Abstract
Selenoproteins are essential in vertebrates because of their crucial role in cellular redox homeostasis, but some invertebrates that lack selenoproteins have recently been identified. Genetic disruption of selenoprotein biosynthesis had no effect on lifespan and oxidative stress resistance of Drosophila melanogaster. In the current study, fruit flies with knock-out of the selenocysteine-specific elongation factor were metabolically labeled with 75Se; they did not incorporate selenium into proteins and had the same lifespan on a chemically defined diet with or without selenium supplementation. These flies were, however, more susceptible to starvation than controls, and this effect could be ascribed to the function of selenoprotein K. We further expressed mouse methionine sulfoxide reductase B1 (MsrB1), a selenoenzyme that catalyzes the reduction of oxidized methionine residues and has protein repair function, in the whole body or the nervous system of fruit flies. This exogenous selenoprotein could only be expressed when the Drosophila selenocysteine insertion sequence element was used, whereas the corresponding mouse element did not support selenoprotein synthesis. Ectopic expression of MsrB1 in the nervous system led to an increase in the resistance against oxidative stress and starvation, but did not affect lifespan and reproduction, whereas ubiquitous MsrB1 expression had no effect. Dietary selenium did not influence lifespan of MsrB1-expressing flies. Thus, in contrast to vertebrates, fruit flies preserve only three selenoproteins, which are not essential and play a role only under certain stress conditions, thereby limiting the use of the micronutrient selenium by these organisms.
Keywords: Aging, Antioxidants, Drosophila Metabolism, Methionine, Oxidation-Reduction, Oxidative Stress, Selenium, Methionine Sulfoxide, Selenocysteine, Selenoprotein
Introduction
Selenium is an important dietary micronutrient in mammals. The major biological form of selenium is the non-canonical amino acid, selenocysteine (Sec).2 The majority of selenoproteins with known type of catalytic activity act as oxidoreductases that use Sec directly for catalysis and maintenance of cellular redox homeostasis. The number of selenoproteins in eukaryotic organisms varies significantly. Higher plants and fungi lack selenoprotein genes, whereas there are many such genes in algae and vertebrates, e.g. 10–57 in algae, 30–37 in fish, and 23–25 in mammals (1). At least five mammalian selenoproteins are essential (2–4). Remarkably, insects possess cysteine-containing homologs or lack all essential mammalian selenoproteins, e.g. thioredoxin reductases and glutathione peroxidases. Moreover, recent studies identified five species of selenoproteinless insects, including the red flour beetle Tribolium castaneum (5), the silkworm Bombyx mori (5), the fly Drosophila willistoni (6), the honey bee Apis mellifera (6), and the wasp Nasonia vitripennis (6). This evolutionary reduction in the use of selenoproteins could be associated with considerable changes in antioxidant defense systems of insects (7–9).
The Sec incorporation machinery is conserved across eukaryotes. Sec is encoded by the UGA codon that usually serves as a termination signal. Decoding of UGA during translation as a codon for Sec insertion requires 1) a unique tRNA with an anticodon to UGA, tRNA[Ser]Sec; 2) a specific stem-loop structure in the 3′-UTR of eukaryotic selenoproteins mRNAs designated selenocysteine insertion sequence (SECIS) element; and 3) Sec-decoding protein factors and enzymes that are required for Sec synthesis on tRNA[Ser]Sec and its insertion into proteins (for review, see Refs. 10–14 and references therein).
The fruit fly Drosophila melanogaster has 3 selenoproteins: selenophosphate synthetase 2 (dSPS2), selenoprotein H (dSelH, also known as BthD), and selenoprotein K (dSelK, also known as G-rich) (15–18). The SPS2 function (synthesis of monoselenophosphate from selenide) is essential for Sec biosynthesis and expression of selenoproteins (19, 20). Thus, the two other Drosophila selenoproteins must be responsible for the biological effects of selenium in fruit flies. Mammalian SelH is a nuclear protein that has a thioredoxin-like fold and possesses glutathione peroxidase activity in vitro (21). This protein up-regulates transcription of genes involved in glutathione synthesis and phase II detoxification (22). In Drosophila, knockdown of the dSelH gene using RNAi technology significantly reduced embryonic viability and caused a decrease in total antioxidant status in Drosophila Schneider 2 (S2) cells (23). However, mutant fruit flies with knock-out of the Sec-specific translational elongation factor dEFsec (24), which failed to decode the UGA codon as a codon for Sec insertion, were viable, fertile, and had the same mean lifespan and oxidative stress resistance as controls (24). It is possible that the phenotypes observed with RNAi against dSelH resulted from off-target effects of siRNA. Drosophila dSelK is a Golgi-resident membrane protein (25). However, two recent studies indicate that mammalian SelK is an ER-resident transmembrane protein (26, 27) that plays an important role in protecting cells from ER stress-induced apoptosis (26) and in immune response (27). To summarize, the specific biological functions of SelH and SelK are not known in Drosophila or in mammals. In this regard, fruit flies offer a useful system to study the functions of these selenoproteins.
Many mammalian selenoproteins are involved in regulation of cell redox homeostasis and could, directly or indirectly, be involved in regulation of longevity. However, there are no published data on the link between overexpression of redox selenoproteins and aging in animals. The fruit fly is a very convenient model organism for such study. Here, we chose a mouse selenoprotein, methionine sulfoxide reductase B1 (MsrB1, also known as SelR or SelX), to characterize a possible connection between overexpression of a selenoenzyme, oxidative stress resistance, and longevity.
MsrB1 is a member of an Msr class of proteins that are responsible for the reduction of methionine sulfoxides to methionine in proteins (for review, see Refs. 28–30 and references therein). Proposed functions of Msrs include protection of cells from oxidative stress through reversible reduction of methionine sulfoxide and repair of oxidatively damaged proteins to preserve their functions (31, 32). Msrs that act on oxidized proteins are classified with respect to their substrate specificity into two types: MsrA that is specific for the reduction of methionine-S-sulfoxide, and MsrB that catalyzes the reduction of methionine-R-sulfoxide. Mammals have one MsrA and three MsrB isozymes, which are targeted to different cellular compartments. MsrB1 is located in the nucleus and cytosol. This protein is remarkable in that it contains an active site Sec in place of cysteine, a common catalytic group for all Msrs (28–30).
Numerous studies provided evidence for an important role of Msrs in aging. It is generally agreed that knock-out of the msrA gene results in accumulation of oxidized and modified proteins (33), increased sensitivity to oxidative stress (34–36), and either a shortened (34, 35, 37) or an unaffected lifespan (36). In contrast, MsrA overexpression led to protection against oxidative stress and increased longevity in yeast and fruit flies (38–40). The effect of MsrB knock-out/overexpression on aging in yeast and flies is less pronounced (37, 41). For example, expression of host MsrA or GFP-fused bovine MsrA in the nervous system increased the median lifespan of Drosophila by ∼20 (39) or 70% (40), respectively. However, overexpression of host MsrB or mouse MsrB2 in the nervous system had no substantial effect on Drosophila lifespan (41).
The different roles of MsrA and MsrB in lifespan regulation could reflect the different biological functions and/or regulatory pathways involving these enzymes. MsrA has broader substrate specificity than MsrB and effectively reduces the free form of methionine-S-sulfoxide to methionine. Thus, MsrA overexpression could change the metabolism of methionine that is important for sulfur and DNA methylation pathways involved in aging (42, 43). In addition, a forkhead transcription factor, FOXO3a, directly activates the human MSRA gene (34) and its homologs (DAF16 in Caenorhabditis elegans and dFOXO in fruit fly) activate worm msrA (34) and Drosophila msrA genes (39). FOXO3a transcription factor is known to up-regulate the expression of genes involved in oxidative stress response and longevity and down-regulate life-shortening genes. FOXO3a target genes include genes involved in cell cycle, stress response, metabolism, and apoptosis. dFOXO overexpression in the adult fatbody of female fruit flies results in increased lifespan (44). Overexpression of Drosophila MsrA caused dFOXO translocation to the nucleus (39), implying that the prolonged lifespan of MsrA-expressing flies may be due not only to the MsrA role in oxidative stress resistance, but also its role in up-regulation of dFOXO. No transcriptional activation of MSRB genes by FOXO3a or its homologs in different organisms was reported thus far.
The fact that the components of Sec biosynthesis and incorporation machineries, as well as selenoproteins themselves, are found in 11 Drosophila species (except for D. willistoni) (6) reflects importance of these proteins for fruit flies; on the other hand, the absence of the evident phenotype of dEFsec knock-out flies and the occurrence of selenoproteinless animals pose an important question. Why are selenoproteins preserved in Drosophila during evolution if these proteins are not important for cellular redox regulation and longevity? In our study, we used dEFsec knock-out flies (24) to address this question. We also used these flies to study the role of selenium as a micronutrient (i.e. not in the form of selenoproteins) in aging. In addition, we used Drosophila as a model organism to clarify the role of mouse MsrB1 in aging. We addressed the following questions. Is it possible to promote resistance to oxidative stress and/or extend lifespan by expressing an additional selenoenzyme with protein repair function? Is it possible to regulate lifespan of these animals by dietary selenium?
EXPERIMENTAL PROCEDURES
Constructs
The ORF of mouse MsrB1 gene was PCR amplified from the pCI-SelR-His construct (45) with 5′-GACTGAATTCATGTCGTTCTGCAGCTTCTTCGGAG-3′ and 5′-GTATGCGGCCGCCTAGTGCCCCTGGGAGGCAGCAGCTTCTTTGC-3′ primers and cloned into the EcoRI/NotI restriction sites of the pUAST vector (46) to yield the pUAST-mMsrB1 construct. The 3′-UTR of Drosophilia selK containing the SECIS element (236 bp after the stop codon) was amplified from total RNA of white mutant (w1118) flies with 5′-GTATGCGGCCGCTAGCGACATCCGGTTCCCAAGACTCTTGG-3′ and 5′-CTACTCTAGAGGAGCTAATAGTTGATAAATGGAACCGACG-3′ primers using the SuperScriptTM II RT kit (Invitrogen) and cloned into NotI/XbaI restriction sites of pUAST-mMsrB1 to yield the pUAST-mMsrB1-SECIS construct. This construct was used for generation of UAS-mMsrB1 transgenic lines.
Transgenic Drosophila Lines
Transgenic flies were obtained using standard techniques for germline transformation and balancing as described (41). Five independent homozygous UAS-mMsrB1 responder lines were obtained and three of them were used in the study. These lines were designated as mMsrB12A, mMsrB13A, and mMsrB13B. The designations of transgenic lines indicate the transgene (i.e. the mouse MsrB1 gene, mMsrB1) followed by the chromosome of insertion (i.e. 2 or 3), followed by a letter (i.e. A or B) showing the independent insertion on that chromosome. None of these insertions influenced viability or development of homozygous UAS-mMsrB1 transgenic lines. Two GAL4-activator lines (drivers) used in this study, whole body da-GAL4 [w*;; P{w+mW.hs = GAL4-da.G32}UH1] (stock number 5460) and nervous system elav-GAL4 [w*, P{w+mW.hs = GawB}elavc155] (stock number 458), were obtained from the Bloomington Drosophila Stock Center at Indiana University. All transgenic lines, including the driver lines, were backcrossed six times to the same isoline of yellow body white eyes flies yw (kindly provided by Dr. R. S. Sohal, University of Southern California) to ensure that the genetic backgrounds were equivalent. dEFsec knock-out flies (KO#24 and KO#46) with impaired selenoproteins synthesis were previously described (24). Briefly, these flies were generated by P-element transformation of mutant dEFsec gene and homologous recombination.
Genetic Crosses and Drosophila Husbandry
The GAL4-UAS binary system (46) was used to drive expression of mMsrB1 in fruit flies. To obtain experimental flies, homozygous mMsrB12A, mMsrB13A, or mMsrB13B males were crossed with virgin females of GAL4-driver. Progeny of crosses between the GAL4-activator line and yw flies (GAL4-activator/yw) or between yw flies and UAS-mMsrB1 lines (yw/UAS-mMsrB1) were used as controls. mMsrB1-expressing or dEFsec KO experimental flies and their controls were obtained and maintained on corn meal food as previously described (41).
Fly Culture Media
Three types of food were used in the lifespan experiments. The first type was corn meal food that is commonly used in our laboratory (41). The second type was a chemically defined medium developed by Martin-Romero et al. (17). This diet contained Grace's insect medium (G-8142, Sigma), 0.01% p-hydroxybenzoic acid methyl ester (Sigma), 2% low melting point agarose (Sigma) and was supplemented with or without sodium selenite (Sigma) at a final concentration of 100 nm. This food was used to examine the role of selenium in aging of dEFsec KO flies. The third diet was another chemically defined medium based on a fruit fly basal mixture developed by Troen et al. (47). This diet contained 62.08 g of Diet TD.04310 (Harlan Teklad), 100 mg of lecitin from soybean (Sigma), 500 mg of ribonucleic acid from Torula yeast (Sigma), 100 g of dextrose, 1.35 g of methionine (Sigma), 20 g of low melting point agarose, 2.85 ml of propionic acid, and 0.255 ml of phosphoric acid (Sigma) per liter of water (47). This food was used to study the effect of selenium during aging of flies expressing Sec-containing mMsrB1. Sodium selenite was added (or was not added) to the food at a final concentration of 10 or 200 nm. The concentration of selenium in the food was measured by the analytical service provided by Oscar E. Olson Biochemistry Laboratories, South Dakota State University. The concentration of selenium in the diets was 13.02 ± 0.05 nm (unsupplemented diet), 19.59 ± 0.03 nm (diet supplemented with 10 nm selenite), and 206.7 ± 2.83 nm (diet supplemented with 200 nm selenite).
Metabolic Labeling of Flies with 75Se
Thirty eight-day-old mated flies were maintained on a Sf-900 II SM serum-free insect medium (Invitrogen) supplemented with 3% dextrose (Sigma), 0.01% p-hydroxybenzoic acid methyl ester, 2% low melting point agarose, and 35 μCi of freshly neutralized [75Se]selenious acid (specific activity 1,000 Ci/mmol, University of Missouri Research Reactor, Columbia, MO) at 25 °C in a 12-h light/dark cycle for 72 h. Whole body homogenates of 35 flies were prepared in 350 μl of PBS buffer containing protease inhibitors (Roche Applied Science). Protein extracts (100 μg) were applied to a NuPAGE® Novex 10% BisTris gel (Invitrogen), electrophoresed, and transferred onto a PVDF membrane (Invitrogen). The 75Se radioactivity pattern on the membrane was visualized using a PhosphorImager system (GE Healthcare).
Lifespan Study
Lifespan studies were performed as described previously (41). Briefly, adult animals were collected within 24 h post-eclosion. In a typical lifespan trial, three-day-old flies were placed in cages (see description in Ref. 41). Three replica cages were used for flies with the same genotype and gender; survivorship curves present the average of those independent replicas. In total, ∼180 mated flies were used for each survivorship curve of dEFsec knock-out flies and 210 virgin flies were used for each survivorship curve of mMsrB1-expressing flies. Fresh food was supplied into cages, and dead flies were removed by aspiration and counted every 3 days. Experimental and control group trials were always performed concurrently.
Stress Resistance and Reproduction Tests
Oxidative resistance test was performed as previously described (41). Flies were starved for 6 h and fed with 5% hydrogen peroxide (Sigma) or freshly prepared 30 mm paraquat (Sigma) in 5% sucrose (Sigma). Eight replicates of 20 mated males (160 flies for each genotype) were used for the hydrogen peroxide resistance test. Nine replicates of 20 animals (180 flies for each genotype) were used for the paraquat resistance test. Results are reported as the means of survived animals in 8 (or 9) replicates ± S.E. for each time point. Starvation resistance tests were performed as previously described (41). Ten groups of 10 animals for dEFsec KO flies (in total 100 flies) or 7 replicates of 20 animals for mMsrB1-expressing flies (in total 140 flies) were used for each genotype and gender. Data are reported as the means of survived animals in 10 replicates (or in 7 replicates) ± S.E. for each time point. All trials were performed concurrently. Age-specific changes in pupa production were determined as previously described (41).
Enzyme Activity Assay
Enzyme activity assays in fly homogenates were performed as previously described (41). Measurements for each sample were performed in triplicate. All data are reported as the mean ± S.E.
Statistical Analysis
Enzyme activities (overexpressor versus control) were compared by unpaired Student's t tests. All average results presented as mean ± S.E. were calculated using Microsoft Excel software. The significance of the difference between the survivors was determined using SAS software version 9.1 (SAS Institute Inc., Cary, NC) as described (41). Nonparametric estimates of the survivor functions by the Kaplan-Meier method were made using the procedure LIFETEST provided by SAS. Survivors were considered statistically different, if statistical parameters (χ2 and p value) of log-rank test were χ2 > 20, p < 0.0001.
RESULTS
Fruit Flies Lacking Selenoproteins Are Susceptible to Starvation
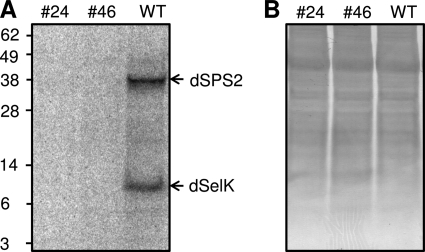
Mice with impaired selenoprotein synthesis die during embryonic development (2). However, fruit flies that lacked dEFsec were viable and fertile (24). These dEFsec knock-out flies did not express dSPS2 and had the same mean lifespan and oxidative stress resistance as wild type flies (24). To examine the presence of selenoproteins in dEFsec knock-out flies, 6-day-old male flies (KO#24 and KO#46) as well as wild type flies were metabolically labeled with [75Se]sodium selenite and selenoproteins in whole body homogenates were detected using a PhosphorImager system (Fig. 1). We observed the absence of any 75Se signal in both knock-out lines, whereas wild type flies showed two selenoprotein bands. Based on the predicted masses of three Drosophila selenoproteins, the lower band was assigned to dSelK, and the higher to dSPS2. dSelH could not be observed as it is specifically expressed during embryonic and larval development and in adult female flies (48). Because the function of dSPS2 is to provide selenium for selenoprotein synthesis, male fruit flies can be used as a unique model for studying dSelK function. Interestingly, we also observed no nonspecific labeling of fruit fly proteins with 75Se, suggesting that selenite does not enter the sulfur pathways for insertion into cellular proteins.
FIGURE 1.
dEFsec knock-out flies do not express selenoproteins. A, selenoprotein pattern in fruit fly homogenates visualized using a PhosphorImager system. Six-day-old male flies were metabolically labeled with 75Se, followed by analysis of proteins by SDS-PAGE. Migration of two Drosophila selenoproteins (dSPS2 and dSelK) is marked by arrows. B, Coomassie Brilliant Blue staining of the same membrane (protein loading control).
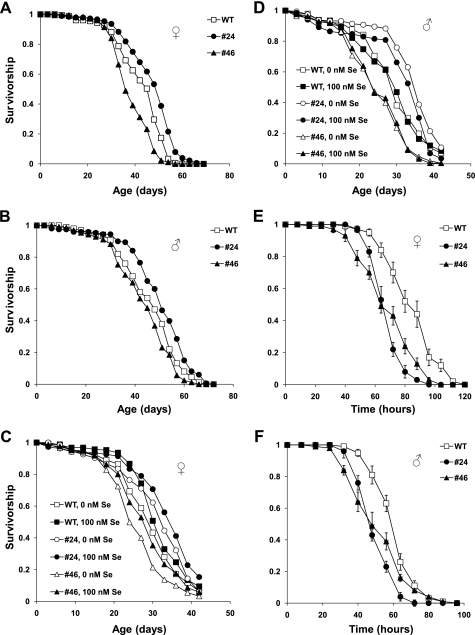
We examined aging of dEFsec KO flies on corn meal food (Fig. 2, A and B). Disruption of selenoprotein synthesis was not critical for Drosophila lifespan, as no significant and reproducible changes in the mean lifespan of dEFsec KO flies were found (Fig. 2, A and B, and Table 1). Although the survivorship data for dEFsec KO female flies were different in comparison with control (Table 1, columns 6 and 7), the mean lifespan varied inconsistently (Table 1, column 4). dEFsec KO male flies had small and inconsistent variations in the mean lifespan (6% increase for KO#24 (χ2 = 10.4, p = 0.0018) and 5% decrease for KO#46 (χ2 = 4.8, p = 0.0279) in comparison with male control). We also confirmed a previous finding (24) that dEFsec knock-out flies had the same resistance to oxidative stress induced by hydrogen peroxide as control flies (data not shown).
FIGURE 2.
Survivorship curves of dEFsec knock-out flies and their controls. A and B, survivorship curves of dEFsec KO (KO#24 and KO#46) and wild type flies fed corn meal food. Mated female and male flies were kept separately during the lifespan study. Each survivor curve represents 180 animals. Genotypes and genders are shown on the plots. All trials were performed concurrently. C and D, survivorship curves of dEFsec KO and wild type flies on the chemically defined food supplemented with or without 100 nm sodium selenite. Mated female and male flies were kept separately during the lifespan study. Genotypes, genders, and selenium concentrations are displayed on the plots. All trials were performed concurrently. E and F, resistance to starvation was studied using 10 replicates of 10 animals (in total, 100 flies were used per genotype). 20-day-old mated female or male flies were kept separately during the test. Data are reported as the means of survived animals in 10 replicates ± S.E. for each time point. Genotypes and sexes are shown on the plot. All trials were performed concurrently.
TABLE 1.
Statistical analysis of survivors for dEFsec knock-out and control animals on corn meal food
Column 1 indicates the letter of the corresponding panel in Fig. 2. The genotypes and genders (female, F; males, M) are shown in columns 2 and 3, respectively. The mean lifespan is shown in column 4. The percent change in the mean lifespan of the experimental flies compared with wild type is displayed in column 5. Comparison of survivors was performed with SAS. Statistics of non-parametrical log rank test (χ2) for comparison with wild type is shown in column 6; p value in column 7.
Flies with disrupted Sec biosynthesis also represent a convenient model to study the role of selenium (as opposed to the role of selenoproteins) as a micronutrient that may regulate aging or could be essential for survival. We examined the lifespan of dEFsec KO flies on a simple chemically defined diet (17), which included Grace's insect medium, low melting point agarose, and p-hydroxybenzoic acid methyl ester, and was supplemented with or without 100 nm sodium selenite. We observed up to 44% reduction in the mean lifespan of flies maintained on this diet compared with that of flies on corn meal food (Fig. 2, C and D, and supplemental Table S1, column 8). In addition, the chemically defined diet did not support the development of flies to adult animals. These findings suggest a nutrient deficiency of the chemically defined diet (17). Survivorship of flies on the diet supplemented with or without 100 nm sodium selenite was identical according to statistical analyses (Fig. 2, C and D, and supplemental Table S1, columns 3–7).
However, we found that dEFsec KO animals were more sensitive to starvation than control flies (Fig. 2, E and F). The median lifespan of female flies of both KO lines was ∼63 versus 84 h in the case of control animals (Fig. 2E). The increased sensitivity of dEFsec KO flies to starvation was also observed for male flies (∼44 h for experimental and 62 h for control flies, Fig. 2F). This effect was substantial and statistically significant.
Expression of a Mammalian Selenoenzyme in Drosophila
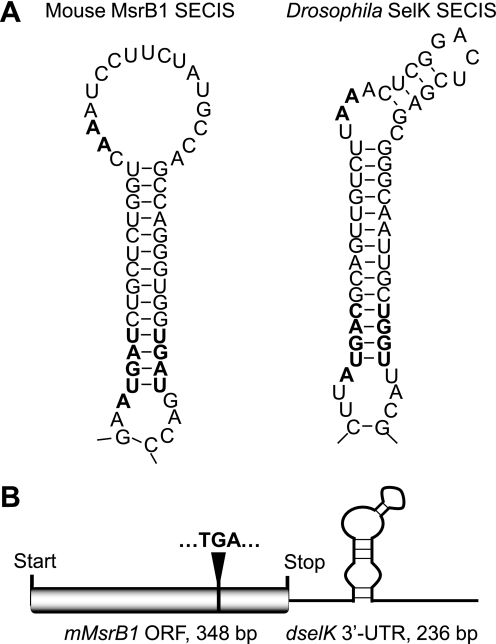
Expression of mouse MsrB1 as a selenoprotein requires the presence of a functional SECIS element in the 3′-UTR. Initially, the mouse gene coding for mMsrB1 (the ORF and the 3′-UTR containing the natural mouse MsrB1 SECIS element) was cloned into pUAST vector (46). Six homozygous lines carrying UAS-mMsrB1 transgenes were generated as described (41). Progeny obtained after crossing these flies with GAL4-activator lines were metabolically labeled with 75Se and selenoprotein expression was analyzed using a PhosphorImager system. Unexpectedly, these flies did not express exogenous mMsrB1 (data not shown). Reverse transcription with primers specific for mouse MsrB1 on total RNA purified from UAS-mMsrB1-responder lines have shown that these flies expressed mMsrB1 mRNA. Based on these data, we hypothesized that expression of exogenous selenoproteins in flies might require a natural Drosophila SECIS element (e.g. SECIS element of dSPS2, dSelH, or dSelK). Indeed, a recent study by Takeuchi et al. (49) has shown that Drosophila SECIS-binding protein 2 exhibits high affinity toward type II SECIS elements. Mouse MsrB1 has a type I SECIS element (Fig. 3A), but all Drosophila selenoprotein genes have type II SECIS elements (Fig. 3A, see Drosophila SelK SECIS element as an example). Type II SECIS element is characterized by an additional minihelix in the apical loop (50).
FIGURE 3.
The MsrB1 coding construct used for generation of transgenic flies. A, secondary structures of mouse MsrB1 SECIS element and Drosophila SelK SECIS element. Conserved nucleotides in the SECIS core and in the apical loop are shown in bold. B, schematic representation of the construct coding for mouse MsrB1 ORF (gray box), Drosophila SelK SECIS element (shown by a black line), and Sec-encoding TGA (shown in bold), which is positioned at 285 bp of the ORF.
Based on this information, we developed a chimeric construct coding for mouse MsrB1 ORF (348 bp) and the Drosophila selK 3′-UTR containing the SECIS element (236 bp) (Fig. 3B). Three of five independent homozygous lines carrying the UAS-mMsrB1 transgene on the second (mMsrB12A) or third (mMsrB13A and mMsrB13B) chromosomes were used in subsequent experiments.
Experimental flies were obtained by crossing flies carrying the UAS-mMsrB1 transgene with the ubiquitous da-GAL4 or neuronal elav-GAL4 driver. The whole body da-GAL4 driver was selected for the study as Drosophila expresses endogenous MsrB ubiquitously. The nervous system driver elav-GAL4 was chosen as the driver with the same genotype used to express MsrA that led to lifespan extension of MsrA-expressing flies (40). Two different types of control flies were used in this study: (i) the driver control flies, i.e. flies carrying the GAL4-transgene (GAL4-activator/yw), and (ii) the responder control flies, i.e. flies carrying the UAS-MsrB transgene (yw/UAS-mMsrB1).
Metabolic labeling of experimental flies showed expression of the mouse selenoprotein in Drosophila whole body (Fig. 4A, lanes 1–6) and the nervous system (Fig. 4B, lanes 1–6). Driver control flies (GAL4-activator/yw) were used as a control (Fig. 4, A and B, lanes 7 and 8). The three previously identified Drosophila selenoproteins, dSPS2, dSelH, and dSelK (16, 17), were clearly discernible in lanes 1, 3, 5, and 7 in Fig. 4A, which corresponded to female flies. The band corresponding to dSelH (approximately 38 kDa) migrated slower than expected (calculated molecular mass of the protein is 28 kDa). The fourth selenoprotein band with the size corresponding to mouse MsrB1 (calculated molecular mass 12.6 kDa) could only be seen in the flies containing the UAS-mMsrB1 transgene (Fig. 4, A and B, lanes 1–6).
FIGURE 4.
Expression of mouse MsrB1 in Drosophila. A and B, selenoprotein pattern in fruit fly homogenates visualized using a PhosphorImager system. Expression of mouse MsrB1 in the whole body (A) or the nervous system (B) was obtained by crossing homozygous UAS-mMsrB1 flies with da-GAL4 activator line or elav-GAL4 activator line, respectively. Panels A and B have the same order of samples: lanes 1 and 2, cross GAL4-activator/mMsrB13A; lanes 3 and 4, cross GAL4-activator/mMsrB12A; lanes 5 and 6, cross GAL4-activator/mMsrB13B; lanes 7 and 8, control cross GAL4-activator/yw. Drosophila selenoproteins (dSPS2, 43 kDa; dSelH, 28 kDa; mMsrB1, 12.6 kDa; and dSelK, 11.5 kDa) are marked on the left side of panel A with arrows. Molecular mass markers are shown on the right side of panel A. C and D, Coomassie Brilliant Blue staining of membranes A and B, respectively (protein loading control). Genders are shown by symbols below panels C and D.
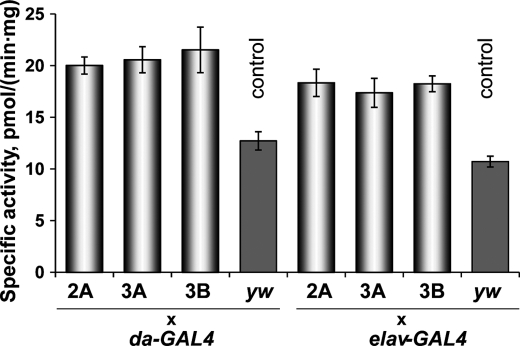
To verify that the expressed Sec-containing mMsrB1 is functional, the specific MsrB activity was measured in the homogenates of transgenic flies. Previously, ubiquitous expression of mouse MsrB2 led to an almost 40-fold increase in MsrB activity in comparison with background activity (due to expression of endogenous fruit fly MsrB) (41). The specific activity of Sec-containing mMsrB1 is ∼4.5-fold higher than the activity of Cys-containing mMsrB2 (51). On the other hand, mMsrB1 expression in fruit flies could be limited by the low levels (or low efficiency) of the Sec insertion machinery. Homogenates prepared from experimental flies showed a 2-fold increase in total MsrB activity in comparison with the homogenates prepared from driver control flies (Fig. 5). Overall, the data showed that expression of catalytically active Sec-containing mouse MsrB1 in Drosophila was achieved.
FIGURE 5.
MsrB activity of homogenates of mMsrB1-expressing male flies. Designations 2A, 3A, 3B, and letters yw refer to mMsrB12A, mMsrB13A, mMsrB13B, and yw lines that were crossed with the indicated GAL4 activator lines. Measurements were performed in triplicate. All data are reported as the mean ± S.E. p < 0.05 was obtained for all combinations of overexpressor versus controls.
Ectopic or Ubiquitous Expression of mMsrB1 in Drosophila Does Not Affect Lifespan
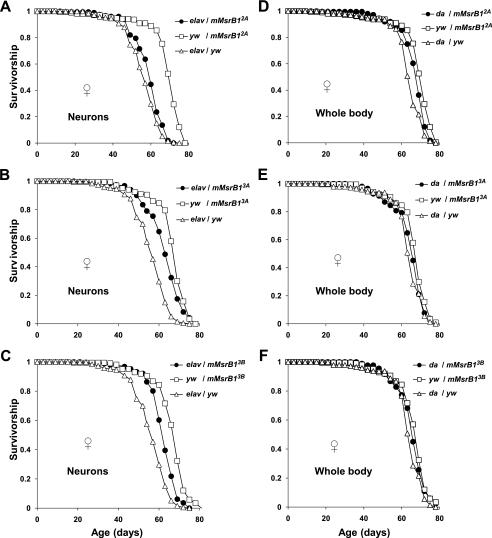
No substantial and reproducible changes (increase or decrease) in the mean lifespan were observed for virgin females expressing mMsrB1 in the nervous system (Fig. 6, A–C, and Table 2, column 4) or in the whole body (Fig. 6, D–F, and Table 2, column 4) in comparison with controls. Both driver and responder control flies were used for each experimental condition. A statistically significant difference (χ2 > 20, p < 0.0001) in survivorship between experimental flies and both types of controls was observed for crosses: elav-GAL4/mMsrB13A and elav-GAL4/mMsrB13B (Table 2, columns 6, 7, 9, and 10). However, the value of the mean lifespan for these flies was between the values of the mean lifespan for two controls. Responder controls had a similar mean lifespan (≈68 days, Table 2, column 4) and their survivorship was undistinguishable by statistical analyses (χ2 = 16.2, p = 0.0011 for comparison yw/mMsrB12A versus yw/mMsrB13B and χ2 = 1.6, p = 0.2069 for comparison yw/mMsrB13A versus yw/mMsrB13B (Table 2, columns 9 and 10)).
FIGURE 6.
Survivorship curves of mMsrB1-expressing virgin females on corn meal food. Expression of mMsrB1 in the nervous system was activated by elav-GAL4 activator line (A–C) and in the whole body by da-GAL4 activator line (D–F). Each survivorship curve represents ∼210 virgin female flies. Genotypes and genders are shown on the plot. All trials were performed concurrently.
TABLE 2.
Statistical analysis of survivors for mMsrB1-expressing flies on corn meal food
Column 1 indicates the letter of the corresponding panel in Fig. 6 or supplemental Fig. S1. The genotypes and genders (female, F; male, M) are shown in columns 2 and 3, respectively. The mean lifespan is shown in column 4. The percent change in the mean lifespan of experimental flies compared with the corresponding driver controls (GAL4-driver/yw) is displayed in column 5 and compared with the corresponding responder control (yw/UAS-mMsrB1) in column 8. Comparison of two types (experimental and control) of survivors was performed with SAS software. Statistics of non-parametrical log rank test (χ2) for comparison with driver control lines is shown in column 6; p value in column 7 and for comparison with responder control lines in columns 9, 10. Asterisks mark a comparison of two responder lines.
Similar data were obtained for virgin male flies (supplemental Fig. S1 and Table 2). Again, these mMsrB1-expressing flies had no substantial and reproducible variation in the mean lifespan in comparison with driver and responder controls (Table 2, columns 5 and 8).
No Effect of Selenium Supplementation of a Chemically Defined Diet on the Mean Lifespan of Experimental Flies
To further examine the role of dietary selenium, we used a recently developed chemically defined diet (47), which supported normal development of flies and provided sufficient nutrients to sustain the lifespan similar to that of flies on the yeast-based food. Using diets with different levels of selenium, we tried to achieve two goals: (i) find conditions of selenium deficiency for flies and observe a possible effect of selenium (as a micronutrient) on lifespan; and (ii) regulate expression of mMsrB1 by selenium in the food and examine the possibility of the life-prolonging effect of this antioxidant enzyme. The diet without selenium supplementation contained 13 nm selenium. The diets supplemented with 10 and 200 nm sodium selenite had 20 and 207 nm selenium, respectively. No effect of selenium supplementation on the mean lifespan of either female or male flies expressing mMsrB1 in the whole body was observed (supplemental Fig. S2 and Table S2). Also, no difference in the mean lifespan for flies of the same genotype and sex kept on the chemically defined diet or on corn meal food was found (supplemental Table S2, column 8).
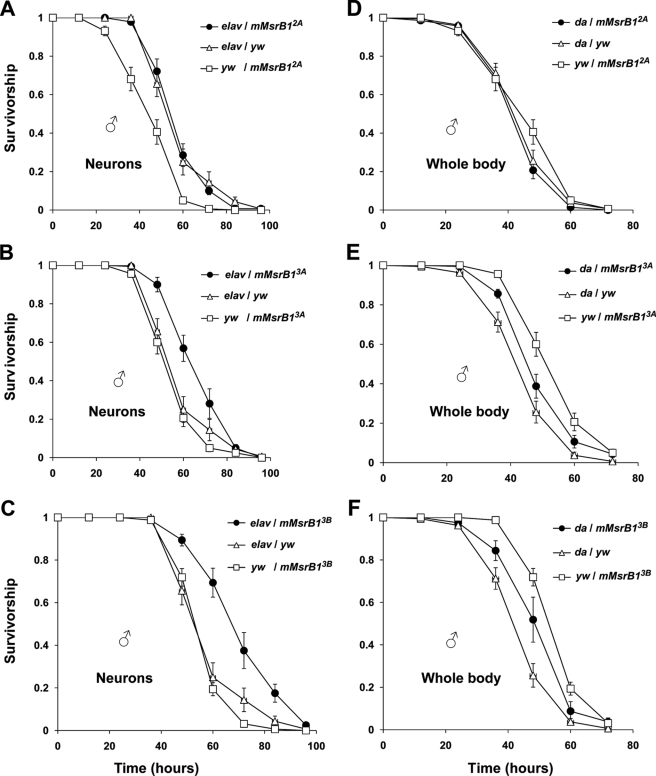
Ectopic Expression of mMsrB1 in Neurons Increased Stress Resistance of Experimental Flies
Extended longevity is often accompanied by the enhanced ability to resist various forms of environmental stress, e.g. oxidative stress, starvation, heat or cold stress (40, 42, 52, 53). However, there are also examples when such a correlation was not observed (52, 54–56). Although mMsrB1 expression had no influence on lifespan, we hypothesized that its antioxidant function could have a more clear effect on resistance against oxidative stress than on such a complex process as aging.
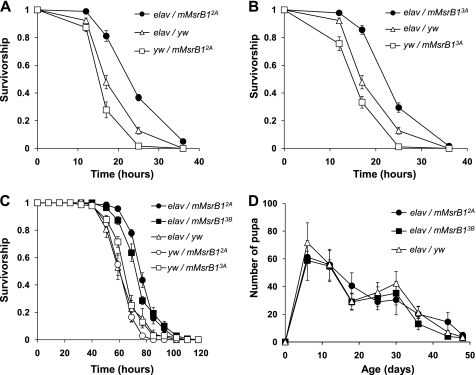
Sixteen-day-old experimental flies and their controls were starved for 6 h to minimize variations in oxidant intake and then fed 5% sucrose and 5% hydrogen peroxide (Fig. 7) or 30 mm paraquat (Fig. 8, A and B). Ectopic expression of mMsrB1 in the nervous system significantly increased survivability of flies on hydrogen peroxide for two crosses elav-GAL4/mMsrB13A and elav-GAL4/mMsrB13B (Fig. 7, B and C). A more pronounced life-prolonging effect was observed in the paraquat resistance test (Fig. 8, A and B). Flies with genotype elav-GAL4/mMsrB12A that did not show enhanced resistance against hydrogen peroxide (Fig. 7A) lived longer on paraquat in comparison with any control (Fig. 8A). Ubiquitous expression of mMsrB1 did not promote survival upon oxidative stress (Fig. 7, D and E). Interestingly, we also observed increased resistance of flies with neuronal mMsrB1 expression against starvation (Fig. 8C).
FIGURE 7.
Resistance of mMsrB1-expressing flies against oxidative stress induced by 5% dietary hydrogen peroxide. Survivorship curves of flies expressing mMsrB1 in the nervous system (A–C) or in the whole body (D–F). 160 male flies (8 replicates of 20 animals) were used for each genotype. Genotypes are shown on the plot. Data for each time point are reported as the means of survived animals in 8 replicates ± S.E. All trials were performed concurrently.
FIGURE 8.
Stress resistance and reproduction of mMsrB1-expressing flies. A and B, survivorship curves of mMsrB1-expressing flies on 30 mm dietary paraquat. 180 male flies (9 replicates of 20 animals) were tested for each genotype. Genotypes are shown on the plot. Data for each time point are reported as the means of survived animals in 9 replicates ± S.E. All trials were performed concurrently. C, starvation resistance of flies expressing mMsrB1 in the nervous system. 140 male flies (7 replicates of 20 animals) were tested for each genotype (shown on the plot). Data are reported as the means of survived animals in 7 replicates ± S.E. for each time point. All trials were performed concurrently. D, age-associated changes in pupa production. Changes in pupa production were determined from counts of pupa developed from eggs laid by five females during 24 h. The y axis represents the number of pupa produced by one female in one vial. 10 replica of each vial were tested. Genotypes are shown on the plot. Data are reported as mean of 10 replica ± S.D.
mMsrB1 Expressing Flies Had the Same Reproduction Vigor as Controls
Previously, we found that flies overexpressing either mouse MsrB2 or Drosophila MsrB had similar physical characteristics and showed no changes in the number of developed pupa in comparison with controls (41). In the current study, we observed that development of mMsrB1-expressing flies from eggs to hatching was the same as in the parental and heterozygous control flies that did not express mMsrB1 (9–10 days, 25 °C). Ectopic expression of mMsrB1 in the nervous system also did not change reproductive vigor (Fig. 8C).
DISCUSSION
dEFsec knock-out flies have a normal lifespan and the same resistance to oxidative stress as the corresponding wild type flies (24). In our study, we have first shown, by 75Se metabolic labeling, that dEFsec KO flies do not express selenoproteins (Fig. 1), whereas three known selenoproteins (dSelK, dSPS2, and dSelH) could be detected in female flies with intact selenoprotein biosynthesis and two of them (dSelK and dSPS2) in male flies (Figs. 1 and 4). These observations support the idea that some insects lost major redox selenoproteins because they no longer provided sufficient benefits, including function in the defense against oxidative stress.
dEFsec KO fly is a convenient model to study the role of selenium as a micronutrient in Drosophila. It is well known that selenoproteins represent the major biological form of selenium. We tested the possibility that another form of selenium may be used by fruit flies, because it is unclear how selenoproteinless animals deal with excess selenium that is present in the environment. Absence of this unknown form of selenium might affect lifespan, and if so, its importance could be revealed using a chemically defined diet supplemented or not with selenium.
Selenophosphate synthetase 1 (SPS1) is a promising candidate that could be responsible for the conversion of sodium selenite to an unknown biological form of selenium. The function of SPS1 is not clear. All selenoproteinless insects have lost SPS2, but all of them have SPS1 (6). Although the two proteins share a common ancestor involved in selenoprotein synthesis, SPS1 cannot be involved in Sec biosynthesis or selenoprotein biosynthesis because (i) it is present in all selenoproteinless insects (5, 6); (ii) SPS1 knock-out leads to Drosophila lethality in the larval/pupal stages (57), whereas selenoproteinless fruit flies are viable; (iii) Drosophila SPS1 is not active in the synthesis of selenophosphate (58); and (iv) knockdown of SPS1 in mammalian cells had no effect on selenoprotein biosynthesis (20). Thus, a possibility that SPS1 is responsible for some other selenium-dependent pathway remains. Using a chemically defined diet supplemented or not with 100 nm sodium selenite, we tested dEFsec KO flies, but did not observe significant changes in the mean lifespan of dEFsec KO flies (Fig. 2, C and D, and supplemental Table S1, columns 5–7). This result suggests that dEFsec KO flies and selenoproteinless insects probably do not utilize selenium. However, the possibility remains that the other biological forms of selenium and selenium-dependent pathways exist in selenoproteinless insects, but they are not essential. Or, alternatively, these pathways are essential, but selenium concentration in the unsupplemented diet is already sufficient to satisfy the needs for this element. Thus, further experiments are needed to shed light on possible new biological forms of selenium or pathways that selenoproteinless insects use to metabolize selenium.
In this work, we found that dEFsec KO male and female flies were more susceptible to starvation than wild type controls (Fig. 2, E and F). dSelK appears to be the only selenoprotein responsible for the biological effects of selenium in male flies (because dSPS2 is involved in Sec biosynthesis), whereas both dSelK and dSelH function in female flies. As we observed decreased resistance to starvation for both genders, it appears that this effect is mediated by dSelK. The specific function of SelK is not known, and ours is the most significant phenotype of SelK knock-out observed in any organism thus far.
The present study is also the first to offer a strategy for expression of exogenous selenoproteins in Drosophila. We expressed mouse mMsrB1 in the whole body and the nervous system of fruit flies (Fig. 4). Consistent with the observation that Drosophila SECIS-binding protein 2 exhibits higher affinity toward the type II SECIS element (46), we found that expression of mMsrB1 required the type II dSelK SECIS element. This information should be useful for further attempts in expressing exogenous selenoproteins in insect cell culture, for example, in Drosophila S2 cells or hundreds of Drosophila cell lines currently available. Preparation of recombinant selenoproteins is difficult in any system. With few exceptions, it is not possible to express eukaryotic selenoproteins in Escherichia coli because the bacterial SECIS element is different from that used in eukaryotes and it is located in the coding region of selenoprotein genes. The yeast system also cannot be used as the fungi lost the Sec insertion machinery and do not have selenoproteins. Selenoproteins can be expressed in mammalian cell culture, but this system has its own limitations and is costly.
MsrA and MsrB are enzymes that catalyze the same biochemical reaction, but their overexpression has different effects on the Drosophila lifespan (39–41). Consistent with our previous study (41), no effect of mMsrB1 expression on lifespan was found (Fig. 6, supplemental Fig. S1, and Table 2). Apparently, MsrA has a biological role distinct from that of MsrB. MsrA effectively reduces the free form of methionine-S-sulfoxide to methionine (MsrB has very low activity with free methionine-R-sulfoxide) that could alter sulfur and DNA methylation pathways involved in aging. Drosophila MsrA activates the dFOXO pathway (no data reported for MsrB) and could indirectly prolong longevity through up-regulation of dFOXO target genes. Thus, the protein repair function (common for MsrA and MsrB) is unlikely to account for the MsrA-induced lifespan extension and other MsrA functions should be considered.
Expression of mMsrB1 in the nervous system significantly increased resistance against oxidative stress induced by hydrogen peroxide and paraquat (Figs. 7, B and C, and 8, A and B). Thus, we conclude that mMsrB1 expression elevated an antioxidant capacity of the cells, but it did not change the mean lifespan of flies. Previously, ectopic expression of catalase (56) or manganese superoxide dismutase (57) in mitochondria of fruit flies enhanced resistance to experimental oxidative stress, but the oxidative stress response was not involved in aging. The nervous system has a high production rate of reactive oxygen species and/or low expression level of antioxidant enzymes, which may cause accumulation of oxidatively damaged molecules (59). Consequently, expression of antioxidant enzymes in the nervous system positively correlates with resistance against oxidative stress (40, 60). Remarkably, flies expressing mMsrB1 in the whole body did not show an increased resistance against oxidative stress (Fig. 7, D–F). We have demonstrated that flies with neuronal expression of mMsrB1 were more resistant to starvation (Fig. 8C). It is possible that mMsrB1 can effectively repair proteins that are important for the regulatory effect of the nervous system on starvation pathways such as lipid biosynthesis or catabolism, whereby promoting survivability of starved flies.
We have attempted to specify the dietary bases of the lifespan of mMsrB1-expressing flies by subjecting animals to a chemically defined diet that supported nutritional needs of fruit flies (supplemental Table S1, column 8) and allowed the selenium levels in the diet to be changed by sodium selenite supplementation. However, variations in selenium levels did not correlate with the mean lifespan (supplemental Fig. S2 and Table S2, columns 5–7). Three explanations are possible: (i) the amount of selenium in the diet did not correlate with selenoprotein expression in flies; (ii) mMsrB1 expression was very low even at the highest selenium concentration used (207 nm); and (iii) bolstering antioxidant levels by mMsrB1 overexpression under elevated selenium levels did not delay the aging process in Drosophila. We were also interested in examining conditions of selenium deficiency and utilized a chemically defined diet without selenium supplementation. However, this diet already had 13 nm selenium that might have been sufficient for normal lifespan (supplemental Fig. S2 and Table S2).
To conclude, we found that selenoproteinless Drosophila are more sensitive to starvation than control flies and ascribed this function to SelK. However, lifespan was not altered by disruption of selenoprotein synthesis, expression of an exogenous selenoprotein, or availability of dietary selenium. Thus, selenoproteins appear to function in insects only under conditions of stress, explaining the observation that several species of insects lost all selenoproteins. This notion is also supported by the observation that Drosophila requires very little dietary selenium and has few selenoproteins that are expressed at a low level. The observed reduction of the Drosophila selenoprotein system could be used for further studies of endogenous selenoproteins (e.g. SelK function in male flies), the function of SPS1, and the role of selenium as a micronutrient. In addition, the Drosophila Sec incorporation machinery is useful for expression of exogenous mammalian selenoproteins. Overall, this study clarified several critical issues related to the use of selenium and selenoproteins in fruit flies.
Supplementary Material
Acknowledgment
We are very grateful to Dr. Daryl A. Travnicek (Department of Statistics, University of Nebraska-Lincoln, Lincoln, NE) for help with statistical analyses.
This work was supported, in whole or in part, by National Institutes of Health Grants AG021518, GM061603 (to V. N. G.), and DK074136 (to L. G. H.), the Intramural Research Program at the Center for Cancer Research, NCI (to D. L. H.), and National Research Foundation of Korea Grant 2010-0001240 (to H. Y. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- Sec
- selenocysteine
- SECIS
- selenocysteine insertion sequence
- tRNA[Ser]Sec
- selenocysteine inserting transfer RNA
- dEFsec
- Drosophila elongation factor for tRNA[Ser]Sec
- Msrs
- methionine sulfoxide reductases
- mMsrB1
- B2, or B3, mouse methionine sulfoxide reductase B1, B2 or B3
- dSelH
- Drosophila selenoprotein H
- dSelK
- Drosophila selenoprotein K
- SPS1
- selenophosphate synthetase 1
- dSPS2
- Drosophila selenophosphate synthetase 2
- UAS
- upstream activation sequence
- GAL4
- gene, encoding the yeast transcription activator protein GAL4
- FOXO3a
- forkhead box O3, subclass of the forkhead family of transcription factors
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- KO
- knock-out.
REFERENCES
- 1. Lobanov A. V., Hatfield D. L., Gladyshev V. N. (2009) Biochim. Biophys. Acta 1790, 1424–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bösl M. R., Takaku K., Oshima M., Nishimura S., Taketo M. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5531–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Conrad M., Bornkamm G. W. (2006) in Selenium, Its Molecular Biology and Role in Human Health (Hatfield D. L., Berry M. J., Gladyshev V. N. eds) pp. 195–206, 2nd Ed., Springer, New York [Google Scholar]
- 4. Yant L. J., Ran Q., Rao L., Van Remmen H., Shibatani T., Belter J. G., Motta L., Richardson A., Prolla T. A. (2003) Free Radic. Biol. Med. 34, 496–502 [DOI] [PubMed] [Google Scholar]
- 5. Lobanov A. V., Hatfield D. L., Gladyshev V. N. (2008) Protein Sci. 17, 176–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chapple C. E., Guigó R. (2008) PLoS ONE 3, e2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kanzok S. M., Fechner A., Bauer H., Ulschmid J. K., Müller H. M., Botella-Munoz J., Schneuwly S., Schirmer R., Becker K. (2001) Science 291, 643–646 [DOI] [PubMed] [Google Scholar]
- 8. Corona M., Robinson G. E. (2006) Insect. Mol. Biol. 15, 687–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Missirlis F., Rahlfs S., Dimopoulos N., Bauer H., Becker K., Hilliker A., Phillips J. P., Jäckle H. (2003) Biol. Chem. 384, 463–472 [DOI] [PubMed] [Google Scholar]
- 10. Papp L. V., Lu J., Holmgren A., Khanna K. K. (2007) Antioxid. Redox Signal. 9, 775–806 [DOI] [PubMed] [Google Scholar]
- 11. Hatfield D. L., Carlson B. A., Xu X. M., Mix H., Gladyshev V. N. (2006) Prog. Nucleic Acids Res. Mol. Biol. 81, 97–142 [DOI] [PubMed] [Google Scholar]
- 12. Gromer S., Eubel J. K., Lee B. L., Jacob J. (2005) Cell. Mol. Life Sci. 62, 2414–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allmang C., Wurth L., Krol A. (2009) Biochim. Biophys. Acta 1790, 1415–1423 [DOI] [PubMed] [Google Scholar]
- 14. Squires J. E., Berry M. J. (2008) IUBMB Life 60, 232–235 [DOI] [PubMed] [Google Scholar]
- 15. Hirosawa-Takamori M., Jäckle H., Vorbrüggen G. (2000) EMBO Rep. 1, 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castellano S., Morozova N., Morey M., Berry M. J., Serras F., Corominas M., Guigó R. (2001) EMBO Rep. 2, 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martin-Romero F. J., Kryukov G. V., Lobanov A. V., Carlson B. A., Lee B. J., Gladyshev V. N., Hatfield D. L. (2001) J. Biol. Chem. 276, 29798–29804 [DOI] [PubMed] [Google Scholar]
- 18. Pallares C., Serras F., Corominas M. (2006) in Selenium, Its Molecular Biology and Role in Human Health (Hatfield D. L., Berry M. J., Gladyshev V. N. eds) pp. 343–353, 2nd Ed., Springer, New York [Google Scholar]
- 19. Xu X. M., Carlson B. A., Mix H., Zhang Y., Saira K., Glass R. S., Berry M. J., Gladyshev V. N., Hatfield D. L. (2007) PLoS Biol. 5, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xu X. M., Carlson B. A., Irons R., Mix H., Zhong N., Gladyshev V. N., Hatfield D. L. (2007) Biochem. J. 404, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Novoselov S. V., Kryukov G. V., Xu X. M., Carlson B. A., Hatfield D. L., Gladyshev V. N. (2007) J. Biol. Chem. 282, 11960–11968 [DOI] [PubMed] [Google Scholar]
- 22. Panee J., Stoytcheva Z. R., Liu W., Berry M. J. (2007) J. Biol. Chem. 282, 23759–23765 [DOI] [PubMed] [Google Scholar]
- 23. Morozova N., Forry E. P., Shahid E., Zavacki A. M., Harney J. W., Kraytsberg Y., Berry M. J. (2003) Genes Cells 8, 963–971 [DOI] [PubMed] [Google Scholar]
- 24. Hirosawa-Takamori M., Chung H. R., Jäckle H. (2004) EMBO Rep. 5, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen C. L., Shim M. S., Chung J., Yoo H. S., Ha J. M., Kim J. Y., Choi J., Zang S. L., Hou X., Carlson B. A., Hatfield D. L., Lee B. J. (2006) Biochem. Biophys. Res. Commun. 348, 1296–1301 [DOI] [PubMed] [Google Scholar]
- 26. Du S., Zhou J., Jia Y., Huang K. (2010) Arch. Biochem. Biophys. 502, 137–143 [DOI] [PubMed] [Google Scholar]
- 27. Verma S., Hoffmann F. W., Kumar M., Huang Z., Roe K., Nguyen-Wu E., Hashimoto A. S., Hoffmann P. R. (2011) J. Immunol. 186, 2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weissbach H., Resnick L., Brot N. (2005) Biochim. Biophys. Acta 1703, 203–212 [DOI] [PubMed] [Google Scholar]
- 29. Moskovitz J. (2005) Biochim. Biophys. Acta 1703, 213–219 [DOI] [PubMed] [Google Scholar]
- 30. Kim H. Y., Gladyshev V. N. (2007) Biochem. J. 407, 321–329 [DOI] [PubMed] [Google Scholar]
- 31. Levine R. L., Mosoni L., Berlett B. S., Stadtman E. R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15036–15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oien D. B., Moskovitz J. (2008) Curr. Top. Dev. Biol. 80, 93–133 [DOI] [PubMed] [Google Scholar]
- 33. Oien D., Moskovitz J. (2007) Amino Acids 32, 603–606 [DOI] [PubMed] [Google Scholar]
- 34. Minniti A. N., Cataldo R., Trigo C., Vasquez L., Mujica P., Leighton F., Inestrosa N. C., Aldunate R. (2009) Aging Cell 8, 690–705 [DOI] [PubMed] [Google Scholar]
- 35. Moskovitz J., Bar-Noy S., Williams W. M., Requena J., Berlett B. S., Stadtman E. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Salmon A. B., Pérez V. I., Bokov A., Jernigan A., Kim G., Zhao H., Levine R. L., Richardson A. (2009) FASEB J. 23, 3601–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Koc A., Gasch A. P., Rutherford J. C., Kim H. Y., Gladyshev V. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moskovitz J., Flescher E., Berlett B. S., Azare J., Poston J. M., Stadtman E. R. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14071–14075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chung H., Kim A. K., Jung S. A., Kim S. W., Yu K., Lee J. H. (2010) FEBS Lett. 584, 3609–3614 [DOI] [PubMed] [Google Scholar]
- 40. Ruan H., Tang X. D., Chen M. L., Joiner M. L., Sun G., Brot N., Weissbach H., Heinemann S. H., Iverson L., Wu C. F., Hoshi T., Chen M. L., Joiner M. A., Heinemann S. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2748–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shchedrina V. A., Vorbrüggen G., Lee B. C., Kim H. Y., Kabil H., Harshman L. G., Gladyshev V. N. (2009) Mech. Ageing Dev. 130, 429–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin M. J., Tang L. Y., Reddy M. N., Shen C. K. (2005) J. Biol. Chem. 280, 861–864 [DOI] [PubMed] [Google Scholar]
- 43. Richardson B. (2003) Ageing Res. Rev. 2, 245–261 [DOI] [PubMed] [Google Scholar]
- 44. Giannakou M. E., Goss M., Jünger M. A., Hafen E., Leevers S. J., Partridge L. (2004) Science 305, 361. [DOI] [PubMed] [Google Scholar]
- 45. Kim H. Y., Gladyshev V. N. (2005) PLoS Biol. 3, e375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Brand A. H., Perrimon N. (1993) Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- 47. Troen A. M., French E. E., Roberts J. F., Selhub J., Ordovas J. M., Parnell L. D., Lai C. Q. (2007) Age 29, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kwon S. Y., Badenhorst P., Martin-Romero F. J., Carlson B. A., Paterson B. M., Gladyshev V. N., Lee B. J., Hatfield D. L. (2003) Mol. Cell. Biol. 23, 8495–8504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takeuchi A., Schmitt D., Chapple C., Babaylova E., Karpova G., Guigo R., Krol A., Allmang C. (2009) Nucleic Acids Res. 37, 2126–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grundner-Culemann E., Martin G. W., 3rd, Harney J. W., Berry M. J. (1999) RNA 5, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim H. Y., Gladyshev V. N. (2004) Mol. Biol. Cell 15, 1055–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vermeulen C. J., Loeschcke V. (2007) Exp. Gerontol. 42, 153–159 [DOI] [PubMed] [Google Scholar]
- 53. Wang H. D., Kazemi-Esfarjani P., Benzer S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12610–12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Andziak B., O'Connor T. P., Qi W., DeWaal E. M., Pierce A., Chaudhuri A. R., Van Remmen H., Buffenstein R. (2006) Aging Cell 5, 463–471 [DOI] [PubMed] [Google Scholar]
- 55. Mockett R. J., Bayne A. C., Kwong L. K., Orr W. C., Sohal R. S. (2003) Free Radic. Biol. Med. 34, 207–217 [DOI] [PubMed] [Google Scholar]
- 56. Mockett R. J., Sohal B. H., Sohal R. S. (2010) Free Radic. Biol. Med. 49, 2028–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alsina B., Serras F., Baguñá J., Corominas M. (1998) Mol. Gen. Genet. 257, 113–123 [DOI] [PubMed] [Google Scholar]
- 58. Persson B. C., Böck A., Jackle H., Vorbrüggen G. (1997) J. Mol. Biol. 274, 174–180 [DOI] [PubMed] [Google Scholar]
- 59. Hamilton M. L., Van Remmen H., Drake J. A., Yang H., Guo Z. M., Kewitt K., Walter C. A., Richardson A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10469–10474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Parkes T. L., Elia A. J., Dickinson D., Hilliker A. J., Phillips J. P., Boulianne G. L. (1998) Nat. Genet. 19, 171–174 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.