Abstract
Cadmium (Cd) is a heavy metal toxic to humans and the accumulation of Cd in the rice grain is a major agricultural problem, particularly in Asia. The role of the iron transporter OsNRAMP1 in Cd uptake and transport in rice was investigated here. An OsNRAMP1:GFP fusion protein was localized to the plasma membrane in onion epidermal cells. The growth of yeast expressing OsNRAMP1 was impaired in the presence of Cd compared with yeast transformed with an empty vector. Moreover, the Cd content of OsNRAMP1-expressing yeast exceeded that of the vector control. The expression of OsNRAMP1 in the roots was higher in a high Cd-accumulating cultivar (Habataki) than a low Cd-accumulating cultivar (Sasanishiki) regardless of the presence of Cd, and the amino acid sequence of OsNRAMP1 showed 100% identity between Sasanishiki and Habataki. Over-expression of OsNRAMP1 in rice increased Cd accumulation in the leaves. These results suggest that OsNRAMP1 participates in cellular Cd uptake and Cd transport within plants, and the higher expression of OsNRAMP1 in the roots could lead to an increase in Cd accumulation in the shoots. Our results indicated that OsNRAMP1 is an important protein in high-level Cd accumulation in rice.
Keywords: Cadmium, metal transporter, OsNRAMP1, phytoremediation, rice
Introduction
Cadmium (Cd) is one of the most toxic heavy metals, and causes serious health problems in humans. Cd accumulates in the human body through the food chain. Accumulation of Cd in rice grain has become a growing threat to agriculture in Asia in recent years because rice is a dietary staple for Asians and a major source of Cd. In Japan, Cd intake from rice accounts for about one-half of the intake from food (Food Safety Commission, 2008). The Codex Alimentarius Commission established a Cd content of 0.4 mg kg−1 polished rice grain as the limit for food safety and human health (CODEX, 2006). Thus, new technologies to reduce the Cd content in rice grains are urgently required to reduce the Cd risk.
One method of reducing Cd in rice grain is to modify the genes involved in Cd uptake or translocation. Although the mechanism underlying the uptake and translocation of Cd in plants is not completely understood, some iron (Fe) transporters take up Cd. Ferrous Fe transporters, OsIRT1 and OsIRT2, take up Cd (Nakanishi et al., 2006), and over-expression of OsIRT1 has been shown to increase Cd accumulation (Lee and An, 2009). Arabidopsis over-expressing AtIRT1 and AtIRT2 also showed increased Cd accumulation (Connolly et al., 2002; Vert et al., 2009).
The natural resistance-associated macrophage proteins (NRAMPs) consist of a large family of integral membrane proteins, and NRAMP genes have been identified in several plant species (Williams et al., 2000; Bereczky et al., 2003; Kaiser et al., 2003; Mizuno et al., 2005; Xiao et al., 2008; Oomen et al., 2009; Wei et al., 2009). The NRAMP family of metal transporters function as Fe transporters. AtNRAMP1 expression is induced by Fe deficiency in the roots and is considered to control Fe homeostasis in plants (Curie et al., 2000). AtNRAMP3 and AtNRAMP4 are also induced by Fe starvation and mobilize vacuolar Fe (Thomine et al., 2000, 2003; Lanquar et al., 2005). Yeast expressing AtNRAMP1, AtNRAMP3, AtNRAMP4, and AtNRAMP6 showed increased sensitivity to Cd, and the over-expression of AtNRAMP3, AtNRAMP4, and At NRAMP6 in Arabidopsis conferred hypersensitivity to Cd (Thomine et al., 2000; Lanquar et al., 2004; Cailliatte et al., 2009). These results suggest that NRAMP proteins also mediate Cd transport. Rice has seven NRAMP genes (Narayanan et al., 2007), and OsNRAMP1 is involved in Fe uptake function in yeast (Curie et al., 2000).
In rice, there is genotypic variation in the Cd levels of grains and shoots, with Cd accumulation in both shoots and grains being generally greater in indica rice cultivars (including Habataki, Jarjan, Anjana Dhan, and Cho-ko-koku) than in japonica cultivars (including Sasanishiki, Nipponbare, and Tsukinohikari; Arao and Ae, 2003; Ishikawa et al., 2005; Uraguchi et al., 2009). Uraguchi et al. (2009) showed that the Cd concentration of xylem sap was higher in indica cultivars. These results suggest that xylem-mediated Cd translocation from roots to shoots is a major factor determining Cd accumulation levels in shoots and grains of rice.
Recently, quantitative trait loci (QTL) for Cd concentration in rice were identified on chromosome 7 (Tezuka et al., 2009; Ueno et al., 2009; Ishikawa et al., 2010). The responsive gene for this QTL of Cho-ko-koku and Anjana Dhan was identified as OsHMA3 (Miyadate et al., 2011; Ueno et al., 2010). Ueno et al. (2010) reported that the amino acid at position 80 in OsHMA3 is critical for the function, and that mutation of this amino acid caused loss of function to sequester Cd into vacuoles in root cells, resulting in the high translocation of Cd from the roots to the shoots in Anjana Dhan. According to this model, Cd concentration in the cytoplasm is higher in the root cells of high Cd-accumulating cultivars than in low Cd-accumulating cultivars. However, the difference in Cd uptake in the root cells between high and low Cd-accumulating cultivars has not been clarified.
In the present study, the role of OsNRAMP1 in Cd uptake and accumulation in rice was investigated using japonica and indica rice cultivars.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa L. cv. Sasanishiki, Nipponbare, Tsukinohikari, Habataki, Jarjan, Anjana Dhan, and Cho-ko-koku) seeds were germinated for 3 d at room temperature on paper soaked with distilled water. After germination, seedlings were transferred to a net floating on distilled water and grown for 7 d at room temperature. Transgenic rice T1 seeds were germinated on MS medium containing 50 mg l−1 hygromycin B and grown for 2 weeks at 28 °C. Seedlings were transferred to a 20 l plastic container and cultivated hydroponically in a greenhouse (30 °C, natural light). The composition of the nutrient solution was described previously by Ishimaru et al. (2006). The nutrient solution was adjusted every day to pH 5.5 with 1 M HCl and changed once per week. For Cd stress treatments, 2-week-old plants were transferred to a nutrient solution containing 0.2 or 1 μM CdCl2 and cultivated for an additional week. For Fe deficiency treatments, 3-week-old plants were transferred to a nutrient solution without Fe and cultivated for another 1 week.
Plant metal content
Harvested plants were washed with tap water and rinsed with distilled water, then blotted using paper towels. The plants were then separated into leaves and roots and dried at 70 °C for 1 week. Sample digestion was performed as described previously by Ishimaru et al. (2010), except that the digestion time and temperature were changed to 20 min at 230 °C. Metal concentrations were measured using inductively coupled plasma atomic emission spectrometry (SPS1200VR; Seiko, Tokyo, Japan). Three biological replicates were used for each treatment.
Cloning of OsNRAMP1 and OsHMA3
Total RNA was extracted from rice using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and then reverse-transcribed using an oligo(dT) primer and SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA). The complete open reading frame (ORF) of OsNRAMP1 was obtained by PCR using the primers HindIII-Forward 5′-CGACTAAGCTTAAGAAGCCGCACTAGTATG-3′ and XbaI-Reverse 5′-CCGGTCTAGAAGGGTACTACACGGGTGGCT-3′, which were designed based on sequence information from the RAP-DB database (http://rapdb.dna.affrc.go.jp). A partial fragment of OsHMA3 was obtained by PCR using forward (5′-CACCATGGCCGGAAAGGATGAGGCG-3′) and reverse (5′-GGCGTGTAGTACTTTGCGCAC-3′) primers.
Subcellular localization of OsNRAMP1
The OsNRAMP1 ORF without the stop codon was cloned into the plant transient expression vector pH7FWG2 (Karimi et al., 2002) using the Gateway system (Invitrogen) to generate a fusion construct with green fluorescent protein (GFP) as an additional C-terminal domain under the control of the CaMV 35S promoter. The OsNRAMP1:GFP fusion construct or an untagged GFP construct was transiently expressed in onion epidermal cells transformed by DNA particle bombardment. After 8 h, fluorescent cells were imaged by confocal microscopy (LSM5Pascal; Carl Zeiss, Göttingen, Germany). To induce cell plasmolysis, onion epidermal cells were soaked in 20% sucrose solution for 20 min before imaging.
Yeast growth conditions and 109Cd uptake analysis
The OsNRAMP1 ORF was inserted into the expression vector pDR195 (Rentsch et al., 1995), then introduced into yeast strain ycf1 using the lithium acetate method. The ycf1 mutant lacks the function for the compartmentalization of Cd into vacuoles (Li et al., 1997). Yeast ycf1 cells transformed with the empty pDR195 vector were used as controls. The transformed yeast cells were grown in synthetic defined (SD) medium and spotted onto SD agar containing 10 μM CdCl2.
For the 109Cd uptake experiment, transformed yeast cells were amplified in SD medium. A 109Cd uptake assay was performed as described previously (Eide et al., 1992), except that 109CdCl2 was substituted for 59FeCl3. Cell samples were collected by centrifugation using Ultrafree MC (pore size 0.45 μm; Millipore, Billerica, MA, USA) and washed twice. The yeast 109Cd content was measured using an AutoWell Gamma System (ARC-300; Aloka, Tokyo, Japan).
Analysis of OsNRAMP1 expression
Total RNA prepared from the shoots or roots of the plants was reverse-transcribed as described above. Quantitative RT-PCR was then performed using the Smart Cycler System (Takara, Shiga, Japan). Amplification was performed using forward (5′-GGATTCTCCTGGGTGCTGGGGTT-3′) and reverse (5′-GCAACAATCTACTCCCATGGGCC-3′) primers with SYBR Premix Ex Taq (Perfect Real Time; Takara); α-tubulin was used as an internal standard (Ishimaru et al., 2006). The products were subcloned using a Zero Blunt TOPO PCR Cloning Kit (Invitrogen). Serially diluted vector was also amplified to calculate the OsNRAMP1 copy number.
Genomic DNA extraction
Leaves were ground thoroughly in liquid nitrogen, and suspended in extraction buffer (200 mM TRIS-HCl, pH 7.5, 250 mM NaCl, 25 mM EDTA, and 0.5% SDS). After incubation for 1 h at room temperature, the supernatant was transferred to a new tube and the DNA was purified by 2-propanol precipitation.
Rice transformation
OsNRAMP1-overexpressing rice was produced using a pH7FWG2-OsNRAMP1 vector described in the section dealing with subcellular localization of OsNRAMP1. Agrobacterium-mediated transformation of rice (cv. Tsukinohikari) was performed as described previously (Higuch et al., 2001). Eight independently transformed over-expressing rice plants (35S-rice) were generated, and two lines showing high levels of OsNRAMP1 expression in the whole plant were selected (see Supplementary Fig. S1 at JXB online).
Results
OsNRAMP1 is localized to the plasma membrane and functions in Cd uptake
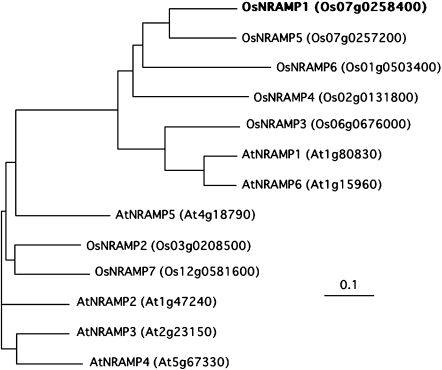
The ORFs of OsNRAMP1 from three rice cultivars (Nipponbare, Sasanishiki, Habataki) were isolated (Fig. 1). There was no difference in nucleotide or amino acid sequence among the three cultivars.
Fig. 1.
Phylogenetic tree of the NRAMP proteins of rice and Arabidopsis. The tree was constructed using Clustal X and the Tree View program with the amino acid sequences of NRAMP.
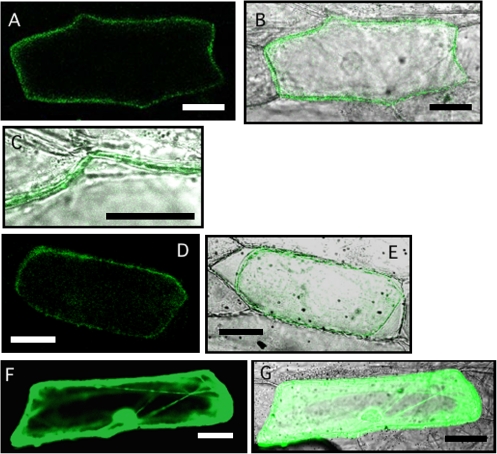
The subcellular localization of OsNRAMP1 was investigated using an OsNRAMP1:GFP fusion protein. Green fluorescence was observed at the periphery of the cells but not inside the cells (Fig. 2A–C). Plasmolysis of the cells was performed to confirm the plasma membrane localization of the protein (Fig. 2D, E).
Fig. 2.
Subcellular localization of OsNRAMP1 in onion epidermal cells. (A) OsNRAMP1 fused to GFP was transiently expressed in onion epidermal cells. (B) Overlay with the transmission image shown in (A). (C) Higher magnification view of (B). (D) OsNRAMP1–GFP expressed in onion epidermal cells under high osmotic conditions. (E) Overlay with the transmission image shown in (D). (F) GFP alone was transiently expressed in onion epidermal cells. (G) Overlay with transmission image shown in (F). Scale bars=50 μm.
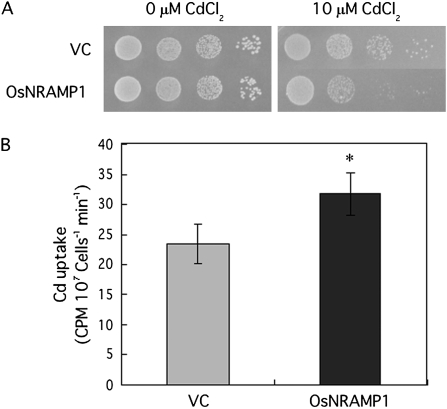
The growth of yeast expressing OsNRAMP1 was impaired compared with that of yeast transformed with the empty vector in the presence of 10 μM Cd (Fig. 3A). Furthermore, with 1 μM Cd treatment for 30 min, the Cd content of the OsNRAMP1-expressing yeast was 1.4 times higher than that of the control (Fig. 3B).
Fig. 3.
(A) Growth of yeast cells expressing OsNRAMP1. A Cd-sensitive mutant, ycf1, was transformed with the empty vector (VC) or with the plasmid containing OsNRAMP1. Yeast cell suspensions were spotted on synthetic defined medium containing 10 μM CdCl2. (B) Cd uptake in yeast. Yeast cells were treated with 1 μM CdCl2 and incubated at 30 °C for 30 min. Non-specific uptake due to cell-surface absorption was determined by preparing parallel assays that were kept on ice for 30 min. The results are presented as the means ±SD of triplicate samples. Asterisks indicate significant difference at P <0.05 by Student's t test.
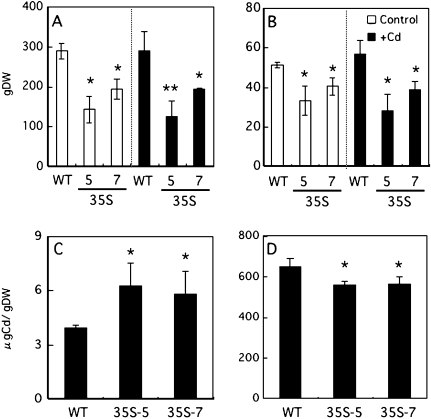
In the presence of 1 μM Cd, the growth of OsNRAMP1-overexpressing rice was significantly inhibited in both shoots and roots (Fig. 4A, B). The growth of OsNRAMP1-overexpressing rice was also inhibited under the control condition (Fig. 4A, B). The Cd concentration in the leaves of 35S-rice was significantly higher than that of non-transformed rice (WT; Fig. 4C). On the other hand, the concentrations of other essential metals (Fe, Zn, Mn, Cu) were not changed in the presence of 1 μM Cd, except for the Fe concentration in 35S-7 (see Supplementary Fig. S2 at JXB online). The Cd concentration in the roots of 35S-rice was lower than that in the wild type (Fig. 4D).
Fig. 4.
Dry weight of the shoots (A) and the roots (B) of 35S-rice in the absence (control) or presence of 1 μM CdCl2. (C) Cd concentration in the leaves of 35S-rice. (D) Cd concentration in the roots of 35S-rice. The results are presented as the means ±SD of triplicate samples. Asterisks indicate significantly different with the wild type at *P <0.05 and **P <0.01 by Student's t test.
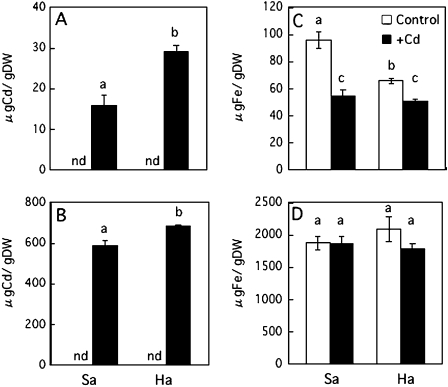
Metal concentrations in the roots and the leaves of Sasanishiki and Habataki
In the presence of 1 μM Cd, the Cd concentrations in the leaves and the roots of Habataki were 1.8 times and 1.2 times higher than those of Sasanishiki, respectively (Fig. 5A, B). Fe concentration in the leaves was decreased in both Sasanishiki and Habataki by Cd treatment, whereas those in the roots remained essentially unchanged (Fig. 5C, D). In the presence of Cd, the concentrations of the other essential metals (Zn, Mn, Cu) also decreased in the leaves and roots of Sasanishiki and Habataki (see Supplementary Fig. S3 at JXB online).
Fig. 5.
Concentrations of Cd (A, B) and Fe (C, D) in the leaves (A, C) or roots (B, D) of Sasanishiki (Sa) and Habataki (Ha) grown in the absence (control) or presence of 1 μM CdCl2. nd, Not detected by ICP-AES. The results are presented as the means ±SD of triplicate samples. Different letters indicate significant differences at P <0.01 by the Tukey–Kramer test.
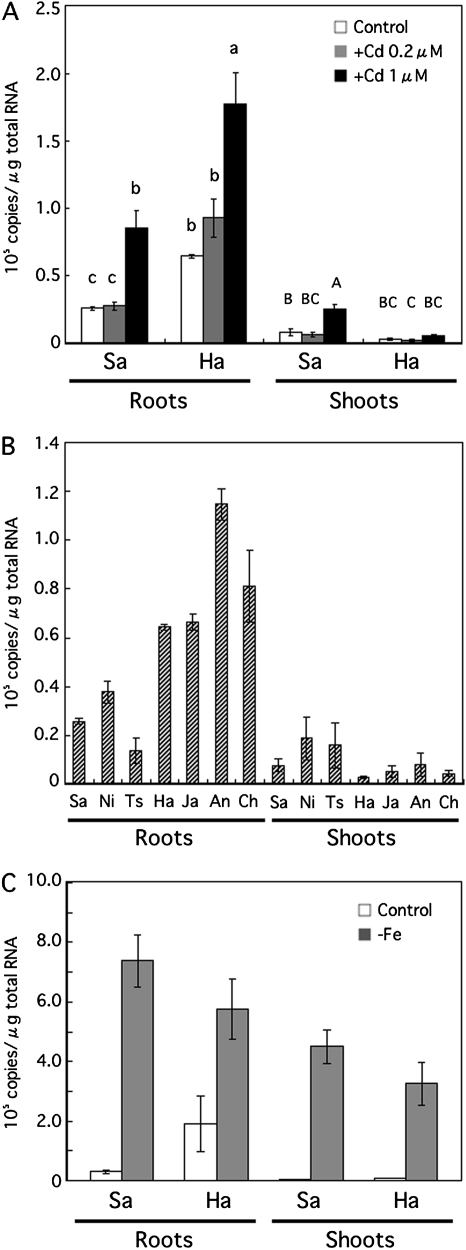
Expression analysis of OsNRAMP1 in different cultivars
The level of OsNRAMP1 expression in the roots was increased in both Sasanishiki and Habataki in the presence of 1 μM Cd (Fig. 6A). The levels of OsNRAMP1 expression in the roots of Habataki were higher than those of Sasanishiki in the presence of 0 (control), 0.2 and 1 μM Cd. In addition, a higher expression of OsNRAMP1 in roots was also observed in other Cd-accumulating indica cultivars, including Jarjan, Anjana Dhan, and Cho-ko-koku, compared with the low Cd-accumulating japonica cultivars, Nipponbare and Tsukinohikari (Fig. 6B). Expression of OsNRAMP1 in shoots was increased only in Sasanishiki in the presence of Cd (Fig. 6A).
Fig. 6.
Quantitative RT-PCR analysis of OsNRAMP1. (A) Total RNA was extracted from roots or shoots of Sasanishiki (Sa) and Habataki (Ha) grown in the absence (control) or presence of 0.2 or 1 μM CdCl2 for 1 week. (B) Total RNA was extracted from the roots or the shoots of Sasanishiki (Sa), Nipponbare (Ni), Tsukinohikari (Ts), Habataki (Ha), Jarjan (Ja), Anjana Dhan (An), and Cho-ko-koku (Ch) grown without Cd. (C) Total RNA was extracted from the roots or shoots of Sasanishiki (Sa) and Habataki (Ha) grown for 1 week under Fe-deficient conditions. The results are presented as the means ±SD of triplicate samples. Different letters indicate significant differences at P <0.01 by the Tukey–Kramer test.
Under conditions of Fe deficiency, the expression of OsNRAMP1 was increased in both the shoots and roots of Sasanishiki and Habataki; the expression levels in the two cultivars were almost identical (Fig. 6C).
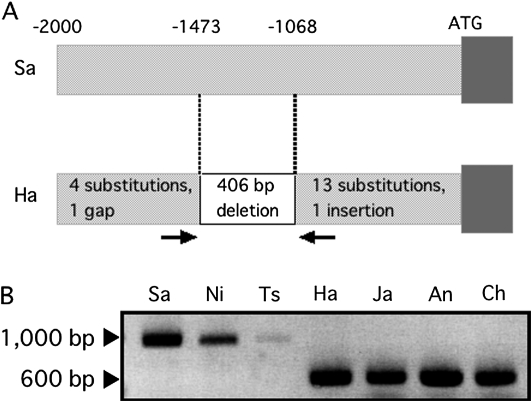
Analysis of the promoter sequence
To gain an insight into the differential expression of OsNRAMP1 among cultivars, the 2 kb 5'-flanking regions of OsNRAMP1 from Sasanishiki and Habataki were isolated and compared. Seventeen nucleotides were substituted, and one nucleotide gap and one nucleotide insertion were found in Habataki (Fig. 7A). Furthermore, 406 bp (−1473 to −1068 bp from the translational initiation codon in Sasanishiki) were missing in Habataki (Fig. 7A). This 400-bp deletion was also found in the 5'-flanking region of other high Cd-accumulating cultivars, including Jarjan, Anjana Dhan, and Cho-ko-koku, whereas it was not found in low Cd-accumulating cultivars, including Nipponbare and Tsukinohikari (Fig. 7B).
Fig. 7.
(A) Structure of the 2 kb 5'-flanking regions of OsNRAMP1 from Sasanishiki (Sa) and Habataki (Ha). (B) Amplification of the 5'-flanking regions of OsNRAMP1 from Sasanishiki (Sa), Nipponbare (Ni), Tsukinohikari (Ts), Habataki (Ha), Jarjan (Ja), Anjana Dhan (An), and Cho-ko-koku (Ch) using the primers indicated by arrows in (A).
Amino acid sequence of OsHMA3 in different cultivars
Part of OsHMA3 was isolated from Sasanishiki and Habataki, and the predicted amino acid sequences were compared (Fig. 8). The substitution from Arg to His at position 80 was observed only in Anjana Dhan and Cho-ko-koku, and the amino acid sequence of OsHMA3 from Habataki was the same as those of the low Cd-accumulating cultivars, Sasanishiki and Nipponbare, at this position.
Fig. 8.
Part of the alignment of OsHMA3 from Sasanishiki (Sa), Habataki (Ha), Nipponbare (Ni), Anjana Dhan (An), and Cho-ko-koku (Ch). Position 80, which is critical for the function of OsHMA3, is indicated by an asterisk.
Discussion
In this study, OsNRAMP1 was isolated and characterized from the low Cd-accumulating rice cultivar Sasanishiki and the high Cd-accumulating cultivar Habataki. OsNRAMP1 was localized to the plasma membrane (Fig. 2), and OsNRAMP1-expressing yeast accumulated larger amounts of Cd than the vector control (Fig. 3B). Furthermore, over-expression of OsNRAMP1 in rice increased Cd accumulation in the leaves (Fig. 4C). These results indicated that OsNRAMP1 participates in cellular Cd uptake and Cd transport within the plant. AtNRAMP1, which shows high homology with OsNRAMP1, has been reported to act as a Mn transporter (Cailliatte et al., 2010). However, OsNRAMP1 did not complement the Mn-deficient phenotype of the yeast smf1 mutant (data not shown), which lacks the Mn2+ uptake function (Supek et al., 1996).
Fe-deficient rice absorbed larger amounts of Cd than Fe-sufficient rice, suggesting that the induced expression of Fe transporters in response to Fe deficiency contributes to Cd uptake in rice (Nakanishi et al., 2006). Among the major Fe transporters induced by Fe deficiency, OsIRT1 and OsIRT2 take up Cd (Nakanishi et al., 2006). Another Fe transporter family, the OsYSLs, has been identified in rice, and OsYSL2 and OsYSL15 were shown to be strongly induced by Fe deficiency (Koike et al., 2004; Inoue et al., 2009). OsYSL2 may not contribute to Cd uptake in the roots. OsYSL2 transports Fe-nicotianamine and Mn-nicotianamine, and is responsible for the internal transport of Fe and Mn (Koike et al., 2004). By contrast, OsYSL15 transports Fe-deoxymugineic acid (DMA) and is responsible for Fe uptake from the soil (Inoue et al., 2009). Nakanishi et al. (2006) reported that the growth of yeast expressing OsYSL15 under Cd stress did not change significantly in the presence of DMA. Meda et al. (2007) reported that the Cd-DMA complex was weak compared with Fe-DMA, and the specificity of Cd-DMA transport by ZmYS1 was very low (Schaaf et al., 2004), suggesting that OsYSL15 is not involved in Cd transport.
OsNRAMP1 could participate in Cd uptake or transport in rice in addition to OsIRT1 and OsIRT2. Cailliatte et al. (2010) discussed the possibility of Mn acquisition in the roots of Arabidopsis by AtIRT1 and AtNRAMP1, and suggested that these transporters have different affinity constants for Mn and act in different tissues in the roots. They also reported that AtIRT1 is expressed exclusively in the external layers of the absorption zone of the root, whereas AtNRAMP1 expression is more widespread along the roots including the inner layers. Thus, it is possible that OsNRAMP1, OsIRT1, and OsIRT2 also function in different tissues in the roots. OsIRT1 was highly expressed in the epidermis and the cortex around the endodermis in the roots, and considered to function in Fe2+ and Cd uptake from the soil (Ishimaru et al., 2006; Nakanishi et al., 2006). The Cd concentration of OsIRT1-overexpressing rice was higher than that of the wild type in both the roots and the shoots (Lee and An, 2009). On the other hand, the Cd concentration in the leaves of OsNRAMP1-overexpressing rice was higher than that of the wild type, whereas the Cd concentration in the roots was lower (Fig. 4C, D). This result suggests that Cd translocation from the roots to the shoots was highly activated in addition to the increase in Cd uptake from the rhizosphere in OsNRAMP1-overexpressing rice. Furthermore, the growth of OsNRAMP1-overexpressing rice was significantly inhibited compared with WT, indicating that ectopic expression of OsNRAMP1 could disturb metal distribution in rice, and suggesting that OsNRAMP1 may play an important role in Cd distribution in rice. It is possible that OsNRAMP1 transports Cd into the cytoplasm in the inner layers of the roots following Cd uptake from the rhizosphere by OsIRT1, and participates in the control of Cd availability for loading into the xylem. Further detailed analysis to determine the tissue localization of OsNRAMP1 will clarify its role in Cd acquisition and translocation.
The level of OsNRAMP1 expression was higher in the roots of high Cd-accumulating indica cultivars than in low Cd-accumulating japonica cultivars (Fig. 6B). As the OsNRAMP1 of Sasanishiki and Habataki showed 100% identity at the amino acid level, the transport ability of OsNRAMP1 does not differ between the low and high Cd-accumulating cultivars. Thus, it is possible that differences in the expression level of OsNRAMP1 in the roots are responsible for the observed differences in Cd accumulation among these cultivars. Comparative analysis of the putative promoter region of OsNRAMP1 revealed a 406 bp deletion in Habataki (Fig. 7A) that was also found in other high Cd-accumulating cultivars (Fig. 7B). Fe deficiency treatment induced expression of OsNRAMP1 in both Sasanishiki and Habataki (Fig. 6C). The core sequences for binding to known transcription factors related to Fe deficiency, including OsIRO2, IDEF1, and IDEF2 (Ogo et al., 2006, 2008; Kobayashi et al., 2007), do not exist within the deleted 406 bp domain, although there are two IDE2 core sequences in the 2 kb 5'-flanking regions of OsNRAMP1 from Sasanishiki and Habataki. Thus, induction by Fe deficiency was unaffected by the deletion of this region. However, it is possible that other cis-acting elements, which do regulate the expression of OsNRAMP1, exist within this 406 bp region. Three putative DNA-binding domains for MYB transcription factors (GGATA, CNGTT) were identified in this region. Some MYB proteins act as negative regulators of gene expression (Chinnusamy et al., 2007; Matsui et al., 2008), and some MYB genes have been reported to be responsive to Cd (Chen et al., 2006). The difference in OsNRAMP1 expression between Sasanishiki and Habataki may be dependent on these transcription factors.
Recently, QTL for Cd accumulation of Habataki was also identified on chromosome 7 (Ishikawa et al., 2010), but the responsive gene for this QTL has yet to be identified. OsNRAMP1 is located in this QTL region, and the amino acid change for the loss of function of OsHMA3 was not observed in Habataki (Fig. 8). Thus, the OsHMA3 mutation is apparently not the reason for Cd accumulation in the shoots of Habataki. The loss of function of OsHMA3 is believed to lead to an elevated Cd concentration in the cytoplasm and higher loading into the xylem. These observations indicate that Cd content in the root cells is an important factor for Cd translocation from the roots to the shoots and Cd accumulation in the shoots. It seems likely that higher levels of OsNRAMP1 expression in the roots of indica cultivars were responsible for higher Cd uptake in the root cells. Our results suggest that OsNRAMP1 is an important protein for Cd higher accumulation in rice.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Expression of OsNRAMP1 in 35S-rice.
Supplementary Fig. S2. Metal concentrations of the roots and leaves of 35S-rice grown in the absence (control) or presence of 1 μM CdCl2.
Supplementary Fig. S3. Metal concentrations of the roots and leaves of Sasanishiki and Habataki grown in the absence (control) or presence of 1 μM CdCl2.
Acknowledgments
This study was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, GMB0001).
References
- Arao T, Ae N. Genotypic variations in cadmium levels of rice grain. Soil Science and Plant Nutrition. 2003;49:473–479. [Google Scholar]
- Bereczky Z, Wang HY, Schubert V, Ganal M, Bauer P. Differential regulation of nramp and irt metal transporter genes in wild type and iron uptake mutants of tomato. Journal of Biological Chemistry. 2003;278:24697–24704. doi: 10.1074/jbc.M301365200. [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Lapeyre B, Briat J-F, Mari S, Curie C. The NRAMP6 metal transporter contributes to cadmium toxicity. Biochemical Journal. 2009;422:217–228. doi: 10.1042/BJ20090655. [DOI] [PubMed] [Google Scholar]
- Cailliatte R, Schikora A, Briat J-F, Mari S, Curie C. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. The Plant Cell. 2010;22:904–917. doi: 10.1105/tpc.109.073023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Yang X, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu J- K. Cold stress regulation of gene expression in plants. Trends in Plant Science. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- CODEX Report of the 38th session of the CODEX committee on Food Additives and Contaminants. Codex Alimentarius Commission. ALINORM 06/29/12: 2006:1–12. [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. The Plant Cell. 2002;14:1347–1357. doi: 10.1105/tpc.001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Jean ML, Ecker JR, Briat J-F. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochemical Journal. 2000;347:749–755. [PMC free article] [PubMed] [Google Scholar]
- Eide D, Davis-Kaplan S, Jordan I, Sipe D, Kaplan J. Regulation of iron uptake in Saccharomyces cerevisiae. Journal of Biological Chemistry. 1992;267:20774–20781. [PubMed] [Google Scholar]
- Food Safety Commission. 2008 Evaluation reports of food contaminants (in Japanese) [Google Scholar]
- Higuch K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S. Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. The Plant Journal. 2001;25:159–167. doi: 10.1046/j.1365-313x.2001.00951.x. [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK. Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. Journal of Biological Chemistry. 2009;284:3470–3479. doi: 10.1074/jbc.M806042200. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Ae N, Sugiyama M, Murakami M, Arao T. Genotypic variation in shoot cadmium concentration in rice and soybean in soils with different levels of cadmium contamination. Soil Science and Plant Nutrition. 2005;51:101–108. [Google Scholar]
- Ishikawa S, Abe T, Kuramata M, Yamaguchi M, Ando T, Yamamoto T, Yano M. A major quantitative trait locus for increasing cadmium-specific concentration in rice grain is located on the short arm of chromosome 7. Journal of Experimental Botany. 2010;61:923–934. doi: 10.1093/jxb/erp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Masuda H, Bashir K, et al. Rice–metal–nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. The Plant Journal. 2010;62:379–390. doi: 10.1111/j.1365-313X.2010.04158.x. [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+ The Plant Journal. 2006;45:335–346. doi: 10.1111/j.1365-313X.2005.02624.x. [DOI] [PubMed] [Google Scholar]
- Kaiser BN, Moreau S, Castelli J, Thomson R, Lambert A, Bogliolo S, Puppo A, Day DA. The soybean NRAMP homologue, GmDMT1, is a symbiotic divalent metal transporter capable of ferrous iron transport. The Plant Journal. 2003;35:295–304. doi: 10.1046/j.1365-313x.2003.01802.x. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Ogo Y, Itai RN, Nakanishi H, Takahashi M, Mori S, Nishizawa NK. The transcription factor IDEF1 regulates the response to and tolerance of iron deficiency in plants. Proceedings of the National Academy of Sciences, USA. 2007;104:19150–19155. doi: 10.1073/pnas.0707010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. OsYSL2 is a rice metal–nicotianamine transporter that is regulated by iron and expressed in the phloem. The Plant Journal. 2004;39:415–424. doi: 10.1111/j.1365-313X.2004.02146.x. [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Barbier-Brygoo H, Thomine S. Regulation and function of AtNRAMP4 metal transporter protein. Soil Science and Plant Nutrition. 2004;50:1141–1150. [Google Scholar]
- Lanquar V, Lelievre F, Bolte S, et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO Journal. 2005;24:4041–4051. doi: 10.1038/sj.emboj.7600864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, An G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant, Cell and Environment. 2009;32:408–416. doi: 10.1111/j.1365-3040.2009.01935.x. [DOI] [PubMed] [Google Scholar]
- Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis (glutathionato) cadmium. Proceedings of the National Academy of Sciences, USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. The Plant Journal. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- Meda AR, Scheuermann EB, Prechsl UE, Erenoglu B, Schaaf G, Hayen H, Weber G, von Wiren N. Iron acquisition by phytosiderophores contributes to cadmium tolerance. Plant Physiology. 2007;143:1761–1773. doi: 10.1104/pp.106.094474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyadate H, Adachi S, Hiraizumi A, et al. OsHMA3, a P1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytologist. 2011;1:190–199. doi: 10.1111/j.1469-8137.2010.03459.x. [DOI] [PubMed] [Google Scholar]
- Mizuno T, Usui K, Horie K, Nosaka S, Mizuno N, Obata H. Cloning of three ZIP/Nramp transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+-transport abilities. Plant Physiology and Biochemistry. 2005;43:793–801. doi: 10.1016/j.plaphy.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Science and Plant Nutrition. 2006;52:464–469. [Google Scholar]
- Narayanan NN, Vasconcelos MW, Grusak MA. Expression profiling of Oryza sativa metal homeostasis genes in different rice cultivars using a cDNA macroarray. Plant Physiology and Biochemistry. 2007;45:277–286. doi: 10.1016/j.plaphy.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Itai RN, Nakanishi H, Inoue H, Kobayashi T, Suzuki M, Takahashi M, Mori S, Nishizawa NK. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. Journal of Experimental Botany. 2006;57:2867–2878. doi: 10.1093/jxb/erl054. [DOI] [PubMed] [Google Scholar]
- Ogo Y, Kobayashi T, Itai RN, Nakanishi H, Kakei Y, Takahashi M, Toki S, Mori S, Nishizawa NK. A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. Journal of Biological Chemistry. 2008;283:13407–13417. doi: 10.1074/jbc.M708732200. [DOI] [PubMed] [Google Scholar]
- Oomen RJFJ, Wu J, Lelievre F, Blanchet S, Richaud P, Barbier-Brygoo H, Aarts MGM, Thomine S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator. Thlaspi caerulescens. New Phytologist. 2009;181:637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. NTR1encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Letters. 1995;370:264–268. doi: 10.1016/0014-5793(95)00853-2. [DOI] [PubMed] [Google Scholar]
- Schaaf G, Ludewig U, Erenoglu BE, Mori S, Kitahara T, Wiren N. ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. Journal of Biological Chemistry. 2004;279:9091–9096. doi: 10.1074/jbc.M311799200. [DOI] [PubMed] [Google Scholar]
- Supek F, Supekova L, Nelson H, Nelson N. A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proceedings of the National Academy of Sciences, USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezuka K, Miyadate H, Katou K, et al. A single recessive gene controls cadmium translocation in the cadmium hyperaccumulating rice cultivar Cho-Ko-Koku. Theoretical and Applied Genetics. 2009;120:1175–1182. doi: 10.1007/s00122-009-1244-6. [DOI] [PubMed] [Google Scholar]
- Thomine S, Lelievre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. The Plant Journal. 2003;34:685–695. doi: 10.1046/j.1365-313x.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proceedings of the National Academy of Sciences, USA. 2000;97:4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno D, Koyama E, Kono I, Ando T, Yano M, Ma JF. Identification of a novel major quantitative trait locus controlling distribution of Cd between roots and shoots in rice. Plant and Cell Physiology. 2009;50:2223–2233. doi: 10.1093/pcp/pcp160. [DOI] [PubMed] [Google Scholar]
- Ueno D, Yamaji N, Kono I, Huang CF, Ando T, Yano M, Ma JF. Gene limiting cadmium accumulation in rice. Proceedings of the National Academy of Sciences, USA. 2010;107:16500–16505. doi: 10.1073/pnas.1005396107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S. Root-to-shoot Cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. Journal of Experimental Botany. 2009;60:2677–2688. doi: 10.1093/jxb/erp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Barberon M, Zelazny E, Seguela M, Briat JF, Curie C. Arabidopsis IRT2 cooperates with the high-affinity iron uptake system to maintain iron homeostasis in root epidermal cells. Planta. 2009;229:1171–1179. doi: 10.1007/s00425-009-0904-8. [DOI] [PubMed] [Google Scholar]
- Wei W, Chai T, Zhang Y, Han L, Xu J, Guan Z. The Thlaspi caerulescens NRAMP homologue TcNRAMP3 is capable of divalent cation transport. Molecular Biotechnology. 2009;41:15–21. doi: 10.1007/s12033-008-9088-x. [DOI] [PubMed] [Google Scholar]
- Williams LE, Pittman JK, Hall JL. Emerging mechanisms for heavy metal transport in plants. Biochimica et Biophysica Acta. 2000;1465:104–126. doi: 10.1016/s0005-2736(00)00133-4. [DOI] [PubMed] [Google Scholar]
- Xiao H, Yin L, Xu X, Li T, Han Z. The iron-regulated transporter, MbNRAMP1, isolated from Malus baccata is involved in Fe, Mn and Cd trafficking. Annals of Botany. 2008;102:881–889. doi: 10.1093/aob/mcn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.