Abstract
The RecA protein is an ATPase that mediates recombination via strand exchange. In strand exchange a single-stranded DNA (ssDNA) bound to RecA binding site I in a RecA/ssDNA filament pairs with one strand of a double-stranded DNA (dsDNA) and forms heteroduplex dsDNA in site I if homology is encountered. Long sequences are exchanged in a dynamic process in which initially unbound dsDNA binds to the leading end of a RecA/ssDNA filament, while heteroduplex dsDNA unbinds from the lagging end via ATP hydrolysis. ATP hydrolysis is required to convert the active RecA conformation, which cannot unbind, to the inactive conformation, which can unbind. If dsDNA extension due to RecA binding increases the dsDNA tension, then RecA unbinding must decrease tension. We show that in the presence of ATP hydrolysis decreases in tension induce decreases in length whereas in the absence of hydrolysis, changes in tension have no systematic effect. These results suggest that decreases in force enhance dissociation by promoting transitions from the active to the inactive RecA conformation. In contrast, increases in tension reduce dissociation. Thus, the changes in tension inherent to strand exchange may couple with ATP hydrolysis to increase the directionality and stringency of strand exchange.
INTRODUCTION
Universally-conserved RecA family proteins mediate DNA recombination and recombinational repair (1–4). RecA presents two binding sites, sites I and II that can each bind to either double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA) (5,6). The binding of either dsDNA or ssDNA to either binding site results in an average extension of ∼0.51 nm/bp (7,8), which is substantially larger than the B-form extension of 0.34 nm/bp. This 0.17 nm/bp change in extension of DNA by RecA protein is believed to play an important role in strand exchange (9). Earlier work on Rad51, a eukaryotic analog of RecA, has suggested that the extension of the dsDNA by the protein results in a tension in the dsDNA that spring loads the filament (10). Such spring loading may promote the unbinding of the protein from the dsDNA-Rad51 filaments.
The initial step in strand exchange is the formation of RecA/ssDNA filaments due to binding of an incoming ssDNA to site I in RecA, which has a higher binding affinity than site II (11,12). The RecA in the filament is in the active state with bound ATP. In strand-exchange the RecA/ssDNA filament searches for homologous dsDNA by binding to dsDNA (13–15). If the dsDNA is not homologous to the ssDNA in the filament, then the RecA/ssDNA filament rapidly unbinds from the dsDNA (14,15). If the dsDNA is homologous, one strand of the dsDNA switches partners and pairs with the incoming ssDNA strand bound in site I. This pairing yields heteroduplex dsDNA in site I and the left-over outgoing ssDNA strand in site II (13–15). Though RecA is an ATPase, the exchange in Watson–Crick pairing that creates the heteroduplex dsDNA during recombination occurs in the absence of ATP hydrolysis (16,17). ATP hydrolysis promotes certain complex RecA-mediated reactions, e.g. progression of strand exchange through short non-sequence matched regions (18,19) and repair of stalled replication forks (20), as well as the release of the heteroduplex dsDNA in site I. Observations of hydrolysis waves have been reported in vitro, but their role in vivo is unclear (21).
The initial binding of the RecA/ssDNA filament to ∼10 bp (22–24) on a homologous dsDNA strand is followed by an extension of the RecA/ssDNA filament along the dsDNA until ∼80 bp (25) or more (26) are incorporated in the RecA/ssDNA–dsDNA complex. This RecA/ssDNA–dsDNA complex is referred to as the strand exchange window. In the presence of ATP hydrolysis, if a longer sequence is participating in strand exchange, then the heteroduplex dsDNA at the lagging end of the strand exchange window is ejected from the filament as dsDNA is bound at the leading edge of the strand exchange window (25). This allows for strand exchange to proceed along the dsDNA for many thousands of base pairs even though fewer base pairs are bound in the strand exchange window at any given time. In contrast, in the absence of hydrolysis, the RecA/ssDNA–dsDNA complex continues to incorporate more dsDNA at the leading end without releasing anything at the lagging end, resulting in RecA/ssDNA–dsDNA complexes that can contain thousands of base pairs (27).
ATP hydrolysis is required for the release of heteroduplex dsDNA from site I of the RecA filament following strand exchange (28) because RecA in the active state cannot unbind from dsDNA (21). Only RecA in the inactive state with bound ADP can unbind from the dsDNA. Thus hydrolysis is required to convert the active form with bound ATP to the inactive form with bound ADP. Earlier work on Rad51 has shown that both the active and inactive forms can be bound simultaneously to the same dsDNA molecule (10,29). It has been suggested that dsDNA bound to Rad51 is spring loaded when it is extended by Rad51 (10). Similarly if dsDNA bound to RecA is spring loaded, then changing the tension on the dsDNA may alter the function of the protein. Earlier work on Rad51 has already shown that the unbinding rate for Rad51 bound to dsDNA is reduced if a tension is applied to the dsDNA indicating that constant tension affects protein function (10). If the bound dsDNA is under tension because the dsDNA is extended, then during strand exchange the tension on the dsDNA must not be constant: the tension at the leading end is increasing as dsDNA is incorporated into the filament, whereas the tension at the lagging end is decreasing as the newly formed heteroduplex dsDNA is released from the strand exchange window. In this work, we consider the possibility that such changes in tension can affect RecA function.
We use our magnetic tweezers apparatus (30,31) to measure the extension of dsDNA as a function of a time-dependent force in the absence and in the presence of free RecA to probe for functional changes in the protein resulting from changes in the tension on the dsDNA. Rather than considering the full complexity of the strand exchange window, we simply consider the effect of tension on dsDNA bound to site I, which is the position of the dsDNA after strand exchange. We note that the unbinding of the dsDNA that occurs at the lagging end of the strand exchange window is the unbinding of dsDNA from site I. In this work, we consider two types of temporal changes in force: stepwise and quasi-continuous. We also consider two different filament states: (i) dsDNA molecules that are completely or nearly completely covered by RecA; and (ii) dsDNA molecules that are only partially covered by RecA, where RecA polymerization and depolymerization may be occurring. In addition, we performed experiments on filaments in the absence of free RecA, so RecA binding from solution is not possible, and in the presence of free RecA in solution.
MATERIALS AND METHODS
Our magnetic tweezers apparatus exerts forces on dsDNA constructs by pulling from labels attached to ssDNA tails at the ends of the dsDNA (30,31). Here, we use dsDNA pulled from the 3′5′-ends of one strand. Each ssDNA tail has six biotin labels which enable one dsDNA end to bind to an extravidin coated 4.5 µm superparamagnetic bead while the other end binds to an extravidin coated glass capillary surface. The beads and 3′5′-DNA constructs are incubated in a solution of RecA buffer pH 7.6 (70 mM Tris, 10 mM MgCl2 and 5 mM dithiothreitol) with 1 µM RecA protein (NEB) and either 1 mM ATP or 1 mM ATPγS for ∼10 min, after which the desired force regime is imposed. In some experiments that examine the effects of reduced hydrolysis, MgCl2 is replaced by CaCl2 to reduce the hydrolysis rate (32). The force exerted on the magnetic beads by a magnet is controlled by the distance between the magnet and the capillary surface. The extension of the dsDNA is measured by monitoring the separation between the surface of the capillary and the surface of the magnetic bead using an inverted optical microscope (30,31).
Flow experiments are performed by connecting the capillary containing the sample to polypropylene tubing and a syringe with buffer. The typical flow rates are 1–3 µl/min and at least one or two full capillary volumes are exchanged in between measurements. One dsDNA molecule was overstretched in the presence of 1 µM RecA and 10 mM ATP until a complete filament was obtained. Then the solution in the capillary was fully exchanged by RecA buffer containing a mixture of 10 mM ATP and 10 µM ATPγS.
RESULTS
Experiments on fully covered dsDNA molecules in the presence of free RecA
Polymerization of RecA in the active state extends the structure of B-DNA by ∼1.5 times (7,8,33); however, the extension of the dsDNA bound to the inactive state is less well known with crystallographic data for RecA/ADP filaments giving values from 1.2× to 1.4× (34). Thus, if the bound RecA is all in the active state; i.e. in the presence of a non-hydrolyzable form of ATP, then the number of active RecA molecules bound to one dsDNA can be determined by measuring, L, the extension of dsDNA single molecules. In contrast, if both active and inactive conformations of RecA are simultaneously bound to dsDNA, then measurements of L are not sufficient to determine the number of bound RecA.
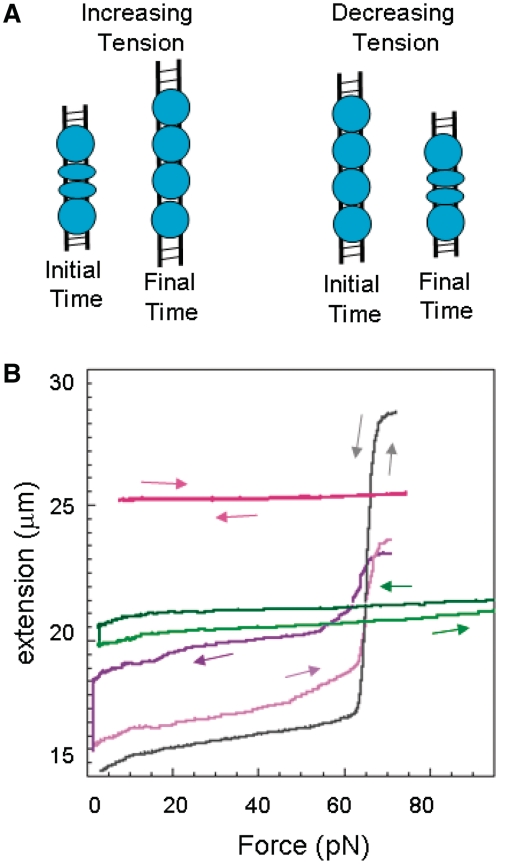
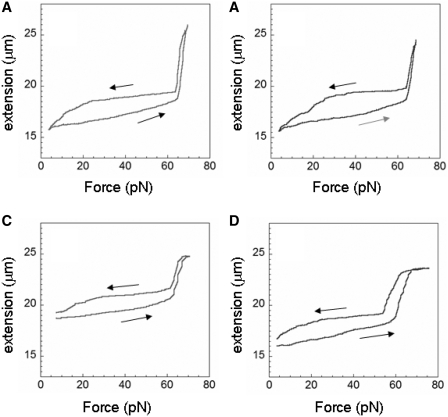
At high ATP concentrations, RecA rapidly binds to stretched dsDNA resulting in complete or nearly complete filaments. Even for a full filament, where all of the RecA remains bound, length changes are possible if the bound RecA makes transitions between the active (circles) and inactive (ovals) states as illustrated in Figure 1A and Supplementary Figure S1. Figure 1B shows the measured extension versus force curves for particular single molecules. The curves are obtained by increasing and decreasing the force at a rate of ∼1 pN/sec. The gray curve corresponds to a control sample in the absence of free RecA, where there is a sudden change in extension at 65 pN resulting in a length increase ΔLoverstretch of ∼11.5 microns, known as the overstretching transition. In contrast, dsDNA completely covered by RecA does not overstretch, as illustrated by the magenta curve which corresponds to a complete RecA filament formed in buffer containing ATPγS. In general, the fraction of dsDNA covered by RecA can be estimated by measuring ΔLoverstretch since B-DNA participates in the overstretching transition, whereas dsDNA covered by RecA does not. We note that at forces below the overstretching transition, the extension of B-dsDNA is shorter than the extension of dsDNA bound to RecA, whereas at forces above the overstretching transition the extension of dsDNA not bound to RecA is longer than the extension of dsDNA bound to RecA.
Figure 1.
Effects of dynamics on dsDNA molecules completely and partially covered by RecA. (A) Schematic representation of dsDNA (black lines) bound to RecA (blue) where the circles represent the ATP-RecA conformation and the ovals the ADP-form. (B) Extension versus force curves for a control in the absence of RecA (gray), a full filament in ATPγS (magenta) and 1 µM RecA, and cycles in buffer containing 4 mM ATP (purple) and 10 mM ATP (green) and 1 µM RecA, where the dark colors show the first half of the cycles. The arrows indicate the direction of the force change.
In ATPγS, conversion to the inactive state is almost negligible; therefore, conformational changes and unbinding do not play a role and the amount of bound RecA at any force can be determined by measuring the extension at that force. In contrast, in buffers where hydrolysis is significant, RecA binding and unbinding and/or conversions between the active and inactive states of bound RecA may play a substantial role in altering the measured extension. Thus, if hydrolysis takes place, at most forces the measured extension does not determine the amount of bound RecA; however, the fraction bound at the overstretching transition can still be determined by measuring ΔLoverstretch.
The purple and green curves in Figure 1B correspond to cycles in buffer containing 4 and 10 mM ATP, respectively, where the arrows indicate the change in force with time and the darker curves correspond to the first half of each cycle. For both curves, the rate of change of the force was ∼1 pN/s except for the several minute pauses at the lowest force. The curve taken in a buffer containing 4 mM ATP shows net RecA unbinding during the first part of the force cycle during which the force is decreasing with time (dark purple) since ΔLoverstretch is larger for the increasing force part of the cycle (light purple). Figure 1B shows that ΔLoverstretch is 4.5 µm during the increasing force part of the cycle versus 2 µm for the decreasing force cycle consistent with bound RecA having been lost when lower forces were applied. Thus, significant RecA unbinding was observed at low forces.
In contrast, the curve taken in a buffer containing 10 mM ATP corresponds to dsDNA completely covered by RecA at 65 pN. Neither the decreasing force cycle (dark green) nor the increasing force cycle (light green) show any overstretching transition suggesting that both curves represent dsDNA fully covered by RecA at 65 pN; however, the light green curve is consistently shorter suggesting that more RecA is in the shorter ADP state. We note that at forces >30 pN, the 10 mM ATP curves match the slope of the magenta curve, which corresponds to a full dsDNA–RecA filament in ATPγS. This similarity between the slopes suggests that at forces >30 pN both curves represent full filaments that are not showing significant net RecA binding or unbinding. In addition, the same filaments are not making significant net transitions between conformations. Finally, in this force range, the change in extension versus force for dsDNA with RecA bound in the inactive state is similar to the extension versus force for dsDNA with RecA bound in the active state. At forces <30 pN there are differences between the slope of the curve in 10 mM ATP and the slope in ATPγS that may indicate conformational changes and/or unbinding. Similar results were obtained for other molecules.
Interpretations of changes in extension with time are complicated if the applied force is also changing with time. Thus, we did experiments in which the force was changed in a step-wise manner. We then observed the extension as a function of time during intervals where the force was held constant. The extension versus time curves for full filaments in the presence of free RecA and 10 mM ATP in solution show increases in extension after force increases and decreases in extension after force decreases, as shown in Supplementary Figure S2A and B. The initial extension changes are very rapid, but the rates quickly decrease as the extension approaches asymptotic values.
Results for dsDNA only partially covered by RecA in the absence of free RecA: filament formation and buffer exchange
The results presented above show that the length of a complete dsDNA/RecA filament depends on the history of the molecule. In particular, decreases in force reduce the length of full filaments. From the data, one cannot tell whether the length increases that accompany increases in force are due to conversions of bound RecA from the inactive state to the active state, to free RecA rebinding from solution or to a combination of both. Thus, we conducted experiments where we created full filaments in ATP and RecA, and then replaced the original buffer to remove any free RecA thus leaving the filament in a buffer solution containing 10 mM ATP and 10 µM ATPγS, which is known to prevent RecA unbinding while preserving full ATPase activity (35).
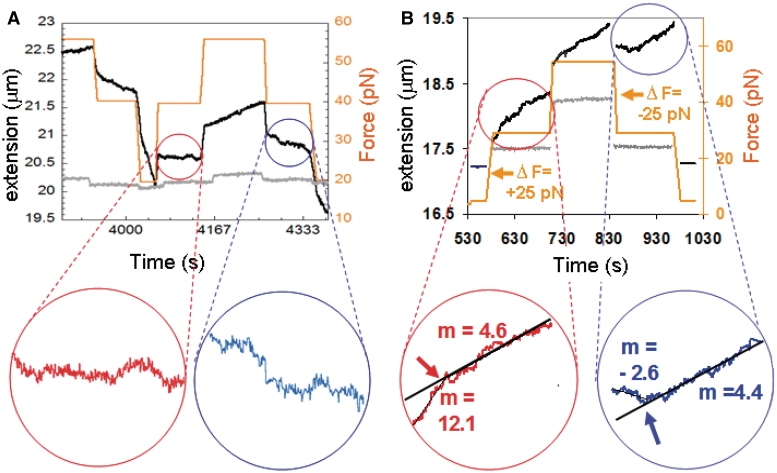
Figure 2A shows additional results for molecules exposed to a stepwise series of constant forces (orange) where the force at each step is varied by at least 10 pN/s. The black curve represents the extension measured after the buffer was exchanged (complete series shown in Supplementary Figure S2C). The black curve corresponds to a partially covered filament since some RecA unbound during the flow exchange.
Figure 2.
Dynamics effects take place during sequences of constant force steps. (A) Extension (black) and force (orange) versus time data in 10 mM ATP and 10 µM ATPγS after exchanging the buffer containing 1 µM RecA and 10 mM ATP using flow, along with a gray curve showing the corresponding extension for a control flow experiment in 1 mM ATPγS. Expanded views are shown in the circles. Linear fits to the curves shown in red and blue are −0.4 ± 0.05 nm/s and −6.4 ± 0.14 nm/s, respectively. The slope during the intervening 120 s is 4.0 nm ± 0.02 nm/s. (B) Plot analogous to that shown in (A) for dsDNA partially covered by RecA in 1 mM ATP and 1 µM RecA. Expanded views are shown in the circles with the y-axis offset between the two views; the black lines show the fits to the slopes during the last 90 s. The measured slopes and the standard deviations in the slopes are 12.1 ± 0.25 nm/s for the first 30 s after the force is increased to 30 pN, and 4.6 ± 0.09 nm/s for the following 90 s. Similarly, the values for the first 30 s and last 90 s after the force is decreased from 55 to 30 pN are −2.6 ± 0.25 and 4.4 ± 0.07 nm/s, respectively. The slopes for the corresponding time interval of the 55 pN second force are 7 ± 0.3 and 4.3 ± 0.06 nm/s.
The complete extension versus time curve for this molecule is shown in Supplementary Figure S2C. For such full filaments that are formed in the presence of RecA and then undergo an exchange of buffer to a buffer without RecA, the extension versus time curves measured in the buffer without free RecA do not approach asymptotic values on the timescale of hundreds of seconds. Thus, we cannot simply compare asymptotic extensions to determine changes in the bound RecA; however, we can compare the extension rate, dL/dt, at a given constant force, Fc, as a function of time after a stepwise force increase (ΔF > 0) with dL/dt after a stepwise force decrease (ΔF < 0). If dynamics plays no role and the amount of bound RecA has no effect, then the two rates should be the same since the applied force is the same, even if the off-rate is force dependent. Instead, we observe that dynamics does play a role and the rates consistently depend on the previous change in force, though they are insensitive to the amount of bound RecA.
Changes in the number of nucleation sites can change the observed extension rates, but comparisons of cases where the number of nucleation sites remains constant show that the observed dynamic effect is not simply a function of a change in the number of nucleation sites. Similarly, in the quasi-continuous protocol we compare the extension rate at a given force Fc when the force is increasing with the extension rate at the same force Fc when the force is decreasing, and for a range of Fc values we observe consistent differences (see below and Supplementary Data).
Figure 2A shows that at ∼4140 s, the force is increased from 40 to 56 pN resulting in a linear extension increase with time at a rate of 4.0 ± 0.02 nm/s. Some RecA that unbound during the low force cycle might subsequently rebind to the dsDNA; however, diffusion should rapidly move unbound RecA away from the dsDNA resulting in a rebinding rate that becomes negligible in less than a second. In contrast, we observe constant extension rates for times >100 s, which strongly suggests that the observed length increase cannot be attributed to the rebinding of RecA that diffused back to the DNA after unbinding. Similar results occur after additional washing cycles, confirming that the results were not due to residual free RecA that remained in solution after the wash. Thus, the observed length increases must be due to a transition in the bound RecA from the inactive state to the active state.
Observed length decreases are more difficult to interpret than length increases since both RecA unbinding and transitions from the active to inactive state produce length decreases; however, we have demonstrated above that a length increase must be due only to a conformational change from the inactive to the active state and not released RecA rebinding. Thus, if such a length increase returns the total extension to a value close to the extension before the previous length decrease, then the previous length decrease must have been dominated by bound RecA making conformational changes from the active state to the inactive state. One example of such a restoration in length is given by the force cycle that begins with the force decrease from 40 to 19 pN that occurs at ∼4030 s for the molecule whose extension versus time curve as shown in Figure 2A.
Checking that there is almost no free RecA in solution after the buffer exchange
The gray curve in Figure 2A corresponds to a control experiment for a partial filament obtained in 1 µM RecA and 1 mM ATPγS after the buffer was replaced by a buffer containing 1 mM ATPγS and no RecA, in order to wash out the RecA protein in solution. When free RecA was present, the ATPγS sample showed increases in extension at constant force, corresponding to the binding of free RecA from solution (see below). In contrast, after removing free RecA with flow, the ATPγS sample showed no change in extension when a constant force was applied even though only a partial filament had been formed. Thus, for molecules in ATPγS, no binding or unbinding is observed after flow confirming that RecA was effectively removed from solution, and no dynamic effects are detected at any force for force changes at rates up to 10 pN/s (see below). This result also shows that in the absence of hydrolysis and free RecA, the observed extension at a given constant force is constant.
Results for dsDNA partially covered by RecA in the presence of free RecA
During strand exchange, dsDNA is only partially covered by RecA. Strand exchange involves both the binding and unbinding of RecA from dsDNA, so it is important to consider the effect of force changes on filaments that are not completely covered in a buffer where the binding of free RecA is possible. In this case, net polymerization and depolymerization can occur as well as changes between bound state conformations.
Figure 2B is analogous to Figure 2A for a single molecule partially covered by RecA in the presence of 1 µM RecA and 1 mM ATP in RecA buffer (black line) and without RecA (gray line). An initial quasi continuous overstretching cycle creates nucleation sites (36–39), and then a step-wise force regime is imposed. The complete series of steps is shown in Supplementary Figure S3. Figure 2B shows a detailed examination of cycle 2, concentrating on Fc = 30 pN with ΔF values of ±25 pN. The negative control in the absence of RecA (gray line) shows force dependent changes in extension, but no significant extension change when the force is constant. In contrast, in the presence of RecA (black line), significant changes in extension occur when the applied force is constant, suggesting that the RecA is binding and unbinding and/or changing conformation. Additionally, Figure 2B shows that the changes in extension depend strongly on the time that has elapsed since the last force change. Thus, unlike the results in the absence of free RecA, the results in the presence of free RecA cannot be characterized by a single extension rate that applies throughout the time during which the force is constant. In order to characterize the results in the presence of free RecA, we divide the 120-s interval after each force change in two intervals: (1) the first 30 s after a force change, ΔF; and (2) the remaining 90 s. We will suggest that for the buffer conditions considered here, dynamics are most important during the first 30 s.
The expanded plots in Figure 2B show that the measured slopes and the standard deviations in the slopes are 12.1 ± 0.25 nm/s for the first 30 s after the force is increased to 30 pN and 4.6 ± 0.09 nm/s for the following 90 s. Similarly, the values for the first 30 s and last 90 s after the force is decreased from 55 to 30 pN are −2.6 ± 0.25 and 4.4 ± 0.07 nm/s, respectively. During the intervening 120-s interval when the applied force was 55 pN, dL/dt = 7 ± 0.3 nm/s for the first 30 s and 4.3 ± 0.06 nm/s for the remaining 90 s. This suggests that in the absence of dynamics the extension rates for all three intervals are the same despite the difference in force. This is consistent with previous results that showed that the extension rate/nucleation site is independent of force (39). Given that earlier work demonstrated a force insensitive RecA binding rate/nucleation site corresponding to length increases of 0.48 ± 0.05 nm/s (39), the data shown in Figure 2B suggests that there were nine nucleation sites present during the entire time shown since the growth rates 30 s after a force change were ∼4.5 nm/s, for both forces during all three time intervals. Thus, though the number of nucleation sites is the same, the extension rates show a significant dependence on previous force changes, indicating that the observed dynamic effect is not due to a change in the number of nucleation sites. In addition, this result also implies that a force dependent off-rate is not playing a significant role in the results since the same extension rates were observed at both 30 and 56 pN. Supplementary Figure S4 shows that the dynamic effect occurs even when filaments are depolymerizing at all forces.
Dynamic effects in dsDNA molecules partially covered by RecA are absent in buffers where hydrolysis is negligible
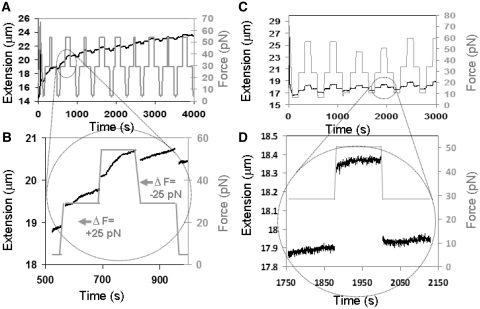
Figure 3A and B is analogous to Figure 2 where the measured force (gray) and extension (black) as a function of time are shown for a molecule in ATPγS, a poorly hydrolysable analog of ATP. In ATPγS, a nearly complete filament is formed after several force cycles, where no decrease in extension is observed even at an applied force of 10 pN. Spontaneous slope changes occur, but they are not correlated with changes in force. In ATPγS, RecA will continue to bind until a full filament is formed. The extensions of full filaments in ATPγS are highly insensitive to changes in applied force as can be seen in Supplementary Figure S5.
Figure 3.
Extension versus time for dsDNA pulled by a sequence of constant forces in conditions where hydrolysis is negligible. (A) Extension (black) and force (gray) as a function of time data in RecA buffer, 1 µM RecA and 1 mM ATPγS. (B) A more detailed sequence during ΔF = +25 pN and ΔF = –25 pN. (C) Extension (black) and force (gray) as a function of time in 1 µM RecA, 10 mM CaCl2 and 1 mM ATP. (D) More detailed sequence during ΔF = +20 pN and ΔF = –20 pN.
Figure 3C and D is also analogous to Figure 2 where the measured force (gray) and extension (black) of dsDNA molecules partially covered by RecA as a function of time in a buffer containing CaCl2 are shown. ATP hydrolysis is known to be suppressed in this buffer (32).
Many measurements of dsDNA partially covered by RecA with different force steps and Fc values in the presence of free RecA
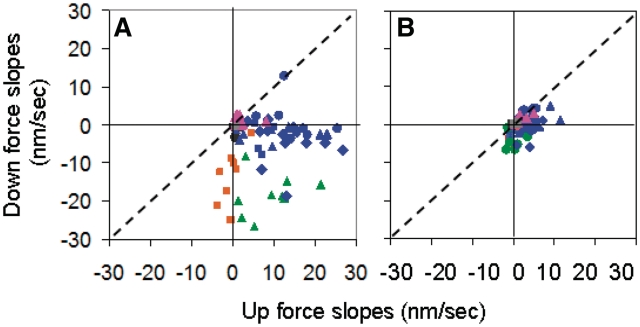
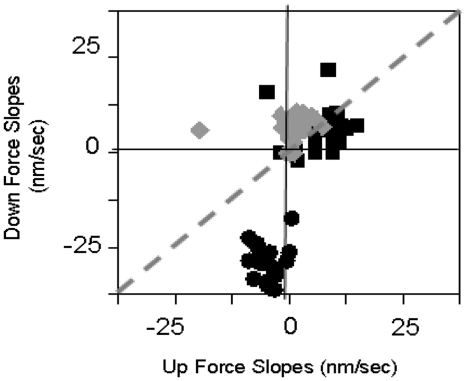
Figure 4 shows the effect of dynamics on the extension rates for many partially covered molecules with Fc values from 10 to 55 pN and |ΔF| values from 10 to 30 pN. Figure 4A and B shows the results for the first 30 s and the remaining 90 s, respectively. Each point in the figure represents two successive measurements made at the same Fc: the x and y coordinates are the measured extension rates after a ΔF > 0 and ΔF < 0, respectively. The negative controls (black points) obtained in RecA buffer and no RecA lie within a ∼1.5 nm/s circle centered at the origin during all time periods as is shown in more detail in Supplementary Figure S6A and B, which are expanded views of Figure 4. If RecA binds but force changes played no role, all the points on the graph would lie along the diagonal x = y line (dotted line), indicating that dL/dt is insensitive to force changes. The ATP results during the first 30 s fall well below the x = y line. In contrast, the rates during the remaining 30–120 s are much smaller and show little clear departure from the x = y line. These results suggest that in most cases the number of nucleation sites was the same for measurements at Fc. Thus, the observed dynamic effect is probably not due to a change in the number of nucleation sites. The observed extension rates during the last 90 s are similar to the rates observed for molecules held at constant force for hundreds of seconds (39), suggesting that the increased slopes observed in the first 30 s are due to transitions between conformations. We note that the effect could not simply be due to a force modulated off- rate since the Fc values are the same. In general, the dynamic effect is more marked at higher cycles.
Figure 4.
Effect of dynamics on many incomplete filaments. (A) Results for 0–30 s. (B) Results for 30–120 s. The x and y coordinates are dL/dt after a ΔF > 0 and after a ΔF < 0, respectively. In ATP, the Fc ranges are orange ≤20 pN, green 20–30 pN and blue >30 pN. All forces for controls, samples in ATPγS and ATP/CaCl2 are shown as black squares, magenta triangles and gray triangles, respectively. The standard deviations for the individual slopes vary, but the average standard deviation is 0.17 nm/s.
In contrast, experiments on incomplete filaments in buffers with suppressed hydrolysis show little or no dynamic effect as illustrated by the magenta and gray points in Supplementary Figure S6A and B, where the magenta and gray points represent data at all forces taken in buffer containing ATPγS and in buffer containing ATP where MgCl2 was replaced by CaCl2, respectively.
Quasi-continuous experiments on dsDNA molecules partially covered by RecA in the presence of free RecA
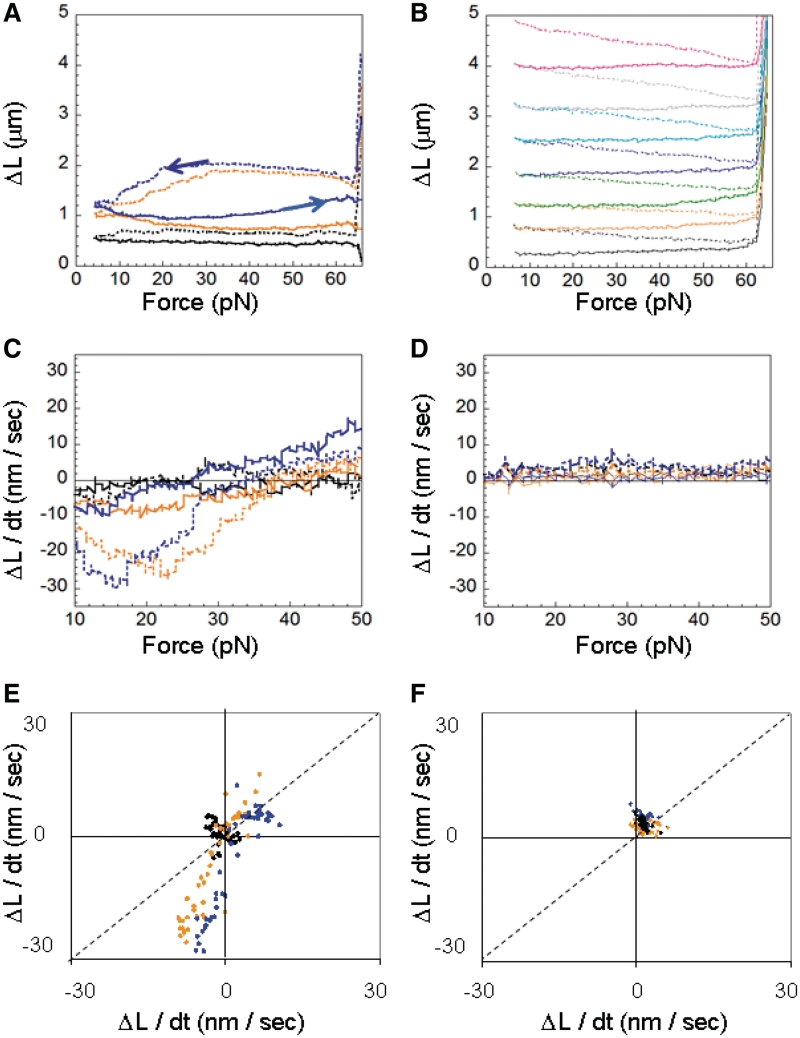
In these experiments each dsDNA molecule is subjected to a force that is cycled in order to measure the change in extension due to RecA binding while the applied force changes at a rate of ∼1 pN/s. Figure 5A and B shows selected cycles in ATP and ATPγS, respectively, from the data shown in Supplementary Figure S7, where ΔL is the measured extension at a given force minus the extension at the same force in the absence of RecA. The black, orange and blue colors represent the first, second and fourth complete cycles, respectively.
Figure 5.
Quasi-continuous measurements. (A) Selected cycles of the complete sequence shown in Supplementary Figure S7 (1 µM RecA and 1 mM ATP), where the extension as a function of force in the absence of RecA has been subtracted. The up cycles are shown with solid lines and the down cycles with dashed lines. (B) Same as (A) but in 1 µM RecA and 1 mM ATPγS. (C) Change in extension as a function of force in 1 mM ATP for the curves shown in (A), where the solid lines correspond to the up cycles and the dashed lines correspond to the down cycles. (D) Same as (C) but in 1 mM ATPγS. (E) Each point corresponds to two successive measurements of the slope at a given force Fc, where the x and y values corresponds to the slopes when the force is increasing and decreasing, respectively, in ATP. (F) Same as (E) in ATPγS. The colors correspond to the cycle number of the complete series of cycles shown in Supplementary Figure S7: black (first), orange (second) and blue (fourth). The variations in slope are ∼±3 nm/s.
Figure 5C and D shows dΔL/dt as a function of force for the data shown in Figure 5A and B, where the up cycles are shown by the solid lines and the down cycles are shown by the dashed lines. In ATP both force directions show significant ΔL increases at forces >30 pN, whereas at forces <30 pN, the down cycles show large ΔL decreases and the up cycles show much smaller changes in ΔL. Some samples eventually reached a condition where successive cycles showed the same force extension behavior as illustrated in Supplementary Figure S8A. In contrast, in ATPγS, Figure 5D shows that dΔL/dt is insensitive to both force and changes in force. In ATPγS, the amount of bound RecA increases with each cycle until a full filament is formed.
We note that even in a buffer containing ATP, if the applied force is constrained to be between 30 and 50 pN, as long as the force is varied at 1 pN/s, dΔL/dt does not change significantly with force or cycle number. In addition, the observed dΔL/dt values are similar to the rates observed in ATPγS for all cycles, suggesting that the number of nucleation sites does not increase much with cycle number and that no significant effects associated with a force dependent off-rate are observed. Finally, consistent with earlier results for full filaments, faster force changes or the application of higher or lower forces do result in dynamic effects and those dynamic effects increase with cycle number.
A different way of analyzing these quasi-continuous results is presented in Figure 5E and F, where the x-coordinate of each point is the increase in ΔL for a cycle when the force is increasing and the y-coordinate is the corresponding ΔL value for a cycle when the force is decreasing, where each point represents a different force value. Again, the dΔL/dt values associated with decreasing forces are less than the dΔL/dt values associated with increasing forces. One can see that in the presence of ATP the dynamics effect was small during the early cycles, but became more prominent as the cycle number increased. The effect really depends on cycle number, not extension as shown in Supplementary Figure S8. In contrast, the dΔL/dt in the presence of ATPγS is similar for all cycles, and no significant dynamic effects are observed in any cycle.
Figure 6 shows the measured extension rates for the quasi-continuous case for forces ramped at ∼1 pN/s. Since the applied force is constant dL/dt = dΔL/dt, Figure 6 is analogous to Figure 4, which showed dL/dt at a constant force after stepwise increases in force. Figure 6 shows two successive measurements of dΔL/dt at the same Fc: the x and y coordinates are the measured dΔL/dt when the force is increasing and when the force is decreasing, respectively. The black circles and squares represent data taken in buffer containing ATP, at forces <30 and ≥30 pN respectively. The gray diamonds represent data taken in ATPγS at all forces.
Figure 6.
Results for quasi continuous force changes. The gray diamonds show data for measurements in 1 µM RecA and 1 mM ATPγS. Black circles and squares correspond to forces <30 pN and ≥30 pN in 1 µM RecA and 1 mM ATP, respectively. The variations in slope are ∼±3 nm/s.
Typical extension versus time results for many cycles of typical molecules are shown in Supplementary Figure S7. We note that at the force minimum there is a large change in dΔL/dt. Hundreds of cycles of tens of molecules have been examined with many different minimum force values, rates of force change and covered filament fractions. If the minimum force is <20 pN, the extension rates at the force minimum show significant negative rates on the decreasing force side and near zero slopes on the increasing force side. For partially covered filaments, if the force is paused at the minimum value, the observed dΔL/dt is much slower than dΔL/dt measured while the force was decreasing (Supplementary Figure S9). Thus, the observed difference in the absolute values dΔL/dt is not dominantly due to an approach to the steady state extension at the minimum force, though such an effect may make a contribution to the observed result.
Figure 7 shows that the dynamic effect occurs when the dsDNA is pulled by both ends or by the 3′3′, 5′5′ and 3′5′-ends. Moreover the effect can be observed when the force is initially increased (3′3′ and 5′5′) or initially decreased (3′5′ and both ends). Thus, the effect seems to depend on the tension on the dsDNA and not on the distribution of the tension. The curves also demonstrate that the dynamic effect is observed whether the increasing force cycle is considered first or the decreasing force cycle is considered first, demonstrating that the temporal order of the cycles is not important.
Figure 7.
Extension versus force curves for different pulling techniques in 1 mM ATP showing significant dynamics effects and large changes in slope at the force minimum. (A) Pulling from 3′3′-ends. (B) 5′5′-ends. (C) 3′5′-ends. (D) Both ends. For these cycles force was initially increased in (A) and (B) and initially decreased in (C) and (D).
Quasi-continuous experiments on dsDNA molecules partially covered by RecA in the absence of free RecA
Quasi-continuous experiments were also done on partial filaments in the absence of free RecA. The results are shown in Supplementary Figure S10. These results are very similar to the results for partial filaments in the absence of RecA shown in Figure 5A, C and E, except that in the absence of free RecA significant length decreases are observed when the force is being decreased from 55 to 30 pN, whereas in the presence of free RecA the extension remains constant until the force is lowered <30 pN. At forces <30 pN, dΔL/dt values of approximately −120 nm/s were observed.
Estimate of the tension on dsDNA inside RecA–dsDNA filaments in the absence of external force
In order to determine whether the results presented above are relevant to the motion of the strand exchange window in vivo, it is important to estimate the tension present inside dsDNA–RecA filaments in the absence of external force. Such an estimate can be made by examining the extension of full filaments as a function of force in ATPγS, such as the graph shown in Supplementary Figure S11. In ATPγS, conformational changes and unbinding are strongly suppressed. Thus, changes in length provide measurements of the elasticity of the filament. If the protein is inextensible, then changes in extension must result from changes in the extension of the dsDNA inside the filament, possibly having an effect at the interfaces between RecA monomers. If the internal tension in the filament is much larger than the tension due to an external force applied on the ends of the dsDNA, then the observed extension should be insensitive to the value of the force once entropic effects are overcome. Supplementary Figure S11 shows no change in extension for forces between 10 and 30 pN. At forces >40 pN, the extension increases linearly with force.
DISCUSSION
The results above suggest that the response of RecA to changes in tension on dsDNA depends on the applied force, the rate of change of the force and the number of previous force cycles experienced by each molecule. The biological relevance of the results depends on whether the experimental conditions are similar to those that occur in vivo in the absence of any external force. In particular, it is important to consider whether the tension and rates of change of tension used in these experiments are similar to those that occur in vivo during strand exchange.
The results shown in Supplementary Figure S11 suggest that the internal tension on dsDNA in a RecA filament is ∼30 pN. As the strand exchange window moves, dsDNA dissociates from the RecA/ssDNA filament at the lagging edge of the strand exchange window. Similarly, the RecA/ssDNA filament binds to new dsDNA at the leading edge of the strand exchange window. Given that the strand exchange window propagates at a rate of of ∼2 nt/s (25) or ∼0.7 RecA/s, the maximum change in dsDNA tension at the end of the filament would be of the order of 20 pN/s. The detailed distribution of tension along the length of the ssDNA/RecA–dsDNA complex is unknown, but the change in tension is probably largest at the ends. Thus, the responses of RecA to changes in dsDNA tension that are described in this work are probably of the order of those observed in vivo during strand exchange.
In a buffer with no free RecA, force changes produce consistent reproducible linear extension rates. At high forces, (>40 pN) the extensions increase slowly and at low forces (<25 pN) the extensions decrease quickly. At ∼40 pN, the extensions decrease after a force decrease and remain approximately constant after a force increase. A given single molecule can be cycled many times with similar results. Thus, changes in the conformation of bound RecA must be the dominant source of the observed extension changes.
This conclusion is also supported by earlier work that showed that the unbinding rate at low force or zero force is of the order of 1 RecA/s (21,35,38), whereas the observed decrease in extension shown in Figure 2A would correspond to 300 RecA/s. It is believed that RecA can only unbind from the ends of filaments (21,35), where each nucleation site results in at most two filament ends. Results presented in this article suggest that under conditions similar to those in Figure 2A there are ∼10 nucleation sites present. Thus, the ∼150 nm/s decreases in extension are probably dominantly due to conformational transitions in the bound RecA rather than RecA unbinding.
In this work, we have shown that if the applied force is varied within the range between 30 and 50 pN at a rate of 1 pN/s, full filaments (Supplementary Figure S12) show no change in ΔL, and partial filaments show no change in dΔL/dt (Figure 5C). At forces <30 pN, the situation is more complicated since unbinding of RecA is observed even in filaments that were initially complete. At 10 mM ATP concentrations in the absence of free RecA (Supplementary Figure S12), the change in extension due to conformational changes is of the order of or larger than the change due to unbinding, so the measured extension changes are indicative of conformational changes. At lower ATP concentrations, length changes at low forces may be dominated by unbinding.
Comparisons of the measured dΔL/dt at given Fc < 30 pN show that decreases in force promote decreases in ΔL and increases in force stabilize ΔL. These observed decreases in ΔL may include transitions in bound RecA from the active conformation to the inactive conformation, as well as unbinding. Transitions from the active state to the inactive state will of course promote future unbinding; therefore, the experimental results presented here suggest that decreases in force promote RecA unbinding and increases in force stabilize RecA binding. This result holds for both stepwise and continuous force changes. The comparison was made between results at the same Fc, so the results are not affected by a force dependence in koff.
In sum, as the strand exchange window progresses along a dsDNA molecule, if the dsDNA tension at the leading end of the strand exchange window is increasing, as suggested by the measurements above that implied that the internal tension is of the order of the tension due to an external force of ∼30 pN, then the tension at the lagging end must be decreasing. The results reported above suggest that increases in dsDNA tension result in conformational changes in the bound RecA from the inactive state to the active state. Similarly, decreases in dsDNA tension result in conformational changes in bound RecA from the active state to the inactive state. Given that RecA in the active state does not unbind from dsDNA, whereas RecA in the inactive state can unbind, the functional response of RecA to changes in dsDNA tension could enhance the motion and directionality of the strand exchange window. This effect would be lost if a non-homologous region resulted in a pause in the motion of the strand exchange window. Such a pause might possibly increase sequence stringency by stalling strand exchange when it reaches a non-homologous region.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Harvard University (to M.P.). N.K.'s research is supported by NIH (grants RO1-GM025326 and R01-GM044794). Funding for open access charge: Harvard University.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- 3.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 4.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 5.Müller B, Koller T, Stasiak A. Characterization of the DNA binding activity of stable RecA-DNA complexes. Interaction between the two DNA binding sites within RecA helical filaments. J. Mol. Biol. 1990;212:97–112. doi: 10.1016/0022-2836(90)90307-8. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi M, Kubista M, Nordén B. Binding stoichiometry and structure of RecA-DNA complexes studied by flow linear dichroism and fluorescence spectroscopy. Evidence for multiple heterogeneous DNA co-ordination. J. Mol. Biol. 1989;205:137–147. doi: 10.1016/0022-2836(89)90371-9. [DOI] [PubMed] [Google Scholar]
- 7.Howard-Flanders P, West SC, Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:17–23. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen Z, Yang H, Pavletich NP. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 9.Shibata T, DasGupta C, Cunningham RP, Radding CM. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc. Natl Acad. Sci. USA. 1979;76:5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Mameren J, Modesti M, Kanaar R, Wyman C, Peterman EJG, Wuite GJL. Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature. 2009;457:745–748. doi: 10.1038/nature07581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazin AV, Kowalczykowski SC. The specificity of the secondary DNA binding site of RecA protein defines its role in DNA strand exchange. Proc. Natl Acad. Sci. USA. 1996;93:10673–10678. doi: 10.1073/pnas.93.20.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazin AV, Kowalczykowski SC. The function of the secondary DNA-binding site of RecA protein during DNA strand exchange. EMBO J. 1998;17:1161–1168. doi: 10.1093/emboj/17.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao J, Singleton SF. Elucidating a key intermediate in homologous DNA strand exchange: structural characterization of the RecA–triple-stranded DNA complex using Fluorescence Resonance Energy Transfer. J. Mol. Biol. 2002;320:529–558. doi: 10.1016/s0022-2836(02)00462-x. [DOI] [PubMed] [Google Scholar]
- 14.Xiao J, Lee AM, Singleton SF. Direct evaluation of a kinetic Model for RecA-mediated DNA-strand exchange: the importance of nucleic acid dynamics and entropy during homologous genetic recombination. ChemBioChem. 2006;7:1265–1278. doi: 10.1002/cbic.200600038. [DOI] [PubMed] [Google Scholar]
- 15.Radding CM. Homologous pairing and strand exchange in genetic recombination. Annu. Rev. Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- 16.Menetski JP, Bear DG, Kowalczykowski SC. Stable DNA heteroduplex formation catalyzed by the Escherichia-coli RecA protein in the absence of ATP hydrolysis. Proc. Natl Acad. Sci. USA. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalczykowski SC, Krupp RA. DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc. Natl Acad. Sci. USA. 1995;92:3478–3482. doi: 10.1073/pnas.92.8.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J-I, Cox M, Inman RB. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem. 1992;267:16438–16443. [PubMed] [Google Scholar]
- 19.Rosselli W, Stasiak A. The ATPase activity of RecA is needed to push the DNA strand exchange through heterologous regions. EMBO J. 1991;10:4391–4396. doi: 10.1002/j.1460-2075.1991.tb05017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robu ME, Inman RB, Cox MM. RecA promotes the regression of stalled replication forks in vitro. Proc. Natl Acad. Sci. USA. 2001;98:8211–8218. doi: 10.1073/pnas.131022698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox JM, Tsodikov OV, Cox MM. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:231–243. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Folta-Stogniew E, O'Malley S, Gupta RC, Anderson KS, Radding CM. Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol. Cell. 2004;15:965–975. doi: 10.1016/j.molcel.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Hsieh P, Camerini-Otero CS, Camerini-Otero RD. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc. Natl Acad. Sci. USA. 1992;89:6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gumbs OH, Shaner SL. Three mechanistic steps detected by FRET after presynaptic filament formation in homologous recombination. ATP hydrolysis required for release of oligonucleotide heteroduplex product from RecA. Biochemistry. 1998;37:11692–11706. doi: 10.1021/bi980646s. [DOI] [PubMed] [Google Scholar]
- 25.van der Heijden T, Modesti M, Hage S, Kanaar R, Wyman C, Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol. Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Stasiak A, Stasiak AZ, Koller T. Visualization of RecA-DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harb. Symp. Quant. Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- 27.Jain SK, Cox MM, Inman RB. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. III. Uni-directional branch migration. J. Biol. Chem. 1994;269:20653–20661. [PubMed] [Google Scholar]
- 28.Cox MM. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 29.Robertson RB, Moses DN, Kwon Y, Chan P, Chi P, Klein H, Sung P, Greene EC. Structural transitions within human Rad51 nucleoprotein filaments. Proc. Natl Acad. Sci. USA. 2009;106:12688–12693. doi: 10.1073/pnas.0811465106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danilowicz C, Coljee VW, Bouzigues C, Lubensky DK, Nelson DR, Prentiss M. DNA unzipped under a constant force exhibits multiple metastable intermediates. Proc. Natl Acad. Sci. USA. 2003;100:1694–1699. doi: 10.1073/pnas.262789199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danilowicz C, Limouse C, Hatch K, Conover A, Coljee VW, Kleckner N, Prentiss M. Demonstration that the force versus extension curves for overstretched DNA depend on which ends are pulled. Proc. Natl Acad. Sci. USA. 2009;106:13196–13201. doi: 10.1073/pnas.0904729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menetski JP, Varghese A, Kowalczykowski SC. Properties of the high-affinity single-stranded-DNA binding state of the Escherichia-coli RecA protein. Biochemistry. 1988;27:1205–1212. doi: 10.1021/bi00404a021. [DOI] [PubMed] [Google Scholar]
- 33.Prévost C, Takahashi M. Geometry of the DNA strands within the RecA nucleofilament: role in homologous recombination. Q. Rev. Biophys. 2004;36:429–453. doi: 10.1017/s0033583504003956. [DOI] [PubMed] [Google Scholar]
- 34.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 35.Lindsley JE, Cox MM. Dissociation pathway for RecA nucleoprotein filaments formed on linear duplex DNA. J. Mol. Biol. 1989;205:695–711. doi: 10.1016/0022-2836(89)90315-x. [DOI] [PubMed] [Google Scholar]
- 36.Leger JF, Robert J, Bourdieu L, Chatenay D, Marko JF. RecA binding to a single double-stranded DNA molecule: a possible role of DNA conformational fluctuations. Proc. Natl Acad. Sci. USA. 1998;95:12295–12299. doi: 10.1073/pnas.95.21.12295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hegner M, Smith SB, Bustamante C. Polymerization and mechanical properties of single RecA–-DNA filaments. Proc. Natl Acad. Sci. USA. 1999;96:10109–10114. doi: 10.1073/pnas.96.18.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shivashankar GV, Feingold M, Krichevsky O, Libchaber A. RecA polymerization on double-stranded DNA by using single-molecule manipulation: the role of ATP hydrolysis. Proc. Natl Acad. Sci. USA. 1999;96:7916–7921. doi: 10.1073/pnas.96.14.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feinstein E, Danilowicz C, Conover A, Gunaratne R, Kleckner N, Prentiss M. Single molecule studies of the stringency factors and rates governing the polymerization of RecA on double stranded DNA. Nucleic Acids Res. 2011;39:3781–3791. doi: 10.1093/nar/gkr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.