Abstract
Type II topoisomerases are ubiquitous enzymes that control the topology and higher order structures of DNA. Type IIA enzymes have the remarkable property to sense locally the global DNA topology. Although many theoretical models have been proposed, the molecular mechanism of chiral discrimination is still unclear. While experimental studies have established that topoisomerases IIA discriminate topology on the basis of crossover geometry, a recent single-molecule experiment has shown that the enzyme has a different processivity on supercoiled DNA of opposite sign. Understanding how cross-over geometry influences enzyme processivity is, therefore, the key to elucidate the mechanism of chiral discrimination. Analysing this question from the DNA side reveals first, that the different stability of chiral DNA cross-overs provides a way to locally sense the global DNA topology. Second, it shows that these enzymes have evolved to recognize the G- and T-segments stably assembled into a right-handed cross-over. Third, it demonstrates how binding right-handed cross-overs across their large angle imposes a different topological link between the topoIIA rings and the plectonemes of opposite sign thus directly affecting the enzyme freedom of motion and processivity. In bridging geometry and kinetic data, this study brings a simple solution for type IIA topoisomerase chiral discrimination.
INTRODUCTION
Type II DNA topoisomerases (topo II) are ubiquitous enzymes that control the topology and the organization of higher order structures of DNA (1–3). They are essential for solving topological problems linked to DNA unwinding during replication and transcription catalysing the passage of one double-stranded DNA segment through a transient-mediated break in another. In addition, growing evidences have shown that they also play multiple cellular roles such as gene regulation and chromosome condensation (2). Remarkably, these enzymes have also the unique property that still remains unexplained, of being able to sense locally the global DNA topology. The present article analyses the type II topoisomerase mechanisms from the DNA point of view and shows how these enzymes may have exploited the intrinsic properties of the double helix and its assembly rules for accomplishing their fascinating cellular roles.
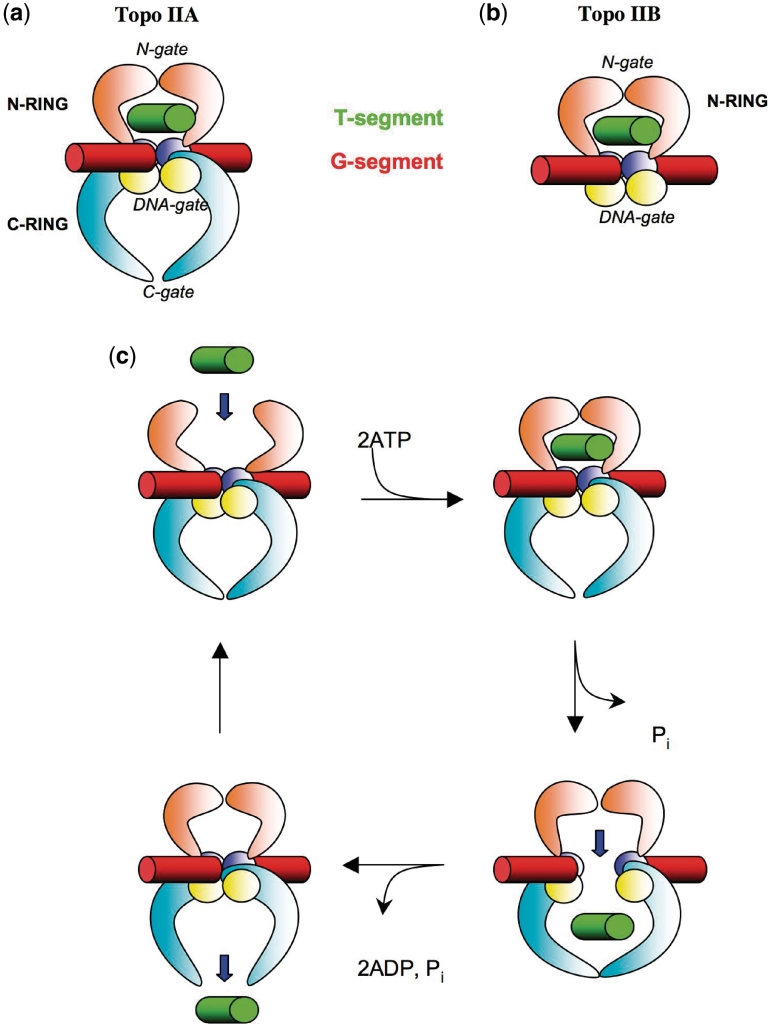
Type II topoisomerases are divided into two subclasses IIA and IIB. Type IIA enzymes are found throughout eubacteria and eukaryotic cells whereas type IIB ones are found in archaea and also in plants (3). Both families share structural analogies, which reveal a complex evolutionary relationship (3–5). They display a modular architecture that contains a series of catalytic modules linked by flexible hinge regions (4,5). While type IIA topoisomerases are composed of two inter-connected rings, type IIB enzymes adopt the overall architecture of a single molecular clamp that contains the two first gates structurally related to the type IIA enzymes (N-ring) (Figure 1a and b). The current model of the DNA strand-passage reaction proposes that both enzyme subclasses bind two DNA segments and catalyse the ATP-dependent transport of one intact DNA double helix (‘T-segment’), through a gate DNA segment that contains the enzyme-mediated transient DNA gate (‘G-segment’) (1,6,7). To accomplish this task, topo IIA acts like two well-synchronized molecular clamps that alternatively capture and release DNA through three gates: the entrance ‘N-gate’, the ‘DNA gate’ and the exit or ‘C-gate’ (8) (Figure 1c). The G-segment is bound to the DNA cleavage/religation domain that contains the catalytic toprim and 5Y-CAP domains (5). Binding of ATP is then coupled to the dimerisation of the ATPase domains that clamp and capture the T-segment into the ‘N-ring’. Cross-talk between ATPase and DNA cleavage domains lead to G-segment cleavage (9). The T-segment is then transported through the open DNA gate for storage into the ‘C-ring’. Finally, opening the C-gate permits the release of the T-segment into the solvent for performing a new catalytic cycle. Recent crystallographic studies of eukaryotic and bacterial type IIA enzymes have unveiled the mode of binding of the G-segment (10–15). In these structures, the G-segment duplex is sharply bent and its central 4 bp located between the two catalytic tyrosines adopt a A-conformation. Since the toprim and CAP domains are organized differently in type IIA and IIB enzymes (16,17), the G-segment binding regions of the two families are structurally distinct to the degree that they generate different DNA-cleavage products (2 bp versus 4 bp overhangs) (5). Consequently, the G-segment might adopt a different conformation and orientation in the two families. Despite these differences, the strand-passage mechanism of both enzyme families is similar, except that in type IIB topoisomerases there is no C-gate and the T-segment is directly released into the solvent after passing the G-gate. A model in which the G-segment is straight and adopts a B-DNA conformation has been proposed for type IIB enzymes (18).
Figure 1.
Type II topoisomerase architecture and catalytic mechanism. (a) Schematic representation of a full-size type IIA topoisomerase. (b) Schematic representation of a full-size type IIB topoisomerase. (c) The strand-passage reaction that transports the T-segment through the transient gate of the G-segment. The T- and G-segments are represented in green and red, respectively. The N- and C-rings are represented in orange and cyan, respectively. The top rim and CAP domains are represented in blue and yellow, respectively.
A functional link between higher order DNA structure and topology
The major tasks of type II topoisomerases are first, the maintenance of topological homeostasis and second, the resolution of the topological problems linked to DNA replication and transcription (19–21). These functions are accomplished by the ATP-dependent strand-passage reaction that enables these enzymes to catalyse supercoil removal or introduction, catenation and decatenation or knotting and unknotting of DNA (1). Bacteria encode gyrase and topoisomerase IV (topo IV). Gyrase is unique in that it introduces negative supercoils into DNA in an ATP-dependent manner to regulate the superhelical density of bacterial DNA (22). Topo IV plays essential roles in untangling daughter chromosomes after DNA replication, removing knots from the genomes and contributing to replication fork progression by relaxing positive supercoils (21). Eukaryotes encode a type II topoisomerase that is homologous to the eubacterial type IIA enzymes and also play critical functions in DNA replication and chromosome segregation (23). Vertebrates express two isoforms, topo IIα and topo IIβ that display similar catalytic properties but differ in their regulation and cellular functions (1). Type II topoisomerases are also involved in the organization of higher order DNA structures in both eukaryotic and prokaryotic cells (2,24). In eukaryotes, these enzymes have been found a long time ago as a major component of chromosomal scaffolds (SAR/MAR) (25–27), and growing evidences show that these enzymes collaborate with condensin thus playing an important role in mitotic chromosome condensation (28–30) and in centromeric function (31). However, their precise role in the building of mitotic chromosome is still controversial and remains to be determined. Consistent with a hierarchic mechanism of chromosome condensation, a recent study reported that type II topoisomerases promote chromatin compaction in vitro, in a manner dependent on histone H1, but independent of ATP (32). It has also been shown that type II topoisomerases participate to the nucleoid organization in prokaryotic cells (33) thus indicating that in both bacteria and eukaryotes, these enzymes establish a functional link between genome architecture and DNA topology. Indeed, although partitioning bacterial sister chromosomes are distinct from chromatid segregation during eukaryotic mitosis (34), the intimate link between the modulation of DNA packaging and topology plays a critical role in gene regulation in both eukaryotic and bacterial cells (24).
Local sensing of global topology
Due to their remarkable ability to sense locally, the global DNA topology type IIA enzymes have been compared to ‘Maxwell Demons’, opening an imaginary door to act on specific DNA topoisomers (more supercoiled, catenated or knotted) and thus reducing the entropy of a system (35). Rybenkov et al. (36) showed that they are able to ‘simplify’ the DNA topology beyond the level expected at the equilibrium. In the presence of ATP, the enzymes generate a steady-state distribution narrower than the equilibrium distribution of topoisomers produced by ATP-independent enzymes such as topoisomerase I. How a small enzyme is able to discriminate the topology of a large DNA molecule still remains a mystery. Several theoretical models have been proposed to explain topology simplification in searching the different possible mechanisms that could increase the probability of the enzyme to select a T-segment belonging to a specific topological state, in a strand-passage reaction (reviewed in 37,38). Some models that include DNA kinking (39), three segment binding (40) and kinetic proofreading (41) suggested that the different probabilities of type II topoisomerase-T-segment interactions could potentially arise via either geometric or kinetic selection. These ‘protein-centric’ models consider that the enzyme is essential in creating new probing information about DNA topology. They contrast with ‘DNA-centric’ models proposing that the topological information is embodied in local juxtaposition geometry and that type II topoisomerases achieve disentanglement by selective segment passages only at pre-existing ‘hooked’ but not free juxtapositions (42–44). However, the computer simulations that account for such effects in both protein- and DNA-centric models are, in general, not sufficient to explain the observed efficiency of local sensing. A recent experimental study has shown that topology simplification is a ubiquitous feature of type IIA topoisomerases (45). The authors concluded that the mechanism of topology simplification might involve both a geometric selection step such as the G-segment bending and a kinetic proofreading process (45). Interestingly, in showing that type IIB topoisomerases are unable to simplify the DNA topology, this study suggested that this property may require the presence of the two enzyme rings (N- and C-ring) and therefore the potential ability to ‘hold’ the T-segment into the C-ring, after the strand-passage reaction (45).
Another kind of local sensing of the global topology of DNA is the ability of some type IIA topoisomerases to efficiently discriminate between knots (46) or supercoiled DNA of opposite signs. For example, bacterial topo IV (47), DNA gyrase (48) and human topoisomerase IIα (49) act preferentially on (+) supercoiled DNA or L-braids. It has been proposed that the C-terminal domain of topo IV acts as a sensor for substrate selection in orienting the T-segment binding (50). In gyrase, this domain is also responsible for the chiral wrapping of DNA around the enzyme tetramer (51,52). A similar role has also been postulated for the eukaryotic C-terminal domains although these domains have no apparent homology with their eubacterial counterparts (49). Similar to the ‘DNA-centric’ models for topology simplification, it has been also suggested that local differences in (+) and (−) supercoiled DNA may be distinguished by the enzymes (53). However, the finding that the C-terminal domains of type IIA topoisomerases play no role in topology simplification suggested that topology simplification and chiral sensing employ different mechanisms (45). Nevertheless, two sets of experimental studies suggest that these two phenomena have in common some mechanistic features probably based on a combination of geometric and kinetic steps of T-segment selection. In the first ones, single-molecules DNA braiding systems have clearly established that type II topoisomerases discern positively and negatively supercoiled DNA on the basis of the local geometry of DNA cross-overs (54–56). Consistent with these finding, chemical cross-linking of topo IV has demonstrated that the enzyme bound to positively supercoiled DNA is in a different conformation from that bound to other forms of DNA (57). Secondly, it has been shown that the relaxation asymmetry is caused by difference in enzyme processivity (58). Indeed, a detailed single-molecule experiment has demonstrated that while Topo IV is highly processive on (+) supercoiled DNA, the enzyme is perfectly distributive on (−) supercoiled DNA. How DNA cross-over geometry does influence the differential mobility of type II topoisomerases on supercoiled DNA of opposite sign represent therefore an interesting conceptual challenge. The detailed knowledge of DNA cross-over properties and their mode of recognition by these enzymes is therefore essential for understanding their puzzling properties and in particular the mode of local sensing of the global topology.
Cross-over DNA topoisomerase II recognition: a complex(’s) picture
A common feature connecting the multiple type II topoisomerase functions and their sense of topology is their binding of DNA cross-overs. As noticed previously, ‘the enzyme’s preference for DNA cross-overs provides an interesting link between its catalytic and structural roles in the cell’ (59). While DNA cross-overs are, at least transiently, an obligate step for the strand-passage reaction (60), they also constitute architectural elements of higher order DNA structures that can be recognized by the proteins condensing DNA (61–63). Indeed, many experimental studies already support the view that type II enzymes bind cross-overs. For example, electron microscopy studies have indicated that they bind preferentially on the helix juxtaposition in the supercoiled or linear DNA (64,65). On another hand, the study of oligonucleotide concentration dependence of DNA cleavage reaction proposed a two-site model in which the transported T-segment must be present for efficient cleavage of G-segment (66). Linking number measurements of plasmid DNA relaxed by vaccinia virus topoisomerase II also concluded to their role in the stabilization of DNA crossings, while suggesting that the stable binding to cross-overs is not directly related to its catalytic cycle (60). Interestingly, it has also been reported that type II topoisomerases bind to particular DNA structures such as four-way-junctions or hairpins to accomplish their multiple functions (67–70). Since type II topoisomerases have presumably multiple DNA binding sites, it is possible that several modes of cross-over binding correspond to distinct functions (71). For example, when the binding of a cross-over occurs before the strand-passage reaction, the G- and T-segment should be both clamped at the interface of the DNA binding and cleavage core and the ATPase domains. Alternatively, a ‘gate-padlock’ model proposes that the enzymes can still hold the two DNA segments after the strand-passage reaction (72). In this model, the enzyme firmly clamps the G- and T-segments trapped into the closed N- and the C-ring, respectively. The formation of this stable complex could participate to the structural role of type II topoisomerases in chromosome architecture (72).
However, from a structural point of view, little is known about the architecture of the ternary complexes as well as the mode of DNA inter-helical interactions within the enzyme cavities. Moreover, the question of cross-over-type II topoisomerase recognition is highly debated, mainly due to the idea that two negatively charged double helices should strongly repel each other, making the formation of such tight ternary complexes improbable. However, the recent crystal structures of two full-size representative enzymes of the topo IIB subclass clearly indicate that the space enclosed between the DNA cleavage–religation domain and the dimerized ATPase domains is suited to accommodate two tightly packed DNA duplexes (16,17). Interestingly, possible candidate for type II enzyme substrates have been observed a long time ago in the crystal structures in which B-DNA double helices pack into tight right-handed cross-overs, thus naturally minimizing electrostatic repulsion (61–63) (Figure 2). It has been suggested that type II topoisomerases and other proteins such as the mismatch repair MutS protein family that recognize either DNA cross-overs or their iso-structural stacked four-way junction (73) may clamp this kind of structures (74). However, it remained to be determined if the close helical approach stabilized in a crystalline environment could also occur in solution. Many more questions are still unsolved: does the simultaneous presence of the G- and T-segments play a role in the structural communication between the ATPase and DNA binding domain? Do DNA–DNA interactions contribute to the catalytic mechanism of strand passage? Are there different cross-over binding modes and do they correspond to distinct functions of the enzymes? And finally, how is cross-over geometry used by the enzyme to sense DNA topology? The present analysis tries to elucidate these questions by dissecting the possible modes of topo IIA and IIB assembly on tight cross-overs.
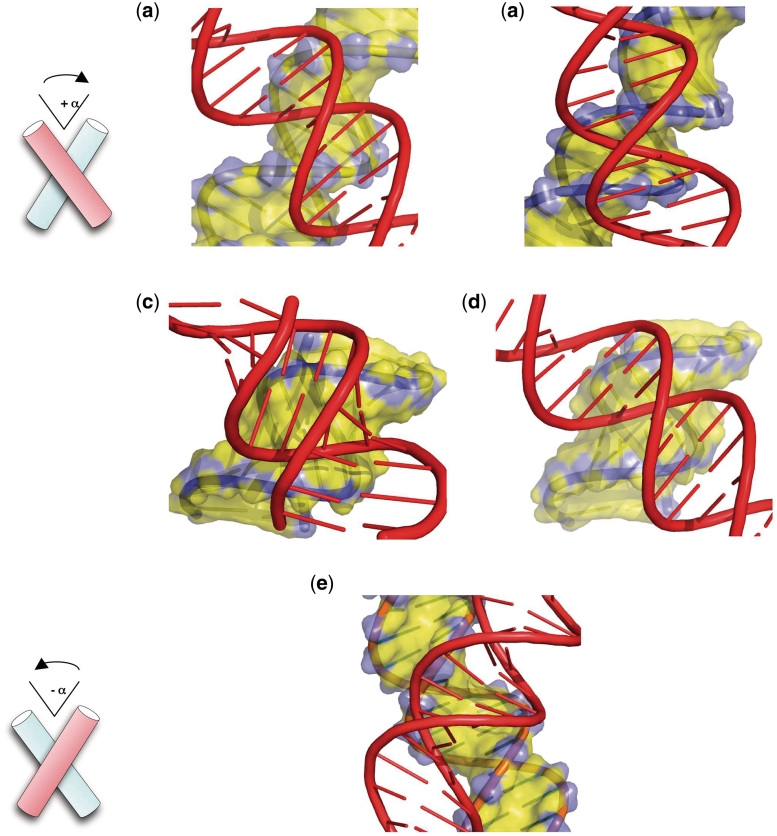
Figure 2.
Modes of inter-helical interactions.Right-handed cross-overs (a–d), left-handed cross-over (e). (a) Major-groove backbone interaction between two B-DNA duplexes. (b) Minor-groove backbone interaction between two B-DNA duplexes. (c) Minor-groove backbone interaction between two A-RNA duplexes. (d) Model of a hybrid cross formed by minor-groove backbone interaction between an A-DNA duplex (yellow) and a B-DNA duplex (red). (e) Major-groove–major groove interaction.
It is proposed herein that the principles governing the geometry and the stability of helical assemblies in nucleic acids may provide useful insights to understand how type II topoisomerases recognize cross-overs and exploit their properties for sensing global topology. Indeed, recent studies have contributed to overcome the conceptual obstacle of electrostatic repulsion. First, the crystallographic structures of large functional assemblies such as ribosomal particles have revealed an extremely dense packing of RNA helices (75–78), similar to the ones observed earlier in crystals of RNA or DNA duplexes, thus supporting the biological relevance of crystal inter-molecular contacts (61). Second, a recent molecular dynamics study has shown that specific helical assemblies observed in crystals can also be stable in solution (79). The detailed knowledge of helical packing modes is therefore essential to understand how cross-over geometry may influence type II topoisomerase functions and mechanisms.
Assembly rules of double helices
The packing of double helices plays essential roles in the architecture and catalysis of nucleic acids. Neutralizing the close approach of negatively charged double helices strictly requires the participation of cations or positively charged amino acids of proteins. Although similar electrostatic rules govern the assembly of RNA and DNA, the helical packing modes of the two molecular cousins differ as a consequence of their distinct secondary structures. Indeed, in addition to adopt a regular A-conformation, the RNA molecules most often fold into more complex structural motifs due to the presence of the 2′-hydroxyl group and extended base pairing rules (80). Thus, RNA structures are characterized by flourishing modes of tertiary interactions, from simple inter-helical interactions to the docking of a wide repertoire of sequence-dependent 3D motifs (81,82). In contrast, DNA molecules mainly form a regular B-DNA double helix stabilized by canonical Watson–Crick base pairing. Following the evolutionary choice of austerity, the building of higher order DNA structures is mainly directed by the B-DNA double helix geometry (62). Crystallographic and theoretical studies have shown that close inter-helical interactions not only minimize the electrostatic repulsion between the negatively charged sugar–phosphate backbones but also optimize the docking between the complementary surfaces of the double helices (83–85). It is also clear that the double helix geometry and handedness play a critical role in the building rules of DNA higher order structures. Thus, the packing of right-handed double helices produces asymmetric DNA cross-overs that differ not only by their inter-molecular contacts but also by their relative stabilities. For example, due to the structural and electrostatic complementarity between the grooves and the sugar–phosphate backbone, right-handed double helices can be mutually fitted by either major-groove or minor-groove backbone interaction into right-handed cross-overs (Figure 2a–d).
From DNA chirality to stable right-handed cross-overs
In the case of B-DNA duplexes, most of the right-handed crosses examined to date from crystallographic data are assembled by major-groove backbone interactions and involve specific cytosine-phosphate group hydrogen bonds (86) (Figure 2a). In solution, a short-range attraction of about −4 kcal mol−1 was observed recently between the duplexes within a right-handed cross-over, in the presence of divalent cations (79). Although less frequent in B-DNA crystals, minor-groove backbone interactions has also been observed (87) (Figure 2b). Interestingly, this mode of interaction has also been observed in a molecular dynamics study of mini-circles of DNA. Under helical tension, monovalent cations seem sufficient to stabilize the neighbouring DNA segments (88). In contrast, in left-handed cross-overs, the B-DNA helices are juxtaposed by groove–groove interactions to minimize their electrostatic repulsion (89) (Figure 2e). This mode of interaction is neither stabilized by sequence-specific contacts between DNA segments, nor by inter-molecular divalent cation bridges. Left-handed cross-overs in ionic conditions favouring the association of right-handed crosses have been found unstable, resulting in a swift dissociation of the helices (79). This study demonstrated that without specific inter-molecular interactions, left-handed helix juxtapositions can only be stable in the crystallographic environment and never in solution.
The A-form double helices differ from the B-form in their modes of inter-molecular recognition (90). As a consequence of a different accessibility and geometry of their grooves, the A-form double helices preferentially self-assemble into right-handed cross-overs formed by minor-groove backbone interactions (Figure 2c). For example, these structures have been observed in the crystal packing of many RNA oligonucleotides (91,92) and are one of the most common elements of the ribosome structure (78). Interestingly, this so called ‘along-groove’ packing motif is thought to play a role in ribosomal function such as tRNA translocation (93,94).
The role of the DNA sequences is also different in the packing of A- and B-DNA helices. A comparison of DNA crystal packing modes revealed that the interactions between A-DNA helices are much less dependent from the DNA sequence than the B-DNA ones (84). Probably because the shallow minor-groove of the A-form provides the opportunity to form many van der Waals and hydrophobic interactions, their stable association has been found less dependent from the formation of specific hydrogen bonds. In consequence, a constant geometry of the A-form assemblies can be maintained for a large variety of sequences. Moreover, as a consequence of a more constraining base stacking scheme in A-form, the sequence-dependent structural variability of the double helix is significantly reduced (95). In summary, despite their differences in sequence requirements, right-handed cross-overs self-fitted by groove backbone interactions constitute a key motif for the stable assembly of both A- and B-form double helices.
From helical interactions to DNA topology
The interplay of local and global properties constitutes a key element in the cellular function of DNA. Factors that affect the local properties of DNA directly influence the global properties of supercoiled DNA and, in turn, changes in superhelicity have repercussions on the local DNA structure and stability (96). For example, it is commonly thought that the underwound form of (−) supercoiled DNA facilitates locally, the strand separation required for transcription or DNA recombination in mesophilic bacteria (97,98). Conversely, the formation of triplex or cruciform structures in specific sequences modulates the rate of encounter and the efficiency of communication between remote sites and may affect transcription through altered global dynamics of supercoiled DNA (99,100). Consequently, local intra- or inter-molecular DNA–DNA interactions play a central role by establishing a link between the two hierarchical levels of structural organization in DNA. In a recent study, it has been shown that the various topological states of the cell are associated with different inter-segmental interactions (101). As seen above, due to the intrinsic helical chirality of DNA, the global topological state of DNA is asymmetrically encoded in the geometry and stability of DNA cross-overs. Indeed, the stable right-handed DNA cross-overs constitute the most probable structure of site juxtaposition in physiological conditions. Thus, right-handed crosses that occur preferentially in (+) supercoiled DNA for geometrical reasons should also be preferentially formed in the absence of superhelical stress, as in relaxed DNA, catenanes or loose knots for electrostatic reasons. This view is fully consistent with the observation that relaxed pBR322 DNA forms positive supercoils in the presence of divalent cations (102). Indeed, the spontaneous formation of right-handed cross-overs is expected to promote the winding of the DNA molecule into a left-handed superhelix, the braiding mode of (+) supercoiled DNA. In contrast, the formation of unstable left-handed DNA crosses is strictly associated with (−) supercoiling which is the normal topological state of mesophilic cells. Consequently, whereas the unstable left-handed cross-overs are exclusively formed in negatively supercoiled DNA, stable right-handed cross-overs constitute the local signature of an unusual topological state in the cell, such as the positively supercoiled or relaxed DNA (see Figure 1 in 101).
As a corollary, the differential stability of cross-overs may be exploited for sensing the global topology of DNA from local interactions. This provides a simple mechanism for the local discrimination of different topological states of DNA and provides new insights for explaining, for example, why negative supercoiling favours decatenation more than positive supercoiling (103). Thus, the different stabilities and geometry of inter-segmental interactions in supercoiled DNA of opposite sign brings a new support for the ‘DNA-juxtaposition-centric’ models (42). Sensing the differential stability and geometry of DNA cross-overs would become the secret of Maxwell’s topological demons (35)?
Type II topoisomerases have evolved to bind stable right-handed DNA cross-overs
It is, therefore, likely that type II topoisomerases have evolved to clamp stable DNA juxtapositions. This paragraph shows indeed that their architecture is designed for selectively bind right-handed cross-overs. The analysis of the available crystal structures of these enzymes shows first that both families have evolved to recognize the G- and T-segments stably assembled into a stable right-handed cross-over. Figure 3a and b reveal indeed that the shape of N-ring in the two class enzymes can only accommodate right-handed crosses. However, due to a different organization of the catalytic domains, types IIA and IIB enzymes recognize DNA cross self-fitted by minor- and major-groove backbone interactions, respectively.
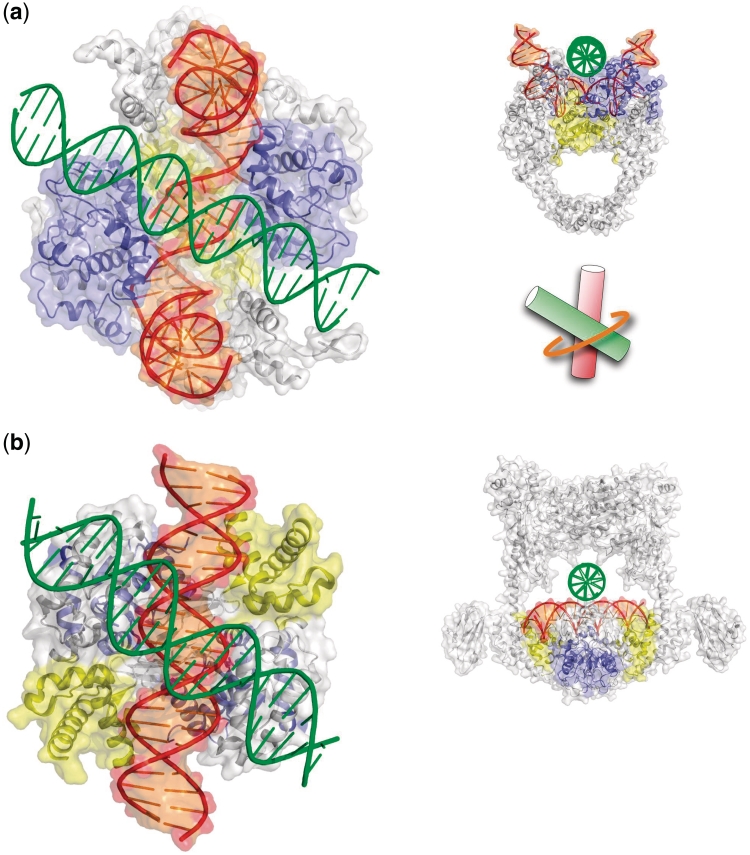
Figure 3.
Type II topoisomerases have evolved to clamp right-handed cross-overs. Model of a topo IIA (a) and of a topo IIB (b) bound to a G- and a T-segment forming a right-handed cross-over. Detailed views of the DNA inter-helical interaction within the enzyme–cross-over complex. The T- and G-segments are assembled by minor-groove backbone interaction within the N-ring of topo IV (a) and by major-groove backbone interaction within the N-ring of topo VI (b). The N-rings (orange) clamp the cross-over across its large angle. The top rim and CAP domains are represented in blue and yellow, respectively. Structural and biochemical data have been brought together for inferring the geometry of cross-overs bound to type II topoisomerases. The available crystallographic coordinates of type II topoisomerases deposited in the PDB and in particular the recent crystal structures of topoII complexed with a G-DNA segment (10–15) have been used for docking DNA cross-overs into the enzymes. The crystal structures of the A-subfragment of the eubacterial Streptococcus pneumonidae topo IV complexed with DNA (12) and the structure of the full-length archaeal methanosarcina mazei topo VI (16) have been considered as two representative models of the topoisomerase IIA and IIB families, respectively. The first one consists into a protein fragment (A-subunit) with a bound G-segment while the other is a full-length enzyme without bound DNA. Since the position of the G-segment is imposed by the protein catalytic domains, the docking problem consists into finding the orientation of T-segment which is dictated by steric and electrostatic constraints imposed by the surface of the protein domains and of the bound G-segment DNA that surround the inner cavity of the N-ring.
Within the type IIB family, a model of the archaeal topo VI core-cleavage domain bound to DNA suggested that the G-segment adopts the B-conformation (18). In this model, the major-groove of the duplex is oriented towards the interior of the N-ring, with the dyad axis of DNA coinciding with the 2-fold axis of the protein dimer. In addition, the crystal structures of two full-size representative enzymes of the topo IIB subclass have shown that the space located between the DNA cleavage–religation domain and the dimerized ATPase domains (N-Ring) is sufficient for fitting two DNA segments (16,17). It has been subsequently shown that these structures can only accommodate right-handed cross-overs self-fitted by groove backbone interactions (Figures 1b and 3b), thus suggesting that type IIB enzymes have evolved to clamp specifically stable right-handed DNA juxtapositions (101).
In the recent structures of type IIA enzymes (10–15), both the bound G-segments and the protein domains shape the N-ring inner surface in such way that the T-segment is also constrained into forming a right-handed cross with the G-segment. However, since the G-segment adopts an A-conformation in its central base pairs and exposes its minor groove towards the incoming T-segment (Figure 3a), the DNA surface available for inter-helix contacts is only compatible with a minor-groove backbone interaction. Consequently, the cross-over clamped by type IIA enzymes is expected to form a hybrid structure composed by a G-segment in the A-conformation and a T-segment in the B-conformation (Figure 2d). Interestingly, the shaping of an enzymatic active site with a DNA duplex has been also observed in DNA polymerases in which the A-form is thought to promote the replication accuracy in buffering the sequence dependent structural variability (95). It is likely that the A-form play a similar role in type IIA topoisomerases in contributing first, to attenuate the sequence-dependent conformational variability of the G-segment in order to keep a constant geometry of the cavity and second, to provide a minor groove surface that can accommodate, without a strict sequence specificity, the backbone of a T-segment (see above).
Thus, independently from their C-terminal domain, the architecture of type IIA and IIB topoisomerases has evolved to clamp selectively stable right-handed cross-overs fitted by groove backbone interaction. An interesting hypothesis is that these enzymes may have exploited the electrostatic properties of cross-overs for their catalytic mechanism of the strand-passage reaction (101). Clamping both the G- and T-segments should be greatly facilitated if there is an attractive interaction between the duplexes (‘pull’). Right-handed crosses are therefore optimal candidates as substrates for the reaction. In contrast, expelling the T-segment from the enzyme would be facilitated by the repulsive interaction between the DNA segments within a left-handed cross-over generated by the reaction (‘push’). Unstable left-handed cross-overs are therefore better candidates for being the product of the strand-passage reaction.
From cross-over geometry to type IIA topoisomerase processivity
Importantly, the ATPase domains that surround the N-ring of both type II topoisomerase families embrace the DNA right-handed cross-overs across their large angle (Figure 3a, right). This mode of binding has then crucial repercussions on the topological relationships between the enzyme rings and the supercoiled DNA of opposite sign, thus directly affecting their ability to move along the DNA chain. This observation provides a simple rational explanation for the chiral discrimination by topo IV that is processive on (+) supercoiled DNA and perfectly distributive on (−) supercoiled DNA. According to Neuman et al. (58), the key step that differentiates the topo IV processivity on (−) and (+) supercoiled DNA is the large conformational changes that accompany the refolding of the DNA, subsequent to the strand-passage reaction. During this step, the DNA chain quickly collapses to restore a plectoneme with a lower energetic state. However, the structural bases explaining the different enzyme behaviour on (−) and (+) supercoiling during this short step remained to be determined.
The present analysis shows that within (+) supercoiled DNA, the enzyme clamps the right-handed cross-overs, in such way that the N-ring is perpendicular to the long supercoiled DNA axis and is not topologically linked to the circular DNA chain (Figure 4a). The strand-passage reaction then transports the T-segment into the C-ring and generates a ‘free’ topological configuration in which, although maintained in their closed forms, the two rings are free to slide and to swivel along the DNA segments. Consequently, similar to a trapeze artist, the enzyme can exploit the large DNA conformational change that occurs during the loop refolding, to correctly reposition itself at the vicinity of another cross-over without dissociating from both DNA segments. The enzyme can then release the T-segment from the C-ring to perform a new catalytic cycle. This provides a geometrical basis that explains the observed processivity of topo IV on (+) supercoiled DNA (58). The C-ring can therefore be viewed as a processivity factor that can slide along the T-segment using the elastic energy of supercoiled DNA after the strand-passage reaction. Consistent with this assumption, is the lack of a corresponding C-ring domain in topoIIB that are unable to sense DNA topology (45). In type IIB enzymes, the T-segment is directly released into the solvent after the strand-passage reaction.
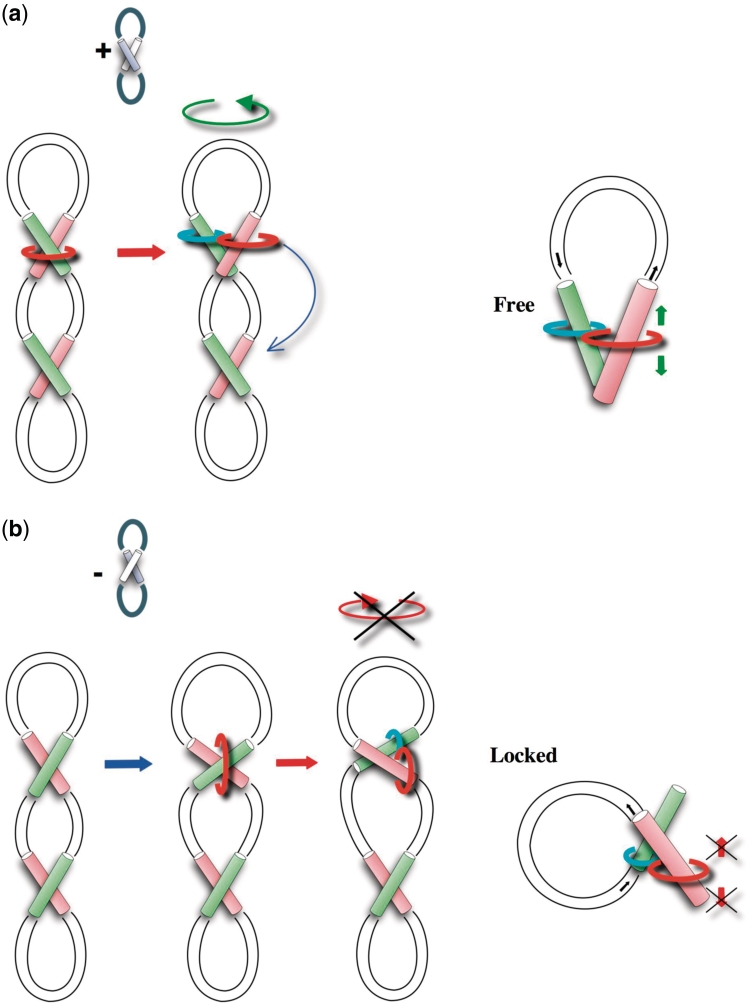
Figure 4.
From cross-over geometry to type IIA topoisomerase processivity. Clamping right-handed cross-overs across their large angles imposes a different topological link between the enzyme’s rings and the plectoneme of opposite signs. (a) In (+) supercoiled DNA, the transport of the T-segment (green) from the N-ring (orange) into the C-ring (cyan) generates a ‘free’ topology compatible with the enzyme motion and processivity along the DNA chain. The enzymes can exploit the loop refolding (green arrow) for finding another cross-over (blue arrow). (b) In (−) supercoiled DNA, the transport of the T-segment into the C-ring generates a ‘locked’ topology incompatible with the enzyme motion along DNA. Consistent with its distributivity on (−) supercoiled DNA, the enzyme is unable to exploit the loop refolding for finding another cross-over.
In contrast, in (−) supercoiled DNA, left-handed cross-overs are the prevalent crossing modes (Figure 4b) (101). For binding its right-handed cross-over substrate, the enzymes must therefore be positioned differently onto the plectoneme. Here, the N-ring that is parallel to the supercoiled DNA axis forms an inter-locked knot and is topologically linked with the circular DNA (the ring passes across the two lobes of the ‘eight’). In consequence, the strand-passage reaction that transports the T-segment into the C-ring generates a ‘locked’ topological configuration that is incompatible with the enzyme mobility along the DNA chain. The set of forces exerted in the opposite direction by the elastic tension of DNA on the two rings not only physically impedes their motion along the DNA but also prevents the loop refolding. This topological situation locks the relaxation process since the enzyme is immobilized on the (−) supercoiled DNA. Consistent with the distributive behaviour of topo IV on (−) supercoiled DNA, the enzyme is unable to exploit the loop refolding for sliding to a next cross-over since the release of the T-segment from the C-ring is required before the loop refolding. In conclusion, the present study brings the missing link between structural and kinetic data and provides an extremely simple explanation for the chiral discrimination by type IIA enzymes. The intrinsic DNA handedness that governs the stable helical associations plays therefore a critical role for the local sensing of DNA topology by type IIA topoisomerases. In conclusion, this analysis suggests that type II topoisomerases have learnt the rules that govern the assembly of DNA molecules in order to perform their multiple functions and to locally sense the global DNA topology.
FUNDING
Funding for open access charge: Centre National de la Recherche Scientifique (CNRS).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The author thanks Chantal Abergel, Jean-Michel Claverie, Pascal Hingamp, Olivier Hyrien, Peter Varnai and Eric Westhof for helpful discussions and the critical reading of the manuscript.
REFERENCES
- 1.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Nittis JL. DNA topoisomerase II and its growing repertoire of biological functions. Nature Rev. Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forterre P, Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 5.Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 6.Bates AD, Maxwell A. The role of ATP in the reactions of type DNA topoisomerases. Biochem. Soc. Trans. 2010;38:438–442. doi: 10.1042/BST0380438. [DOI] [PubMed] [Google Scholar]
- 7.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl Acad. Sci. USA. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roca J. The path of DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 2004;279:25783–25788. doi: 10.1074/jbc.M402555200. [DOI] [PubMed] [Google Scholar]
- 9.Mueller-Planiz F, Hershlag D. Coupling between ATP binding and DNA cleavage by DNA topoisomerase II. A unifying kinetic and structural mechanism. J. Biol. Chem. 2008;283:17463–17476. doi: 10.1074/jbc.M710014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1206. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature. 2010;465:641–644. doi: 10.1038/nature08974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laponogov I, Sohi MK, Veselkov DA, Pan XS, Sawhney R, Thompson AW, McAuley KE, Fisher LM, Sanderson MR. Structural insight into the quinolone-DNA cleavage complex of type IIA topoisomerases. Nat. Struct. Mol. Biol. 2009;16:667–669. doi: 10.1038/nsmb.1604. [DOI] [PubMed] [Google Scholar]
- 13.Laponogov I, Pan XS, Veselkov DA, McAuley KE, Fisher LM, Sanderson MR. Structural basis of gate-DNA breakage and resealing by type II topoisomerases. PLoS One. 2010 doi: 10.1371/journal.pone.0011338. 2010 5:e11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bax BD, Chan PF, Eggleston DS, Fosberry A, Gentry DR, Gorrec F, Giordano I, Hann MM, Hennessy A, Hibbs M, et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature. 2010;466:935–940. doi: 10.1038/nature09197. [DOI] [PubMed] [Google Scholar]
- 15.Wohlkonig A, Chan PF, Fosberry AP, Homes P, Huang J, Kranz M, Leydon VR, Miles TJ, Pearson ND, Perera RL, et al. Structural basis of quinolone inhibition of type IIA topoisomerases and target-mediated resistance. Nat. Struct. Mol. Biol. 2010;17:1152–1153. doi: 10.1038/nsmb.1892. [DOI] [PubMed] [Google Scholar]
- 16.Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- 17.Graille M, Cladière L, Durand D, Lecointe F, Gadelle D, Quevillon-Cheruel S, Vachette P, Forterre P, van Tilbeurgh H. Crystal structure of an intact type II DNA topoisomerase: insight into DNA transfer mechanisms. Structure. 2008;16:360–370. doi: 10.1016/j.str.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 18.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc. Natl Acad. Sci. USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stupina V, Wang JC. DNA axial rotation and the merge of oppositely supercoiled DNA domains in Escherichia coli: Effects of DNA bends. Proc. Natl Acad. Sci. USA. 2004;101:8608–8613. doi: 10.1073/pnas.0402849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol. Microbiol. 2004;52:925–931. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- 22.Gellert M, Mizuuchi K, O’Dea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3878. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20:6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travers A, Muskhelishvili G. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 2007;8:147–151. doi: 10.1038/sj.embor.7400898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi Y, Käs E, Laemmli UK. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989;8:3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berrios M, Osheroff N, Fisher PA. In situ localization of DNA topoisomerase II, a major polypeptide component of the Drosophila nuclear matrix fraction. Proc. Natl Acad. Sci. USA. 1985;82:4142–4146. doi: 10.1073/pnas.82.12.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasser SM, Laemmli UK. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 28.Belmont A. Mitotic chromosome structure and condensation Curr. Opin. Cell. Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 29.Haering CH, Nasmyth K. Building and breaking bridges between sister chromatids. Bioessays. 2003;25:1178–1191. doi: 10.1002/bies.10361. [DOI] [PubMed] [Google Scholar]
- 30.Swedlow JR, Hirano T. The making of the mitotic chromosomes: modern insights into classical questions. Mol. Cell. 2003;11:557–569. doi: 10.1016/s1097-2765(03)00103-5. [DOI] [PubMed] [Google Scholar]
- 31.Porter CG, Farr CJ. Topoisomerase II: untangling its contribution at the centromere. Chrom. Res. 2004;12:569–583. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- 32.Hizume K, Araki S, Yoshikawa K, Takeyasu K. Topoisomerase II, scaffold component, promotes chromatin compaction in vitro in a linker-histone H1-dependent manner. Nucleic Acids Res. 2007;35:2787–2799. doi: 10.1093/nar/gkm116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu YH, Chung MW, Li TK. Distribution of gyrase and topoisomerase IV on bacterial nucleoid: implications for nucleoid organization. Nucleic Acids Res. 2006;34:3128–3138. doi: 10.1093/nar/gkl392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dasgupta S, Maisnier-Patin S, Nordström K. New genes with old modus operandi. The connection between supercoiling and partitioning of DNA in Escherichia coli. EMBO Rep. 2000;1:323–327. doi: 10.1093/embo-reports/kvd077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulleyblank DE. Of Topo and Maxwell’s dream. Science. 1997;277:648–649. doi: 10.1126/science.277.5326.648. [DOI] [PubMed] [Google Scholar]
- 36.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 37.Vologodskii A. Theoretical models of topology simplification by type IIA DNA topoisomerases. Nucleic Acid Res. 2009;37:3125–3133. doi: 10.1093/nar/gkp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z, Deibler RW, Chan HS, Zechiedrich L. The why and how DNA unlinking. Nucleic Acid Res. 2009;37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vologodskii AV, Zhang W, Rybenkov V, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerase. Proc. Natl Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trigueros S, Salceda J, Bermudez I, Fernandez X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J. Mol. Biol. 2004;335:723–731. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 41.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Mann JK, Zechiedrich EL, Chan HS. Topological information embodied in local juxtaposition geometry provides a statistical mechanical basis for unknotting by type-2 DNA topoisomerases. J. Mol. Biol. 2006;361:268–285. doi: 10.1016/j.jmb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 43.Buck GR, Zechiedrich EL. DNA Disentangling by Type-2 Topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 44.Randall GL, Pettitt BM, Buck GR, Zechiedriech EL. Electrostatics of DNA–DNA juxtapositions: consequences for type II topoisomerase function. J. Phys. Condens. Matter. 2006;18:S173–S185. doi: 10.1088/0953-8984/18/14/S03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stuchinskaya T, Mitchenall LA, Schoeffler AJ, Corbett KD, Berger JM, Bates AD, Maxwell A. How do type II topoisomerases use ATP hydrolysis to simplify DNA topology beyond equilibrium? Investigating the relaxation reaction of non supercoiling type II topoisomerases. J. Mol. Biol. 2009;385:1397–1408. doi: 10.1016/j.jmb.2008.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw S, Wang JC. Chirality of DNA trefoils: implications in intramolecular synapsis of distant DNA segments. Proc. Natl Acad. Sci. USA. 1997;94:1692–1697. doi: 10.1073/pnas.94.5.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Gene Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nöllmann N, Stone MD, Bryant Z, Gore J, Crisona NJ, Hong SC, Mitelheiser S, Maxwell A, Bustamante C, Cozzarelli NR. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 49.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIα rapidly relaxes positively supercoiled DNA. Implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 50.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JB. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Kampranis SC, Maxwell A. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl Acad. Sci. USA. 1996;93:14416–14421. doi: 10.1073/pnas.93.25.14416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruthenburg AJ, Graybosch DM, Huetsch JC, Verdine GL. A Superhelical Spiral in the Escherichia coli DNA Gyrase A C-terminal Domain Imparts Unidirectional Supercoiling Bias. Proc. Natl Acad. Sci. USA. 2005;280:26177–26184. doi: 10.1074/jbc.M502838200. [DOI] [PubMed] [Google Scholar]
- 53.Fogg JM, Catanese DJ, Randall GL, Swick MC, Zechiedrich LE. Differences between positively and negatively supercoiled DNA that topoisomerases may distinguish. In Mathematics of DNA Structure, Function and Interactions. The IMA Volumes in Mathematics and its Applications; 2009. 2009, 150, 73–121. [Google Scholar]
- 54.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamente C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Charvin G, Strick TR, Bensimon D, Croquette V. Tracking topoisomerase activity at the single-molecule level. Annu. Rev. Biophys. Biomol. Struct. 2005;34:201–219. doi: 10.1146/annurev.biophys.34.040204.144433. [DOI] [PubMed] [Google Scholar]
- 57.Crisona NJ, Cozzarelli NR. Alteration of Escherichia coli topoisomerase IV conformation upon enzyme binding to positively supercoiled DNA. J. Biol. Chem. 2006;281:18927–18932. doi: 10.1074/jbc.M603068200. [DOI] [PubMed] [Google Scholar]
- 58.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Osheroff N, Zechiedrich EL, Gale K. Catalytic function of topoisomerase II. Bioessays. 1990;13:269–275. doi: 10.1002/bies.950130603. [DOI] [PubMed] [Google Scholar]
- 60.Roca J, Berger JM, Wang JC. On the simultaneous binding of eukaryotic DNA topoisomerase II to a pair of double-stranded DNA helices. J. Biol. Chem. 1993;268:14250–14255. [PubMed] [Google Scholar]
- 61.Timsit Y, Moras D. Groove-Backbone Interaction in B-DNA. Implication for DNA condensation and recombination. J. Mol. Biol. 1991;221:919–940. doi: 10.1016/0022-2836(91)80184-v. [DOI] [PubMed] [Google Scholar]
- 62.Timsit Y, Moras D. DNA self-fitting: the double helix directs the geometry of its supramolecular assembly. EMBO J. 1994;13:2737–2746. doi: 10.1002/j.1460-2075.1994.tb06567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Timsit Y, Moras D. Cruciform structures and functions. Q. Rev. Biophys. 1996;29:279–307. doi: 10.1017/s0033583500005862. [DOI] [PubMed] [Google Scholar]
- 64.Zechiedrich EL, Osheroff N. Eukaryotic topoisomerases recognize nucleic acid topology by preferentially interacting with DNA crossovers. EMBO J. 1990;9:4555–4562. doi: 10.1002/j.1460-2075.1990.tb07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howard MT, Lee MP, Hsieh T-S, Griffith JD. Drosophila topoisomerase II-DNA interactions are affected by DNA structure. J. Mol. Biol. 1991;217:53–62. doi: 10.1016/0022-2836(91)90610-i. [DOI] [PubMed] [Google Scholar]
- 66.Corbett AH, Zechiedrich EL, Osheroff N. A role for the passage helix in the cleavage reaction of eukaryotic topoisomerase II. A two-site model for enzyme mediated DNA cleavage. J. Biol. Chem. 1992;267:683–686. [PubMed] [Google Scholar]
- 67.West K, Austin CA. Human DNA topoisomerase IIβ binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999;27:984–992. doi: 10.1093/nar/27.4.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Froelich-Ammon SJ, Gale K, Osheroff N. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 1994;269:7719–7725. [PubMed] [Google Scholar]
- 69.Jonstrup AT, Thomsen T, Wang Y, Knudsen B, Koch J, Andersen AH. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIα. Nucleic Acids Res. 2008;36:6165–6174. doi: 10.1093/nar/gkn640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.René B, Fermandjian S, Mauffret O. Does topoisomerase II recognize and cleave hairpins, cruciforms and crossovers of DNA?. (2007) Biochimie. 89:508–515. doi: 10.1016/j.biochi.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 71.Timsit Y, Duplantier B, Jannink G, Sikorav JL. Symmetry and chirality in topoisomerase II-DNA crossover recognition. J. Mol. Biol. 1998;284:1289–1299. doi: 10.1006/jmbi.1998.2281. [DOI] [PubMed] [Google Scholar]
- 72.Roca J. Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 2009;37:721–730. doi: 10.1093/nar/gkn994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eichman BF, Ortiz-Lombardia M, Aymami J, Coll M, Ho PS. The inherent properties of DNA four-way junctions: comparing the crystal structures of Holliday junctions. J. Mol. Biol. 2002;320:1037–1051. doi: 10.1016/s0022-2836(02)00540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Timsit Y. Convergent evolution of MutS and Topoisomerase II for clamping DNA crossovers and stacked Holliday junctions. J. Biomol. Struct. Dyn. 2001;19:215–218. doi: 10.1080/07391102.2001.10506733. [DOI] [PubMed] [Google Scholar]
- 75.Ban N, Nissen P, Hansen J, Moore PB, Steitz A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 76.Wimberly BT, Brodersen DE, Clemons W, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartschk T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 77.Harms J, Schluenzen F, Zarivach R, Basham A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 78.Nissen P, Ippolito JA, Ban N, Moore P, Steitz T. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varnai P, Timsit Y. Differential stability of chiral DNA crossovers mediated by divalent cations. Nucleic Acid Res. 2010;38:4163–4172. doi: 10.1093/nar/gkq150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Leontis NB, Lescoute A, Westhof E. The buidling blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 83.Srinivasan AR, Olson WK. DNA associations: packing calculation in A-, B- and Z-DNA structures. Biophys. Chem. 1992;43:279–310. doi: 10.1016/0301-4622(92)85027-2. [DOI] [PubMed] [Google Scholar]
- 84.Timsit Y, Moras D. Crystallization of DNA. Meth. Enzym. 1992;211:409429. doi: 10.1016/0076-6879(92)11022-b. [DOI] [PubMed] [Google Scholar]
- 85.Murthy VL, Rose GD. Is counterion delocalization responsible for collapse in RNA folding. Biochemistry. 2000;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- 86.Timsit Y, Varnai P. Cytosine, the double helix and DNA self-assembly. J. Mol. Recognit. 2011;24:137–138. doi: 10.1002/jmr.1082. [DOI] [PubMed] [Google Scholar]
- 87.Wood AA, Nunn CM, Trent JO, Neidle S. Sequence-dependent crossed helix packing in the crystal structure of a B-DNA decamer yields a detailed model for the Holliday junction. J. Mol. Biol. 1997;269:827–841. doi: 10.1006/jmbi.1997.1089. [DOI] [PubMed] [Google Scholar]
- 88.Mitchell JS, Laughton CA, Harris SA. Atomistic simulations reveal bubbles, kinks and wrinkles in supercoiled DNA. Nucleic Acids Res. 2011;39:3928–3938. doi: 10.1093/nar/gkq1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Timsit Y, Shatzky-Schwartz M, Shakked Z. Left-handed DNA crossovers. Implications for DNA-DNA recognition and structural alterations. J. Biomol. Struct. Dyn. 1999;16:775–785. doi: 10.1080/07391102.1999.10508292. [DOI] [PubMed] [Google Scholar]
- 90.Lu X-J, Shakked Z, Olson W. A-form conformational motifs in ligand-bound DNA structures. J. Mol. Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 91.Schindelin H, Zhang M, Bald R, Fürste J-P, Erdmann VA, Heinemann U. Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol. 1995;249:595–603. doi: 10.1006/jmbi.1995.0321. [DOI] [PubMed] [Google Scholar]
- 92.Baeyens K, De Bondt HL, Holbrook SR. Structure of an RNA double helix including uracil-uracil base pairs in an internal loop. Nat. Struct. Biol. 1996;2:56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- 93.Gagnon M, Steinberg S. GU receptors of double helices mediate tRNA movement in the ribosome. RNA. 2002;8:873–877. doi: 10.1017/s135583820202602x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gagnon MG, Mukhopadhyay A, Steinberg SV. Close packing of helices 3 and 12 of 16S rRNA is required for the normal ribosome function. J. Biol. Chem. 2006;281:39349–39357. doi: 10.1074/jbc.M607725200. [DOI] [PubMed] [Google Scholar]
- 95.Timsit Y. DNA structure and polymerase fidelity. J. Mol. Biol. 1999;293:835–853. doi: 10.1006/jmbi.1999.3199. [DOI] [PubMed] [Google Scholar]
- 96.Minsky A. Information content and complexity in the higher organisation of DNA. Annu. Rev. Biophys. Biomol. Struct. 2004;33:317–342. doi: 10.1146/annurev.biophys.33.110502.133328. [DOI] [PubMed] [Google Scholar]
- 97.Benham CJ, Mielke SP. DNA mechanics. Annu. Rev. Biomed. Eng. 2005;7:21–53. doi: 10.1146/annurev.bioeng.6.062403.132016. [DOI] [PubMed] [Google Scholar]
- 98.Vologodskii AV, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- 99.Shlyakhtenko LS, Hsieh P, Grigoriev M, Potaman VN, Sinden R, Lyubchenko YL. A cruciform structural transition provides a molecular switch for chromosome structure and dynamics. J. Mol. Biol. 2000;296:1169–1173. doi: 10.1006/jmbi.2000.3542. [DOI] [PubMed] [Google Scholar]
- 100.Vetcher AA, Napierala M, Wells RD. Sticky DNA: effect of the polypurine. polypyrimidine sequence. J. Biol. Chem. 2002;277:39228–39234. doi: 10.1074/jbc.M205210200. [DOI] [PubMed] [Google Scholar]
- 101.Timsit Y, Varnai P. Helical chirality: a link between local interactions and global topology in DNA. PLos ONE. 2010;5:e9326. doi: 10.1371/journal.pone.0009326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu Y-C, Bremer H. Winding of the DNA helix by divalent metal ions. Nucleic Acids Res. 1997;25:4067–4071. doi: 10.1093/nar/25.20.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roca J. Varying levels of positive and negative supercoiling differently affect the efficiency with which topoisomerase II catenates and decatenates DNA. J. Mol. Biol. 2001;305:441–450. doi: 10.1006/jmbi.2000.4307. [DOI] [PubMed] [Google Scholar]