Abstract
In this study, we constructed and evaluated a target-specific, salt-resistant antimicrobial peptide (AMP) that selectively targeted Streptococcus mutans, a leading cariogenic pathogen. The rationale for creating such a peptide was based on the addition of a targeting domain of S. mutans ComC signaling peptide pheromone (CSP) to a killing domain consisting of a portion of the marine-derived, broad-spectrum AMP pleurocidin to generate a target-specific AMP. Here, we report the results of our assessment of such fusion peptides against S. mutans and two closely related species. The results showed that nearly 95% of S. mutans cells lost viability following exposure to fusion peptide IMB-2 (5.65 μM) for 15 min. In contrast, only 20% of S. sanguinis or S. gordonii cells were killed following the same exposure. Similar results were also observed in dual-species mixed cultures of S. mutans with S. sanguinis or S. gordonii. The peptide-guided killing was further confirmed in S. mutans biofilms and was shown to be dose dependent. An S. mutans mutant defective in the CSP receptor retained 60% survival following exposure to IMB-2, suggesting that the targeted peptide predominantly bound to the CSP receptor to mediate killing in the wild-type strain. Our work confirmed that IMB-2 retained its activity in the presence of physiological or higher salt concentrations. In particular, the fusion peptide showed a synergistic killing effect on S. mutans with a preventive dose of NaF. In addition, IMB-2 was relatively stable in the presence of saliva containing 1 mM EDTA and did not cause any hemolysis. We also found that replacement of serine-14 by histidine improved its activity at lower pH. Because of its effectiveness, salt resistance, and minimal toxicity to host cells, this novel target-specific peptide shows promise for future development as an anticaries agent.
INTRODUCTION
Streptococcus mutans is a Gram-positive bacterium that depends on a “biofilm lifestyle” for survival and persistence in dental plaque, a multispecies biofilm community characterized by vast diversity (>700 species) and high cell density (1011 cells/g wet weight) (1, 14). Under appropriate environmental conditions, the organism can rapidly produce acids from fermentable dietary carbohydrates, resulting in demineralization of the tooth surface or dental caries (1, 2). Although hundreds of species of microorganisms are isolated from dental biofilms, decades of studies in epidemiology, microbiology, biochemistry, and animal models have strongly implicated S. mutans as the primary etiological agent of dental caries worldwide (1, 2, 13). This is because S. mutans is highly efficient in expressing several virulence-associated traits, which provide it with ecological advantages for its succession and persistence in dental biofilm, resulting in its emergence in significant numbers in dental biofilms that may initiate caries (13, 17, 21). Although much effort has been invested in treatment and prevention, dental caries remains a “silent epidemic,” left untreated in many undeveloped regions and countries around the world (2, 11). Moreover, S. mutans can be a cause of subacute infective endocarditis (1, 13, 21). Diagnosing, treating, and re-treating caries and caries-related diseases is still a major financial burden to public health systems worldwide (2, 11, 20). Additionally, eliminating S. mutans from dental biofilms using broad-spectrum antimicrobial agents is not practical, because these agents inevitably result in indiscriminate killing of the resident microflora, leading to ecological disruption and other negative consequences. Therefore, prevention of S. mutans infection and its associated diseases requires the development of new strategies that specifically target S. mutans without significant disruption of the resident oral flora.
Antimicrobial peptides (AMPs) comprise a wide range of short, cationic peptide antibiotics that constitute the first line of innate immune defense against infectious agents (12, 35). Over the last 2 decades, AMPs have been rigorously investigated as alternatives to small-molecule antibiotics and as potential solutions to the growing crisis of antibiotic-resistant bacterial infections (3, 12, 35). AMPs can be found in virtually all forms of life, including microorganisms, insects, invertebrates, amphibians, plants, birds, and mammals (12, 35). For example, many AMPs have been isolated from a variety of organisms in the oceans, which cover over 70% of the earth's surface and represent a rich source of antimicrobial agents (24, 33). In particular, the high salinity (3.3% or 565 mM) of the marine environment makes it likely that AMPs isolated from marine organisms naturally possess greater salt resistance than those derived from other sources (23, 24). This feature may allow marine-derived AMPs to maintain their activities better in relatively high-salt environments, such as in saliva, gastrointestinal fluid, serum, or other body fluids (12, 23, 24). Unlike small-molecule antimicrobial compounds, which may lose activity when their basic structures are modified, many AMPs can be optimized to enhance their effectiveness and stability through modification of their primary sequences, making them good templates for the development of therapeutic agents (3, 12, 35). Although many reports have explored the potential of AMPs as antimicrobial agents in vitro, there are considerable challenges in their clinical application. These include doubts about the ability to achieve high antimicrobial activities under physiological salt, pH, and serum conditions; the susceptibility of peptides to proteolytic degradation; the lack of information about potential toxicities in vivo; and comparatively high costs associated with peptide production (12, 35).
In this study, we aimed to construct a small, target-specific, salt-resistant AMP that selectively killed S. mutans. The rationale for constructing such a peptide was based on the addition of a targeting domain of the S. mutans ComC signaling peptide (CSP) to a killing domain consisting of the active portion of a marine-derived AMP to generate a target-specific fusion peptide against S. mutans strains. To achieve this goal, we initially screened 26 pleurocidin variants (see Table S1 in the supplemental material), a group of AMPs isolated from skin mucous secretions of the marine flatfish (5, 6, 25). From this peptide library, we identified one pleurocidin variant, NRC-4, which was highly effective against all S. mutans strains. We then designed peptides containing the targeting domain of S. mutans CSP and various domains of NRC-4 based on the rationale developed from the structure-activity analyses of NRC-4 (30) and S. mutans CSP (31, 32). Here, we report the results of our assessment of these peptides against S. mutans and two closely related oral streptococci.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study included S. mutans wild-type strains (UA159, NG8, and GS5); a comD deletion mutant (SmΔcomD) that was constructed from UA159 and that is defective in the CSP receptor (31); and two closely related species, S. sanguinis SK108 and S. gordonii DL1. We also included two lacZ transcriptional reporter strains, SmdC-PcomDE and SmdC-PnlmAB, to assay transcriptional activation of S. mutans comDE and nlmAB genes, respectively, in response to the fusion peptides, since these genes were directly controlled by the ComCDE quorum-sensing system (17, 18). All strains were maintained on Todd-Hewitt yeast extract (THYE) medium or THYE supplemented with appropriate antibiotics.
Design and synthesis of fusion peptides.
We initially designed and synthesized three fusion peptides, IMB-1, IMB-2, and IMB-3, based on the structure-activity relationships of a pleurocidin variant, NRC-4 (31); S. mutans CSP (32); and a GG linker (7). The new fusion peptides consisted of the targeting domain of CSP, a GG linker, and either the full length of NRC-4 (IMB-1), the N-terminal portion of NRC-4 (IMB-2), or its C-terminal portion (IMB-3) (Table 1). To facilitate microscopic analysis of peptide binding, we also synthesized two peptides, IMB2-Biot and IMB2-H/S14, with N-terminal biotinylation. All peptides were commercially synthesized and purified by reversed-phase high-pressure liquid chromatography with purities of >80%. Their molecular masses were confirmed by mass spectrometry (BioBasic Inc., Markham, Ontario, Canada). With the exception of CSP, all the peptides were C-terminally amidated to reduce the end charge and increase stability (25). The peptides were dissolved in sterile distilled water as stock solutions (5.12 mg/ml) and further diluted to final concentrations for individual experiments.
Table 1.
Sequences and antimicrobial activities of targeted peptides against S. mutans
| Peptidea | Targeting domain-linker-killing domainb | MICc (μM) |
|---|---|---|
| CSP | SGSLSTFFRLFNRSFTQALGK | NA |
| NRC-4 | GWGSFFKKAAHVGKHVGKAALTHYL·NH2 | 1.0 |
| IMB-1 | TFFRLFNR-GG-GWGSFFKKAAHVGKHVGKAALTHYL·NH2 | 2.2 |
| IMB-2 | TFFRLFNR-GG-GWGSFFKKAAHVGKL·NH2 | 2.8 |
| IMB-3 | TFFRLFNR-GGG-HVGKHVGKAALTHYL·NH2 | 44 |
| IMB2-Biot | Biot-TFFRLFNR-GG-GWGSFFKKAAHVGKL·NH2 | 2.8 |
| IMB2-H/S14 | Biot-TFFRLFNR-GG-GWGHFFKKAAHVGKL·NH2 | 2.8 |
CSP and NRC-4 are listed as the references for their original sequences. All the peptides were chemically synthesized and amidated at their C termini, except CSP. H/S indicates a His-to-Ser substitution (boldface) in the killing domain.
The minimal targeting domain of S. mutans CSP is underlined. -GG- or -GGG- indicates a linker sequence. Biot- indicates biotinylated peptides at the N terminus.
The MICs of the peptides are based on the results from triplicate experiments against S. mutans UA159. NA, not applicable.
Structural analysis of fusion peptides by NMR spectroscopy.
To understand structure-activity relationships, we determined the structures of three newly synthesized fusion peptides (IMB-1, -2, and -3) in dodecylphosphocholine (DPC) by nuclear magnetic resonance (NMR) spectroscopy. Sample preparation, NMR analysis, data calculation, and structural determination of the peptides were conducted by methods described previously (31, 32; see the supplemental material). In addition, we analyzed the effects of a single Ser-to-His substitution at position 14 of IMB-2 (IMB2-H/S14) at acidic pH on the structure of IMB2-H/S14 in DPC micelles.
MIC and MBC tests.
A modified broth dilution assay was used to determine the MICs of all the peptides against each bacterial strain based on methods described previously (22, 23, 25). Briefly, an aliquot of mid-log-phase cells (∼105 CFU/ml) was inoculated into 100 μl/well of THYE containing 2-fold serial dilutions of a peptide in a 96-well microtiter plate. Erythromycin (10 μg/ml) was used as a positive control, while THYE without addition of the peptide was used as a negative control. The cultures were incubated anaerobically at 37°C for 20 h. The optical density was read at 590 nm (OD590) with a microplate reader (Synergy HT; Biotek). To determine the minimal bactericidal concentration (MBC) of these peptides, samples were also taken from the exposed cultures, serially diluted, and spread on THYE plates for viable-cell counts.
Selective-killing assay.
To determine the killing kinetics and selectivity of the fusion peptides, we performed time-kill experiments against S. mutans strains, S. sanguinis SK108, and S. gordonii DL1 in both single- and dual-species cultures using methods described previously (19, 23, 26). Briefly, mid-log-phase cultures of these strains were first adjusted to the same cell density at OD590. The killing was initiated by adding an aliquot of cell suspension (∼105 CFU/ml) to 100 μl/well of THYE medium containing a killing concentration of a peptide in a 96-well microtiter plate. A one-to-one ratio of cell suspensions of two species was added in dual-species cultures. Aliquots of samples were taken at 1, 5, and 15 min for viable-cell counts, while samples without the peptide were used as controls. Viable-cell counts were obtained by growing cells either on THYE agar plates (single species) or on blood agar plates (dual species). S. mutans could be distinguished from S. sanguinis or S. gordonii by examining the colony morphology and hemolysis on blood agar plates. Survival percentages were determined as the number of surviving CFU per milliliter divided by total CFU per milliliter in control samples. The targeted killing of S. mutans cells by the peptides was also examined under a fluorescence microscope (Leica AF7000) with a Live/Dead BacLight bacterial-viability kit (Invitrogen) using the protocol recommended by the manufacturer.
Bactericidal activity against biofilms.
To examine the killing activity of the fusion peptide against biofilm, single-species S. mutans biofilm was initiated by inoculating an aliquot of mid-log-phase cells (∼106 CFU/ml) into 4×-diluted THYE plus 5 mM sucrose in a 96-well microtiter plate. The culture was incubated anaerobically at 37°C for 6 h. Planktonic cells were removed and replaced carefully with the same volume of the fresh medium containing increasing concentrations of a peptide. Biofilm samples were taken at 15 min by washing cells from the surfaces of wells with 10 mM phosphate buffer. The cell suspensions were serially diluted and spread on THYE agar plates to determine survival percentages as described above. A biotinylated peptide, IMB2-Biot, was used to examine peptide binding and distribution in biofilms. In this case, biofilms were developed on coverslips immersed in the medium in a 24-well microtiter plate. The culture medium was replaced with fresh medium containing the peptide and incubated for 5 min, and then a fluorescent dye, Alexa Fluor 546 conjugated to streptavidin (Invitrogen), was added. The cultures were incubated in the dark for an additional 5 min before the coverslips were removed for examination under a fluorescence microscope (Leica AF7000). Samples were also stained with 0.1% crystal violet and immediately examined under a phase-contrast microscope.
Bactericidal activity in the presence of NaCl, NaF, or acidic pH.
The killing assay in the presence of increasing concentrations of NaCl or a preventive dose of NaF or at acidic pH was performed using the same procedures described above but in THYE medium, which already contained 34 mM NaCl commercially, supplemented either with increasing concentrations of NaCl (25, 50, 100, 150, or 200 mM) or with 10 ppm NaF or in THYE medium adjusted to pH 6.0 or 5.5.
Assay for the stability of the fusion peptide in human saliva.
Unstimulated whole saliva was collected from five healthy volunteers and centrifuged at 5,000 × g at 4°C to remove food debris. The supernatants were passed through 0.2-μm filters, pooled, and stored at −20°C. We first examined the bactericidal activity of IMB-2 in the presence of saliva with or without 1 mM EDTA by transferring S. mutans log-phase cells into 1:4-diluted saliva-THYE medium. Viable-cell counts were performed to determine the percent survival using the same method as described above. The assay for peptide degradation in saliva was initiated by incubating aliquots (15 μg) of the peptide in 100 μl of saliva with or without 1 mM EDTA in a 37°C water bath. Aliquots of sample were taken at 0, 5, 10, and 15 min; boiled immediately; and analyzed with a 16% SDS-PAGE gel (Bio-Rad). A peptide sample without saliva was included as a control. The gels were then stained with Coomassie blue, and the peptide bands were quantitated by densitometry for subsequent calculation of percent degradation compared to the 0-min samples.
Hemolytic assay.
To assess the effects of the peptides on red blood cells (RBCs), blood from a healthy volunteer was collected in 5 ml EDTA-containing vacutainers. RBCs were isolated by gentle centrifugation, washed with 150 mM NaCl and then with 100 mM PBS, and diluted in PBS to ∼5 × 108 RBCs/ml. To initiate hemolysis, 100 μl of the cells was added to 100 μl of a 2-fold serially diluted peptide in PBS in a 96-well flat-bottom microtiter plate (4). Wells without peptide were used as negative controls, while wells containing 1% Triton X-100 were used as positive controls. The plate was incubated at 37°C for 1 h and centrifuged at 3,000 × g for 10 min. An aliquot (120 μl) of supernatant from each well was transferred to a new plate to read the absorbance at 541 nm using a microtiter plate reader. The percentage of hemolysis was calculated by the following equation: (A541 of the peptide-treated sample − A541 of buffer-treated sample)/(A541 of Triton X-100-treated sample − A541 of buffer-treated sample) × 100% (4).
Assay for activation of the ComDE transduction pathway by IMB-2.
The transcriptional activation of the comDE and nlmAB genes in response to a sublethal concentration (0.56 μM) of IMB-2 was performed using the lacZ transcriptional reporter strains, SmdC-PcomDE and SmdC-PnlmAB, and methods described previously (32). Briefly, an early mid-log-phase culture was dispensed into three tubes, the first containing IMB-2, the second (positive control) containing CSP (50 nM), and the third (negative control) containing water. Aliquots of samples were taken from the cultures at 0, 5, and 15 min to collect cell pellets by centrifugation at 4°C. The cells were permeabilized by glass beads after being resuspended in 1 ml of 50 mM Tris-HCl buffer (pH 7.5) containing 0.27% (vol/vol) β-mercaptoethanol, 50 μl chloroform, and 20 μl of 0.1% SDS. Aliquots of the supernatant were taken and incubated for 60 min with 80 μM O-nitrophenyl-d-galactopyranoside (ONPG). lacZ reporter (β-galactosidase [β-Gal]) activity was assayed by optical density readings at 420 nm, as described previously (31, 32). The data on β-Gal activity were normalized against negative controls, and specific β-Gal activities (A420 min/ml/mg protein) were calculated from triplicate assays.
Statistical analysis.
All killing assays were performed in triplicate in two independent experiments. The results were analyzed by Student's t test, and P values of ≤0.05 were considered statistically significant.
RESULTS
Structural analysis of the fusion peptides.
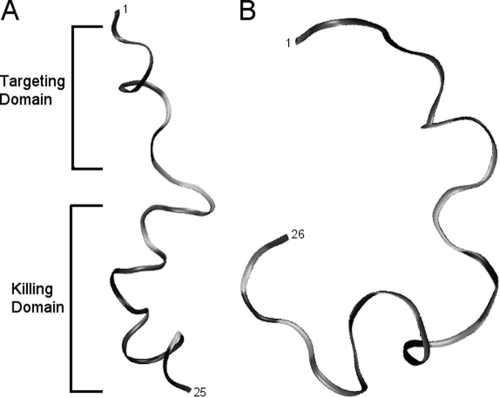
NMR spectroscopy showed that both IMB-1 and IMB-2 formed an α-helical structure within the CSP domain, a flexible unstructured linker, and an amphipathic α-helix for the NRC-4 N-terminal portion in DPC micelles (Fig. 1A). IMB-1 had a structure similar to that of IMB-2 with the exception of an unstructured C terminus (see Fig. S1 to S3 in the supplemental material). NMR diffusion measurements confirmed that these two peptides were strongly associated with DPC micelles. However, IMB-3 was relatively unstructured in the presence of DPC micelles and did not strongly associate with the micelles, based on diffusion measurements (Fig. 1B).
Fig. 1.
NMR-derived structures of IMB-2 (A) and IMB-3 (B) determined in DPC micelles (peptide/DPC ratio, 1:70 [wt/wt]). The lowest-energy structure of IMB-2 is an amphipathic peptide with α-helical character, whereas IMB-3 is a random coil.
Antimicrobial and bactericidal activities of the fusion peptides.
Both IMB-1 and IMB-2 showed good antimicrobial activity (MIC, 2 to 3 μM) against UA159 (Table 1) and other S. mutans strains (see Table S2 in the supplemental material), while IMB-3, which lacks the N-terminal portion of NRC-4, showed much lower activity (44 μM) against these strains. IMB-1 and IMB-2 had MBCs of 4.5 and 5.65 μM, respectively, which were only 2-fold higher than their MICs, and their killing activities were dose dependent. Interestingly, nearly 4-fold-higher concentrations were required to achieve the MBCs of these peptides against S. sanguinis SK108 and S. gordonii DL1, although their MICs were just slightly higher than those of the same peptides against S. mutans strains (see Table S2 in the supplemental material). Since IMB-2 was the shortest peptide effective against S. mutans strains, we selected this fusion peptide for most experiments unless otherwise specified.
IMB-2 selectively targets and kills S. mutans cells.
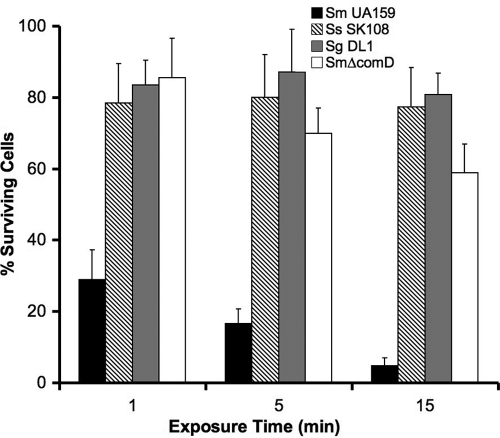
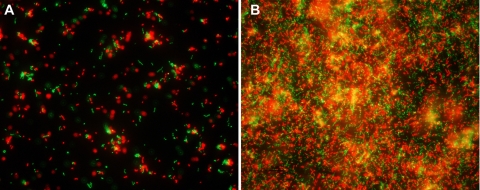
We next examined the killing kinetics and selectivity of IMB-2 against S. mutans strains and two closely related oral streptococci, S. sanguinis SK108 and S. gordonii DL1, in single- and dual-species planktonic cultures. As shown in Fig. 2, only 30% of S. mutans cells survived following exposure to IMB-2 (5.65 μM) for 1 min, and less than 5% of the cells survived following exposure to the peptide for 15 min. Similar results were observed when the viability of S. mutans was monitored with a Live/Dead BacLight bacterial-viability assay with fluorescence microscopy (Fig. 3). In contrast, less than 20% of S. sanguinis and S. gordonii cells were killed by exposure to the same concentration of IMB-2 for 15 min, indicating that IMB-2 selectively targeted S. mutans cells for killing. To explore the mechanism of the selectivity of the peptide against S. mutans, we examined the killing activity of IMB-2 against an S. mutans comD mutant (SmΔcomD) that was defective in the CSP receptor. Interestingly, we found that nearly 60% of the mutant cells retained their viability even after exposure to the peptide for 15 min (Fig. 2), suggesting that IMB-2 bound predominantly to the CSP receptor to trigger killing in the wild-type strains of S. mutans. To determine whether binding of IMB-2 to the CSP receptor activated the ComDE signal transduction pathway, we examined the transcriptional activation of comDE and nlmAB promoters in response to a sublethal concentration (0.56 μM) of the peptide by using two lacZ transcriptional reporter strains, SmdC-PcomED and SmdC-PnlmAB (31, 32). The results showed little detectable lacZ reporter activity of these genes (data not shown), suggesting that the fusion peptide did not activate the ComDE signal transduction pathway or its regulated genes.
Fig. 2.
Selective killing activity of IMB-2 (5.65 μM) against planktonic cells of S. mutans UA159 (Sm UA159), S. sanguinis SK108 (Ss SK108), S. gordonii DL1 (Sg DL1), and an S. mutans UA159 comD mutant (SmΔcomD) defective in the CSP receptor (ComD protein). The error bars indicate standard deviations.
Fig. 3.
Fluorescence microscopy of the targeted killing of S. mutans planktonic cells (A) and biofilm cells (B) following exposure to IMB-2 (5.65 μM) for 5 min. The cells are labeled with Live/Dead BacLight bacterial-viability dye, in which viable cells are green and dead cells are red.
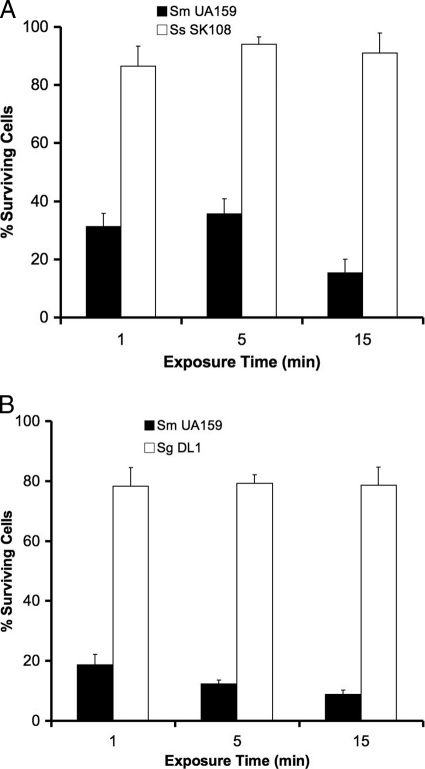
To further confirm the selectivity of the fusion peptide, we examined the bactericidal activity of IMB-2 (5.65 μM) against S. mutans cells in dual-species planktonic cultures, which contained a one-to-one ratio of mid-log-phase cells of S. mutans UA159 with either S. sanguinis SK108 or S. gordonii DL1. In the mixed culture of S. mutans and S. sanguinis, 65 to 85% of S. mutans cells were killed by exposure of up to 15 min, but only 10% of S. sanguinis cells were killed (Fig. 4A). Similarly, IMB-2 in the mixed culture of S. mutans and S. gordonii killed 80 to 90% of S. mutans cells but killed only 20% of S. gordonii cells during the same exposure (Fig. 4B). These results confirmed that IMB-2 selectively targeted S. mutans for killing in dual-species mixed cultures.
Fig. 4.
Selective killing activity of IMB-2 (5.65 μM) against S. mutans in dual-species cultures of S. mutans UA159 with S. sanguinis SK108 (A) or S. gordonii DL1 (B). S. mutans was distinguished from S. sanguinis and S. gordonii by colony morphology and hemolysis by bacterial cells grown on blood agar plates. The error bars indicate standard deviations.
IMB-2 is also active against S. mutans biofilms.
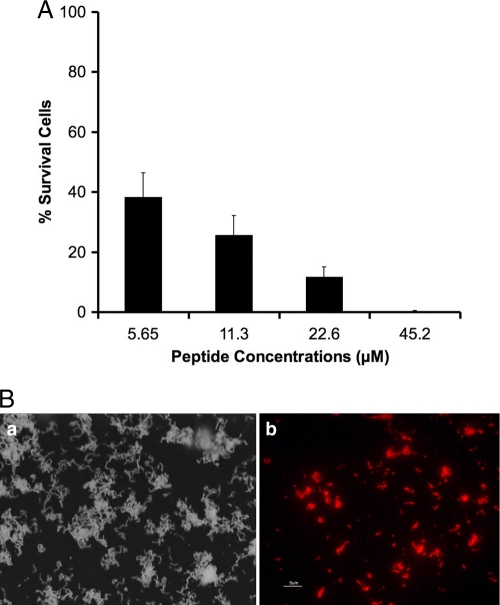
The killing assay of S. mutans biofilms showed that approximately 60% of biofilm cells were killed following a 15-min exposure to IMB-2 at the same concentration (5.65 μM) as that in planktonic culture (Fig. 5A). At higher concentrations (22.6 μM), nearly 90% of biofilm cells were killed, and at 45.2 μM, all the biofilm cells were completely killed. Consistent with these results, fluorescence microscopy also showed that biotin-labeled peptide, IMB2-Biot, clearly bound to the biofilm (Fig. 5B).
Fig. 5.
(A) Killing activities of different concentrations of IMB-2 against S. mutans biofilms (the error bars indicate standard deviations). (B) Microscopic images of treated cells. (a) Example of the same biofilm under a phase-contrast microscope. (b) Binding of a biotin-labeled peptide, IMB2-Biot, to the biofilm under a fluorescence microscope.
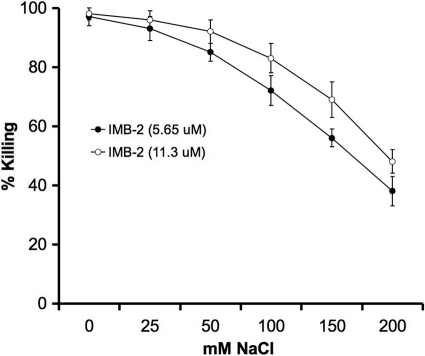
Bactericidal activities in different salt concentrations.
To determine the effects of increasing concentrations of salt on the fusion peptide, we examined the killing activity of IMB-2 against S. mutans grown in THYE medium with different concentrations of NaCl. At salt concentrations ranging from 59 to 134 mM, IMB-2 retained 70 to 90% of the killing activity at a concentration of 5.65 μM and 85 to 95% of the activity at a concentration of 11.2 μM (Fig. 6). The results confirmed that the fusion peptide retained killing activity against S. mutans under physiological salt conditions (45 to 80 mM) found in human saliva (29). However, further increases in salt concentrations to 184 to 234 mM resulted in decreased killing activity.
Fig. 6.
Killing activity of IMB-2 against S. mutans UA159 at 5.65 and 11.3 μM in the presence of different concentrations of NaCl, which is added to THYE medium that already contained 34 mM NaCl. The error bars indicate standard deviations.
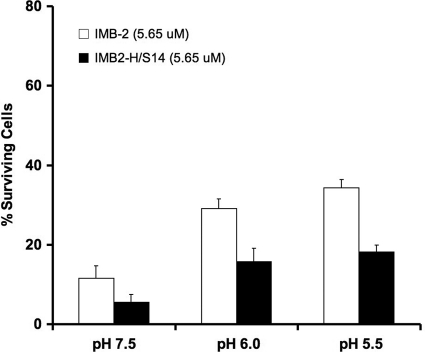
Bactericidal activity at acidic pH.
To determine whether the fusion peptide retained its activity at lower pH, a condition that was frequently encountered in dental biofilms (20, 27), we examined the killing activity of IMB-2 against S. mutans grown at different pHs. At pH 6.0, approximately 70% of S. mutans cells were killed by IMB-2 (5.65 μM), and at pH 5.5, about 65% of the cells were killed, indicating that the killing activity of the peptide was reduced by 20 to 25% compared to that at pH 7.5 (Fig. 7). Interestingly, we found that a single Ser-to-His substitution in the peptide, IMB2-H/S14, improved the activity of the peptide 2-fold at all three pHs (Fig. 7). To further explore the effect of this substitution on the peptide, we examined the NMR spectra of IMB2-H/S14 in DPC micelles at pH 5.0, 6.6, and 7.0. At all pHs, the proton resonances of the peptide were similar to that of IMB-2 (see Fig. S4 in the supplemental material), suggesting that IMB2-H/S14 has a structure similar to that of IMB-2 at the lower pHs. This was consistent with the improved killing activity of the peptide at acidic pH.
Fig. 7.
Percent survival of S. mutans UA159 treated with IMB-2 (5.65 μM) and IMB2-H/S14 (5.65 μM) in THYE broth at different pHs. The percentages of surviving cells following exposure to the peptides for 15 min are shown. The error bars indicate standard deviations.
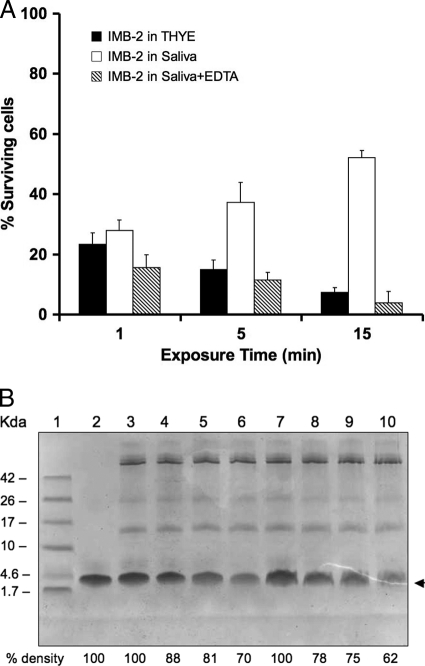
Bactericidal activity in the presence of saliva.
The killing assay of IMB-2 in the presence of saliva revealed little change in the activity of the peptide during a 1-min exposure to the saliva (Fig. 8A), although this was decreased by 20% after 5 min and by approximate 50% after 15 min. Addition of 1 mM EDTA to the culture effectively prevented the saliva-induced loss of the killing activity, although 1 mM EDTA alone did not significantly affect the growth of S. mutans (data not shown). Incubating an aliquot of IMB-2 in saliva followed by SDS-PAGE analysis revealed progressive degradation of the peptide over 15 min (Fig. 8B, lanes 7 to 10). Consistent with the results of the killing assay, addition of 1 mM EDTA to the saliva reduced the degradation of the peptide (lanes 3 to 6). Increasing the concentration of EDTA (5 mM) provided further protection against degradation (data not shown).
Fig. 8.
Killing activity and stability of IMB-2 (5.65 μM) in the presence of saliva. (A) Survival of cells in saliva exposed to IMB-2 with or without addition of 1 mM EDTA compared to no saliva. (B) SDS-PAGE analysis of peptide degradation in the presence of saliva with or without addition of 1 mM EDTA. The peptide bands were quantitated by densitometry to calculate percent degradation compared to the 0-min samples. Lane 1, low-molecular-weight markers; lane 2, IMB-2 only (∼4.5 kDa) as a blank control; lanes 3 to 6, IMB-2 exposed to saliva with 1 mM EDTA for 0, 5, 10, and 15 min; lanes 7 to 10, IMB-2 exposed to saliva without EDTA for 0, 5, 10, and 15 min. The arrowhead indicates IMB-2.
Synergistic killing of IMB-2 with 10 ppm NaF.
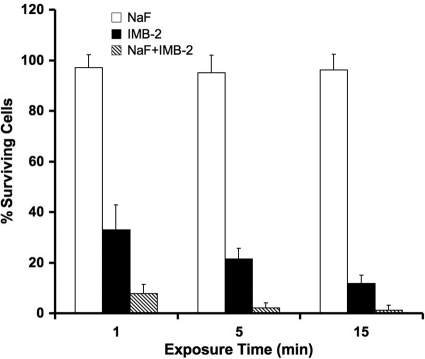
The killing assay of IMB-2 in the presence of NaF revealed that exposure of S. mutans UA159 to IMB-2 (5.65 μM) and NaF (10 ppm) resulted in synergistic killing of S. mutans, although NaF alone at 10 ppm did not affect the growth of S. mutans (Fig. 9).
Fig. 9.
Killing activity of IMB-2 (5.65 μM) against S. mutans in the presence of 10 ppm NaF. This concentration of NaF alone does not significantly affect the survival of S. mutans cells. The error bars indicate standard deviations.
The fusion peptide shows little cytotoxicity.
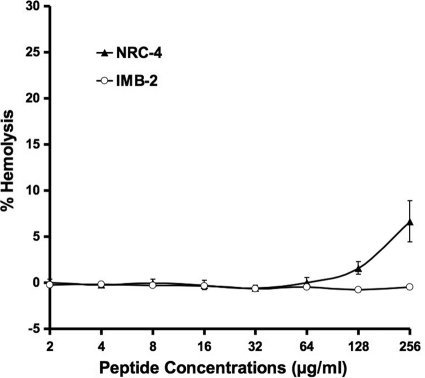
To assess the safety of the fusion peptide toward eukaryotic cells, we examined the hemolytic activity of IMB-2 and NRC-4 against human red blood cells. As shown in Fig. 10, IMB-2 at concentrations as high as 256 μg/ml (90.5 μM) caused little detectable hemolysis, although its parent peptide, NRC-4, showed a very low level (7%) of hemolysis at this concentration.
Fig. 10.
Hemolysis caused by peptide IMB-2 and its parent peptide, NRC-4. The percentages of hemolysis were calculated by the following equation: (A541 of the peptide-treated sample − A541 of buffer-treated sample)/(A541 of the Triton X-100-treated sample − A541 of the buffer-treated sample) × 100%.
DISCUSSION
A major challenge in using antimicrobial agents or antibiotics to treat an infectious disease is that these antimicrobial agents may indiscriminately kill the resident organisms, resulting in ecological disruption of the natural flora and other negative clinical consequences (21). To circumvent this problem, an innovative approach has recently been developed to design a new class of antimicrobials, called “specifically targeted antimicrobial peptides” (STAMPs), which selectively target a specific organism with little effect on the other members of the resident flora (7, 10, 26). The design of such target-specific AMPs requires the identification of two functionally independent peptide components, a killing moiety comprised of a nonspecific AMP that can rapidly kill bacterial cells and a targeting moiety consisting of an organism-specific, high-affinity binding peptide (7, 26). The two moieties are then joined via a small linker to generate a fusion AMP without detrimental changes in their independent activities. The major advantage of such an AMP is that the targeting moiety can guide the fusion peptide to bind selectively to the target organism, allowing peptide-guided killing (7, 26). It is also desirable that such a fusion peptide should be relatively stable and retain its bactericidal activity under a range of physiological conditions, such as different salt concentrations, ionic strengths, pHs, and other factors in body fluids (8, 15, 34). By studying marine-derived pleurocidin variants (25, 30) and an S. mutans quorum-sensing signaling peptide pheromone or CSP (17, 18, 31) we have determined the structure-activity relationships and mechanisms of action of these two types of biologically active peptides. By taking advantage of our studies and those of others (7, 10, 26), we have designed several fusion peptides that consist of a short targeting domain of S. mutans CSP fused to a killing domain from a selected pleurocidin, NRC-4, via a small linker (7). In addition, the fusion peptides are amidated at their C termini to eliminate the end charge, since peptides amidated at their C termini frequently show better antimicrobial activities than their unamidated parent peptides (23, 25). Our work demonstrates that all the fusion peptides, with the exception of IMB-3, show strong bactericidal activity against S. mutans strains. In particular, the fusion peptide IMB-2 selectively targets S. mutans in the presence of closely related species found in dental biofilms under the same conditions. Structural analysis using NMR spectroscopy also confirms that the active targeted peptide forms an amphipathic, α-helical character, which appears to favor its movement in liquid and interaction with the receptor in the cell membrane, whereas the inactive peptide (IMB-3) adopts a relatively unstructured, random-coil form. It is predicted that IMB-1 and IMB-2 behave in a manner similar to that of NRC-4 in its active state, based on the amphipathic α-helical character of the NRC-4 N-terminal portion of these fusion peptides (30).
The evidence from this study suggests that the binding of IMB-2 to S. mutans cells is species specific, since enhanced killing activity against S. mutans cells was clearly observed in both monocultures and mixed cultures consisting of either of two closely related species, S. sanguinis and S. gordonii. Deletion of the comD gene that encodes the CSP receptor results in significant reduction in the killing of S. mutans, suggesting that the targeting domain of CSP is sufficient to guide the fusion peptide to bind selectively to its receptor on S. mutans cells. Our result is consistent with the findings of Eckert et al. (7), who showed that the targeted killing of S. mutans by a STAMP peptide, C16G2, was also species specific but involved CSP receptor-independent binding. The reason for this divergence is unclear, but one possibility is that the bulky carboxyfluorescein-labeled C16G2 used by the researchers may favor the binding of the peptide to components other than the CSP receptor. Alternatively, the difference may be due to the different bacterial strains used in the two studies, since S. mutans UA140, used by Eckert et al. (7) in the binding assay, was reported to differ somewhat from UA159 in terms of variations in genomic sequences and some phenotypes (1, 36). Therefore, one cannot completely rule out the possibility that there might be a structural variation in the CSP receptor between the two strains. However, our work does not exclude the possibility that the IMB-2 fusion peptide may involve an additional, unknown binding site on the surfaces of S. mutans cells, since IMB-2 also killed 20 to 40% of the ComD mutant cells following the same exposure. We speculate that, because of its amphipathic nature, IMB-2 may interact mainly with the CSP receptor and to a lesser extent with the membranes themselves. The fact that IMB-2 also kills about 20% of S. sangunis or S. gordonii cells appears to support this hypothesis. No matter which binding mechanism is involved, the targeted peptides tested both in our study and in the study by Eckert et al. (7) showed significantly enhanced killing of S. mutans cells but did not activate the signal transduction pathway or its regulated genes, such as comDE and nlmAB (18, 31, 32). This may be because all the fusion peptides lack the C-terminal structural motif of CSP, which is known to activate the signal transduction pathway (31) and which was replaced in our study by the killing domain of the AMP NRC-4.
It is well known that some AMPs are sensitive to the physiological salt concentration or the ionic strength, pH, and proteolytic activity in body fluids (12, 15). Thus, the ability to resist physiological salts, pH fluctuation, or proteolytic degradation is crucial for AMPs to function in body fluids, such as saliva (8, 12). Human whole saliva is a mixed fluid that contains 99% water and 1% organic and inorganic molecules with an average pH 6.7 (29). Total electrolytes in saliva largely account for ionic strength or salt concentrations (45 to 80 mM), which are normally lower than those in serum and vary extremely with saliva flow and health status (29). In addition, whole saliva contains a number of components, such as antimicrobial compounds, proteins, and glycoproteins from epithelial cells, host defense cells, and bacteria, which account for most of its proteolytic activity (29, 34). The evidence from our study indicates that IMB-2 is capable of maintaining killing activity in physiological or even higher concentrations of salt, particularly for the short time (15 min) that is sufficient for the peptide to exhibit full killing activity. This result is not surprising, because IMB-2 has a structure, an amphipathic α-helix, similar to that of its parent NRC-4, a pleurocidin variant that is active in high-salt environments (5, 25). Previous studies showed that the pleurocidin isolated from the winter flounder, which migrates from its habitat of brackish water (0.8% salinity) to seawater (3.3% salinity), was naturally resistant to high concentrations of salt (5, 25). Importantly, the pleurocidin did not depend on high salt concentrations for killing activity (5, 28). The evidence from our study suggests that IMB-2 retains a level of salinity resistance similar to that of its ancestor peptide.
In dental biofilms, S. mutans can produce acids during ingestion of dietary fermentable carbohydrates and lower the environmental pH (20, 27), which is a key factor for the organism to initiate dental caries (1, 2, 13). By evaluating the killing activity of the fusion peptide at low pH, we found that IMB-2 could kill 70 to 75% of S. mutans cells at pH 5.5 to 6.0 during a 15-min exposure, which was about 20 to 25% lower than that at pH 7.5. This suggests that acidic pH affects the activity of the fusion peptide to some degree, although the remaining activity at the lower pH is still considered to be good. It was previously reported that clavanin, a histidine-rich α-helical AMP isolated from leukocytes of a tunicate (Styela clava), exerted substantial antimicrobial activity at pH 5.5 (15). Histidine residues in clavanin were suggested to be responsible for the resistance to low pH. However, the antimicrobial activity of clavanin was pH dependent, and the activity appeared to be insignificant at neutral pH (15). Li et al. also constructed a histidine-rich AMP, which showed resistance to low pH but also needed acidic pH for activity (16). To explore this characteristic of histidine in the activity of our AMPs, we designed a new peptide, IMB2-H/S14, replacing serine-14 with a histidine residue. We found that this Ser-to-His substitution improved the killing activity of the peptide at the lower pHs by 10 to 20%. Interestingly, IMB-2H/S14 did not depend on low pH for its activity, since the killing activity of the peptide was also improved at pH 7.5. The NMR spectra of IMB2-H/S14 in DPC micelles confirmed that the peptide retained a similar structure at lower pH. These results support the notion that histidine plays an important role in maintaining the activity and stability of an AMP at acidic pH. This also suggests that IMB-2H/S14 might be more desirable for use under conditions with frequent pH fluctuations.
Many AMPs are sensitive to degradation by proteases existing in body fluids, such as serum, saliva, or gastrointestinal fluid, resulting in a decrease or loss of activity (12, 34). It is also known that chelating agents, such as EDTA, or protease inhibitors can prevent the degradation of AMPs (34). In this study, we assessed the activity and stability of the fusion peptide in the presence of human saliva. We found that IMB-2 was capable of retaining its killing activity for a short time (5 to 10 min) in the presence of saliva, but longer exposure (15 min or longer) resulted in the loss of about 50% of the activity. Since a 1-min exposure was sufficient to attain 80% of the killing, the bactericidal activity of IMB-2 in the presence of saliva is still considered to be reasonably good. In particular, using 1 mM EDTA can effectively reduce the degradation and improve the killing activity. The higher concentration (5 mM) of EDTA confers even better protection against degradation, suggesting that it should be an advantage to use the fusion peptide combined with EDTA in the oral cavity or in the presence of potential proteases. It has been reported that proteases recognize only l-form amino acids of peptides from natural sources and that peptide degradation can be decreased or prevented by replacement of l-form amino acids with their d-form amino acids (12, 34). Thus, it will be interesting to construct and assess d-form fusion peptides for killing activity against S. mutans in the presence of saliva.
Fluoride has been widely used as an anticaries agent for years and is considered safe at a concentration of 1 to 2 ppm in drinking water (9, 20). Clinically, up to 1,000-fold-higher concentrations of fluoride may be used as a topical procedure during dental cleaning or treatments (9, 20, 21). Sodium fluoride (NaF) is the most commonly used form of fluoride (20). An interesting finding from this study was that the fusion peptide, together with use of a preventive dose (10 ppm) of NaF, resulted in a synergistic killing effect on S. mutans cells, although this concentration of NaF alone did not significantly affect the growth of S. mutans (20). The mechanism behind this synergy is unclear; however, fluoride is known to repress enzymatic activities in carbohydrate metabolism, resulting in inhibition of glycolysis, especially at higher concentrations or at lower pH (9, 20). Because of its weak-acid property (pKa = 3.15), fluoride also acts as a transmembrane proton conductor that increases the proton permeability of bacterial cells, resulting in an increased demand for ATP to move protons out of the cells (20). It is possible that the killing activity of the fusion peptide is enhanced in the presence of fluoride due to the increased burden of the bacterial cells in dealing with antibiotic stress, proton movement, and energy demand through the above mechanisms.
In summary, we have successfully designed and tested a novel target-specific AMP, IMB-2, which selectively targets and kills S. mutans. Our work confirms that the peptide retains good killing activity in the presence of physiological or higher salt concentrations. Furthermore, it shows a synergistic killing effect on S. mutans with a preventive dose of NaF. We have also found that replacement of serine 14 by histidine can improve the stability of the peptide (IMB2-H/S14) at acidic pH. In addition, IMB-2 is relatively stable and retains the killing activity in the presence of saliva with a low concentration of EDTA. Because of its effectiveness, salt resistance, and minimal toxicity toward host cells, this novel target-specific peptide shows good potential for further study and development as an anticaries agent. We suggest that such an agent could be an additional strategy for topical use to prevent caries in high-risk populations, such as young children who have “nursing” or rampant caries, caries in chemotherapy or diabetes patients, and individuals whose immune system is compromised or who have dry mouth.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NSERC discovery grant RGPIN-311682-07 to Y.-H.L. J.M. was the recipient of a Ph.D. scholarship from the China Scholarship Council (CSC). Y.-H.L. was the recipient of a Nova Scotia-CIHR Regional Partnership Program New Investigator Award.
The NMR data were collected at the Biomolecular Magnetic Resonance Facility (BMRF) housed within NRC-IMB. The He-cooled probe for the 700-MHz spectrometer at the NRC-IMB was provided by Dalhousie University through an ACOA Grant. We thank Tong-Jun Lin in the Department of Microbiology and Immunology at Dalhousie University for access to the fluorescence microscope.
This is NRC publication 2011-54057.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
Published ahead of print on 15 August 2011.
REFERENCES
- 1. Ajdić D., et al. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bowen W. H. 2002. Do we need to be concerned about dental caries in the coming millennium? Crit. Rev. Oral Biol. Med. 13:126–131 [DOI] [PubMed] [Google Scholar]
- 3. Brogden K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238–250 [DOI] [PubMed] [Google Scholar]
- 4. Bulmus V., et al. 2003. A new pH-response and glutathione-reactive, endosomal membrane-disruptive polymeric carrier for intracellular delivery of biomolecular drugs. J. Control Release 93:105–120 [DOI] [PubMed] [Google Scholar]
- 5. Cole A. M., Weis P., Diamond G. 1997. Isolation and characterization of pleuricidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 272:12008–12013 [DOI] [PubMed] [Google Scholar]
- 6. Douglas S. E., Gallant J. W., Gong Z., Hew C. 2001. Cloning and developmental expression of a family of pleurocidin-like antimicrobial peptides from winter flounder, Pleuronectes americanus. Dev. Comp. Immunol. 25:137–147 [DOI] [PubMed] [Google Scholar]
- 7. Eckert R., et al. 2006. Targeted killing of Streptococcus mutans by a pheromone-guided “smart” antimicrobial peptide. Antimicrob. Agents Chemother. 50:3651–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedrich C., Scott M. G., Karunaratne N., Yan H., Hancock R. E. W. 1999. Salt-resistant alpha-helical cationic antimicrobial peptides. Antimicrob. Agents Chemother. 43:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffin S. O., Regnier E. P. M., Huntley V. 2007. Effectiveness of fluoride in preventing caries in adults. J. Dent. Res. 86:410–415 [DOI] [PubMed] [Google Scholar]
- 10. He J., et al. 2007. Novel synthetic antimicrobial peptides against Streptococcus mutans. Antimicrob. Agents Chemother. 51:1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Health Canada 2009. Canadian task force on the periodic health examination. Canadian guide to clinical preventive health care; Ottawa, Canada: http://www.fptdwg.ca/assets/PDF/CHMS/CHMS-E-summ.pdf [Google Scholar]
- 12. Jenssen H., Hamill P., Hancock R. E. W. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuramitsu H. K. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historic perspective. Crit. Rev. Oral Biol. Med. 14:331–344 [DOI] [PubMed] [Google Scholar]
- 14. Kuramitsu H. K., He X., Lux R., Anderson M. H., Shi W. 2007. Interspecies interactions within oral microbial communities. Microbiol. Mol. Biol. Rev. 71:653–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee I. H., Cho Y., Lehrer R. I. 1997. Effects of pH and salinity on the antimicrobial properties of clavanins. Infect. Immun. 65:2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li L., et al. 2010. Design and characterization of an acid-activated antimicrobial peptide. Chem. Biol. Drug Des. 75:127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y. H., Lau P. C. Y., Lee J. H., Ellen R. P., Cvitkovitch D. G. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y. H., et al. 2001. A quorum sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 183:6875–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marquis R. E., Clock S. A., Mota-Meira M. 2003. Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev. 26:493–510 [DOI] [PubMed] [Google Scholar]
- 20. Marsh P. D., Bradshaw D. J. 1997. Physiological approaches to the control of oral biofilms. Adv. Dent. Res. 11:176–185 [DOI] [PubMed] [Google Scholar]
- 21. Mitchell T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1:219–230 [DOI] [PubMed] [Google Scholar]
- 22. Ouyang J., Tian X. L., Versey J., Wishart A., Li Y. H. 2010. The BceABRS four-component system regulates bacitracin-induced cell envelope stress response in Streptococcus mutans. Antimicrob. Agents Chemother. 54:3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Patrzykat A., Friedrich C. L., Zhang L., Mendoza V., Hancock R. E. W. 2002. Sublethal concentrations of pleurocidin-derived antimicrobial peptides inhibit macromolecular synthesis in Escherichia coli. Antimicrob. Agents Chemother. 46:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patrzykat A., Douglas S. E. 2003. Gone gene fishing: how to catch novel marine antimicrobials. Trends Biotechnol. 21:362–369 [DOI] [PubMed] [Google Scholar]
- 25. Patrzykat A., Gallant J. W., Seo J.-K., Pytyck J., Douglas S. E. 2003. Novel antimicrobial peptides derived from flatfish genes. Antimicrob. Agents Chemother. 47:2464–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qiu X. Q., et al. 2003. An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat. Biotechnol. 21:1480–1485 [DOI] [PubMed] [Google Scholar]
- 27. Quivey R. Q., Jr., Kuhnert W. L., Hahn K. 2001. Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12:301–314 [DOI] [PubMed] [Google Scholar]
- 28. Saint N., Cadiou H., Bessin Y., Molle G. 2002. Antimicrobial peptide pleurocidin forms ion channels in planar lipid bilayers. Biochim. Biophys. Acta 1564:359–364 [DOI] [PubMed] [Google Scholar]
- 29. Syeebny L. M., et al. 1992. Saliva: its role in health and disease. Int. Dent. J. 42(Suppl. 2):291–304 [PubMed] [Google Scholar]
- 30. Syvitski R. T., Burton I., Mattatall N. R., Douglas S. E., Jakeman D. L. 2005. Structural characterization of the antimicrobial peptide pleurocidin from winter flounder. Biochemistry 44:7282–7293 [DOI] [PubMed] [Google Scholar]
- 31. Syvitski R. T., et al. 2007. Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J. Bacteriol. 189:1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tian X. L., et al. 2009. A method for structure-activity analysis of quorum-sensing signaling peptides from naturally transformable streptococci. Biol. Proced. Online 11:207–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tincu J. A., Taylor S. W. 2004. Antimicrobial peptides from marine invertebrates. Antimicrob. Agents Chemother. 48:3645–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei G. X., Bobek L. A. 2005. Human salivary mucin MUC7 12-mer-L and 12-mer-D peptides: antifungal activity in saliva, enhancement of activity with protease inhibitor cocktail or EDTA, and cytotoxicity to human cells. Antimicrob. Agents Chemother. 49:2336–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wiesner J., Vilcinskas A. 2010. Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464 [DOI] [PubMed] [Google Scholar]
- 36. Wu C., et al. 2010. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl. Environ. Microbiol. 76:5815–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.