Abstract
The MyD88-independent pathway, one of the two crucial TLR signaling routes, is thought to be a vertebrate innovation. However, a novel Toll/interleukin-1 receptor (TIR) adaptor, designated bbtTICAM, which was identified in the basal chordate amphioxus, links this pathway to invertebrates. The protein architecture of bbtTICAM is similar to that of vertebrate TICAM1 (TIR-containing adaptor molecule-1, also known as TRIF), while phylogenetic analysis based on the TIR domain indicated that bbtTICAM is the oldest ortholog of vertebrate TICAM1 and TICAM2 (TIR-containing adaptor molecule-2, also known as TRAM). Similar to human TICAM1, bbtTICAM activates NF-κB in a MyD88-independent manner by interacting with receptor interacting protein (RIP) via its RHIM motif. Such activation requires bbtTICAM to form homodimers in endosomes, and it may be negatively regulated by amphioxus SARM (sterile α and armadillo motif-containing protein) and TRAF2. However, bbtTICAM did not induce the production of type I interferon. Thus, our study not only presents the ancestral features of vertebrate TICAM1 and TICAM2, but also reveals the evolutionary origin of the MyD88-independent pathway from basal chordates, which will aid in understanding the development of the vertebrate TLR network.
Keywords: TLR, TICAM, MyD88-independent pathway, innate immunity, evolution
Introduction
Toll-like receptors (TLRs) play a critical role in innate immunity by detecting invading pathogens 1. Downstream signaling of mammalian TLRs involves 5 TIR (Toll/interleukin-1 receptor) domain-containing adaptors, including MyD88, MAL (MyD88-adaptor-like, also known as TIRPA), TICAM1 (TIR-containing adaptor molecule-1, also known as TRIF), TICAM2 (TIR-containing adaptor molecule-2, also known as TRAM), and SARM (sterile α and armadillo motif-containing protein) 2. MyD88 mediates a universal pathway for all vertebrate TLRs except TLR3, whereas MAL functions as a partner for MyD88 when it is used by TLR4 3, 4. TICAM1 mediates a MyD88-independent pathway specifically utilized by TLR3 5. When coupled with TICAM2, TICAM1 can also be recruited by TLR4, leading to the activation of nuclear factor-κB (NF-κB) and the induction of type I interferon (IFN) 6.
MyD88, the first-identified TIR adaptor in mammals, is also present in poriferans 7, cnidarians 8, Drosophila 9, sea urchin 10, 11, and amphioxus 12. The direct interaction of MyD88 with TLRs in Drosophila and amphioxus suggests that this pathway is functionally conserved during evolution 13. Unlike MyD88, no homolog of TICAM1 or TICAM2 has been identified in non-chordates such as Drosophila, Cnidaria 8, or sea urchin 10, 11. Although TICAM1 orthologs could be found in the early vertebrates lamprey 14 and zebrafish 15, 16, 17, 18, 19, they do not function in the same manner as those found in mammals. For example, human TICAM1 can induce the secretion of type I interferon by activating transcriptional factors interferon-regulatory factor (IRF) 3 and IRF7 via interaction with tumor necrosis factor receptor-associated factor 6 (TRAF6) 20, while zebrafish TICAM1, localized on the Golgi apparatus 18, induces production of zebrafish IFN in a IRF3/7-independent manner and activates NF-κB via interaction with RIP, but not with TRAF6 17. Zebrafish TICAM1 is novel and exhibits unique structural and functional features, which are not observed in mammalian TICAM1 17.
The genomic comparison of mammalian TICAM1 and 2 genes indicates that TICAM1 and 2 were duplicated from a common ancestor 17. However, unlike orthologs of mammalian TICAM1 in early vertebrates, homologs of TICAM2 have not been identified in other vertebrates besides mammals and shark 21, such as chicken, Xenopus, zebrafish, and lamprey 14, 17, 22, 23. TICAM2 specifically acts as a bridge for TICAM1 to be successfully recruited when TLR4 recognizes LPS by coupling with CD14 and MD-2 5, 24, 25. Since endotoxin recognition by TLR4 is absent in lamprey 14, jawfish 19, and Xenopus 22, it was previously believed that the endotoxin recognition complex and the downstream signaling pathway in mammals arose after the divergence of fish and tetrapods 19. Thus, the absence of TICAM2 in bony fish and the rapid evolution from tetrapods to mammals may have been associated with the development of the functional requirement for the formation of the endotoxin recognition complex.
Analysis of the lamprey TLR system has identified two TICAM homologs and suggests that pmTICAM-1b is the ortholog of jawed vertebrate TICAM-1 14. In addition, a TICAM homolog has been identified in the genome of the tunicate Ciona, but its function has been not studied 17. These studies indicate that a common ancestor of vertebrate TICAM1 and TICAM2 may have existed in species that were more primitive than Ciona. Since amphioxus occupies a crucial evolutionary position as a basal chordate, and its genome displays the ancestral features of present-day vertebrates, we performed a search for TICAM-like genes in amphioxus in order to investigate whether the TICAM homolog emerged in basal chordates and how it functioned. Functional studies of this novel molecule will not only help to understand the original formation of the MyD88-independent pathway in basal chordates, but also assist in further understanding the functional divergence of TICAM1 and TICAM2 in vertebrates.
Results
Sequence and structural analysis of bbtTICAM
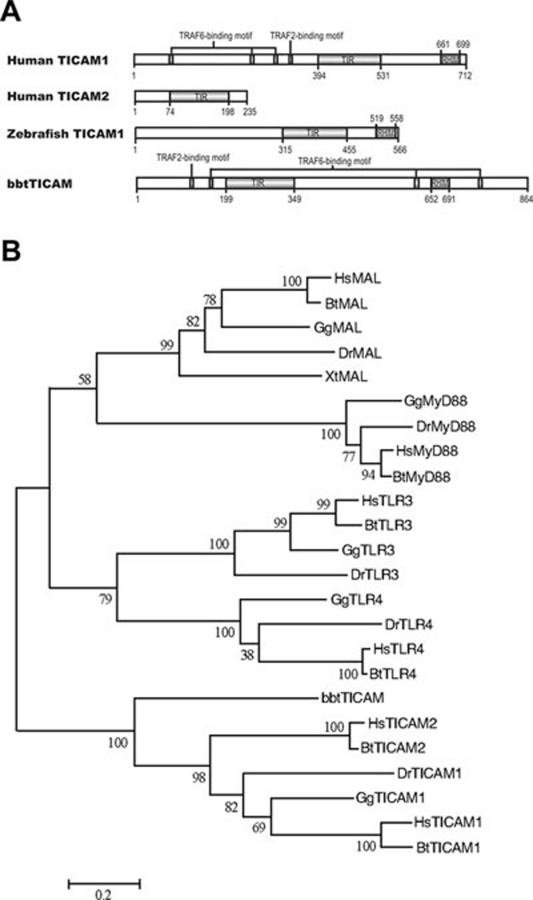
A full-length cDNA of 2 592 bp, similar to mammalian TICAM1, was isolated from a Chinese amphioxus (Branchiostoma belcheri tsingtauense) intestine cDNA library and designated bbtTICAM. bbtTICAM encodes a polypeptide of 864 amino acids with only one highly conserved protein structure, the TIR domain. BLAST analysis of the TIR domain showed 29%-32% amino-acid identity between bbtTICAM and mammalian TICAM2s. However, analysis of the entire sequence revealed one TRAF6-binding motif and one TRAF2-binding motif in the N terminus and two TRAF6-binding motifs and one RHIM motif in the C terminus 26, 27, which is structurally similar to mammalian TICAM1 (Figure 1A). A phylogenetic tree based on TIR domains of TLR3, TLR4, MyD88, MAL, TICAM1 and TICAM2 from human, cow, chicken, Xenopus, zebrafish, and amphioxus was constructed. This phylogenetic tree suggests that the TIR domains of TICAM1 and TICAM2 are different from those of TLR3, TLR4, MyD88 and MAL. The phylogenetic tree also shows that bbtTICAM is the ortholog of vertebrate TICAM1 and TICAM2 (Figure 1B). Amphioxus is a basal chordate, and no TICAM ortholog is present in any non-chordate based on current genomic analysis. Therefore, bbtTICAM is the earliest ortholog of TICAM molecules. In other words, mammalian TICAM1 and TICAM2 may have originated from this ancestral molecule of the basal chordate through gene duplication and function refinement.
Figure 1.
Sequence analysis of bbtTICAM. (A) Domain topology of bbtTICAM compared with humanTICAM1, humanTICAM2 and zebrafishTICAM1. The TIR domains were predicted by the SMART website (http://smart.embl-heidelberg.de/smart/set_mode.cgi?NORMAL=1), whereas the TRAF6-binding motif (PxExx[Ac/Ar], x-represents any residue), TRAF2-binding motif (PxQxS, x-represents any residue), and RHIM motif were manually predicted by sequence alignment according to the literature. (B) Neighbor-joining tree of bbtTICAM with TLR3, TLR4, MAL, MyD88, TICAM1, and TICAM2 was constructed using protein sequences of TIR domain. The numbers at nodes indicated bootstrap values. Clustal W 1.83 was used in multiple alignments. Alignment was edited and refined with GeneDoc software. The phylogenetic tree was built with Mega v3.1. The neighbor-joining method was used to calculate the trees, with 1 000 bootstrap tests and handling gaps with pair-wise deletion. Hs: Homo sapiens; Bt: Bos taurus, cow; Gg: Gallus gallus, chicken; Xt: Xenopus tropicalis, frog; Dr: Danio rerio, zebrafish.
Analysis of bbtTICAM expression pattern
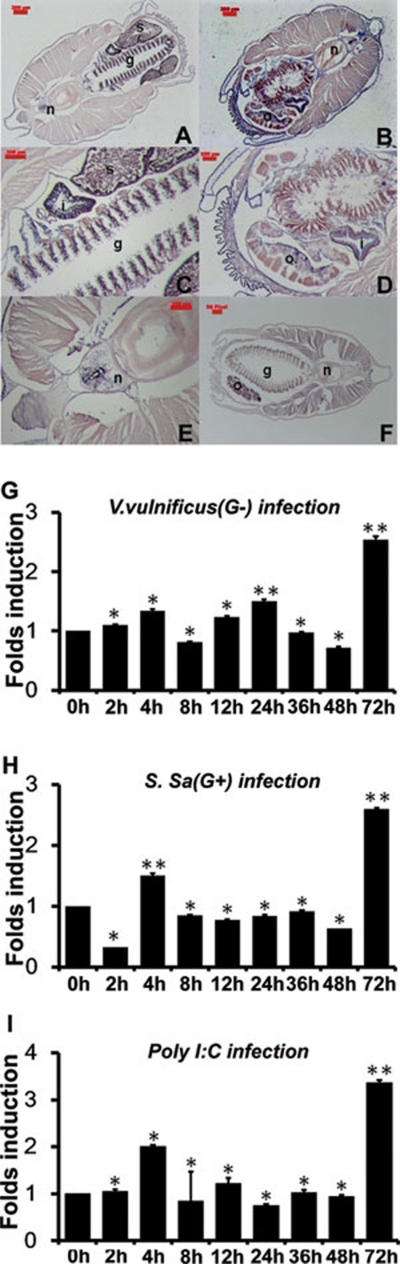
To describe expression pattern and possible function of bbtTICAM in adult amphioxus, in situ hybridization was conducted. In adult amphioxus, transcripts of bbtTICAM were strongly detected in the epithelial cells of intestine, gill, and skin (Figure 2A-2D). There is almost no signal in the ovary and spermatozoa (Figure 2A-2D). A weak signal was found in notochord (Figure 2E). As the skin, gill, and digestive system are presumed to be the primary line of immune defense in amphioxus, bbtTICAM should be immune relevant. To further study the immunological significance of bbtTICAM, real-time PCR analysis was performed. Figure 2G and 2H show that the transcription of bbtTICAM in intestines was upregulated after challenge of Gram-negative (G−) bacterium V. vulnificus and Gram-positive (G+) bacteria SSA. The response of bbtTICAM to poly I:C was also shown (Figure 2I). Although it was not dramatically upregulated, the abundance in immune tissue and the response to challenge suggested that bbtTICAM is multi-functional in amphioxus immunity.
Figure 2.
The tissue distribution and expression pattern of bbtTICAM after challenge with bacteria and polyI:C. (A, B) Section in situ hybridization analysis of bbtTICAM anti-sense probe showed predominant expression in intestine, gill, and skin. bbtTICAM was not abundant in notochord. The magnification factor is 45. (C-E) Macroscopic view of the hybridization signals of bbtTICAM in gill, intestine, and notochord. The magnification factor is 150. A, C and E show the slice from a male adult, while B and D show the slice from a female adult. (F) Section in situ hybridization analysis of bbtTICAM using sense probe as negative control. n = notochord, i = intestine, g = gill, o = ovary, s = spermatozoa. The scale bars are shown in red. Blue indicates strong hybridization, and dark brown means a weak signal. (G-I) Quantitative real-time PCR (RT-PCR) analysis of the expression of bbtTICAM after challenge with gram-negative bacteria (G), gram-positive bacteria (H) and PolyI:C (I). Results were presented as fold induction of mRNA expression in triplicate from two parallel experiments, using 2-ΔΔCt method. Endogenous control for quantification was cytoplasmic β-actin. Values were considered to be significant at P < 0.05. Student's t-test (two-tailed distribution, two-sample unequal variance) was used for the calculation of all P-values. *P < 0.05 and **P < 0.01.
bbtTICAM overexpression specifically activates NF-κB signal pathway in a MyD88-independent fashion
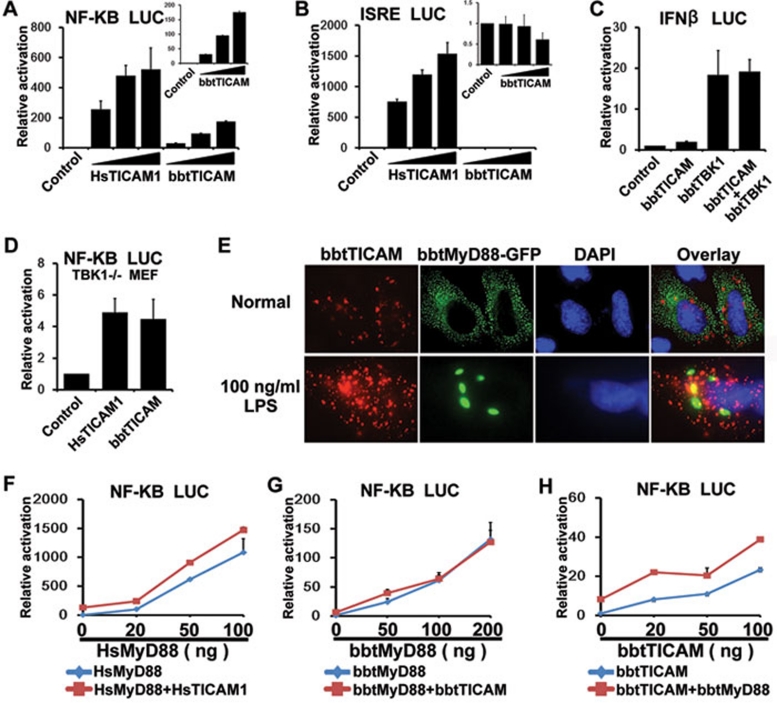
As both TICAM1 and TICAM2 are critical for the MyD88-independent pathway via activation of NF-κB or IRFs in mammals 5, 6, 25, luciferase assays were performed to reveal whether bbtTICAM has similar activities. Due to the absence of amphioxus cells currently, human cell lines were chosen to express amphioxus genes. Overexpression of bbtTICAM specifically activated NF-κB in a dose-dependent manner, but had no effect on interferon-stimulated response element (ISRE) (Figure 3A, 3B and Supplementary information, Figure S1A and S1B). In vertebrates, TICAM1 interacts with TBK1 to activate IRF3 and induces type I interferon. Thus, we further carry out co-expression of bbtTICAM with bbtTBK1, which could perform the function of human TBK1 in the TBK1−/− MEF cells (Supplementary information, Figure S2). Reporter assays showed that, although the possible downstream molecules of bbtTICAM in amphioxus were compensated in human cells, bbtTICAM did not induce the production of type I interferon (Figure 3C). In addition, with overexpressed bbtTICAM in TBK1−/− MEF cells, the luciferase assay showed that bbtTICAM could induce activation of NF-κB without human TBK1 as human TICAM1 (Figure 3D). Thus, we can assume that the primitive role of bbtTICAM is involved in NF-κB activation, but not in the induction of type I IFN.
Figure 3.
bbtTICAM overexpression specifically activates a MyD88-independent NF-κB signal pathway. (A) bbtTICAM stimulates NF-κB signal pathway in a dose-dependent manner in the HEK239T cell line. (B) bbtTICAM has no effect on ISRE promoter. In A and B, HsTICAM1 (human TICAM1/TRIF) has been used as a positive control. The small panels of the bar graph show the relative activation of luciferase reporter by bbtTICAM. (C) Co-transfected HEK293T cells with bbtTICAM and bbtTBK1, IFN-β promoter was not co-stimulated. bbtTBK1 is the possible downstream molecule of bbtTICAM in amphioxus. (D) The result shows that bbtTICAM can induce the production of NF-κB as HsTICAM in TBK1−/− MEF cells. (E) bbtTICAM and bbtMyD88 have different subcellular distributions and do not co-localize in Hela cell line. At 24 h after transfection, 100 ng/ml LPS was added into the culture medium of cells for 2 h. After stimulation by LPS, the subcellular distribution of bbtTICAM is unchanged, while bbtMyD88 become larger spots and still do not co-localize with bbtTICAM. (F) HsTICAM1 and HsMyD88 (human MyD88) do not co-stimulate the signal pathway, as is the case with the controls. The HsMyD88 expression vector was titrated into transfections in HEK293T cells in the absence (blue curve) and presence (red curve) of 5 ng of the HsTICAM1 expression vector, 100 ng of the NF-κB response promoter luciferase reporter and 5 ng Rellina luciferase reporter plasmid. (G) bbtTICAM and bbtMyD88 do not co-stimulate the signal pathway. The bbtMyD88 expression vector was titrated into transfections in HEK293T cells in the absence (blue curve) and presence (red curve) of 10 ng of the bbtTICAM1 expression vector and 100 ng of the NF-κB response promoter luciferase reporter and 5 ng Rellina luciferase reporter plasmid. (H) bbtTICAM and bbtMyD88 do not co-stimulate the signal pathway. The bbtTICAM expression vector was titrated into transfections in HEK293T cells in the absence (blue curve) and presence (red curve) of 5 ng of the bbtMyD88 expression vector and 100 ng of the NF-κB response promoter luciferase reporter and 5 ng Rellina luciferase reporter plasmid. Experiments of subcellular localization were conducted in the Hela cell line. Reporter assays were done in HEK293T cell line in triplicate and repeated at least twice in all cases. Data were expressed as 'Relative activation' (mean ± sd) relative to control induction for a representative experiment.
bbtMyD88, identified in our previous study, is a conserved molecule that induces NF-κB activation, suggesting the existence of a MyD88-dependent pathway in amphioxus 13. As mammalian TICAMs mediate a MyD88-independent pathway, it is important to assess whether activation of bbtTICAM is independent of MyD88. The co-localization of bbtTICAM and bbtMyD88 showed that these two molecules were not overlapping, although they were both distributed in the cytosol, appearing as spots of differing sizes (Figure 3E). To test if the subcellular distribution is altered with stimulation, 100 ng/ml LPS was added into the culture medium of cells at 24 h after transfection. After the stimulation of LPS for 2 h, bbtTICAM and bbtMyD88 did not co-localize, although the spots of bbtMyD88 became larger in vision (Figure 3E). Luciferase assays showed that, similar to human TICAM1/TRIF, bbtTICAM had no effect on the activation of NF-κB induced by bbtMyD88 (Figure 3F-3H). Thus, the activation of NF-κB by bbtTICAM is MyD88-independent, indicating that the MyD88-independent pathway emerged at the basal chordate level.
Activity of bbtTICAM depends on its TIR domain and C terminus, which determine its localization on endosomes and formation of homodimers
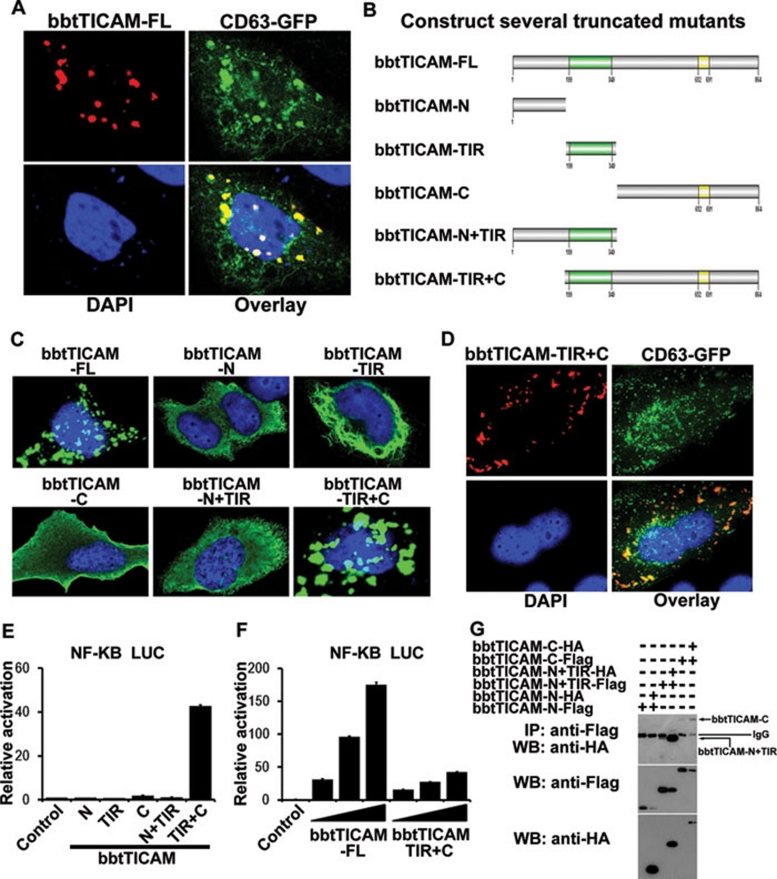
In mammals, the delivery of TICAM2 to endosomes is a requisite step in the TLR4-TICAM2-TICAM1 pathway 28. To further investigate the mechanism of bbtTICAM, and to discover if its subcellular localization is similar to that of mammalian TICAM1 and TICAM2, Hela cell line were co-transfected with bbtTICAM and the endosome marker, CD63. Signal analysis by confocal microscopy showed that bbtTICAM co-localized with endosomes when overexpressed in Hela cells (Figure 4A). To find which domain of bbtTICAM determines its endosome localization, five truncated mutants were constructed for confocal and reporter assays (Figure 4B) 29. The N-terminal domain of bbtTICAM was localized diffusely in the cytosol, while the TIR domain was critical for the accumulation and formation of the fiber-like structure (Figure 4C). Both the TIR domain and the C-terminal domain appeared as a speckled formation and the truncated mutant without the N terminus showed a similar subcellular localization to bbtTICAM-FL (Figure 4C). Further investigation indicated that both the TIR domain and the C-terminal region were involved in the subcellular localization of bbtTICAM-FL, as co-localization of the endosome marker and bbtTICAM-TIR+C mutant was observed (Figure 4D). To assess whether the endosome localization of bbtTICAM was indispensable for its signaling activity, luciferase assays were performed. The mutant construct of bbtTICAM-TIR+C affected NF-κB activation in a dose-dependent manner, while the other truncated mutants showed no effect on the activation of NF-κB (Figure 4E and 4F). When bbtTICAM was overexpressed in Hela cells, two expression bands were detected by western blot. To investigate whether bbtTICAM forms homodimers for its function, co-IP assays were conducted. The results indicate that both the TIR domain and C terminal domain were involved in the formation of homodimers (Figure 4G). Taken together with the confocal analysis, we suggest that the subcellular localization on the endosome and the formation of homodimers are crucial for bbtTICAM to initiate the NF-κB signaling.
Figure 4.
The endosomal localization is important for the function of bbtTICAM. (A) The full-length of bbtTICAM co-localizes with endosome marker (CD63). (B) Name of the truncated mutants used in this study. (C) The subcellular localization of full-length and all the truncated mutants. (D) The truncated mutant bbtTICAM-TIR+C co-localizes with endosome marker (CD63). (E, F) bbtTICAM-TIR+C is a unique mutant that induces NF-κB activation in a dose-dependent manner, although the activity is much lower than the full length. Other truncated mutants have no effect. (G) The co-IP results show that both truncated mutants bbtTICAM-N+TIR and bbtTICAM-C can form homodimers.
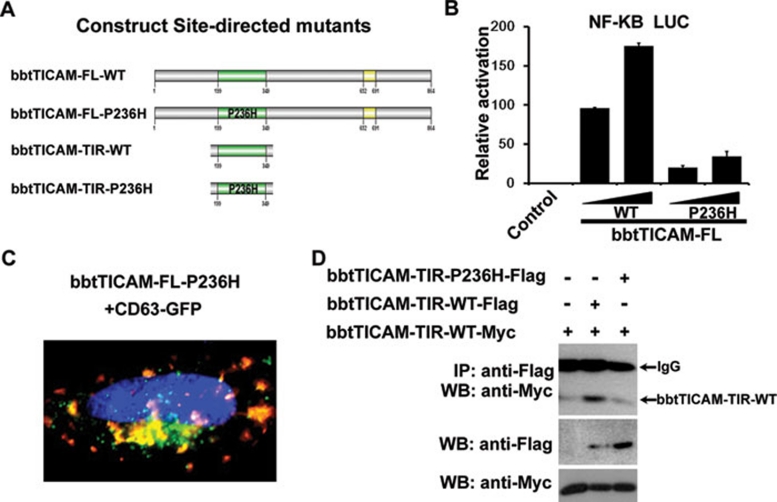
The Pro236 in TIR domain is required for full activation of bbtTICAM
Interaction among TIR-containing proteins requires their respective BB loops, which contain a conserved proline residue necessary for successful interaction with other TIR domains. Sequence analysis revealed a conserved proline residue at 236aa. Subsequently, site-directed mutagenesis by substitution of Pro236 for histidine (p263H) was conducted (Figure 5A and Supplementary information, Figure S3) and the luciferase assay showed that the P236H mutant attenuated activation of NF-κB by 20-30% compared with the wild type (Figure 5B). Further, confocal assays indicated that the P263H mutant maintained co-localization with endosomes (Figure 5C), indicating that Pro236 in TIR domain does not participate in the subcellular localization of bbtTICAM to endosomes. Co-IP assays showed that the mutation in TIR domain (P236H) could weaken the formation of the homodimer (Figure 5D). Therefore, the Pro236 in TIR domain is considered to be important for interaction between TIR domains.
Figure 5.
The Pro236 in TIR domain is required for full activation of bbtTICAM and the formation of homodimer. (A) Name of the site-directed mutants used in this study. P236H-Pro236 is substituted for His. (B) The mutant bbtTICAM-FL-P236H can attenuate the signal, which has 20-30% activity compared with the wild type. (C) The site-directed mutant, bbtTICAM-FL-P236H, co-localizes with endosome marker (CD63). (D) Co-IP assay shows that the TIR domain of bbtTICAM can form homodimers. With mutation in TIR domain (P236H), the formation of homodimer becomes weak.
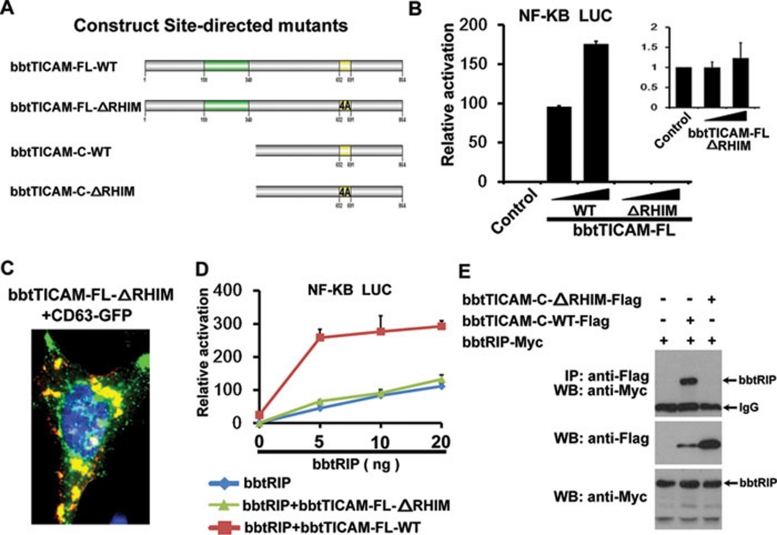
The C-terminal RHIM is indispensable for the function of bbtTICAM
As the RIP homotypic interaction motif (RHIM) in human TICAM1 is responsible for its interaction with RIP1 and RIP3 to activate the NF-κB pathway, a second site-directed mutagenesis was conducted by replacing the four conserved residues 652IQIG691 within the RHIM domain with four Ala for confocal and luciferase reporter assays (Figure 6A). When 652IQIG691 within the RHIM was replaced with four Ala, the activation of the NF-κB promoter by bbtTICAM was completely eliminated (Figure 6B). Further confocal assays indicated that the site-directed mutant maintained its co-localization with endosomes (Figure 6C), indicating that the RHIM did not participate in the subcellular localization of bbtTICAM to endosomes. Accordingly, the RHIM may be in charge of contact with other RHIM-containing proteins, and such interaction is critical for the full function of bbtTICAM.
Figure 6.
C-terminal RHIM is indispensable for the function of bbtTICAM. (A) Name of the site-directed mutants used in this study. ΔRHIM indicates that four conserved residues within RHIM domain 652IQIG691 are replaced with continuous four Ala. (B) With mutation of RHIM, the activation of NF-κB promoter is eliminated. (C) bbtTICAM-FL-ΔRHIM co-localizes with endosome marker (CD63). (D) bbtRIP and bbtTICAM-FL-WT co-stimulate the NF-κB signal pathway. The bbtRIP expression vector was titrated into transfections in HEK293T cells in the absence (blue curve) and presence (red curve) of 10 ng of the bbtTICAM-FL-WT expression vector, 100 ng of the NF-κB response promoter luciferase reporter, and 5 ng Rellina luciferase reporter plasmid. bbtRIP and the mutant bbtTICAM-FL-ΔRHIM do not co-stimulate the NF-κB signal pathway. The bbtRIP expression vector was titrated into transfections in HEK293T cells in the absence (blue curve) and presence (green curve) of 10 ng of the bbtTICAM-FL-ΔRHIM expression vector, 100 ng of the NF-κB response promoter luciferase reporter, and 5 ng Rellina luciferase reporter plasmid. (E) Co-IP results show that bbtTICAM-C directly interacts with bbtRIP1, and such interaction is abolished with the mutation of RHIM. This indicates that bbtRIP interacts with bbtTICAM via the RHIM within the C-terminus. The bands of bbtRIP have been indicated in the figure, and other bands are unspecific.
As human and zebrafish TICAM1 also contain an RHIM for their direct interaction with RIP1, we investigated whether bbtTICAM can associate with bbtRIP1, the ortholog of human RIP1 in amphioxus. Our results indicated that bbtRIP1 co-activated the NF-κB signal pathway with bbtTICAM (Figure 6D). Co-IP assays showed that bbtTICAM can directly interact with bbtRIP1, and such interaction was abolished when the RHIM of bbtTICAM was replaced with four Ala, indicating that the RHIM is indispensable for bbtTICAM to interact with the downstream bbtRIP1 for the activation of NF-κB (Figure 6E).
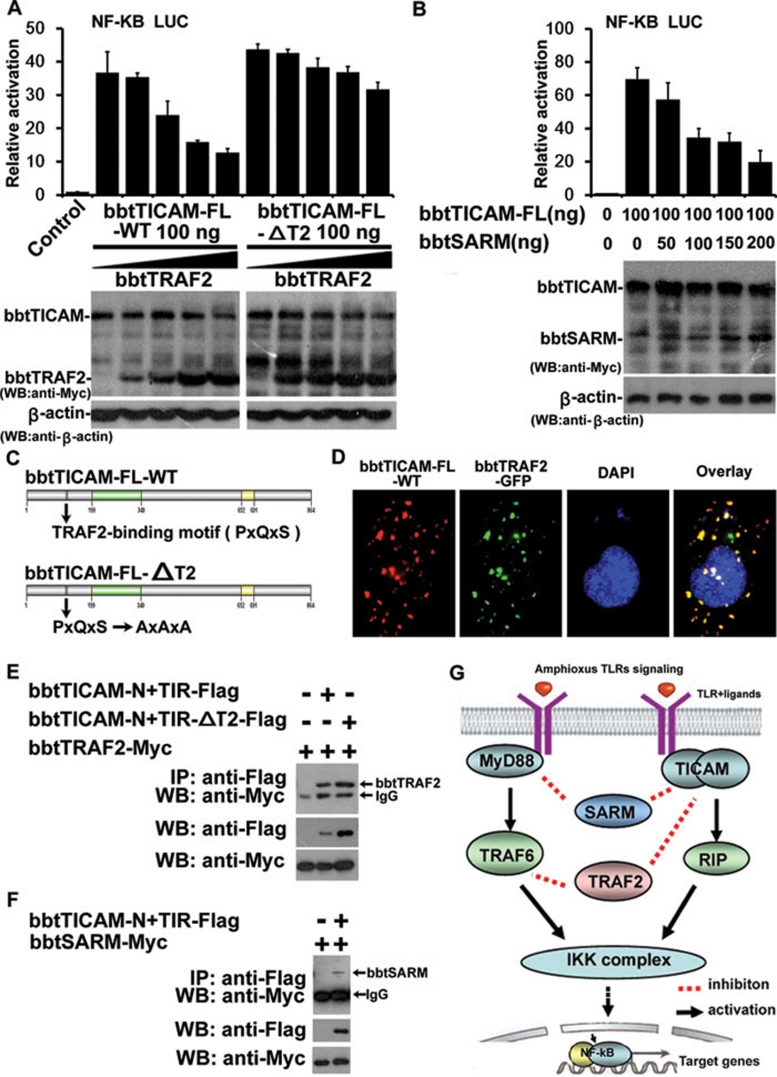
bbtTICAM failed to interact with bbtTRAF3, 4, and 6, but associated with bbtRIP and is negatively regulated by bbtSARM and bbtTRAF2
In mammals, TRAF2 and TRAF6 bind TICAM1 directly to participate in the activation of the TLR3/4-NF-κB pathway 26, 30, 31, 32. TRAF3 and TICAM1 form the complex to induce IFN-β via TRAF2/6. Conversely, TRAF1, TRAF4 and SARM act as inhibitors in TICAM1-dependent signaling 33, 34, 35, 36. As amphioxus TRAF2, TRAF3, TRAF4, TRAF6, and SARM were cloned by our group in other studies 37, 38, 39, we investigated the relationship between these molecules and bbtTICAM. The results showed that bbtTRAF3, bbtTRAF4, and bbtTRAF6 had no effect on the NF-κB activation mediated by bbtTICAM (Supplementary information, Figure S5A-S5C), while bbtTRAF2 and bbtSARM attenuated the activity of bbtTICAM in a dose-dependent manner (Figure 7A and 7B). Furthermore, confocal assay and co-IP tests showed that bbtTICAM and bbtTRAF2 co-localized and interacted with each other (Figure 7D and 7E), while bbtTRAF3, 4, and 6 did not (Supplementary information, Figure S5D and S5E). As a PxQxS TRAF2-binding motif was identified in bbtTICAM, site-directed mutagenesis was also conducted by changing PxQxS to AxAxA (Figure 7C). The AxAxA mutant activated the signal pathway as wild type without the inhibitory effect of bbtTRAF2 (Figure 7A). Further, co-IP confirmed that bbtTICAM directly interacts with bbtTRAF2 and bbtSARM (Figure 7E and 7F). However, when the TRAF2-binding motif was mutated, the interaction was attenuated but not completely abolished, indicating that other unidentified TRAF-binding motifs may participate in the interaction, as such a motif is short and not always conserved (Figure 7E). Therefore, the NF-κB activation mediated by bbtTICAM could be negatively regulated by bbtTRAF2 and bbtSARM, suggesting similarities and differences in the regulatory mechanism between human TICAM1 and bbtTICAM.
Figure 7.
The relationship of bbtTICAM with other adaptors. (A) bbtTRAF2 attenuates the signal pathway induced by bbtTICAM-FL-WT in a dose-dependent manner. bbtTICAM-FL-ΔT2 is not affected by bbtTRAF2 as bbtTICAM-FL-WT. The pictures from western blot below the bar graph show expression of relevant proteins, including bbtTICAM, bbtTRAF2, and β-actin. (B) bbtTICAM-FL can be negatively regulated by bbtSARM in a dose-dependent manner. The pictures from western blot below the bar graph show expression of relevant proteins, including bbtTICAM, bbtSARM and β-actin. (C) Site-directed mutation of TRAF2-binding motif. PxQxS (TRAF2-binding site) is substituted for AxAxA. (D) bbtTICAM-FL-WT co-localizes with bbtTRAF2 in Hela cell line. (E) Co-IP results show that the truncated mutant bbtTICAM-N+TIR can interact directly with bbtTRAF2. When the TRAF2-binding motif was mutated, the interaction was attenuated. (F) Co-IP results show that the truncated mutant bbtTICAM-N+TIR can interact with bbtSARM directly. (G) Signal transduction and regulation of amphioxus MyD88-dependent and -independent pathways, setting up the foundation of vertebrate TLR signaling network.
Discussion
bbtTICAM is the earliest ortholog of vertebrate TICAM1 and TICAM2; TICAM2 evolved with the development of endotoxin recognition complex
Amphioxus occupies a basal position in the Chordata and hence is an important point of reference for the evolution of vertebrate immunity, particularly that of early chordates. Analysis of immune-related genes in the amphioxus genome identified a TICAM2-like molecule 12. However, the full-length sequence revealed that the protein architecture of bbtTICAM is more similar to vertebrate TICAM1, including its length, position of the TIR domain, the RHIM in the C terminus, and the potential TRAF2-binding motif in the N terminus. The phylogenetic tree, based on the TIR domain, suggested that the molecule can be regarded as the ortholog of vertebrate TICAM1 and TICAM2. To date, no homolog for TICAM-1 or TICAM-2 has been reported in Cnidaria 8, sea urchin 10, 11 or other non-chordates, further suggesting that TICAM first emerged in the basal chordates and bbtTICAM might have evolved as the earliest molecule of the ancestral vertebrate TICAM1 and TICAM2.
In the early vertebrates, TICAM1 is evolutionarily conserved and easily identified 40. Homologs of TICAM2 have not been identified in teleosts 15, 16, 40, Xenopus, 22 or chickens 17, 23, except in mammals and sharks 21. As two TICAM homologs were identified in lamprey 14, Iliev et al. 40 assumed that a TICAM2 ortholog existed in the common predecessor of fish and tetrapods. Sullivan et al. confirmed this hypothesis and went further by suggesting that TICAM2 was probably lost after the divergence of rayfin and lobefin fishes 450 million years ago 17. The LPS-CD14/MD2/TLR4 endotoxin-recognition complex is absent in Takifugu and zebrafish 41, indicating that this recognition mechanism may have emerged after the divergence of fish and tetrapods 15, 16, 19, 42, 43, 44. The function of TICAM2 in the LPS-TLR4 pathway, together with the functional characterization of bbtTICAM presented here, suggests the following evolutionary process of TICAM molecules: bbtTICAM was duplicated in a basal chordate and TICAM2 was subsequently lost in teleosts, amphibians, reptiles, and birds, and then evolved rapidly with TLR4 until the endotoxin recognition complex was formed and stabilized in mammals.
Gradual adaptation of the MyD88-independent pathway with the evolution of the vertebrate anti-virus mechanism
There are two MyD88-like proteins and two atypical TLRs in Hydra 8, one MyD88 homolog and one TLR-1-like protein in Nematostella 8, four MyD88-like and more than two hundred TLRs in the sea urchin 10, 11, and one MyD88 ortholog and three TLRs in Ciona 45. The identification of both TLR and MyD88 in different evolutionary stages and the functional interaction of amphioxus TLR with MyD88 13 suggest that the MyD88-dependent pathway is conserved and crucial for invertebrate TLR signaling in detecting invading pathogens. The characterization of bbtTICAM indicated that it is unrelated to amphioxus MyD88, although it showed a functional effect on NF-κB activation. Thus, our study of bbtTICAM revealed that the MyD88-independent pathway was not a vertebrate innovation, but appeared in the basal chordates.
Mammalian TICAM1 plays an important role in the induction of type I IFN anti-viral response by activating the transcription factors IRF3 and IRF7 5. Zebrafish and Takifugu TICAM1 have been demonstrated to interact with fish TLR3 located on the endosome, and recognize the dsRNA virus 42, indicating that the RNA sensing pathway mediated by TICAM1 is conserved in vertebrates. In this study, the real-time assay shows transcription of bbtTICAM was upregulated after polyI:C challenge. Transcripts of zebrafish TICAM1 are altered after polyI:C challenge in a similar fashion 18, indicating that bbtTICAM may participate in an anti-virus response. However, bbtTICAM specifically activates NF-κB, but does not induce the production of type I IFN. Moreover, genome annotation indicated that amphioxus lacks orthologs of IRF3, IRF7, or IFN 12. Thus, it is inferred that the antiviral mechanism of bbtTICAM may not be fully established or may be completely different from vertebrates.
Research has shown that in vertebrates, TICAM-1 localizes diffusely in the cytosol of resting cells. Once TLR3 is activated by dsRNA, TICAM-1 transiently co-localizes with TLR3 on the cell surface or on the endosomes 46. The localization of TICAM-2 on endosomes is both necessary and sufficient to induce TLR4 signaling and support a model whereby LPS induces the internalization of TLR4 into endosomes when the TRAM-TRIF-dependent signaling pathway is activated 28. For amphioxus, the crucial formation of homodimer in endosomes, which is determined by both the TIR domain and C-terminal domain, is important for its function, indicating that the primitive TICAM path in amphioxus has set up the functional foundation for the endosomal TLRs, which chiefly participate in cellular virus detection through induction of IFNs. To date, no IFNs (specifically type I IFNs), the cytokines that bridge the innate and adaptive immunity soon after the recognition of pathogen-associated molecular patterns, have been reported in invertebrates. The adaptive immunity mediated primarily by lymphocytes bearing a unique (somatically diversified) antigen receptor is also considered vertebrate specific. Thus, the co-emergence of IFNs and adaptive immunity may indicate that the TICAM1-mediated anti-virus pathway evolved from the primitive MyD88-independent path by developing along with the emergence of adaptive immunity.
The regulatory mechanism of bbtTICAM on NF-κB activation sheds light on the foundation of vertebrate TLRs
Mammalian TICAM1 comprises a proline-rich N-terminal domain, a central TIR domain, and a C-terminal domain with RHIM motif 17. The N-terminal domain recognizes TRAF6 to activate both NF-κB and IRF3/7 pathways, while the central TIR domain is essential for interaction with other TIR domains. The RHIM motif within the C terminus interacts with RIP1/3 for the induction of mitochondria-independent apoptosis via formation of an RIP/FADD/caspase-8 complex 26, 30, 31. Conversely, TICAM1-mediated signaling can be negatively regulated by direct interaction with TRAF1 and TRAF4 via its N-terminal domain and through interaction of SARM via its TIR domain 33, 34, 35. Zebrafish TICAM1, co-localized with Golgi apparatus 18, stimulates NF-κB activation via RIP and TBK1, but not with TRAF6, due to the lack of TRAF6-binding motifs in zebrafish TICAM1 17. The newly identified TICAM1 homologs in lamprey resemble zebrafish TICAM1, lacking the TRAF6-binding motif, but with the conserved RHIM-like domain 14.
The relationship between the RHIM motif in bbtTICAM and bbtRIP is similar to that in mammals, zebrafish, and lamprey. However, bbtTICAM1 alone can specifically stimulate the NF-κB signal pathway, but not induce type I IFN production. Thus, the TICAM-RIP-NF-κB signal pathway seems to be the most primitive MyD88-independent pathway in amphioxus. It is noteworthy that the TRAF domain of human TRAF6 can attenuate the signal pathway induced by bbtTICAM in a dose-dependent manner (Supplementary information, Figure S5F). Moreover, the activation of NF-κB by bbtTICAM is TBK1 independent, indicating that the IKK complex should participate in the downstream signaling for bbtTICAM, although the mechanism among RIP1, TRAF6, and the IKK complex still requires further study. Similar to human TICAM1, activity of bbtTICAM is negatively regulated by amphioxus SARM through direct TIR-TIR interaction. Although bbtTRAF3, bbtTRAF4, and bbtTRAF6 have no relationship with bbtTICAM, bbtTRAF2 acts as an inhibitor of the function of bbtTICAM. Thus, the amphioxus signaling activity and the interaction network are different from vertebrate TICAM1 in certain aspects, although the protein architecture of bbtTICAM is similar to that of mammalian TICAM1. These relationships indicate that the more primitive regulation via interaction with other TIR adaptors and TRAF family members was developed when the MyD88-independent pathway was established in basal chordates. This aids in understanding the functional formation and divergence of the TLR signaling network at its earliest stage during chordate evolution.
Conclusion
The present study not only presents the primitive roles of the ancestral molecule with the combined structure and function of vertebrate TICAM1 and TICAM2 in amphioxus, but also sheds light on the MyD88-independent TLR signaling in invertebrates, providing evidence for the evolution of TICAM molecules and the formation of the endotoxin recognition complex. The functional analysis of the crucial relationships among bbtTICAM, TRAFs, RIP, and SARM will also contribute to interpreting the evolving function of TICAM molecules and the development of the TLR system in vertebrates, particularly in humans.
Materials and Methods
Animals and cells
Adult Chinese amphioxus Branchiostoma belcheri tsingtauense were obtained from Qingdao, China. Human embryonic kidney (HEK) 293T and Hela cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics. Wild-type MEF and TBK−/− MEF was kindly provided by Dr Tang's laboratory (Institute of Biophysics, Chinese Academy of Sciences).
Cloning of bbtTICAM cDNAs
A mammalian TICAM2 ortholog was identified in the B. floridae genome. Based on this sequence, a partial sequence of bbtTICAM (TICAM from B. belcheri tsingtauense) was cloned from Chinese amphioxus intestinal cDNAs by a specific primer pair derived from bfTICAM (TICAM from B. floridae). Subsequently, 5′-RACE and 3′-RACE were performed according to the manufacturer's protocol using a GeneRACE Kit (Invitrogen) for full-length sequence cloning.
Section in situ hybridization
Digoxigenin-labeled sense and anti-sense probes for bbtTICAM were synthesized with DIG RNA Labeling Kit (Roche). Section in situ hybridization was performed following the procedure described previously 13.
Acute immune challenges of adult amphioxus and real-time PCR
The challenged and un-challenged amphioxus were cultured in separate tanks. Intestines from five individuals were collected at 2, 4, 8, 12, 24, 36, 48, and 72 h post-injection as a single sample. Intestines from five PBS-injected animals were collected concurrently as non-challenged controls. After total RNA was prepared, reverse transcription was performed following the manufacturer's instructions (TOYOBO). Real-time PCRs were run using the conditions: 2 min at 95 °C followed by 40 cycles of 30 s at 95 °C, 15 s at 60 °C, and 30 s at 72 °C. Data were quantified using the 2-ΔΔCt method based on Ct values of bbtTICAM and β-actin from two parallel experiments done in triplicate. Levels of expression following challenge were normalized to the expression in PBS-injected animals. Values were considered to be significant at P < 0.05.
Expression plasmids
For the expression of bbtTICAM in HEK293T cells, PCR fragments encoding for amino acids 1-188, 189-359, 360-864, 1-359, 189-864, and 1-864 of bbtTICAM fused with 3′Flag tag or 3′HA tag were inserted into pcDNA3.0 vector (Invitrogen) and designated as bbtTICAM-N, bbtTICAM-TIR, bbtTICAM-C, bbtTICAM-N+TIR, bbtTICAM-TIR+C, and bbtTICAM-FL, respectively (Figure 4B). For the study of subcellular localization, full-length bbtMyD88 and bbtTRAFs were inserted into pEGFP-N1 (Clontech). For the luciferase assay and the Co-IP test, the full lengths of bbtTRAF2, bbtSARM, and bbtRIP were inserted into pCMV-Myc (Clontech) fused with Myc-tag. For the endosomal marker, PCR fragment encoding for the endosomal marker protein CD63 was inserted into pEGFP-N1 vector fused with GFP-tag. Vectors containing full-length human MyD88 and human TICAM1 fused with Flag-tag were provided by Dr Tang's laboratory (Institute of Biophysics, Chinese Academy of Science). The site-directed mutants were constructed according to the QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene).
Immunofluorescence imaging
Seeded Hela cells on coverslips (10 mm × 10 mm) in a 24-well plate were transfected with 400 ng indicated expression plasmids by jetPEI (PolyPlus-transfection) according to the manufacturer's instructions. After 20-24 h, cells were fixed for 15 min in a 4% formaldehyde solution, washed three times in PBST (0.05% Tween-20 in PBS) and blocked with 5% BSA in PBST at room temperature for 1 h. They were then incubated with 1 μg/ml anti-Flag mAb for 1 h. The cells were washed three times in PBST, and incubated with the second antibody for 1 h. Following triple washing in PBS, cells were labeled with 0.2 μg/ml DAPIs in PBS for 5 min, and then washed three times in PBS. Cells were mounted in MOWIOL R4-88 Reagent (Calbiochem) and photographed with a ZESSI Axio vision 4 microscope.
Transient transfection and luciferase reporter assay
HEK293T cells were plated in 48-well plates and, 14-20 h later, transfected with 500 ng/well DNA. The mixed DNA contained the indicated amount of expression vectors, 5 ng/well of Renilla luciferase reporter plasmid (Promega), to allow normalization of data for transfection efficiency and 100 ng/well of the NF-κB or IFN response promoter luciferase reporter and the addition of empty vectors. Samples were measured by a dual-luciferase reporter assay system (Promega). Luciferase activities were normalized to Renilla luciferase activity and expressed as the fold stimulation relative to that measured in cells transfected with the empty vector. Each experiment was performed at least in triplicate and repeated at least twice in all cases. Data were expressed as 'relative activation' (mean ± sd) relative to control induction for a representative experiment.
Coimmunoprecipitation
HEK293T cells in six-well dishes were transfected with 6 μg DNA plasmids (3 μg/expression vector). At 20-36 h post-transfection, the whole-cell extracts were prepared in IP lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholic acid sodium salt and cocktail protease inhibitor (Roche)) and incubated with primary antibodies at 4 °C for 4 h, then incubated with Protein G Sepharose (Roche) at 4 °C overnight. On the second day, the mix was washed three times with lysis buffer. Analysis was conducted using SDS-PAGE followed by western blot, using the ECL protocol (Amersham Pharmacia). The monoclonal antibody to HA (1:5 000), FLAG (1:1 000), MYC (1:7 000) and the anti-mouse secondary antibody (1:5 000) were purchased from Sigma.
Acknowledgments
This work was supported by grants from the National Basic Research Program (2007CB815800 and 2011CB946101), the State High-Tech Development Project (2008AA092603), International S&T Cooperation Program from the Ministry of Science and Technology of China (2007DFA30840), the Ministry of Education (0107), and the National Natural Science Foundation of China (30901305, 30730089), and the Fundamental Research Funds for the Central Universities (11lgzd16). Anlong Xu is recipient of “Outstanding Young Scientist Award” from the National Natural Science Foundation of China.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
BbtTICAM has no effect on IFNα4 and IFNβ promoters in HEK239T cell-line.
BbtTBK1 could substitute for human TBK1 in the TBK−/− MEF cells.
The conserved three boxes and BB loop in the TIR domains.
The sub-cellular distribution of three site directed mutants: bbtTICAM-FL-P236H, bbtTICAM-FL-ΔRHIM, and bbtTICAM-FL-P236H+ΔRHIM.
References
- Roach JC, Glusman G, Rowen L, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny EF, O'Neill LA. Signalling adaptors used by Toll-like receptors: an update. Cytokine. 2008;43:342–349. doi: 10.1016/j.cyto.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Jenkins KA, Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine. 2010;49:237–244. doi: 10.1016/j.cyto.2009.01.009. [DOI] [PubMed] [Google Scholar]
- O'Neill LA. The role of MyD88-like adapters in Toll-like receptor signal transduction. Biochem Soc Trans. 2003;31:643–647. doi: 10.1042/bst0310643. [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- Seya T, Oshiumi H, Sasai M, Akazawa T, Matsumoto M. TICAM-1 and TICAM-2: toll-like receptor adapters that participate in induction of type 1 interferons. Int J Biochem Cell Biol. 2005;37:524–529. doi: 10.1016/j.biocel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Wiens M, Korzhev M, Perovic-Ottstadt S, et al. Toll-like receptors are part of the innate immune defense system of sponges (demospongiae: Porifera) Mol Biol Evol. 2007;24:792–804. doi: 10.1093/molbev/msl208. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Hemmrich G, Ball EE, et al. The innate immune repertoire in cnidaria--ancestral complexity and stochastic gene loss. Genome Biol. 2007;8:R59. doi: 10.1186/gb-2007-8-4-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilak H, Tauszig-Delamasure S, Imler JL. Toll and Toll-like receptors in Drosophila. Biochem Soc Trans. 2003;31:648–651. doi: 10.1042/bst0310648. [DOI] [PubMed] [Google Scholar]
- Rast JP, Smith LC, Loza-Coll M, Hibino T, Litman GW. Genomic insights into the immune system of the sea urchin. Science. 2006;314:952–956. doi: 10.1126/science.1134301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Loza-Coll M, Messier C, et al. The immune gene repertoire encoded in the purple sea urchin genome. Dev Biol. 2006;300:349–365. doi: 10.1016/j.ydbio.2006.08.065. [DOI] [PubMed] [Google Scholar]
- Huang S, Yuan S, Guo L, et al. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity. Genome Res. 2008;18:1112–1126. doi: 10.1101/gr.069674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Huang S, Zhang W, et al. An amphioxus TLR with dynamic embryonic expression pattern responses to pathogens and activates NF-kappaB pathway via MyD88. Mol Immunol. 2009;46:2348–2356. doi: 10.1016/j.molimm.2009.03.022. [DOI] [PubMed] [Google Scholar]
- Kasamatsu J, Oshiumi H, Matsumoto M, Kasahara M, Seya T. Phylogenetic and expression analysis of lamprey toll-like receptors. Dev Comp Immunol. 2010;34:855–865. doi: 10.1016/j.dci.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Jault C, Pichon L, Chluba J. Toll-like receptor gene family and TIR-domain adapters in Danio rerio. Mol Immunol. 2004;40:759–771. doi: 10.1016/j.molimm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Meijer AH, Gabby Krens SF, Medina Rodriguez IA, et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Sullivan C, Postlethwait JH, Lage CR, Millard PJ, Kim CH. Evidence for evolving Toll-IL-1 receptor-containing adaptor molecule function in vertebrates. J Immunol. 2007;178:4517–4527. doi: 10.4049/jimmunol.178.7.4517. [DOI] [PubMed] [Google Scholar]
- Fan S, Chen S, Liu Y, et al. Zebrafish TRIF, a Golgi-localized protein, participates in IFN induction and NF-kappaB activation. J Immunol. 2008;180:5373–5383. doi: 10.4049/jimmunol.180.8.5373. [DOI] [PubMed] [Google Scholar]
- Sepulcre MP, Alcaraz-Perez F, Lopez-Munoz A, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- Sato S, Sugiyama M, Yamamoto M, et al. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J Immunol. 2003;171:4304–4310. doi: 10.4049/jimmunol.171.8.4304. [DOI] [PubMed] [Google Scholar]
- Wu B, Xin B, Jin M, Wei T, Bai Z. Comparative and phylogenetic analyses of three TIR domain-containing adaptors in metazoans: implications for evolution of TLR signaling pathways. Dev Comp Immunol. 2011;35:764–773. doi: 10.1016/j.dci.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Ishii A, Kawasaki M, Matsumoto M, Tochinai S, Seya T. Phylogenetic and expression analysis of amphibian Xenopus Toll-like receptors. Immunogenetics. 2007;59:281–293. doi: 10.1007/s00251-007-0193-y. [DOI] [PubMed] [Google Scholar]
- Keestra AM, van Putten JP. Unique properties of the chicken TLR4/MD-2 complex: selective lipopolysaccharide activation of the MyD88-dependent pathway. J Immunol. 2008;181:4354–4362. doi: 10.4049/jimmunol.181.6.4354. [DOI] [PubMed] [Google Scholar]
- Fitzgerald KA, Rowe DC, Barnes BJ, et al. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198:1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Sasai M, Shida K, Fujita T, Matsumoto M, Seya T. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278:49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- Sasai M, Tatematsu M, Oshiumi H, et al. Direct binding of TRAF2 and TRAF6 to TICAM-1/TRIF adaptor participates in activation of the Toll-like receptor 3/4 pathway. Mol Immunol. 2010;47:1283–1291. doi: 10.1016/j.molimm.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Meylan E, Burns K, Hofmann K, et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5:503–507. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat Immunol. 2008;9:361–368. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funami K, Sasai M, Oshiumi H, Seya T, Matsumoto M. Homo-oligomerization is essential for Toll/interleukin-1 receptor domain-containing adaptor molecule-1-mediated NF-kappaB and interferon regulatory factor-3 activation. J Biol Chem. 2008;283:18283–18291. doi: 10.1074/jbc.M801013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KJ, Su X, Xu LG, Bin LH, Zhang J, Shu HB. Mechanisms of the TRIF-induced interferon-stimulated response element and NF-kappaB activation and apoptosis pathways. J Biol Chem. 2004;279:15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Offermann MK. Apoptosis induced by the toll-like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol. 2005;174:4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- Cusson-Hermance N, Khurana S, Lee TH, Fitzgerald KA, Kelliher MA. Rip1 mediates the Trif-dependent toll-like receptor 3- and 4-induced NF-{kappa}B activation but does not contribute to interferon regulatory factor 3 activation. J Biol Chem. 2005;280:36560–36566. doi: 10.1074/jbc.M506831200. [DOI] [PubMed] [Google Scholar]
- Su X, Li S, Meng M, et al. TNF receptor-associated factor-1 (TRAF1) negatively regulates Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF)-mediated signaling. Eur J Immunol. 2006;36:199–206. doi: 10.1002/eji.200535415. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Ishii KJ, Kobiyama K, et al. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur J Immunol. 2005;35:2477–2485. doi: 10.1002/eji.200526151. [DOI] [PubMed] [Google Scholar]
- Carty M, Goodbody R, Schroder M, Stack J, Moynagh PN, Bowie AG. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006;7:1074–1081. doi: 10.1038/ni1382. [DOI] [PubMed] [Google Scholar]
- Peng J, Yuan Q, Lin B, et al. SARM inhibits both TRIF- and MyD88-mediated AP-1 activation. Eur J Immunol. 2010;40:1738–1747. doi: 10.1002/eji.200940034. [DOI] [PubMed] [Google Scholar]
- Yuan S, Liu T, Huang S, et al. Genomic and functional uniqueness of the TNF receptor-associated factor gene family in amphioxus, the basal chordate. J Immunol. 2009;183:4560–4568. doi: 10.4049/jimmunol.0901537. [DOI] [PubMed] [Google Scholar]
- Yuan S, Wu K, Yang M, et al. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J Immunol. 2010;184:6874–6881. doi: 10.4049/jimmunol.0903675. [DOI] [PubMed] [Google Scholar]
- Yuan S, Liu H, Gu M, et al. Characterization of the extrinsic apoptotic pathway in the basal chordate amphioxus. Sci Signal. 2010;3:ra66. doi: 10.1126/scisignal.2000906. [DOI] [PubMed] [Google Scholar]
- Iliev DB, Roach JC, Mackenzie S, Planas JV, Goetz FW. Endotoxin recognition: in fish or not in fish. FEBS Lett. 2005;579:6519–6528. doi: 10.1016/j.febslet.2005.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H, Tsujita T, Shida K, Matsumoto M, Ikeo K, Seya T. Prediction of the prototype of the human Toll-like receptor gene family from the pufferfish, Fugu rubripes, genome. Immunogenetics. 2003;54:791–800. doi: 10.1007/s00251-002-0519-8. [DOI] [PubMed] [Google Scholar]
- Baoprasertkul P, Peatman E, Somridhivej B, Liu Z. Toll-like receptor 3 and TICAM genes in catfish: species-specific expression profiles following infection with Edwardsiella ictaluri. Immunogenetics. 2006;58:817–830. doi: 10.1007/s00251-006-0144-z. [DOI] [PubMed] [Google Scholar]
- Purcell MK, Smith KD, Hood L, Winton JR, Roach JC. Conservation of Toll-Like receptor signaling pathways in teleost fish. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:77–88. doi: 10.1016/j.cbd.2005.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebl A, Goldammer T, Seyfert HM. Toll-like receptor signaling in bony fish. Vet Immunol Immunopathol. 2010;134:139–150. doi: 10.1016/j.vetimm.2009.09.021. [DOI] [PubMed] [Google Scholar]
- Azumi K, De Santis R, De Tomaso A, et al. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: “waiting for Godot”. Immunogenetics. 2003;55:570–581. doi: 10.1007/s00251-003-0606-5. [DOI] [PubMed] [Google Scholar]
- Funami K, Sasai M, Ohba Y, Oshiumi H, Seya T, Matsumoto M. Spatiotemporal mobilization of Toll/IL-1 receptor domain-containing adaptor molecule-1 in response to dsRNA. J Immunol. 2007;179:6867–6872. doi: 10.4049/jimmunol.179.10.6867. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BbtTICAM has no effect on IFNα4 and IFNβ promoters in HEK239T cell-line.
BbtTBK1 could substitute for human TBK1 in the TBK−/− MEF cells.
The conserved three boxes and BB loop in the TIR domains.
The sub-cellular distribution of three site directed mutants: bbtTICAM-FL-P236H, bbtTICAM-FL-ΔRHIM, and bbtTICAM-FL-P236H+ΔRHIM.