Background: Tumor stem cells contribute to tumor recurrence after chemotherapy.
Results: Reactivation of antioxidant enzymes through Nrf2-p21 signaling contributes to stem cell enrichment.
Conclusion: Nrf2 stabilization through reduced 26 S proteasome activity generates cells with tumor stem cell-like properties.
Significance: Nrf2, 26 S proteasome, and p21 are the key players in the emergence of drug-resistant tumor stem cells after chemotherapy.
Keywords: ABC Transporter, Anticancer Drug, Apoptosis, Cancer Therapy, Reactive Oxygen Species (ROS), Stem Cells, Senescence, Tumor Stem Cells
Abstract
Tumor recurrence after chemotherapy or radiation remains a major obstacle to successful cancer treatment. A subset of cancer cells, termed cancer stem cells, can elude conventional treatments and eventually regenerate a tumor that is more aggressive. Despite the large number of studies, molecular events that govern the emergence of aggressive therapy-resistant cells with stem cell properties after chemotherapy are poorly defined. The present study provides evidence for the rare escape of tumor cells from drug-induced cell death, after an intermediate stay in a non-cycling senescent stage followed by unstable multiplication characterized by spontaneous cell death. However, some cells appear to escape and generate stable colonies with an aggressive tumor stem cell-like phenotype. These cells displayed higher CD133 and Oct-4 expression. Notably, the drug-selected cells that contained low levels of reactive oxygen species (ROS) also showed an increase in antioxidant enzymes. Consistent with this in vitro experimental data, we observed lower levels of ROS in breast tumors obtained after neoadjuvant chemotherapy compared with samples that did not receive preoperative chemotherapy. These latter tissues also expressed enhanced levels of ROS defenses with enhanced expression of superoxide dismutase. Higher levels of Oct-4 and CD133 were also observed in tumors obtained after neoadjuvant chemotherapy. Further studies provided evidence for the stabilization of Nrf2 due to reduced 26 S proteasome activity and increased p21 association as the driving signaling event that contributes to the transition from a high ROS quiescent state to a low ROS proliferating stage in drug-induced tumor stem cell enrichment.
Introduction
Adjuvant and neoadjuvant chemotherapy continue to play a critical role in the management of breast cancer. The primary hurdle for effective chemotherapy is the inherent or acquired resistance of cancer cells to a variety of anticancer agents. Thus, chemotherapy in many patients can only delay recurrence and very often is accompanied by rapid, untreatable progression. Understanding the mechanisms of how such chemoresistance is generated is a critical clinical requirement. An emerging concept in tumor biology is the cancer stem cell (CSC)3 hypothesis, which postulates that in bulk tumors, the potential to grow and propagate is restricted to a rare population of cells called CSCs that can reinitiate the tumor even after a successful elimination of the primary tumor mass by surgery, chemotherapy, or radiotherapy. Such cellular and molecularly heterogeneous subsets have been identified in hematopoietic as well as solid malignancies (1–3). Previous studies suggest that in breast cancer, tumorigenic breast cancer stem cells are enriched in cells with a CD44+/CD24− phenotype (1, 3). Side population cells defined by Hoechst 33342 dye exclusion have also been found to be enriched in tumor stem cells due to high expression of drug efflux transporter proteins (4). These subpopulations appear to be more resistant to most antitumor agents due to efficient DNA repair capacity, inherent resistance to apoptosis, property of clonal expansion, and asymmetric division (5–9). Against this background, we employed a live cell tool to monitor caspase-mediated cell death or survival after in vitro drug treatment to investigate events associated with enrichment of CSCs following drug treatment. With this tool, we could track dying or surviving cells after chemotherapy for longer time periods. Drug-selected cells were isolated from human breast cancer cell lines treated with different antitumor agents for further characterization. Among multiple drugs used, we observed that emergence of drug-resistant cells is associated with a transient appearance of a quiescent senescent population of cells. The postsenescent, drug-surviving cells were enriched with dye efflux cells and embryonic stem cell markers. Senescent cells generated during the drug treatment showed a significantly higher level of ROS, but the senescence-escaped stable colonies were enriched with cells of very low ROS subsequent to reactivation of antioxidant machinery.
Our study indicates that oxidative stress caused by chemotherapeutic agents will generate senescent-like cells from which the emergence of a population with enriched stem cell activity occurs, and this may well be one of the mechanisms by which CSCs evade chemotherapy and repopulate the tumor. Moreover, the study identified reactivation of the antioxidant system through stabilization of the master regulator Nrf2 that culminates in enrichment of cells with low 26 S proteasome activity, low ROS, and high Oct-4.
EXPERIMENTAL PROCEDURES
Cell Lines and Primary Tumors
Human breast tumor cell lines MCF-7, MDA MB231, and T47D were obtained from NCI, National Institutes of Health (Bethesda, MD). All cancer cell lines were maintained in RPMI medium supplemented with 1% penicillin/streptomycin and 10% fetal bovine serum. All cell lines were incubated in a humidified incubator at 37 °C supplied with 5% carbon dioxide.
Tumor specimens were obtained after informed consent from patients who underwent surgical resection at the Division of Surgical Oncology, Regional Cancer Centre, Thiruvananthapuram. The study was approved by the Human Ethics Committee. Fresh tumor samples obtained immediately after surgery were rinsed, mechanically minced, and digested for 3 h at 37 °C in a shaking incubator with 0.2% collagenase type IV (Invitrogen) in DMEM. After red blood cell lysis, the single cell suspension was washed twice with DMEM/F-12 supplemented with 20% FBS.
Side Population (SP) Analysis
The cell suspensions were labeled with Hoechst 33342 dye (Invitrogen) and analyzed for side population as described previously (4). Briefly, cells were resuspended at 1 × 106/ml in prewarmed DMEM with 2% FBS and 10 mm HEPES buffer containing 5 μg/ml of Hoechst 33342 in the presence or absence of verapamil at 37 °C for 90 min. At the end of the incubation, cells were washed and resuspended in ice-cold Hanks' balanced salt solution containing 2% FBS and 10 mm HEPES. Propidium iodide at a final concentration of 2 μg/ml was added to the cells to gate viable cells. Side population analyses and sorting were done on a FACSAria I flow cytometer (BD Biosciences).
Drug Treatment
Cells were seeded at a density of 2 × 105 cells/well in 12-well plates containing DMEM supplemented with 10% FBS. After overnight incubation, different drugs were added to each cell line and maintained at 37 °C in a water-saturated atmosphere containing 5% CO2. Medium replacement was done every fourth day with fresh drug-containing medium. The duration of exposure to each drug was about 2–3 weeks. A cross-resistance assay was performed by seeding 2000 cells in 100 μl of growth medium in 96-well plates. After overnight incubation, cells were incubated in the presence of chemotherapeutic drugs at 37 °C in 5% CO2 for 24 h. Drugs and dose used in the assay are given in supplemental Table 1.
Measurement of Intracellular ROS
For intracellular ROS analysis, cells were incubated in the dark with 10 μm dichlorodihydrofluorescein diacetate (Invitrogen) for 30 min at 37 °C and immediately analyzed by flow cytometry on a FACSAria I flow cytometer (BD Biosciences).
Senescence-associated β-Galactosidase (SA-β-gal) Staining
SA-β-gal staining was performed as described previously (10). In brief, cells or tissues (5-μm-thick frozen sections of breast tissue) were fixed for 5 min in 3% formaldehyde, washed, and incubated at 37 °C with 5-bromo-4-chloro- 3-indolyl β-d-galactopyranoside (1 mg/ml), dissolved in a solution containing 40 mm citric acid (pH 6.5), 5 mm potassium ferrocyanide, 5 mm potassium ferricyanide, 150 mm NaCl, and 2 mm MgCl2. After overnight incubation at 37 °C, tissues or cells were visualized by microscopy.
Rhodamine 123 Dye Efflux
For analysis of rhodamine 123 efflux activity, cells were resuspended in DMEM supplemented with 2% FBS containing 0.1 μg/ml rhodamine 123 (Calbiochem) for 30 min. Cells were then washed with cold PBS and reincubated at 37 °C for 45 min in DMEM supplemented with 10% FBS to allow dye efflux. After the efflux period, cells were washed and analyzed using a FACSAria I flow cytometer (BD Biosciences).
Apoptosis Analysis
After treatment with different drugs, the cells were stained with Hoechst 33342 dye (1 μg/ml) and incubated for 10 min at 37 °C before imaging under an epifluorescent microscope (TE-2000E, Nikon, Japan) using the UV2A filter. Cells with condensed or fragmented nuclei were taken as apoptotic and counted against the total number of cells in the field and plotted graphically with the percentage of apoptotic populations against the treatments.
Western Blot Analysis and Immunoprecipitation
Cells were harvested, washed three times in PBS, and lysed in radioimmune precipitation assay buffer (50 mm Tris-Cl (pH 7.4), 1% Nonidet P-40, 40 mm NaF, 10 mm NaCl, 10 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride, 10 mm dithiothreitol, and 1 μg/ml each leupeptin and aprotinin). To extract total protein from tissue, the frozen tissue was crushed with a mortar and pestle in liquid nitrogen and lysed in radioimmune precipitation assay buffer. For immunoprecipitation, cell lysate prepared from parental and drug-escaped cells in ice-cold immunoprecipitation lysis buffer containing 50 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm Na3VO4, 1 mm dithiothreitol, 1 mm PMSF, 5 mm EDTA, and protease inhibitor mixture. After centrifugation, lysates were incubated with p21 antibody for 4 h at 4 °C followed by the addition of protein A-Sepharose beads for another 2 h. Western blot was performed with Nrf2 antibody. Total protein concentration was measured by Bradford assay according to the manufacturer's instructions (Sigma). 50 μg of protein was resolved by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad) by the wet transfer method. The membranes were blocked and then probed with the primary antibodies overnight at 4 °C (supplemental Table 2). After incubation, membranes were washed with TBS-Tween 0.2% and subjected for incubation with horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h. The membranes were washed again and developed by enhanced chemiluminescence (Amersham Biosciences).
In Vitro Invasion Assay
Cellular potential for invasiveness of drug-resistant cells was determined using a BD BioCoat FluoroBlok 24-multiwell insert system (BD Biosciences). Cells were seeded into upper inserts at 1 × 105 cells/insert in serum-free DMEM. Outer wells were filled with DMEM containing 10% FBS as chemoattractant. Cells were incubated at 37 °C with 5% CO2 for 48 h. After 48 h, cells that migrated through the pores of the membrane to the bottom chamber were stained with 4 μg/ml Calcein AM (BD Biosciences) in Hanks' balanced salt solution for 60 min at 37 °C and viewed under a fluorescent microscope. Cells in four random optical fields were counted to determine the number of invaded cells.
Mammosphere and Soft Agar Colony Assay
For the mammosphere formation assay, parental and drug-escaped cells were seeded on ultralow attachment 96-well plates (Corning Glass) in serum-free mammary epithelial basal medium (Lonza) up to 12 days. The mammosphere growth was analyzed under a phase-contrast microscope with a ×10 objective and counted from multiple wells. For determining anchorage-independent growth potential of the drug-escaped cells, a soft agar colony assay was performed as per the standard protocol. Briefly, 5000 cells/well were prepared in 0.3% agarose and layered on the bottom layer of 1% agar in a 24-well plate and allowed to grow for 21 days. The colonies were stained with 0.05% crystal violet and photographed using × 4 objective.
Immunophenotyping
For immunophenotyping by flow cytometry, cells were trypsinized into a single cell suspension, washed with phosphate-buffered saline, and stained with antibodies specific for human cell surface markers: CD24-PE and CD44-FITC (Invitrogen). A total of 25,000 cells were incubated with antibodies for 1 h at room temperature. Unbound antibody was washed off, and cells were analyzed using FACSAria I flow cytometer (BD Biosciences).
Reporter Gene Assay
Cells were trypsinized and transfected with 2 μg of NQO1-antioxidant response element (ARE) luciferase reporter plasmid and 0.2 μg of PRL −TK control plasmid using neon transfection system as per the instruction manual (Invitrogen). The electroporated cells were seeded on 6-well plates and allowed to grow for 48 h. After 48 h, the cells were lysed to determine the Renilla and firefly luciferase activities using a dual luciferase reporter system with a luminometer (Turner).
RNAi Silencing
Nrf2 (sc-37030), Oct-4 (sc-36123), p21 (sc-29427), and negative control scrambled (sc-37007) small interfering RNAs were procured from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). RNAi duplex was transfected into drug-resistant cells by a neon transfection system as per the instructions. After 24 h of transfection, cells were trypsinized and seeded onto 12-well plates for 24 h before treatment with different drugs. For stable knockdown of p21, the shRNA plasmid targeting the human p21 gene was used as described earlier (11). Briefly, the cells were transfected with shRNAp21 vector or control vector (shRNA Luc) using Lipofectamine followed by selection in 800 μg/ml G418.
26 S Proteasome Activity Assay
Chymotryptic, tryptic, and caspase-like proteasome activities were determined using fluorogenic proteasome substrates, Suc-LLVY-AMC (chymotryptic substrates), benzyloxycarbonyl-AAR-AMC (tryptic substrates), and benzyloxycarbonyl-LLE-AMC (caspase-like substrates) as described (12). All substrates were procured from Calbiochem. Briefly, cells grown on 6-well plates were scraped, washed, and lysed in lysis buffer (25 mm Tris, pH 7.5, 100 mm NaCl, 5 mm ATP, 0.2% Nonidet P-40, 20% glycerol) by sonication. To measure 26 S proteasome activities, 100 μg of protein from each sample was diluted in assay buffer containing 80 μm respective fluorogenic substrates in triplicate. Proteolytic activities were continuously monitored for 2 h at 37 °C by measuring the release of the free AMC using a fluorescent plate reader (Tecan-200) with excitation and emission wavelengths of 380 and 460 nm, respectively.
Detection of Caspase-3 Activation in Live Cells Using Fluorescence Resonance Energy Transfer (FRET)
For live cell visualization of caspase activation, the caspase sensor expression vector, SCAT3.1, which consists of ECFP and EYFP (Venus), separated by the caspase cleavage site, DEVD, was employed to generate stable clones as described above. Stable clones were expanded and sorted further for cells enriched with high FRET using FACSAria I flow cytometer (BD Biosciences). For FRET imaging by microscopy, cells were seeded on a chambered coverglass (Nunc International) and maintained in a live cell incubation chamber (Tokai-Hit) at 37 °C with 5% CO2 for the indicated time periods. Images were collected using a CARVII unit mounted on an epifluorescent microscope TE-2000E (Nikon, Japan) at regular intervals. Single excitations of ECFP at 440/20 and dual emission of ECFP at 480/20 and FRET channel at 542/27 were collected using an automated excitation and emission filter wheel controlled by IP Lab software (BD Biosciences). For long term analysis of cell fate after drug treatment, the FRET probe-expressing stable cells were seeded on 96-well glass bottom imaging plates and exposed to different drugs for the indicated time periods. The cells were imaged using a 96-well plate BD Pathway Bio imager (BD Biosciences) configured with the donor and FRET filter sets (Semrock) in ratio mode using a ×20 objective in 3 × 3 or 6 × 6 montage capture, as described (13).
Assessment of Cell Cycle Progression in Live Cells
For live cell visualization of cell cycle progression, a G1-specific fluorescent component of FUCCI, the Cdt1-Kusabira Orange (KO) fusion construct was employed to generate stable clones. Cells were transfected with the expression vector Cdt1-KO followed by selection in G418. Stable integration and cell cycle-specific fluorescence were evaluated by live cell imaging after staining the cells with Hoechst 33342 (1 μg/ml) for 5 min at 37 °C. The cells were seeded on chambered coverglass for live cell imaging as described above, employing Texas Red filter sets.
RESULTS
Breast Cancer Cell Lines and Primary Tumors Contain SP Cells and Their Enrichment by Chemotherapy
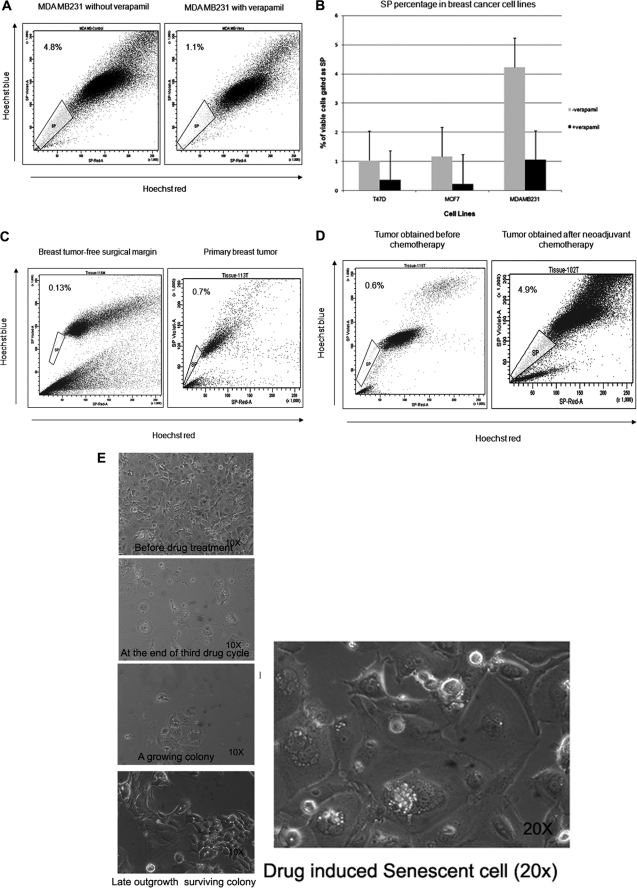
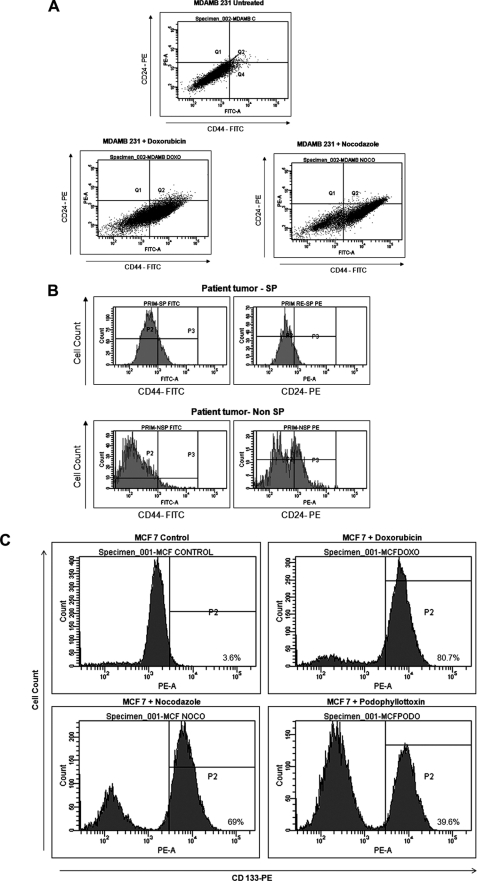
We first examined the existence of SP cells in four human breast cancer cell lines by staining them with Hoechst 33342 dye to generate a Hoechst blue-red profile. SP gate was defined as the diminished region in the presence of verapamil. The staining profile of MDA MB231 is shown as an example (Fig. 1A). All of the breast cancer cell lines contained a distinct fraction of SP cells, ranging from 1 to 4.8% of gated cells, which decreased significantly in the presence of verapamil (Fig. 1B). To see if primary tumors also contain SP, we used single cell suspension from surgical resections from breast cancer patients. Lymphocytes were excluded using forward and side scatter signals. The SP fraction was present in tumor-free surgical margins as well as tumors (Fig. 1C). Among 12 tumor specimens obtained after neoadjuvant chemotherapy, eight samples showed an increased prevalence of SP cells (Fig. 1D) relative to the basal percentage of 1% SP in prechemotherapy samples. Postsorting culture was performed to ensure that SP and non-SP fractions analyzed were enriched with epithelial cells (supplemental Fig. 1A).
FIGURE 1.
SP cells were present in breast cancer cell lines and breast tumor samples. A, MDA MB231 cells were stained with Hoechst 33342 dye in the presence (right) or absence (left) of 50 μmol/liter verapamil and analyzed by flow cytometry. B, percentage of SP cells gated with or without the addition of verapamil in listed cancer cell lines from three representative experiments. C, a representative scatter plot of SP analysis from tumor-free surgical margin (left) and primary tumor (right). D, SP analysis from a tumor obtained after neoadjuvant chemotherapy (right) and a tumor that had not received preoperative chemotherapy (left). Significantly higher proportions of SP cells were present in tumors after chemotherapy. E, different stages of clonal expansion of doxorubicin-treated MCF-7 cells observed by phase-contrast microscopy. A ×20 magnified image of senescent culture is also shown in the left panel. Error bars, S.D.
Because tumor samples collected after neoadjuvant chemotherapy showed a higher percentage of SP cells, we decided to characterize the enrichment of the drug-resistant population after in vitro drug treatment. MDA MB 231, T47D, SKBR3, and MCF7 and the respective EGFP-expressing cells were treated with a panel of anti-cancer drugs as described earlier. A concentration of drug that kills more than 60% of cells at 24–48 h was used for all experiments (supplemental Table 2). The majority of cells died by the fifth day. Over the next 10 days few enlarged cells were seen with flattened, senescent-like morphology. The cells were refed with fresh medium containing drugs every 4 days for two more cycles. After 3–4 weeks, small, spindle-shaped cells were seen with occasional visible small colonies (Fig. 1E and supplemental Fig. 1B). Such late outgrowth was observed in all of the four cell lines treated with 5-fluorouracil, paclitaxel, doxorubicin, colchicine, vincristine, podophyllotoxin, and nocodazole. However, in MG132, thapsigargin, and staurosporine treated sets, no surviving cells were seen or any indication of senescent-like cells observed, indicating a close association of senescence and emergence of late outgrowth colonies.
Experimental Model to Track the Emergence of Drug-resistant Colonies
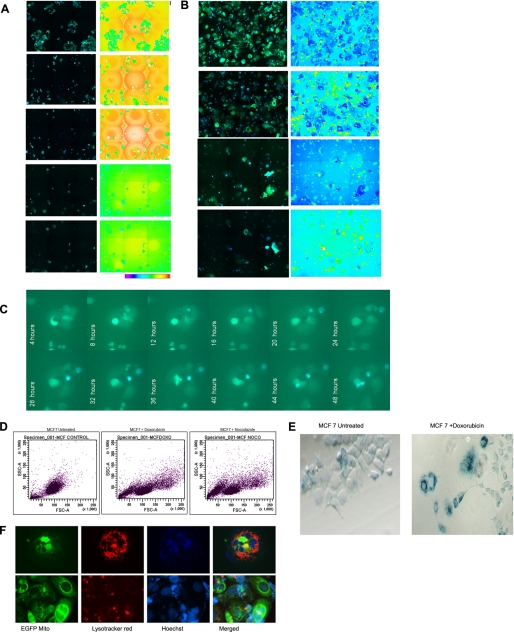
Most antitumor agents trigger apoptosis with activation of caspases. It is possible that some rare cells escape from anticancer drug-induced caspase activation, leading to generation of caspase-resistant cells that may contribute to drug resistance and therapy failure. To track the emergence of rare outgrowth colonies from drug-induced caspase activation, we generated stable breast cancer cell lines expressing caspase-specific FRET probe ECFP-DEVD-EYFP. With this tool, caspase activation can be monitored in live cells on ratio image, and the remaining resistant cells at the end of drug treatment can be identified by live cell FRET imaging as described under “Experimental Procedures.” Because our initial studies indicate that the emergence of drug-resistant clones is a rare event, we configured the 96-well BD Pathway Bio imager to perform ratio imaging so that the large area of same wells could be repeatedly imaged for tracking the cells that escape caspase activation. Representative images of MCF7-SCAT3.1 cells treated with different drugs are given in Fig. 2, A and B, and supplemental Fig. 2A. During prolonged in vitro drug treatment, it was seen that after an initial phase of massive cell death by caspase activation, a few cells enter a noncycling quiescent-like state with large, flattened morphology, a characteristic feature of senescent cells. Most senescent-like cells underwent spontaneous caspase activation and cell death. However, rare senescent cells showed evidence for unstable cell division in a delayed manner that is also associated with spontaneous cell death. A representative live cell image of senescent cells generated after vincristine treatment and its cell proliferation and spontaneous caspase activation for 0–48 h is given in Fig. 2C. We observed the emergence of rare outgrowth colonies from some of the wells in a delayed manner at around 3–4 weeks, indicating that some cells may escape spontaneous cell death and give rise to a drug-tolerant population.
FIGURE 2.
Drug-induced senescence and emergence of multidrug-resistant cells. A, MCF-7 cells stably expressing FRET probe SCAT3 consisting of fusion of ECFP-DEVD-EYFP developed as described. The cells were exposed to doxorubicin. ECFP/EYFP FRET ratio imaging was carried out using a 96-well plate, Bio-Imager, pathway 435 at 6 × 6 montage at the indicated days after drug treatment. The merged image of the ECFP channels and the EYFP FRET channel is shown. Loss of FRET upon caspase activation enhances ECFP signal, leading to an increase in blue color in the real merged image. Ratio image and ratio scale are also given. B, the above cell lines were treated with nocodazole and imaged as described earlier. C, MCF-7 SCAT3 stable cells were treated with vincristine for three cycles as described. The remaining senescent cells were trypsinized and seeded on a chambered coverglass. FRET live cell imaging was done using a BD-CARV Bio-Imager for 48 h as described. The merged ECFP and EYFP FRET channel at the indicated time point is shown. Occasional cell division and spontaneous cell death by caspase activation are evident in the image. D, flow cytometry analysis of the side scatter (y) and forward scatter (x) factors (i.e. granularity and size, respectively) of MCF-7 cells after a 30-day treatment with paclitaxel. Highly heterogeneous populations with three different scatter factor values emerged after drug exposure. E, expression of SA-β-gal in MCF-7 cells following prolonged exposure to doxorubicin. The enlarged, flat cells showed strong cytoplasmic positivity, whereas the newly emerged cells were negative for SA-β-gal. F, MCF-7 cells expressing Mito-EYFP were treated with vincristine for three cycles. The cells were stained with LysoTracker Red and nuclear dye Hoechst and imaged under a fluorescent microscope to visualize mitochondria (green), lysosomes (red), and nuclei (blue). Two representative images of cells showing senescent cells with high lysosomal and mitochondrial density are shown. In the lower panel, few cells of small size with less lysosomal mass are evident.
The late outgrowth cells were small in size and always observed near the senescent cells. Extensive microscopic follow up examinations of in vitro drug-treated cultures suggested their possible emergence from senescent cells. The scatter plot analysis by FACS showed two heterogeneous populations with great variability in the forward (size) and side (granularity) scatter factors compared with the parental cell line (Fig. 2D). The emerged clones were stained for SA-β-gal activity. We observed that the large, flat cells stained positive for SA-β-gal activity, whereas most of the smaller cells were negative for SA-β-gal, suggesting that the late outgrowth cells were senescence-escaped cells (Fig. 2E). To confirm additional features associated with senescence, such as an increase in acidic organelles, cells expressing mitochondrial EYFP were stained with LysoTracker Red. Microscopic analysis showed that the enlarged flattened cells had a more profuse lysosomal network (Fig. 2F).
Senescent Cells Had High Levels of ROS, whereas Drug-selected Cells Had Low ROS
Most anti-cancer drugs are known to act through oxidative stress (i.e. by increased production of superoxide free radicals in the cells), resulting in irreversible cell injury, ultimately leading to cancer cell death (14, 15). Previous studies have also shown that human and murine breast CSCs, similar to their normal tissue counterparts, maintain low levels of ROS that afford them radioprotection (16). Hence, we examined ROS levels in the late outgrowth cells by flow cytometry. A significant subset of cells displayed a low ROS phenotype, whereas the subpopulation with higher forward and side scatter factor values showed high ROS levels (supplemental Fig. 3A). Quite interestingly, drug-tolerant cells always showed a very low level of ROS compared with the parent untreated population, indicating that this phenotype is secondarily acquired rather than the selection of previously existing low ROS cells.
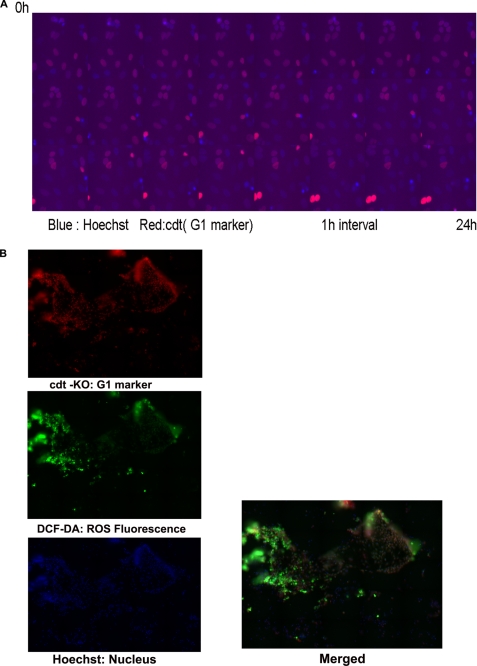
Results from the above studies indicate the possibility of generation of low ROS cells from senescent cells that contain high ROS by occasional reentry into the cell cycle. Further, to track the cell cycle progression of drug-treated cells, time lapse imaging was done in MDAMB 231 cells transfected with Cdt1-KO, a live cell marker for G1 cell cycle (17). The stable integration of Cdt1 and its remarkable property to indicate cell cycle transition was analyzed by live cell imaging after Hoechst staining. Cells in the G1 phase show red color in the nucleus with disappearance of nuclear red upon entry into S phase (24-h live cell imaging is shown in Fig. 3A and supplemental Video 1). The same cells were exposed to nocodazole, and resistant clones were developed as described. A representative well containing both senescent and resistant clones was stained with dichlorodihydrofluorescein diacetate and imaged using a BD Pathway imager to cover a large area in a well. It was confirmed that the senescence-evaded cells were in an actively dividing state with both red and blue nuclei. The newly emerged cells also showed low ROS levels (Fig. 3B). The image shows that multiple clones with low and high levels of ROS emerge after drug treatment in the transition stage. The observation suggests that occasionally some cells with low ROS are generated from the high ROS senescent population.
FIGURE 3.
Analysis of ROS in breast cancer cell lines and cell cycle status. A, MDAMB 231 cells were transfected with Cdt1-KO, a live cell marker for the G1 cell cycle phase. The stably expressing cells were generated as described. The stable integration of Cdt1 was validated by live cell imaging after Hoechst staining. Imaging was carried out for 24 h at an interval of 30 min. Cells in the G1 phase show red color in the nucleus with gradual disappearance of nuclear red upon entry into S phase and complete loss at G2 phase. Non-red blue nuclei in the merged image represent G2 phase cells. A representative merged image is shown. B, MDAMB 231 stably expressing Cdt1-KO cells grown on 96 imaging plates were treated with nocodazole as described. A representative well containing both senescent and growing clone was stained with dichlorodihydrofluorescein diacetate as described, counterstained with Hoechst. Imaging was carried out by a 96-well plate Bio-Imager with 5 × 5 montages to cover a large area of view. Multiple clones with varying level of ROS are seen. The red and non-red nuclei indicate the cycling status of cells.
Drug-surviving Cells Are Resistant to Drugs and Enriched with Tumor Stem Cell-like Properties
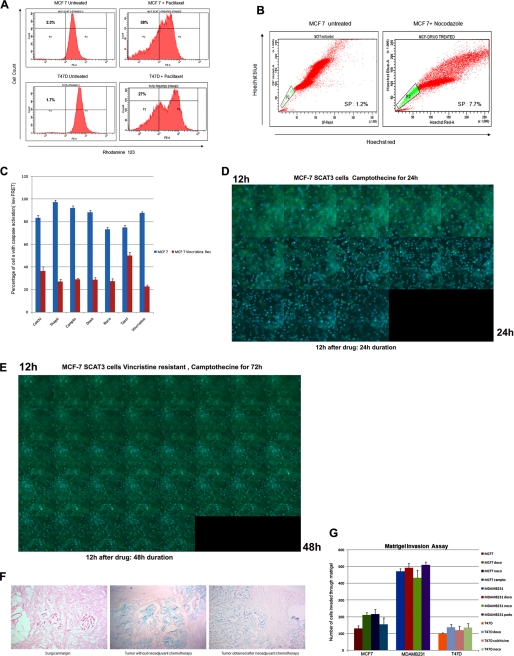
To examine whether drug surviving cells displayed higher drug efflux capacity, we performed an SP analysis and rhodamine 123 efflux assay. There was an enrichment of SP cells and Rholow population in the drug-selected cells (Fig. 4, A and B). The high proportion of Hoechst- or rhodamine-excluding cells could be associated with chemotherapeutic resistance, and this resistance may be partly due to the overexpression of ABC transporter proteins, which efflux drugs. Drug-selected cells exhibited high levels of cross-resistance to other structurally unrelated drugs (data not shown). The live cell FRET assay was done in drug-selected MCF-7 cells transfected with SCAT3.1. Very few cells showed caspase activation (low FRET). The percentage of parental MCF-7 and vincristine-resistant cells with loss of FRET for each drug is shown in Fig. 4C. Representative time lapse images of MCF-7 SCAT3.1 vincristine-resistant cells and parental cells exposed to camptothecin are given in Fig. 4, D and E. By 24 h of drug treatment, most parent cells showed an increase in ECFP fluorescence, indicating caspase activation (blue cells), the percentage of cells with FRET loss is significantly less in drug-resistant clone even at 48 h of camptothecin treatment (supplemental Videos 2 and 3).
FIGURE 4.
Characteristics of drug-selected cells. A, MCF-7 and T47D cells and drug-escaped clones from a paclitaxel-treated population were stained for the rhodamine 123 dye efflux assay as described. Drug efflux was analyzed by FACS. Drug-selected cells generated a significant rhodamine 123low fraction when compared with the parental cell line. B, MCF-7 and MCF-7 drug-escaped cells after treatment with nocodazole were analyzed for side population. An increase in side population cells is evident in the surviving clones. C, MCF-7 SCAT3 cells and MCF-7 SCAT3 vincristine-resistant clones were exposed to different drugs and imaged by FRET microscopy. The percentage of cells with loss of FRET is calculated for each drug, and the histogram is shown. D, MCF-7 SCAT3 cells and MCF-7 SCAT3 vincristine-resistant clones were exposed to camptothecin. After 12 h of drug treatment, the cells were placed in a live cell incubation chamber, and FRET imaging was carried out for an additional 12 h for MCF-SCAT3 cells and 48 h for vincristine-resistant cells at an interval of 10 min. Representative time lapse images of MCF7 SCAT3 cells (D) and vincristine-resistant cells (E) are shown. By 24 h of drug treatment, most parent cells showed an increase in ECFP fluorescence, indicating caspase activation (blue cells); the percentage of FRET lost cells is significantly less in drug-resistant clone even at 48 h of camptothecin treatment. F, breast tissue from surgical margin and from tumor with and without adjuvant chemotherapy was stained for SA-β-gal as described. Representative images are given. Strong positive staining for SA-β-gal is evident in the tumor samples. G, the respective parental cells and respective drug-surviving cells were evaluated for an in vitro Matrigel invasion assay as described. Each experiment was performed in triplicate, and the number of cells invaded across the membrane is presented. Error bars, S.D.
Our results indicate the possible role of drug-induced senescence in tumor recurrence through clonal expansion of multidrug-resistant cells with low ROS. We also saw increased SP in tumors from patients who received neoadjuvant chemotherapy. Upon analysis for the presence of senescent cells in the tumor samples, strong positive staining for SA-β-gal was evident (Fig. 4F). To investigate possible differences in invasion between postsenescent cells and parental cells, an in vitro Matrigel invasion assay was done. Fig. 4G shows that MCF-7 cells that evaded senescence were significantly more invasive than the parental cells.
Reactivation of Antioxidant Enzymes and Stem Cell Markers in Drug-escaped Cells
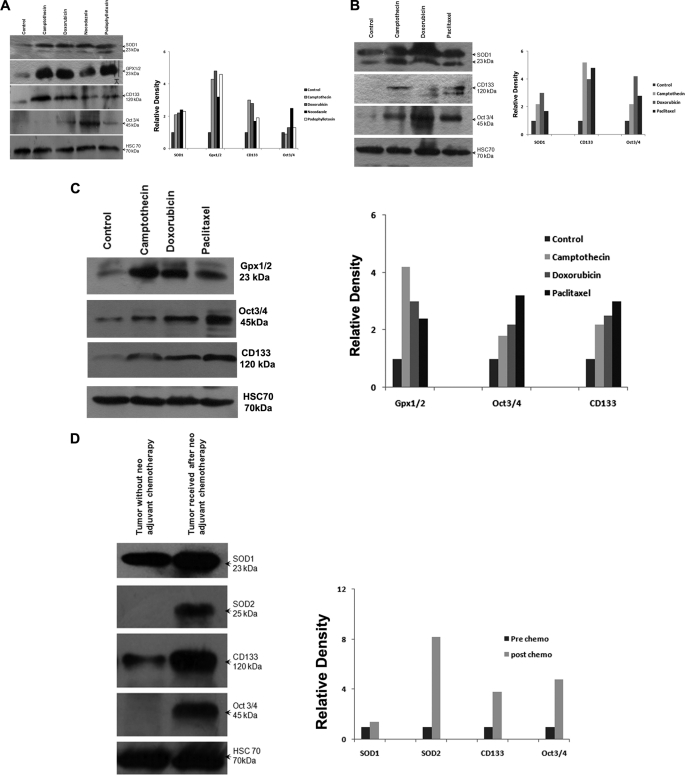
Previous studies have reported that normal stem cells as well as tumor stem cells are characterized by low intracellular ROS that often allows them to survive adverse conditions (6, 13). We observed very low levels of ROS in drug-selected cells compared with the parent untreated cells. This could be a secondary consequence that was acquired during drug treatment. Antioxidant enzymes such as superoxide dismutase (SOD) and glutathione peroxidase are closely associated with regulation of cellular ROS (18–20). Hence, to understand the relationship between antioxidant enzymes and acquired drug resistance in breast cancer, Western blot analysis for levels of antioxidant enzymes in the drug-surviving cells was done. Superoxide Dismutase 1 (SOD1) and glutathione peroxidase were more strongly expressed in the drug-surviving cells (Fig. 5, A–C). Consistent with in vitro data, analysis of the expression of key antioxidant enzymes, SOD1 and SOD2, in primary breast tumors indicated higher levels of expression in cases that received neoadjuvant chemotherapy compared with those that did not receive preoperative chemotherapy (Fig. 5D). Among 12 postchemotherapy samples analyzed, nine showed increased expression compared with samples without chemotherapy. Higher expression of the embryonic stem cell marker Oct-4 was also found in drug-surviving cells (Fig. 5, A–C).
FIGURE 5.
Analysis of Oct-4 and antioxidant enzymes in drug-selected cells and primary tumors. A, Western blot analysis of total SOD1, glutathione peroxidase, CD133, and Oct-4 levels from whole cell lysates of MCF-7 control and MCF-7 drug-selected cells (50 μg of protein/lane). Hsc70 served as loading control. Densitometry measurements of bands were quantitated using ImageJ software and shown as a graph. B, Western blot analysis of total SOD1, CD133, and Oct-4 levels from whole cell lysates of MDA MB231 control and MDA MB231 drug-selected cells (50 μg of protein/lane). Hsc70 was used as loading control. Densitometry measurements of bands were quantitated using ImageJ software and shown as a graph. C, Western blot analysis of total glutathione peroxidase, Oct-4, and CD133 levels from whole cell lysates of T47D control and T47D drug-selected cells (50 μg of protein/lane). Hsc70 was used as loading control. Densitometry measurements of bands were quantitated and are shown as relative density. D, Western blots of tissue extracts from primary breast tumors probed for SOD1, SOD2, CD133, and Oct-4 (50 μg of protein/lane). Hsc70 was used as loading control. Relative density of the bands analyzed by ImageJ software is shown as a graph.
Analysis of CSC Markers in Drug-selected Cells and Primary Tumors
To determine whether the drug-surviving cells have other intrinsic properties of stem cells (such as preferential expression of stem cell markers), flow cytometric analyses for expression of CSC and embryonic stem cell markers were performed. The drug-selected cells were enriched with a CD44+/CD24−/low subpopulation, which corresponds to the previously identified phenotype of breast CSCs (Fig. 6A). To find a correlation between CD44/CD24 staining profile and SP phenotype, immunophenotypic analysis of sorted SP and non-SP fractions from primary breast tumors was also performed by flow cytometry. We observed that SP cells possessed a significant CD44+/CD24−/low subset, suggesting that drug-resistant cells are more frequent in the tumor stem cell compartment (Fig. 6B). Drug-selected cells expressed higher levels of CD133 as compared with the parental cells (Figs. 5 (A–C) and 6C).
FIGURE 6.
Analysis of CSC markers in drug-selected cells and primary tumors. A, MDAMB 231 and drug-selected cells were stained using CD44-FITC and CD24-PE as described. Scatter plot of CD44+/CD24−/low subpopulation in MDAMB 231 drug-selected cells by flow cytometry is indicated. Cells in Q4 correspond to CD44+/CD24−/low cells. B, FACS analysis of candidate surface markers for breast cancer stem cells CD44 and CD24 in the SP and non-SP fractions from a primary breast tumor. The majority of SP cells showed the stem cell phenotype CD44+/CD24−/low. C, MCF-7 parental cells and resistant clones generated after the indicated drug treatment were stained with CD133 PE antibody. Respective FACS histograms are shown.
Reduced 26 S Proteasome Activity Contributes to Nrf2 Stabilization in Drug-escaped Cells
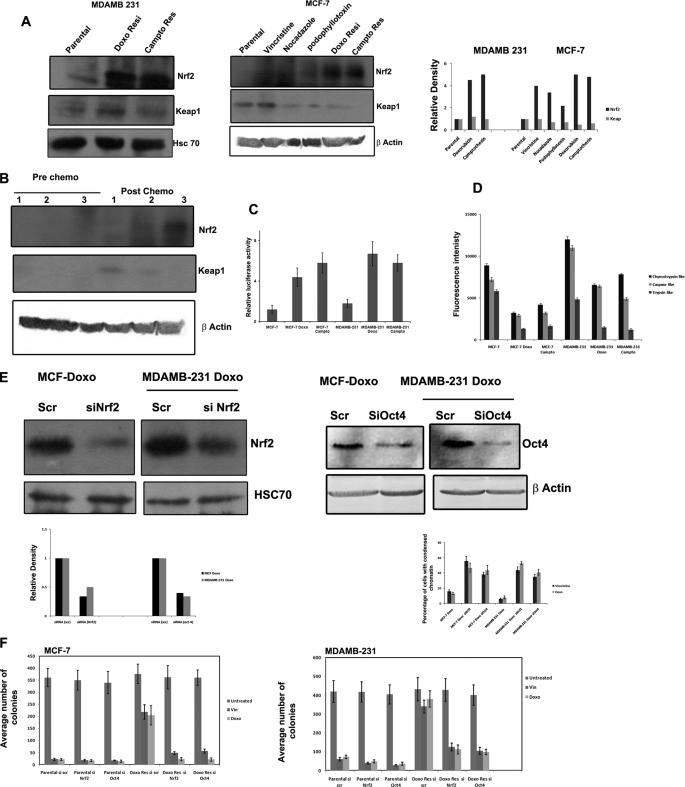
The above described results indicate that reactivation of antioxidant signaling is a critical step in tumor stem cell enrichment after drug exposure. Most of the cytoprotective antioxidant genes like SOD, GPX, etc. are under the control of the master regulator, transcription factor Nrf2, which is negatively regulated by Keap1 (21). In unstressed cells, the Keap1 targets its substrate, Nrf2, for ubiquitination and subsequent degradation by the 26 S proteasome. Inhibition of Keap1-dependent ubiquitination of Nrf2 increases the steady-state level of Nrf2 and enables activation of cytoprotective Nrf2-dependent genes. We analyzed the steady state level of Nrf2 in drug-escaped and parental cells by Western blot. Interestingly, all of the drug-escaped cells showed an increased constitutive Nrf2 level compared with untreated cells. However, the Western blot failed to show a concomitant decrease in Keap1 level in all of the drug-escaped cells (Fig. 7A). Immunofluorescent microscopy also supported an increase in the level of Nrf2 with occasional nuclear positivity in drug-escaped cells compared with parental cells (supplemental Fig. 4). Consistent with the increase in Nrf2 protein, antioxidant response reporter gene activity was also enhanced in drug-escaped cells, as seen from the NQO1-ARE Luc activity (Fig. 7C). The results indicate that Nrf2 stabilization and its transcriptional activity contribute to drug resistance and induction of antioxidant genes, such as SOD, GPX, and Oct-4. Because the Nrf2 induction is not associated with Keap1 down-regulation, we further analyzed whether 26 S proteasome activity essential for the degradation of Nrf2 is compromised in drug-escaped cells. As shown in Fig. 7D, chymotryptic, tryptic, and caspase-like 26 S proteasome activity were reduced in drug escaped cells compared with the parental population. The results indicate loss of 26 S proteasome activity to be the reason for increased Nrf2 activity in drug-escaped populations. We also used breast cancer tissue collected after and before chemotherapy to understand whether Nrf2 is induced after chemotherapy. Western blot analysis showed increased Nrf2 activity in postchemotherapy samples compared with prechemotherapy tissue samples (Fig. 7B). Because we observed increases in Oct-4 and Nrf2 consistently in all drug-escaped cells, RNAi silencing of both of the proteins was carried out to see whether acquired resistance can be reversed by knocking down these targets. As shown in Fig. 7E, the drug-escaped cells showed increased sensitivity to both vincristine and doxorubicin once Nrf2 or Oct-4 is silenced rather than the scrambled RNAi-transfected cells. Similarly, long term colony assay also substantiated that Nrf2 and Oct-4 silencing sensitizes the drug-resistant clones to drugs more than the parental cells (Fig. 7F and supplemental Fig. 4B).
FIGURE 7.
Nrf2 signaling in drug-escaped cells and possible signaling of CSC induction. A, the whole cell extracts prepared from MDAMB 231, MCF-7, and the indicated drug-resistant cells were separated on SDS-PAGE and blotted using antibodies against Nrf2, Keap1, Hsc70, and β-actin. B, whole cell extract prepared from three prechemotherapy and three postchemotherapy breast cancer patients were probed using antibodies against Nrf2 and Keap1. β-Actin served as loading control. C, MCF-7 and MDAMB 231 and the indicated drug-escaped cells were transfected with NQO1-ARE luciferase reporter plasmid and PRL −TK as control plasmid. 48 h after transfections, luciferase activity was measured as described. D, chymotryptic, tryptic, and caspase-like 26 S proteasome activities were determined using protein extract prepared from the indicated cell lines by spectrofluorimetry as described. Mean fluorescence intensity representing the corresponding enzymatic activity is shown (n = 3). E, doxorubicin-resistant MCF-7 and MDAMB 231 cells were transfected with either scrambled siRNA or siRNA against Oct-4 or Nrf2. The Western blot of Nrf2 and Oct-4 in transfected cells is shown to confirm the silencing. Densitometry measurements of bands were quantitated using ImageJ software and shown as graph. 48 h after transfection, the cells were treated with vincristine or doxorubicin for 24 h. The cell death was quantified after staining the cells with Hoechst dye to calculate the percentage of cells with condensed chromatin (n = 3). F, doxorubicin-resistant MCF-7 and MDAMB 231 cells as well as the respective parental cells were transfected with either scrambled siRNA or siRNA against Oct-4 or Nrf2. The cells were treated with vincristine or doxorubicin as above for 72 h. A long term colony assay was carried out as described. The average number of colonies counted for each group is represented as a graph (n = 3). Error bars, S.D.
p21-mediated Stabilization of Nrf2 Contributes to Tumor Stem Cell Enrichment
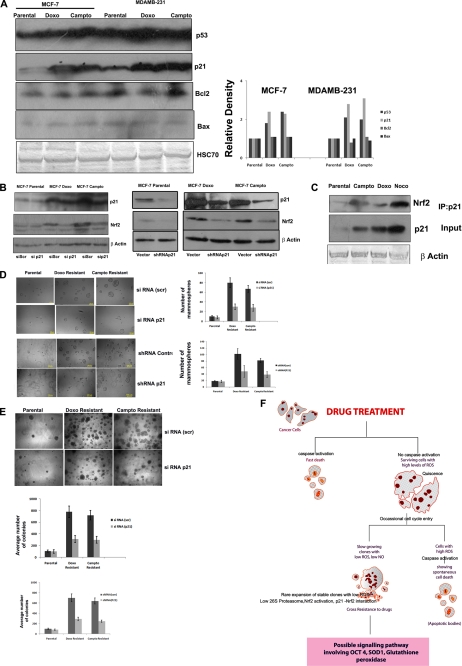
Genotoxic stress often induces the master regulator p53, promoting expression of many proteins associated with apoptosis and cell cycle arrest to counteract the stressors causing DNA damage. On the contrary, p53 is a direct target of 26 S proteasome. Because 26 S proteasome activity can regulate both pro- and antiapoptotic proteins, the status of p53 and its important targets was analyzed in drug-resistant clones. An increase in p53 was noticed in drug-escaped cells without significant change in the proapoptotic Bax or antiapoptotic Bcl2 proteins. However, a significant increase in p21 was noticed in all of the drug-resistant clones compared with control (Fig. 8A). A Keap1 independent regulation of Nrf2 was reported previously that involves its direct interaction with cyclin-dependent kinase inhibitor, p21 (22). Further, to understand whether the stabilization of Nrf2 in the drug-escaped cells also involves its direct interaction with p21 in addition to reduced 26 S proteasome, we immunoprecipitated the p21 from parental and drug-resistant clones, followed by probing with Nrf2 antibody. Consistent with the increase in p21 expression, an increased association of Nrf2 and p21 was noticed in drug-resistant clones compared with parental MCF-7 cells (Fig. 8C). Again, RNAi silencing of p21 reduced the expression of Nrf2 in drug-resistant clones, substantiating the importance of this interaction for the stabilization of Nrf2 (Fig. 8B).
FIGURE 8.
p21-mediated stabilization of Nrf2 contributes to tumor stem cell enrichment. A, the whole cell extract prepared from the indicated cells was probed with p53, p21, Bax, and Bcl2 as described. HSC70 served as loading control. Densitometry measurements of bands were quantitated using ImageJ software and shown as a graph. B, parental MCF-7 cells and doxorubicin-resistant and camptothecin-resistant clones were transfected with RNAi p21 and blotted for p21 and Nrf2. The left panel represents the indicated cells stably transfected with shRNA against p21 or with control vector, probed with p21, Nrf2, and β-actin antibodies. C, p21 was immunoprecipitated from parental MCF-7 cells and doxorubicin-resistant and camptothecin-resistant clones. Probed with Nrf2 antibody. The whole cell extract used for immunoprecipitation (IP) is probed with p21 and β-actin antibody (Input). D, parental MCF-7 cells and doxorubicin-resistant and camptothecin-resistant clones after indicated gene silencing were seeded on 96-well plates for mammosphere culture as described. The mammospheres formed are imaged with ×10 objective. Representative images are shown. The average number of mammospheres from three different experiments is used for plotting the graph. E, parental MCF-7 cells and doxorubicin-resistant and camptothecin-resistant clones after the indicated gene silencing were seeded on 24-well plates for a soft agar colony assay. Representative images and the graph of average colonies counted in each group are shown (n = 3). F, the possible signaling involved in stem cell enrichment after chemotherapy is presented. Anticancer drugs eliminate most of the cells through classical caspase-mediated cell death, leaving some quiescent senescent-like cells. These senescent cells are characterized by high ROS, and they rarely enter into the cell cycle, which often culminates in death at mitosis by a caspase-mediated mechanism. Some cells appear to escape spontaneous cell death through the reactivation antioxidant system by stabilizing Nrf2 subsequent to reduced 26 S proteasome activity and by direct interaction with p21.
The ability to form mammosphere in low attachment growth conditions is a good index of the tumor-forming potential of stem cells. We have analyzed the mammosphere formation in the drug-resistant clones as well as the parental MCF-7 cells. As shown in Fig. 8D, compared with parental cells, the number and size of mammospheres formed are greater in drug-escaped cells. Interestingly, the silencing of p21 reduced the mammosphere-forming potential. The soft agar colony assay also showed similar results both in transient and stable silencing of p21 (Fig. 8E). Overall, the results substantiated that apart from reduced 26 S proteasome activity, p21-mediated stabilization of Nrf2 plays a critical role in the emergence of drug-resistant cells with reduced ROS.
DISCUSSION
Recent research has provided compelling evidence for presence of cancer stem cells in a variety of tumors and their potential to reinitiate the whole tumor with more aggressiveness. Conventional treatments fail to target such cells with an aggressive phenotype that eventually return as stronger and therapy-resistant tumors. In this study, we provide further evidence for clonal expansion of cells with a stemlike phenotype after drug treatment that is often preceded with an intermediate senescence-like phenotype.
Previously, several studies have reported the generation of drug-resistant cells with or without stem cells characteristic after initial drug treatment. In a remarkable study by Sharma et al. (23), a unique reversible drug-tolerant state was observed as an acute response of non-small cell lung cancer cells after a lethal exposure of epidermal growth factor receptor-tyrosine kinase inhibitor. They also observed that drug-tolerant cells were enriched with the putative progenitor marker CD133. In the current study, we employed some widely used antitumor agents to test the nature of drug-tolerant cells and found that resistant cells were more often generated with a drug that transiently induces a senescent-like phenotype and not by other drugs, such as kinase inhibitors or proteasome inhibitors. Only a few compounds were capable of inducing a senescent-like phenotype that is always coupled with an initial phase of massive cell death by caspase activation. The senescent cells most often showed resistance against a second phase of drug treatment. Gradual spontaneous death was seen in some subsets of postsenescent cells. Time lapse imaging and high throughput imaging of caspase sensor-expressing cells indirectly revealed that senescent cells may reenter the cell cycle, leading to the generation of drug-sensitive and drug-resistant fractions.
Cellular senescence is often described as the response of normal cells undergoing limited replication before entering into a terminally growth-arrested state due to telomere attrition (24–26). Tumor cells are often thought of as immortal with an irreversible loss of senescence response. Tumor cells also retain the capacity to undergo senescence in response to genotoxic stress, radiotherapy, and chemotherapy (27, 28). This accelerated senescence or pseudosenescence is also considered as a strategy to limit tumor growth. The presence of senescent cells has been observed in primary tumors undergoing radio- and chemotherapy. Cells with premature senescent phenotype are also abundant in premalignant lesions. Senescent cells are generally resistant to apoptosis and can serve as reservoirs of secreted factors for mitogenic and angiogenic activity, leading to tumor recurrence (29).
Preceding studies indicate that drug-induced senescent cells rarely reenter cell cycle (30–32). It has also been reported that paracrine factors produced by senescent cells contribute to growth and survival of tumor cells (33). Previous studies have also suggested that drug-induced senescent cells can bypass the senescence plateau and give rise to highly tumorigenic cells (32, 34). A study involving p21-induced senescence provided evidence for reentry of cells into mitosis after prolonged cell cycle arrest but with grossly abnormal mitotic figures (35). Recent studies have also suggested that the drug-tolerant phenotype might be acquired through chromatin alterations that occur due to stress-induced mutagenesis (23). It is possible that drug-induced quiescence in senescent cells is regulated by excessive cytosolic ROS. However, some epigenetic changes may induce the cells to undergo asymmetric division leading to generation of cells with high ROS and low ROS, resulting in enrichment of stemlike cells with low ROS. Further studies are required to experimentally validate this hypothesis.
Recent studies have shown that tumor stem cells retain low levels of ROS, which contribute to less DNA damage and to protection from radiation injury (16). Our study favors the concept that drug-induced senescence contributes to selective expansion of cells with low ROS exhibiting high drug efflux capacity and invasiveness. However, currently, the molecular switch that triggers senescent cells with high ROS to enter into the cell cycle, leading to the generation of cells with two distinct phenotypes, is not clear. In fact, several studies have reported that low levels of ROS can activate cell proliferation and gene expression by inactivating phosphatases and activating kinases (36). Thus, ROS can contribute to the transcriptional activation of drug transporter proteins and matrix metalloproteases that enhance drug resistance as well as invasiveness (33). It is also possible that occasional reactivation of the antioxidant system by senescent cells help them to escape senescence and enter the cell cycle.
Quite surprisingly, the results demonstrate that drug-selected cells and tumors obtained after preoperative chemotherapy contain increased levels of Oct-4, a transcription factor seen in embryonic stem cells. Recent studies have shown that overexpression of Oct-4 enhanced whereas Oct-4 knockdown reduced liver cancer cell resistance to chemotherapeutic drugs in vitro and in xenograft tumors (37). It has also been reported that Oct-4 contributes to the stem cell-like phenotypes found in many tumors (38–40). Expression of CD133 and Oct-4 was also associated with poor clinical response to chemotherapy and worse survival in patients in stage IV colorectal cancer (41). Recent studies have also shown that the DNA-binding activity of Oct-4 is sensitive to oxidation. Along these lines, our results indicate that Oct-4 expression may be a key element in breast cancer in the development of a chemoresistant phenotype.
Drug-selected cells possessed an increased CD44+/CD24low subpopulation, which has been identified as breast cancer-initiating cells (1). It has been reported that breast tumors from patients treated with chemotherapy contained a higher proportion of CD44+/CD24low cells with CSC properties compared with breast tumors from untreated patients. The stem/progenitor cell phenotype in our drug-selected cells was further confirmed by the expression of CD133, a putative CSC marker. CD133 expression has been reported in CSCs of several different cancers, including brain, prostate, and lung (2, 42–44). CD133 expression has also been found to be associated with chemoresistance in Ewing sarcoma cells (45).
The study identified a role for Nrf2 in determining the induction of stem cell-like properties and reactivation of antioxidant signaling after drug exposure. Nrf2, being a master regulator of antioxidant signaling, regulates its downstream target genes by binding to the AREs in the promoters of many genes involved in detoxification and cell survival(21). Several recent studies implicate Nrf2-dependent antioxidant signaling to play a key role in cellular protection against many detrimental effects and drug-induced toxicity. The activity of Nrf2 is tightly controlled primarily at the protein level by a negative regulator Keap1, an adaptor for a Cul3-based E3 ligase that promotes constitutive proteasome-mediated Nrf2 degradation. Quite interestingly, we observed an enhanced constitutive expression of Nrf2 in drug-escaped cells as well as in postchemotherapy breast cancer tissues. However, the expression of its inhibitor, Keap1, was not significantly altered in these cells, indicating a Keap1 independent regulation of Nrf2 for the transition. In fact, Keap1 sequesters Nrf2 in the cytoplasm, thereby promoting its ubiquitination and degradation by 26 S proteasome complex. A recent study using Nrf2 mutant lacking the Keap1-interacting motif reported its ability to accumulate rapidly in the presence of MG132, suggesting Keap1-independent proteasome-mediated degradation of Nrf2 (46). We analyzed the most important component of Nrf2 degradation, 26 S proteasome activity using fluorescent substrates that showed marked reduction of 26 S proteasome activity in drug-resistant cells compared with parental populations. Recent studies also indicated reduced 26 S proteasome activity in tumor stem cell-like compartment, although the mechanism behind this is currently not known (47). Reduced proteasome activity coincided with the expression of stem cell markers and lack of differentiation markers. Our observation is also consistent with previous data that showed that, unlike Keap1, Nrf2 degradation is highly dependent on 26 S proteasome activities (46). Further studies were done to address involvement of any other signaling apart from the reduced 26 S proteasome in stabilization of Nrf2 because both pro- and antiapoptotic proteins are under the regulative control of 26 S proteasome. Quite interestingly, drug-escaped cells showed enhanced expression of cyclin-dependent kinase inhibitor p21. Consistent with the induction of p21, its close association with Nrf2 appears to play a critical role in the stabilization of Nrf2. It is interesting to note that p21 is also an important target of 26 S proteasome. The importance of the interaction of p21 and Nrf2 in the drug resistance and tumor recurrence was further substantiated by the reduction of mammosphere formation and soft agar colony formation of the drug-escaped cells concomitant with RNAi silencing of p21. The increased expression of Oct-4 also appears to be contributed by reduced 26 S proteasome activity because Oct-4 regulation is known to be regulated by ubiquitin-mediated degradation (48). Quite interestingly, silencing of both Nrf2 and Oct-4 sensitized the drug-resistant cells to anticancer drugs, opening a therapeutic window to address acquired resistance.
Most of the drugs used as chemotherapeutics induce genomic instability on tumor cells. A recent study implicated genomic instability as a cause for tumor stem cell induction (49). DNA damage and ROS generation may significantly induce genomic instability, leading to massive cell death. It is possible that the emergence of drug resistant stem cell-like population is augmented by genomic instability-mediated heterogeneity, where induction of Nrf2 helps the survival of cells from ROS-mediated genomic instability. In support of the antioxidant system, we observed a significant increase in the expression of primary antioxidant enzymes, such as SOD1 and glutathione peroxidase, in the drug-selected cells. Higher levels of antioxidant enzymes were also found in breast tumors from patients who received neoadjuvant therapy. Previous studies have shown that in tumor tissues, total and mitochondrial SOD activities were increased compared with noncancerous tissues, and this increase was dependent upon overexpression of the enzymes (50). It has also been shown that lower ROS levels in breast CSCs are associated with increased expression of free radical scavenging systems (16). This suggests that total antioxidant capacity may be a critical determinant for the transition of high ROS to low ROS levels (Fig. 7F). Increased expression of antioxidant molecules may be an effective adaptive mechanism to keep ROS levels within the range that allows cancer cells to escape severe oxidative damage and survive under stress and ROS-mediated mutations.
In short, our studies suggest that drug-induced senescence contributes to sustenance of tumor stem cell-like cells with low ROS, providing additional deleterious effects apart from its indirect contribution of tumorigenicity by soluble factors. It is also quite interesting to note that the senescence-inducing potential and generation of drug-tolerant cells appear to be restricted to certain groups of drugs and not all. To conclude, we believe that our studies for the first time define molecular events associated with the emergence of drug-resistant stem cell-like cells after chemotherapy and identify Nrf2, p21, and 26 S proteasome as key players for the transition from high ROS to low ROS state involving reactivation of antioxidant defense signaling.
Acknowledgments
We thank the Flow Cytometry Technical Support Team (Rajiv Gandhi Centre for Biotechnology) for FACS experiments. We also thank Dr. Atsushi Miyawaki (RIKEN, Japan) for the generous gift of the expression vectors SCAT3 and Cdt-KO, Dr. Masayuki Yamamoto (University of Tsukuba) for NQO1-Luc vector, and Dr. Ben Ho Park (Johns Hopkins University School of Medicine) for shRNAp21 vector.
This work was supported by Department of Biotechnology, Government of India, Grant BT/PR/7793/MED/14/1112/2006 (to M. R. P.) and Innovative Young Biotechnologist Award Grant BT/B1/12/040/2005 (to T. R. S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2, Figs. 1–4, and Videos 1–3.
- CSC
- cancer stem cell
- SOD
- superoxide dismutase
- ROS
- reactive oxygen species
- SP
- side population
- AMC
- 7-amido-4-methylcoumarin
- ECFP
- enhanced cyan fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- KO
- Kusabira Orange
- SA-β-gal
- senescence-associated β-galactosidase
- ARE
- antioxidant response element.
REFERENCES
- 1. Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 3. Ponti D., Costa A., Zaffaroni N., Pratesi G., Petrangolini G., Coradini D., Pilotti S., Pierotti M. A., Daidone M. G. (2005) Cancer Res. 65, 5506–5511 [DOI] [PubMed] [Google Scholar]
- 4. Goodell M. A., McKinney-Freeman S., Camargo F. D. (2005) Methods Mol. Biol. 290, 343–352 [DOI] [PubMed] [Google Scholar]
- 5. Ho M. M., Ng A. V., Lam S., Hung J. Y. (2007) Cancer Res. 67, 4827–4833 [DOI] [PubMed] [Google Scholar]
- 6. Chen M. F., Lin C. T., Chen W. C., Yang C. T., Chen C. C., Liao S. K., Liu J. M., Lu C. H., Lee K. D. (2006) Int. J. Radiat. Oncol. Biol. Phys. 66, 244–253 [DOI] [PubMed] [Google Scholar]
- 7. Fillmore C. M., Kuperwasser C. (2008) Breast Cancer Res. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li X., Lewis M. T., Huang J., Gutierrez C., Osborne C. K., Wu M. F., Hilsenbeck S. G., Pavlick A., Zhang X., Chamness G. C., Wong H., Rosen J., Chang J. C. (2008) J. Natl. Cancer Inst. 100, 672–679 [DOI] [PubMed] [Google Scholar]
- 9. Hu L., McArthur C., Jaffe R. B. (2010) Br. J. Cancer 102, 1276–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dimri G. P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E. E., Linskens M., Rubelj I., Pereira-Smith O. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karakas B., Weeraratna A. T., Abukhdeir A. M., Konishi H., Gustin J. P., Vitolo M. I., Bachman K. E., Park B. H. (2007) Cancer Biol. Ther. 6, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vlashi E., Kim K., Lagadec C., Donna L. D., McDonald J. T., Eghbali M., Sayre J. W., Stefani E., McBride W., Pajonk F. (2009) J. Natl. Cancer Inst. 101, 350–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joseph J., Seervi M., Sobhan P. K., Retnabai S. T. (2011) PLoS One 6, e20114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vogler M., Weber K., Dinsdale D., Schmitz I., Schulze-Osthoff K., Dyer M. J., Cohen G. M. (2009) Cell Death Differ. 16, 1030–1039 [DOI] [PubMed] [Google Scholar]
- 15. Ozben T. (2007) J. Pharm. Sci. 96, 2181–2196 [DOI] [PubMed] [Google Scholar]
- 16. Diehn M., Cho R. W., Lobo N. A., Kalisky T., Dorie M. J., Kulp A. N., Qian D., Lam J. S., Ailles L. E., Wong M., Joshua B., Kaplan M. J., Wapnir I., Dirbas F. M., Somlo G., Garberoglio C., Paz B., Shen J., Lau S. K., Quake S. R., Brown J. M., Weissman I. L., Clarke M. F. (2009) Nature 458, 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., Imamura T., Ogawa M., Masai H., Miyawaki A. (2008) Cell 132, 487–498 [DOI] [PubMed] [Google Scholar]
- 18. Portakal O., Ozkaya O., Erden Inal M., Bozan B., Koan M., Sayek I. (2000) Clin. Biochem. 33, 279–284 [DOI] [PubMed] [Google Scholar]
- 19. Ramanathan B., Jan K. Y., Chen C. H., Hour T. C., Yu H. J., Pu Y. S. (2005) Cancer Res. 65, 8455–8460 [DOI] [PubMed] [Google Scholar]
- 20. Rao A. K., Ziegler Y. S., McLeod I. X., Yates J. R., Nardulli A. M. (2008) Mol. Endocrinol. 22, 1113–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwak M. K., Kensler T. W. (2010) Toxicol. Appl. Pharmacol. 244, 66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen W., Sun Z., Wang X. J., Jiang T., Huang Z., Fang D., Zhang D. D. (2009) Mol Cell 34, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharma S. V., Lee D. Y., Li B., Quinlan M. P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M. A., Wong K. K., Brandstetter K., Wittner B., Ramaswamy S., Classon M., Settleman J. (2010) Cell 141, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim N. W., Piatyszek M. A., Prowse K. R., Harley C. B., West M. D., Ho P. L., Coviello G. M., Wright W. E., Weinrich S. L., Shay J. W. (1994) Science 266, 2011–2015 [DOI] [PubMed] [Google Scholar]
- 25. Chen Q., Fischer A., Reagan J. D., Yan L. J., Ames B. N. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4337–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sahin E., Depinho R. A. (2010) Nature 464, 520–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chang B. D., Xuan Y., Broude E. V., Zhu H., Schott B., Fang J., Roninson I. B. (1999) Oncogene 18, 4808–4818 [DOI] [PubMed] [Google Scholar]
- 28. te Poele R. H., Okorokov A. L., Jardine L., Cummings J., Joel S. P. (2002) Cancer Res. 62, 1876–1883 [PubMed] [Google Scholar]
- 29. Zheng X., Chou P. M., Mirkin B. L., Rebbaa A. (2004) Cancer Res. 64, 1773–1780 [DOI] [PubMed] [Google Scholar]
- 30. Romanov S. R., Kozakiewicz B. K., Holst C. R., Stampfer M. R., Haupt L. M., Tlsty T. D. (2001) Nature 409, 633–637 [DOI] [PubMed] [Google Scholar]
- 31. Sundaram M., Guernsey D. L., Rajaraman M. M., Rajaraman R. (2004) Cancer Biol. Ther. 3, 207–218 [DOI] [PubMed] [Google Scholar]
- 32. Karimi-Busheri F., Rasouli-Nia A., Mackey J. R., Weinfeld M. (2010) Breast Cancer Res. 12, R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Radisky D. C., Levy D. D., Littlepage L. E., Liu H., Nelson C. M., Fata J. E., Leake D., Godden E. L., Albertson D. G., Nieto M. A., Werb Z., Bissell M. J. (2005) Nature 436, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Levina V., Marrangoni A. M., DeMarco R., Gorelik E., Lokshin A. E. (2008) PLoS One 3, e3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang B. D., Watanabe K., Broude E. V., Fang J., Poole J. C., Kalinichenko T. V., Roninson I. B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4291–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hancock J. T., Desikan R., Neill S. J. (2001) Biochem. Soc. Trans. 29, 345–350 [DOI] [PubMed] [Google Scholar]
- 37. Wang X. Q., Ongkeko W. M., Chen L., Yang Z. F., Lu P., Chen K. K., Lopez J. P., Poon R. T., Fan S. T. (2010) Hepatology 52, 528–539 [DOI] [PubMed] [Google Scholar]
- 38. Guo Y., Einhorn L., Kelley M., Hirota K., Yodoi J., Reinbold R., Scholer H., Ramsey H., Hromas R. (2004) Stem Cells 22, 259–264 [DOI] [PubMed] [Google Scholar]
- 39. Chen Y. C., Hsu H. S., Chen Y. W., Tsai T. H., How C. K., Wang C. Y., Hung S. C., Chang Y. L., Tsai M. L., Lee Y. Y., Ku H. H., Chiou S. H. (2008) PLoS One 3, e2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wong D. J., Liu H., Ridky T. W., Cassarino D., Segal E., Chang H. Y. (2008) Cell Stem Cell 2, 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ong C. W., Kim L. G., Kong H. H., Low L. Y., Iacopetta B., Soong R., Salto-Tellez M. (2010) Mod. Pathol. 23, 450–457 [DOI] [PubMed] [Google Scholar]
- 42. Collins A. T., Maitland N. J. (2006) Eur. J. Cancer 42, 1213–1218 [DOI] [PubMed] [Google Scholar]
- 43. Miki J., Furusato B., Li H., Gu Y., Takahashi H., Egawa S., Sesterhenn I. A., McLeod D. G., Srivastava S., Rhim J. S. (2007) Cancer Res. 67, 3153–3161 [DOI] [PubMed] [Google Scholar]
- 44. Eramo A., Lotti F., Sette G., Pilozzi E., Biffoni M., Di Virgilio A., Conticello C., Ruco L., Peschle C., De Maria R. (2008) Cell Death Differ. 15, 504–514 [DOI] [PubMed] [Google Scholar]
- 45. Jiang X., Gwye Y., Russell D., Cao C., Douglas D., Hung L., Kovar H., Triche T. J., Lawlor E. R. (2010) BMC Cancer 10, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McMahon M., Itoh K., Yamamoto M., Hayes J. D. (2003) J. Biol. Chem. 278, 21592–21600 [DOI] [PubMed] [Google Scholar]
- 47. Pan J., Zhang Q., Wang Y., You M. (2010) PLoS One 5, e13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xu H., Wang W., Li C., Yu H., Yang A., Wang B., Jin Y. (2009) Cell Res. 19, 561–573 [DOI] [PubMed] [Google Scholar]
- 49. Liang Y., Zhong Z., Huang Y., Deng W., Cao J., Tsao G., Liu Q., Pei D., Kang T., Zeng Y. X. (2010) J. Biol. Chem. 285, 4931–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhong W., Oberley L. W., Oberley T. D., St Clair D. K. (1997) Oncogene 14, 481–490 [DOI] [PubMed] [Google Scholar]