Abstract
Medicago truncatula is a model for investigating legume genetics, including the genetics and evolution of legume–rhizobia symbiosis. We used whole-genome sequence data to identify and characterize sequence polymorphisms and linkage disequilibrium (LD) in a diverse collection of 26 M. truncatula accessions. Our analyses reveal that M. truncatula harbors both higher diversity and less LD than soybean (Glycine max) and exhibits patterns of LD and recombination similar to Arabidopsis thaliana. The population-scaled recombination rate is approximately one-third of the mutation rate, consistent with expectations for a species with a high selfing rate. Linkage disequilibrium, however, is not extensive, and therefore, the low recombination rate is likely not a major constraint to adaptation. Nucleotide diversity in 100-kb windows was negatively correlated with gene density, which is expected if diversity is shaped by selection acting against slightly deleterious mutations. Among putative coding regions, members of four gene families harbor significantly higher diversity than the genome-wide average. Three of these families are involved in resistance against pathogens; one of these families, the nodule-specific, cysteine-rich gene family, is specific to the galegoid legumes and is involved in control of rhizobial differentiation. The more than 3 million SNPs that we detected, approximately one-half of which are present in more than one accession, are a valuable resource for genome-wide association mapping of genes responsible for phenotypic diversity in legumes, especially traits associated with symbiosis and nodulation.
Keywords: association genetics, population genomics, selection scan, haplotype map
Legumes comprise a highly diverse plant family that is the second most important crop family in the world. Among cultivated plants, legumes are unique in their ability to fix atmospheric nitrogen through their symbiotic relationship with rhizobia bacteria. Symbiotic nitrogen fixation contributes nearly 90 billion kg nitrogen/y to the global ecosystem (1). Because legumes are not limited for nitrogen, they have remarkably high levels of protein, a property that is both biologically and agriculturally significant. Nearly 33% of all human nutritional requirement for nitrogen comes from legumes, and in many developing countries, legumes serve as the most important source of protein for people and livestock (2).
Medicago truncatula, a diploid, predominantly self-fertilizing close relative of alfalfa (M. sativa), serves as a model for investigating the genetics and evolution of legume–rhizobia symbiosis (3–5), legume genetics, and genome evolution (6) as well as the genetics and evolution of plant–mycorrhizal symbiosis (7), a symbiosis that is common among land plants but not found in the primary plant genetic model, Arabidopsis thaliana. The utility of M. truncatula as a model is built on a modest genome size of about 500 million bp (Mbp) (6), short seed to seed generation time (3–4 mo), excellent collections of tagged mutants (8), and large collections of diverse ecotypes (9). Moreover, a BAC-based, high-quality genome sequence for M. truncatula covering most of its euchromatin has recently become available (www.medicago.org).
In addition to facilitating gene discovery and comparative genomics, the M. truncatula reference genome enables alignment of genome-scale sequencing using next generation approaches, allowing for genome-scale analyses of nucleotide diversity as well as inferences on the evolutionary and demographic forces that shape that diversity (10–18). In particular, the scale of sampling achieved by whole-genome sequencing allows for robust descriptions of how nucleotide diversity varies along chromosomes, the importance of both background and positive selection in shaping that diversity, the extent of linkage disequilibrium or evolutionary independence of genes in different chromosomal regions, and the relative importance of recombination and mutation in introducing variation.
In addition to the insights that can be gained into the evolutionary forces that shape genomic diversity, whole-genome sequence data from a population sample allow for the development of tools needed for genome-wide association studies (GWAS). The use of GWAS for identifying genetic variants in complex traits remains challenging, especially in humans (19, 20). However, in plant species (for which phenotypic data can be collected in highly replicated experiments with low environmental variation), both candidate gene and GWAS seem to be a powerful approach for identifying genes underlying naturally occurring variation [e.g., maize (21), A. thaliana (22), and rice (23)].
We used Illumina next generation DNA sequencing technology to sequence 26 M. truncatula accessions to ∼15× average mapped coverage. We used these data to characterize genome-wide patterns of nucleotide diversity, recombination, and linkage disequilibrium and identify genomic regions that may have evolved in response to recent and strong bouts of selection. These data also yield in excess of 3 million SNPs that will be a robust foundation for future SNP-based GWAS of phenotypic diversity in legumes, especially traits associated with symbiosis and nodulation.
Results and Discussion
Diversity.
We aligned to the reference genome (A17) an average of 82 million 90-base paired end reads (∼32× coverage) from each of the 26 M. truncatula lines. Approximately 50% of these reads aligned to a unique position in the reference genome and were used for SNP calling (mean unique coverage was ∼15×). The distributions of both aligned and uniquely aligned reads were, however, skewed to lower coverage; the among-line average of median coverage was 9.0, and the median unique coverage was 7.9 (Table S1).
Focusing on regions covered by reads in at least 20 of 26 genomes, we can confidently probe 53% of the 257 Mbp comprising the reference M. truncatula genome (ignoring gaps). We detected 3,063,923 SNPs resulting in genome-wide estimates of nucleotide diversity (θw = 0.0063 and θπ = 0.0043 bp−1) (Table 1), approximately three times more diversity than is found in genome-scale estimates of diversity in the economically important legume Glycine max (θW cultivated = 0.0017 bp−1 and θW wild = 0.0023 bp−1) (24). SNPs bp−1 were approximately three times more frequent at synonymous sites in coding regions than at replacement sites (θWREP:θWSYN = 0.41 and θπREP:θπSYN = 0.39) (Fig. 1 and Table 1). Both lower diversity and a higher proportion of low-frequency segregating SNPs at replacement than synonymous sites (Fig. 2) are consistent with the expectation of stronger purifying selection acting at replacement sites (25).
Table 1.
Coverage and diversity statistics by nucleotide class
| Covered bases (Mbp) | Total bases (%) | Polymorphic sites | π bp−1 | θw bp−1 | |
| Total | 138 | — | 3,063,923 | 0.0043 | 0.0063 |
| Coding | 28.1 | 0.20 | 447,496 | 0.0032 | 0.0044 |
| Synonymous | 4.85 | 0.04 | 168,529 | 0.0072 | 0.0093 |
| Replacement | 18.4 | 0.13 | 264,903 | 0.0028 | 0.0038 |
| Introns | 34.2 | 0.25 | 656,266 | 0.0038 | 0.0054 |
| Intergenic | 66.4 | 0.48 | 1,797,133 | 0.0053 | 0.0077 |
| UTR 5′ | 4.55 | 0.03 | 73,824 | 0.0030 | 0.0046 |
| UTR 3′ | 4.82 | 0.03 | 89,204 | 0.0035 | 0.0052 |
Synonymous and replacement data are presented only for genes for which there are data from >80% of coding regions from ≥20 accessions. UTRs of genes 5′ and 3′ of the coding sequence are designated UTR 5′ and UTR 3′, respectively.
Fig. 1.
Distributions of nucleotide diversity (θW) found in 100-kb sliding windows and replacement (gray) and synonymous (red) sites among 30,768 gene models.
Fig. 2.
MAF at replacement, synonymous, intergenic, intron, and UTR sites.
The minor allele frequency (MAF) spectrum of polymorphic sites (Fig. 2) shows that the frequencies of rare variants (present in only one accession) were similar in introns, intergenic regions, and replacement sites, with rare polymorphisms more frequent at each of these three classes than at synonymous sites. This pattern is similar to that found in A. thaliana (18). Moreover, nucleotide diversity was higher at synonymous sites in coding regions than in either introns or intergenic regions (Table 1), similar to patterns recently reported for A. thaliana (26), Populus balsamifera (27), and Drosophila melanogaster (28). These findings are not consistent with the traditional view that intergenic, intron, and synonymous sites are all equally selectively neutral, but rather, they suggest that introns and intergenic regions may experience stronger selective constraints than synonymous coding sites (29). We caution, however, that higher synonymous than intergenic and intron diversity may be an artifact of the difficulties in aligning noncoding regions—coding regions with highly diverse synonymous sites will be easier to align to the reference genome, because the highly diverse synonymous sites are interspersed with less variable replacement sites. Regardless, similar MAF spectrums at intergenic and replacement sites should not be viewed as evidence of these sites experiencing equal selective constraint; SNPs were far less frequent at replacement than intergenic sites.
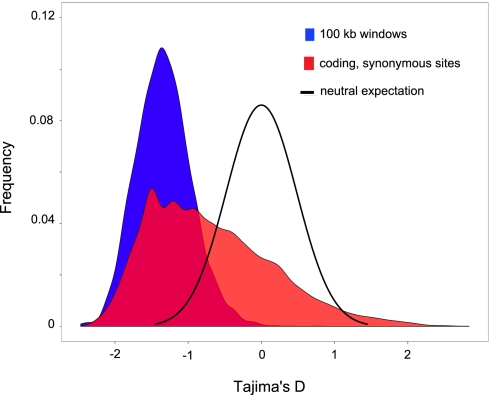
Low-frequency SNPs are much more common in M. truncatula than expected under a standard neutral model (SNM), which was reflected in strongly negatively skewed distributions of Tajima's D (DT) for both sliding windows and synonymous sites in putative coding regions (mean DT = −1.34 and mean DT = −0.95, respectively) (Fig. 3). The skewed distributions may reflect a recent population expansion (3); however, sequencing error as well as population structure also may contribute to that pattern. Because we were interested in capturing species-wide diversity, we sampled a single individual from multiple subpopulations throughout a large part of the species range. This sampling scheme is similar to the approach used in initial genome-wide surveys in A. thaliana (11), maize (13), and soybean (24). Sampling a single individual from multiple equally related subpopulations is not expected to cause the SNP MAFS to deviate from expectations of an SNM (30). However, most plant species, including M. truncatula (31, 32), likely are comprised of subpopulations of unequal relatedness, and sampling a single individual from multiple unequally related subpopulations can result in SNP frequency distributions that deviate considerably from SNM expectations (30, 33, 34).
Fig. 3.
Distributions of Tajima's D statistic for 100-kb sliding windows (blue) and synonymous sites found in the 23,468 gene models with more than or equal to two segregating sites. The black line shows the expected distribution of D with no selection in a panmictic population of constant size.
Selection Candidates.
In the absence of strong selection shaping diversity, we would expect not to find contiguous windows exhibiting either low diversity or an excess of low-frequency variants (low DT). By contrast, we detected three chromosomal regions where contiguous 100-kb windows harbor low diversity (lowest 1% of empirical distribution), including four contiguous windows of low diversity at a telomeric end of chromosome 1 (base pairs 1–400,000) (Tables S2 and S3). If windows are independent, the probability of finding four contiguous windows by chance is extremely low (P < 1 × 10−8). Four regions contained two or more contiguous windows with DT estimates that were among the most negative 1%, including three windows at a telomeric end of chromosome 8, two windows on chromosome 5, and two pairs of contiguous windows on chromosome 3 (Tables S2 and S3). One of the pairs of windows on chromosome 3 was embedded within five contiguous windows with DT values that were among the lowest 2% of genome-wide values.
The large regions of low diversity (θW) or very negative DT are obvious a posteriori regions to search for targets of recent species-wide selective sweeps. With a couple of exceptions of genes that may be involved in defense against pathogens [a gene with an NB-ARC (nucleotide binding adaptor shared by APAF-1, R proteins, and CED-4) domain located on the top of chromosome 1 and a leucine-rich repeat (LRR) located in a window of chromosome 3—both in windows that are among the lowest in diversity and DT] (Dataset S1), these regions do not harbor identified genes with putative functions (e.g., pathogen defense) that make them obvious targets of strong selection. However, one of seven windows that fell into the lowest 1% of the distribution of both DT and θW contains an early nodulin gene (ENOD93) (5), suggesting that this episode of selection may have been associated with host–rhizobia interactions. Genes previously identified as targets of selection in M. truncatula, including DMI1, a nodulation-related gene (3), as well as genes identified as possible targets of adaptation to saline environments in Tunisian populations of M. truncatula (35) were not located in chromosomal regions harboring unusual levels of diversity or skewed frequency distributions (i.e., DT) in our sample. The lack of correspondence between this study and previous studies may not be surprising—the studies by De Mita et al. (3) and Friesen et al. (35) both sampled from geographically restricted locations, whereas our range-wide sample may be powerful for detecting species-wide sweeps but poorly suited for identifying genes involved in local adaptation to biotic or abiotic conditions.
Correlates of Diversity.
Nucleotide diversity (θw silent) decreased from centromeric to telomeric regions of the euchromatin-rich reference genome (Fig. 4 and Table 2), with the distance from the centromere accounting for ∼13% of the variance in θw silent. The strength of the correlation, however, differed significantly among chromosomes, with chromosomal position accounting for ∼5% of the variance on chromosome 7 and >40% of the variance on chromosome 2. Negative correlations between nucleotide diversity and distance from the centromere are also seen in A. thaliana (11). By contrast, nucleotide diversity increases with increasing distance from the centromeric regions in Zea mays (13), D. simulans (10), and humans (36). In M. truncatula, we also do not see noticeable reductions in diversity or recombination (Fig. 4) most close to and far from the centromeres, which is seen in Z. mays (13) and Drosophila (10). Not finding reduced diversity or recombination near the centromeres and telomeres may be related to heterochromatic regions that are missing from the M. truncatula reference genome and may not reflect fundamental differences in the forces shaping nucleotide diversity.
Fig. 4.
Sliding window analyses from chromosome 5 showing (A) the relationship between total polymorphism (blue, θW) and population-scaled recombination rate (red, ρ), (B) the ratio of ρ to θW (dashed line marks a ratio of one), and (C) gene density. The location of the centromere is marked by the vertical black (A) or red line (B and C). Data for other chromosomes are in Fig. S1.
Table 2.
Correlations between genome-wide estimates (from 2,533 100-kb windows) of silent nucleotide diversity (θW silent), map-based recombination rate (R), population-scaled recombination rate (ρ), gene density (proportion of the window containing the coding sequence), and distance from centromere (proportional distance to the tip of each chromosome arm)
| Raw correlations | θW silent | R | ρ | Gene density | Distance from centromere |
| θW silent | — | 0.18 (P < 0.05) | 0.44 (P < 0.01) | −0.24 (P < 0.01) | −0.36 (P < 0.01) |
| R | — | 0.13 (P = 0.06) | −0.11 (P = 0.12) | −0.27 (P = 0.3) | |
| ρ | — | −0.17 (P < 0.02) | −0.29 (P = 0.14) | ||
| Gene density | — | 0.18 (P < 0.05) | |||
| Distance from centromere | — |
To account for the spatial autocorrelation of 100-kb windows, significance (P values; shown in parentheses) was evaluated using a permutation test that kept the linear order of estimates intact.
Nucleotide diversity (θw silent) was also negatively correlated with gene density estimated through either physical distance (r = −0.24) (Table 2) or the proportion of genic regions per centimorgan, (r = −0.22) and positively correlated with map-based estimates of recombination R (Table 2). The negative correlation between θw silent and gene density is consistent with either background selection or genetic hitchhiking with sites that have experienced selective sweeps (37–39). Similar to A. thaliana, two aspects of the M. truncatula data suggest that the negative correlation between diversity and gene density is more likely because of background selection than hitchhiking; selective sweeps are expected to cause negative values of DT (40), but we find no correlation between DT and gene density (r = 0.01) and M. truncatula harbors a significant load of deleterious mutations, which is reflected in the excess of rare replacement relative to synonymous mutations (Fig. 2) (39).
Nucleotide diversity of coding regions differed significantly among both gene annotation classes (Fig. S2) and groups of genes that share similar protein domains (Fig. 5). Among annotation groups, genes supported by full-length or expressed sequence matches (18,926 gene models covering >70% of putative gene length; θWSYN = 0.010) harbored <70% of diversity found at genes identified on the basis of protein homology (8,855 gene models; θWSYN = 0.014) or ab initio or low-confidence gene calls (2,987 gene models; θWSYN = 0.015). The higher diversity at homology-based as well as low-confidence gene calls may reflect weaker selective constraints on genes that are expressed either infrequently or at lower levels (and thus, not detected among either full-length or expressed genes), pseudogenes, or annotation error.
Fig. 5.
Average replacement and synonymous site diversity (θW bp−1) for the 51 gene families represented by ≥50 members (red) and 1,000 randomly selected groups of 50 genes (gray) selected from all gene models that had been assigned as a member of a gene family. The four groups harboring greater diversity than the highest 2.5% of resampled groups are labeled.
Among putative functional groups, four classes of genes—toll interleukin repeat, NB-ARC, LRR, and nodule-specific cysteine rich—harbor significantly higher replacement and synonymous site diversity relative to other gene families (Fig. 5). The members of three of these gene families (toll interleukin repeat, NB-ARC, and LRR) play well-established roles in the activation of the resistance response against pathogens. By contrast, the nodule-specific cysteine rich gene family, which is found only in the galegoid lineage of legumes, contains members with direct antimicrobial properties as well as members involved in controlling the terminal differentiation of the nitrogen-fixing rhizobial bacteroids inside of nodules (41). High average diversity of the members of large, defense-related gene families, which has also been found in genome-wide surveys of A. thaliana (11, 42), likely reflects both frequency-dependent selection favoring rare alleles (43) as well as relaxed selective constraint acting on nonfunctional gene copies (44).
Recombination and Linkage Disequilibrium.
M. truncatula is a predominantly selfing species, and thus, we expected to find extended linkage disequilibrium and low rates of effective recombination. We found that, within our broad geographic sample, mean r2 between pairs of SNPs fell to approximately one-half of the initial value within ∼3 kb and <0.3 within ∼5 kb, although linkage disequilibrium (LD) can be extremely variable and estimates of LD span the entire range of values (i.e., from absence of to complete LD) from ∼1- to 10-kb distances (Fig. 6). Moreover, ∼65% of the more than 1 million SNPs present at frequency >0.2 are not in complete LD with an adjacent SNP. The population-scaled recombination rate, ρ = 4Ner (where Ne is the effective population size and r is the effective recombination rate), calculated on 100-kb windows varied from 0.05 to 32 kb−1 with a genome-wide average of ρ = 1.8 kb−1. Genome-wide, the ratio of population recombination rate to the effective mutation rate (ρ/θ) is equal to 0.29 (Fig. 4), indicating that mutations occur approximately three to four times more frequently than recombination events. These estimates of the relative importance of recombination and mutation are for a range-wide sample. Because of the high selfing rate in M. truncatula, the LD within local populations may be more extensive than the LD found in our range-wide sample; as such, the relative importance of recombination to mutation in generating diversity may be lower within local populations. Moreover, the extent of LD within local populations may be more important than range-wide LD when considering the effects of linkage on the efficacy of selection.
Fig. 6.
Mean LD decay (red line) as measured by pairwise r2, with the 50% and 90% ranges of values shown in light and dark gray, respectively.
We find two genome regions, each of approximately 0.5 Mbp, with contiguous 100-kb windows with very low levels of estimated recombination (ρ). Chromosome 3 contains a region from 42.6 to 43.3 Mbp comprised of seven contiguous windows with estimates of ρ that are among the lowest 5%, including three contiguous windows in the lowest 0.5%. Chromosome 1 (32.3–32.9 Mbp) contains six contiguous windows in the lowest 2.5% of genome-wide estimates. These two regions of very low recombination may contain chromosomal inversions or perhaps large insertions or other structural polymorphisms segregating within M. truncatula.
The population-scaled recombination rate estimated on 100-kb windows is only weakly correlated with the map-based recombination estimates (r = 0.13) (Table 2). This weak correlation may be because of population structure, double cross-over events not captured by the distantly spaced markers used to construct the genetic map, changes in recombination through time, or genetic distance between lines used to generate the genetic map. The line used to generate the M. truncatula reference genome and used as a parent for the map-based recombination rate estimates has a large-scale rearrangement involving chromosomes 4 and 8 (45). This rearrangement does not, however, seem to explain the lack of a strong correlation between population- and map-based estimates of recombination; all correlations were similar when these two chromosomes were removed from the dataset.
At the genome scale, recombination and LD of M. truncatula look very similar to recombination and LD of A. thaliana, in which r2 among a diverse set of genotypes drops to less than one-half of its initial value in 3–4 kb, LD blocks are short, approximately one-third of SNPs are not in LD with an adjacent SNP, ρ = 0.8/kb, and ρ/θ = 0.05 (18, 39). The ratios of recombination to diversity in both M. truncatula and A. thaliana are consistent with the expectation that the evolutionary transition from outcrossing to selfing will have much greater effects on recombination than mutation (46). Neither these genomes nor estimates from the highly selfing wild barley (47), however, are consistent with the hypothesis that selfing species have extensive LD that would act as a major constraint to adaptive evolution (48). By contrast, domesticated selfing species, including indica rice (Oryza sativa ssp. inidica) and soybean (G. max), show LD extending >50 kb (24, 49). The difference between domesticated compared with nondomesticated selfing taxa suggests that the bottleneck that accompanied domestication may contribute strongly to the extensive LD found in these taxa.
Implications for GWAS.
Based on our analyses and the current costs of whole-genome resequencing, a tagged SNP approach for conducting GWAS to identify genetic variants responsible for naturally occurring phenotypic variation does not provide a clear advantage over whole-genome resequencing. In particular, a tag SNP approach designed to assay all common SNPs (MAF ≥ 0.2) detected in our survey would require more than 800,000 tag SNPs (i.e., the number of SNPs not in complete LD with an adjacent SNP plus the number of complete LD blocks). Moreover such a strategy would entail substantial ascertainment bias and impede assaying low-frequency SNPs, which may increase the probability of identifying potentially misleading synthetic associations (19) while decreasing the power to correctly identify causal variants and characterize the genetic architecture of complex traits.
Methods
Data Collection.
We sequenced 26 M. truncatula accessions sampled from geographically distinct populations (Fig. S3) that were chosen, because they capture the range of simple-sequence repeat (SSR) diversity found among naturally occurring lines (9) or are parents of biparental recombinant inbred line (RIL) mapping populations (Table S1). Each accession was self-fertilized for a minimum of three generations before growing seedlings for DNA extraction. Total DNA was extracted from a pool of ∼30-d-old dark-grown seedlings using a modified CTAB extraction.
Alignments and SNP discovery described here are based on the Mt3.0 version of the M. truncatula genome sequence (www.medicago.org) as a reference. This assembly is a BAC-based assembly for M. truncatula accession A17 (hereafter referred to as HM101) that covers ∼70% of the euchromatin. The Mt3.0 version consists of essentially the same sequence data found in the more recent Mt3.5 version, except that the order/orientation of scaffolds in Mt3.0 was based on genetic map anchoring, whereas the assembly of Mt3.5 was based on newer optical map results (50). Although the Mt3.0/Mt3.5 assemblies have been supplemented by Illumina-based whole-genome sequencing to capture missing portions of the genome (www.medicago.org), we did not use these supplemental sequences for alignment or SNP discovery.
Genomic paired-end Illumina sequencing libraries were prepared for sequencing by synthesis according to standard methods (51). Insert sizes (not including the adapters) ranged from ∼200 to 450 nt. Libraries were sequenced using GAII or GAIIx Illumina sequencing instruments to yield paired 90- or 151-mer reads. The latter were subsequently trimmed back to 90 oligomers for this analysis. The Illumina image analysis pipeline with default parameters was used for image analysis, base calling, and read filtering. Additional filtering was done on later runs to remove adapter and PhiX contamination based on blast alignment (pairs with ≥14 nt aligned at ≥98% were removed). All Illumina sequence data have been deposited in the National Center for Biotechnology Information short-read archive, and Sanger-sequenced PCR products have been deposited in GenBank (short-read archive project SRP001874). Coverage data and called SNPs are available at www.medicago.org.
All reads that passed the initial quality control filter were aligned to the HM101 reference genome using the Genomic Short-Read Nucleotide Alignment Program (GSNAP) (52). Only reads ≥91% identical to a region in the reference genome and aligned to fewer than five locations were included in the alignment output file. We required that four additional criteria be met before identifying polymorphisms: (i) a read align to only one position in the reference genome, meaning that it does not align equally well to any other region of the genome, (ii) more than or equal to two reads cover that nucleotide position, (iii) the variant nucleotide was called by >70% of the reads that covered that site, and (iv) each of the nucleotides that called an alternate allele was required to have an Illumina quality score ≥10 (results from analyses on data requiring a quality score ≥20 were very similar; e.g., the correlation between θπ per 100-kb window from the two datasets was very high at R2 > 0.99) (Dataset S2). The >70% requirement means that we identified no heterozygous sites, although we expect this finding will have minor effects on our data given that there should be minimal residual heterozygosity because of high selfing rates in natural populations (>95%) (31, 32) and more than or equal to three generations of selfing before DNA extraction.
The alignment criteria were chosen after preliminary analyses of three genomes that covered the range of diversity in our sample: the reference genome HM101, HM005 (also known as DZA315-16), and M. tricycla (HM029). Illumina DNA sequence reads from these genomes were aligned to the reference at three levels of stringency (95%, 93%, and 91% identity), and SNPs were called requiring one or two reads with a minimum of 30%, 50%, or 70% of reads calling the base (total of 18 parameter combinations per genome). The quality of SNP calls for each of these conditions was evaluated by comparing the aligned sequences to 100 randomly selected regions that had been PCR-amplified and then Sanger-sequenced (roughly 60 kb/genome).
To evaluate quality of our called SNPs, we compared our SNP calls for 47 genomic regions, ranging from 190 to 2,956 bp (45,565 total bp), that had been PCR-amplified and Sanger-sequenced (53) from each of 16 of the same M. truncatula lines that we used in this study. Among the 16 lines, we confirmed 2,843 nonreference base calls (i.e., a variant relative to the reference was identified in both GSNAP-aligned Illumina data and Sanger sequence) and 102 variants that were identified in GSNAP-aligned Illumina data but not verified by Sanger resequencing.
Nucleotide Diversity.
We characterized nucleotide diversity using two standard estimates of the scaled mutation rate θw = 4Neμ, the proportion of segregating sites (54), and θπ, the average pairwise nucleotide diversity (55). The frequency distribution of segregating sites was summarized using Tajima's D statistic, DT (55). All summary statistics were calculated along all eight chromosomes using nonoverlapping sliding windows of 100 kb (Dataset S2). Summary statistics were also calculated for each of 30,768 for which we had sufficient sequence coverage of ∼51,000 gene models identified by the International Medicago Genome Annotation Group (Dataset S3). Putative genes were included in analyses only if resequence data covered ≥80% of the putative coding sequences from ≥20 accessions. Similarly, for sliding window analyses, we included only those sites for which we had data from ≥20 accessions. Windows were truncated at gaps in the reference genome. New windows were opened after the gap, and windows with <10 kb of covered sites were excluded from analyses (2,538 windows, with an average of 54,084 covered bases per window, were included in analyses). For coding regions, we calculated DT only for genes with more than two polymorphic sites to avoid biasing the distribution of the statistic. Analyses were conducted using C++ code available in the libsequence software library (56), available R codes, or custom R or PERL scripts (written by AB or PZ). For coding regions, we calculated summary statistics for replacement sites, synonymous sites, and total coding region, with site identity based on International Medicago Genome Annotation Group annotation.
Recombination and Linkage Disequilibrium.
Population-scaled recombination rates (ρ = 4Ner) along each of the eight chromosomes were estimated using the program interval in the LDhat (57) package using standard methods (58, 59). In brief, we ran the MCMC algorithm implemented in LDhat interval on 100-kb sliding windows for 1,000,000 generations sampling every 1,000 generations, including only SNPs that were at an MAF of >0.1 at sites covered in >20 genomes. As with sliding window analyses of summary statistics, windows were truncated at gaps in the reference genome. In addition to calculating ρ, we estimated the rate of LD decay by calculating pairwise r2 between 50 randomly selected SNPs within 200-kb windows that were sliding every 100 kb. For this analysis, we used an MAF of >0.2 to minimize the effects of rare variants.
Map-based recombination distances were estimated using data from a cross between the M. truncatula line used for developing the reference genome A17 (HM101) and line A20 (HM018) using genetic markers that had been mapped to the physical genome (60). Line HM018, although traditionally treated as M. truncatula, is more closely related to M. littoralis and M. tricycla (Fig. S2) and therefore, was not included in other analyses. To translate map-based estimates of recombination to 100-kb windows at which we estimated population genetic parameters, we used the average physical location of markers that had identical map distances and linearly interpolated the recombination rate between adjacent markers.
We used Pearson correlations to examine the linear relationship between estimates of diversity, recombination, gene density, and distance from the centromere. Because 100-kb window estimates of these variables are autocorrelated (61), we estimated the statistical significance of correlations by 1,000 permutations in which the chromosomal order of observations were kept intact (39).
Supplementary Material
Acknowledgments
We thank Stephen Keller, Maren Friesen, and Sergey Nuzhdin for discussions, Jean-Marie Prosperi and Magalie Delalande for the development and management of the M. truncatula germplasm collection (seeds available at http://www1.montpellier.inra.fr/BRC-MTR/), and Thierry Huguet and M. El Arbi for development of some M. truncatula germplasm. This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute and was funded by National Science Foundation Grant 0820005.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the NCBI Sequence Read Archive (accession no. SRP001874).
See Author Summary on page 17253.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104032108/-/DCSupplemental.
References
- 1.Kinzig AP, Socolow RH. Human impacts on the nitrogen cycle. Phys Today. 1994;47:24–35. [Google Scholar]
- 2.Graham PH, Vance CP. Legumes: Importance and constraints to greater use. Plant Physiol. 2003;131:872–877. doi: 10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Mita S, et al. Investigation of the demographic and selective forces shaping the nucleotide diversity of genes involved in nod factor signaling in Medicago truncatula. Genetics. 2007;177:2123–2133. doi: 10.1534/genetics.107.076943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heath K, Tiffin P. Context dependence in the coevolution of plant and rhizobial mutualists. Proc R Soc Lond B Biol Sci. 2007;274:1905–1912. doi: 10.1098/rspb.2007.0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stacey G, Libault M, Brechenmacher L, Wan JR, May GD. Genetics and functional genomics of legume nodulation. Curr Opin Plant Biol. 2006;9:110–121. doi: 10.1016/j.pbi.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Young ND, Udvardi M. Translating Medicago truncatula genomics to crop legumes. Curr Opin Plant Biol. 2009;12:193–201. doi: 10.1016/j.pbi.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Harrison MJ. Signaling in the arbuscular mycorrhizal symbiosis. Annu Rev Microbiol. 2005;59:19–42. doi: 10.1146/annurev.micro.58.030603.123749. [DOI] [PubMed] [Google Scholar]
- 8.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 9.Ronfort J, et al. Microsatellite diversity and broad scale geographic structure in a model legume: Building a set of nested core collection for studying naturally occurring variation in Medicago truncatula. BMC Plant Biol. 2006;6:28. doi: 10.1186/1471-2229-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begun DJ, et al. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 12.McNally KL, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 14.Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3:1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williamson SH, et al. Localizing recent adaptive evolution in the human genome. PLoS Genet. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian F, Stevens NM, Buckler ES., 4th Tracking footprints of maize domestication and evidence for a massive selective sweep on chromosome 10. Proc Natl Acad Sci USA. 2009;106:9979–9986. doi: 10.1073/pnas.0901122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordborg M, et al. The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2002;30:190–193. doi: 10.1038/ng813. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]
- 19.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt A, Vilhjálmsson BJ, Nordborg M. Conditions under which genome-wide association studies will be positively misleading. Genetics. 2010;186:1045–1052. doi: 10.1534/genetics.110.121665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 22.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 24.Lam HM, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42:1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218. doi: 10.1146/annurev.genet.39.073003.112420. [DOI] [PubMed] [Google Scholar]
- 26.DeRose-Wilson LJ, Gaut BS. Transcription-related mutations and GC content drive variation in nucleotide substitution rates across the genomes of Arabidopsis thaliana and Arabidopsis lyrata. BMC Evol Biol. 2007;7:66. doi: 10.1186/1471-2148-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson MS, et al. Nucleotide diversity and linkage disequilibrium in balsam poplar (Populus balsamifera) New Phytol. 2010;186:526–536. doi: 10.1111/j.1469-8137.2009.03174.x. [DOI] [PubMed] [Google Scholar]
- 28.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 29.Wright SI, Andolfatto P. The impact of natural selection on the genome: Emerging patterns in Drosophila and Arabidopsis. Annu Rev Ecol Evol Syst. 2008;39:193–213. [Google Scholar]
- 30.Nordborg M. Structured coalescent processes on different time scales. Genetics. 1997;146:1501–1514. doi: 10.1093/genetics/146.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonnin I, Ronfort J, Wozniak F, Olivieri I. Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae) Mol Ecol. 2001;10:1371–1383. doi: 10.1046/j.1365-294x.2001.01278.x. [DOI] [PubMed] [Google Scholar]
- 32.Siol M, Prosperi JM, Bonnin I, Ronfort J. How multilocus genotypic pattern helps to understand the history of selfing populations: A case study in Medicago truncatula. Heredity. 2008;100:517–525. doi: 10.1038/hdy.2008.5. [DOI] [PubMed] [Google Scholar]
- 33.Moeller DA, Tenaillon MI, Tiffin P. Population structure and its effects on patterns of nucleotide polymorphism in teosinte (Zea mays ssp. parviglumis) Genetics. 2007;176:1799–1809. doi: 10.1534/genetics.107.070631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arunyawat U, Stephan W, Städler T. Using multilocus sequence data to assess population structure, natural selection, and linkage disequilibrium in wild tomatoes. Mol Biol Evol. 2007;24:2310–2322. doi: 10.1093/molbev/msm162. [DOI] [PubMed] [Google Scholar]
- 35.Friesen ML, et al. Population genomic analysis of Tunisian Medicago truncatula reveals candidates for local adaptation. Plant J. 2010;63:623–635. doi: 10.1111/j.1365-313X.2010.04267.x. [DOI] [PubMed] [Google Scholar]
- 36.Hellmann I, et al. Population genetic analysis of shotgun assemblies of genomic sequences from multiple individuals. Genome Res. 2008;18:1020–1029. doi: 10.1101/gr.074187.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephan W. Genetic hitchhiking versus background selection: The controversy and its implications. Philos Trans R Soc Lond B Biol Sci. 2010;365:1245–1253. doi: 10.1098/rstb.2009.0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordborg M, et al. The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 2005;3:e196. doi: 10.1371/journal.pbio.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charlesworth D, Charlesworth B, Morgan MT. The pattern of neutral molecular variation under the background selection model. Genetics. 1995;141:1619–1632. doi: 10.1093/genetics/141.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 42.Borevitz JO, et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rose LE, et al. The maintenance of extreme amino acid diversity at the disease resistance gene, RPP13, in Arabidopsis thaliana. Genetics. 2004;166:1517–1527. doi: 10.1534/genetics.166.3.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gos G, Wright SI. Conditional neutrality at two adjacent NBS-LRR disease resistance loci in natural populations of Arabidopsis lyrata. Mol Ecol. 2008;17:4953–4962. doi: 10.1111/j.1365-294X.2008.03968.x. [DOI] [PubMed] [Google Scholar]
- 45.Kamphuis LG, et al. The Medicago truncatula reference accession A17 has an aberrant chromosomal configuration. New Phytol. 2007;174:299–303. doi: 10.1111/j.1469-8137.2007.02039.x. [DOI] [PubMed] [Google Scholar]
- 46.Nordborg M. Linkage disequilibrium, gene trees and selfing: An ancestral recombination graph with partial self-fertilization. Genetics. 2000;154:923–929. doi: 10.1093/genetics/154.2.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morrell PL, Toleno DM, Lundy KE, Clegg MT. Low levels of linkage disequilibrium in wild barley (Hordeum vulgare ssp. spontaneum) despite high rates of self-fertilization. Proc Natl Acad Sci USA. 2005;102:2442–2447. doi: 10.1073/pnas.0409804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takebayashi N, Morrell PL. Is self-fertilization an evolutionary dead end? Revisiting an old hypothesis with genetic theories and a macroevolutionary approach. Am J Bot. 2001;88:1143–1150. [PubMed] [Google Scholar]
- 49.Hyten DL, et al. Highly variable patterns of linkage disequilibrium in multiple soybean populations. Genetics. 2007;175:1937–1944. doi: 10.1534/genetics.106.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S, et al. A single molecule scaffold for the maize genome. PLoS Genet. 2009;5:e1000711. doi: 10.1371/journal.pgen.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Mita S, Chantret N, Loridon K, Ronfort J, Bataillon T. Molecular adaptation in flowering and symbiotic recognition pathways: Insights from patterns of polymorphism in the legume Medicago truncatula. BMC Evol Biol. 2011;11:229. doi: 10.1186/1471-2148-11-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 55.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thornton K. Libsequence: A C++ class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- 57.McVean GA, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 60.Mun JH, et al. Distribution of microsatellites in the genome of Medicago truncatula: A resource of genetic markers that integrate genetic and physical maps. Genetics. 2006;172:2541–2555. doi: 10.1534/genetics.105.054791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hahn MW, SMBE Tri-National Young Investigators Proceedings of the SMBE Tri-National Young Investigators’ Workshop 2005. Accurate inference and estimation in population genomics. Mol Biol Evol. 2006;23:911–918. doi: 10.1093/molbev/msj094. [DOI] [PubMed] [Google Scholar]