Abstract
HIV-1 infections lead to a progressive depletion of CD4 cells culminating in AIDS. The coreceptor usage by HIV varies from CCR5 (R5) tropic early in infection to CXCR4 (X4) tropic in later infections. Although the coreceptor switch from R5 to X4 tropic HIV is well associated with progression to AIDS, the role of CCR5 in disease progression especially in patients infected exclusively with R5 isolates throughout the disease remains enigmatic. To better understand the role of CCR5 and R5 tropic HIV envelope in AIDS pathogenesis, we asked whether the levels of CCR5 and/or HIV Env-mediated fusion determine apoptosis of bystander cells. We generated CD4+ T cell lines expressing varying levels of CCR5 on the cell surface to show that CCR5 expression levels correlate with bystander apoptosis induction. The mechanism of apoptosis involved caspase-3 activation and mitochondrial depolarization and was dependent on gp41 fusion activity as confirmed by fusion-restricted gp41 point mutants and use of the fusion inhibitor T20. Interestingly, lower levels of CCR5 were able to support virus replication in the absence of bystander apoptosis. Our findings suggest that R5 HIV-1-mediated bystander apoptosis is dependent on both CCR5 expression levels as well as fusogenic activity of the Env glycoprotein.
Keywords: AIDS, Chemokines, Pathogenesis, Virology, Virus Entry
Introduction
For viral entry, HIV-1 utilizes CD4 as receptor and one of the chemokine receptors as coreceptors, the most common ones being CXCR4 (X4) and CCR5 (R5) (1–3). It is well known that R5 tropic viruses predominate during the initial establishment of infection as they are transmitted with greater efficiency (4–6), whereas the X4 viruses emerge later during the disease and are associated with rapid progression to AIDS (7, 8). The progressive increase in virulence in vivo during the late stages of disease is attributed in many cases to a coreceptor switch from CCR5 tropic to CXCR4 tropic HIV (9–11). Although in approximately half of the HIV infections the virus switches coreceptor usage to CXCR4, and there are several hypotheses to explain this phenomenon (12–15), the mechanism by which R5 tropic HIV isolates lead to AIDS remains poorly defined especially in patients that remain solely infected with R5 isolates throughout the disease.
Although HIV selectively infects CD4+ cells, the relatively few infected cells in vivo do not account for the extensive depletion of CD4 cells. This has led to the idea that the virus is able to kill uninfected bystander CD4+ cells (16) via proteins like Env2 (17), Nef (18, 19), Tat (20, 21), and Vpr (22, 23). There is growing interest in the role of the Envelope (Env) glycoprotein in bystander apoptosis due to the fact that it is expressed on the surface of infected cells and interacts with CD4 and a coreceptor (CXCR4/CCR5), thereby initiating apoptotic signaling in uninfected bystander T cells. However, in vitro studies show that although the binding of the Env gp120 subunit to CD4 and coreceptor are required for apoptosis induction, these interactions are not sufficient (24, 25). In support of this observation, we and others showed that the fusion process mediated by the gp41 subunit of HIV envelope may be critical in bystander killing (26–28). Furthermore, through mutagenesis studies, we demonstrated that hemifusion (incomplete fusion accompanied by partial mixing of apposing cell membranes) induced by gp41 subunit is the major mechanism for HIV Env-mediated bystander T cell apoptosis (29).
The fusion of biological membranes mediated by HIV Env, specifically in the case of R5 tropic viruses, is dependent on the expression level of CCR5 coreceptor on cells. Surface CCR5 expression levels in turn are dependent on the CCR5 gene (30) and promoter polymorphism (31). Platt et al. (32) demonstrated that different levels of CCR5 expression in HeLa cells can affect fusion mediated by different R5 isolates. The importance of CCR5 expression levels is further emphasized by its effects (within physiological limits) on Env-mediated fusion and virus replication (32, 33). However, whether increased surface expression of CCR5 accounts for higher Env-mediated fusion and apoptosis in certain R5 virus-infected patients remains to be determined. Previous studies observed that CCR5Δ32 heterozygous (CCR5Δ32+/−) individuals, while susceptible to HIV infection, show a slower progression to AIDS (34–36), possibly because of reduced surface expression of CCR5 (30). On the contrary, CCR5Δ32 homozygous populations resist HIV infection with R5 tropic viruses (37). Similarly, SCID-hu mice reconstituted with CCR5Δ32+/− thymus grafts were resistant to CCR5 virus-mediated CD4 cell loss even in the presence of virus replication (38). These studies demonstrate the importance and complexity of CCR5 expression levels on HIV pathogenesis.
In this study we asked what was the role of CCR5 cell surface expression as well as Env fusion activity on bystander apoptosis induction by HIV-1 YU-2 Env, a CCR5 isolate. We engineered SupT1 cells to express either low, medium, or high levels of CCR5 on the surface or used R5 Env mutants with different fusogenic activities to address these questions. We show here that R5 Env-mediated bystander apoptosis is a function of both CCR5 expression levels and Env fusogenic activity. However, this bystander apoptosis induction is independent of virus replication, supporting the idea that individuals with lower levels of CCR5, as in the case of CCR5Δ32+/−, may be able to support virus replication without bystander apoptosis and consequently slower progression to AIDS. To the best of our knowledge, this is the first study showing that CCR5 cell surface levels determine susceptibility to bystander apoptosis by HIV-1 Env independent of virus replication.
MATERIALS AND METHODS
Cells and Reagents
SupT1 cells were maintained in RPMI 1640 media supplemented with 10% FBS and penicillin/streptomycin (5000 units/ml). HeLa and TZM cells (National Institutes of Health AIDS Research and Reference Reagent Program) were maintained in Dulbecco's modified Eagles medium supplemented with 10% FBS and penicillin/streptomycin (5000 units/ml). Fusion inhibitor T20 (Enfuvirtide) was provided by National Institutes of Health AIDS Research and Reference Reagent Program. Pan caspase inhibitor Z-VAD-fmk and mitochondrial potential sensor dye DiOC6 were obtained from Calbiochem.
Plasmid Constructs
Molecular clones of HIV-1 YU-2 and NL4-3 and Env clone QH0692.42 (39) were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. EcoRI-XhoI fragments from the YU-2 molecular clone were cloned into pcDNA 3.1 expression vector (Invitrogen) making the wild type (WT) YU-2 Env construct. The fragment contains open reading frames of env, tat, and rev genes. Point mutations were introduced into the WT Env construct using QuikChange site-directed mutagenesis kit (Stratagene). The mutants were numbered based on Env sequence of HXB-2 reference strain. All mutant and WT YU-2 Env regions were also introduced into the NL4-3 molecular clone using the EcoRI-XhoI fragments to generate infectious molecular clones containing YU-2 env and henceforth referred to as NL-YU-2.
Expression of Env Glycoprotein
HeLa cells were transfected in 6-well plates with the HIV-1 Env vectors using ExGen 500 (Fermentas Life Science) transfection reagent as per the manufacturer's instructions. 24 h post-transfection, the cells were collected and used for either cell to cell fusion assay or bystander apoptosis induction in coculture experiments with target cells.
Generation of CCR5-expressing Cell Lines
For the generation of cell lines expressing different levels of CCR5, first a lentiviral vector was generated. CCR5 gene was amplified from plasmid pSFF-CCR5 a kind gift from Dr. David Kabat (32). The PCR-amplified region was cloned into pLENTi 6.3 vector (Invitrogen) using a TOPO cloning approach to generate pLenti-CCR5. The insert was sequenced for confirmation. Lentiviral particles were generated in 293T cells by transfecting with pLenti-CCR5 in combination with the helper construct and VSVG as per the manufacturer's instructions (Invitrogen). The lentivirus particles were used for transduction of SupT1 cells followed by two-step flow cytometric sorting using FACS ARIA II system (BD Biosciences). Individual clones were selected and expanded in blasticidin media and characterized for CD4 as well as CCR5 expression. The resulting CCR5-expressing cell lines referred to as SupT-R5 clones, H6 (high CCR5), M10 (medium CCR5), and L23 (low CCR5) were selected for further analysis and used in all subsequent experiments.
Apoptosis Induction
For induction of apoptosis, coculture between Env-transfected HeLa cells and SupT-R5 cell lines was performed. HeLa cells transfected with either WT or mutant Env constructs were seeded in 24-well plates at 105 cells/well. The cells were allowed to adhere for 4–6 h. Subsequently, the media was removed, and SupT-R5 cells at 0.5–1 × 106 cells were added. Different inhibitors were added at the time of coculture. The cells were cocultured for 24 h following which the suspension cells were collected, stained with annexin V (BD Biosciences), and analyzed by flow cytometry using a FACS CANTO-II (BD Biosciences). For some assays, apoptosis was detected by staining with mitochondrial potential sensitive dye DiOC6 (10 μm) (Calbiochem) followed by flow cytometry. At least 10,000 events were collected and analyzed using FACS DIVA (BD Biosciences) or Tree Star FlowJo software.
Dye Transfer Assay
Env-transfected HeLa cells were labeled with either cytoplasmic dye CMTMR (10 μm) or lipophilic membrane dye DiI (Vybrant cell labeling kit, Molecular Probes). The cells were washed and plated in 24-well plates at 105 cells/well. After 4–6 h, unlabeled SupT-R5 cells (H6, M10, or L23) were added at 5 × 105 cells/well and cocultured for 24 h. Subsequently, the nonadherent target cells were collected and stained with annexin V for detection of apoptosis and analyzed by flow cytometry. Transfer of dye (cytoplasmic or membrane) was acquired in the red channel (FL-2), and the green channel (FL1) was used for apoptosis detection via annexin V FITC. This assay determined whether the apoptotic target cells seen in our system had taken up cytoplasmic or membrane dyes from the effector cells.
In Vitro Infection and Apoptosis Detection
SupT-R5 cell lines (H6, M10, and L23) were infected with equal reverse transcriptase (RT) activity units of NL-YU-2 virus and cultured for indicated times. The cultures were split 1:3 every 2nd or 3rd day, and culture supernatants were harvested for determination of RT activity. Cells were collected at day 7 post-infection, fixed, permeabilized, and stained with anti-p24 RD-1 antibody clone KC57 (Beckman Coulter) for detection of virus infection and with activated caspase indicator, Z-VAD-FITC (Promega), for apoptosis. Flow cytometry was performed on the samples using a FACS CANTO-II flow cytometer. Data were analyzed using FACS DIVA software with at least 20,000 events acquired for each sample. At day 3, 5, 7, and 10 post-infection, cells were also assayed for viability using the Cell Titer Glo (Promega) viability assay that is based on measuring cellular ATP levels. Supernatants from the cultures were collected at different time points and assayed for virus replication using RT assay as described previously (40).
Cell to Cell Fusion Assay
HeLa cells transfected with different YU-2 Env expression vectors (which also express HIV-1 Tat) were seeded in 96-well plates at 2 × 104 cells/well. TZM cells expressing CD4 and CXCR4 as well as Tat-dependent luciferase reporter gene were added at the same concentration. The cells were cocultured for 6 h following which the luciferase activity was measured by BriteLite luciferase substrate (PerkinElmer Life Sciences). For fusion with SupT-R5 cell lines, a dye transfer assay was performed as described by Gallo et al. (41) with minor modifications. Briefly, HeLa cells transfected with YU-2 Env WT were labeled with a red fluorescent dye, CMTMR (10 μm). SupT-R5 cell lines were labeled with a green fluorescent dye, CMFDA (10 μm). The two cell lines were mixed, and fusion was detected by fluorescent microscopy 24 h post coculture using a Nikon Ti Eclipse microscope. Fusion was calculated as percent of area double-positive for red and green dye versus total red area using NIS Elements software (Nikon).
RESULTS
Establishment of Cell Lines Expressing Different Levels of CCR5
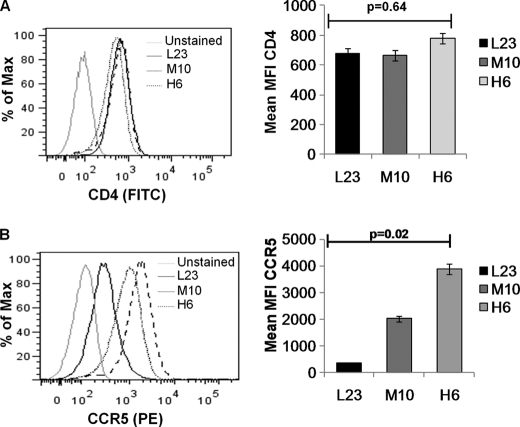
The surface levels of CCR5 vary considerably among the human population because of polymorphisms both in the CCR5 gene (42) and promoter region (43). This differential surface expression of CCR5 has been correlated with progression to AIDS in HIV-infected individuals (14, 31), although the mechanism behind this phenomenon remains undetermined. To better understand the correlation between cell surface CCR5 levels and AIDS pathogenesis, we established CD4+ T cell lines that express different levels of CCR5. SupT1 cells that naturally lack CCR5 expression were transduced with a lentiviral vector expressing CCR5. Single clones were sorted by flow cytometry and selected using blasticidin resistance. After initial characterization of clones for CD4 and CCR5 levels, three cell lines representing low (L23), medium (M10), and high (H6) CCR5 expression were selected for further studies based on stable CD4 and CCR5 expression levels. As seen in Fig. 1A, the expression of CD4 in all the cell lines was comparable. However, the cell lines showed progressively higher CCR5 expression levels (Fig. 1B) starting with L23 followed by M10 and the highest expressing H6 cell lines based on mean fluorescent intensity of CCR5 staining. The expression of CD4 and CCR5 was stable in the cell lines over several weeks of culture in the presence of blasticidin. This is depicted in Fig. 1, A, right panels, and B, where mean fluorescent intensity of CD4 and CCR5 surface staining is from three independent time points in culture.
FIGURE 1.
Characterization of SupT1 cell lines expressing different levels of CCR5. SupT1 cell line transduced with a lentiviral vector expressing CCR5 was sorted into single clones. Three selected clones based on surface CD4 and CCR5 (high H6, medium M10 or low L23) expression levels were stained using either anti-CD4 (A) or anti-CCR5 (B) antibodies and analyzed by flow cytometry. Mean fluorescence intensity for selected cell clones H6, M10, and L23 is shown on the right. The bar graphs represent mean fluorescence intensity (MFI) of CD4 and CCR5 surface staining for each clone from three independent time points in culture.
Susceptibility to Bystander Apoptosis in CCR5-expressing T Cells Is Dependent on the Cell Surface Expression Levels of CCR5
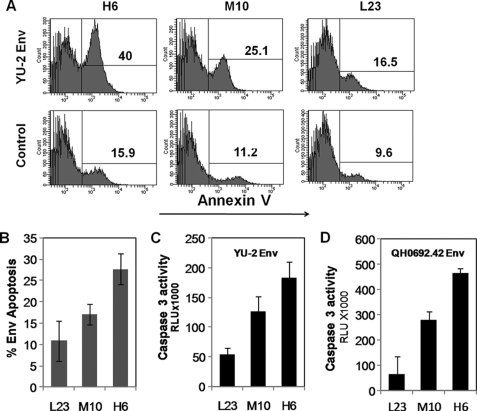
HIV envelope-mediated apoptosis of uninfected bystander T cells is one of the mechanisms of CD4 T cell depletion during progression to AIDS (44). We previously showed that an X4 tropic isolate of HIV causes bystander T cell apoptosis via gp41-mediated fusion/hemifusion (27, 29). In this study, we asked whether this was true for an R5 tropic HIV envelope and if bystander apoptosis mediated by an R5 HIV envelope correlated with surface CCR5 expression levels. We used a coculture assay whereby HeLa cells transfected with a R5 Env expression vector (YU-2 Env) were cocultured with SupT-R5 cell lines H6, M10, or L23. Apoptosis was determined 24 h post coculture using annexin V staining as a marker of apoptosis. As seen in Fig. 2A, apoptosis induction was highest for H6 (high CCR5 expressing) cell line followed by M10 (medium CCR5) and L23 (low CCR5 expression). Env-specific apoptosis was calculated for each cell line by subtracting corresponding background apoptosis derived using control vector-transfected cells (Fig. 2B). We further confirmed the differential induction of bystander apoptosis in the cell lines by determining caspase 3 activity in the cocultures (Fig. 2C). This was proportional to the amount of Env-specific apoptosis in corresponding cultures. That this differential bystander apoptosis was not specific for YU-2 Env was confirmed by using an R5 tropic primary Env clone QH0692.42 (39) from the reference panel of B subtype Envs (Fig. 2D). Hence, there was a good correlation between CCR5 expression and bystander apoptosis induction suggesting that the bystander apoptosis induction capacity of R5 HIV Env is dependent on the level of CCR5 cell surface expression.
FIGURE 2.
CCR5 tropic Env-mediated apoptosis is dependent on cell surface CCR5 expression. HeLa cells transfected with the YU-2 Env expression vector were cocultured with the SupT-R5 cell lines H6, M10, and L23. Apoptosis was determined 24 h later via annexin V staining. A, representative histograms are shown. B, Env-specific apoptosis for each cell line was determined by subtracting background apoptosis. Data are means ± S.D. from three independent experiments. Apoptosis was also determined by caspase 3 activity assay using caspase Glo 3/7 assay with YU-2 Env (C) or QHO692.42 Env (D). Data are mean ± S.D. from three independent experiments.
Expression Levels of CCR5 Determine Cell to Cell Fusion Capacity of HIV Env Glycoprotein
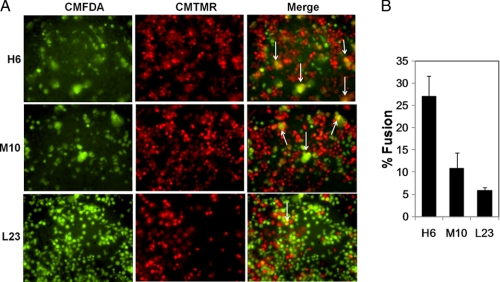
Given that Env glycoprotein-mediated fusion depends on CCR5 levels (32) and that such resultant cell-cell fusion determines bystander apoptosis induction (27), we wanted to determine whether CCR5 expression levels affected cell to cell fusion capacity in our cell lines. We conducted a coculture assay similar to the one used for apoptosis induction described above. However, in this case the effector (HeLa-expressing Env) and the target (H6, M10, or L23) cells were labeled with a green (CMFDA) and red (CMTMR) fluorescent dye, respectively. As seen in Fig. 3A, the high CCR5-expressing H6 cell line underwent extensive fusion with effector cells showing numerous large syncytia (white arrows) followed by the medium CCR5-expressing cell line, M10, whereas only limited and small syncytia were seen with the low CCR5-expressing L23 cell line. Quantitation of fusion for each cell line shows clear differences in the level of fusion with the highest fusion with H6 followed by M10 and L23 (Fig. 3B). This suggests that the cell lines undergo differential cell to cell fusion mediated by HIV Env glycoprotein that correlates with CCR5 expression levels. These data also support the correlation between cell to cell fusion and bystander apoptosis induction by R5 tropic HIV Env.
FIGURE 3.
Cell to cell fusion mediated by YU-2 Env in the cell lines expressing different levels of CCR5. A, HeLa cells transfected with YU-2 Env vector were labeled with the red fluorescent dye CMTMR. SupT-R5 cell lines H6, M10, and L23 were labeled with green fluorescent dye CMFDA. The cells were cocultured for 24 h following which fluorescence images were collected using Nikon Eclipse Ti microscope. Single channel images and merge are shown. Syncytia are marked with white arrows. B, quantitation of fusion was done by calculating the percent of area positive for both red and green dye compared with total red area in the image using NIS elements software. Data are mean ± S.D. from triplicate observations.
Low Levels of CCR5 Expression Support Virus Replication in the Absence of Bystander Apoptosis
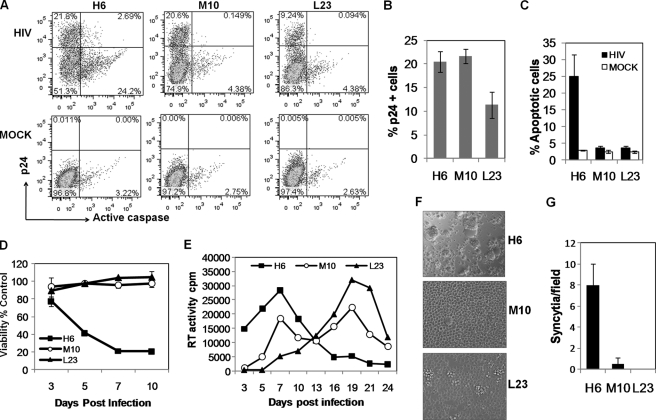
Although the levels of CCR5 vary considerably in HIV-infected individuals, which in many cases correlates with differences in disease progression (45), there seems to be a relatively smaller effect of cellular CCR5 expression levels on susceptibility to viral infection and/or viremia (46). Hence, we asked whether the cell lines expressing lower levels of CCR5 could support HIV-1 replication and whether this infection was associated with bystander apoptosis. We infected the SupT-R5 clones H6, M10, and L23 with NL-YU-2 virus that contains the R5 Env region of YU-2 in the NL4-3 backbone, and we monitored the cultures for virus replication and bystander apoptosis induction over a period of several days. The number of infected cells was determined by staining for intracellular HIV-1 Gag p24 antigen using specific antibody, whereas apoptosis was measured via caspase activation using Z-VAD-FITC. As seen in Fig. 4A, at day 7 post-infection p24+ cells were seen in all three cell lines infected with NL-YU-2 virus. Interestingly, apoptosis was largely restricted to the H6 cell line that expresses the highest levels of CCR5. In essence, the number of p24-positive cells in H6 and M10 cell lines was similar (Fig. 4B); however, there was a marked absence of apoptotic cells in M10-infected cultures (Fig. 4C). Moreover, in the infected H6 cell line, the caspase-positive cells were largely p24-negative suggesting that these were uninfected cells that were probably in contact with infected cells and undergoing bystander apoptosis consistent with our previous findings (47) and those by Holm et al. (48). We also observed that cell to cell fusion leading to syncytia formation was largely restricted to H6 cells (Fig. 4, F and G) at the peak of virus replication at day 7 (Fig. 4E). The lack of apoptosis in the M10 and L23 cell lines was further supported by the loss of viability in the H6 cell line but not the other cell lines expressing lower levels of CCR5 (Fig. 4D). While following the cells for virus replication over a period of time, we consistently observed that the peak virus replication occurred at day 7 in H6 cells followed by a sharp decline associated with cell death most likely due to bystander killing. In contrast, peak virus replication in M10 cells occurred at day 7 followed by a plateau that lasted until day 19 (Fig. 4E). The prolonged replication of virus in M10 cells is supportive of the lack of bystander apoptosis allowing for more viable cells for virus replication. Another possibility is that the rate of cell killing due to direct infection was lower than the cell division allowing for a broader replication curve. Conversely, virus in L23 cells replicated slowly up to day 19 (Fig. 4E) consistent with slower kinetics also seen in the p24 staining experiments. In physiological settings, this cell line may represent the prolonged lower viremia seen in some individuals with CCR5 polymorphisms. These findings suggest that although lower levels of CCR5 can support virus replication, with altered kinetics, there is a marked difference in the cytopathic effects specifically bystander apoptosis, which is highly dependent on the levels of CCR5.
FIGURE 4.
Cell lines with lower levels of CCR5 support virus replication in the absence of bystander apoptosis. A, SupT-R5 cell lines H6, M10, and L23 were infected with NL-YU-2 virus and subsequently stained with anti-p24 Ab to detect virus infection and Z-VAD-FITC to detect apoptosis induction on day 7 post-infection. Cumulative data representing virus infection (B) and apoptosis (C) on day 7 postinfection is shown. D, cell viability in the cultures was determined by measuring ATP levels in cells using cell titer Glo assay at the indicated time points. E, virus replication in the cultures was determined by measuring RT activity in culture supernatants every 2–3 days post-infection. F, syncytia formation was recorded in the cultures on day 7. G, number of syncytia per field were counted for at least four images for each cell line. All error bars show mean ± S.D. of triplicate observations.
Apoptosis Mediated by CCR5 Env Is Dependent on Caspase Activation and Mitochondrial Depolarization
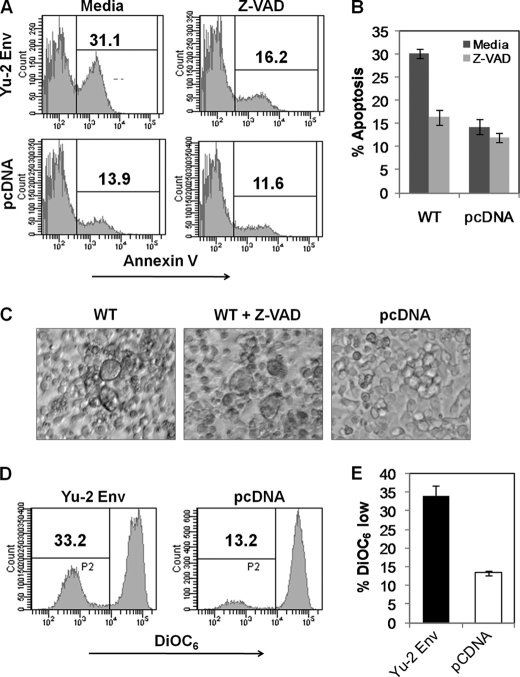
We have previously shown that the signaling pathway involved in X4 tropic HIV Env glycoprotein-mediated apoptosis involves caspase activation and mitochondrial depolarization. However, the apoptotic signaling pathway for R5 viruses remains undetermined. Hence, we asked whether apoptosis mediated in bystander cells by R5 tropic Env under our experimental conditions is dependent on caspase activation and/or involves mitochondrial depolarization. The requirement of caspase cascade for apoptosis induction can be determined by inhibition of apoptosis by pan caspase inhibitor Z-VAD-fmk. As seen in Fig. 5A, addition of Z-VAD-fmk at 40 μm inhibited apoptosis induced by Env glycoprotein in bystander cells. The decrease was significant only for the Env-expressing cocultures with little effect on vector control transfected cells (Fig. 5B). Interestingly, inhibition of apoptosis by Z-VAD-fmk did not alter the fusion process as syncytia formation was evident with or without Z-VAD-fmk addition (Fig. 5C). Further analysis of mechanism of apoptosis by R5 Env was done using the membrane potential sensor dye DiOC6 and suggested that R5 Env-mediated apoptosis pathway involved mitochondrial membrane depolarization (Fig. 5D). YU-2 Env-expressing cocultures exhibited a significantly higher percentage of cells displaying mitochondrial depolarization (DiOC6 low) when compared with control vector-expressing cultures (Fig. 5E). These findings suggest that R5 Env-mediated apoptosis involves the classical apoptotic pathway involving mitochondrial depolarization and caspase activation similar to X4 viruses reported previously (27).
FIGURE 5.
Apoptosis mediated by R5 Env in high CCR5-expressing cell line is dependent on caspase activation and mitochondrial depolarization. HeLa cells transfected with YU-2 Env were cocultured with SupT-R5-H6 cell line. Apoptosis inhibitor Z-VAD-fmk (40 μm) was added at the time of coculture. Apoptosis was detected 24 h later using annexin V staining. A, representative histograms are shown. B, apoptosis inhibition by Z-VAD-fmk in high CCR5-expressing cell line H6 using data from three independent (mean ± S.D.) experiments. C, syncytia formation in cocultures in the presence or absence of Z-VAD-fmk. D, apoptosis was also determined by DiOC6 labeling. E, YU-2 Env causes increased mitochondrial depolarization in bystander cells as determined by DiOC6 staining. Data are mean ± S.D. from three independent experiments.
Apoptosis Mediated by CCR5 Env Is Dependent on Env Fusogenic Activity
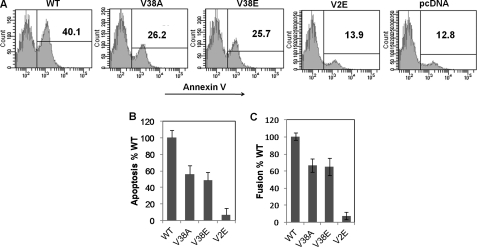
Binding of HIV Env glycoprotein to its receptor CD4 and a coreceptor, either CXCR4 or CCR5, has been implicated in bystander apoptosis induction (49). The fact that higher levels of CCR5 on cells result in greater bystander apoptosis induction naturally raises the possibility that signaling via CCR5 may be critical for apoptosis induction. However, recent data with X4 tropic viruses suggest that fusogenic activity of the gp41 subunit of the Env glycoprotein determines bystander apoptosis induction (26, 29). In case of X4 viruses, bystander apoptosis can be inhibited by abolishing gp41 function either by using peptide inhibitors (T20/Enfuvirtide) or mutations in different regions of gp41 that abrogate/attenuate fusion activity and consequently apoptosis (29). However, whether the same phenomenon holds true for R5 tropic Env in our cell line expressing high levels of CCR5 is not known. To address these issues, we generated several mutations in the gp41 region of the YU-2 Env vector. Mutations V38A and V38E in the heptad repeat 1 region of gp41 are associated with a reduction in fusogenic activity (50). Conversely, the mutation V2E in the second amino acid of gp41 severely restricts fusogenic activity of the Env glycoprotein (51). Using these mutants and the SupT-R5 cell line H6, we found that for R5 Env, bystander apoptosis was dependent on gp41-mediated fusogenic activity as measured by annexin V staining (Fig. 6A). More interestingly, the bystander apoptosis inducing activity (Fig. 6B) correlated with cell to cell fusion capacity (Fig. 6C) of the mutants. Taken together, these data suggest that bystander apoptosis induced by R5 Env in cells expressing high levels of CCR5 is dependent on gp41 function.
FIGURE 6.
Apoptosis induction correlates with cell to cell fusion activity of different Env mutants. HIV YU-2 Env gp41 mutants were tested for bystander apoptosis induction in cocultures between HeLa cells transfected with WT, V38A, V38E, or V2E Env-expressing vectors after coculture with high CCR5-expressing cell line SupT-R5-H6. A, apoptosis was detected 24 h later by annexin V staining. B, percent Env-specific apoptosis normalized to WT Env is shown. C, cell to cell fusion mediated by HeLa cells expressing WT, V38A, V38E, or V2E Env in TZM cell coculture. Percent fusion normalized to WT Env is shown. B and C, data represent mean ± S.D. from three independent experiments.
Apoptosis Induction in Bystander Cells Is a Result of gp41-mediated Hemifusion
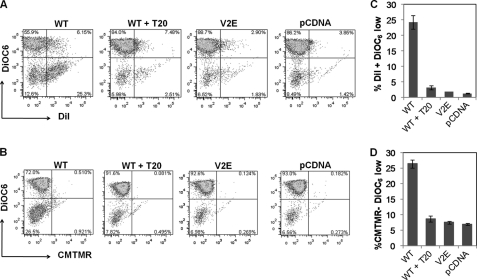
In HIV-1 infections, Env fusogenic activity correlates with pathogenesis and bystander apoptosis both in vitro (26, 44, 52) and in vivo (28, 53, 54). However, the fact that there are few multinucleated syncytia seen in vivo remains a controversial issue. This observation has led to the hypothesis that HIV Env-mediated apoptosis in bystander cells is a process mediated by hemifusion rather than complete fusion in target cells (26, 27). Hemifusion is an intermediate step in the fusion process characterized by the merger of the outer membrane leaflets of two biological membranes without the formation of a fusion pore or mixing of the inner leaflets (55). This phenomenon can be studied by the use of cytoplasmic versus membrane dye labeling of cells. We (27, 29) and others (26) have used dye transfer assays to study the phenomenon of hemifusion and its correlation with bystander apoptosis mediated by X4 HIV Env. Using the same technique, we next asked whether bystander apoptosis induced by R5 Env in CCR5 high expressing cell line (SupT-R5-H6) was hemifusion-dependent. For this purpose, we labeled HeLa cells transfected with YU-2 Env with either a cytoplasmic dye CMTMR or a lipophilic dye DiI. These cells were then cocultured with unlabeled SupT-R5-H6 cell line. The nonadherent cells were collected 24 h post coculture, and apoptosis was detected by either DiOC6 or annexin V staining followed by flow cytometry. The transfer of cytoplasmic dye (an indicator of complete cell fusion leading to the mixing of cytoplasmic contents) or membrane dye (an indicator of membrane mixing or partial fusion/hemifusion) was detected in the red channel, whereas apoptosis was detected in the green channel. As seen in Fig. 7A, the apoptotic cells (DiOC6 low) were also DiI-positive indicative of exchange of membrane components characteristic of hemifusion. However, as evident in Fig. 7B, the apoptotic cells characterized by low DiOC6 staining were negative for the cytoplasmic dye (CMTMR) suggesting that there was no mixing of cellular contents. That this process was specific to HIV gp41 was further validated by inhibition of both bystander apoptosis and DiI transfer by using the gp41 inhibitor T20 or the mutation V2E that abolishes fusion (Fig. 7, C and D). Similar results were seen using annexin V as a marker for apoptosis (supplemental Fig. 1). Taken together, these findings suggest that the process of bystander apoptosis by R5 Env is dependent on gp41 function and more specifically hemifusion mediated by the Env glycoprotein.
FIGURE 7.
Apoptosis induction in bystander cells mediated by R5 Env in high CCR5-expressing cells is gp41-induced hemifusion-dependent. HeLa Env expressing cells were labeled either with lipophilic dye DiI (A) or cytoplasmic dye CMTMR (B) and cocultured with unlabeled SupT-R5-H6 cells. Suspension cells were collected and analyzed for dye transfer (x axis) and apoptosis by DiOC6 staining (y axis) by flow cytometry. C, Env-mediated apoptosis is restricted to hemifused cells characterized by DiI-positive and DiOC6 low labeling. D, apoptotic cells fail to take up cytoplasmic dye characterized as CMTMR-negative and DiOC6 low labeling. Data are mean ± S.D. of triplicate observations. All experiments were repeated three times with similar results.
DISCUSSION
The role of a specific type of coreceptor in HIV pathogenesis remains much debated (reviewed by Schuitemaker et al. (56)). However, it is evident that the switch of coreceptor usage by HIV from CCR5 to CXCR4 precedes the rapid decline in CD4 cells and AIDS development in a number of cases (57). The differences in CXCR4 and CCR5 expression on CD4+ T cells may be related to the relative pathogenesis of X4 versus R5 viruses (58). Although the correlation between CCR5 polymorphism and HIV-1 disease progression has been extensively studied, it is still unclear as to why individuals with CCR5 polymorphisms like the CCR5Δ32 heterozygous mutation in its coding region or CCR5–2459 A/G mutation in the promoter show slower progression to AIDS. It has been reported previously that CCR5Δ32 heterozygous mutation as well as CCR5–2459A/G affect CCR5 expression levels (45); however, the functional consequence of this phenomenon and how it affects progression to AIDS remain unknown. Also, the mechanism via which levels of CCR5 play a role in disease progression in patients infected exclusively with R5 tropic viruses remains undetermined.
In an animal model (SCID-hu mice), Scoggins et al. (38) demonstrated that thymus grafts from CCR5Δ32+/− individuals can support virus replication in the absence of CD4 depletion. Whether this was a result of reduced replication or cytopathic effect is not known. That viremia per se does not lead to AIDS is further supported by the absence of AIDS in simian immunodeficiency virus infection of African green monkeys and Sooty Mangabeys in the wild despite high levels of virus replication (59, 60). We therefore hypothesized that although lower levels of CCR5 can support virus replication, they may be insufficient for bystander apoptosis mediated by HIV Env glycoprotein. Consistent with this hypothesis, our study establishes a correlation between CCR5 levels and bystander apoptosis and provides mechanistic insights into this phenomenon.
The increased fusogenicity of X4 viruses is suggested to be a contributing factor in the enhanced pathogenesis of these viruses (61, 62). However, in 50% of patients there is no switch in coreceptor usage, and although the presence of X4 tropic virus has been associated with poor prognosis in patients, it is evident that the coreceptor switch is not absolutely essential for progression to AIDS (63). Nevertheless, the selection of more pathogenic CCR5 utilizing HIV has been documented. In fact, Olivieri et al. (64) showed that CCR5 tropic viruses isolated from patients early during disease (pre-AIDS) are different from those isolated at the development of AIDS with a specific change in the glycosylation site at position Asn-362. More importantly, this change is associated with increased Env fusion activity suggesting that Env fusion and CD4 loss are correlative for R5 viruses as well. The role of Env glycoprotein fusion activity in apoptosis by CCR5 tropic viruses is supported by other studies as well. Blanco et al. (52) used fusion inhibitor T20 to show that the apoptotic mechanism was similar between X4 and R5 viruses and required gp41 function. In another study, Espert et al. (65) showed that autophagy is a phenomenon that is regulated by HIV Env in a gp41-dependent manner. However, in these studies the role of CCR5 expression levels in regulating bystander cell death mediated by HIV Env and whether it is hemifusion-dependent was not determined.
Our study addresses two important questions. 1) Is there a role for CCR5 expression levels on bystander apoptosis induction by the R5 tropic HIV Env? 2) Is R5 Env-mediated apoptosis dependent upon gp41-mediated hemifusion reaction? Our study demonstrates that bystander apoptosis mediated by R5 HIV Env correlates with surface expression of CCR5. Next, the phenomenon of bystander apoptosis mediated by CCR5 tropic Env glycoprotein is dependent on hemifusion mediated by the gp41 domain of Env glycoprotein. Taken together, the fact that CCR5 levels determine Env-mediated fusion, which in turn correlates with bystander apoptosis, suggests that the three phenomenon are likely interdependent.
Another important question that remains unanswered in HIV pathogenesis is the phenomenon of coreceptor switch that only occurs in some individuals. The answer may once again lie in the differences in the levels of CCR5 cell surface expression. In fact in vitro evolution of R5 viruses to CXCR4 usage can be achieved by culturing in the presence of limiting amounts of CCR5 and high levels of CXCR4 (66, 67). Hence, individuals with lower levels of CCR5 may facilitate the virus to switch coreceptors due to lower levels of virus replication. This is also supported by the observation that the likelihood of finding CXCR4 tropic virus in CCR5Δ32+/− individuals is higher. Conversely, recent studies by Schuitemaker et al. (56) suggest that all viruses eventually evolve to CXCR4 usage. However, in some individuals terminal disease is reached before conversion to X4 tropism. Our data suggest that higher levels of CCR5 not only support better virus replication but also result in enhanced pathogenesis via HIV Env-mediated bystander apoptosis and consequently progression to AIDS without or prior to coreceptor switch. However, in this case, subtype differences may play a role in combination with CCR5 expression levels in determining rate of disease progression and/or coreceptor switch.
The gamut of CCR5 expression levels in individuals makes CCR5 an important and interesting variable in HIV pathogenesis (68). Our establishment of T cell lines with varied levels of CCR5 should provide valuable reagents for studying both virus replication and pathogenesis of other clinically relevant R5 isolates. In our preliminary study using these cell lines, we saw some interesting phenomena with regard to virus replication. For example, the H6 (high CCR5) cell line showed quick virus replication followed by extensive cytopathic effects resulting in a decline in virus after the peak. However, lower levels of CCR5 in cells allowed for prolonged replication of virus in the M10 cell line in the absence of cytopathic effects. Taken together, our findings may reflect the outcome of R5 viruses in individuals with lower levels of CCR5 where virus replication is seen in the absence of cytopathic effects. Nevertheless, many reports suggest that in case of CCR5 polymorphism lower viral set points and replication are correlative with disease progression (69). However, lower virus replication alone cannot explain the reduced rate of CD4 decline in CCR5Δ32+/− individuals as suggested by Hunt and Carrington (46). Even after correction for virus replication, there is a protective effect of CCR5 polymorphism on AIDS progression (70). Hence, the virus replication independent effects of CCR5 expression level on disease progression may be explained by our findings.
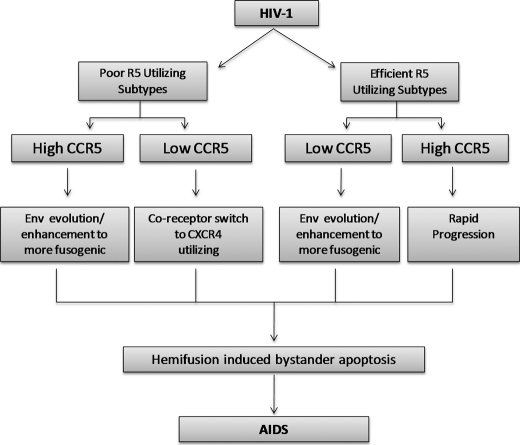
Our study suggests that bystander apoptosis in HIV infection is possibly a result of a combination of Env fusion activity and CCR5 expression levels. The subtype and quasi-species variability in HIV Env are likely to play a marked role in pathogenesis by regulating the efficiency with which the Env can use CCR5 for fusion/hemifusion and bystander apoptosis. Based on this, we propose a model for the interdependence of these factors (Fig. 8). In our hypothetical model, infections with certain subtypes that are relatively inefficient at using CCR5 combined with lower levels of CCR5 on the cell surface drives coreceptor change to X4 tropism and pathogenic consequence. However, with subtypes that efficiently utilize CCR5, the coreceptor tropism is maintained as the virus evolves toward increased fusion activity and pathogenesis while maintaining CCR5 usage. In each case the enhancement of Env-mediated fusogenic activity results in increased hemifusion-induced bystander apoptosis resulting in progression to AIDS.
FIGURE 8.
Model for interdependence of surface CCR5 expression levels and Env fusogenic activity in AIDS pathogenesis. Infections with certain HIV-1 subtypes that are relatively inefficient at using CCR5 combined with lower levels of CCR5 on cell surface may drive coreceptor change to X4 tropism. However, for subtypes that efficiently utilize CCR5, the coreceptor tropism is maintained as the virus evolves toward increased Env fusion activity and pathogenesis while maintaining CCR5 usage.
Although our study is a first step in studying the role of CCR5 cell surface levels in bystander apoptosis, the results of our in vitro study need to be interpreted with caution. Our findings are only indicative of the association between CCR5 expression levels and bystander apoptosis and consequently with AIDS progression in patients infected with HIV-1 R5 isolates. Further studies utilizing primary cells and a larger panel of R5 tropic Envs will be needed to determine whether this actually holds true in vivo.
Supplementary Material
Acknowledgments
We are grateful to the National Institutes of Health AIDS Research and Reference Reagent Program for supplying valuable reagents used in this study. Cell sorting was done at the cytometry core facility at Texas Tech University, Department of Biomedical Sciences.
This work was supported in part by a Seed Grant from Texas Tech University Health Sciences Center (to H. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- Env
- envelope
- Z-
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- DiOC6
- 3,3′dihexyloxacarbocyanine iodide
- DiI
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- CMTMR
- (5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine)
- CMFDA
- 5-chloromethylfluorescein diacetate.
REFERENCES
- 1. Albright A. V., Shieh J. T., Itoh T., Lee B., Pleasure D., O'Connor M. J., Doms R. W., González-Scarano F. (1999) J. Virol. 73, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bazan H. A., Alkhatib G., Broder C. C., Berger E. A. (1998) J. Virol. 72, 4485–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rucker J., Edinger A. L., Sharron M., Samson M., Lee B., Berson J. F., Yi Y., Margulies B., Collman R. G., Doranz B. J., Parmentier M., Doms R. W. (1997) J. Virol. 71, 8999–9007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Keele B. F., Giorgi E. E., Salazar-Gonzalez J. F., Decker J. M., Pham K. T., Salazar M. G., Sun C., Grayson T., Wang S., Li H., Wei X., Jiang C., Kirchherr J. L., Gao F., Anderson J. A., Ping L. H., Swanstrom R., Tomaras G. D., Blattner W. A., Goepfert P. A., Kilby J. M., Saag M. S., Delwart E. L., Busch M. P., Cohen M. S., Montefiori D. C., Haynes B. F., Gaschen B., Athreya G. S., Lee H. Y., Wood N., Seoighe C., Perelson A. S., Bhattacharya T., Korber B. T., Hahn B. H., Shaw G. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salazar-Gonzalez J. F., Bailes E., Pham K. T., Salazar M. G., Guffey M. B., Keele B. F., Derdeyn C. A., Farmer P., Hunter E., Allen S., Manigart O., Mulenga J., Anderson J. A., Swanstrom R., Haynes B. F., Athreya G. S., Korber B. T., Sharp P. M., Shaw G. M., Hahn B. H. (2008) J. Virol. 82, 3952–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van't Wout A. B., Kootstra N. A., Mulder-Kampinga G. A., Albrecht-van Lent N., Scherpbier H. J., Veenstra J., Boer K., Coutinho R. A., Miedema F., Schuitemaker H. (1994) J. Clin. Invest. 94, 2060–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schuitemaker H., Koot M., Kootstra N. A., Dercksen M. W., de Goede R. E., van Steenwijk R. P., Lange J. M., Schattenkerk J. K., Miedema F., Tersmette M. (1992) J. Virol. 66, 1354–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van 't Wout A. B., Blaak H., Ran L. J., Brouwer M., Kuiken C., Schuitemaker H. (1998) J. Virol. 72, 5099–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Camerini D., Su H. P., Gamez-Torre G., Johnson M. L., Zack J. A., Chen I. S. (2000) J. Virol. 74, 3196–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Connor R. I., Sheridan K. E., Ceradini D., Choe S., Landau N. R. (1997) J. Exp. Med. 185, 621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kwa D., Boeser-Nunnink B., Schuitemaker H. (2003) AIDS 17, 759–761 [DOI] [PubMed] [Google Scholar]
- 12. van Rij R. P., Worobey M., Visser J. A., Schuitemaker H. (2003) Virology 314, 451–459 [DOI] [PubMed] [Google Scholar]
- 13. Bleul C. C., Wu L., Hoxie J. A., Springer T. A., Mackay C. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ostrowski M. A., Justement S. J., Catanzaro A., Hallahan C. A., Ehler L. A., Mizell S. B., Kumar P. N., Mican J. A., Chun T. W., Fauci A. S. (1998) J. Immunol. 161, 3195–3201 [PubMed] [Google Scholar]
- 15. Kitchen S. G., Zack J. A. (1999) AIDS Res. Hum. Retroviruses 15, 143–148 [DOI] [PubMed] [Google Scholar]
- 16. Gougeon M. L., Montagnier L. (1993) Science 260, 1269–1270 [DOI] [PubMed] [Google Scholar]
- 17. Perfettini J. L., Castedo M., Roumier T., Andreau K., Nardacci R., Piacentini M., Kroemer G. (2005) Cell Death Differ. 12, Suppl. 1, 916–923 [DOI] [PubMed] [Google Scholar]
- 18. Priceputu E., Rodrigue I., Chrobak P., Poudrier J., Mak T. W., Hanna Z., Hu C., Kay D. G., Jolicoeur P. (2005) J. Virol. 79, 6377–6391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zauli G., Gibellini D., Secchiero P., Dutartre H., Olive D., Capitani S., Collette Y. (1999) Blood 93, 1000–1010 [PubMed] [Google Scholar]
- 20. Li C. J., Friedman D. J., Wang C., Metelev V., Pardee A. B. (1995) Science 268, 429–431 [DOI] [PubMed] [Google Scholar]
- 21. Purvis S. F., Jacobberger J. W., Sramkoski R. M., Patki A. H., Lederman M. M. (1995) AIDS Res. Hum. Retroviruses 11, 443–450 [DOI] [PubMed] [Google Scholar]
- 22. Jian H., Zhao L. J. (2003) J. Biol. Chem. 278, 44326–44330 [DOI] [PubMed] [Google Scholar]
- 23. Muthumani K., Choo A. Y., Hwang D. S., Chattergoon M. A., Dayes N. N., Zhang D., Lee M. D., Duvvuri U., Weiner D. B. (2003) Biochem. Biophys. Res. Commun. 304, 583–592 [DOI] [PubMed] [Google Scholar]
- 24. Scheller C., Jassoy C. (2001) Virology 282, 48–55 [DOI] [PubMed] [Google Scholar]
- 25. Blanco J., Jacotot E., Cabrera C., Cardona A., Clotet B., De Clercq E., Esté J. A. (1999) AIDS 13, 909–917 [DOI] [PubMed] [Google Scholar]
- 26. Blanco J., Barretina J., Ferri K. F., Jacotot E., Gutiérrez A., Armand-Ugón M., Cabrera C., Kroemer G., Clotet B., Esté J. A. (2003) Virology 305, 318–329 [DOI] [PubMed] [Google Scholar]
- 27. Garg H., Blumenthal R. (2006) J. Leukocyte Biol. 79, 351–362 [DOI] [PubMed] [Google Scholar]
- 28. Etemad-Moghadam B., Rhone D., Steenbeke T., Sun Y., Manola J., Gelman R., Fanton J. W., Racz P., Tenner-Racz K., Axthelm M. K., Letvin N. L., Sodroski J. (2001) J. Virol. 75, 5646–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Garg H., Joshi A., Freed E. O., Blumenthal R. (2007) J. Biol. Chem. 282, 16899–16906 [DOI] [PubMed] [Google Scholar]
- 30. Wu L., Paxton W. A., Kassam N., Ruffing N., Rottman J. B., Sullivan N., Choe H., Sodroski J., Newman W., Koup R. A., Mackay C. R. (1997) J. Exp. Med. 185, 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salkowitz J. R., Bruse S. E., Meyerson H., Valdez H., Mosier D. E., Harding C. V., Zimmerman P. A., Lederman M. M. (2003) Clin. Immunol. 108, 234–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. (1998) J. Virol. 72, 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Platt E. J., Durnin J. P., Kabat D. (2005) J. Virol. 79, 4347–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Michael N. L., Chang G., Louie L. G., Mascola J. R., Dondero D., Birx D. L., Sheppard H. W. (1997) Nat. Med. 3, 338–340 [DOI] [PubMed] [Google Scholar]
- 35. Zimmerman P. A., Buckler-White A., Alkhatib G., Spalding T., Kubofcik J., Combadiere C., Weissman D., Cohen O., Rubbert A., Lam G., Vaccarezza M., Kennedy P. E., Kumaraswami V., Giorgi J. V., Detels R., Hunter J., Chopek M., Berger E. A., Fauci A. S., Nutman T. B., Murphy P. M. (1997) Mol. Med. 3, 23–36 [PMC free article] [PubMed] [Google Scholar]
- 36. Dean M., Carrington M., Winkler C., Huttley G. A., Smith M. W., Allikmets R., Goedert J. J., Buchbinder S. P., Vittinghoff E., Gomperts E., Donfield S., Vlahov D., Kaslow R., Saah A., Rinaldo C., Detels R., O'Brien S. J. (1996) Science 273, 1856–1862 [DOI] [PubMed] [Google Scholar]
- 37. Liu R., Paxton W. A., Choe S., Ceradini D., Martin S. R., Horuk R., MacDonald M. E., Stuhlmann H., Koup R. A., Landau N. R. (1996) Cell 86, 367–377 [DOI] [PubMed] [Google Scholar]
- 38. Scoggins R. M., Taylor J. R., Jr., Patrie J., van't Wout A. B., Schuitemaker H., Camerini D. (2000) J. Virol. 74, 3205–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C. (2005) J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Joshi A., Nagashima K., Freed E. O. (2006) J. Virol. 80, 7939–7951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gallo S. A., Reeves J. D., Garg H., Foley B., Doms R. W., Blumenthal R. (2006) Retrovirology 3, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Roda Husman A. M., Koot M., Cornelissen M., Keet I. P., Brouwer M., Broersen S. M., Bakker M., Roos M. T., Prins M., de Wolf F., Coutinho R. A., Miedema F., Goudsmit J., Schuitemaker H. (1997) Ann. Intern. Med. 127, 882–890 [DOI] [PubMed] [Google Scholar]
- 43. McDermott D. H., Zimmerman P. A., Guignard F., Kleeberger C. A., Leitman S. F., Murphy P. M. (1998) Lancet 352, 866–870 [DOI] [PubMed] [Google Scholar]
- 44. Garg H., Blumenthal R. (2008) Cell. Mol. Life Sci. 65, 3134–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shieh B., Liau Y. E., Hsieh P. S., Yan Y. P., Wang S. T., Li C. (2000) Int. Immunol. 12, 1311–1318 [DOI] [PubMed] [Google Scholar]
- 46. Hunt P. W., Carrington M. (2008) Curr. Opin HIV AIDS 3, 342–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garg H., Joshi A., Ye C., Shankar P., Manjunath N. (2011) Virol. J. 8, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holm G. H., Zhang C., Gorry P. R., Peden K., Schols D., De Clercq E., Gabuzda D. (2004) J. Virol. 78, 4541–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahr B., Robert-Hebmann V., Devaux C., Biard-Piechaczyk M. (2004) Retrovirology 1, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garg H., Joshi A., Blumenthal R. (2009) AIDS Res. Hum. Retroviruses 25, 811–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Freed E. O., Delwart E. L., Buchschacher G. L., Jr., Panganiban A. T. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blanco J., Barretina J., Clotet B., Esté J. A. (2004) J. Leukocyte Biol. 76, 804–811 [DOI] [PubMed] [Google Scholar]
- 53. Karlsson G. B., Halloran M., Schenten D., Lee J., Racz P., Tenner-Racz K., Manola J., Gelman R., Etemad-Moghadam B., Desjardins E., Wyatt R., Gerard N. P., Marcon L., Margolin D., Fanton J., Axthelm M. K., Letvin N. L., Sodroski J. (1998) J. Exp. Med. 188, 1159–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reimann K. A., Li J. T., Veazey R., Halloran M., Park I. W., Karlsson G. B., Sodroski J., Letvin N. L. (1996) J. Virol. 70, 6922–6928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chernomordik L. V., Kozlov M. M. (2005) Cell 123, 375–382 [DOI] [PubMed] [Google Scholar]
- 56. Schuitemaker H., van 't Wout A. B., Lusso P. (2011) J. Transl. Med. 9, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koot M., van 't Wout A. B., Kootstra N. A., de Goede R. E., Tersmette M., Schuitemaker H. (1996) J. Infect. Dis. 173, 349–354 [DOI] [PubMed] [Google Scholar]
- 58. Jekle A., Keppler O. T., De Clercq E., Schols D., Weinstein M., Goldsmith M. A. (2003) J. Virol. 77, 5846–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Silvestri G., Paiardini M., Pandrea I., Lederman M. M., Sodora D. L. (2007) J. Clin. Invest. 117, 3148–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Silvestri G., Sodora D. L., Koup R. A., Paiardini M., O'Neil S. P., McClure H. M., Staprans S. I., Feinberg M. B. (2003) Immunity 18, 441–452 [DOI] [PubMed] [Google Scholar]
- 61. Spijkerman I., de Wolf F., Langendam M., Schuitemaker H., Coutinho R. (1998) J. Infect. Dis. 178, 397–403 [DOI] [PubMed] [Google Scholar]
- 62. Penn M. L., Grivel J. C., Schramm B., Goldsmith M. A., Margolis L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karlsson I., Grivel J. C., Chen S. S., Karlsson A., Albert J., Fenyö E. M., Margolis L. B. (2005) J. Virol. 79, 11151–11160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Olivieri K., Scoggins R. M., Bor Y. C., Matthews A., Mark D., Taylor J. R., Jr., Chernauskas D., Hammarskjöld M. L., Rekosh D., Camerini D. (2007) Virology 358, 23–38 [DOI] [PubMed] [Google Scholar]
- 65. Espert L., Varbanov M., Robert-Hebmann V., Sagnier S., Robbins I., Sanchez F., Lafont V., Biard-Piechaczyk M. (2009) PLoS One 4, e5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Edo-Matas D., van Dort K. A., Setiawan L. C., Schuitemaker H., Kootstra N. A. (2011) Virology 412, 269–277 [DOI] [PubMed] [Google Scholar]
- 67. Pastore C., Ramos A., Mosier D. E. (2004) J. Virol. 78, 7565–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Reiche E. M., Bonametti A. M., Voltarelli J. C., Morimoto H. K., Watanabe M. A. (2007) Curr. Med. Chem. 14, 1325–1334 [DOI] [PubMed] [Google Scholar]
- 69. Reynes J., Portales P., Segondy M., Baillat V., André P., Réant B., Avinens O., Couderc G., Benkirane M., Clot J., Eliaou J. F., Corbeau P. (2000) J. Infect. Dis. 181, 927–932 [DOI] [PubMed] [Google Scholar]
- 70. Ioannidis J. P., Rosenberg P. S., Goedert J. J., Ashton L. J., Benfield T. L., Buchbinder S. P., Coutinho R. A., Eugen-Olsen J., Gallart T., Katzenstein T. L., Kostrikis L. G., Kuipers H., Louie L. G., Mallal S. A., Margolick J. B., Martinez O. P., Meyer L., Michael N. L., Operskalski E., Pantaleo G., Rizzardi G. P., Schuitemaker H., Sheppard H. W., Stewart G. J., Theodorou I. D., Ullum H., Vicenzi E., Vlahov D., Wilkinson D., Workman C., Zagury J. F., O'Brien T. R. (2001) Ann. Intern. Med. 135, 782–795 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.