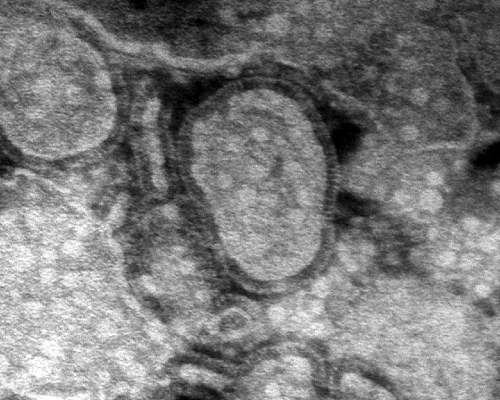Abstract
Summary: It has been 10 years since human metapneumovirus (HMPV) was identified as a causative agent of respiratory illness in humans. Since then, numerous studies have contributed to a substantial body of knowledge on many aspects of HMPV. This review summarizes our current knowledge on HMPV, HMPV disease pathogenesis, and disease intervention strategies and identifies a number of areas with key questions to be addressed in the future.
INTRODUCTION
Respiratory tract infections (RTIs) are a leading cause of morbidity and mortality worldwide. For children under the age of 5 years old, RTIs are ranked as the second leading cause of death, regardless of geographical location (172). In children, respiratory syncytial virus (RSV), parainfluenza viruses (PIVs), and influenza virus are known major causes of bronchiolitis and lower respiratory tract illnesses. In 2001, a previously unknown virus, human metapneumovirus (HMPV), was added to this list. In this review, the current knowledge on HMPV is summarized.
HMPV BIOLOGY
Description of the Agent
Human metapneumovirus was first detected upon inoculation of tertiary monkey kidney cells with respiratory specimens collected from children with RTIs for which the etiological agent could not be identified using diagnostic assays for known respiratory viruses. Cytopathic effects morphologically indistinguishable from those induced by RSV were observed. Virus-infected cell supernatants revealed pleomorphic particles measuring 150 to 600 nm, with short envelope projections of 13 to 17 nm, by electron microscopy (Fig. 1). These supernatants did not display hemagglutinating activity, and virus propagation was found to be dependent on trypsin (231). PCR and sequence analysis revealed a viral genome with close resemblance to that of avian metapneumovirus (AMPV), a virus causing swollen head syndrome and rhinotracheitis in chickens and turkeys (37). AMPVs have been classified into four subgroups, subgroups A through D (18), among which AMPV-C is most closely related to HMPV. However, the highly variable attachment (G) protein and small hydrophobic (SH) protein of AMPV and HMPV share only 20 to 30% amino acid sequence identity, while the percent sequence identity is ∼80% for the other structural proteins (230).
Fig. 1.
Electron micrograph of HMPV particles. Virions concentrated from infected cell culture supernatants were visualized by negative-contrast electron microscopy after phosphotungstic acid staining. Magnification, ×92,000.
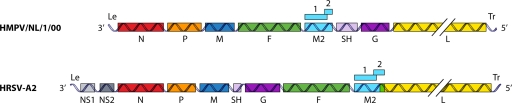
Similar to other pneumoviruses, HMPV virions contain a lipid membrane envelope surrounding the matrix (M) protein and three transmembrane surface glycoproteins, the fusion (F), G, and SH proteins. Within the envelope lies a helical ribonucleoprotein (RNP) complex, which consists of nucleoprotein (N), phosphoprotein (P), large polymerase protein (L), and the nonsegmented single-stranded negative-sense RNA genome. The genome size of HMPV ranges in length from 13,280 to 13,378 nucleotides and contains at least 8 genes and 9 open reading frames (ORFs). Beyond the genes for the proteins mentioned above, the HMPV genome, similar to the RSV genome, contains the M2 gene, from which the M2-1 and M2-2 proteins are expressed. However, distinct from RSV, the HMPV genome lacks nonstructural genes (NS1 and NS2), and the order of genes between M and L is different (in RSV, the order is SH-G-F-M2, and in HMPV, the order is F-M2-SH-G) (230, 231) (Fig. 2).
Fig. 2.
Genomic maps of HMPV and RSV showing the important differences between the two viruses. Compared to HMPV, RSV expresses two extra proteins, NS1 and NS2, the positions of the SH and G proteins differ, and the reading frames for M2 and L overlap in RSV. The double diagonal lines crossing the L ORF indicate the shortened representation of the L gene. Le, leader; N, nucleoprotein; P, phosphoprotein; M, matrix protein; F, fusion protein; SH, small hydrophobic protein; G, attachment protein; L, large polymerase protein; Tr, trailer; NS1 and NS2, nonstructural proteins 1 and 2.
Taxonomy.
Human metapneumovirus belongs to the order Mononegavirales, in the family Paramyxoviridae. HMPV was classified as the first human member of the Metapneumovirus genus, in the subfamily Pneumovirinae of the family Paramyxoviridae (231).
HMPV Replication
The HMPV replication cycle begins with attachment of the virus to the host cell, which is thought to be directed by the G protein (136). The G protein is the most variable protein among HMPV isolates (236). The deduced amino acid sequence of the G protein contains a single hydrophobic region that is located near the N terminus and is thought to serve as both an uncleaved signal peptide and a membrane anchor. The C-terminal three-fourths of the molecule are thought to be extracellular. The HMPV G protein has a high content of serine and threonine residues, which are potential acceptor sites for O-linked glycosylation, and a high content of proline residues (230)—features shared with heavily glycosylated mucin-like structures. The predicted structural features of the G protein were confirmed by analyses of the biosynthesis, glycosylation, intracellular transport, and cell surface expression of the G protein (144). It has been suggested that cellular glycosaminoglycans, including heparin sulfate-like molecules, are involved in the binding of the G protein to the host cell (224). Recombinant viruses lacking the G protein are able to replicate both in vitro and in vivo, indicating that attachment via the G protein is not required for subsequent steps in the replication cycle (22, 25).
Fusion of the viral membrane with the host cell membrane is mediated by the F protein. The structural organization of the HMPV F protein is similar to that of other viral class I fusion proteins, where the F protein is synthesized as an F0 precursor protein that requires cleavage by proteases to yield the activated disulfide-linked F1 and F2 subunits (reviewed in reference 135). Although the cleavage motif of the HMPV F protein (RQSR↓F) contains a minimal furin cleavage site that is typical for most paramyxoviruses (RXXR↓F), the HMPV F protein appears to require exogenous protease activation, as it is not cleaved intracellularly (reviewed in reference 212). In vitro, cleavage of the HMPV F protein is facilitated by the addition of trypsin to the cell culture medium. However, some laboratory strains have been shown to replicate in the absence of exogenous trypsin, likely due to a change in the cleavage site (RQPR↓F) (198), though this altered protease cleavability does not affect virulence (23). For most paramyxoviruses, fusion depends on an interaction between the F protein and its cognate attachment protein (reviewed in reference 136). However, the F proteins of members of the Pneumovirinae subfamily do not depend upon their cognate G proteins for fusion and are processed efficiently and correctly into a biologically active form when expressed in the absence of other viral proteins (114, 125, 178, 203, 220). This is consistent with the observation that HMPV lacking the G protein/gene remains replication competent in vitro and in vivo (22). Recently, it was shown that the HMPV F protein can engage in binding to host cells via integrin αvβ1 via a conserved Arg-Gly-Asp (RGD) motif, providing evidence for a role of the F protein in attachment in addition to membrane fusion (47).
Membrane fusion promoted by paramyxovirus F proteins generally occurs at the plasma membrane of the host cell at neutral pH (134, 195), which contrasts with the case for viruses gaining entry via a pH-dependent endocytic route. Interestingly, it has been shown that syncytium formation for HMPV strain Can97-83 is promoted by the HMPV F protein at low pH, suggesting a unique mechanism of triggering fusion among the paramyxovirus F proteins (203). However, the low-pH dependency is not a general phenomenon for HMPV F proteins and appears to be restricted to a few laboratory strains that contain an E294G substitution in the F protein (114). As for most paramyxoviruses, the trigger that leads to the membrane fusion event remains unknown, although it is tempting to speculate that binding to integrin αvβ1 may provide such a trigger (47). By generating chimeric viruses between HMPV and AMPV, it has been shown that the F protein is an important determinant of metapneumovirus host range (57).
Besides the F and G proteins, HMPV harbors a third putative transmembrane surface glycoprotein, SH. Hydrophilicity profiles of AMPV, HMPV, and RSV SH proteins were found to be similar, although the RSV SH protein appears to be truncated compared to the SH proteins of AMPV and HMPV (233). The SH protein of HMPV has a high threonine/serine content of ∼22% and contains 10 cysteine residues (230). Recombinant HMPV lacking only the SH gene was not attenuated or showed only marginal attenuation in vitro and in animal models (12, 22, 58). Infection of mice with HMPV lacking the SH gene resulted in enhanced secretion of proinflammatory cytokines compared to that in mice infected with wild-type virus (12). However, similar analyses using human lung epithelial cells infected with wild-type HMPV or the mutant virus lacking the SH gene did not reveal differential expression of genes or proteins (58). Moreover, the latter study did not reveal any effects of the SH protein on replication kinetics, cytopathic effects, or plaque formation of HMPV. Thus, the function of the HMPV SH protein remains elusive.
Following membrane fusion, the viral RNP containing the negative-sense viral RNA (vRNA) genome is released into the cytoplasm, where it serves as a template for the synthesis of mRNA and antigenomic cRNA. Most of the knowledge on HMPV transcription is inferred from the knowledge of RSV and other paramyxoviruses (136). The HMPV genomic termini contain leader and trailer sequences that are partially complementary and act as promoters to direct the transcription of mRNA and cRNA or vRNA, respectively. The leader and trailer of HMPV are less complementary than those of related paramyxoviruses. Their functions were confirmed in minigenome assays showing that the leader and trailer sequences were sufficient to drive replication and expression of heterologous genes when HMPV polymerase complex proteins were present (110). Using minigenome assays and recombinant virus rescue, it was further shown that the polymerase complex proteins and genomic termini of HMPV and AMPV are interchangeable, in agreement with the close genetic relationship between these two viruses (54). Nevertheless, viruses with HMPV-AMPV chimeric polymerase complexes were found to be attenuated in animal models and may thus represent useful live vaccine candidates (54, 184). Similar minigenome and reverse genetics studies further showed that while the L, P, and N proteins were absolutely required for minigenome expression or recombinant virus rescue, the M2-1 protein was dispensable (110). The M2-1 protein of RSV has been shown to promote transcription processivity (46, 78), but such a role was not observed for M2-1 of HMPV (35, 110).
In addition to the M2-1 ORF, the M2 gene of pneumoviruses also contains a second ORF, the M2-2 ORF. Expression of the M2-2 protein is achieved via a process of coupled translation, a mechanism of translational initiation in which the ribosomes that translate the first ORF move a short distance upstream after termination and reinitiate translation from the second overlapping ORF (94). For RSV, the M2-2 protein is thought to play a role in shifting the balance of RNA synthesis from mRNA to vRNA (20). Recent evidence suggests that M2-2 of HMPV regulates RNA synthesis in a similar fashion (35) and potentially also affects the fidelity of the polymerase complex (197). More work is needed to gain detailed knowledge of the HMPV polymerase complexes and the roles of M2-1 and M2-2 during virus replication.
The genome of HMPV further contains noncoding regions between each ORF that range in size from 23 to 209 nucleotides and contain gene end signals, intergenic regions, and gene start signals, with little overall sequence identity with the noncoding regions of RSV and AMPV. The role of these noncoding regions is likely the same as that for other paramyxoviruses, in which the gene end and gene start sequences control transcription termination and reinitiation, leading to a gradient of mRNA abundance that decreases from the 3′ end of the genome (N gene) toward the 5′ end (L gene) (136). The gene start consensus sequence of HMPV is CCCUGUUU/CA, and the start codon of each ORF is found 4 nucleotides downstream of this sequence (230). Using this knowledge, several noncoding regions have been duplicated in the HMPV genome to facilitate expression of green fluorescent protein and other heterologous ORFs, providing wonderful tools for HMPV research (24, 55).
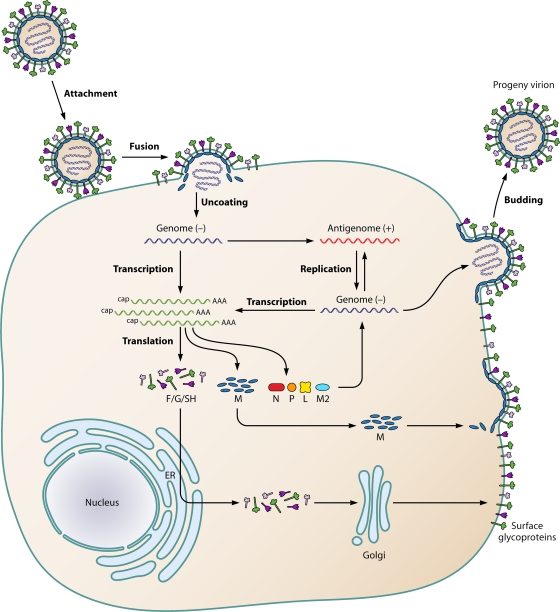
Steps in the HMPV replication cycle after the synthesis of RNA and viral proteins have not been investigated extensively. At present, as for other paramyxoviruses, it is assumed that virus assembly and budding occur through similar mechanisms (136) (Fig. 3).
Fig. 3.
Schematic representation of the HMPV life cycle. After attachment of the virion to the plasma membrane, the viral and plasma membranes fuse, resulting in uncoating of the virion and release of the RNP (containing the negative-sense viral RNA) into the cytoplasm. After primary transcription, the genome is replicated to produce the antigenome. The antigenome is used to synthesize genomic RNA, which is used to produce additional antigenomes for incorporation into progeny virions or as a template for secondary transcription. After translation, M proteins and RNPs are transported intracellularly to the plasma membrane and the viral glycoproteins F (fusion), G (glycoprotein), and SH (small hydrophobic) are transported from the endoplasmic reticulum (ER) to the Golgi apparatus and then the plasma membrane. Finally, new virions are assembled and are subsequently released from the plasma membrane by a budding process.
EPIDEMIOLOGY
Molecular Epidemiology and Virus Evolution
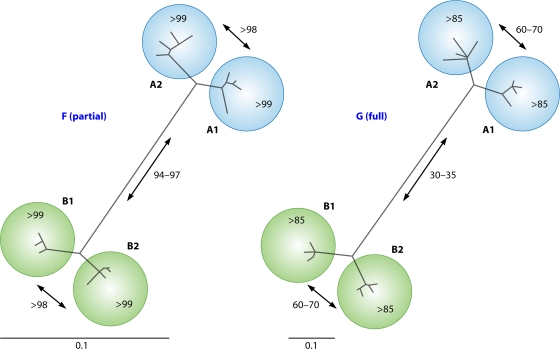
When HMPV was first described as the causative agent of RTIs in children, it was immediately recognized that at least two genetic lineages of HMPV were circulating in humans (231). Subsequent phylogenetic analysis of additional sequences obtained for the F and G genes revealed that each of these main lineages, A and B, can be divided into two sublineages: A1 and -2 and B1 and -2 (Fig. 4) (231). The maximum percent amino acid sequence identity between the F proteins of viruses belonging to lineages A and B was 95 to 97%; in contrast, there was only 30 to 35% identity for G protein sequences. The full-length genome sequences of prototypes of each of these lineages have become available in GenBank (under accession numbers AF371337 [A1], FJ168779 [A2], AY525843 [B1], and FJ168778 [B2]) (54). The similarities between HMPV strains of different lineages are in the same range as that described for the subgroups of AMPV and RSV (204, 215). The circulation of the 4 genetic lineages of HMPV was confirmed in studies throughout the world, most notably in long-term retrospective studies conducted in the United States from 1981 to 2001 (248, 253). From these studies, it can be concluded that (i) the prevalence of particular lineages is not restricted to certain locations and times and (ii) multiple lineages can circulate in the same period at a given location.
Fig. 4.
Phylogenetic trees for fusion (F) and attachment (G) genes of selected HMPV isolates. For each of the four genetic lineages (233), four representative isolates were selected, and maximum likelihood trees were generated for the G gene (right) and for 451 nucleotides of the F gene (left). Numbers in trees represent percent amino acid identities between virus isolates.
In 2004, a new variant of HMPV that was distantly related to previously described HMPV strains was detected in Germany (200). Unfortunately, the virus could not be isolated in cell culture and was therefore not characterized further, and other groups have not confirmed its detection. However, several other groups have subsequently reported the detection of newly emerging sublineages of lineages A and B (2, 9, 41, 44, 70, 118, 146, 242). In some of these studies, the available genetic information was limited to only small fragments of the HMPV genome, potentially giving rise to misclassifications. Nevertheless, it has become clear that sublineages of HMPV do not persist and that old lineages may be replaced with newly emerging variants. For instance, while lineage A1 circulated extensively in humans from 1982 to 2003 (233, 253), it has rarely been detected since 2004. In numerous other studies, it has been shown that the predominant circulating strains may vary by year and that predominant strains may be replaced, on average, every 1 to 3 years (2, 9, 40, 44, 70, 118, 140, 146, 152, 154, 166, 177, 185, 188, 242).
Although antibody responses elicited against the highly conserved F protein of HMPV may provide significant cross-protection against different HMPV lineages in animal studies (210), it has been postulated that antigenic variation may provide a plausible explanation for the cocirculation of multiple genetic sublineages of HMPV in humans (233). Virus neutralization assays performed with lineage-specific ferret antisera demonstrated that homologous virus neutralizing titers were significantly higher than titers against other HMPV lineages (233). Likewise, in reciprocal cross-neutralizing assays with sera from infected Syrian golden hamsters, the antigenic relatedness between viruses from two genetic lineages was relatively low (155). Based on these observations, as well as robust reinfections of cynomolgus macaques (232) and humans (66–68, 183, 253) with genetically distinct HMPV strains, it is possible that the cocirculation of multiple lineages of HMPV is facilitated by the limited cross-protection induced by HMPV. In this scenario, antigenic variation of the G and SH proteins, which may vary by as much as 70% at the amino acid level, may be sufficient to explain a selective advantage for heterologous virus lineages during subsequent epidemics, even in the presence of broadly cross-reactive anti-F humoral immunity in the population.
With the advent of a rapidly increasing number of HMPV (and AMPV) gene sequences available in public databases, the evolutionary history and dynamics of HMPV have been studied. Investigations of the evolutionary dynamics of HMPV and AMPV-C, using G, F, and N nucleotide sequences, demonstrated higher substitution rates for the G gene (3.5 × 10−3 nucleotide substitution per site per year) than for the N (9 × 10−4 nucleotide substitution per site per year) and F (7.1 × 10−4 to 8.5 × 10−4 nucleotide substitution per site per year) genes (56, 259). Such high evolutionary rates are not uncommon for RNA viruses (120). In both studies (56, 259), a limited number of positively selected sites were found in the F gene, and none were found in the N gene. Mutations in the G gene were likely to be either neutral or positively selected. For the G protein of RSV, a strong association between neutralizing epitopes and positively selected sites has been reported (263). In contrast to the case for other paramyxoviruses, such as RSV, the HMPV G protein is not a major neutralizing or protective antigen (209). Presently, there is limited knowledge about the locations of neutralizing epitopes in the F protein of HMPV. It is possible that there is a correlation between positive selection of epitopes and neutralizing epitopes in the HMPV F protein, because the F protein represents the major neutralization and protective antigen. However, in contrast to those of other paramyxoviruses, the HMPV F gene does not display substantial evolutionary progressive drift (233).
Extensive phylogenetic analyses have provided approximate calculations of the time of the most recent common ancestor of HMPV and AMPV-C. These analyses indicate that HMPV diverged from AMPV-C around 200 years ago (an average of 180 to 269 years, depending on the study and gene under investigation). The current genetic diversity of HMPV appears to have come about in the last ∼100 years (97 years in reference 259 and 133 years in reference 56). Each of the main genetic lineages (A and B) appears to be 34 to 51 years old, while each of the sublineages (A1, A2, B1, and B2) appears to be less than 30 years of age (56, 259). Thus, the genetic diversity within the four sublineages is of extremely recent origin, with several lineage diversifications occurring at approximately the same time.
Clinical Epidemiology
Since the first description of HMPV in 2001, the virus has been discovered worldwide on all continents and independent of the economic situations of different countries (1, 3, 5, 6, 17, 41, 44, 45, 49, 64, 67, 70, 83, 84, 93, 96, 97, 116, 132, 140, 145, 147, 148, 152, 154, 161, 165, 167, 170, 174, 181, 182, 186, 211, 219, 223, 228, 238, 243). HMPV infections can occur throughout the year, but seasonality has been described in several studies, with the epidemiological peak occurring 1 to 2 months later than that observed for RSV epidemics (2, 3, 107, 157, 193, 245). The intriguing question of whether different HMPV lineages are associated with differences in clinical courses of disease has so far remained unresolved. Several groups have suggested that HMPV lineage A might be associated with more severe clinical disease (9, 126, 162, 240). However, others reported that lineage B may cause more severe illness (72, 185), while other groups found no evidence for differential severity caused by different HMPV lineages (4, 140, 160, 258). A better understanding of the host response to HMPV lineages is needed to understand the mechanisms that may contribute to differences in clinical severity.
HMPV infections are observed in all age groups, with a high prevalence in pediatric patients (Table 1). The first HMPV infection appears to take place at 6 months of life, after which infections may occur repeatedly and frequently. The elderly represent the second group of patients that are severely affected by HMPV, and severe HMPV infections in the elderly occur despite high seroprevalence rates and independent of immunosenescence (62, 153). Reports on HMPV infections in otherwise healthy adults are relatively rare. Seroprevalence studies indicated that all adults have been infected with HMPV by the age of 25, with very high seroprevalence rates beginning from 5 years of age (68, 147, 150, 153, 255, 261, 262). The nosocomial impact of HMPV is estimated to be as high as that for RSV. In an HMPV outbreak in Japan, 34.8% of elderly patients who shared the same day care room in a hospital were infected with HMPV (116).
Table 1.
Overview of selected recent clinical studies on the epidemiology and diagnostics of HMPV infectionsa
| Age group (yr) in study | No. of HMPV-positive patients/total no. of patients (% positive) | Sample type | Detection method | Study period | Genotypes detected | Country (reference) |
|---|---|---|---|---|---|---|
| ≤2 | 18/217 (8.3) | NPA | RT-PCR of N gene | May 2006–November 2007 | A2, B1, B2 | Uruguay (186) |
| Children | 198/3,934 (5) | NPA | RT-PCR of N gene, polymerase gene, and NL-N genes | October 2004–April 2008 | ND | Switzerland (107) |
| Age groups are named A to K but are not specified in more detail | 142/7,091 and 118/4,282 | Respiratory samples | Pooling of clinical samples, nested RT-PCR | July 2006–June 2008 | A2, B1, B2 | Scotland (90) |
| 0–15 | 9/237 (3.8) | Nasopharyngeal swabs or aspirates | RT-PCR of nucleoprotein | October 2006–April 2007 | ND | Italy (73) |
| ≤5 | 191/1,670 (11.4) | NPA | GeneScan, RT-PCR of F gene | January 2003–December 2006 | B1 (2004–2005), A2, B2 | Brazil (177) |
| 0–91 | 143/4,989 (2.8) | NPA | RT-PCR of L gene | 2002–2006 | A (96%), B (4%) | Sweden (188) |
| ≤1 | 18/111 (16.2) | NPA | Nested RT-PCR of matrix gene | October 2003–April 2004 | ND | Spain (179) |
| 0–16 | 617/12,299 (5) | NPA | Multiplex RT-PCR, ELISA | 2002–2006 | ND | Germany (244) |
| ≤5 | 48/347 (13.8) | Nasal washes | RT-PCR of F gene | October 2005–April 2007 | A2, B1, B2 | Italy (39) |
| ≤5 | 68/516 (13.2) | Nasal washes | RT-PCR of L gene | November 2001–October 2002 | ND | Israel (254) |
| ≤58 | 65/3,858 (1.7) | NPA | Multiplex PCR/RT-PCR | 2007–2009 | B2, A2 | Cambodia (9) |
| ≤14 | 8/137 (5.8) | NPA | RT-PCR of polymerase gene | June 2002–August 2002 | ND | South Africa (79) |
| ≤2 | 10/182 (5.5) | NPA | RT-PCR of N gene | 2003 | ND | Chile (151) |
| ≤3 | 19/111 (17.1) | Nasopharyngeal secretions | RT-PCR of M, F, and N genes | April–May 2002 | ND | Brazil (49) |
| ≤3 | 95/865 (11) | Nasopharyngeal swabs | Singleplex RT-PCR | October 2005–September 2007 | ND | Alaska (208) |
| ≤5 | 14/497 (2.8) | NPA | RT-PCR of N gene | April 2000–December 2007 | A1, A2, B1, B2 | Brazil (40) |
| ≤2 | 109/1,612 (6.8) | NPA | RT-PCR of F gene | October 2000–October 2007 | ND | Austria (3) |
| ≤2 | 101/1,322 (7.6) | NPA | RT-PCR of M and L genes | October 2000–June 2005 | A1, A2, B1, B2 | Spain (88) |
| 0–89 | 12/420 (2.9) | Pharyngeal swab | RT-PCR of N and F genes | January 2002–November 2003 | B1, B2 | Peru (97) |
| 64–102 | 3/16 (18.8) | Throat swab | Direct IF RT-PCR | December 2007–January 2008 | ND | Australia (180) |
| ≤14 | 45/661 (6.8) | NPA | RT-PCR of M gene | December 2006–November 2008 | A2, A1, B1 | China (258) |
| Pediatric patients | 50/808 (6.2) | NPA | RT-PCR of N and F genes | December 2001–November 2004 | A1, A2, B1, B2 | Italy (91) |
| 0–≥40 | 34/1,294 (2.6) | Respiratory specimen (75% were nasal washes) | RT-PCR of F and N genes | October 2001–May 2004 | A1, A2, B1, B2 | USA (96) |
| <3 and children of >3 | 248/2,009 (1.4) | Nasal washing | Hexamer PCR, RT-PCR, and cell culture | 1976–2001 | ND | USA (248) |
ND, not detected; NPA, nasopharyngeal aspirates; RT-PCR, reverse transcription-PCR; N, nucleocapsid protein/gene; F, fusion protein/gene; IF, immunofluorescence.
CLINICAL FEATURES OF HMPV INFECTION
Symptoms and Pathology
HMPV is associated with a variety of symptoms and diagnoses localized to the respiratory tract (Table 2). Children with HMPV infection most commonly exhibit upper respiratory symptoms such as rhinorrhea, cough, or fever. Conjunctivitis, vomiting, diarrhea, and rash have been reported but are not frequent (234). The duration of symptoms prior to medical evaluation is usually less than a week, and limited data suggest that children shed virus for 1 to 2 weeks (67, 235, 248). Only one study has detected HMPV in serum by reverse transcription-PCR (RT-PCR) (159), suggesting that HMPV infection is usually limited to the respiratory tract. Studies of rodents and nonhuman primates have failed to detect HMPV in tissues outside the respiratory tract (7, 106, 131, 251, 257). The lower respiratory illnesses most frequently caused by HMPV are bronchiolitis, pneumonia, croup, and asthma exacerbation. Clinical signs and symptoms of HMPV infection overlap with those for other common respiratory viruses, and reliable distinctions cannot be made. Although symptoms usually overlap, differences in clinical presentation can occur. It has been reported that fever is more frequent in HMPV-infected patients, while rhinorrhea is observed more often in RSV-infected patients (19). Reinfection with HMPV occurs, although repeated infections are more likely to be limited to the upper respiratory tract in otherwise healthy children (248, 253). Some data suggest that dominant HMPV strains vary by season, presumably to avoid herd immunity, and thus reinfection may be more likely with heterologous viruses (2, 3). Further studies are needed to clarify the importance of antigenic variation of HMPV in human populations with respect to the clinical course of infection. HMPV is associated with acute otitis media, and viral RNA can be detected in middle ear fluid (199, 217, 252).
Table 2.
Symptoms and clinical diagnoses associated with human metapneumovirus infection of children
| Parameter | Valuea in reference: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 29 | 235 | 72 | 248 | 253 | 173 | 63 | 19 | |
| No. of patients | 12 | 25 | 53 | 49 | 118 | 26 | 50 | 26 |
| % of patients with symptom or diagnosis | ||||||||
| Fever | 67 | 61 | 77 | 52 | 54 | 73 | 44 | 13 |
| Rhinorrhea | 92 | 80 | 64 | 88 | 82 | 77 | 90 | 2 |
| Cough | 100 | 72 | 68 | 90 | 66 | 92 | 90 | 16 |
| Wheezing | 83 | 24 | 51 | 52 | * | * | 56 | 6 |
| Vomiting | 25 | * | * | 10 | 20 | * | 36 | 4 |
| Diarrhea | 8 | * | * | 17 | 14 | * | 14 | 1 |
| Rash | 0 | * | * | 4 | 3 | * | 2 | 5 |
| Abnormal chest radiograph | * | 62 | 56 | 50 | * | 85 | 67 | * |
| Bronchiolitis | 67 | * | * | 59 | * | 23 | 48 | * |
| Pneumonia | 17 | * | * | 8 | * | 23 | 34 | 9 |
| Croup | 0 | * | * | 18 | * | * | 4 | * |
| Asthma | * | * | * | 14 | * | 27 | * | * |
| Acute otitis media | 50 | * | * | 37 | 50 | 15 | 6 | * |
*, not reported.
Whether there is an association between HMPV infection and asthma is not clear. An Australian study of outpatient children with asthma did not identify an association between HMPV and asthma exacerbations (189), while a related study of outpatient children found a significant association between HMPV and the diagnosis of acute asthma (248). Studies of children hospitalized for wheezing and adults hospitalized with asthma exacerbations detected HMPV in a substantial number of these admissions (247, 250). Measurements of cytokines implicated in asthma pathogenesis in nasal washes of HMPV-infected infants have yielded conflicting results (119, 133). One retrospective study found a strong association between HMPV infection during infancy and the subsequent diagnosis of asthma (86). A major challenge for studies of respiratory viruses and asthma is the difficulty in firmly establishing a diagnosis of asthma during infancy, when acute wheezing associated with viral infections is common. However, the available data and analogy with RSV and human rhinoviruses suggest an association between HMPV and asthma exacerbations.
One study reported histopathologic changes during HMPV infection of young patients. In this study, bronchoalveolar lavage (BAL) fluid samples and lung biopsy specimens from HMPV-positive children displayed epithelial cell degeneration or necrosis with detached ciliary tufts and round red cytoplasmic inclusions, hemosiderin-laden macrophages, frequent neutrophils, and mucus (237). However, these samples were obtained from patients with underlying disease, and similar studies of otherwise healthy children infected with HMPV have not been done. Studies of nonhuman primates as well as small animals have demonstrated that HMPV infections remain restricted to the respiratory tract and do not spread to other internal organs. Histopathology studies of infected macaques and infected cotton rats have shown that infection is associated with a disruption of the epithelial architecture, sloughing of epithelial cells, loss of ciliation, and the presence of inflammatory infiltrates in the lungs (106, 131, 251, 257). HMPV-infected mice have been shown to develop parenchymal pneumonia and neutrophilic infiltrates during infection (50, 106). HMPV appears to exhibit a primary tropism limited to respiratory epithelia, as shown in immunohistochemistry studies of infected cynomolgus macaques, mice, and cotton rats. Viral expression was found in the epithelial cells of nasal tissue, all the way down to cells in the bronchioles, and was found less frequently in type I pneumocytes and alveolar macrophages (106, 131, 251).
Risk Groups Other than Children
Populations at risk of HMPV infection are children, the immunocompromised, and the elderly. Most studies of the elderly were performed with study groups where the elderly were defined as persons above the age of 65 years (http://www.who.int/healthinfo/survey/ageingdefnolder/en/index.html). However, this definition is dependent on the geographic and social background of the population of the elderly. HMPV infection may be more severe in patients with underlying medical conditions. It has been shown that 30 to 85% of children hospitalized with HMPV have chronic conditions, such as asthma, chronic lung disease due to prematurity, congenital heart disease, or cancer (29, 63, 72, 168, 173, 225, 234). Hospitalization of adults or children for HMPV-associated lower respiratory tract infections is more likely for patients with underlying conditions such as asthma, chronic obstructive pulmonary disease (COPD), HIV infection, immunocompromised status, or prematurity (34, 66, 75, 78, 106, 122, 163, 168, 187, 193, 217, 229, 242, 253, 254).
Immunosuppressed individuals.
HMPV is capable of causing severe infections in immunocompromised hosts, a phenomenon that has been well described for most respiratory viruses, including influenza virus, RSV, and PIV. There are reports of fatal infection attributed to HMPV in cancer patients, and several studies suggest that HMPV is a relatively common cause of acute respiratory infection in children and adults with malignancy or hematopoietic stem cell transplants (27, 38, 80, 183, 249). The basis for the increased severity of disease in these different groups is likely related to a reduced capacity to control virus replication, but the mechanisms are not well understood. Long-term prospective studies are needed to better characterize disease due to HMPV infection in immunocompromised hosts.
Older adults.
Infections in adults are probably underreported, as many hospitals do not routinely screen adults for HMPV. The yearly incidence of HMPV infection in adults has been reported to be 4 to 11% (78, 247), but the incidence varies depending on the group studied, e.g., adults, high-risk patients, elderly adults, and residents of a long-term care facility. Infections in older adults are detected mostly in late winter and early spring, and viral coinfections are observed mainly with RSV; up to 22.9% of infected elderly patients have been shown to be affected by at least one additional pathogen (78).
HMPV is a significant cause of acute respiratory disease in older adults (>65 years) and adults with comorbid illness, such as COPD, asthma, cancer, or lung transplantation (30, 75, 103, 163, 229, 239, 241, 247). Given this, HMPV is responsible for many hospitalized cases (28, 30, 43, 241), and infections in the elderly can result in death (28, 30, 43). Pneumonia has been documented for 40% of HMPV-infected frail elderly subjects (28, 117, 149). It has been reported that the most frequent admission diagnoses for HMPV infection in the elderly are acute bronchitis, COPD exacerbation, pneumonia, and congestive heart failure (241). Twenty-seven percent of the hospitalized patients in this study had substantial airway infiltrates observed by chest radiographs, with an average length of hospitalization of 9 days, but the duration was twice as long for the high-risk group, reaching 34 days in the most severe cases, with 13.2% requiring intensive care. High fatality rates have been reported for elderly patients with underlying disease who died of general respiratory failure (33, 182, 247). In one study, a case of HMPV-induced fatal pneumonia in an old woman who had no medical condition other than advanced age was presented (30), illustrating that HMPV infection can cause severe pneumonia leading to death in otherwise healthy elderly individuals. In addition, a single case of severe acute pericarditis associated with HMPV was reported for an otherwise healthy 62-year-old woman (52); thus, complications may occur that might be linked to HMPV infection.
The reasons for the higher morbidity in older adults have not been determined and require attention. Increased morbidity as well as a delay in clearance of symptoms of virus infections has been reported for the elderly, a feature consistent with the impairment of innate and adaptive immunity commonly associated with aging. Aging causes both qualitative and quantitative alterations, and intrinsic defects in the T cells of aged mice have been shown to contribute to decreased virus-specific T cell responses (33, 121–123, 144, 187). Although defects in the early events after virus infection may also influence the age-associated delay in clearance of the virus, it is possible that one reason for the clinical aggravation observed in the elderly could be an exaggerated immune response with inflammation, as opposed to a declining immune response (214). In the context of virus infection, these outcomes may be affected differently by the strain or HMPV subtype. It is clear that additional studies are required to determine the nature of the age-related defects during HMPV infection, and appropriate animal models are essential for these investigations.
Healthy adults suffer from asymptomatic infection, colds, and influenza-like illnesses (102). Asymptomatic infections are more common for HMPV infections than for RSV or influenza A virus infections. Asymptomatic infection in adults was also reported in a survey study of immunocompromised bone marrow transplant recipients (53). Both asthma (75, 247) and COPD (6, 9, 17, 20, 74, 103, 115, 118a, 163, 179, 194, 214) are exacerbated in immunocompromised adults. Serious and prolonged respiratory infections are associated with mortality in adults with underlying disease or hematological malignancies (247) and following hematopoietic stem cell or lung transplantation (139, 191, 216). Elderly subjects frequently suffer from bronchitis and pneumonia, and HMPV is responsible for many hospitalized cases in this population (17, 115, 128).
Coinfections
Many studies evaluating HMPV infection have tested for other viruses by using sensitive methods and have detected RSV in 5 to 17% of patients infected with HMPV (10, 17, 29, 31, 38, 49, 63, 67, 72, 81, 84, 119, 133, 159, 168, 173, 182, 189, 225, 235, 238, 248, 253). Most studies have not described exacerbated disease in patients with codetection of multiple viruses. Note that highly sensitive RT-PCR techniques may detect viruses for several weeks after an acute infection. A few studies of hospitalized patients have described much higher coinfection rates (30 to 60%) (87, 119, 130, 238), raising the question of whether HMPV infections are more severe if another virus is present. One group addressed this question by using a nested RT-PCR assay to test BAL fluids from 30 intubated infants with RSV infection, and they detected HMPV in 21/30 (70%) infants (98). They subsequently used the same nested PCR assay to test specimens from children admitted to the intensive care unit and the general wards. HMPV and RSV coinfections were detected in 18/25 (72%) intensive care patients and in 15/171 (9%) general ward patients, leading the authors to conclude that dual infection with RSV and HMPV was associated with severe bronchiolitis (205). However, a study of 46 inpatients with either mild or severe RSV disease found no coinfections with HMPV (141). Also, a Dutch study did not find any HMPV-RSV coinfections in children with severe RSV bronchiolitis (236). Whether these conflicting findings are due to methodological differences or to variability in circulating viruses is unknown. Further studies are needed to clarify the nature of disease associated with codetections; however, it is clear that the majority of HMPV infections are not associated with other viruses and that HMPV is a primary respiratory pathogen.
Treatment Options
The majority of children infected with HMPV can be managed with supportive care. For infants and children who require hospitalization, the primary therapies are supplementary oxygen and intravenous hydration. Bronchodilators and corticosteroids have been used empirically, but there are no controlled trials of these medications for HMPV and no data to support or refute their efficacy. One animal study suggested that there were benefits of bronchodilator and corticosteroid treatment of experimental HMPV infection in cotton rats (104). The only currently licensed antiviral drug for a related virus, i.e., RSV, is ribavirin, a nucleoside analogue administered by aerosol. Both ribavirin and polyclonal human immunoglobulin exhibit in vitro neutralizing activity against HMPV equivalent to their activity against RSV (256). There are no published animal or human data for these interventions, although they may be worthy of consideration for severely immunocompromised hosts with lower respiratory infections due to HMPV. Ribavirin has been used in severely immunocompromised patients, such as hematopoietic stem cell transplant recipients, often in conjunction with RSV-specific immunoglobulin, with some evidence of efficacy (60).
IMMUNE RESPONSES IN HUMANS AND ANIMALS
Innate Immunity
Acute viral infection is known to induce an early innate immune response characterized by induction of an antiviral type I interferon (IFN) response that involves JAK/STAT signaling, leading to the production of a variety of antiviral proteins, particularly IFN-stimulated gene products (218). Toll-like receptors (TLRs) and RNA helicases (RIG-I and MDA-5) are pattern recognition receptors most commonly activated by viral infections, and their activation contributes to a signaling cascade that governs the expression of proinflammatory and immune mediators. The TLRs, some of which are located in the endosomal cellular compartment, operate mainly in primary antigen-presenting and dendritic cells, while RIG-I and MDA-5 are cytoplasmic sensors and have been identified as being essential for IFN induction by several viruses. Mitochondrial antiviral signaling protein (MAVS) is an adaptor protein linking both RIG-I and MDA-5 to downstream activation of IRF3 and NF-κB, ultimately leading to type I IFN and IFN-stimulated gene expression (127, 206). RNA viruses use a large number and variety of mechanisms to subvert innate antiviral host defenses, and many of these mechanisms involve evasion of IFN and IFN-regulated responses (89).
Limited information is available on the interaction of HMPV and the innate immune system. In cultured human epithelial cells, HMPV was shown to be a strong inducer of the type I IFN pathway (14), and HMPV sensing and subsequent activation of the IFN pathway appeared to occur through the RIG-MAVS pathway, with no involvement of MDA-5 (143). Different HMPV proteins have been suggested to subvert the antiviral IFN pathway. Mutant HMPV lacking the G gene was shown to induce higher levels of chemokines and type I IFN in infected airway epithelial cells than those induced by the wild-type virus (12, 13, 15). The phosphoprotein in HMPV genotype B viruses was shown to be an IFN antagonist, but interestingly, the phosphoprotein in genotype A HMPV lacked this activity (95). In this study, it appeared that RIG-I signaling was not blocked by the phosphoprotein, while other studies suggested that the G protein binds to RIG-I to inhibit the IFN pathway (12, 13, 15). Two studies have shown that HMPV blocks double-stranded RNA (dsRNA)-induced IFN expression (100, 101), and in cultured epithelial cells, HMPV prevented IFN-induced phosphorylation and subsequent nuclear translocation of STAT1 (61). These findings contradict a report showing that HMPV infection of human A549 cells induces STAT1 phosphorylation (14). The contradictory results might relate to differences in the strains of HMPV used in the studies, as accumulating evidence suggests that there are strain differences in the host response to HMPV infection.
Limited data are available on the innate immune response to HMPV infection in humans; however, the available data suggest that differences may exist between HMPV and RSV. A Finnish study reported increased levels of interleukin-8 (IL-8) and decreased levels of CCL5 (RANTES) in nasal secretions of HMPV-infected children compared with levels from RSV-infected children (119). In contrast, a prospective study of infants infected with HMPV revealed significantly lower levels of IL-1β, IL-6, IL-8, IL-12, and tumor necrosis factor alpha (TNF-α) in nasal washes than those in RSV- or influenza virus-infected infants (133). Experimental HMPV infection of human dendritic cells ex vivo also induced cytokine profiles that differed significantly from responses evoked by RSV (100). In addition, a study with HMPV-infected BALB/c mice demonstrated significantly larger amounts of IFN-α than those in mice infected with RSV (99). These findings suggest that HMPV may induce a distinct host response, perhaps characterized by potent innate responses to infection.
Humoral Response
The prevalence of HMPV antibodies in persons between the ages of 6 months and 1 year, as measured by immunofluorescence assay, is 25%, and this prevalence increases to 55% for those between 1 and 2 years, to 70% for 2- to 5-year-olds, and to 100% for those over 5 years of age. Neutralizing antibody titers are lower for younger than for older children, with proportions of 25% for 6- to 12-month-olds, 31% for 1- to 2-year-olds, 38% for 2- to 5-year-olds, and 75 to 100% for those over 5 years of age (231). Thus, while all persons have been infected by age 5, a substantial proportion of those who are seropositive by immunofluorescence assay do not exhibit high neutralizing antibody titers. Consistent with these findings, several studies have shown that high HMPV seropositivity is associated with age and that high overall rates of seropositivity occur after the age of 5 years (68, 142, 181, 255). In general, it appears that despite protective antibody titers in adults, reinfections with homotypic and heterotypic HMPV strains occur in both healthy and immunocompromised humans (66, 157, 183, 248, 253). It is not known whether this phenomenon is due to limited cross-protective immunity between strains of HMPV, but these findings have led to the hypothesis that humoral immune responses may have only a minor role in the clearance of HMPV. However, this hypothesis is weakened by the observation that all permissive animal models, i.e., those that can be infected with HMPV and suffer from HMPV-associated disease, have been shown to develop serum neutralizing antibodies upon experimental HMPV infection (7, 106, 155, 232, 251, 257). Furthermore, passive antibody transfer alone, either polyclonal or monoclonal, has been shown to protect against HMPV replication and disease (7, 227, 246). Several different experimental approaches have shown that the HMPV F protein is a primary target of neutralizing antibodies (25, 171, 196, 209). However, since virtually all humans are seropositive yet severe infection occurs, antibody-mediated protection is probably not sufficient to prevent disease pathogenesis. One study using macaques showed waning immunity at 12 weeks and complete loss of protection 8 months following experimental infection (232). A poor HMPV antibody response was observed in aged mice, and this was not due to less viral replication, as the virus load in the lung at day 3 was shown to be greater in the older mice than in younger mice (51). This impaired antibody response was reflected in the IgG2a isotype, where the production of neutralizing antibodies was also impaired. It is possible that the HMPV-specific antibody response may contribute to protection against the severity of illness. This hypothesis is consistent with the finding that passive transfer of HMPV-immune serum protects naïve BALB/c mice from challenge and with results showing that a neutralizing monoclonal antibody to the F protein confers protection against challenge (8, 246).
Cellular Immunity
Reports of severe and fatal infections in immunocompromised populations suggest that T cell immunity is important for virus clearance and resolution of illness (38, 69, 139, 164, 183, 249). Many of these reports identified lymphopenia and cytotoxic therapy as important risk factors for severe HMPV disease, emphasizing a likely role for T cells in protection. Consistent with these findings, HMPV infection has been shown to be more severe in immunodeficient HIV-infected persons (85, 118a, 156, 157). HLA class I-restricted cytotoxic T lymphocyte (CTL) responses have been evaluated ex vivo in cells from HMPV-infected humans by enzyme-linked immunosorbent spot (ELISPOT) assay and chromium release assays (109). However, the contributions of T cells during primary and secondary infections of humans are unclear. In mice, depletion of NK cells was associated with significantly increased HMPV titers in the lungs, suggesting that cytotoxic cell types may contribute to HMPV immune surveillance and control (7). The contribution of T cells to HMPV immunity has been investigated to a limited extent by use of animal models. CD4+, CD8+, or CD4+ CD8+ T cell-depleted mice exhibited diminished weight loss and lung histopathology, suggesting that host immunity, while required for virus clearance, contributes to disease (129). However, if secondary virus challenge was performed in T cell-depleted mice, no difference in titer occurred between depleted and control groups of mice at the single time point examined, despite an absence of antibody in the CD4+ T cell-depleted mice. Therefore, the contributions of CD4+ and CD8+ T cells to protection or pathology remain poorly defined.
Murine major histocompatibility complex (MHC) class I-restricted epitopes for HMPV have been described (108, 169). One group used algorithms to identify candidate epitopes and tested predicted epitopes by peptide immunization (108). This group found that a few of these epitopes induced peptide-specific CTL responses against the same immunizing peptide and resulted in modest protection against virus challenge. However, the peptides were not screened against CTLs induced by virus infection, and thus it is not clear that these are true epitopes presented during natural infection. A similar bioinformatic approach was used to identify potential H-2d-restricted epitopes, i.e., epitopes recognized by murine MHC II, the analog of the human leukocyte antigen (HLA) complex (169). Predicted epitopes were screened as synthetic peptides against HMPV-immune splenocytes from BALB/c mice by ELISPOT assay. These experiments identified H-2d-restricted dominant (M2181–189) and subdominant (N307–315) epitopes. Primary T cell lines generated ex vivo by stimulation with these peptides were CD8+, IFN-γ secreting, and functional for lytic activity. Adoptive transfer of these cell lines into Rag−/− mice provided a modest reduction in virus titer, but lung pathology or signs of disease were not determined. These studies suggest that T cells contribute to protective immunity.
ANIMAL MODELS OF HMPV INFECTION
Several animal models have been developed to study HMPV infections. These animal models include mice, cotton rats, hamsters, guinea pigs, and ferrets, as well as nonhuman primates such as chimpanzees, macaques, and African green monkeys (155, 201). Each animal model has unique features that allow for investigations into the mechanisms of immunity and disease pathogenesis. The following sections summarize the roles of animal models and their contributions to our understanding of the biology of HMPV infection, replication, and disease intervention strategies.
Mouse Model
Experimental HMPV infection of mice (Mus musculus) has been studied with several mouse strains, but the majority of these studies were conducted with the BALB/c strain, as numerous studies have demonstrated robust HMPV replication in the lungs of BALB/c mice, with peak titers from days 5 to 7 (7, 8, 51, 213). In an early study, HMPV was shown to exhibit biphasic growth kinetics in the lungs of BALB/c mice, with peak titers occurring at days 7 and 14 postinfection (p.i.), while infectious HMPV could be recovered from the lungs until day 60 p.i. and HMPV RNA could be detected at ≥180 days p.i. (7). These results suggested the ability of HMPV to establish persistence following infection and are consistent with a more recent publication showing persistence of HMPV RNA in the lungs of mice, with associated pulmonary inflammation, out to day 154 p.i. (105). However, similar results were not reported in other studies showing that HMPV induces a self-limiting infection. The observed biphasic growth kinetics might reflect the specifics of the virus strain used or methods employed. The level of illness associated with weight loss has been linked with the level of virus replication and ranges from 5 to 20% weight loss, depending on initial viral inoculums (7, 8, 213). No viral RNA or infectious virus has been detected in the serum, spleen, kidneys, heart, trachea, or brain tissue (7). However, it has also been reported that HMPV has limited replication in mice (155). In these studies, BALB/c mice infected intranasally with HMPV/NL/1/00 were shown to be semipermissive for HMPV infection.
The disease pathogenesis associated with HMPV infection has been well characterized in mice. In general, following intranasal HMPV infection, lung histopathology reveals a prevalent mononuclear cell infiltration in the interstitium, beginning at day 2 p.i. and decreasing by day 14 p.i., and this has been associated with airway remodeling, increased mucus production, and airway hyperresponsiveness (AHR) (7, 129). In the BALB/c mouse model of HMPV infection, infection has been associated with an indolent inflammatory response characterized by innate immune and CD4+ T cell trafficking to the lungs, low-level IFN expression, induction of IL-10 expression at later stages of infection, and delayed CTL activity that coincides with persistent virus replication in the lungs (8). In addition, one study showed age-associated aggravation of HMPV clinical disease in BALB/c mice (51). Young and aged mice showed respiratory dysfunction, weight loss, and similar histopathological abnormalities. However, aged mice were far more susceptible than young mice to HMPV infection. It was suggested that this increased susceptibility was linked to the loss of cellular immune responses that are important for controlling HMPV infection. The virus and host mechanisms that contribute to pathogenesis are not fully understood, but several studies have addressed these. In an intranasal inoculation model, it was shown that HMPV-infected BALB/c mice developed parameters of clinical disease, including airway obstruction and hyperresponsiveness, and that this was mediated predominantly by CD4 T cells in comparison to CD8 T cells (129).
Cotton Rat Model
The cotton rat (Sigmodon hispidus and Sigmodon fulviventer) has become an accepted model for studying respiratory virus infections. Numerous studies have used the cotton rat model to evaluate HMPV infection, vaccine candidate efficacy, and disease pathogenesis (25, 35, 111, 155, 184, 197, 209, 210, 222, 251). In cotton rats infected intranasally with HMPV, peak levels of virus replication in the lung generally occur between days 4 and 5 p.i. Clinical symptoms are generally not observed, although infection has been associated with significant inflammation and histopathological changes associated with the development of peribronchial inflammatory infiltrates in the lower respiratory tract (251). No significant peripheral tissue pathology was evident in any of multiple tissues examined, and immunostaining revealed the presence of antigen at the apical surface of respiratory epithelial cells, suggesting a tropism for respiratory epithelial cells. Differences in susceptibility to different HMPV strains have been examined. In one study, cotton rats were inoculated intranasally with one of three strains, i.e., HMPV-26575, HMPV-26583, or RL Bx (257). Genetic analysis indicates that HMPV-26575 belongs to genotype B, while HMPV-26583 and RL Bx belong to genotype A (257). Interestingly, the genotype A viruses had similar replication kinetics and grew to high titers; however, the type B virus failed to replicate to high levels in the lungs of cotton rats. Increasing evidence demonstrates that the results obtained were more likely to be due to strain differences than genotype differences.
Young adult cotton rats are good models for evaluating disease intervention strategies for HMPV, because changes seen in cotton rats appear to be similar to those reported to occur in nonhuman primates following HMPV infection (131). In both species, HMPV infection results in inflammation in and around the bronchi and bronchioles, with markedly increased numbers of leukocytes surrounding these regions. Virus-specific antibody staining revealed HMPV antigen predominantly on the apical surface of the columnar cells, and peak viral titers in the lungs generally occurred around day 5 p.i. (201). Infection is accompanied by an inflammatory response characterized by upregulation of several chemokines and cytokines in the lungs, including IFN-α, CCL5 (RANTES), CCL2 (MCP-1), CCL3 (MIP-1α), and IL-2 mRNAs (32). HMPV infection in cotton rats results in a partially protective immune response, a reduced level of virus replication in the lungs following challenge, and a virus-specific serum neutralizing antibody response (32).
Hamster and Ferret Models
Several studies have evaluated HMPV infection and vaccine efficacy in hamsters (Mesocricetus auratus). HMPV replication in the lungs of hamsters has been shown to be high (105 PFU/g lung tissue) and similar to levels observed in cotton rats (155). This feature makes the hamster an ideal model for evaluating vaccine antigenicity and efficacy (113). There has been little done in the ferret model of HMPV infection. In one study that evaluated a panel of small animal models for HMPV, ferrets were infected intranasally with 106 PFU of HMPV, and at day 4 p.i., levels of virus in the turbinates and lung tissues were determined (155). The results showed that like hamsters, ferrets support HMPV replication in the respiratory tract to high titers, e.g., 4 to 4.7 log10 PFU/g tissue. Interestingly, there was no evidence of fever in the ferrets, which were monitored daily, and no clinical signs of illness. However, both hamsters and ferrets developed neutralizing HMPV antibodies. Given these limited features, the higher costs associated with ferrets, and the limited availability of immunological reagents for ferrets, it is likely that few studies will emerge using the ferret model.
HMPV Infection in Nonhuman Primates
To date, several nonhuman primate species have been evaluated for HMPV replication and viral pathogenesis, including rhesus macaques, cynomolgus macaques, African green monkeys, and chimpanzees (22, 155, 210, 221). In chimpanzees screened for the presence of neutralizing anti-HMPV antibodies in serum, 61% (19 of 31 animals) of animals were seropositive for either genotype A or genotype B HMPV strains (210), i.e., HMPV circulated within the chimps' group or was transmitted by their keepers. When chimpanzees were infected experimentally with HMPV, they demonstrated signs of clinical disease, including nasal discharge, thick mucus, and decreased appetites; however, despite these clinical changes, viral titers reached only 1.8 to 3.2 log10 in either the lower or upper respiratory tract (210). Rhesus macaques infected with 105.2 50% tissue culture infective doses (TCID50) of HMPV demonstrated only low levels of viral replication, with peak titers ranging from 0.9 to 2.6 log10 TCID50 in nasopharyngeal swabs and tracheal lavage fluid. In contrast, rhesus macaques infected with a similar inoculum yielded viral titers of 2.2 to 3.7 and 4.9 to 5.0 log10 TCID50 from the upper and lower respiratory tracts, respectively, and developed serum neutralizing antibody titers ranging from 9.1 to 10.9 log2, with initial infection conferring resistance against subsequent infections with both homologous and heterologous HMPV strains (155, 221). The African green monkey model has been utilized to investigate the generation of novel HMPV-based vaccines.
VACCINE DEVELOPMENT
A number of vaccines against HMPV have been investigated, including those prepared from chimeric viruses, live-attenuated viruses, and subunits of the virus. One of the first vaccine studies evaluated the efficacy of a chimeric virus vector consisting of bovine parainfluenza virus type 3 (PIV3) containing the F and HN genes of human PIV3 and expressing the HMPV F protein (222). In this study, vaccination of hamsters or African green monkeys induced HMPV-specific neutralizing antibodies that protected against HMPV challenge (221, 222). Other chimeric vaccine candidates were evaluated, such as a virus where the nucleoprotein or phosphoprotein of HMPV was replaced with that of AMPV type C. High levels of protective neutralizing antibodies were observed following intranasal vaccination, and upon challenge, lung virus titers were reduced substantially compared to those of controls at 3 days postchallenge (184). A study examining a chimeric virus vaccine based on alphavirus replicon particles encoding the G or F protein of HMPV demonstrated that the virus harboring the F protein of HMPV induced protective immunity against subsequent challenge. The latter observation made for HMPV is in contrast to the case for alphavirus harboring the G protein, which did not appear to be immunogenic and protective (170).
A live-attenuated HMPV vaccine candidate was generated by repeated passages of HMPV at low temperature in Vero cells, resulting in the accumulation of mutations in the viral genome (111). These mutations were reverse engineered into a wild-type HMPV backbone and resulted in a number of viruses with a temperature-sensitive phenotype. Replication of these temperature-sensitive HMPV vaccine candidates was reduced in the upper respiratory tract of hamsters and undetectable in the lower respiratory tract, but it was sufficient to induce high titers of protective HMPV-specific antibodies (111). Other live-attenuated vaccine candidates were generated by deleting the SH and/or G gene or the second ORF of the M2 gene. These vaccine candidates were shown to be attenuated in hamsters and nonhuman primates but were immunogenic, as they protected animals from subsequent challenge (25, 35, 197). However, a mutant virus lacking the M2-1 gene did not induce protective immunity, indicating that M2-1 is essential for virus replication.
The F protein has been used to develop several subunit vaccine candidates. Vaccination of hamsters with adjuvanted soluble F protein was shown to induce protective immunity against HMPVs of both genetic lineages (112). In addition, DNA vaccination with plasmid DNA carrying the F gene or vaccination with a purified soluble F protein lacking the transmembrane domain induced protective immunity (48). Moreover, the use of CTL peptide epitopes has been tested as a potential vaccination strategy in BALB/c mice: vaccination with such peptides was shown to reduce viral loads and immune pathology in the lungs of HMPV-challenged mice, an effect linked to enhanced expression of Th1 cytokines, including IFN-γ and IL-12 (108). Venezuelan equine encephalitis virus replicon particles encoding the HMPV F or G protein have also been evaluated for immunogenicity and protective efficacy in mice (171). Although several vaccine strategies have been developed, caution is needed, based on the disastrous outcome of formalin-inactivated RSV (FI-RSV) vaccines. FI-RSV vaccines administered to young children in the late 1960s sensitized vaccinees for vaccine-enhanced illness upon natural RSV infection (26). Similar FI-HMPV vaccines have been examined in mice, and the results show that HMPV challenge of FI-HMPV-vaccinated mice also leads to vaccine-enhanced disease (59, 260).
DIAGNOSTICS
Molecular Diagnostics
Currently, there exists neither a “gold standard” nor a consensus assay for the detection of HMPV in clinical samples. RT-PCR-based techniques are generally the methods of choice for the detection of HMPV (29, 71, 76, 98, 158, 182, 230); however, other assays, such as isothermal real-time nucleic acid sequence-based amplification (NASBA), have been used (92). Most PCR protocols detect all HMPV genotypes and rely on conserved and essential regions within the N gene or the F gene. Commercial singleplex assays are available from several diagnostic companies. These assays include both RT-PCR (e.g., assays by GenProbe/Prodesse, San Diego, CA) (82) and NASBA (bioMérieux, Marcy l'Etoile, France) (92) assays. Unfortunately, data on the utility of those assays are rare, despite the fact that NASBA was shown to be as sensitive as PCR-based detection methods (92). Recent studies used multiplex assays to evaluate large cohorts of patients coinfected with two or more pathogens (21, 190). In some patients, up to five pathogens were detected simultaneously. Most multiplex assays for the detection of respiratory viruses also include HMPV detection reagents. The xTAG respiratory virus panel (RVP; Luminex, Toronto, Canada) was shown to have superior negative predictive values, with acceptable positive predictive values that were marginally lower than the positive predictive value of the ResPlex II test by Qiagen (Hilden, Germany) (11). The ResPlex II assay has a broader pathogen range and detects up to 19 different viruses, compared to 17 viruses detected by the xTAG RVP assay (11). With respect to HMPV, the xTAG RVP, ResPlex II, and MultiCode-PLx assays have similar detection efficiencies (11). In addition, the MultiCode-PLx assay (EraGen Biosciences, Madison, WI) (175) has an identical negative predictive value to that of the Qiagen assay, but both values are lower than that for the Luminex assay (11). Thus, the xTag assay has received FDA approval, which is a prerequisite for its favored use in routine laboratory diagnostics in the United States, as in-house validation would otherwise be required. Another FDA-approved multiplex assay is the FilmArray respiratory panel by Idaho Technology Inc. (Salt Lake City, UT). This assay is a fully automated cassette that contains all necessary reagents, starting from extraction via a combined multiplex RT-PCR followed by singleplex second-stage PCRs to the final detection via endpoint melting curve. The assay is designed to detect up to 15 different agents in a single sample and has a very high specificity (99.2% for HMPV). An alternative multiplex detection method for respiratory pathogens is the RespiFinder technology (Pathofinder, Maastricht, The Netherlands), which is based on multiplex PCRs that are analyzed by subsequent capillary gel electrophoresis (192). This technology make use of multiplex ligation-dependent probe amplification (MLPA) and employs two probes which ligate exclusively in the presence of target-specific complementary sequences (34, 192). This assay was shown to have high sensitivity (98.2%) and specificity (100%) for HMPV in a study that investigated 144 clinical samples (192). Unfortunately, this assay is restricted in its use to those laboratories that are equipped with a capillary sequencing unit. The first version of this assay detected up to 15 pathogens simultaneously, but the second version, i.e., RespFinder19, is able to detect 19 pathogens simultaneously, including 15 respiratory viruses. Recent studies have shown that the RespiFinder assay (34) and the Seeplex RV15 ACE assay (Seegene, Eschborn, Germany) (65) are more sensitive than cell culture but comparable to singleplex real-time RT-PCR. There are currently other novel technologies being developed for respiratory virus diagnostics. Some examples include microfluid chip-compatible assays (36), RT-PCR coupled to electrospray ionization mass spectrometry (42), and surface-enhanced Raman spectroscopy assays (207), which may improve the sensitivity and range of pathogen detection.
Fluorescent-Antibody Staining and ELISA
Diagnosis of HMPV infection is based on the direct detection of viral components (protein, particles, or RNA) rather than indirect detection of antiviral antibodies in patient sera; however, there is value in evaluating antibody levels, particularly for understanding vaccine efficacy. Several studies have reported high seroprevalences based on enzyme-linked immunosorbent assays (ELISAs) (68, 145, 176, 262) or microneutralization assays (77, 153). ELISA detection of HMPV is generally performed with in-house assays, because assay kits are currently not commercially available. As an alternative method, cytospin-assisted direct immunofluorescence assay (DFA) is used occasionally for HMPV diagnosis in Europe and has become quite standard in the United States, as it allows for rapid detection of viral proteins from clinical samples, with acceptable sensitivities (124, 137). Commonly used antibodies are those from Chemicon/Millipore (Chemicon International, Temecula, CA) (137, 138); these monoclonal antibodies are offered as complete kits, e.g., the Light Diagnostics human metapneumovirus (HMPV) direct immunofluoresence assay, or as the SimulFluor HMPV/RSV reagent (both from Millipore, Billerica, MA). Also, Diagnostic Hybrids (Athens, OH) has developed and launched FDA-cleared assays, namely, the D3 DFA metapneumovirus identification kit for the detection of HMPV and the D3FastPoint L-DFA respiratory virus identification kit, which allows the identification of 8 different viruses, including HMPV. The latter test is claimed to be as sensitive and accurate as DFA, but a recent study showed that it is less sensitive than PCR and DFA; however, the time for multiplex detection is shorter (M. Barger, D. Vestal, M. Nye, and B. A. Body, presented at the 26th Clinical Virology Symposium, 25 to 28 April 2010, Daytona Beach, FL).
Cell Culture
HMPV can be cultured in several cell lines, including tertiary monkey kidney cells (231), Vero cells (231), LLC-MK2-cells, BEAS-2B cells (226), A549 cells (14), and HepG2 cells (202). These cell culture models facilitate HMPV research; however, there are caveats related to the different envelopes the viruses acquire as they bud from the cells, and these different envelopes may modify immune responses and may interfere with some assays.
FUTURE PERSPECTIVES
Although a substantial amount of knowledge on HMPV has been gained during the last decade, many issues remain unsolved. Despite extensive efforts to understand the molecular basis of the HMPV life cycle, the functions of several HMPV proteins need to be investigated further. For instance, studies regarding the F protein have shown that, as for most paramyxoviruses, the trigger that leads to the membrane fusion event remains unknown. The binding of F protein to integrin αvβ1 may provide such a trigger and is an intriguing area for further investigation. In addition, the function of the SH protein of paramyxoviruses has remained elusive, and more work is needed to gain detailed knowledge of the HMPV polymerase complexes and the roles of M2-1 and M2-2 during virus replication.
As for the clinical impact of HMPV, long-term studies are needed to characterize disease pathogenesis and to understand immunity in very young, old, or immunocompromised hosts responding to HMPV infection. There is also a need to optimize commercial diagnostic reagents and methods for detection of HMPV infection. Although a plethora of studies have been done on the clinical impact of HMPV, more detailed studies are needed to clarify the importance of antigenic variation of HMPV in human populations. This is important for the development of a cross-protective vaccine and for understanding antiviral approaches. Moreover, there is a need to develop safe and effective vaccines that induce protective immunity and perhaps give cross-protection against related viruses, such as RSV. To achieve these goals, a better understanding of the host response is needed at the genome level as well as the immune level, and the mechanisms of innate immunity that facilitate adaptive immunity need to be determined.
ACKNOWLEDGMENTS
V.S., O.S., C.M., and R.A.T. have no conflicts of interest to declare. B.V.D.H. is named inventor on several patents related to HMPV. These patents have been licensed to MedImmune USA. R.F. is named inventor on several patents related to HMPV. These patents have been licensed to MedImmune USA. R.F. is also a holder of certificates for 1% of the shares in ViroClinics Biosciences B.V. ViroClinics Biosciences is a contract research organization providing an array of services to the biopharmaceutical industry. To avoid any possible conflict of interests, Erasmus MC policy dictates that the shares as such are held by the Stichting Administratiekantoor Erasmus Personeelsparticipaties. The board of this foundation is appointed by the Board of Governors of the Erasmus MC and exercises all voting rights with regard to these shares. J.W. served as a consultant to MedImmune and Novartis and also serves on the Scientific Advisory Board of Quidel. R.A. is an employee of the U.S. Government. This work was prepared as part of official duties.
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, the Department of Defense, or the U.S. Government.
Biographies
 Verena Schildgen studied biology at the University of Cologne. Her diploma thesis was on recombinant T cell receptors and their utility as therapeutic molecules. She performed her Ph.D. research at the University of Düsseldorf and specialized in the molecular mechanisms of myelodysplastic syndromes, which are age-related diseases with assumed mitochondrial defects. Since 2007, her research interest has been on age-related aspects of viral infections and comorbidities that contribute to severe infections.
Verena Schildgen studied biology at the University of Cologne. Her diploma thesis was on recombinant T cell receptors and their utility as therapeutic molecules. She performed her Ph.D. research at the University of Düsseldorf and specialized in the molecular mechanisms of myelodysplastic syndromes, which are age-related diseases with assumed mitochondrial defects. Since 2007, her research interest has been on age-related aspects of viral infections and comorbidities that contribute to severe infections.
 Bernadette van den Hoogen studied plant biology at Rijks Hogere Agrarische School (RHAS), Wageningen, The Netherlands. She moved to the field of virology as a research assistant in the Department of Microbiology at the University of Pennsylvania School of Medicine in Philadelphia (under S. R. Ross). She was involved in identifying the receptor for the mouse mammary tumor virus. After accepting a position at Erasmus MC in 1999, she received a Ph.D. in Medicine from Erasmus University in 2004, for her studies on the discovery and characterization of the human metapneumovirus (under R. Fouchier and A. D. Osterhaus). As a senior postdoctoral fellow in the same department, she received a VENI grant from the Dutch Organisation for Scientific Research (NWO) to start her own research line. Her research focuses on the interaction of paramyxoviruses and the innate immune system and the development of therapies against HMPV. She is a frequent reviewer for journals in the field of virology and a member of the editorial board of the Journal of Clinical Virology.
Bernadette van den Hoogen studied plant biology at Rijks Hogere Agrarische School (RHAS), Wageningen, The Netherlands. She moved to the field of virology as a research assistant in the Department of Microbiology at the University of Pennsylvania School of Medicine in Philadelphia (under S. R. Ross). She was involved in identifying the receptor for the mouse mammary tumor virus. After accepting a position at Erasmus MC in 1999, she received a Ph.D. in Medicine from Erasmus University in 2004, for her studies on the discovery and characterization of the human metapneumovirus (under R. Fouchier and A. D. Osterhaus). As a senior postdoctoral fellow in the same department, she received a VENI grant from the Dutch Organisation for Scientific Research (NWO) to start her own research line. Her research focuses on the interaction of paramyxoviruses and the innate immune system and the development of therapies against HMPV. She is a frequent reviewer for journals in the field of virology and a member of the editorial board of the Journal of Clinical Virology.
 Ron Fouchier studied microbiology at the RHAS in Wageningen, The Netherlands, and received a Ph.D. in Medicine from the University of Amsterdam in 1995. He was a postdoctoral fellow at the Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, PA, from 1995 to 1998, in the laboratory of M. Malim. Achievements of his team include the identification and characterization of several “new” viruses: human metapneumovirus (HMPV), a human coronavirus (hCoV-NL), the severe acute respiratory syndrome coronavirus (SARS-CoV), and a new influenza A virus subtype (H16). He is part of the NIH/NIAID-funded Centers of Excellence in Influenza Research and Pathogenesis and received a prestigious VICI grant from the Dutch Organisation for Scientific Research (NWO) in 2009. He was elected a member of the “Young Academy” of the Royal Dutch Academy of Sciences (2005–2010) and is a member of the Council for Medical Sciences (RMW) and the Central Committee on Animal Experimentation (CCD) to advise the ministry of VWS, Chairman of the Medical and Veterinary Subcommittee of the Committee for Genetic Modification (COGEM), and Vice-Chairman of the COGEM board to advise the ministry of VROM. Dr. Fouchier is active in several WHO and FAO working groups and serves on numerous international advisory boards and on the editorial boards of leading journals. Dr. Fouchier received the Heine-Medin Award in Virology in 2007 and is currently Professor of Molecular Virology at Erasmus MC Rotterdam.
Ron Fouchier studied microbiology at the RHAS in Wageningen, The Netherlands, and received a Ph.D. in Medicine from the University of Amsterdam in 1995. He was a postdoctoral fellow at the Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, Philadelphia, PA, from 1995 to 1998, in the laboratory of M. Malim. Achievements of his team include the identification and characterization of several “new” viruses: human metapneumovirus (HMPV), a human coronavirus (hCoV-NL), the severe acute respiratory syndrome coronavirus (SARS-CoV), and a new influenza A virus subtype (H16). He is part of the NIH/NIAID-funded Centers of Excellence in Influenza Research and Pathogenesis and received a prestigious VICI grant from the Dutch Organisation for Scientific Research (NWO) in 2009. He was elected a member of the “Young Academy” of the Royal Dutch Academy of Sciences (2005–2010) and is a member of the Council for Medical Sciences (RMW) and the Central Committee on Animal Experimentation (CCD) to advise the ministry of VWS, Chairman of the Medical and Veterinary Subcommittee of the Committee for Genetic Modification (COGEM), and Vice-Chairman of the COGEM board to advise the ministry of VROM. Dr. Fouchier is active in several WHO and FAO working groups and serves on numerous international advisory boards and on the editorial boards of leading journals. Dr. Fouchier received the Heine-Medin Award in Virology in 2007 and is currently Professor of Molecular Virology at Erasmus MC Rotterdam.
 Ralph Tripp is a Professor and holds a Georgia Research Alliance Chair in Vaccine Studies. He was trained in the field of viral immunity under the tutelage of Linda Gooding at Emory University and then with Peter C. Doherty at St. Jude Children's Research Hospital. Following these postdoctoral programs, he led a research team in vaccine studies for important human viral diseases, particularly RSV, in the Respiratory and Enteric Viruses Branch at the CDC in Atlanta, GA. Professor Tripp now oversees research activities at the Animal Health Research Center (AHRC) at the University of Georgia, which is a biosafety level 2 (BSL2)/BSL3 biocontainment facility where his laboratory develops platform-enabling technologies in pathogen biosensing (using nanotechnology-based approaches), antiviral drugs, and vaccines, using state-of-the-art technologies with an in-house GMP vaccine facility. He is an Editor for Virology and Cellular Immunology and an ad hoc reviewer for several journals. He has received several awards for his work on respiratory viruses.
Ralph Tripp is a Professor and holds a Georgia Research Alliance Chair in Vaccine Studies. He was trained in the field of viral immunity under the tutelage of Linda Gooding at Emory University and then with Peter C. Doherty at St. Jude Children's Research Hospital. Following these postdoctoral programs, he led a research team in vaccine studies for important human viral diseases, particularly RSV, in the Respiratory and Enteric Viruses Branch at the CDC in Atlanta, GA. Professor Tripp now oversees research activities at the Animal Health Research Center (AHRC) at the University of Georgia, which is a biosafety level 2 (BSL2)/BSL3 biocontainment facility where his laboratory develops platform-enabling technologies in pathogen biosensing (using nanotechnology-based approaches), antiviral drugs, and vaccines, using state-of-the-art technologies with an in-house GMP vaccine facility. He is an Editor for Virology and Cellular Immunology and an ad hoc reviewer for several journals. He has received several awards for his work on respiratory viruses.
 Rene Alvarez graduated with a degree in biology and received his Ph.D. in microbiology and immunology. During his scientific career, he was a postdoctoral fellow in the Laboratory of the Agricultural Research Service, Athens, GA, and at Southeast Poultry Research and was a research fellow at the Centers for Disease Control and Prevention, National Centers for Infectious Diseases, Division of Respiratory and Enteric Viruses, Atlanta, GA. Between 2004 and 2006, he was Research Assistant Professor at the University of Georgia College of Veterinary Medicine and took various positions up to Associate Director at Alnylam Pharmaceuticals, Cambridge, MA. Currently, he is Supervisory Research Biologist, Principal Investigator, and Head of the Applied Laboratory Science Division, Naval Medical Research Unit, San Antonio, TX. During his career, he has received multiple national scientific awards.
Rene Alvarez graduated with a degree in biology and received his Ph.D. in microbiology and immunology. During his scientific career, he was a postdoctoral fellow in the Laboratory of the Agricultural Research Service, Athens, GA, and at Southeast Poultry Research and was a research fellow at the Centers for Disease Control and Prevention, National Centers for Infectious Diseases, Division of Respiratory and Enteric Viruses, Atlanta, GA. Between 2004 and 2006, he was Research Assistant Professor at the University of Georgia College of Veterinary Medicine and took various positions up to Associate Director at Alnylam Pharmaceuticals, Cambridge, MA. Currently, he is Supervisory Research Biologist, Principal Investigator, and Head of the Applied Laboratory Science Division, Naval Medical Research Unit, San Antonio, TX. During his career, he has received multiple national scientific awards.
 Catherine Manoha-Bourgeois obtained her Ph.D. in Microbiology-Virology in 1991. In 1992, she became Hospital and University Assistant in the Laboratory of Virology, CHU, Dijon, France. Her research activity focused on respiratory syncytial virus. Since 1998, as the Attachée Scientifique, she has led a small research group that is working on the pathogenesis of respiratory viruses, focusing on metapneumovirus in more recent years. She also directs clinical research on respiratory viruses in the Laboratory of Virology, CHU, Dijon, France.
Catherine Manoha-Bourgeois obtained her Ph.D. in Microbiology-Virology in 1991. In 1992, she became Hospital and University Assistant in the Laboratory of Virology, CHU, Dijon, France. Her research activity focused on respiratory syncytial virus. Since 1998, as the Attachée Scientifique, she has led a small research group that is working on the pathogenesis of respiratory viruses, focusing on metapneumovirus in more recent years. She also directs clinical research on respiratory viruses in the Laboratory of Virology, CHU, Dijon, France.
 John Williams received his B.A. with distinction in biology from the University of Virginia and his M.D. from the Medical College of Virginia, where he was elected to the Alpha Omega Alpha medical honor society. He completed a residency in pediatrics at the Children's Hospital in Pittsburgh and postdoctoral training in pediatric infectious diseases at Vanderbilt University Medical Center. His postdoctoral training was supported by a Pasteur Merieux Connaught award from the Infectious Diseases Society of America. He joined the Department of Pediatrics at Vanderbilt University as an Assistant Professor in 2003. Dr. Williams was awarded the Pediatric Infectious Diseases Society Young Investigator award in 2007.
John Williams received his B.A. with distinction in biology from the University of Virginia and his M.D. from the Medical College of Virginia, where he was elected to the Alpha Omega Alpha medical honor society. He completed a residency in pediatrics at the Children's Hospital in Pittsburgh and postdoctoral training in pediatric infectious diseases at Vanderbilt University Medical Center. His postdoctoral training was supported by a Pasteur Merieux Connaught award from the Infectious Diseases Society of America. He joined the Department of Pediatrics at Vanderbilt University as an Assistant Professor in 2003. Dr. Williams was awarded the Pediatric Infectious Diseases Society Young Investigator award in 2007.
 Oliver Schildgen studied biology at the University of Cologne. His diploma thesis was on the pathogensis of the picornavirus ECHO9. Following his diploma thesis, Oliver Schildgen worked at the University of Cologne's Institute for Genetics and the Max Planck Institute for Neurological Research in Cologne, Germany, and studied the baculovirus model before performing his Ph.D. thesis work on the woodchuck hepatitis virus in the lab of M. Roggendorf at the University of Essen. Between January 2002 and May 2009, he worked at the University of Bonn with an independent research group that focuses on the epidemiology of new respiratory pathogens. Since June 2009, the group has been based at the Institute for Pathology at the Hospital of the Private University of Witten/Herdecke in Cologne-Merheim. He has received several national and international awards for his work on newly discovered respiratory viruses.
Oliver Schildgen studied biology at the University of Cologne. His diploma thesis was on the pathogensis of the picornavirus ECHO9. Following his diploma thesis, Oliver Schildgen worked at the University of Cologne's Institute for Genetics and the Max Planck Institute for Neurological Research in Cologne, Germany, and studied the baculovirus model before performing his Ph.D. thesis work on the woodchuck hepatitis virus in the lab of M. Roggendorf at the University of Essen. Between January 2002 and May 2009, he worked at the University of Bonn with an independent research group that focuses on the epidemiology of new respiratory pathogens. Since June 2009, the group has been based at the Institute for Pathology at the Hospital of the Private University of Witten/Herdecke in Cologne-Merheim. He has received several national and international awards for his work on newly discovered respiratory viruses.
REFERENCES
- 1. Abdullah Brooks W., et al. 2007. Human metapneumovirus infection among children, Bangladesh. Emerg. Infect. Dis. 13:1611–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aberle J. H., Aberle S. W., Redlberger-Fritz M., Sandhofer M. J., Popow-Kraupp T. 2010. Human metapneumovirus subgroup changes and seasonality during epidemics. Pediatr. Infect. Dis. J. 29:1016–1018 [DOI] [PubMed] [Google Scholar]
- 3. Aberle S. W., Aberle J. H., Sandhofer M. J., Pracher E., Popow-Kraupp T. 2008. Biennial spring activity of human metapneumovirus in Austria. Pediatr. Infect. Dis. J. 27:1065–1068 [DOI] [PubMed] [Google Scholar]
- 4. Agapov E., Sumino K. C., Gaudreault-Keener M., Storch G. A., Holtzman M. J. 2006. Genetic variability of human metapneumovirus infection: evidence of a shift in viral genotype without a change in illness. J. Infect. Dis. 193:396–403 [DOI] [PubMed] [Google Scholar]
- 5. Albuquerque M. C., et al. 2009. Novel respiratory virus infections in children, Brazil. Emerg. Infect. Dis. 15:806–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ali S. A. Human metapneumovirus in hospitalized children in Amman, Jordan. J. Med. Virol. 82:1012–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alvarez R., Harrod K. S., Shieh W. J., Zaki S., Tripp R. A. 2004. Human metapneumovirus persists in BALB/c mice despite the presence of neutralizing antibodies. J. Virol. 78:14003–14011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alvarez R., Tripp R. A. 2005. The immune response to human metapneumovirus is associated with aberrant immunity and impaired virus clearance in BALB/c mice. J. Virol. 79:5971–5978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arnott A., et al. 1 February 2011. Genetic variability of human metapneumovirus amongst an all ages population in Cambodia between 2007 and 2009. Infect. Genet. Evol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bach N., et al. 2004. Acute respiratory tract infections due to a human metapneumovirus in children: descriptive study and comparison with respiratory syncytial virus infections. Arch. Pediatr. 11:212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balada-Llasat J. M., LaRue H., Kelly C., Rigali L., Pancholi P. 2011. Evaluation of commercial ResPlex II v2.0, MultiCode-PLx, and xTAG respiratory viral panels for the diagnosis of respiratory viral infections in adults. J. Clin. Virol. 50:42–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bao X., et al. 2008. Human metapneumovirus small hydrophobic protein inhibits NF-kappaB transcriptional activity. J. Virol. 82:8224–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bao X., et al. 2008. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS Pathog. 4:e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bao X., et al. 2007. Airway epithelial cell response to human metapneumovirus infection. Virology 368:91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bao X., et al. 2008. Identification of human metapneumovirus-induced gene networks in airway epithelial cells by microarray analysis. Virology 374:114–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reference deleted.
- 17. Bastien N., et al. 2003. Human metapneumovirus infection in the Canadian population. J. Clin. Microbiol. 41:4642–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bäyon-Auboyer M. H., Arnauld C. T. D., Eterradossi N. 2000. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J. Gen. Virol. 81:2723–2733 [DOI] [PubMed] [Google Scholar]
- 19. Beneri C., Ginocchio C. C., Manji R., Sood S. 2009. Comparison of clinical features of pediatric respiratory syncytial virus and human metapneumovirus infections. Infect. Control Hosp. Epidemiol. 30:1240–1241 [DOI] [PubMed] [Google Scholar]
- 20. Bermingham A., Collins P. L. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. U. S. A. 96:11259–11264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bharaj P., et al. 2009. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol. J. 6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Biacchesi S., et al. 2005. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 79:12608–12613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Biacchesi S., et al. 2006. Modification of the trypsin-dependent cleavage activation site of the human metapneumovirus fusion protein to be trypsin independent does not increase replication or spread in rodents or nonhuman primates. J. Virol. 80:5798–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biacchesi S., et al. 2004. Recovery of human metapneumovirus from cDNA: optimization of growth in vitro and expression of additional genes. Virology 321:247–259 [DOI] [PubMed] [Google Scholar]
- 25. Biacchesi S., et al. 2004. Recombinant human metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. J. Virol. 78:12877–12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blanco J. C., Boukhvalova M. S., Shirey K. A., Prince G. A., Vogel S. N. New insights for development of a safe and protective RSV vaccine. Hum. Vaccin. 6:482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boeckh M., Erard V., Zerr D., Englund J. 2005. Emerging viral infections after hematopoietic cell transplantation. Pediatr. Transplant. 9(Suppl. 7):48–54 [DOI] [PubMed] [Google Scholar]
- 28. Boivin G., et al. 2002. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J. Infect. Dis. 186:1330–1334 [DOI] [PubMed] [Google Scholar]
- 29. Boivin G., et al. 2003. Human metapneumovirus infections in hospitalized children. Emerg. Infect. Dis. 9:634–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boivin G., et al. 2007. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin. Infect. Dis. 44:1152–1158 [DOI] [PubMed] [Google Scholar]
- 31. Bosis S., et al. 2005. Impact of human metapneumovirus in childhood: comparison with respiratory syncytial virus and influenza viruses. J. Med. Virol. 75:101–104 [DOI] [PubMed] [Google Scholar]
- 32. Boukhvalova M. S., Prince G. A., Blanco J. C. 2009. The cotton rat model of respiratory viral infections. Biologicals 37:152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brien J., Uhrlaub J. L., Hirsch A., Wiley C. A., Nikolich-Zugich J. 2009. Key role of T cell defects in age-related vulnerability to West Nile virus. J. Exp. Med. 206:2735–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bruijnesteijn van Coppenraet L. E., et al. 2010. Comparison of two commercial molecular assays for simultaneous detection of respiratory viruses in clinical samples using two automatic electrophoresis detection systems. J. Virol. Methods 169:188–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buchholz U. J., et al. 2005. Deletion of M2 gene open reading frames 1 and 2 of human metapneumovirus: effects on RNA synthesis, attenuation, and immunogenicity. J. Virol. 79:6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burton R. E. W., et al. 2010. A microfluid chip-compatible bioassay based on single-molecule detection with high sensitivity and multiplexing. Lab Chip 10:843–851 [DOI] [PubMed] [Google Scholar]
- 37. Byus S. B., et al. 1989. Swollen head syndrome in chicken: a preliminary report on the isolation of a possible aetiological agent. J. S. Afr. Vet. Assoc. 60:221–222 [PubMed] [Google Scholar]
- 38. Cane P. A., van den Hoogen B. G., Chakrabarti S., Fegan C. D., Osterhaus A. D. 2003. Human metapneumovirus in a haematopoietic stem cell transplant recipient with fatal lower respiratory tract disease. Bone Marrow Transplant. 31:309–310 [DOI] [PubMed] [Google Scholar]
- 39. Caracciolo S., et al. 2008. Human metapneumovirus infection in young children hospitalized with acute respiratory tract disease: virologic and clinical features. Pediatr. Infect. Dis. J. 27:406–412 [DOI] [PubMed] [Google Scholar]
- 40. Carneiro B. M., et al. 2009. Detection of all four human metapneumovirus subtypes in nasopharyngeal specimens from children with respiratory disease in Uberlandia, Brazil. J. Med. Virol. 81:1814–1818 [DOI] [PubMed] [Google Scholar]
- 41. Carr M. J., et al. 2008. Molecular epidemiology of human metapneumovirus in Ireland. J. Med. Virol. 80:510–516 [DOI] [PubMed] [Google Scholar]
- 42. Chen K. F., et al. 2011. Reverse transcription polymerase chain reaction and electrospray ionization mass spectrometry for identifying acute viral upper respiratory tract infections. Diagn. Microbiol. Infect. Dis. 69:179–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chiu C. Y., et al. 2007. Diagnosis of a critical respiratory illness caused by human metapneumovirus by use of a pan-virus microarray. J. Clin. Microbiol. 45:2340–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chung J. Y., Han T. H., Kim S. W., Hwang E. S. 2008. Genotype variability of human metapneumovirus, South Korea. J. Med. Virol. 80:902–905 [DOI] [PubMed] [Google Scholar]
- 45. Chung J. Y., Han T. H., Kim S. W., Hwang E. S. 2007. Respiratory picornavirus infections in Korean children with lower respiratory tract infections. Scand. J. Infect. Dis. 39:250–254 [DOI] [PubMed] [Google Scholar]
- 46. Collins P. L., Hill M. G., Cristina J., Grosfeld H. 1996. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc. Natl. Acad. Sci. U. S. A. 93:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cseke G., et al. 2009. Integrin alphavbeta1 promotes infection by human metapneumovirus. Proc. Natl. Acad. Sci. U. S. A. 106:1566–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cseke G., et al. 2007. Human metapneumovirus fusion protein vaccines that are immunogenic and protective in cotton rats. J. Virol. 81:698–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cuevas L. E., et al. 2003. Human metapneumovirus and respiratory syncytial virus, Brazil. Emerg. Infect. Dis. 9:1626–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Darniot M., Petrella T., Aho S., Pothier P., Manoha C. 2005. Immune response and alteration of pulmonary function after primary human metapneumovirus (hMPV) infection of BALB/c mice. Vaccine 23:4473–4480 [DOI] [PubMed] [Google Scholar]
- 51. Darniot M., et al. 2009. Age-associated aggravation of clinical disease after primary metapneumovirus infection of BALB/c mice. J. Virol. 83:3323–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. David P., Hirschwerk D., Goldberg S., Ginocchio C. 2009. Acute pericarditis caused by human metapneumovirus. Infect. Dis. Clin. Pract. 17:283–285 [Google Scholar]
- 53. Debiaggi M., et al. 2006. Persistent symptomless human metapneumovirus infection in hematopoietic stem cell transplant recipients. J. Infect. Dis. 194:474–478 [DOI] [PubMed] [Google Scholar]
- 54. de Graaf M., et al. 2008. Specificity and functional interaction of the polymerase complex proteins of human and avian metapneumoviruses. J. Gen. Virol. 89:975–983 [DOI] [PubMed] [Google Scholar]
- 55. de Graaf M., et al. 2007. An improved plaque reduction virus neutralization assay for human metapneumovirus. J. Virol. Methods 143:169–174 [DOI] [PubMed] [Google Scholar]
- 56. de Graaf M., Osterhaus A. D., Fouchier R. A., Holmes E. C. 2008. Evolutionary dynamics of human and avian metapneumoviruses. J. Gen. Virol. 89:2933–2942 [DOI] [PubMed] [Google Scholar]
- 57. de Graaf M., et al. 2009. Fusion protein is the main determinant of metapneumovirus host tropism. J. Gen. Virol. 90:1408–1416 [DOI] [PubMed] [Google Scholar]
- 58. de Graaf M. H. S., et al. 2009. Metapneumovirus; determinants of host range and replication. Erasmus University Rotterdam, Rotterdam, The Netherlands [Google Scholar]
- 59. de Swart R. L., et al. 2007. Immunization of macaques with formalin-inactivated human metapneumovirus induces hypersensitivity to hMPV infection. Vaccine 25:8518–8528 [DOI] [PubMed] [Google Scholar]
- 60. DeVincenzo J. P., Hirsch R. L., Fuentes R. J., Top F. H., Jr 2000. Respiratory syncytial virus immune globulin treatment of lower respiratory tract infection in pediatric patients undergoing bone marrow transplantation—a compassionate use experience. Bone Marrow Transplant. 25:161–165 [DOI] [PubMed] [Google Scholar]
- 61. Dinwiddie D. L., Harrod K. S. 2008. Human metapneumovirus inhibits IFN-alpha signaling through inhibition of STAT1 phosphorylation. Am. J. Respir. Cell Mol. Biol. 38:661–670 [DOI] [PubMed] [Google Scholar]
- 62. Ditt V., Lusebrink J., Tillmann R. L., Schildgen V., Schildgen O. 2011. Respiratory infections by HMPV and RSV are clinically indistinguishable but induce different host response in aged individuals. PLoS One 6:e16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dollner H., Risnes K., Radtke A., Nordbo S. A. 2004. Outbreak of human metapneumovirus infection in Norwegian children. Pediatr. Infect. Dis. J. 23:436–440 [DOI] [PubMed] [Google Scholar]
- 64. Don M., Korppi M., Valent F., Vainionpaa R., Canciani M. 2008. Human metapneumovirus pneumonia in children: results of an Italian study and mini-review. Scand. J. Infect. Dis. 40:821–826 [DOI] [PubMed] [Google Scholar]
- 65. Drews S. J., et al. 2008. Use of the Seeplex RV detection kit for surveillance of respiratory viral outbreaks in Toronto, Ontario, Canada. Ann. Clin. Lab. Sci. 38:376–379 [PubMed] [Google Scholar]
- 66. Ebihara T., et al. 2004. Early reinfection with human metapneumovirus in an infant. J. Clin. Microbiol. 42:5944–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ebihara T., et al. 2004. Human metapneumovirus infection in Japanese children. J. Clin. Microbiol. 42:126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ebihara T., et al. 2004. Comparison of the seroprevalence of human metapneumovirus and human respiratory syncytial virus. J. Med. Virol. 72:304–306 [DOI] [PubMed] [Google Scholar]
- 69. Englund J. A., et al. 2006. Brief communication: fatal human metapneumovirus infection in stem-cell transplant recipients. Ann. Intern. Med. 144:344–349 [DOI] [PubMed] [Google Scholar]
- 70. Escobar C., et al. 2009. Genetic variability of human metapneumovirus isolated from Chilean children, 2003-2004. J. Med. Virol. 81:340–344 [DOI] [PubMed] [Google Scholar]
- 71. Esper F., Boucher D., Weibel C., Martinello R. A., Kahn J. S. 2003. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111:1407–1410 [DOI] [PubMed] [Google Scholar]
- 72. Esper F., et al. 2004. A 1-year experience with human metapneumovirus in children aged <5 years. J. Infect. Dis. 189:1388–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fabbiani M., et al. 2009. Epidemiological and clinical study of viral respiratory tract infections in children from Italy. J. Med. Virol. 81:750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Falsey A. R., Criddle M. C., Walsh E. E. 2006. Detection of respiratory syncytial virus and human metapneumovirus by reverse transcription polymerase chain reaction in adults with and without respiratory illness. J. Clin. Virol. 35:46–50 [DOI] [PubMed] [Google Scholar]
- 75. Falsey A. R., Erdman D., Anderson L. J., Walsh E. E. 2003. Human metapneumovirus infections in young and elderly adults. J. Infect. Dis. 187:785–790 [DOI] [PubMed] [Google Scholar]
- 76. Falsey A. R., Formica M. A., Treanor J. J., Walsh E. E. 2003. Comparison of quantitative reverse transcription-PCR to viral culture for assessment of respiratory syncytial virus shedding. J. Clin. Microbiol. 41:4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Falsey A. R., Formica M. A., Walsh E. E. 2009. Microneutralization assay for the measurement of neutralizing antibodies to human metapneumovirus. J. Clin. Virol. 46:314–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fearns R., Collins P. L. 1999. Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Reference deleted.
- 80. Franquet T., et al. 2005. Human metapneumovirus infection in hematopoietic stem cell transplant recipients: high-resolution computed tomography findings. J. Comput. Assist. Tomogr. 29:223–227 [DOI] [PubMed] [Google Scholar]
- 81. Freymouth F., et al. 2003. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr. Infect. Dis. J. 22:92–94 [DOI] [PubMed] [Google Scholar]
- 82. Fuenzalida L., et al. 2010. Usefulness of two new methods for diagnosing metapneumovirus infections in children. Clin. Microbiol. Infect. 16:1663–1668 [DOI] [PubMed] [Google Scholar]
- 83. Furuse Y. Detection of novel respiratory viruses from influenza-like illness in the Philippines. J. Med. Virol. 82:1071–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Galiano M., et al. 2004. Evidence of human metapneumovirus in children in Argentina. J. Med. Virol. 72:299–303 [DOI] [PubMed] [Google Scholar]
- 85. Garbino J., et al. 2008. Respiratory viruses in HIV-infected patients with suspected respiratory opportunistic infection. AIDS 22:701–705 [DOI] [PubMed] [Google Scholar]
- 86. Garcia-Garcia M. L., et al. 2007. Human metapneumovirus bronchiolitis in infancy is an important risk factor for asthma at age 5. Pediatr. Pulmonol. 42:458–464 [DOI] [PubMed] [Google Scholar]
- 87. Garcia-Garcia M. L., et al. 2006. Human metapneumovirus infections in hospitalised infants in Spain. Arch. Dis. Child. 91:290–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Garcia-Garcia M. L., et al. 2006. Prevalence and clinical characteristics of human metapneumovirus infections in hospitalized infants in Spain. Pediatr. Pulmonol. 41:863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Garcia-Sastre A. 2006. Antiviral response in pandemic influenza viruses. Emerg. Infect. Dis. 12:44–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Gaunt E., McWilliam-Leitch E. C., Templeton K., Simmonds P. 2009. Incidence, molecular epidemiology and clinical presentations of human metapneumovirus; assessment of its importance as a diagnostic screening target. J. Clin. Virol. 46:318–324 [DOI] [PubMed] [Google Scholar]
- 91. Gerna G., et al. 2005. Changing circulation rate of human metapneumovirus strains and types among hospitalized pediatric patients during three consecutive winter-spring seasons. Brief report. Arch. Virol. 150:2365–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ginocchio C. C., Manji R., Lotlikar M., Zhang F. 2008. Clinical evaluation of NucliSENS magnetic extraction and NucliSENS analyte-specific reagents for real-time detection of human metapneumovirus in pediatric respiratory specimens. J. Clin. Microbiol. 46:1274–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gioula G., Chatzidimitriou D., Melidou A., Exindari M., Kyriazopoulou-Dalaina V. 2010. Contribution of human metapneumovirus to influenza-like infections in North Greece, 2005-2008. Euro. Surveill. 15:19499. [DOI] [PubMed] [Google Scholar]
- 94. Gould P. S., Easton A. J. 2007. Coupled translation of the second open reading frame of M2 mRNA is sequence dependent and differs significantly within the subfamily Pneumovirinae. J. Virol. 81:8488–8496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Goutagny N., et al. 2010. Cell type-specific recognition of human metapneumoviruses (HMPVs) by retinoic acid-inducible gene I (RIG-I) and TLR7 and viral interference of RIG-I ligand recognition by HMPV-B1 phosphoprotein. J. Immunol. 184:1168–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gray G. C., et al. 2006. Multi-year study of human metapneumovirus infection at a large US midwestern medical referral center. J. Clin. Virol. 37:269–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gray G. C., et al. 2006. Human metapneumovirus, Peru. Emerg. Infect. Dis. 12:347–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Greensill J., et al. 2003. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg. Infect. Dis. 9:372–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guerrero-Plata A., et al. 2005. Activity and regulation of alpha interferon in respiratory syncytial virus and human metapneumovirus experimental infections. J. Virol. 79:10190–10199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Guerrero-Plata A., et al. 2006. Differential response of dendritic cells to human metapneumovirus and respiratory syncytial virus. Am. J. Respir. Cell Mol. Biol. 34:320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Guerrero-Plata A., Kolli D., Hong C., Casola A., Garofalo R. P. 2009. Subversion of pulmonary dendritic cell function by paramyxovirus infections. J. Immunol. 182:3072–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hamelin M. E., Abed Y., Boivin G. 2004. Human metapneumovirus: a new player among respiratory viruses. Clin. Infect. Dis. 38:983–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hamelin M. E., et al. 2005. Human metapneumovirus infection in adults with community-acquired pneumonia and exacerbation of chronic obstructive pulmonary disease. Clin. Infect. Dis. 41:498–502 [DOI] [PubMed] [Google Scholar]
- 104. Hamelin M. E., Prince G. A., Boivin G. 2006. Effect of ribavirin and glucocorticoid treatment in a mouse model of human metapneumovirus infection. Antimicrob. Agents Chemother. 50:774–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hamelin M. E., Prince G. A., Gomez A. M., Kinkead R., Boivin G. 2006. Human metapneumovirus infection induces long-term pulmonary inflammation associated with airway obstruction and hyperresponsiveness in mice. J. Infect. Dis. 193:1634–1642 [DOI] [PubMed] [Google Scholar]
- 106. Hamelin M. E., et al. 2005. Pathogenesis of human metapneumovirus lung infection in BALB/c mice and cotton rats. J. Virol. 79:8894–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Heininger U., Kruker A. T., Bonhoeffer J., Schaad U. B. 2009. Human metapneumovirus infections—biannual epidemics and clinical findings in children in the region of Basel, Switzerland. Eur. J. Pediatr. 168:1455–1460 [DOI] [PubMed] [Google Scholar]
- 108. Herd K. A., et al. 2006. Cytotoxic T-lymphocyte epitope vaccination protects against human metapneumovirus infection and disease in mice. J. Virol. 80:2034–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Herd K. A., Nissen M. D., Hopkins P. M., Sloots T. P., Tindle R. W. 2008. Major histocompatibility complex class I cytotoxic T lymphocyte immunity to human metapneumovirus (hMPV) in individuals with previous hMPV infection and respiratory disease. J. Infect. Dis. 197:584–592 [DOI] [PubMed] [Google Scholar]
- 110. Herfst S., et al. 2004. Recovery of human metapneumovirus genetic lineages A and B from cloned cDNA. J. Virol. 78:8264–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Herfst S., et al. 2008. Generation of temperature-sensitive human metapneumovirus strains that provide protective immunity in hamsters. J. Gen. Virol. 89:1553–1562 [DOI] [PubMed] [Google Scholar]
- 112. Herfst S., et al. 2007. Immunization of Syrian golden hamsters with F subunit vaccine of human metapneumovirus induces protection against challenge with homologous or heterologous strains. J. Gen. Virol. 88:2702–2709 [DOI] [PubMed] [Google Scholar]
- 113. Herfst S., Fouchier R. A. 2008. Vaccination approaches to combat human metapneumovirus lower respiratory tract infections. J. Clin. Virol. 41:49–52 [DOI] [PubMed] [Google Scholar]
- 114. Herfst S., et al. 2008. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J. Virol. 82:8891–8895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. High K. P. 2005. Pneumonia in older adults. New categories add complexity to diagnosis and care. Postgrad. Med. 118:18–20,25-28. [DOI] [PubMed] [Google Scholar]
- 116. Honda H., et al. 2006. Outbreak of human metapneumovirus infection in elderly inpatients in Japan. J. Am. Geriatr. Soc. 54:177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hopkins P., et al. 2008. Human metapneumovirus in lung transplant recipients and comparison to respiratory syncitial virus. Am. J. Respir. Crit. Care Med. 178:876–881 [DOI] [PubMed] [Google Scholar]
- 118. Huck B., et al. 2006. Novel human metapneumovirus sublineage. Emerg. Infect. Dis. 12:147–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118a. IJpma F. F., et al. 2004. Human metapneumovirus infection in hospital referred South African children. J. Med. Virol. 73:486–493 [DOI] [PubMed] [Google Scholar]
- 119. Jartti T., van den Hoogen B., Garofalo R. P., Osterhaus A. D., Ruuskanen O. 2002. Metapneumovirus and acute wheezing in children. Lancet 360:1393–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jenkins G. M. R., et al. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:156–165 [DOI] [PubMed] [Google Scholar]
- 121. Jiang J., et al. 2009. Limited expansion of virus-specific CD8 T cells in the aged environment. Mech. Ageing Dev. 130:713–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Jiang T., et al. 2009. CpG oligodeoxynucleotides protect against the H1N1 pandemic influenza virus infection in a murine model. Antiviral Res. 89:124–126 [DOI] [PubMed] [Google Scholar]
- 123. Jiang T. J., et al. 2009. Preferential loss of Th17 cells is associated with CD4 T cell activation in patients with pandemic H1N1 swine-origin influenza A infection. Clin. Immunol. 137:303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Jokela P., Piiparinen H., Luiro K., Lappalainen M. 2010. Detection of human metapneumovirus and respiratory syncytial virus by duplex real-time RT-PCR assay in comparison with direct fluorescent assay. Clin. Microbiol. Infect. 16:1568–1573 [DOI] [PubMed] [Google Scholar]
- 125. Kahn J. S., Schnell M. J., Buonocore L., Rose J. K. 1999. Recombinant vesicular stomatitis virus expressing respiratory syncytial virus (RSV) glycoproteins: RSV fusion protein can mediate infection and cell fusion. Virology 254:81–91 [DOI] [PubMed] [Google Scholar]
- 126. Kaida A., et al. 2006. Seasonal distribution and phylogenetic analysis of human metapneumovirus among children in Osaka City, Japan. J. Clin. Virol. 35:394–399 [DOI] [PubMed] [Google Scholar]
- 127. Kawai T. A. S. 2005. Pathogen recognotion with Toll-like receptors. Curr. Opin. Immunol. 17:338–344 [DOI] [PubMed] [Google Scholar]
- 128. Kaye M., Skidmore S., Osman H., Weinbren M., Warren R. 2006. Surveillance of respiratory virus infections in adult hospital admissions using rapid methods. Epidemiol. Infect. 134:792–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kolli D., et al. 2008. T lymphocytes contribute to antiviral immunity and pathogenesis in experimental human metapneumovirus infection. J. Virol. 82:8560–8569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Konig B., et al. 2004. Prospective study of human metapneumovirus infection in children less than 3 years of age. J. Clin. Microbiol. 42:4632–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kuiken T., et al. 2004. Experimental human metapneumovirus infection of cynomolgus macaques (Macaca fascicularis) results in virus replication in ciliated epithelial cells and pneumocytes with associated lesions throughout the respiratory tract. Am. J. Pathol. 164:1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Laguna-Torres V. A., et al. 2011. Influenza and other respiratory viruses in three Central American countries. Influenza Other Respi. Viruses 5:123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Laham F. R., et al. 2004. Differential production of inflammatory cytokines in primary infection with human metapneumovirus and with other common respiratory viruses of infancy. J. Infect. Dis. 189:2047–2056 [DOI] [PubMed] [Google Scholar]
- 134. Lamb R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1–11 [DOI] [PubMed] [Google Scholar]
- 135. Lamb R. A., Jardetzky T. S. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Lamb R. A. P., Parks G. D. 2007. Paramyxoviridae: the viruses and their replication, p. 1449–1456 In Knipe D. M., et al. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 137. Landry M. L., Cohen S., Ferguson D. 2008. Prospective study of human metapneumovirus detection in clinical samples by use of Light Diagnostics direct immunofluorescence reagent and real-time PCR. J. Clin. Microbiol. 46:1098–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Landry M. L., Ferguson D., Cohen S., Peret T. C., Erdman D. D. 2005. Detection of human metapneumovirus in clinical samples by immunofluorescence staining of shell vial centrifugation cultures prepared from three different cell lines. J. Clin. Microbiol. 43:1950–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Larcher C., et al. 2005. Human metapneumovirus infection in lung transplant recipients: clinical presentation and epidemiology. J. Heart Lung Transplant. 24:1891–1901 [DOI] [PubMed] [Google Scholar]
- 140. Larcher C., et al. 2008. Comparison of human metapneumovirus genotypes from the province of Bolzano in northern Italy with strains from surrounding regions in Italy and Austria. Jpn. J. Infect. Dis. 61:154–156 [PubMed] [Google Scholar]
- 141. Lazar I., et al. 2004. Human metapneumovirus and severity of respiratory syncytial virus disease. Emerg. Infect. Dis. 10:1318–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Leung J., Esper F., Weibel C., Kahn J. S. 2005. Seroepidemiology of human metapneumovirus (hMPV) on the basis of a novel enzyme-linked immunosorbent assay utilizing hMPV fusion protein expressed in recombinant vesicular stomatitis virus. J. Clin. Microbiol. 43:1213–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Liao S., et al. 2008. Role of retinoic acid inducible gene-I in human metapneumovirus-induced cellular signalling. J. Gen. Virol. 89:1978–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Liu L., Bastien N., Li Y. 2007. Intracellular processing, glycosylation, and cell surface expression of human metapneumovirus attachment glycoprotein. J. Virol. 81:13435–13443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Liu L., Bastien N., Sidaway F., Chan E., Li Y. 2007. Seroprevalence of human metapneumovirus (hMPV) in the Canadian province of Saskatchewan analyzed by a recombinant nucleocapsid protein-based enzyme-linked immunosorbent assay. J. Med. Virol. 79:308–313 [DOI] [PubMed] [Google Scholar]
- 146. Ljubin-Sternak S., et al. 2008. Detection of genetic lineages of human metapneumovirus in Croatia during the winter season 2005/2006. J. Med. Virol. 80:1282–1287 [DOI] [PubMed] [Google Scholar]
- 147. Ljubin Sternak S., Vilibic Cavlek T., Falsey A. R., Walsh E. E., Mlinaric Galinovic G. 2006. Serosurvey of human metapneumovirus infection in Croatia. Croat. Med. J. 47:878–881 [PMC free article] [PubMed] [Google Scholar]
- 148. Loo L. H., et al. 2007. Human metapneumovirus in children, Singapore. Emerg. Infect. Dis. 13:1396–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Louie J. K., et al. 2007. A summer outbreak of human metapneumovirus infection in a long-term-care facility. J. Infect. Dis. 196:705–708 [DOI] [PubMed] [Google Scholar]
- 150. Lu G. Large-scale seroprevalence analysis of human metapneumovirus and human respiratory syncytial virus infections in Beijing, China. Virol. J. 8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Luchsinger V., Escobar C., Avendano L. F. 2005. Detection of human metapneumovirus in children hospitalized for acute lower respiratory infection in Santiago, Chile. Rev. Med. Chil. 133:1059–1064 [DOI] [PubMed] [Google Scholar]
- 152. Ludewick H. P., et al. 2005. Human metapneumovirus genetic variability, South Africa. Emerg. Infect. Dis. 11:1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Lusebrink J., et al. High seroprevalence of neutralizing capacity against human metapneumovirus in all age groups studied in Bonn, Germany. Clin. Vaccine Immunol. 17:481–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Mackay I. M., et al. 2006. Genetic diversity of human metapneumovirus over 4 consecutive years in Australia. J. Infect. Dis. 193:1630–1633 [DOI] [PubMed] [Google Scholar]
- 155. MacPhail M., et al. 2004. Identification of small-animal and primate models for evaluation of vaccine candidates for human metapneumovirus (hMPV) and implications for hMPV vaccine design. J. Gen. Virol. 85:1655–1663 [DOI] [PubMed] [Google Scholar]
- 156. Madhi S. A., Ludewick H., Abed Y., Klugman K. P., Boivin G. 2003. Human metapneumovirus-associated lower respiratory tract infections among hospitalized human immunodeficiency virus type 1 (HIV-1)-infected and HIV-1-uninfected African infants. Clin. Infect. Dis. 37:1705–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Madhi S. A., et al. 2007. Seasonality, incidence, and repeat human metapneumovirus lower respiratory tract infections in an area with a high prevalence of human immunodeficiency virus type-1 infection. Pediatr. Infect. Dis. J. 26:693–699 [DOI] [PubMed] [Google Scholar]
- 158. Maertzdorf J., et al. 2004. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J. Clin. Microbiol. 42:981–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Maggi F., et al. 2003. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J. Clin. Microbiol. 41:2987–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Manoha C., Espinosa S., Aho S. L., Huet F., Pothier P. 2007. Epidemiological and clinical features of hMPV, RSV and RVs infections in young children. J. Clin. Virol. 38:221–226 [DOI] [PubMed] [Google Scholar]
- 161. Mao H. W., Yang X. Q., Zhao X. D. 2008. Characterization of human metapneumoviruses isolated in Chongqing, China. Chin. Med. J. (Engl.) 121:2254–2257 [PubMed] [Google Scholar]
- 162. Martinello R. A., Chen M. D., Weibel C., Kahn J. S. 2002. Correlation between respiratory syncytial virus genotype and severity of illness. J. Infect. Dis. 186:839–842 [DOI] [PubMed] [Google Scholar]
- 163. Martinello R. A., et al. 2006. Human metapneumovirus and exacerbations of chronic obstructive pulmonary disease. J. Infect. 53:248–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Martino R., et al. 2005. Prospective study of the incidence, clinical features, and outcome of symptomatic upper and lower respiratory tract infections by respiratory viruses in adult recipients of hematopoietic stem cell transplants for hematologic malignancies. Biol. Blood Marrow Transplant. 11:781–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Mathisen M., et al. 2009. RNA viruses in community-acquired childhood pneumonia in semi-urban Nepal; a cross-sectional study. BMC Med. 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Matsuzaki Y., et al. 2008. Clinical impact of human metapneumovirus genotypes and genotype-specific seroprevalence in Yamagata, Japan. J. Med. Virol. 80:1084–1089 [DOI] [PubMed] [Google Scholar]
- 167. Mazhul L. A., Schildgen O., Isaeva E. I., Viazov S. O. 2007. Human metapneumovirus as a common cause of respiratory tract disease. Vopr. Virusol. 52:4–8 [PubMed] [Google Scholar]
- 168. McAdam A. J., et al. 2004. Human metapneumovirus in children tested at a tertiary-care hospital. J. Infect. Dis. 190:20–26 [DOI] [PubMed] [Google Scholar]
- 169. Melendi G. A., et al. 2007. Mapping and characterization of the primary and anamnestic H-2(d)-restricted cytotoxic T-lymphocyte response in mice against human metapneumovirus. J. Virol. 81:11461–11467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Mirazo S., Ruchansky D., Blanc A., Arbiza J. 2005. Serologic evidence of human metapneumovirus circulation in Uruguay. Mem. Inst. Oswaldo Cruz 100:715–718 [DOI] [PubMed] [Google Scholar]
- 171. Mok H., et al. 2008. An alphavirus replicon-based human metapneumovirus vaccine is immunogenic and protective in mice and cotton rats. J. Virol. 82:11410–11418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Monto A. S. 2002. Epidemiology of viral respiratory infections. Am. J. Med. 112(Suppl. 6A):4S–12S [DOI] [PubMed] [Google Scholar]
- 173. Mullins J. A., et al. 2004. Human metapneumovirus infection among children hospitalized with acute respiratory illness. Emerg. Infect. Dis. 10:700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Nicholson K. G., McNally T., Silverman M., Simons P., Zambon M. C. 2003. Influenza-related hospitalizations among young children in Leicestershire. Pediatr. Infect. Dis. J. 22:S228–S230 [DOI] [PubMed] [Google Scholar]
- 175. Nolte F. S., et al. 2007. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J. Clin. Microbiol. 45:2779–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Okamoto M., et al. 2010. Development and evaluation of a whole virus-based enzyme-linked immunosorbent assay for the detection of human metapneumovirus antibodies in human sera. J. Virol. Methods 164:24–29 [DOI] [PubMed] [Google Scholar]
- 177. Oliveira D. B., et al. 2009. Epidemiology and genetic variability of human metapneumovirus during a 4-year-long study in Southeastern Brazil. J. Med. Virol. 81:915–921 [DOI] [PubMed] [Google Scholar]
- 178. Olmsted R. A., et al. 1986. Expression of the F glycoprotein of respiratory syncytial virus by a recombinant vaccinia virus: comparison of the individual contributions of the F and G glycoproteins to host immunity. Proc. Natl. Acad. Sci. U. S. A. 83:7462–7466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Ordas J., et al. 2006. Role of metapneumovirus in viral respiratory infections in young children. J. Clin. Microbiol. 44:2739–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Osbourn M., McPhie K. A., Ratnamohan V. M., Dwyer D. E., Durrheim D. N. 2009. Outbreak of human metapneumovirus infection in a residential aged care facility. Commun. Dis. Intell. 33:38–40 [PubMed] [Google Scholar]
- 181. Pavlin J. A., et al. 2008. Human metapneumovirus reinfection among children in Thailand determined by ELISA using purified soluble fusion protein. J. Infect. Dis. 198:836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Peiris J. S., et al. 2003. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg. Infect. Dis. 9:628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Pelletier G., Dery P., Abed Y., Boivin G. 2002. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg. Infect. Dis. 8:976–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Pham Q. N., et al. 2005. Chimeric recombinant human metapneumoviruses with the nucleoprotein or phosphoprotein open reading frame replaced by that of avian metapneumovirus exhibit improved growth in vitro and attenuation in vivo. J. Virol. 79:15114–15122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Pitoiset C., et al. 2010. Human metapneumovirus genotypes and severity of disease in young children (n = 100) during a 7-year study in Dijon hospital, France. J. Med. Virol. 82:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Pizzorno A., et al. Molecular detection and genetic variability of human metapneumovirus in Uruguay. J. Med. Virol. 82:861–865 [DOI] [PubMed] [Google Scholar]
- 187. Po J. L., Gardner E. M., Anaraki F., Katsikis P. D., Murasko D. 2002. Age-associated decrease in virus-specific CD8+ T lymphocytes during primary influenza infection. Mech. Ageing Dev. 123:1167–1181 [DOI] [PubMed] [Google Scholar]
- 188. Rafiefard F., Yun Z., Orvell C. 2008. Epidemiologic characteristics and seasonal distribution of human metapneumovirus infections in five epidemic seasons in Stockholm, Sweden, 2002-2006. J. Med. Virol. 80:1631–1638 [DOI] [PubMed] [Google Scholar]
- 189. Rawlinson W. D., et al. 2003. Asthma exacerbations in children associated with rhinovirus but not human metapneumovirus infection. J. Infect. Dis. 187:1314–1318 [DOI] [PubMed] [Google Scholar]
- 190. Raymond F., et al. 2009. Comparison of automated microarray detection with real-time PCR assays for detection of respiratory viruses in specimens obtained from children. J. Clin. Microbiol. 47:743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Raza K., et al. 2007. Successful outcome of human metapneumovirus (hMPV) pneumonia in a lung transplant recipient treated with intravenous ribavirin. J. Heart Lung Transplant. 26:862–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Reijans M., et al. 2008. RespiFinder: a new multiparameter test to differentially identify fifteen respiratory viruses. J. Clin. Microbiol. 46:1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Robinson J. L., Lee B. E., Bastien N., Li Y. 2005. Seasonality and clinical features of human metapneumovirus infection in children in Northern Alberta. J. Med. Virol. 76:98–105 [DOI] [PubMed] [Google Scholar]
- 194. Rohde G., et al. 2005. Relevance of human metapneumovirus in exacerbations of COPD. Respir. Res. 6:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Russell C. J., Luque L. E. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol. 14:243–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Ryder A. B., Tollefson S. J., Podsiad A. B., Johnson J. E., Williams J. V. 2010. Soluble recombinant human metapneumovirus G protein is immunogenic but not protective. Vaccine 28:4145–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Schickli J. H., et al. 2008. Deletion of human metapneumovirus M2-2 increases mutation frequency and attenuates growth in hamsters. Virol. J. 5:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Schickli J. H., Kaur J., Ulbrandt N., Spaete R. R., Tang R. S. 2005. An S101P substitution in the putative cleavage motif of the human metapneumovirus fusion protein is a major determinant for trypsin-independent growth in Vero cells and does not alter tissue tropism in hamsters. J. Virol. 79:10678–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Schildgen O., Geikowski T., Glatzel T., Schuster J., Simon A. 2005. Frequency of human metapneumovirus in the upper respiratory tract of children with symptoms of an acute otitis media. Eur. J. Pediatr. 164:400–401 [DOI] [PubMed] [Google Scholar]
- 200. Schildgen O., et al. 2004. New variant of the human metapneumovirus (HMPV) associated with an acute and severe exacerbation of asthma bronchiale. J. Clin. Virol. 31:283–288 [DOI] [PubMed] [Google Scholar]
- 201. Schildgen O., Simon A., Williams J. 2007. Animal models for human metapneumovirus (HMPV) infections. Vet. Res. 38:117–126 [DOI] [PubMed] [Google Scholar]
- 202. Schildgen V., et al. 2010. Human HepG2 cells support respiratory syncytial virus and human metapneumovirus replication. J. Virol. Methods 163:74–81 [DOI] [PubMed] [Google Scholar]
- 203. Schowalter R. M., Smith S. E., Dutch R. E. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 80:10931–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Seal B. S. S., et al. 2000. Fusion protein predicted amino acid sequence of the first US avian pneumovirus isolate and lack of heterogeneity among other US isolates. Virus Res. 66:139–147 [DOI] [PubMed] [Google Scholar]
- 205. Semple M. G., et al. 2005. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J. Infect. Dis. 191:382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Seth R. B. S., et al. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signalling protein that actrivates NF-kappa B and IRF-3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 207. Shanmukh S., et al. 2008. Identification and classification of respiratory syncytial virus (RSV) strains by surface-enhanced Raman spectroscopy and multivariate statistical techniques. Anal. Bioanal. Chem. 390:1551–1555 [DOI] [PubMed] [Google Scholar]
- 208. Singleton R. J., et al. 2010. Viral respiratory infections in hospitalized and community control children in Alaska. J. Med. Virol. 82:1282–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Skiadopoulos M. H., et al. 2006. Individual contributions of the human metapneumovirus F, G, and SH surface glycoproteins to the induction of neutralizing antibodies and protective immunity. Virology 345:492–501 [DOI] [PubMed] [Google Scholar]
- 210. Skiadopoulos M. H., et al. 2004. The two major human metapneumovirus genetic lineages are highly related antigenically, and the fusion (F) protein is a major contributor to this antigenic relatedness. J. Virol. 78:6927–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Sloots T. P., et al. 2006. Human metapneumovirus, Australia, 2001-2004. Emerg. Infect. Dis. 12:1263–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Smith E. C., Popa A., Chang A., Masante C., Dutch R. E. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 276:7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Spetch L., Bowlin T. L., Casola A. 2008. Effect of NMSO3 treatment in a murine model of human metapneumovirus infection. J. Gen. Virol. 89:2709–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Stout-Delgado H. W., Du W., Shirali A. C., Booth C. J., Goldstein D. 2009. Aging promotes neutrophil-induced mortality by augmenting IL-17 production during viral infection. Cell Host Microbe 6:446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Sullender W. M. 2000. Respiratory syncytial virus genetic and antigenic diversity. Clin. Microbiol. Rev. 13:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Sumino K. C., et al. 2005. Detection of severe human metapneumovirus infection by real-time polymerase chain reaction and histopathological assessment. J. Infect. Dis. 192:1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Suzuki A., et al. 2005. Detection of human metapneumovirus from children with acute otitis media. Pediatr. Infect. Dis. J. 24:655–657 [DOI] [PubMed] [Google Scholar]
- 218. Takaoka A. Y. H. 2006. Interferon signalling network in innate defense. Cell. Microbiol. 8:907–922 [DOI] [PubMed] [Google Scholar]
- 219. Talavera G. A., Mezquita N. E. 2007. Human metapneumovirus in children with influenza-like illness in Yucatan, Mexico. Am. J. Trop. Med. Hyg. 76:182–183 [PubMed] [Google Scholar]
- 220. Tang R. S., et al. 2004. Parainfluenza virus type 3 expressing the native or soluble fusion (F) protein of respiratory syncytial virus (RSV) confers protection from RSV infection in African green monkeys. J. Virol. 78:11198–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Tang R. S., et al. 2005. A host-range restricted parainfluenza virus type 3 (PIV3) expressing the human metapneumovirus (hMPV) fusion protein elicits protective immunity in African green monkeys. Vaccine 23:1657–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Tang R. S., et al. 2003. Effects of human metapneumovirus and respiratory syncytial virus antigen insertion in two 3′ proximal genome positions of bovine/human parainfluenza virus type 3 on virus replication and immunogenicity. J. Virol. 77:10819–10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Tecu C., et al. 2007. First detection of human metapneumovirus in children with respiratory infections in Romania. Roum. Arch. Microbiol. Immunol. 66:37–40 [PubMed] [Google Scholar]
- 224. Thammawat S., Sadlon T. A., Hallsworth P. G., Gordon D. L. 2008. Role of cellular glycosaminoglycans and charged regions of viral G protein in human metapneumovirus infection. J. Virol. 82:11767–11774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Thanasugarn W., et al. 2003. Human metapneumovirus infection in Thai children. Scand. J. Infect. Dis. 35:754–756 [DOI] [PubMed] [Google Scholar]
- 226. Tollefson S. J., Cox R. G., Williams J. V. 2010. Studies of culture conditions and environmental stability of human metapneumovirus. Virus Res. 151:54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Ulbrandt N. D., et al. 2006. Isolation and characterization of monoclonal antibodies which neutralize human metapneumovirus in vitro and in vivo. J. Virol. 80:7799–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Ulloa-Gutierrez R., et al. 2009. Human metapneumovirus in Costa Rica. Pediatr. Infect. Dis. J. 28:452–453 [DOI] [PubMed] [Google Scholar]
- 229. van den Hoogen B. G. 2007. Respiratory tract infection due to human metapneumovirus among elderly patients. Clin. Infect. Dis. 44:1159–1160 [DOI] [PubMed] [Google Scholar]
- 230. van den Hoogen B. G., Bestebroer T. M., Osterhaus A. D., Fouchier R. A. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119–132 [DOI] [PubMed] [Google Scholar]
- 231. van den Hoogen B. G., et al. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7:719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. van den Hoogen B. G., et al. 2007. Experimental infection of macaques with human metapneumovirus induces transient protective immunity. J. Gen. Virol. 88:1251–1259 [DOI] [PubMed] [Google Scholar]
- 233. van den Hoogen B. G., et al. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg. Infect. Dis. 10:658–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. van den Hoogen B. G., Osterhaus D. M., Fouchier R. A. 2004. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr. Infect. Dis. J. 23:S25–S32 [DOI] [PubMed] [Google Scholar]
- 235. van den Hoogen B. G., et al. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J. Infect. Dis. 188:1571–1577 [DOI] [PubMed] [Google Scholar]
- 236. van Woensel J. B., Bos A. P., Lutter R., Rossen J. W., Schuurman R. 2006. Absence of human metapneumovirus co-infection in cases of severe respiratory syncytial virus infection. Pediatr. Pulmonol. 41:872–874 [DOI] [PubMed] [Google Scholar]
- 237. Vargas S. O., Kozakewich H. P., Perez-Atayde A. R., McAdam A. J. 2004. Pathology of human metapneumovirus infection: insights into the pathogenesis of a newly identified respiratory virus. Pediatr. Dev. Pathol. 7:478–486 [DOI] [PubMed] [Google Scholar]
- 238. Viazov S., Ratjen F., Scheidhauer R., Fiedler M., Roggendorf M. 2003. High prevalence of human metapneumovirus infection in young children and genetic heterogeneity of the viral isolates. J. Clin. Microbiol. 41:3043–3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Vicente D., Montes M., Cilla G., Perez-Trallero E. 2004. Human metapneumovirus and chronic obstructive pulmonary disease. Emerg. Infect. Dis. 10:1338–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Vicente D., Montes M., Cilla G., Perez-Yarza E. G., Perez-Trallero E. 2006. Differences in clinical severity between genotype A and genotype B human metapneumovirus infection in children. Clin. Infect. Dis. 42:e111–e113 [DOI] [PubMed] [Google Scholar]
- 241. Walsh E. E., Peterson D. R., Falsey A. R. 2008. Human metapneumovirus infections in adults: another piece of the puzzle. Arch. Intern. Med. 168:2489–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Wang H. C., et al. 2008. Co-circulating genetically divergent A2 human metapneumovirus strains among children in southern Taiwan. Arch. Virol. 153:2207–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Wang S. M., Liu C. C., Wang H. C., Su I. J., Wang J. R. 2006. Human metapneumovirus infection among children in Taiwan: a comparison of clinical manifestations with other virus-associated respiratory tract infections. Clin. Microbiol. Infect. 12:1221–1224 [DOI] [PubMed] [Google Scholar]
- 244. Weigl J. A., et al. 2007. Ten years' experience with year-round active surveillance of up to 19 respiratory pathogens in children. Eur. J. Pediatr. 166:957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Wilkesmann A., et al. 2006. Human metapneumovirus infections cause similar symptoms and clinical severity as respiratory syncytial virus infections. Eur. J. Pediatr. 165:467–475 [DOI] [PubMed] [Google Scholar]
- 246. Williams J. V., et al. 2007. A recombinant human monoclonal antibody to human metapneumovirus fusion protein that neutralizes virus in vitro and is effective therapeutically in vivo. J. Virol. 81:8315–8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Williams J. V., et al. 2005. Human metapneumovirus infection plays an etiologic role in acute asthma exacerbations requiring hospitalization in adults. J. Infect. Dis. 192:1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248. Williams J. V., et al. 2004. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N. Engl. J. Med. 350:443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Williams J. V., et al. 2005. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J. Infect. Dis. 192:1061–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Williams J. V., et al. 2005. Human metapneumovirus infection in children hospitalized for wheezing. J. Allergy Clin. Immunol. 115:1311–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Williams J. V., Tollefson S. J., Johnson J. E., Crowe J. E., Jr 2005. The cotton rat (Sigmodon hispidus) is a permissive small animal model of human metapneumovirus infection, pathogenesis, and protective immunity. J. Virol. 79:10944–10951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Williams J. V., Tollefson S. J., Nair S., Chonmaitree T. 2006. Association of human metapneumovirus with acute otitis media. Int. J. Pediatr. Otorhinolaryngol. 70:1189–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Williams J. V., et al. 2006. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J. Infect. Dis. 193:387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254. Wolf D. G., et al. 2006. Comparison of human metapneumovirus, respiratory syncytial virus and influenza A virus lower respiratory tract infections in hospitalized young children. Pediatr. Infect. Dis. J. 25:320–324 [DOI] [PubMed] [Google Scholar]
- 255. Wolf D. G., Zakay-Rones Z., Fadeela A., Greenberg D., Dagan R. 2003. High seroprevalence of human metapneumovirus among young children in Israel. J. Infect. Dis. 188:1865–1867 [DOI] [PubMed] [Google Scholar]
- 256. Wyde P. R., Chetty S. N., Jewell A. M., Boivin G., Piedra P. A. 2003. Comparison of the inhibition of human metapneumovirus and respiratory syncytial virus by ribavirin and immune serum globulin in vitro. Antiviral Res. 60:51–59 [DOI] [PubMed] [Google Scholar]
- 257. Wyde P. R., Chetty S. N., Jewell A. M., Schoonover S. L., Piedra P. A. 2005. Development of a cotton rat-human metapneumovirus (hMPV) model for identifying and evaluating potential hMPV antivirals and vaccines. Antiviral Res. 66:57–66 [DOI] [PubMed] [Google Scholar]
- 258. Xiao N. G., et al. 2010. Prevalence and clinical and molecular characterization of human metapneumovirus in children with acute respiratory infection in China. Pediatr. Infect. Dis. J. 29:131–134 [DOI] [PubMed] [Google Scholar]
- 259. Yang C. F., et al. 2009. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virol. J. 6:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Yim K. C., et al. 2007. Human metapneumovirus: enhanced pulmonary disease in cotton rats immunized with formalin-inactivated virus vaccine and challenged. Vaccine 25:5034–5040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Zeng L., et al. 2011. Association between human metapneumovirus seroprevalence and hypertension in elderly subjects in a long-term care facility. Hypertens. Res. 34:474–478 [DOI] [PubMed] [Google Scholar]
- 262. Zhang Q., Yang X. Q., Zhao Y., Zhao X. D. 2008. High seroprevalence of human metapneumovirus infection in children in Chongqing, China. Chin. Med. J. (Engl.) 121:2162–2166 [PubMed] [Google Scholar]
- 263. Zlateva K. T., Lemey P., Vandamme A. M., Van Ranst M. 2004. Molecular evolution and circulation patterns of human respiratory syncytial virus subgroup A: positively selected sites in the attachment G glycoprotein. J. Virol. 78:4675–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]