Abstract
The p53 family member, p73, has been characterized as a tumor suppressor and functions in a similar manner as p53 to induce cellular death. The phosphatase and tensin homolog (PTEN) can function as a dual specificity lipid/protein phosphatase. However, recent data have described multiple roles for nuclear PTEN independent of its lipid phosphatase activity. PTEN can directly or indirectly activate p53 to promote apoptosis. We examined whether PTEN would interact and regulate p73 independent of p53. Co-localization in the nucleus and complex formation of p73/PTEN were observed after DNA damage. Furthermore, we also demonstrate that p73α/PTEN proteins directly bind one another. Both overexpressed and endogenous p73-PTEN interactions were determined to be the strongest in the nuclear fraction after DNA damage, which suggested formation of a transcriptional complex. We employed chromatin immunoprecipitation (ChIP) and found that p73 and PTEN were associated with the PUMA promoter after genotoxic stress in TP53-null cells. We found that another p73 target, BAX, had an increased expression in the presence of p73 and PTEN. In addition, in virus-transduced cell lines stably expressing p73, PTEN, or both p73/PTEN, we found that the p73/PTEN cells were more sensitive to genotoxic stress and cellular death as measured by increased poly(ADP-ribose) polymerase cleavage and PUMA/Bax induction. Conversely, knockdown of PTEN dramatically reduced Bax and PUMA levels. Thus, a p73-PTEN protein complex is engaged to induce apoptosis independent of p53 in response to DNA damage.
Keywords: Apoptosis, p73, Phosphatase, Transcription Factors, Tumor Suppressor Gene, PTEN, PUMA, bax
Introduction
p73 and p63 belong to the p53 tumor suppressor family (1–4). Genetically, both p73 and p63 are components of a p53-dependent network to induce apoptosis (5). All p53 family members can have multiple isoforms as follows: ΔN (no transactivation domain) or TA (containing transactivation domain), which result from alternative promoter usage (6). As for TAp73, it is capable of transactivating a number of genes including, but not limited to, p21 and GADD45 involved in cell cycle arrest and BAX, NOXA, and 14-3-3 involved in apoptosis (7). In response to apoptotic stimuli, the PUMA gene (p53 up-regulated modulator of apoptosis) is induced by TAp73, which triggers Bax mitochondrial translocation and release of cytochrome c to activate the caspase cascade. The ΔNp73 isoform can repress the caspase cascade by acting as a dominant negative to both p73 and p53 (8). Recently, a ubiquitin ligase named p73-induced ring protein 2 (PIR2) has been demonstrated to be induced by TAp73, which leads to an increase in the ratio of TAp73/ΔNp73 with preferential ubiquitin-mediated degradation of ΔNp73 (9). The regulation by this ubiquitin ligase supports the pro-apoptotic function of TAp73. Therefore, p73 induces apoptosis in a similar fashion to p53, and isoform-specific regulation of p73 dramatically affects the balance between cell survival and programmed cell death.
The PTEN2 tumor suppressor has been extensively investigated with respect to somatic mutations associated with inherited human genetic diseases and post-translational modifications, which have defined the role of PTEN in cell polarity, genomic maintenance, and regulating survival signaling (10–15). Thus, PTEN is a multifaceted protein involved in tumor suppression networks (16). PTEN functions as a dual specificity phosphatase whose activity has been shown to dephosphorylate phosphatidylinositol 3,4,5-triphosphate and some proteins (17–20). The loss of phosphatidylinositol 3,4,5-triphosphate opposes Akt function through inhibition of phosphatidylinositol 3-kinase (PI3K) for regulation of cellular migration and cell cycle and proliferation and apoptotic events (21–24). PTEN may also undergo nuclear translocation, although its function in the nucleus remains unclear, it seems to be involved in genomic regulation.
The PTEN gene is also a transcriptional target of p53 in response to DNA damage (25), and at the biochemical level PTEN can regulate the tumor suppressor p53 by a direct protein-protein interaction or indirectly regulating the p53 antagonist Mdm2 by blocking nuclear localization (26–28). PTEN can form a direct protein interaction with p53 and has been mapped to the C2 domain amino acids 186–351 on PTEN and on the C-terminal negative regulatory region of p53 (29). Although PTEN traditionally functions as a lipid phosphatase in the cytoplasmic fraction of the cell, it has been reported to enter the nucleus. Interestingly, PTEN lacks classical nuclear localization signals and nuclear export signals, yet contains motifs that appear to promote its nuclear entry (30).
Here, we demonstrate in response to genotoxic stress that human p73 and PTEN integrate into a common pathway to activate apoptotic genes. In response to DNA damage, p73 and PTEN protein levels are increased and both proteins co-localize to the nucleus. We found that the TAp73α isoform had the highest affinity for binding to PTEN. Co-immunoprecipitation experiments using both endogenous and overexpressed p73 and PTEN were found to have increased interaction post-DNA damage in nuclear fractions. This complex was found associated with the PUMA promoter after genotoxic stress. The subsequent increase in apoptotic mediators, PUMA and Bax, corresponded with increased PARP cleavage. Knockdown of PTEN dramatically reduced levels of Bax and PUMA. Our work demonstrates that independent of p53, a p73-PTEN complex can induce apoptosis.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
The p53-null human non-small cell lung carcinoma cell line H1299, human kidney epithelial cell line 293T, and human foreskin fibroblast immortalized with hTert BJ-ERT and p53-null derivative Shp53 BJ-ERT described previously (31) were all cultured at 37 °C in a humidified incubator with 5% CO2. The H1299 cell line was virally transduced with PTEN (H1299 (PTEN)) and p73α (H1299 (p73α)), and p73α and PTEN (H1299 (p73α/PTEN)) were constructed from insertion of full-length human p73α or PTEN into a pLNCX2 vector. Knockdown of PTEN (ShPTEN) or control (ShGFP) was made in H1299 cells in a pLVTHM vector using previously described sequences (31). Virus was produced as described previously (32). All cell lines were maintained in Dulbecco's modified Eagle's medium with high glucose (Invitrogen) supplemented with 10% fetal bovine serum (Atlanta Biological), 50 units/ml penicillin, and 50 μg/ml streptomycin sulfate (Invitrogen). For transfection of 293T cells, cells were plated overnight and transfected at ∼75% confluency in 6-well plates with 1–2 μg of DNA of pcDNA3.1 HA-p73α, HA-p73β, myc-p73ϵ, myc-p73γ (a generous gift from Dr. Makoto Hijikata), or pCMV5-FLAG PTEN using 3 μl each of Lipofectamine and Plus Reagent (Invitrogen) in 1 ml of serum- and antibiotic-free Iscove's modified Dulbecco's medium overnight.
Antibodies
The antibodies used for detection were as follows: anti-p73 and PTEN polyclonal antibodies (Bethyl Laboratories); N-terminal PTEN monoclonal antibody (Abgent); p73 mouse monoclonal antibody (BD Biosciences); mouse monoclonal PTEN (A2B1), anti-Bax (2D2), anti-ARF, and anti-GST (1E5) antibodies (Santa Cruz Biotechnology); anti-FLAG (M2) monoclonal antibody and anti-PUMA C-terminal polyclonal antibody (Sigma); anti-hemagglutinin (HA) 12CA5 clone (Roche Diagnostics), and mouse monoclonal anti-His tag (EZ BioLabs Inc.).
Protein Expression and Purification
The PTEN(186–351) construct was a generous gift from Dr. C. Eng and was subcloned into pGEX4T3 vector. p73α was subcloned from a pcDNA3.1 expression plasmid into a pRSET vector to generate His-tagged p73α. Wild-type PTEN and mutant C124S were subcloned from a pCMV5 expression plasmid into pGEX4T3 to generate GST-tagged PTEN and C124S versions. All recombinant proteins were expressed and induced with isopropyl 1-thio-β-d-galactopyranoside in Escherichia coli. His-tagged proteins were purified over a 2-ml nickel-nitrilotriacetic acid column or over glutathione for GST-tagged proteins.
Immunoprecipitation, Cytoplasmic/Nuclear Fractionation, and Western Blotting
Cells for both whole cell lysates and immunoprecipitates (unless indicated) were solubilized in lysis buffer: 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 1 mm EDTA, 1 mm EGTA, 1% (octylphenoxy)polyethoxyethanol (IGEPAL), 1 mm phenylmethylsulfonyl fluoride (PMSF), 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm sodium orthovanadate (Na3VO4), and 10 mm sodium fluoride (NaF). 0.25–1 mg of protein (as indicated in the figure legends) was immunoprecipitated with either 1 μg of α-PTEN (A2B1) monoclonal antibody or epidermal growth factor (Santa Cruz Biotechnology), α-p73 monoclonal antibody (BD Biosciences), or normal mouse IgG (Calbiochem). Immunocomplexes were absorbed onto 20 μl of A/G-Sepharose beads (Santa Cruz Biotechnology) for 5 h at 4 °C. The immunoprecipitates were washed three times with 1 ml of lysis buffer and then boiled in 30 μl of 2× Laemmli buffer prior to Western blotting analysis. Cytoplasmic and nuclear extracts were made as described previously with immunoprecipitation from these extracts as described above (33).
Confocal Microscopy
H1299, BJ-ERT, and Shp53 BJ-ERT cells were fixed in 3% paraformaldehyde in PBS for 15 min, washed with PBS, and permeabilized in 1% Triton X-100 in PBS for 15 min. Slides were blocked with 5% fetal bovine serum in PBS/Tween and incubated with a rabbit polyclonal antibody to p73 (Bethyl Laboratories), followed by an anti-rabbit Cy3 secondary antibody (The Jackson Laboratory). Additionally, slides were blocked and then incubated with a rabbit polyclonal antibody to PTEN (Bethyl Laboratories) followed by an anti-rabbit Cy5 secondary antibody (The Jackson Laboratory). Slides were washed with PBS-T following primary antibody and secondary antibody incubation. Nuclei were stained with DAPI, and slides were mounted with nonfading gel-mount prior to visualization on a Zeiss multiphoton microscope.
ReporterAssay/(ChIP) Assay
The PTEN-luc reporter has been previously described, and reporter assay methods have been previously described (28). The ChIP assay to detect p73 and PTEN protein-DNA interactions in vivo were performed as described previously (34). Immunoprecipitations were performed overnight with specific antibodies α-p73 (Bethyl Laboratories) or a mixture of antibodies against PTEN as follows: α-PTEN (A2B1) (Santa Cruz Biotechnology), α-PTEN (Bethyl Laboratories), and α-PTEN (N-terminal) (Abgent) or with secondary α-mouse or α-rabbit antibodies (Santa Cruz Biotechnology) as negative controls. Complexes were then incubated with protein G-Sepharose beads (Pierce). Immunoprecipitates were washed with TE and eluted by incubation at 65 °C overnight in 1% SDS (in 0.1 m NaHCO3 at 65 °C for 4 h). DNA was purified using a PCR purification kit (Roche Diagnostics) according to the manufacturer's instructions and eluted with TE; templates were used in PCRs to detect PUMA promoter regions bound by specific proteins. PCR primer sequences used were as follows: PUMA, forward 5′-CTGTGGCCTTGTGTCTGTGAGTAC-3′ and reverse 5′-CCTAGCCCAAGGCAAGGAGGAC-3′. PCR products were resolved on a 2% agarose gel and visualized by ethidium bromide staining.
RESULTS
Increased Levels of p73 and PTEN in Response to DNA Damage
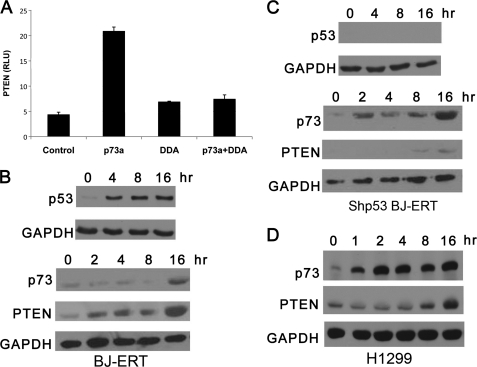
To determine whether there was an integrated pathway that connected p73 and PTEN, which was independent of p53, we transfected p73 and human PTEN promoter fused to a luciferase construct into p53-null H1299 cells. The ΔNp73α isoform has been reported to bind the PTEN promoter and negatively repress its activity in thyroid cancer cells (35). We observed that p73α robustly stimulated the human PTEN promoter (Fig. 1A). The dominant negative p73 DDA abrogated the ability of p73α to stimulate the PTEN promoter (Fig. 1A). Thus, PTEN can be induced by the TAp73α isoform. The fact that TAp73 activated the PTEN promoter and the ΔNp73 isoform repressed this activity is in agreement with these two isoforms differentially regulating promoters (35).
FIGURE 1.
p73 and PTEN induction in response to DNA damage. A, H1299 were transfected with human PTEN promoter coupled to the luciferase gene RSV-β-gal (control), p73α, or p73α-DDA. Each sample was done in triplicate and expressed as relative luciferase units (RLU). B, BJ-ERT cells were treated with 2 μm doxorubicin for the indicated times, and whole cell lysates were prepared for Western blotting. Blots were probed separately with polyclonal antibodies to p73 and PTEN and monoclonal to GAPDH for protein loading (bottom panels) or p53 (top panel). C, BJ-ERT (shp53) cells were treated and probed with antibodies as in B. D, H1299 cells were treated and probed with antibodies as in C.
To examine if p73 and PTEN protein levels change in response to DNA-damaging agents, a time course in response to doxorubicin was conducted in multiple cell lines. Normal human diploid BJ fibroblasts immortalized with E1a, H-Ras-V12, and hTERT (BJ-ERT cells) and BJ-ERT cells transduced with control ShRNA or ShRNA to p53 (Shp53 BJ-ERT) were used (31). We treated both Shp53 and ShControl BJ-ERT cell lines with 2 μm doxorubicin. The protein levels of p53 were analyzed to ensure the cells were responsive to genotoxic stress. An increase in p53 levels was observed in BJ-ERT at 4 h with levels maintained through 16 h (Fig. 1B, top panel) but not in Shp53 BJ-ERT (Fig. 1C, top panel). p73 and PTEN protein levels were increased in response to DNA damage in BJ-ERT cells (Fig. 1B, lower panel). Shp53 BJ-ERT cells had a rapid induction of p73 protein levels following doxorubicin treatment beginning at 2 h, although PTEN induction occurred at 8–16 h (Fig. 1C, lower panel). A similar response for p73 and PTEN was detected in H1299 cells, a p53 null cell line (Fig. 1D). Thus, in both immortalized fibroblasts devoid of p53 and a p53-null cancer cell line, we observed that DNA damage results in an increase in p73 and PTEN protein levels.
Because the levels of p73 and PTEN were increased, we next examined the cellular localization of p73 and PTEN in response to DNA damage. A time course of ShControl BJ-ERT cells untreated or with 2 μm doxorubicin over a 16-h time course and was used to detect p73 and PTEN by confocal microscopy. In response to DNA damage, both p73 and PTEN localized to the nucleus (Fig. 2A). Merged images revealed that p73 and PTEN co-localized at 4 h. To show that the p73/PTEN co-localization was not dependent on p53, Shp53 BJ-ERT cells were subjected to DNA damage followed by confocal imaging. Once again in response to DNA damage, p73 and PTEN were found in the nucleus, which was independent of p53 (Fig. 2B).
FIGURE 2.
p73 and PTEN co-localize in response to DNA damage. A, DNA damage in transformed fibroblast cell lines leads to co-localization of p73 and PTEN. DNA damage was induced with 2 μm doxorubicin for the indicated times in BJ-ERT fibroblasts that have wild-type p53, which were then prepared for confocal microscopy. Merged images show co-localization of p73 and PTEN in the nucleus. Nuclei are stained with DAPI. B, Shp53 BJ-ERT fibroblasts devoid of p53 were treated and prepared as described above.
Complex Formation of PTEN and p73
Because we observed both p73 and PTEN nuclear co-localization with similar kinetics, we tested if there was a direct protein-protein interaction. Purified His-p73α and GST-PTEN and a lipid phosphatase mutant, GST-C124S, were all produced from bacteria from which GST pulldown assays were performed. TAp73α was pulled down with both GST tagged PTEN and C124S but not GST (Fig. 3A). This indicates that both wild-type and lipid phosphatase mutant forms of PTEN directly bind TAp73α protein.
FIGURE 3.
p73 and PTEN form an interaction in vivo and in vitro. A, p73 and PTEN form a direct interaction in vitro. Recombinant GST-PTEN, GST-C124S (PTEN lipid phosphatase mutant), and His-p73α were expressed and purified from E. coli and used in GST-pulldown assays. Interactions were detected with anti-His, and pulldown efficiency was counter-blotted with anti-GST. B, p73 and PTEN interact in vivo. FLAG-PTEN and HA-tagged p73α or p73β were overexpressed in 293T cells (left panel), and immunoprecipitation (I.P.) with PTEN antibodies was performed. Blots were probed for HA and then stripped and re-probed with PTEN. FLAG-PTEN and Myc-tagged p73ϵ or p73γ were overexpressed (right panel) and immunoprecipitated with PTEN antibodies as described for the previous panel and then stripped and re-probed with Myc antibodies. C, schematic diagram of p73 isoforms. Specific protein domains are labeled throughout the diagram as follows: transactivation domain, DNA binding domain, oligomerization, and the C terminus. Positive results from PTEN binding to various p73 isoforms is denoted by plus symbols. D, PTEN domain mapping with p73. An in vitro GST pulldown assay was performed with GST-PTEN and GST(186–351) after addition of His-p73α. A Western blot was probed for anti-His and counter-blotted for anti-GST, and positive results of p73 interaction with PTEN are displayed as plus symbols.
We next tested if the interaction was TAp73 isoform-specific. An alignment of TAp73 isoforms α, β, ϵ, and γ are depicted (Fig. 3C). p73 shares extensive homology to p53 in their transactivation domain, DNA binding domain, and oligomerization domain. To test which p73 isoforms would interact with PTEN, we overexpressed multiple TAp73 isoforms and PTEN in 293T cells and examined binding interactions by immunoprecipitation. HA-tagged p73α and PTEN interaction was evident, and the PTEN-p73β was not (Fig. 3B, left panel). We also tested the ability of p73ϵ and p73γ isoforms and found a marginal interaction with PTEN (Fig. 3B, right panel). A summary of the various isoforms of TAp73 and binding to PTEN is depicted in Fig. 3C which illustrates that the C-terminal domain is important for binding to PTEN.
Previous reports have demonstrated that p53 and PTEN form a direct protein-protein interaction (29). To test where p73 binds PTEN, recombinant GST-PTEN and GST(186–351) were expressed and purified from E. coli and then mixed with His-p73α with a GST pulldown performed. As depicted in Fig. 3D, PTEN C-terminal domain bound to p73α. This is in agreement with full-length PTEN and the 186–351 fragment increasing p53 transactivation (36).
Based on the observation that p73 and PTEN bind directly and DNA damage appears to enhance cellular compartmentalization at a similar time course, we determined if DNA damage enhanced this interaction. To test if p73 and PTEN complex formation would increase with DNA damage, whole cell lysates from H1299 cells that were transduced with both TAp73α and PTEN and treated with cisplatin (CDDP) were prepared. Analysis of p73 protein levels increased at 8 and 16 h, although PTEN levels remained unchanged (Fig. 4A, bottom panel). When cells were treated with CDDP and p73 was immunoprecipitated, an increase in PTEN binding was observed at 8 and 16 h (Fig. 4A, top panel). Thus, the interaction of p73 and PTEN was increased upon platinum treatment because levels of both proteins were elevated after DNA damage.
FIGURE 4.
Interaction between overexpressed p73 and PTEN is enhanced with DNA damage. A, Western blot of immunoprecipitated p73 and PTEN from overexpressing cell lines. H1299 (p73/PTEN) cells were treated with 10 μm cisplatin for the indicated times. Cell extracts were immunoprecipitated (IP) with polyclonal p73 antibodies (top panel), and the blot was probed for PTEN (A2B1) and stripped and re-probed with anti-p73. Whole cell extracts (bottom panel) were probed with monoclonal p73 and PTEN (A2B1) antibodies and GAPDH for loading controls. B, cytoplasmic (C)/nuclear (N) fractionation of p73 and PTEN in response to DNA damage. H1299 (p73/PTEN) cells were treated as in A, and cytoplasmic and nuclear fractions were made from whole cell lysates. Blot was probed with monoclonal antibodies against p73 and PTEN (A2B1). GAPDH and PARP were probed for cytoplasmic and nuclear fractionation controls. C, immunoprecipitations were performed from cytoplasmic and nuclear extracts from B with anti-p73 (monoclonal) or normal mouse IgG as control. The Western blot was probed with polyclonal antibodies to PTEN (denoted by arrow) to detect co-precipitation and then stripped and re-probed with monoclonal p73 antibodies.
Next, we examined cellular fractionation on p73 and PTEN protein levels after platinum-induced DNA damage. H1299 (p73/PTEN) cells were treated with CDDP over a 16-h time course. Cytoplasmic and nuclear fractions were prepared, and a Western blot was used to analyze p73 and PTEN localization. In response to CDDP, we observed an increase in nuclear p73 levels. In addition, the overall levels of p73 levels were increased in response to DNA damage (Fig. 4B). PTEN levels did not appear to change after damage in the cytoplasmic fraction; however, a robust increase in nuclear PTEN occurred at 16 h (Fig. 4B). The data from H1299 cells is in agreement with the confocal data results in Fig. 2 from Shp53 BJ-ERT cells.
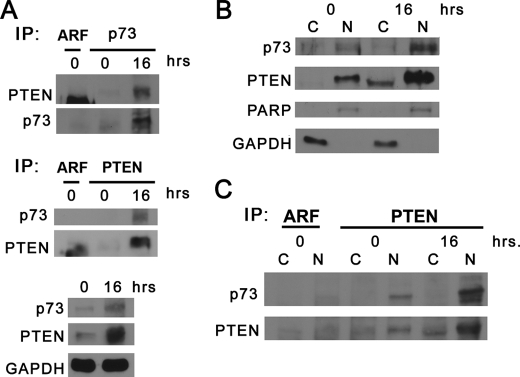
Based on our immunoprecipitation results where the interaction of p73 and PTEN was more abundant after DNA damage, we wanted to see if this occurred in the nuclear fraction. H1299 cells were used to immunoprecipitate endogenous p73 from cytoplasmic and nuclear fractions after CDDP treatment. Mock immunoprecipitation with IgG antibodies did not co-purify p73. However, PTEN was detected at 16 h after genotoxic stress, as denoted by the arrow in Fig. 4C. Nuclear p73 versus cytoplasmic p73 increased with DNA damage (Fig. 4C). PTEN was detected with p73 in all nuclear fractions compared with the cytoplasmic fractions at 16 h (Fig. 4C). To examine if endogenous p73 would co-precipitate with PTEN, whole cell extracts were prepared from Shp53 BJ-ERT cells after 16 h of CDDP treatment. Immunoprecipitation with p73 antibody led to an increased amount of PTEN co-purified with p73 after CDDP treatment (Fig. 5A, top panel). In addition, immunoprecipitation of PTEN detected endogenous p73 under DNA damage conditions with little detection of p73 with no treatment (Fig. 5A, middle panel). Counter-blotting revealed an enhanced level of PTEN protein immunoprecipitated with CDDP treatment versus control, which is in agreement with levels observed in lysates (Fig. 5A, bottom panel). These results demonstrate that an endogenous PTEN-p73 complex forms after DNA damage, which likely stems from higher levels of both proteins being present. To further define the endogenous localization of these tumor suppressors, we made cytoplasmic/nuclear extracts from H1299 cells after treatment with DNA damage. After 16 h of CDDP treatment, levels of nuclear PTEN and p73 increased compared with cytoplasmic fractions (Fig. 5B), which is in agreement with confocal microscopy and lysates from cells overexpressing p73 and PTEN in Figs. 2, A and B, and 4B, respectively. Both H1299 cytoplasmic and nuclear extracts were immunoprecipitated with PTEN or ARF (control) antibodies and then probed for reactivity to p73 (Fig. 5C). Results show a clear increase in the amount of endogenous p73 that co-precipitates with PTEN after 16 h of CDDP treatment in nuclear extracts. This immunoprecipitation with PTEN antibodies used in other experiments mirrors the results observed in Fig. 4C (immunoprecipitated p73 in overexpressed cells), where in nuclear extracts post-CDDP treatment increased the p73-PTEN complex. Thus, DNA damage levels clearly increase the levels of nuclear p73 and PTEN proteins, which forms a p73-PTEN complex.
FIGURE 5.
Endogenous p73-PTEN interactions are enhanced in the nuclear compartment in response to DNA damage. A, Western blot of immunoprecipitated (IP) endogenous p73 and PTEN in response to DNA damage. Shp53 BJ-ERT cells were treated with 20 μm cisplatin for the indicated times. Whole cell extracts were immunoprecipitated with polyclonal p73 antibodies or ARF as a control (top panel), and the blot was probed for PTEN (monoclonal) and stripped and re-probed with anti-p73. Whole cell extracts were immunoprecipitated with monoclonal PTEN antibodies or ARF for control (middle panel), and the blot was probed for p73 (polyclonal) and stripped and re-probed with anti-PTEN. Lysates (bottom panel) were probed with polyclonal p73, PTEN (A2B1) antibodies, and GAPDH. B, cytoplasmic (C)/nuclear (N) fractionation of endogenous p73 and PTEN in response to DNA damage. H1299 cells were treated as in A, and cytoplasmic and nuclear fractions were made from whole cell lysates. Blot was probed with monoclonal antibodies against p73 and PTEN (A2B1), PARP, and GAPDH. C, immunoprecipitations were performed from cytoplasmic and nuclear extracts from B with anti-PTEN (A2B1) or ARF as a control. The Western blot was probed with p73 monoclonal antibodies to detect co-precipitation and then stripped and re-probed with PTEN polyclonal antibodies for counter blot.
Induction of Apoptotic Genes by p73/PTEN
p73 will induce BAX and PUMA gene expression independent of p53 (8). To test if a p73-PTEN complex would influence BAX expression, the human BAX promoter upstream of luciferase was used. Control or H1299 cells stably overexpressing PTEN (H1299-PTEN)) were transiently transfected with RSV-βgal, BAX reporter, and either control TAp73α or p73α+DDA constructs. In Fig. 6A, overexpression of TAp73α increases BAX luciferase activity compared with the control plasmid. This activity was increased 2-fold in H1299 (PTEN) cells versus control H1299. This effect was abrogated by p73α DDA (dominant negative) for the induction of BAX in either cell line (Fig. 6A). Next we examined if Bax protein was induced in response to DNA damage. A time course with 2 μm doxorubicin treatment revealed an increase in Bax levels at 8–16 h (Fig. 6B). Also, another p73 target, PUMA, was also elevated at 8–16 h (Fig. 6B). The increase of Bax and PUMA protein levels coincides with the co-localization of p73/PTEN in the nucleus (Fig. 2, A and B) and protein complex formation (Figs. 4C and 5C).
FIGURE 6.
p73/PTEN enhances BAX apoptotic promoter activity and PUMA promoter binding. A, H1299 or cells overexpressing PTEN, H1299 (PTEN), were transfected with human BAX promoter coupled to the luciferase gene, RSV-β-gal (control), p73α, or p73α-DDA. Each sample was done in triplicate and expressed as relative luciferase units (RLU). B, H1299 cells were treated with 5 μm doxorubicin for the indicated times, and Western blots were probed with anti-Bax or PUMA antibodies and GAPDH for loading controls. C, PTEN and p73 bind the human PUMA promoter. Shp53 BJ-ERT or H1299 cells were treated with 5 μm doxorubicin for the indicated times, and then chromatin immunoprecipitation (ChIP) was performed with antibodies against p73 or PTEN as described under “Experimental Procedures.” Input was nonimmunoprecipitated (I.P.) chromosomal DNA. Bar graphs are the average of two independent experiments with no DNA control subtracted and represented as the percent change in PUMA promoter binding. A representative ChIP experiment from Shp53 BJ-ERT cells is depicted below the graphs.
Because Bax and PUMA levels are increased with the complex formation of nuclear p73 and PTEN, we next determined if p73 and PTEN were associated with the PUMA genomic promoter using a ChIP assay. p73 will bind to apoptotic promoters in response to genotoxic stress (37). However, PTEN has never been shown to be in a complex on a promoter element. We analyzed in Shp53 BJ-ERT and H1299 cancer cells. These cells were treated with 5 μm doxorubicin for 24 h followed by covalent cross-linking of proteins to DNA and immunoprecipitation of either p73 or PTEN. We observed an increase in p73 binding to the PUMA promoter after 24 h in response to DNA damage in Shp53 BJ-ERT cells (Fig. 6C, left graph). A representative ChIP assay from Shp53 BJ-ERT cells depicting both immunoprecipitations with p73 and PTEN antibodies is shown below the graphs in Fig. 6C. PTEN was also found associated with the PUMA promoter at the 24-h time point (Fig. 6C, left graph). These experiments were replicated in H1299 cells, and a similar trend was observed (Fig. 6C, right graph) with each cell line data coming from at least two independent experiments.
In response to genotoxic stress, the elevation of p73/PTEN leading to induction of Bax and PUMA would be predictive of an activated caspase cascade and consequently the initiation of apoptosis. One of the downstream targets of caspase activation is the cleavage of PARP. To test if PARP cleavage is dependent on the levels of p73 and PTEN, H1299, H1299 (p73α), H1299 (PTEN), and H1299 (p73α/PTEN) were treated with doxorubicin. A Western blot was made from whole cell lysates from the various cells lines to show overexpression of PTEN and p73 (Fig. 7A). Next, H1299 cells were treated in a dose response to find the optimal dose for PARP cleavage. A Western blot for cleaved PARP revealed that 10 μm doxorubicin was the maximal dose after 20 h (Fig. 7B). Moreover, PARP cleavage was initiated at a lower dose of 2.5 μm doxorubicin in cells overexpressing both p73/PTEN. A comparison of all cell lines was performed after treatment with 10 μm doxorubicin at both 8 and 16 h. At 8 h the p73/PTEN-overexpressing cell line showed maximal PARP cleavage that was sustained through 16 h. All cell lines showed some level of PARP cleavage at 16 h. The levels of PARP cleavage were not as robust in other cell lines (PTEN, p73, or control), which is not surprising as H1299 cells are very resistant to genotoxic stress (Fig. 7C). To test if overexpression of p73/PTEN led to an induction in Bax protein levels, cell extracts were made after 16 h of CDDP treatment or no treatment. p73 protein levels in cells overexpressing p73 alone and cells overexpressing both p73/PTEN were increased in response to CDDP (Fig. 7D). Bax levels are already elevated in cells overexpressing both p73/PTEN but not each individual tumor suppressor protein (Fig. 7D). However, in response to CDDP, the levels of Bax are increased in cells overexpressing PTEN and to the highest level in the cell line overexpressing both p73/PTEN (Fig. 7D). Thus, overexpression of both p73/PTEN leads to maximal activation of the pro-apoptotic protein Bax in cells independent of p53. To validate the role of PTEN in stimulation of apoptosis, we silenced PTEN levels in H1299 cells with ShRNA to PTEN or GFP (control). Levels of PTEN were decreased ∼90%, and levels do not increase in response to genotoxic stress. ShGFP control cells showed a predictable increase in Bax (Fig. 7E), whereas ShPTEN cells showed a decrease in Bax induction. Puma levels were not detectable in ShPTEN cells and were not induced following genotoxic stress (Fig. 7E). Thus, the presence of both p73 and PTEN tumor suppressors facilitates optimal induction of apoptotic genes after DNA damage. Our model shows a p73-PTEN interaction enhanced in the nuclear compartment (Fig. 7F). These tumor suppressors are capable of binding to and activating the BAX and PUMA promoters independent of p53.
FIGURE 7.
Cooperative induction of apoptosis through p73 and PTEN. A, H1299 and cell line derivatives overexpressing p73, PTEN, or both were lysed, and whole cell extracts were probed with p73, PTEN, and GAPDH antibodies. B, H1299, H1299 (p73α), H1299 (PTEN), and H1299 (p73α/PTEN) cell lines were treated with 0–10 μm doxorubicin (dox) as indicated for 20 h. Cells were harvested, and whole cell lysates were probed for cleaved PARP and GAPDH. C, H1299, H1299 (p73α), H1299 (PTEN), and H1299 (p73α/PTEN) cell lines were treated with 10 μm doxorubicin for 8 and 16 h, and Western blots were probed as in B. Lane C, control. D, H1299 cell line derivatives (described in C) were treated with 10 μm cisplatin for 16 h, and lysates were probed with the indicated antibodies. E, H1299 stable cell lines for ShGFP or ShPTEN were treated with 10 μm cisplatin for 16 h, and lysates were probed with the indicated antibodies. F, model for p73-PTEN interactions stimulating p53-independent apoptosis. PTEN and p73 are present in the cytoplasmic fraction and have limited binding with each other. DNA damage signals nuclear p73/PTEN levels, which lead to an enhancement in p73-PTEN complex formation. The end result is recruitment of PTEN binding to the PUMA promoter by transcriptional activity of p73, which in turn enhances BAX activity.
DISCUSSION
The responsiveness of cancer cells to chemotherapeutic agents depends on intact tumor suppressor pathways. The degree that p53 regulates apoptosis without contribution from p63 and p73 remains understudied (5, 38). Although there are clear similarities and possible overlapping functions and regulation of p53 and p73, there are distinct differences. For instance, Mdm2 represents the predominant E3 ubiquitin ligase for proteasomal degradation of p53, although p73 is not a target of Mdm2 (39). The NEDD4-like E3 ligase Itch has been demonstrated to interact with p73 and mediate its degradation through ubiquitination, yet this enzyme does not influence p53 (40). Additionally, other proteins that are known to bind p53 such as human papillomavirus E6, adenovirus E1B 55K, and simian virus 40 T do not associate with p73α and p73β isoforms (41). Although there are distinct differences in p73 and p53 protein-binding partners and how these proteins are degraded, there are functional overlaps in transcriptional gene targets. p73α and p73β have both been shown to be competent in transcription of p53-responsive genes such as BAX, GADD45, and others, but they differ in the degree of transcription between p73 isoforms and with respect to p53 (7). Thus, there are clear delineations of p73 and p53 in the manner in which they respond to DNA damage and protein binding partners.
A mouse phenotype where complete p73 isoform knock-out (Trp73−/−), including both TA and ΔN, has been performed reveals a variety of neurological and inflammatory defects. However, spontaneous tumors were not observed, which serves as a hallmark for identification of a tumor-suppressor protein (42). Recently, to delineate the differences between TAp73 and ΔNp73, separate mouse knockouts were performed. The TAp73 null mouse (TAp73−/−) was found to have a higher incidence of carcinogen-induced and spontaneous tumors along with infertility, genomic instability, and aneuploidy (43). A knock-in mouse model of the ΔNp73 isoforms (ΔNp73−/−) is sensitive to chemotherapeutic agents and has increased p53-dependent apoptosis (44). Thus, TAp73 appears to be a tumor suppressor like p53 capable of transactivating genes involved in apoptosis, whereas the ΔNp73 opposes this effect by interfering with DNA damage signaling pathways.
PTEN represents a very important arm of cellular tumor suppression by negatively regulating PI3K/Akt activity. The PTEN gene is frequently lost or mutated in human cancers affirming its role as a primary tumor suppressor (14). We observed a robust induction of PTEN protein levels in response to DNA damage in the absence of p53 (Figs. 1, C and D, and 3A). PTEN levels followed a similar time course profile for induction of protein levels to p73 with (Fig. 1, C and D). This is consistent with p73 activation in response to DNA damage and its ability to impair chemoresistance in cancer cells (45).
Our data shows that PTEN can bind both in vitro and in vivo to the p73α isoform (Fig. 3, A and B). p73α is the only isoform that contains a complete C-terminal domain that includes the sterile α-motif (Fig. 3C). The sterile α-motif domain in p73α is dispensable for membrane binding to phospholipids yet functions as an autoinhibitory domain for its transcriptional activity, which prevents the interaction with p300/CBP (46). PTEN binding to this region on p73α could compete for p73 binding to membranes and make it more transcriptionally competent by relieving this inhibition. We also observed that in cells expressing p73 and PTEN, the overall levels of p73 were elevated (Fig. 7A). This may implicate a role in PTEN in altering the stability of p73 by protecting it from ubiquitin-mediated proteolysis. Further investigation to understand this mechanism is currently underway. In addition, PTEN is known to facilitate p53 acetylation by p300 (47). Interestingly, p73 transcriptional activity can also be inhibited by mutant p53, because the mutant rather than wild-type p53 can bind to p73 (7). The selective pressure of cancer cells to favor mutant p53 presents an interesting problem for determining how to activate p73. Therefore, there must be other mechanisms present to alleviate the negative repression on p73 from mutant p53.
Previous reports have hinted that a p73-PTEN interaction may exist. 1) A PTEN-regulated pathway, mammalian target of rapamycin in the PI3K signaling pathway, is an upstream regulator of p73 (48). 2) ΔNp73 has shown to inhibit PTEN expression in thyroid cancer cells (35). We have demonstrated that TAp73α can stimulate the PTEN promoter, which can be inhibited with the addition of the p73 DDA construct (Fig. 1A), which is in agreement with Ref. 35. An additional luciferase reporter assay was performed which showed PTEN overexpression with p73α enhanced BAX transcription (Fig. 6A). Our novel finding of PTEN (direct or indirect DNA binding) on an apoptotic promoter further validates its role as a tumor suppressor, especially in cells devoid of p53 (Fig. 6C).
Opposing the tumor suppressor functions of p73 and p53 is the murine double minute (Mdm2) oncoprotein. Additionally, there is further interplay with Mdm2 in conjunction with PTEN and p53 which is reviewed in Ref. 16. A strong link between signaling in the PI3K pathway and Mdm2 has been demonstrated. PTEN impedes Akt activity to promote Mdm2 nuclear entry through phosphorylation of serine 166 and 186 on Mdm2 (26). This allows for cytoplasmic retention and destabilization of Mdm2, thereby keeping p53 active. Furthermore, a functional connectivity between PTEN and p53 has been established, whereby cancer cells are more sensitive to DNA-damaging agents when both tumor suppressors are present (27). Perhaps this is the same case for p73 and PTEN in the absence of p53. In agreement is our observation that DNA damage increases the levels of both p73 and PTEN protein. From our co-precipitation experiments utilizing both overexpressed and endogenous p73 and PTEN, we detected enhanced interactions under conditions of DNA damage (Figs. 4, A and C, and 5, A, C, and D). The p73-PTEN interaction is substantially increased in endogenous nuclear fractions after DNA damage (Fig. 5C). However, our experiments are not able to discern if the enhanced p73-PTEN nuclear protein interaction is directly stimulated by DNA damage or if this is merely a secondary effect due to the induction of both proteins after damage. Some data links PTEN nucleoplasmic shuttling to the major vault protein through Ca+2 signaling (49). Precisely, how PTEN nuclear levels would increase after DNA damage and required upstream signaling events clearly warrants further investigation.
In support of our observation that a p73-PTEN protein complex acts as a co-activator of apoptosis, is the observation that cells overexpressing both tumor suppressors induce an overall higher level of PARP cleavage in cells (Fig. 7B). Furthermore, overexpression of both p73/PTEN leads to the greatest level of induction of Bax (Fig. 7D), whereas silencing PTEN completely reduces levels of PUMA and substantially decreases Bax in the background of p73 activation by DNA damage (Fig. 7E). These results provide interesting evidence to the degree of the importance of PTEN in activating downstream apoptotic targets in response to p73 activation after DNA damage. These experiments substantiate our observations that the p73 and PTEN tumor suppressors interact together to enhance apoptosis. Mdm2 expression is also regulated by PTEN independent of p53. Cells null for PTEN expressed higher levels of the Mdm2 P1 promoter, and consequently PTEN through its lipid phosphatase activity negatively regulates expression of the p90 Mdm2 (50). Interestingly, a p53-PTEN-Mdm2 complex could not be co-precipitated from cells (29). Therefore, a fine-tuning mechanism between PTEN, p53, p73, and Mdm2 is in place in human cells to mediate cellular apoptosis in response to DNA damage. To this end, Mdm2 represents a viable target for drug design and chemotherapeutic intervention (51–53).
Thus, our data reveal an important role for nuclear PTEN in facilitating p73 binding to apoptotic promoters and inducing their expression. This is the first demonstration that PTEN can be found on an apoptotic promoter, which further substantiates its role as a tumor suppressor. Given the high incidence of tumors with mutant or null p53, stimulation of the p73/PTEN axis to enhance apoptosis is an evolving area for further investigation.
This work was supported, in whole or in part, by National Institutes of Health Grant CA109262 from NCI (to L. D. M.) and NRSA T32 CA 111198 (to J. A. L.).
- PTEN
- phosphatase and tensin homolog
- PARP
- poly(ADP-ribose) polymerase
- ARF
- ADP ribosylation factor
- CDDP
- cisplatin
- DDA
- dominant negative.
REFERENCES
- 1. Li Y., Prives C. (2007) Oncogene 26, 2220–2225 [DOI] [PubMed] [Google Scholar]
- 2. Deyoung M. P., Ellisen L. W. (2007) Oncogene 26, 5169–5183 [DOI] [PubMed] [Google Scholar]
- 3. Tomasini R., Mak T. W., Melino G. (2008) Trends Cell Biol. 18, 244–252 [DOI] [PubMed] [Google Scholar]
- 4. Rosenbluth J. M., Pietenpol J. A. (2008) Genes Dev. 22, 2591–2595 [DOI] [PubMed] [Google Scholar]
- 5. Flores E. R., Tsai K. Y., Crowley D., Sengupta S., Yang A., McKeon F., Jacks T. (2002) Nature 416, 560–564 [DOI] [PubMed] [Google Scholar]
- 6. Dötsch V., Bernassola F., Coutandin D., Candi E., Melino G. (2010) Cold Spring Harbor Perspect. Biol. 2, a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Como C. J., Gaiddon C., Prives C. (1999) Mol. Cell. Biol. 19, 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Melino G., Bernassola F., Ranalli M., Yee K., Zong W. X., Corazzari M., Knight R. A., Green D. R., Thompson C., Vousden K. H. (2004) J. Biol. Chem. 279, 8076–8083 [DOI] [PubMed] [Google Scholar]
- 9. Sayan B. S., Yang A. L., Conforti F., Tucci P., Piro M. C., Browne G. J., Agostini M., Bernardini S., Knight R. A., Mak T. W., Melino G. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 12877–12882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Orloff M. S., Eng C. (2008) Oncogene 27, 5387–5397 [DOI] [PubMed] [Google Scholar]
- 11. Leslie N. R., Batty I. H., Maccario H., Davidson L., Downes C. P. (2008) Oncogene 27, 5464–5476 [DOI] [PubMed] [Google Scholar]
- 12. Yin Y., Shen W. H. (2008) Oncogene 27, 5443–5453 [DOI] [PubMed] [Google Scholar]
- 13. Wang X., Jiang X. (2008) Oncogene 27, 5454–5463 [DOI] [PubMed] [Google Scholar]
- 14. Keniry M., Parsons R. (2008) Oncogene 27, 5477–5485 [DOI] [PubMed] [Google Scholar]
- 15. Carracedo A., Pandolfi P. P. (2008) Oncogene 27, 5527–5541 [DOI] [PubMed] [Google Scholar]
- 16. Mayo L. D., Donner D. B. (2002) Trends Biochem. Sci. 27, 462–467 [DOI] [PubMed] [Google Scholar]
- 17. Stambolic V., Suzuki A., de la Pompa J. L., Brothers G. M., Mirtsos C., Sasaki T., Ruland J., Penninger J. M., Siderovski D. P., Mak T. W. (1998) Cell 95, 29–39 [DOI] [PubMed] [Google Scholar]
- 18. Maehama T., Dixon J. E. (1998) J. Biol. Chem. 273, 13375–13378 [DOI] [PubMed] [Google Scholar]
- 19. Myers M. P., Pass I., Batty I. H., Van der Kaay J., Stolarov J. P., Hemmings B. A., Wigler M. H., Downes C. P., Tonks N. K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13513–13518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leslie N. R., Maccario H., Spinelli L., Davidson L. (2009) Adv. Enzyme Regul. 49, 190–196 [DOI] [PubMed] [Google Scholar]
- 21. Sun H., Lesche R., Li D. M., Liliental J., Zhang H., Gao J., Gavrilova N., Mueller B., Liu X., Wu H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 6199–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tamura M., Gu J., Matsumoto K., Aota S., Parsons R., Yamada K. M. (1998) Science 280, 1614–1617 [DOI] [PubMed] [Google Scholar]
- 23. Ramaswamy S., Nakamura N., Vazquez F., Batt D. B., Perera S., Roberts T. M., Sellers W. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li D. M., Sun H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stambolic V., MacPherson D., Sas D., Lin Y., Snow B., Jang Y., Benchimol S., Mak T. W. (2001) Mol. Cell 8, 317–325 [DOI] [PubMed] [Google Scholar]
- 26. Mayo L. D., Donner D. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mayo L. D., Dixon J. E., Durden D. L., Tonks N. K., Donner D. B. (2002) J. Biol. Chem. 277, 5484–5489 [DOI] [PubMed] [Google Scholar]
- 28. Mayo L. D., Seo Y. R., Jackson M. W., Smith M. L., Rivera Guzman J., Korgaonkar C. K., Donner D. B. (2005) J. Biol. Chem. 280, 25953–25959 [DOI] [PubMed] [Google Scholar]
- 29. Freeman D. J., Li A. G., Wei G., Li H. H., Kertesz N., Lesche R., Whale A. D., Martinez-Diaz H., Rozengurt N., Cardiff R. D., Liu X., Wu H. (2003) Cancer Cell 3, 117–130 [DOI] [PubMed] [Google Scholar]
- 30. Planchon S. M., Waite K. A., Eng C. (2008) J. Cell Sci. 121, 249–253 [DOI] [PubMed] [Google Scholar]
- 31. Cipriano R., Patton J. T., Mayo L. D., Jackson M. W. (2010) Cell Cycle 7, 1373–1379 [DOI] [PubMed] [Google Scholar]
- 32. Patton J. T., Mayo L. D., Singhi A. D., Gudkov A. V., Stark G. R., Jackson M. W. (2006) Cancer Res. 66, 3169–3176 [DOI] [PubMed] [Google Scholar]
- 33. Jackson M. W., Patt L. E., LaRusch G. A., Donner D. B., Stark G. R., Mayo L. D. (2006) J. Biol. Chem. 281, 16814–16820 [DOI] [PubMed] [Google Scholar]
- 34. Araki S., Eitel J. A., Batuello C. N., Bijangi-Vishehsaraei K., Xie X. J., Danielpour D., Pollok K. E., Boothman D. A., Mayo L. D. (2010) J. Clin. Invest. 120, 290–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vella V., Puppin C., Damante G., Vigneri R., Sanfilippo M., Vigneri P., Tell G., Frasca F. (2009) Int. J. Cancer 124, 2539–2548 [DOI] [PubMed] [Google Scholar]
- 36. Tang Y., Eng C. (2006) Cancer Res. 66, 736–742 [DOI] [PubMed] [Google Scholar]
- 37. Mantovani F., Piazza S., Gostissa M., Strano S., Zacchi P., Mantovani R., Blandino G., Del Sal G. (2004) Mol. Cell 14, 625–636 [DOI] [PubMed] [Google Scholar]
- 38. Flores E. R., Sengupta S., Miller J. B., Newman J. J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. (2005) Cancer Cell 7, 363–373 [DOI] [PubMed] [Google Scholar]
- 39. Bálint E., Bates S., Vousden K. H. (1999) Oncogene 18, 3923–3929 [DOI] [PubMed] [Google Scholar]
- 40. Rossi M., De Laurenzi V., Munarriz E., Green D. R., Liu Y. C., Vousden K. H., Cesareni G., Melino G. (2005) EMBO J. 24, 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Marin M. C., Jost C. A., Irwin M. S., DeCaprio J. A., Caput D., Kaelin W. G., Jr. (1998) Mol. Cell. Biol. 18, 6316–6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang A., Walker N., Bronson R., Kaghad M., Oosterwegel M., Bonnin J., Vagner C., Bonnet H., Dikkes P., Sharpe A., McKeon F., Caput D. (2000) Nature 404, 99–103 [DOI] [PubMed] [Google Scholar]
- 43. Tomasini R., Tsuchihara K., Wilhelm M., Fujitani M., Rufini A., Cheung C. C., Khan F., Itie-Youten A., Wakeham A., Tsao M. S., Iovanna J. L., Squire J., Jurisica I., Kaplan D., Melino G., Jurisicova A., Mak T. W. (2008) Genes Dev. 22, 2677–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wilhelm M. T., Rufini A., Wetzel M. K., Tsuchihara K., Inoue S., Tomasini R., Itie-Youten A., Wakeham A., Arsenian-Henriksson M., Melino G., Kaplan D. R., Miller F. D., Mak T. W. (2010) Genes Dev. 24, 549–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Irwin M. S., Kondo K., Marin M. C., Cheng L. S., Hahn W. C., Kaelin W. G., Jr. (2003) Cancer Cell 3, 403–410 [DOI] [PubMed] [Google Scholar]
- 46. Liu G., Chen X. (2005) J. Biol. Chem. 280, 20111–20119 [DOI] [PubMed] [Google Scholar]
- 47. Li A. G., Piluso L. G., Cai X., Wei G., Sellers W. R., Liu X. (2006) Mol. Cell 23, 575–587 [DOI] [PubMed] [Google Scholar]
- 48. Rosenbluth J. M., Mays D. J., Pino M. F., Tang L. J., Pietenpol J. A. (2008) Mol. Cell. Biol. 28, 5951–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Minaguchi T., Waite K. A., Eng C. (2006) Cancer Res. 66, 11677–11682 [DOI] [PubMed] [Google Scholar]
- 50. Chang C. J., Freeman D. J., Wu H. (2004) J. Biol. Chem. 279, 29841–29848 [DOI] [PubMed] [Google Scholar]
- 51. Lehman J. A., Eitel J. A., Batuello C. N., Mayo L. D. (2008) Expert Opin. Drug Discov. 3, 1309–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Waning D. L., Lehman J. A., Batuello C. N., Mayo L. D. (2010) Pharmaceuticals 3, 1576–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marine J. C., Lozano G. (2010) Cell Death Differ. 17, 93–102 [DOI] [PubMed] [Google Scholar]