001; Pavri et al., 2010; Robbiani et al., 2009; Shen et al., 1998; Yamane et al., 2011). While non-Ig gene mutation frequencies are low, it has been estimated that AID mutates as many as 25% of all genes expressed in germinal center B cells (Liu et al., 2008).
The full spectrum of potential AID targets was revealed by AID-chromatin immunoprecipitation studies, which showed AID occupancy at more than 5,000 gene promoters bearing stalled RNA polymerase II (Yamane et al., 2011). AID is targeted to these genes through its interaction with Spt5, an RNA polymerase stalling factor (Pavri et al., 2010). Consistent with its genome-wide distribution, mice that over-express AID exhibit chromosomal instability and develop translocation-associated lymphomas (Okazaki et al., 2003; Robbiani et al., 2009). Yet, c-myc is the only gene conclusively shown to translocate as a result of AID-induced DSBs (Ramiro et al., 2007; Robbiani et al., 2008). It has been estimated that up to 5% of activated primary B lymphocytes carry IgH fusions to unidentified partners which may or may not be selected during transformation (Franco et al., 2006; Jankovic et al., 2010; Ramiro et al., 2006; Robbiani et al., 2009; Wang et al., 2009; Yan et al., 2007). Additionally, recent deep-sequencing studies have revealed hundreds of genomic rearrangements within human cancers and documented their propensity to involve genes (Campbell et al., 2008; Pleasance et al., 2010a; Pleasance et al., 2010b; Stephens et al., 2009) However, the role of selection or other physiologic constraints in the genesis of these events is unclear because methods for mapping chromosomal translocations in primary cells do not yet exist.
Here we describe a novel, genome-wide strategy to document primary chromosomal rearrangements. We provide insight into the effects of genomic position and transcription on the genesis of chromosomal rearrangements and DSB resolution. Our data also reveal the extent of recurrent AID-mediated translocations in activated B cells.
Results
Translocation Capture Sequencing
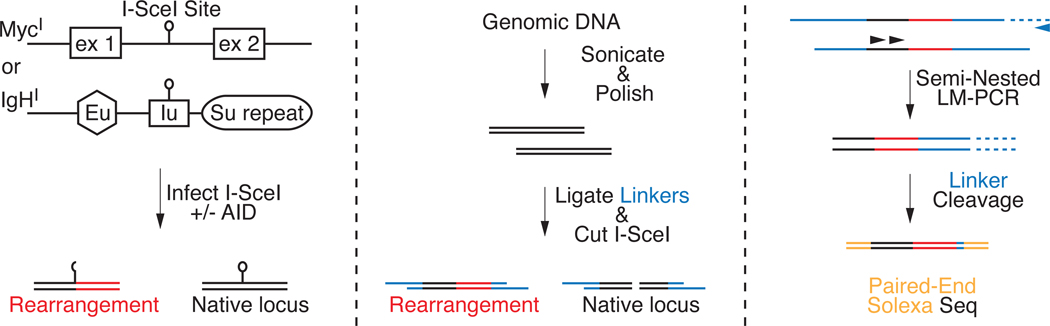
To discover the extent and nature of chromosomal rearrangements in activated B lymphocytes we developed an assay to capture and sequence rearranged genomic DNA (TC-Seq). In this system, DSBs are induced at the c-myc (chromosome 15) or IgH (chromosome 12) loci, which were engineered to harbor the I-SceI meganuclease target sequence (Robbiani et al., 2008). c-mycI-SceI/I-SceI or IgHI-SceI/I-SceI (hereafter referred to as MycI and IgHI) B cells were stimulated and infected with a retrovirus expressing I-SceI, in the presence or absence of AID. Rearrangements to I-SceI sites were recovered by semi-nested ligation-mediated PCR from genomic DNA that had been fragmented, A-tailed (to prevent intra-molecular ligation) and ligated to asymmetric DNA linkers (Figure 1). Site-specific primers were placed at least 150bp from the I-SceI site allowing for the capture of rearrangements involving moderate end-processing. PCR products were submitted for high-throughput paired-end sequencing and reads were aligned to the mouse genome. Identical reads were clustered as single events. Since sonication generates unique linker ligation points in each cell, this method allows for the study of independent events without sequencing through rearrangement breakpoints.
Figure 1. TC-Seq schematic.
IgHI or MycI primary B cells are infected with retroviruses encoding I-SceI with or without AID. Genomic DNA is fragmented, blunted, A-tailed, ligated to T-tailed asymmetric linkers and native loci are eliminated by I-SceI digestion. Rearrangements are amplified by semi-nested ligation-mediated PCR followed by linker cleavage and paired-end deep sequencing.
AID-independent translocations
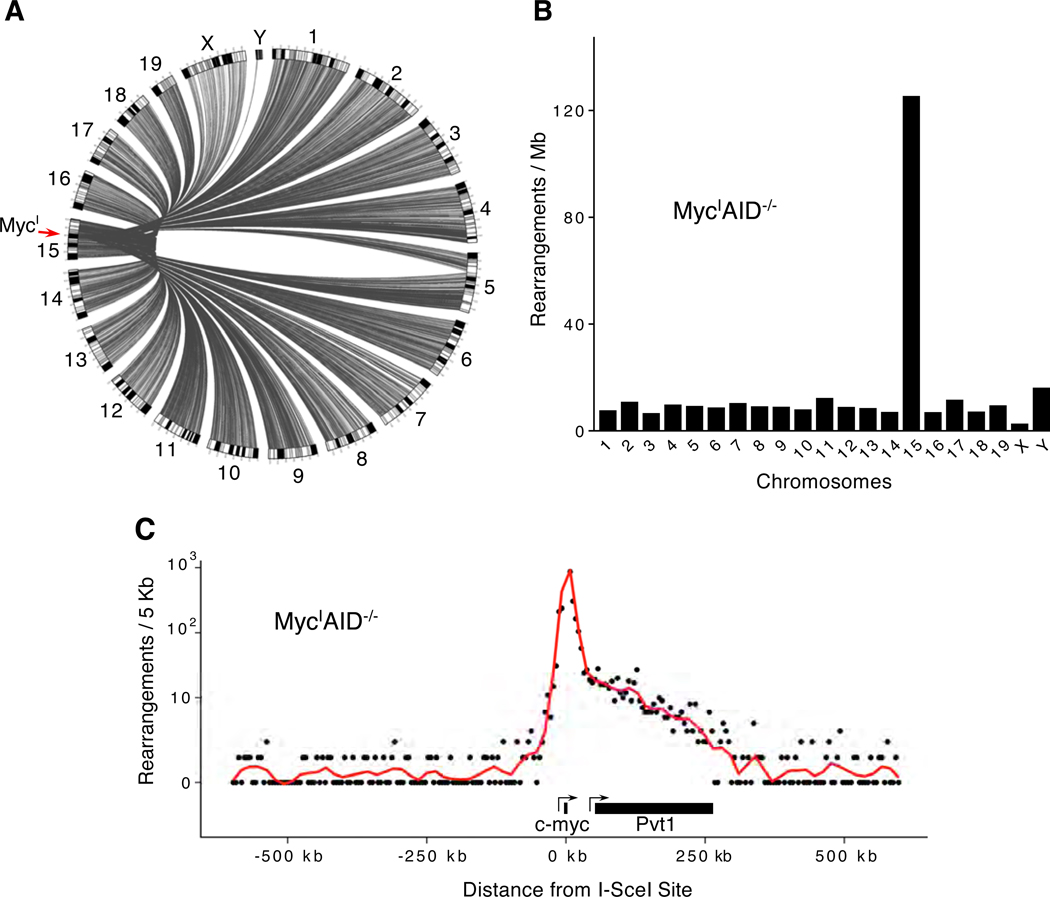
In the absence of AID, DSBs arise as by-products of normal cellular metabolism including transcription and DNA replication (Branzei and Foiani, 2010). Consistent with a global distribution of DSBs, we mapped 28,548 unique rearrangements between the I-SceI site and every chromosome in MycIAID−/− B cells (100 million cells assayed, Figure 2A). To determine whether there is a genome-wide bias for rearrangement, these events were characterized based on location, transcription and histone modification of the locus.
Figure 2. Rearrangements to a DSB site documented by TC-Seq.
(A) Genome-wide view of rearrangements to MycI in AID−/− B cells. (B) Rearrangements per mappable megabase to each chromosome in MycIAID−/− B cells. (C) Profile of rearrangements around the I-SceI site in 5 kb intervals.
We found a marked enrichment of intra-chromosomal rearrangements on chromosome 15, with approximately 125 events per mappable megabase (11,066 rearrangements), or ~40% of all events (Figure 2B). Translocations between MycI and other chromosomes were evenly distributed throughout the genome (Figure 2B and Table S1). Notably, 86.7% (9,591 of 11,066) of all intra-chromosomal rearrangements were localized within a 350 kb domain surrounding the I-SceI site (from −50 kb to +300 kb; Figure 2C). This is consistent with the observation that 92% of intra-chromosomal rearrangements in the breast cancer genome involve aberrant joining of DSBs within 2 Mb of each other (Stephens et al., 2009), and that 87% of RAG-mediated intra-chromosomal rearrangements in Abl-transformed pre-B cells lie within 200 kb of a recombination substrate (Mahowald et al., 2009). The asymmetrical distribution of events in the direction of c-myc transcription and the adjacent Pvt1 gene is also consistent with the idea that gene expression facilitates rearrangement (Thomas and Rothstein, 1989). I-SceI-proximal events may be the result of either resection and rejoining of I-SceI breaks, bona fide rearrangements between I-SceI and random DSBs, or a combination of DNA end resection and balanced translocations. Regardless of the precise molecular mechanism, the abundance of these events reveals a strong preference for DSBs to be resolved by ligation to a proximal sequence, a DNA repair strategy that may minimize gross genomic alterations.
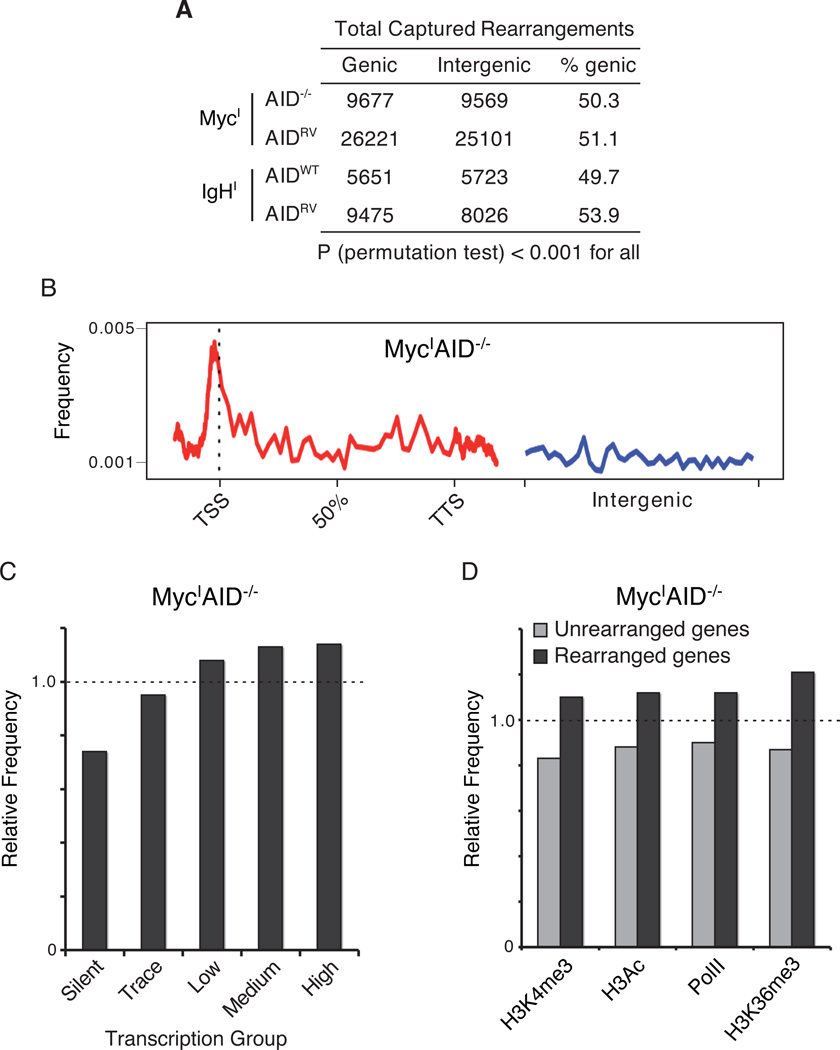
Recent cancer genome sequencing experiments uncovered a modest but highly significant preference for cancer-associated rearrangements to occur in genes, which compose only 41% of the human genome. For example, in 24 sequenced breast cancer genomes, 50% of all rearrangements involved genes (Stephens et al., 2009). Whether this bias resulted from selection or some inherent feature of DSB formation and repair specific to cancer cells could not be determined. To ascertain whether a similar bias is seen in primary cells in short term cultures, AID-independent rearrangements in MycIAID−/− B lymphocytes (excluding 1 Mb of DNA around the I-SceI site) were classified as genic or intergenic. Consistent with the human tumor studies, 51% (9,677 of 19,246) of the events were associated with genes (Figure 3A). Because only 40% of the mouse genome is genic, this represents a small (1.25-fold) but significant difference (permutation test P < 0.001) relative to intergenic regions. Moreover, the genic rearrangements were particularly enriched at transcription start sites (Figure 3B).
Figure 3. Rearrangements to MycI occur near the TSSs of actively transcribed genes.
(A) Rearrangements, excluding the 1 Mb around I-SceI, were categorized as genic or intergenic. (B) Composite density profile of genomic rearrangements from MycI to genes (red line) and intergenic (blue line) regions. TSS = transcription start site, TTS = transcription termination site. (C) Relative frequency (fe) of rearrangements in genes that are either silent or display trace, low, medium or high levels of transcription in activated B cells as determined by RNA-Seq (Figure S1). Dashed line indicates the expected rearrangement frequency based on a random model. P < 0.001 for all (permutation test). (D) Relative frequency of rearrangements in PolII- and activating histone mark-associated gene groups (Yamane et al., 2011). Dashed line indicates expected frequency based on a random model. P < 0.001 for all samples (permutation test). Also see Figure S1.
Consistent with the preference for genic rearrangements, we also observed a bias to transcribed genes. Fewer rearrangements than expected occurred at silent (fe = 0.74, P < 0.001) and trace (fe = 0.95, P < 0.001) transcribed genes, while more than expected occurred at low (fe = 1.08 P < 0.001), medium (fe = 1.13, P < 0.001), and highly (fe = 1.14, P < 0.001) transcribed genes (Figures 3C and S1). Additionally, rearrangements were enriched in genes bearing PolII and activating histone marks such as H3K4 trimethylation, H3 acetylation, and H3K36 trimethylation (P < 0.001, Figure 3D). Thus, there is a propensity for a DSB to recombine with gene rich regions of the genome and more specifically to transcription start sites of actively transcribed genes.
AID mediated lesions captured by TC-Seq
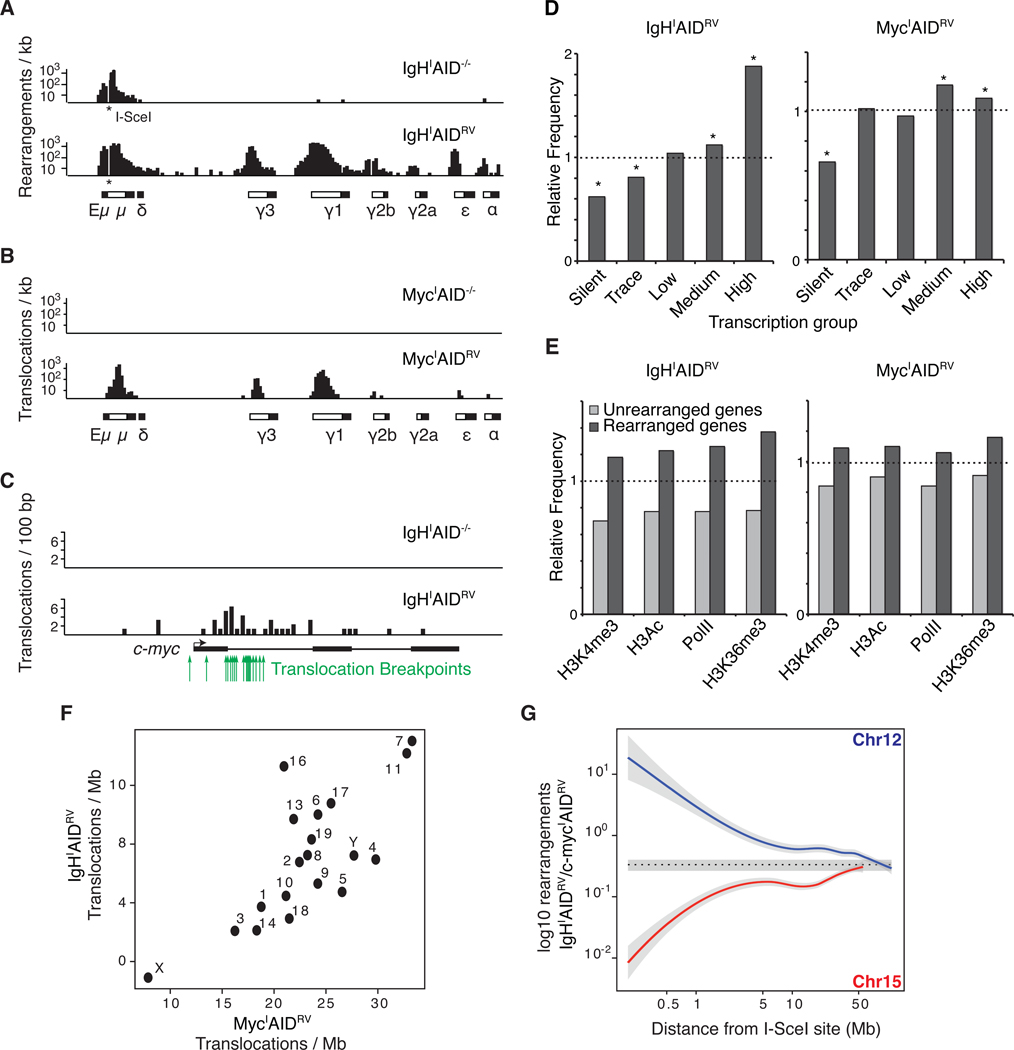
Processing of AID induced U:G mismatches can result in DSBs in Ig and non-Ig genes such as c-myc (Robbiani et al., 2008). To determine whether AID-mediated DSBs can be captured by TC-Seq we examined the IgH and c-myc loci in B cells expressing retrovirally encoded AID (IgHIAIDRV or MycIAIDRV). IgHI B cells expressing both I-SceI and AID showed extensive AID-dependent rearrangement between the I-SceI site and downstream switch (S) regions (Figure 4A). The frequency of rearrangements resembled the pattern of AID-mediated CSR in LPS+IL-4 cultures (e.g. IgG1>>IgG3>IgE), with 18,686 mapping to Sγ1, 3,192 to Sγ3, and 1,433 to Sε (Table S2). Furthermore, translocations between c-myc and IgH were entirely dependent on AID (Figures 4B and 4C). In two biological replicate samples totaling 100 million B cells, we observed 45 translocations from IgHI to c-myc (the I-SceI DSB was in IgH), and 3,463 from MycI to IgH (the I-SceI DSB was in c-myc) (Table S2). Additionally, TC-Seq tags mapping to c-myc from IgHI correlate well with c-myc/IgH translocation breakpoints sequenced from primary B cells (Figure 4C) (Robbiani et al., 2008). This suggests that TC-Seq reads are an accurate proxy for breakpoints. Furthermore, the data corroborate previous findings showing that AID induced breaks at c-myc are rate limiting for c-myc/IgH translocations (Robbiani et al., 2008) and suggest that AID-dependent IgH breaks are two orders of magnitude more frequent than those at c-myc. We conclude that TC-Seq captures rearrangements and translocations between DSBs in IgHI or MycI and known AID targets.
Figure 4. Rearrangements to IgHI or MycI in primary B cells expressing AID.
(A) Rearrangements per kb to I-SceI sites (indicated with an asterisk) in IgH or (B) c-myc in AID−/− (top panel) or AIDRV cells (bottom panel). White boxes in the schematics below each graph represent Ig switch domains while black boxes depict constant regions. (C) Translocations per 100 bp from IgHI to c-myc in AID−/− (top panel) or AIDRV cells (bottom panel). Green arrows indicate c-myc/IgH translocation breakpoints sequenced from primary B cells (Robbiani et al., 2008). (D) Relative frequency of rearrangements in transcription-level gene groups (Figure S1), dashed line indicates expected frequency based on a random model. Asterisks highlight values with a P < 0.001 (permutation test). (E) Relative frequency of rearrangements in PolII-associated or activating histone mark-associated gene groups (Yamane et al., 2011). Dashed line indicates the expected frequency based on a random model. P < 0.001 for all samples (permutation test). (F) Graph comparing the number of translocations per mappable megabase to each chromosome from IgHI (y axis) or MycI (x axis). (G) Ratio of IgHI captured to MycI captured events in 500 kb bins moving away from the I-SceI capture site (both directions combined). Dotted line represents the average trans-chromosomal joining rate computed on all chromosomes other than 12 or 15. Gray areas show 2 standard deviations around the mean. Also see Figure S2.
As was the case for AID deficient samples, MycIAIDRV and IgHIAIDRV libraries were enriched in intra-chromosomal rearrangements: 17% (10,633 of 63,772 total events) for MycI and 70% (36,019 of 51,312) for IgHI (Table S1 and Figures S2A and S2B). The difference in enrichment between the two was mostly the result of AID activity on chromosome 12, which generated a large number of rearrangements to IgH variable and constant domains (Figure 4A). Expression of AID did not alter the distribution of events around MycI, with 72.5% (7,707 of 10,633) mapping within −50 kb to 300 kb of the break (Figure S2C). A notable exception was an additional cluster of rearrangements associated with Pvt1 exon 5 (Figure S2C). These events coincided precisely with documented chromosomal translocations isolated from AID sufficient mouse plasmacytomas (Cory et al., 1985; Huppi et al., 1990) and likely represent an AID hotspot.
In agreement with the MycIAID−/− samples, translocations between IgHI or MycI and other chromosomes were evenly distributed throughout the genome, except for the MycI capture sample, which displayed a marked bias for chromosome 12 due to creation of DSBs at the IgH locus by AID (Figure S2A). Similar to MycIAID−/− samples, rearrangements in both cases were more likely to occur in regions that are genic, transcriptionally active, recruiting PolII, and associated with activating histone marks (Figures 3A, 4D and 4E). Furthermore, intragenic rearrangements were enriched at transcription start sites of genes (Figure S2D). In contrast to recent studies that used Nbs1 as an indirect marker of AID mediated damage (Staszewski et al., 2011), we found little or no difference in rearrangements to genomic repeats in the presence of AID (Table S3). Thus, AID does not dramatically alter the general profile of rearrangements.
Next, we examined whether IgHI and MycI capture DSB targets at similar rates. Indeed, in AID sufficient samples, total translocations from a given chromosome to IgHI or MycI occurred at roughly similar frequencies (Figure 4F). This similarity could be explained by the close physical proximity of IgH and c-myc, as suggested by studies with EBV-transformed B lymphoblastoid cells (Roix et al., 2003). Alternatively, the correlation in trans-chromosomal joining might represent random ligation between I-SceI DSBs in IgH or c-myc and DSBs on other chromosomes. We conclude that extra-chromosomal DSBs ligate DSBs in IgHI and c-mycI at similar rates.
To examine the nature of the intra-chromosomal rearrangement bias we calculated the ratio of IgHI to MycI captured events for each 500 kb segment of the genome and compared the values for chromosome 12 and 15 to the trans-chromosomal average (Figures 4G and S2E). This analysis revealed that DSBs are preferentially captured intra-chromosomally and this effect diminishes at a rate inversely proportional to the distance from the I-SceI site (d−1.29) (Figure S2F). This effect was most prominent locally but was evident at up to ~50 Mb away from the I-SceI break. We conclude that paired DSBs are preferentially joined intra-chromosomally and that the magnitude of this effect decreases with increasing distance between the two lesions.
Translocation hotspots
To determine whether there are hotspots for rearrangement, we searched the B cell genome for local accumulations of reads in AID deficient and sufficient samples. TC-Seq hotspots were defined as a localized enrichment of rearrangements above what is expected from a uniform genomic distribution. We removed likely artifacts; namely hotspots containing >80% of reads within DNA repeats, and those with footprints of <100nt (because translocations are amplified from randomly sonicated DNA (Figure 1), deep-sequence tags associated with bona fide rearrangements are unlikely to map within a small region).
We identified 34 hotspots captured by MycI in the absence of AID (Table S4). There were 31 hotspots in 17 genes and 3 in non-genic regions (Table S4). 17 of the hotspots were in Pvt1, within 500 kb of the I-SceI site. In addition, 2 hotspots occurred within 5 kb of cryptic I-SceI sites (each bearing one mismatch to the 18-base pair recognition sequence). For example, one such hotspot at chr15:16219195-16219312 containing a 1-off I-SceI recognition sequence bore 8 rearrangements (Figure S3). A genome-wide search for rearrangements within 5 kb of cryptic I-SceI sites (83 within the mouse genome with 1 or 2 mismatches to the canonical I-SceI recognition sequence) yielded 57 events, 7 times more than expected in a random distribution model. When allowing up to 6 mismatches we find a total of 5 out of 17 AID-independent hotspots near putative cryptic I-SceI sites. Although I-SceI has been used to generate a unique DSB in gene targeting and DNA repair experiments, our data suggest that DNA recognition by I-SceI can be promiscuous in the mouse genome, as demonstrated for other yeast endonucleases (Argast et al., 1998).
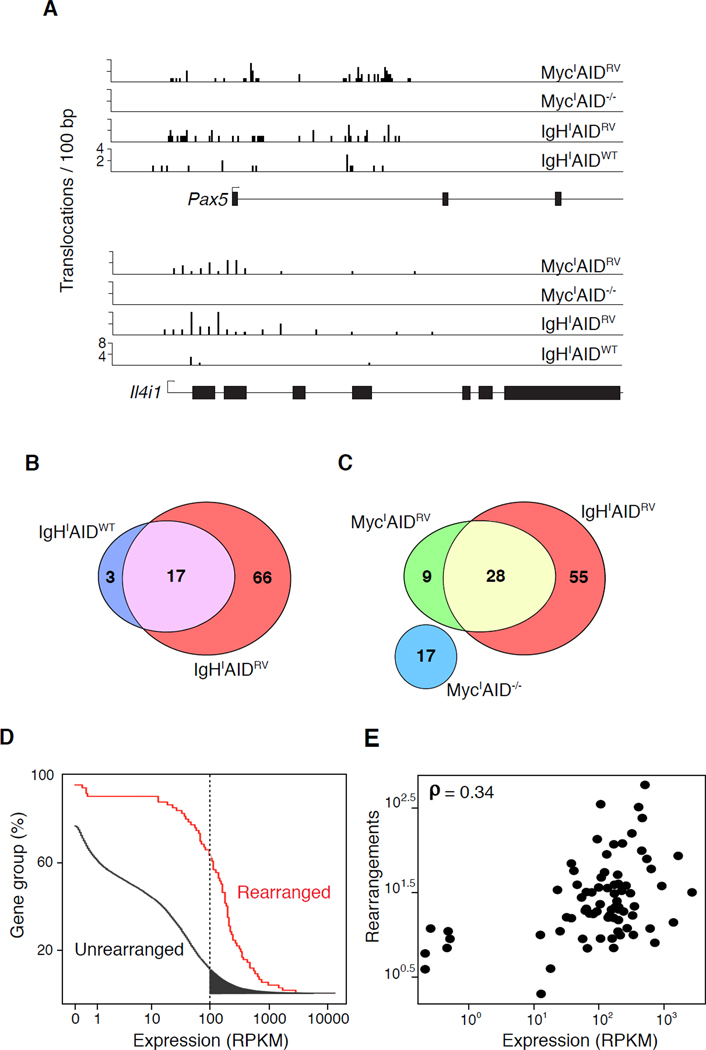
In contrast to AID−/−, we found 157 hotspots in 83 genes captured by IgHIAIDRV and 60 hotspots in 37 genes by MycIAIDRV in 100 million B cells. (Table S4). 80% of the hotspots captured by c-myc and 90% of those captured by IgH were within genes. For example, we found robust AID-dependent hotspots on Il4i1 and Pax5 (a recurring IgH translocation partner in lymphoplasmacytoid lymphoma (Kuppers, 2005)) (Figure 5A and Table S4). AID-dependent hotspots were similar for IgHI B cells expressing wild type levels (WT) or retrovirally over-expressed (RV) AID, however the number of TC-Seq captured events per hotspot was decreased in the former (Figures 5A, 5B, and Table S4). Therefore, translocations to AID targets occur in cells expressing physiological levels of AID and hotspots are not dependent on AID over-expression. We conclude that AID produces substrates for translocations in a number of discreet sites throughout the genome, and these sites are mainly in genes.
Figure 5. AID-dependent rearrangement hotspots.
(A) Screenshots of translocations per 100 bp present at Il4i1 and Pax5 genes in all samples. (B) Overlap of AID-dependent hotspot-bearing genes in IgHIAIDRV and IgHIAIDWT experiments. (C) Overlap of AID-dependent hotspot-bearing genes in IgHIAIDRV, MycIAID−/− and MycIAIDRV experiments. (D) Empirical cumulative distribution showing transcript abundance in genes displaying (red) or lacking (black) rearrangement hotspots. Filled-in gray slice represents ~2000 highly transcribed unrearranged genes. (E) Total rearrangements as a function of gene expression (RNA-Seq) (Yamane et al., 2011) in genes bearing rearrangement hotspots. Also see Figure S3.
Genes containing AID-dependent hotspots overlapped between IgHIAIDRV and MycIAIDRV samples (Figure 5C). Consistent with the similar capture rates observed for trans-chromosomal targets (Figure 4F), we found that 28 of the frequently translocated targets were shared (Table S5). In contrast, we found a number of unique intra-chromosomal AID-dependent hotspots. For example, rearrangements to Inf2 on chromosome 12 (~850 kb from IgHI) were only found by IgHI capture while rearrangements near Pvt1 on chromosome 15 (up to ~350 kb from MycI) were only found by MycI capture (Table S5). Thus, there is a bias towards recombination between I-SceI breaks and AID hotspots within the same chromosome. Additionally, the finding that some hotspots are only captured in cis indicates that TC-Seq underestimates the number of AID mediated DSBs in the genome and suggests that we have not reached saturation.
Combined analysis of the IgHI and MycI TC-Seq data sets shows that AID-dependent hotspots are primarily found in transcribed genes (Figure 5D). However, although nearly all of the translocated genes are actively transcribed, there is no clear correlation between transcript abundance and rearrangement frequency (Figure 5E). Furthermore, ~2000 highly transcribed genes are not rearranged (Figure 5D, shaded area). Therefore transcription is necessary but not sufficient for AID targeting, and transcription levels alone cannot account for AID-dependent DSBs.
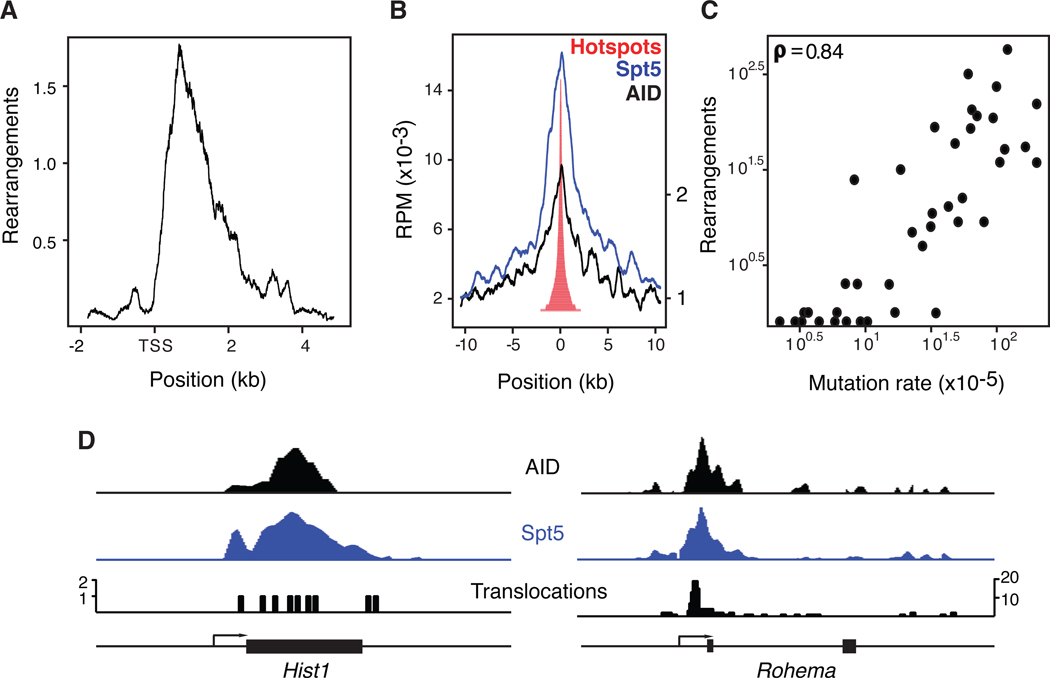
AID-dependent hotspots are biased to the region around the transcription start site (Figure 6A). This finding is consistent with the accumulation of AID and Spt5 around the promoters of stalled genes and the distribution of somatic hypermutation (Pavri et al., 2010; Yamane et al., 2011). Indeed, AID-dependent TC-Seq hotspots overlap with regions of AID (Figure S4A) and Spt5 accumulation (Figures 6B and 6D). This correlation prompted us to explore the relationship between AID activity and accumulation of chromosomal translocations by measuring somatic hypermutation at TC-Seq captured AID targets ((Yamane et al., 2011) and Table S6). We found a positive correlation (Spearman coefficient = 0.84) between hypermutation and rearrangement frequency (Figure 6C). All genes analyzed with a mutation rate over 10×10−5 bear rearrangements, and all genes with AID-dependent TC-Seq hotspots show mutations (Figure 6C). Rearrangements were only seen rarely in genes with lower rates of mutation (Figure 6C). This suggests that the rate of hypermutation and the frequency of AID-induced DSBs are directly proportional. We conclude that AID-dependent TC-Seq hotspots occur on stalled genes that accumulate Spt5, AID, and high rates of hypermutation.
Figure 6. Characterization of AID-dependent hotspots.
(A) Composite density graph showing the distribution of rearrangements in genes associated with AID-dependent hotspots relative to the TSS. (B) Spt5 and AID recruitment at genomic sites associated with translocation hotspots (Pavri et al., 2010; Yamane et al., 2011). (C) Somatic hypermutation frequency versus number of rearrangements in genes bearing AID-dependent translocation hotspots. (D) Distribution of AID, Spt5 and translocations in Rohema and Hist1 genes. Also see Figure S4.
Among AID-dependent hotspot containing genes we find several that are translocated or deleted in mature B cell lymphoma. These include Pax5/IgH, Pim1/Bcl6, Il21r/Bcl6, Gas5/Bcl6 and Ddx6/IgH translocations and Junb and Socs1 deletions in diffuse large B cell lymphoma, Birc3/Malt1 translocation in MALT lymphoma, Ccnd2/IgK translocation and Bcl2l11 deletion in mantle cell lymphoma, Aff3/Bcl2 and Grhpr/Bcl6 translocations in follicular lymphoma, mir142/c-myc translocation in B cell prolymphocytic leukemia as well as c-myc/IgH and Pvt1/IgK translocations in Burkitt’s lymphoma (Table 1). Interestingly, we find that AID is capable of inducing DSBs in Fli1 (Table S4), which is translocated to EWS in 90% of Ewing’s sarcomas, a malignant tumor of uncertain origin (Riggi and Stamenkovic, 2007). We conclude that in addition to mutating many genes, AID also initiates DSBs in numerous non-Ig genes. These genes serve as substrates for translocations associated with mature B-cell lymphoma, strongly implicating AID as a source of genomic instability in these cancers.
Table 1. AID-dependent translocations in human B cell lymphoma.
Genes bearing AID-dependent TC-Seq hotspots are shown with the associated translocation or deletion observed in human mature B cell lymphoma (BCL). BCL types are abbreviated as follows: MALT (mucosa-associated lymphoid tissue lymphoma), DLBCL (diffuse large B cell lymphoma), FL (follicular lymphoma), MCL (mantle cell lymphoma), BL (Burkitt’s lymphoma), B-PLL (B cell prolymphocytic leukemia). Asterisk indicates a review article.
| TC-Seq Gene | Translocation in Mature BCL | Mature BCL Type |
Reference |
|---|---|---|---|
| Birc3 (Api2) | t(11;18)(q21;q21) | MALT | (Rosebeck et al., 2011) |
| Il21r | t(3;16)(q27;p11) | DLBCL | (Ueda et al., 2002) |
| Pax5 | t(9;14)(p13;q32) | DBLCL | (Iida et al., 1999) |
| Pim1 | t(3;6)(q27;p21.2) | DLBCL | (Yoshida et al., 1999) |
| Aff3 | t(2;18)(q11.2;q21) | FL | (Impera et al., 2008) |
| Gas5 | t(1;3)(q25;q27) | DLBCL | (Nakamura et al., 2008) |
| Ccnd2 | t(2;12)(p12;p13) | MCL | (Gesk et al., 2006) |
| c-myc | t(8;14)(q23;q32) | BL | (Kuppers, 2005) |
| Ddx6 (Rck) | t(11;14)(q23;q32) | DLBCL | (Lu and Yunis, 1992) |
| Grhpr | t(3;9)(q27;p11) | FL | (Akasaka et al., 2003) |
| Bcl2l11 | Deleted | MCL | (Bea et al., 2009) |
| Socs1 | Deleted | DLBCL | (Mottok et al., 2009) |
| Junb | Deleted | DLBCL | (Mao et al., 2002) |
| mir142 | t(8;17) | B-PLL | (Gauwerky et al., 1989) |
| Pvt1 | t(2;8)(p11.2;q24.1) | BL | (Einerson et al., 2006) |
| IgH | several | several | (Kuppers, 2005) |
| IgK | several | several | (Kuppers, 2005) |
| IgL | several | several | (Kuppers, 2005) |
Akasaka, T., Lossos, I.S., and Levy, R. (2003). BCL6 gene translocation in follicular lymphoma: a harbinger of eventual transformation to diffuse aggressive lymphoma. Blood 102, 1443–1448.
Bea, S., Salaverria, I., Armengol, L., Pinyol, M., Fernandez, V., Hartmann, E.M., Jares, P., Amador, V., Hernandez, L., Navarro, A., et al. (2009). Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood 113, 3059–3069.
Einerson, R.R., Law, M.E., Blair, H.E., Kurtin, P.J., McClure, R.F., Ketterling, R.P., Flynn, H.C., Dogan, A., and Remstein, E.D. (2006). Novel FISH probes designed to detect IGK-MYC and IGL-MYC rearrangements in B-cell lineage malignancy identify a new breakpoint cluster region designated BVR2. Leukemia 20, 1790–1799.
Gauwerky, C.E., Huebner, K., Isobe, M., Nowell, P.C., and Croce, C.M. (1989). Activation of MYC in a masked t(8;17) translocation results in an aggressive B-cell leukemia. Proc Natl Acad Sci U S A 86, 8867–8871.
Gesk, S., Klapper, W., Martin-Subero, J.I., Nagel, I., Harder, L., Fu, K., Bernd, H.W., Weisenburger, D.D., Parwaresch, R., and Siebert, R. (2006). A chromosomal translocation in cyclin D1-negative/cyclin D2-positive mantle cell lymphoma fuses the CCND2 gene to the IGK locus. Blood 108, 1109–1110.
Iida, S., Rao, P.H., Ueda, R., Chaganti, R.S., and Dalla-Favera, R. (1999). Chromosomal rearrangement of the PAX-5 locus in lymphoplasmacytic lymphoma with t(9;14)(p13;q32). Leuk Lymphoma 34, 25–33.
Impera, L., Albano, F., Lo Cunsolo, C., Funes, S., Iuzzolino, P., Laveder, F., Panagopoulos, I., Rocchi, M., and Storlazzi, C.T. (2008). A novel fusion 5'AFF3/3'BCL2 originated from a t(2;18)(q11.2;q21.33) translocation in follicular lymphoma. Oncogene 27, 6187–6190.
Kuppers, R. (2005). Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer 5, 251–262.
Lu, D., and Yunis, J.J. (1992). Cloning, expression and localization of an RNA helicase gene from a human lymphoid cell line with chromosomal breakpoint 11q23.3. Nucleic Acids Res 20, 1967–1972.
Mao, X., Lillington, D., Child, F., Russell-Jones, R., Young, B., and Whittaker, S. (2002). Comparative genomic hybridization analysis of primary cutaneous B-cell lymphomas: identification of common genomic alterations in disease pathogenesis. Genes Chromosomes Cancer 35, 144–155.
Mottok, A., Renne, C., Seifert, M., Oppermann, E., Bechstein, W., Hansmann, M.L., Kuppers, R., and Brauninger, A. (2009). Inactivating SOCS1 mutations are caused by aberrant somatic hypermutation and restricted to a subset of B-cell lymphoma entities. Blood 114, 4503–4506.
Nakamura, Y., Takahashi, N., Kakegawa, E., Yoshida, K., Ito, Y., Kayano, H., Niitsu, N., Jinnai, I., and Bessho, M. (2008). The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet Cytogenet 182, 144–149.
Rosebeck, S., Madden, L., Jin, X., Gu, S., Apel, I.J., Appert, A., Hamoudi, R.A., Noels, H., Sagaert, X., Van Loo, P., et al. (2011). Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-kappaB activation. Science 331, 468–472.
Ueda, C., Akasaka, T., Kurata, M., Maesako, Y., Nishikori, M., Ichinohasama, R., Imada, K., Uchiyama, T., and Ohno, H. (2002). The gene for interleukin-21 receptor is the partner of BCL6 in t(3;16)(q27;p11), which is recurrently observed in diffuse large B-cell lymphoma. Oncogene 21, 368–376.
Yoshida, S., Kaneita, Y., Aoki, Y., Seto, M., Mori, S., and Moriyama, M. (1999). Identification of heterologous translocation partner genes fused to the BCL6 gene in diffuse large B-cell lymphomas: 5'-RACE and LA - PCR analyses of biopsy samples. Oncogene 18, 7994–7999.
Discussion
To date, the study of chromosomal aberrations has been primarily limited to events identified in tumors and tumor cell lines. Although we have learned a great deal about the importance of genomic rearrangements in cancer, it has not been possible to develop an understanding of the cellular and molecular requirements that govern their genesis. To examine genomic rearrangements in primary cells in short term cultures, we developed a technique to catalog these events by deep sequencing, TC-seq. Our results and analysis reveal the importance of transcription and physical proximity in recombinogenesis, and identifies hotspots for AID-mediated translocations in mature B cells.
Nuclear Proximity and Chromosomal Position
The existence of chromosome territories, regions in which individual chromosomes segregate, has been long proposed (Cremer and Cremer, 2001) and recently shown to be a key feature of genome organization (Lieberman-Aiden et al., 2009). Our analysis provides evidence that physical proximity and chromosome territories are partial determinants for joining of specific rearrangement partners. The effects of physical proximity are most evident in the 350 kb region around the DSB. In the absence of AID the plurality rearrangements fall in this region. This observation is consistent with the analysis of rearrangements in the breast cancer genome and suggests that the abundance of these events is independent of cancer specific selection (Stephens et al., 2009). Additionally, a preference for DSB repair within 350 kb matches the range of gamma-H2AX spreading from a DSB (Bothmer et al., 2011). This is consistent with the idea that the DNA damage response facilitates proximal rearrangement, a phenomenon most prominent at the IgH locus during CSR.
The magnitude of the effect of chromosome territories on rearrangement is far less prominent than proximal joining, but is consistent with recent genome mapping data obtained by high-throughput chromosome conformation capture (Hi-C) (Lieberman-Aiden et al., 2009). Intra-chromosomal joining bias is evident in the preferential joining of AID hotspots and non-hotspots on Chr12 and Chr15 with their respective I-SceI breaks. When compared to trans-chromosomal joining, the bias to intra-chromosomal rearrangements is evident even when DSBs are separated by as much as 50 Mb. In mouse, the mean autosome size is ~130 Mb, so a 50 Mb preference for intra-chromosomal joining on either side of a DSB will encompass nearly the entire average chromosome. We conclude that intra-chromosomal joining is preferred to trans-chromosomal joining.
Since this effect diminishes with distance, it is mediated by proximity, a likely consequence of local chromosome packing and nuclear chromosomal territories. A strong preference for proximal intra-chromosomal rearrangement minimizes gross genomic alterations. We propose that this may be an important feature of DSB repair regulation that maintains genomic integrity.
Transcription
Transcription is associated with increased rates of DNA damage and genome instability; these effects are likely mediated by a number of different mechanisms (Gottipati and Helleday, 2009). Transcription may expose ssDNA, which is susceptible to chemical or oxidative damage (Aguilera, 2002). Additionally, head-on collision of the replication and transcription machinery has been implicated in fork stalling and genomic instability (Takeuchi et al., 2003). Consistent with these ideas, TC-Seq reveals that transcription facilitates DNA rearrangement. In the case of the c-myc locus, transcription increases the size of the local area around a DSB that is available for recombination from 50 kb to 300 kb. Moreover, I-SceI breaks rearrange predominantly to transcribed genes genome-wide and more specifically to the TSS. Thus, exposed ssDNA may serve as a primary source of genomic instability. AID expression further reinforces this phenomenon by creating U:G mismatches in ssDNA at sites of PolII stalling downstream of the TSS (Pavri et al., 2010).
A bias for rearrangement between genic regions was also reported in recent studies of the cancer genome, but the role of transcription, transformation or selection in these events could not be evaluated (Stephens et al., 2009). Our experiments demonstrate that transcribed genic regions are over-represented in chromosomal rearrangements in primary cells in short-term cultures. In addition to being more susceptible to damage, this effect may be due to the increased physical proximity of transcribed regions to each other in the nucleus (Lieberman-Aiden et al., 2009). We speculate that this phenomenon may have consequences for tumorigenesis. The rearrangement of proto-oncogenes to transcribed regions may lead to their deregulation or produce hybrid entities that alter cellular metabolism.
AID and Chromosome Translocation
AID initiates SHM, CSR, and chromosome translocation by deaminating cytosine residues in ssDNA exposed by transcription (Chaudhuri and Alt, 2004; Di Noia and Neuberger, 2007; Nussenzweig and Nussenzweig, 2010; Peled et al., 2008; Stavnezer et al., 2008). AID targets the IgH locus and the TSSs of stalled genes through direct interaction with Spt5, a PolII stalling factor (Pavri et al., 2010), resulting in widespread somatic mutations (Yamane et al., 2011). Additionally, AID has been shown to initiate DSBs in non-Ig targets such as c-myc, and generates diverse translocations and chromosome breaks (Robbiani et al., 2008; Robbiani et al., 2009). However the precise relationships between AID and Spt5 occupancy, mutation, and translocations have not previously been investigated.
By capturing and sequencing chromosomal rearrangements, a readout for aberrantly resolved DSBs, we have gained insight into the mechanisms by which AID targets DNA for chromosomal rearrangement. First, we show that AID targets discreet sites in the genome for DSB. These sites are predominantly genic and actively transcribed. A recent study using Nbs1-ChIP as a surrogate for DNA damage suggested that AID targets repeat rich sequences (Staszewski et al., 2011). In contrast, we find no AID-dependent increase in rearrangements to repeats. Moreover, AID-dependent rearrangement hotspots predominantly occur in genes, not in or near repeat regions that are not transcribed. Hotspots that do fall in repeats (Figure S4B), are not AID-dependent and do not suffer somatic hypermutation (Table S6). While it is difficult to map short reads to repetitive sequences, these data suggest that rearrangements to repeats may be from AID-independent DSB.
While genes rearranged by AID are largely transcribed, expression and PolII accumulation do not correlate directly with rearrangement frequency suggesting that transcription is necessary but not rate-limiting for rearrangement. Reflecting the distribution of AID and its co-factor Spt5 in the genome (Pavri et al., 2010; Yamane et al., 2011), AID-dependent rearrangements occur mainly on transcription start sites of stalled genes that carry high levels of the PolII stalling factor Spt5. In addition, we find a strong and direct correlation between hypermutation and rearrangements, suggesting that genes susceptible to AID mediated recombinogenesis are a subset of the most highly mutated genes in the genome. Consistent with this notion, we show that Pax5, Il21r, Gas5, Ddx6, Birc3, Ccnd2, Aff3, Grhpr, c-myc, Pvt1, Bcl2l11, Socs1, mir142, Junb and Pim1, which are translocated or deleted in mature B cell lymphomas (Table 1) are among the more highly mutated AID targets and bear AID-dependent translocation hotspots. Our experiments were performed on in vitro stimulated B cells. Germinal center B cells will have an alternate gene expression profile that might influence the number and position of AID target sites. We conclude that in addition to hypermutation, AID is also a source of genomic instability in mature B cell lymphomas.
Finally, we note that TC-seq can be adapted for use in other cell types to study translocation biology in any tissue.
Experimental Procedures
B cell cultures, infections and sorting
Resting B lymphocytes were isolated from mouse spleens by immunomagnetic depletion with anti-CD43 MicroBeads (Miltenyi Biotech) and cultured at 0.5 × 106 cells/ml in RPMI supplemented with L-glutamine, sodium pyruvate, antibiotic/antimycotic, HEPES, 50 µM 2-mercaptoethanol (all from GIBCO-BRL), and 10% fetal calf serum (Hyclone). B cells were stimulated in the presence of 500ng/ml RP105 (BD Pharmingen), 25 µg/ml lipopolysaccharide (LPS) (Sigma) and 5 ng/ml mouse recombinant IL-4 (Sigma). Retroviral supernatants were prepared by cotransfection of BOSC23 cells with pCL-Eco and pMX-IRES-GFP-derived plasmids encoding for I-SceI-mCherry or AID-GFP with Fugene 6, 72 hr before infection. At 20 and 44 hr of lymphocyte culture, retroviral supernatants were added, and B cells were spinoculated at 1150 g for 1.5 hr in the presence of 10 µg/ml polybrene. For dual infection, separately prepared retroviral supernatants were added simultaneously on both days. After 4 hr at 37°C, supernatants were replaced with LPS and IL-4 in supplemented RPMI. At 96 hr from the beginning of their culture, singly infected B cells were collected and frozen in 10 million cell pellets at −80C. Dually infected B cells were sorted for double positive cells with a FACSAria instrument (Becton Dickson) then frozen down.
Translocation capture sequencing (TC-Seq)
Genomic DNA library preparation
5×10 million B cell aliquots were lysed in Proteinase K buffer (100mM Tris pH8, 0.2% SDS, 200mM NaCl, 5mM EDTA) and 50ul of 20mg/ml Proteinase K. Genomic DNA was extracted by phenol chloroform precipitation and fragmented by sonication (Bioruptor - Diagenode) to yield a 500–1350 bp distribution of DNA fragments. DNA was divided into (5ug) aliquots in 1.5mL eppendorf tubes. Each experiment consisted of genomic DNA from 50 million B cells in 50 × 5ug aliquots for a total of 250ug of fragmented genomic DNA per experiment. Subsequent reactions were performed individually on 5ug aliquots. DNA was blunted by End-It DNA Repair Kit (Epicentre), purified, then adenosine-tailed by Klenow fragment 3->5’ exo− (NEB) and purified. Fragments were ligated to 200pmol of annealed linkers (pLT + pLB) and unrearranged loci were eliminated by I-SceI digestion. Reactions were purified and pooled.
Rearrangement amplification
Pooled linker-ligated DNA was divided into 2 equal parts for semi-nested ligation-mediated PCR using either forward or reverse primers (to capture rearrangements to either side of the I-SceI break). All PCRs were performed using the Phusion Polymerase system (NEB). DNA was divided into 1ug aliquots and subjected to single-primer PCR with biotinylated pMycF1, pMycR1, pIghF1 or pIghR1 [1×(98C-1min) 12×(98C-15sec, 65C-30sec, 72C-45sec) 1×(72C-1min)]. Each reaction was spiked with pLinker and subjected to additional cycles of PCR [1×(98C-1min) 35×(98C-15sec, 65C-30sec, 72C-45sec) 1×(72C-5min)]. Forward and reverse PCR reactions were pooled separately. Higher molecular weight products were isolated by agarose gel electrophoresis and magnetic streptavidin bead purification. Semi-nested PCR was performed on the magnetic beads with pMycF2, pMycR2, pIghF2 or pIghR2 and pLinker [1×(98C-1min) 35×(98C-10sec, 65C-30sec, 72C-40sec) 1×(72C-5min)]. Higher molecular weight products were isolated by agarose gel electrophoresis.
Paired-end library preparation
Linkers were removed by AscI digestion. Fragments were blunted by End-It DNA Repair Kit (Epicentre), purified, adenosine-tailed and ligated to Illumina paired-end adapters. Higher molecular weight products were isolated by agarose gel electrophoresis and adapter-ligated fragments were enriched by 25 cycles of PCR with Illumina primers PE1.0 and PE2.0. Forward and reverse libraries for the same sample were mixed in equimolar ratios and sequenced by 36×36 or 54×54 paired end deep sequencing on an Illumina GAII.
TC-Seq Computational analysis
Read Alignment
Each end of the paired end sequences was matched against the relevant bait primer plus genomic sequences allowing up to two mismatches with bowtie (V 0.12.5, cite <PMID 19261174>; command line options: –v2). For read pairs longer than 2 × 36 nts, 10 nts were trimmed of the 3’ end of each read. Each read pair with a single match to one of the primers was then checked for a perfect match to the linker on the second arm. If the linker was present, this arm was designated a target arm, linker sequence was trimmed, and the remainder was aligned against the mouse genome (NCBI 37/mm9) with bowtie allowing up to 2 mismatches and requiring unique alignments in the best alignment stratum (command line options: -v2 --all --best --strata -m1). Exactly identical alignments (same position, same strand) were combined into a single putative translocation event and events supported by a single alignment were not considered in any analyses. We also removed putative translocation events closer than 1 kb to their respective bait. For hotspot analyses the exclusion limit was increased to 50 kb. Translocation positions were given as the position of the 5’ end of the read in the alignment. Data from technical and biological repeats were pooled to increase saturation (Table S7).
Mapping of translocation hotspots
A translocation hotspot was defined as a localized enrichment of translocation events above what is expected from the null hypothesis of uniform distribution of translocation events along the genome. To identify such hotspots, candidate regions were defined as locations containing consecutive translocations with distances shorter than expected from the mappable size of the mm9 genome assembly (P < 0.01 each as determined by a negative binomial test). For a candidate region to be called a hotspot it had to (1) have more than 3 translocations and (2) have at least one read from each of the two sides of the bait and (3) have at least 10% of the translocations come from each side of the bait and (4) have a combined P value less than 10−9 given the number of translocations and length of the region as determined by a negative binomial test. Hotspots with a large degree (>80%) of overlap with repeat regions, small footprints (<100nt) or less than 10-fold enrichment over the AID−/− control were removed. Analyses of RNA-Seq, chromatin modifications, AID-, PolII-, and Spt5-ChIP as well as the identification of cryptic I-SceI sites, TSSs, genic and intergenic domains were carried out in R (http://www.R-project.org).
Hypermutation analysis
CD43− splenocytes from IgkAID-Ung−/− or Aicda−/− mice were cultured at 0.1 × 106 cells/ml with LPS+IL-4, and 0.5 mg/ml of aCD180 (RP105) antibody (RP/14, BD Pharmingen). At 72 hrs cells were diluted 1:4 and cultured for another 48 hs. 50 ng of genomic DNA was amplified for 30 cycles with Phusion DNA polymerase (New England Biolabs) and specific primers. For nested PCR, two-20 cycle amplifications were performed with DMSO. The amplicon was cloned using PCR Zero blunt (Invitrogen) and sequenced.
Highlights.
A new method to map chromosomal rearrangements genome-wide in primary cells.
Transcription favors chromosome rearrangement.
Rearrangements reveal chromosome territories.
AID generates rearrangements to many genes, including proto-oncogenes.
Supplementary Material
Acknowledgements
We thank all the members of the Nussenzweig and Casellas labs for valuable input and advice, Klara Velinzon and Svetlana Mazel for FACSorting and David Bosque and Thomas Eisenreich for animal management. We also thank Scott Dewell of the Rockefeller Genomics Resource Center and Gustavo Gutierrez of the NIAMS genome facility for high-throughput sequencing and guidance, as well as Christopher Mason of the Weill Cornell Medical College for assistance with data analysis. I.A.K. was supported by NIH MSTP grant GM07739, and is a Cancer Research Institute Predoctoral Fellow and a William Randolph Hearst Foundation Fellow. A.B. is a Cancer Research Institute Predoctoral Fellow. This work was supported by NIH grant #AI037526 to M.C.N., NYSTEM #C023046 and the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health. M.C.N. is an HHMI investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers.
The TC-Seq datasets are deposited in SRA (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA039959.
I.A.K. designed and performed experiments and analysis and wrote the manuscript. W.R. designed and performed data analysis. M.J. performed TC-Seq experiments. T.O. designed and performed data analysis. A.Y. and H.N. performed hypermutation sequencing and analysis. M.D.V. and A.B. assisted with TC-Seq experiments. D.F.R. assisted with TC-Seq experiments and contributed mice. A.N. made suggestions on the manuscript. R.C. and M.C.N. designed experiments and analysis and wrote the manuscript.
References
- Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argast GM, Stephens KM, Emond MJ, Monnat RJ., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J Mol Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, et al. Regulation of DNA End Joining, Resection, and Immunoglobulin Class Switch Recombination by 53BP1. Mol Cell. 2011;42:319–329. doi: 10.1016/j.molcel.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- Cory S, Graham M, Webb E, Corcoran L, Adams JM. Variant (6;15) translocations in murine plasmacytomas involve a chromosome 15 locus at least 72 kb from the c-myc oncogene. EMBO J. 1985;4:675–681. doi: 10.1002/j.1460-2075.1985.tb03682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- Franco S, Gostissa M, Zha S, Lombard DB, Murphy MM, Zarrin AA, Yan C, Tepsuporn S, Morales JC, Adams MM, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Goldman JM, Melo JV. Chronic myeloid leukemia--advances in biology and new approaches to treatment. N Engl J Med. 2003;349:1451–1464. doi: 10.1056/NEJMra020777. [DOI] [PubMed] [Google Scholar]
- Gordon MS, Kanegai CM, Doerr JR, Wall R. Somatic hypermutation of the B cell receptor genes B29 (Igbeta, CD79b) and mb1 (Igalpha, CD79a) Proc Natl Acad Sci U S A. 2003;100:4126–4131. doi: 10.1073/pnas.0735266100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostissa M, Yan CT, Bianco JM, Cogne M, Pinaud E, Alt FW. Long-range oncogenic activation of Igh-c-myc translocations by the Igh 3' regulatory region. Nature. 2009;462:803–807. doi: 10.1038/nature08633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipati P, Helleday T. Transcription-associated recombination in eukaryotes: link between transcription, replication and recombination. Mutagenesis. 2009;24:203–210. doi: 10.1093/mutage/gen072. [DOI] [PubMed] [Google Scholar]
- Honjo T. Does AID need another aid? Nat Immunol. 2002;3:800–801. doi: 10.1038/ni0902-800. [DOI] [PubMed] [Google Scholar]
- Huppi K, Siwarski D, Skurla R, Klinman D, Mushinski JF. Pvt-1 transcripts are found in normal tissues and are altered by reciprocal(6;15) translocations in mouse plasmacytomas. Proc Natl Acad Sci U S A. 1990;87:6964–6968. doi: 10.1073/pnas.87.18.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic M, Robbiani DF, Dorsett Y, Eisenreich T, Xu Y, Tarakhovsky A, Nussenzweig A, Nussenzweig MC. Role of the translocation partner in protection against AID-dependent chromosomal translocations. Proc Natl Acad Sci U S A. 2010;107:187–192. doi: 10.1073/pnas.0908946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppers R. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Mahowald GK, Baron JM, Mahowald MA, Kulkarni S, Bredemeyer AL, Bassing CH, Sleckman BP. Aberrantly resolved RAG-mediated DNA breaks in Atm-deficient lymphocytes target chromosomal breakpoints in cis. Proc Natl Acad Sci U S A. 2009;106:18339–18344. doi: 10.1073/pnas.0902545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, Klein I, Ansarah-Sobrinho C, Resch W, Yamane A, Reina San-Martin B, Barreto V, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–133. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010a;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010b;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M. Neoplastic development in plasma cells. Immunol Rev. 2003;194:177–195. doi: 10.1034/j.1600-065x.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Rabbitts TH. Commonality but diversity in cancer gene fusions. Cell. 2009;137:391–395. doi: 10.1016/j.cell.2009.04.034. [DOI] [PubMed] [Google Scholar]
- Ramiro A, Reina San-Martin B, McBride K, Jankovic M, Barreto V, Nussenzweig A, Nussenzweig MC. The role of activation-induced deaminase in antibody diversification and chromosome translocations. Adv Immunol. 2007;94:75–107. doi: 10.1016/S0065-2776(06)94003-6. [DOI] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- Riggi N, Stamenkovic I. The Biology of Ewing sarcoma. Cancer Lett. 2007;254:1–10. doi: 10.1016/j.canlet.2006.12.009. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet. 2003;34:287–291. doi: 10.1038/ng1177. [DOI] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Staszewski O, Baker RE, Ucher AJ, Martier R, Stavnezer J, Guikema JE. Activation-induced cytidine deaminase induces reproducible DNA breaks at many non-Ig Loci in activated B cells. Mol Cell. 2011;41:232–242. doi: 10.1016/j.molcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storb U, Shen HM, Longerich S, Ratnam S, Tanaka A, Bozek G, Pylawka S. Targeting of AID to immunoglobulin genes. Adv Exp Med Biol. 2007;596:83–91. doi: 10.1007/0-387-46530-8_8. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tsai AG, Lieber MR. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics. 2010;11 Suppl 1:S1. doi: 10.1186/1471-2164-11-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AG, Lu H, Raghavan SC, Muschen M, Hsieh CL, Lieber MR. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135:1130–1142. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- Yamane A, Resch W, Kuo N, Kuchen S, Li Z, Sun HW, Robbiani DF, McBride K, Nussenzweig MC, Casellas R. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–69. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, Lewis S, Alt FW. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.