Abstract
Production of reactive oxygen species represents a fundamental innate defense against microbes in a diversity of host organisms. Oxidative stress, amongst others, converts peptidyl and free methionine to a mixture of methionine-S- (Met-S-SO) and methionine-R-sulfoxides (Met-R-SO). To cope with such oxidative damage, methionine sulfoxide reductases MsrA and MsrB are known to reduce MetSOs, the former being specific for the S-form and the latter being specific for the R-form. However, at present the role of methionine sulfoxide reductases in the pathogenesis of intracellular bacterial pathogens has not been fully detailed. Here we show that deletion of msrA in the facultative intracellular pathogen Salmonella (S.) enterica serovar Typhimurium increased susceptibility to exogenous H2O2, and reduced bacterial replication inside activated macrophages, and in mice. In contrast, a ΔmsrB mutant showed the wild type phenotype. Recombinant MsrA was active against free and peptidyl Met-S-SO, whereas recombinant MsrB was only weakly active and specific for peptidyl Met-R-SO. This raised the question of whether an additional Met-R-SO reductase could play a role in the oxidative stress response of S. Typhimurium. MsrC is a methionine sulfoxide reductase previously shown to be specific for free Met-R-SO in Escherichia (E.) coli. We tested a ΔmsrC single mutant and a ΔmsrBΔmsrC double mutant under various stress conditions, and found that MsrC is essential for survival of S. Typhimurium following exposure to H2O2, as well as for growth in macrophages, and in mice. Hence, this study demonstrates that all three methionine sulfoxide reductases, MsrA, MsrB and MsrC, facilitate growth of a canonical intracellular pathogen during infection. Interestingly MsrC is specific for the repair of free methionine sulfoxide, pointing to an important role of this pathway in the oxidative stress response of Salmonella Typhimurium.
Introduction
Salmonella affects humans and animals causing a variety of diseases including gastroenteritis, septicemia, and typhoid fever. Salmonella is a facultative intracellular pathogen. Inside macrophages, the pathogen is exposed to oxidative stress [1]. Individuals with chronic granulomatous disease, who suffer from a genetic defect of the phagocyte NADPH oxidase gene, show a severely impaired production of reactive oxygen intermediates (ROI) due to the lack of superoxide anion O2 -, and are more susceptible to infections with Salmonella [2]. Likewise, susceptibility to Salmonella is increased in macrophages from mice with diminished NADPH oxidase activity [3]. Pathogens counteract oxidative stress by detoxifying ROI and repairing damage caused by ROI. For detoxification Salmonella possesses two classes of enzymes, catalases and peroxiredoxin-type peroxidases (peroxiredoxins).Three members of the catalase family, KatG, KatE, and KatN, and three members of the peroxiredoxin family, AhpC, TsaA, and Tpx are present in Salmonella, pointing to the multiple enzymes evolved in Salmonella for this route [4]–[6].
Despite detoxification, repair is an essential part of oxidative stress response in most bacteria. Exposure to ROI particularly affects sulfur residues, due to their high susceptibility to oxidation [7]. Sulfur containing amino acids such as methionine can be easily repaired to their native form by specific enzymes [7]. It has been suggested that reduction of methionine not only repairs damage afflicted by oxidative defense of the host, but even provides an advantage, as it could scavenge oxidants and thereby preventing other, less readily reversible reactions [8]. Methionine sulfoxide, whether in protein or as free amino acid, exists in two epimers, methionine-(S)-sulfoxide (Met-S-SO) and methionine-(R)-sulfoxide (Met-R-SO) [7]. Two sulfoxide reductases encoded by msrA and msrB, mediate reduction of Met-S-SO and Met-R-SO, respectively [7], [9]. In E. coli MsrA acts on both free and peptidyl Met-S-SO whereas MsrB acts on peptidyl Met-R-SO and shows little activity for free Met-R-SO [10]. Repair of free Met-R-SO has been shown to be mediated by a third methionine sulfoxide reductase encoded by msrC [10]–[12].
In bacteria such as Mycobacterium (M.) smegmatis [13], Staphylococcus (S.) aureus [14], and E. coli [15] mutants of msrA were hypersensitive to ROI. In Campylobacter (C.) jejuni [16], and Enterococcus (E.) faecalis [17], mutants of msrA and msrB showed hypersensitivity towards ROI, and the effect was much more pronounced in the msrA mutant than in the msrB mutant. In Helicobacter (H.) pylori and Neisseria (N.) gonorrhoeae msrA and msrB are fused together. Deletion of the fusion protein increased sensitivity towards ROI in both species [18], [19]. However, none of these bacterial species are typical intracellular pathogens. In Mycobacterium (M.) tuberculosis deletion in either msrA or msrB did not affect resistance to ROI in vitro [20]. Thus at present the role of MsrA and MsrB for pathogenesis of intracellular pathogens has not been fully understood. There are even less data available on the role of MsrC in bacterial pathogenesis. Recently its ability to be stereo and size specific exclusively for the reduction of free Met-R-SO has been described in S. aureus, E. coli, N. meningitides and in Saccharomyces (S.) cerevisiae [10], [11], [21], [22]. However, except for S. cerevisiae a role of MsrC in oxidative response has not been reported in these organisms.
Here we generated single and double mutants of msrA and msrB and showed that msrA is essential for survival of S. Typhimurium exposed to exogenous H2O2, and growth of the pathogen in activated macrophages and in mice. In contrast, a ΔmsrB mutant showed the wild type phenotype. Further experiments showed that MsrB has weak reductive activity towards peptidyl Met-R-SO and no activity towards free Met-R-SO. Repair of free Met-R-SO was carried out by MsrC. ΔmsrC single and ΔmsrBΔmsrC double mutants were more susceptible to exogenous H2O2, showed reduced growth in activated macrophages, and the ΔmsrBΔmsrC double mutant was attenuated in mice. These results not only suggest that the methionine sulfoxide reductase pathway plays a key role in oxidative stress response of S. Typhimurium, but also indicates that the repair of free oxidized amino acids contributes significantly to resistance of the pathogen towards oxidative stress.
Results
In vitro growth and survival of ΔmsrA and ΔmsrB mutants following exposure to exogenous H2O2
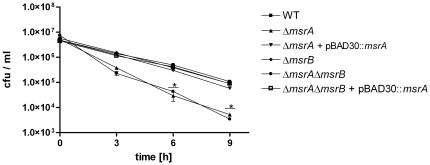
STM4408 was annotated as msrA on the chromosome of S. Typhimurium, with 89% identity (amino acids) to MsrA (AAC77176) of E. coli. STM1291 was annotated as yeaA on the chromosome of S. Typhimurium, with 86% identity (amino acids) to yeaA (AAC74848) of E. coli (coliBASE) [23]. Subsequently, yeaA of E. coli was renamed to msrB [24]. For this present study, we initially generated mutants in S. Typhimurium with deletions in msrA (STM4408), and msrB (STM1291), and double mutants with deletions in both genes. Neither mutant revealed any general growth defect under normal aerobic culture conditions in LB broth (Fig. S1). The impact of the mutations on the susceptibility of S. Typhimurium towards exogenous H2O2 was assessed in vitro. In the presence of 2 mM exogenous H2O2 the ΔmsrA mutant was more susceptible than the wild type after 6 and 9 hours of exposure to H2O2 (Fig. 1). The wild type phenotype could be restored by expressing the wild type msrA on a plasmid under the control of its own promoter. In contrast, survival of the ΔmsrB mutant was not affected in the presence of 2 mM H2O2 in vitro after 6 and 9 hours of exposure to H2O2. A ΔmsrAΔmsrB mutant was constructed to test whether the double mutant has a stronger phenotype than the msrA single mutant in the presence of H2O2. The results shown in Figure 1 indicate that the phenotype of the single ΔmsrA and the double ΔmsrAΔmsrB mutant do not differ from each other. This is in line with the fact that the single msrA from wild type was sufficient to restore the phenotype of the ΔmsrAΔmsrB double mutant (Fig. 1).
Figure 1. Susceptibility of S. Typhimurium ΔmsrA, ΔmsrB, ΔmsrAΔmsrB, ΔmsrA + pBAD30::msrA, and ΔmsrAΔmsrB+ pBAD30::msrA towards exogenous H2O2.
Wild type and mutant strains ΔmsrA, ΔmsrB, ΔmsrAΔmsrB, ΔmsrA + pBAD30::msrA and ΔmsrAΔmsrB + pBAD30::msrA were exposed to 2 mM exogenous H2O2 in LB medium, and plated for colony forming units at the indicated time points. Analysis was done in duplicates, and is a representative of three independent experiments. Asterisks indicate P-values <0.05.
Growth of ΔmsrA and ΔmsrB mutants inside activated macrophages
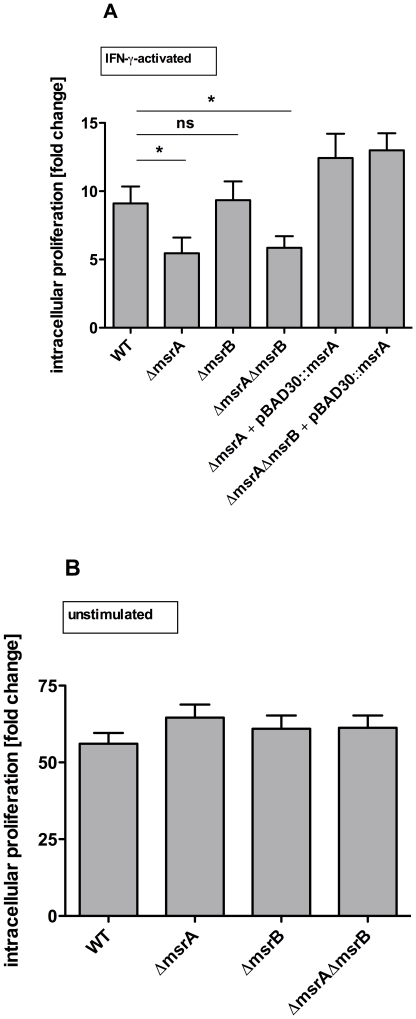
To elucidate the role of msrA and msrB during intracellular growth of S. Typhimurium, the murine cell-line RAW 264.7 was infected with wild type and mutant strains. Intracellular bacteria were enumerated 2 and 16 hours (h) post infection (p.i.) by plating for colony forming units. Proliferation was calculated by dividing the bacterial numbers 16 h p.i. by the number of bacteria 2 h p.i.. In IFN-γ-activated RAW 264.7 macrophages the single ΔmsrA and the ΔmsrAΔmsrB double mutant were attenuated to the same extent, while growth of the ΔmsrB mutant was not affected (Fig. 2A). Complementation of the ΔmsrAΔmsrB double mutant with the wild type msrA was sufficient to restore the wild type phenotype. Similar studies addressing the role of Msr in E. faecalis, Actinobacillus (A.) actinomycetemcomitans and M. smegmatis used non-stimulated cells as controls [13], [17], [25]. Thus we tested the strains in non-stimulated RAW 264.7 cells, and found that neither the ΔmsrA and ΔmsrB single mutants, nor the ΔmsrAΔmsrB double mutant were affected in their intracellular growth in comparison to the wild type (Fig. 2B).
Figure 2. Intracellular proliferation of S. Typhimurium wild type, ΔmsrA, ΔmsrB, ΔmsrAΔmsrB in RAW 264.7 cells.
A. S. Typhimurium wild type, ΔmsrA, ΔmsrB, ΔmsrAΔmsrB, ΔmsrA + pBAD30::msrA and ΔmsrAΔmsrB + pBAD30::msrA were tested in IFN-γ-activated RAW 264.7 cells. B. S. Typhimurium wild type, ΔmsrA, ΔmsrB, and ΔmsrAΔmsrB, were tested in non-stimulated RAW 264.7 cells. Intracellular proliferation was determined in a gentamicin protection assay. Number of bacteria 16 h post infection was divided by the number of bacteria 2 h post infection. Results shown are the means ± standard error of the mean for three independent experiments, each in triplicate. P-values <0.05 were considered to be significant as indicated by asterisks. ns: not significant.
In vivo susceptibility of ΔmsrA and ΔmsrB mutants in Balb/cJ mice
To test whether the deletion of msrA or msrB affects virulence of S. Typhimurium in mice, the fitness of the mutants was directly compared to that of the wild type in a competition experiment [26]. For each mutant and time point groups of 5 female Balb/cJ mice were infected intraperitoneally with a mixture of two bacterial strains in a ratio of 1∶1 and bacterial burden in liver and spleen was enumerated 1 and 3 days after infection. Competitive indices (CI) were calculated as described elsewhere [26] and are given for bacterial loads of various strains in spleen and liver 1 day and 3 days after infection (Table 1 and 2). The ΔmsrA mutant was significantly attenuated in the liver and spleen 1 and 3 days after infection. In contrast, the ΔmsrB mutant was not attenuated in the mouse model of infection in the liver and spleen. The ΔmsrAΔmsrB double mutant was also attenuated both in the liver and the spleen.
Table 1. Competitive indices (CI) of ΔmsrA, ΔmsrB and ΔmsrAΔmsrB versus wild type on day 1 post infection.
| Strains | CI liver 1 day p.i. | P-value* | CI spleen 1 day p.i. | P-value* |
| ΔmsrA vs. WT | 0.69±0.11 | <0.05 | 0.81±0.10 | <0.05 |
| ΔmsrB vs. WT | 1.01±0.27 | >0.05 | 0.93±0.27 | >0.05 |
| ΔmsrAΔmsrB vs. WT | 0.72±0.21 | <0.05 | 0.62±0.11 | <0.05 |
Table 2. Competitive indices (CI) of ΔmsrA, ΔmsrB and ΔmsrAΔmsrB versus wild type on day 3 post infection.
| Strains | CI liver 3 days p.i. | P-value* | CI spleen 3 days p.i. | P-value* |
| ΔmsrA vs. WT | 0.16±0.03 | <0.05 | 0.24±0.07 | <0.05 |
| ΔmsrB vs. WT | 1.13±0.06 | <0.05 | 1.12±0.09 | >0.05 |
| ΔmsrAΔmsrB vs. WT | 0.23±0.09 | <0.05 | 0.13±0.05 | <0.05 |
*P-values were calculated using the Wilcoxon Signed Rank Test to test whether the actual median differs significantly from 1 (theoretical median). CIs are shown as medians ± standard deviation.
Biochemical characterization of MsrA and MsrB
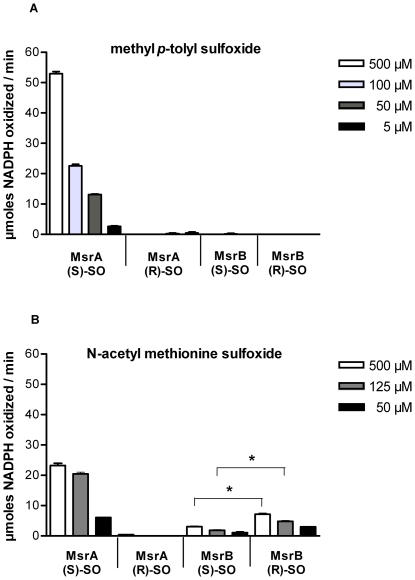
To investigate the enzymatic activity of MsrA and MsrB we overexpressed both proteins in E. coli followed by purification of recombinant MsrA and MsrB. Enzymatic activities of MsrA and MsrB were tested initially with methyl p-tolyl sulfoxide, as both the R- and the S-form are commercially available. In a test system coupled to NADPH oxidation, thioredoxin (Trx) was used as an electron donor for Msr. MsrA was active against the S-epimer of methyl p-tolyl sulfoxide but not the R-epimer. MsrB displayed no activity against either epimer of methyl p-tolyl sulfoxide (Fig. 3A). N-acetyl methionine sulfoxide has been used as a surrogate for peptidyl MetSO [20]. We chemically synthesized N-acetyl Met-R-SO and N-acetyl Met-S-SO and tested activities of both MsrA and MsrB. MsrA and MsrB each were active against N-acetyl MetSO in a stereospecific manner, because MsrA reduced the S-form of N-acetyl MetSO, while MsrB reduced the R-form of N-acetyl MetSO (Fig. 3B). It seems that MsrB was active against N-acetyl Met-S-SO, too. But an NMR-analysis of this substrate showed, that N-acetyl Met-S-SO was not pure but contaminated with N-acetyl Met-R-SO. In fact, purity of N-acetyl Met-S-SO as determined by 1H-NMR was 80%. Thus the observed activity is due to this contamination.
Figure 3. NADPH-linked reductase activity assay of recombinant MsrA and MsrB from S. Typhimurium.
A. The graph shows the amount of oxidized NADPH per minute for the reduction of 500, 100, 50 and 5 µM methyl p-tolyl sulfoxide by MsrA and MsrB, respectively. B. The graph shows the amount of oxidized NADPH per minute for the reduction 500, 125 and 50 µM N-acetyl methionine sulfoxide by MsrA and MsrB, respectively. The decrease of absorbance at 340 nm due to NADPH oxidation was measured in the presence of the S- or R-isomer of methyl p-tolyl sulfoxide (A), and in the presence of the S- or R-isomere of N-acetyl methionine sulfoxide (B) using purified His-tagged MsrA or MsrB. Besides 10 µM purified MsrA or MsrB, the test system contained 10 µM TrxR, 10 µM TrxB, 450 µM NADPH, and various concentrations of S- or R-methyl p-tolyl sulfoxide (Sigma), and N-acetyl methionine sulfoxide (MOLISA), respectively. Reactions were filled up to a final volume of 500 µl with a buffer containing 50 mM HEPES and 1 mM EDTA (pH 7.4). Reactions were carried out at 25°C and started after 10 min pre-incubation by addition of the oxidized methionine derivatives.
Utilization of methionine sulfoxide by a ΔmsrB and a ΔmsrC mutant
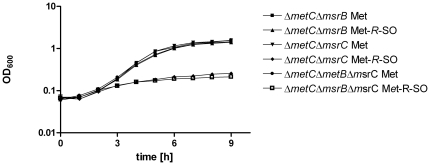
The wild type phenotype of the mutant with a deletion in STM1291 (msrB) prompted us to perform a more detailed analysis of methionine sulfoxide reductases that specifically act on the Met-R-SO epimer. We included MsrC (STM1847) in our analysis, which had been shown to encode a methionine sulfoxide reductase that reduces free but not peptidyl Met-R-SO in E. coli [10]. The open reading frame STM1847 was annotated on the chromosome of S. Typhimurium, with 85% identity (amino acids) to fRMsr (AAC77176) of E. coli (coliBASE) [23]. Eventually fRMsr was named MsrC (EcoGene database at http://ecogene.org/ ), and this latter designation was used throughout this study. An auxotrophic mutant that is unable to synthesize methionine and deficient for methionine sulfoxide reductases fails to grow on MetSO, because MetSO cannot be used by acylate tRNAmet as a substrate [27]. Thus methionine auxotrophic mutants can be used to test the function of methionine sulfoxide reductases. Using the ΔmetC background, a ΔmsrB mutant was generated. The ΔmetCΔmsrB double mutant could grow on Met-R-SO (Fig. 4), suggesting that lack of MsrB does not alter the ability of the mutant to reduce free Met-R-SO, which is in line with the enzyme activity data of MsrB. In contrast, the ΔmetCΔmsrC double mutant and the ΔmetCΔmsrBΔmsrC triple mutant were unable to grow on Met-R-SO. Taken together these results suggest that MsrC repairs free Met-R-SO in S. Typhimurium.
Figure 4. Growth of methionine auxotrophic mutants with deletions in msrB and msrC on methionine and free Met-R-SO.
Bacteria were grown overnight in LB, harvested, washed and cultivated in M9 minimal medium supplemented with 13 µg ml−1methionine (Met) or methionine-R-sulfoxide (Met-R-SO) (MOLISA), respectively, started at an optical density (OD600) of 0.05. Growth of ΔmetCΔmsrB, ΔmetCΔmsrC and ΔmetCΔmsrBΔmsrC was observed by measurement of OD600 every hour for nine hours of cultivation. Experiments were done in triplicates. Means are shown ± standard deviation.
In vitro growth and survival of a ΔmsrC mutant following exposure to exogenous H2O2
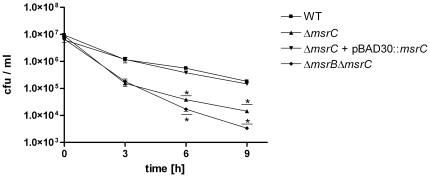
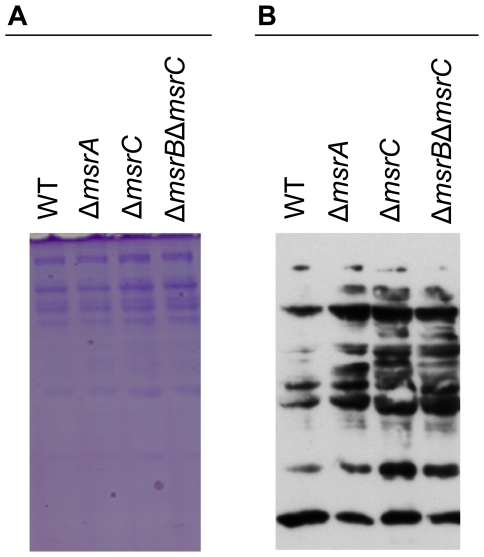
To clarify the role of MsrC in oxidative stress response we generated mutants with a single deletion in msrC and a double mutant with deletions in both genes msrC and msrB in the S. Typhimurium wild type background. The mutants did not show a general growth defect under normal aerobic culture conditions in LB broth (Fig. S2). In the presence of 2 mM exogenous H2O2 the ΔmsrC mutant was more susceptible than the wild type after incubation of 6 and 9 hours (Fig. 5). The wild type phenotype could be restored by expressing msrC on a plasmid under control of its own promoter. Interestingly, the survival of the ΔmsrBΔmsrC double mutant was more reduced than the ΔmsrC single mutant in the presence of 2 mM H2O2 in vitro after 6 and 9 hours (Fig. 5) of incubation. Since the ΔmsrA, ΔmsrC, and ΔmsrBΔmsrC mutants are more susceptible to H2O2 in vitro, we sought to characterize the oxidized protein profiles of these mutants in comparison to the wild type following exposure to H2O2. Accumulation of oxidized proteins was visualized using the oxyblot method. Signals of immunoblots of the mutant strains clearly differed from those of the wild type, indicating a higher level of oxidation in the absence of msrA, msrC, or msrB and msrC (Fig. 6B).
Figure 5. Susceptibility of S. Typhimurium ΔmsrC, ΔmsrC + pBAD30::msrC and ΔmsrB .
ΔmsrC towards exogenous H2O2. Wild type and mutant strains ΔmsrC, ΔmsrC + pBAD30::msrC and ΔmsrBΔmsrC were treated with 2 mM exogenous H2O2 in LB medium and plated for colony forming units at the indicated time points. The data shown are representatives and were performed at least three times with identical results. Experiments were done in duplicates and means are shown ± standard deviation. Asterisks indicate P-values <0.05.
Figure 6. Oxyblot of whole cell extracts of S. Typhimurium wild type, ΔmsrA, ΔmsrC, ΔmsrBΔmsrC.
Cells grown to OD600 1 were incubated in 0.75 mM H2O2 for 3 h. A. SDS-PAGE stained with Coomassie. B. Oxyblot film exposed for 2 min.
Growth of a ΔmsrC mutant inside activated macrophages
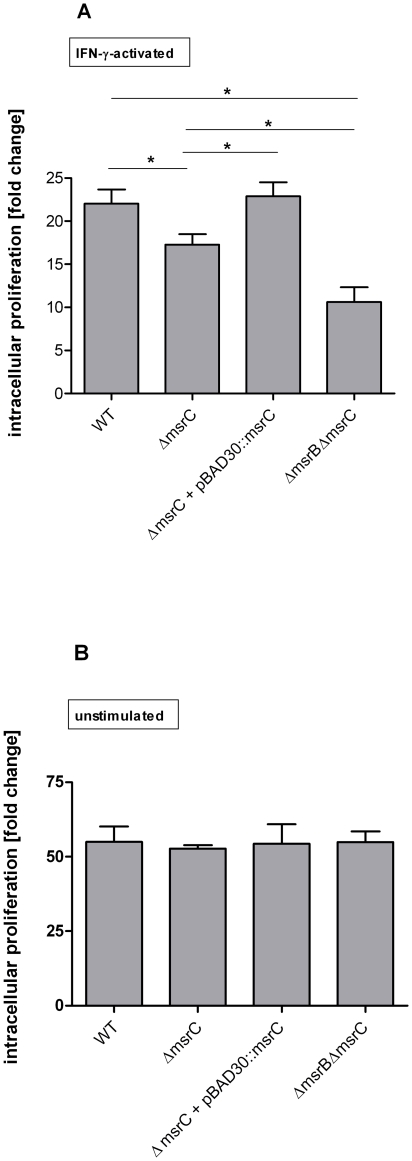
To elucidate the role of MsrC during intracellular growth of S. Typhimurium, RAW 264.7 macrophages were infected with wild type and mutant strains. The experiment was performed as described above. In IFN-γ-activated RAW 264.7 cells the single ΔmsrC mutant was attenuated compared to the wild type. The growth of the ΔmsrBΔmsrC mutant was significantly more affected than the ΔmsrC mutant (Fig. 7A), which is in line with the higher susceptibility of the ΔmsrBΔmsrC double mutant compared to the ΔmsrC single mutant towards exogenous H2O2. When we tested the strains in unstimulated RAW 264.7 cells, we found that the mutants were not affected in their intracellular growth in comparison to the wild type (Fig. 7,B).
Figure 7. Intracellular proliferation of S. Typhimurium wild type, ΔmsrC, ΔmsrB .
ΔmsrC in RAW 264.7 cells. S. Typhimurium wild type, ΔmsrC, ΔmsrC + pBAD30::msrC and ΔmsrBΔmsrC were tested A. in IFN-γ-activated, and B. in non-stimulated RAW 264.7 cells. Intracellular proliferation was determined in a gentamicin protection assay. Number of bacteria 16 h post infection was divided by the number of bacteria 2 h post infection. Results shown are the means ± standard error of the mean for three independent experiments, each in triplicate. P-values <0.05 were considered to be significant as indicated by asterisks.
In vivo susceptibility of a ΔmsrC mutant in Balb/cJ mice
To test whether MsrC or MsrB and MsrC together affect virulence of S. Typhimurium in mice, the fitness of the ΔmsrC single mutant and the ΔmsrBΔmsrC double mutant were directly compared to that of the wild type in a competition experiment [26]. The experiments were performed as described above. The ΔmsrC mutant was not attenuated in the liver and in the spleen 1 and 3 days after infection (Table 3 and 4). The ΔmsrBΔmsrC mutant was attenuated in the mouse model of infection in the liver and in the spleen 3 days after infection (Table 4). These data are consistent with the higher susceptibility of the ΔmsrBΔmsrC double mutants compared to the ΔmsrC mutant towards exogenous H2O2 and the more severe attenuation of the ΔmsrBΔmsrC double mutant compared to the ΔmsrC mutant in IFN-γ-activated RAW 264.7 cells. Taken together these results suggest that beside the repair of protein bound MetSO by MsrA and MsrB, the repair of free MetSO is also crucial for resistance of S. Typhimurium towards oxidative stress in vitro and in vivo.
Table 3. Competitive indices (CI) of ΔmsrC and ΔmsrBΔmsrC versus wild type on day 1 post infection.
| Strains | CI liver 1 day p.i. | P-value* | CI spleen 1 day p.i. | P-value* |
| ΔmsrC vs. WT | 0.91±0.21 | >0.05 | 1.07±0.25 | >0.05 |
| ΔmsrBΔmsrC vs. WT | 0.67±0.15 | <0.05 | 0.95±0.46 | >0.05 |
Table 4. Competitive indices (CI) of ΔmsrC and ΔmsrBΔmsrC versus wild type on day 3 post infection.
| Strains | CI liver 3 days p.i. | P-value* | CI spleen 3 days p.i. | P-value* |
| ΔmsrC vs. WT | 0.77±0.13 | >0.05 | 0.82±0.31 | >0.05 |
| ΔmsrBΔmsrC vs. WT | 0.57±0.15 | <0.05 | 0.46±0.06 | <0.05 |
*P-values were calculated using the Wilcoxon Signed Rank Test to test whether the actual median differs significantly from 1 (theoretical median). CIs are shown as medians ± standard deviation.
Discussion
Previously MsrA and MsrB were identified as the two principle methionine sulfoxide reductases that play a role in protecting bacteria against oxidative damage [28]. The genetic organization of msrA and msrB varies between organisms [29], [30]. In S. Typhimurium and other pathogens such as E. coli, C. jejuni, S. aureus, E. faecalis, M. smegmatis and M. tuberculosis msrA and msrB are transcribed independently as single genes [13]–[17], [20], [31]–[33]. Of these, only S. Typhimurium and M. tuberculosis are considered facultative intracellular pathogens, and they are able to replicate inside macrophages in the host. In M. tuberculosis deletion of msrA did not show a phenotype when challenged with exogenous H2O2, in activated macrophages, or in mice. Thus of the two intracellular pathogens in which the msr-system has been analyzed in detail, only in Salmonella MsrA is highly efficient in mediating resistance to oxidative stress.
In contrast to the ΔmsrA mutant, the ΔmsrB single mutant of S. Typhimurium showed the wild type phenotype at all conditions tested. Our results further showed that recombinant MsrA reduced both free and peptidyl Met-S-SO, whereas MsrB was specific only for peptidyl Met-R-SO. In this respect, our in vitro data concur with the substrate specificities of MsrA and MsrB in M. tuberculosis. In M. tuberculosis the purified MsrA is active against both free and peptidyl Met-S-SO, whereas MsrB showed weak activity for peptidyl Met-R-SO, and was inactive towards free Met-R-SO [20].
We speculated that an explanation for the phenotypic difference of the ΔmsrA and ΔmsrB mutant of S. Typhimurium during oxidative stress could be the limited substrate range of MsrB. In contrast to MsrA which is active against both peptidyl and free Met-S-SO; MsrB is only active against the peptidyl Met-R-SO. This idea implied that yet another methionine sulfoxide reductase for the R-form exists that could play a role in oxidative stress response of S. Typhimurium. A free methionine-R-sulfoxide reductase (MsrC) was first characterized by Lin and colleagues in E. coli [10], and was subsequently purified from N. meningitidis and from S. aureus. High substrate specificity for free Met-R-SO was reported in all three bacterial organisms, and detailed analysis with respect to biochemical properties and structural composition was carried out [10], [21], [22]. Lee and Gladyshev described MsrC as a highly specific Msr for free Met-R-SO that does not reduce peptidyl methionine sulfoxides, and compensates for the low or missing activity of MsrB with respect to free Met-R-SO [34].
However, none of the studies performed experiments to elucidate the role of MsrC in bacterial pathogenesis. In S. cerevisiae the role of MsrC in reducing free Met-R-SO was also clearly demonstrated [11]. Here the authors showed that MsrC mediates resistance to S. cerevisiae when challenged with H2O2 in vitro [11]. The influence of MsrC for growth or survival inside macrophages or in mice could not be studied in S. cerevisae, as the organism is non-pathogenic. The present work demonstrates that the methionine sulfoxide reductase MsrC is essential for growth and survival of an intracellular pathogen in macrophages, and, in combination with MsrB, in mice. We also showed by growth experiments with a msrC deficient mutant in a methionine auxotrophic background that in S. Typhimurium MsrC repairs free Met-R-SO. In all biochemical studies available at present, MsrC has been shown to be specific for free Met-R-SO, and there is no experimental evidence that MsrC can use peptidyl Met-R-SO as a substrate. In addition, we performed computational analysis and homology modelling using the available structural information of MsrC from E. coli (pdb: 1vhm) [10], and predicted the binding of free Met-R-SO but not protein based Met-R-SO as substrates for MsrC due to the conformation of its substrate binding pocket (data not shown).
It has been suggested that the cycle of oxidation of methionine and enzymatic reduction serves as a sink for oxidants [8]. Within this cycle MsrA, MsrB and MsrC could act together, to completely reduce free and protein based methionine sulfoxides [34]. Our data showed that MsrB has a function in reduction of protein bound Met-R-SO. The lack of activity towards free Met-R-SO can be compensated by MsrC [34]. Thus in contrast to Met-S-SO the repair of Met-R-SO requires two enzymes, one that is specific for the free amino acid and another for the protein-based form. In the ΔmsrC mutant, Met-R-SO within proteins can still be reduced by MsrB explaining its moderate phenotype. The synergistic effect of the msrB and msrC double mutation in S. Typhimurium following exposure to H2O2, in activated RAW 264.7 macrophages, and in mice, is due to the inability of the mutant to repair both free Met-R-SO and peptidyl Met-R-SO, causing a more dramatic decline of the overall scavenger capacity of methionine in the cell. It is noteworthy that a ΔmsrA mutant, which typically acts on both free and peptidyl-Met-S-SO shows a strong phenotype in Salmonella (this study) and in most other bacteria [13], [16], [19], [32], [35]. Thus free methionine plays a more important role during oxidative stress in bacteria than predicted so far.
Finally MsrC has been proposed to act as a signaling molecule in oxidative stress response as the protein contains a GAF domain [10], [22], and it is tempting to speculate that alteration of signaling contributes to the phenotype of the ΔmsrC mutant in S. Typhimurium. The GAF domain within the MsrC molecule was first described by Lin and coworkers, who also solved the structure of the protein. Based on the identification of the GAF domain it was speculated that the binding and reduction of free methionine sulfoxide could mediate cell signaling in response to oxidative stress [10]. An alternative hypothesis for the role of the GAF domain, however, challenged the idea of MsrC being a signaling molecule, and suggested that MsrC is an unusual example for a GAF fold that simply acts as an enzyme without a modular function [22].
Thus in summary, we showed that mutants of S. Typhimurium that lack components of the methionine sulfoxide reductase pathway are attenuated in vitro when exposed to H2O2, inside activated macrophages and in mice. Previously, MsrA and MsrB were considered to be the principle enzymes of the msr-system that play a role in bacterial resistance to oxidative stress. Here we showed that in addition MsrC contributes significantly to counteract the damage that is afflicted to bacterial pathogens by oxidative stress.
Materials and Methods
Ethics Statement
The mouse experiments were approved by Stockholm Norra Djurförsöksetiska Nämnd, Stockholms Tingsrätt, Box 8307, 104 20 Stockholm. The mouse experiments were performed in the animal facility at the Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet (Stockholm, Sweden) in accordance with both institutional and national guidelines (ethical permit N345/08).
Bacterial strains and plasmids
Bacterial strains used in this study are shown in Table 5. Salmonella enterica serovar Typhimurium ATCC14028 (S. Typhimurium) was used as wild type and all mutants were generated in this genetic background. Salmonella was grown overnight in LB broth in a “roller drum” under aerobic conditions at 37°C with antibiotics if necessary (ampicillin 100 µg ml−1, chloramphenicol 10 µg ml−1, kanamycin 25 µg ml−1). E. coli was grown under shaking conditions at 37°C in LB medium with appropriate antibiotics.
Table 5. Strains used in this study.
| Strain | characteristics | selective marker | reference |
| E. coli HB101 | F–, thi-1, hsdS20 (rB –, mB –), supE44, recA13, ara-14, leuB6, proA2, lacY1, galK2, rpsL20 (strr), xyl-5, mtl-1. | Promega | |
| T7 lysY Express | MiniF, lysY (CamR) / fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::miniTn10--TetS )2 [dcm] R(zgb-210::Tn10--TetS ) endA1 Δ(mcrC-mrr)114::IS10 | chloramphenicol | NEB |
| VK1 | E. coli T7 lysY Express transformed with pET28c::msrA | kanamycin | This study |
| VK2 | E. coli T7 lysY Express transformed with pET28c::msrB | kanamycin | This study |
| S. Typhimurium | wild type | - | ATCC 14028 |
| SHO11 | ΔmsrA | chloramphenicol | This study |
| SHO18 | ΔmsrAΔmsrB | chloramphenicol, kanamycin | This study |
| SHO19 | ΔmsrB | kanamycin | This study |
| SHO35 | ΔmsrA transformed with pBAD30::msrA | chloramphenicol, ampicillin | This study |
| SHO36 | ΔmsrAΔmsrB transformed with pBAD30::msrA | chloramphenicol, kanamycin, ampicillin | This study |
| LD24 | ΔmetCΔmsrB | kanamycin | This study |
| LD40 | ΔmetCΔmsrC | chloramphenicol | This study |
| LD41 | ΔmetCΔmsrBΔmsrC | chloramphenicol, kanamycin | This study |
| LD39 | ΔmsrC | chloramphenicol | This study |
| LD49 | ΔmsrBΔmsrC | chloramphenicol | This study |
| LD55 | ΔmsrC transformed with pBAD30::msrC | chloramphenicol, ampicillin | This study |
Construction of deletion mutants in S. Typhimurium 14028 and complementation
DNA sequences of S. Typhimurium were obtained from the database coliBASE (strain LT2). Generation of deletion mutants for the open reading frames msrA (STM4408), msrB (STM1291), metC (STM3161) and msrC (STM1847) was performed using the one-step inactivation via homologous recombination [36]. Primers and plasmids are listed in Table 6 and 7.
Table 6. Primers used in this study.
| Primer | Sequence (5′→3′) | Purpose |
| msrA1 | ATG CGT TAC CAG GAC GCA ACA CCC CGA TGC GAT CGC CAC TCT GCG TAG GCT GGA GCT GCT TC | Deletion of msrA |
| msrA2 | GCG ATG GTC TCC GGC GGC GGT CAT TGC CGA CTG AAA GCG CTC GCG TAT GAA TAT CCT CCT TAG | Deletion of msrA |
| msrA ST fwd | CTGGTTACTCAAGCAGATGCG | Confirmation of msrA deletion |
| msrA ST rvs | CAGGGCTAGTATAGCGTAAGC | Confirmation of msrA deletion |
| msrB1 | GTG TTA ATG TTT TGT TAG AAT CGG TCA GGC AAT GTG AGC ACG GTA GGC TGG AGC TGC TTC | Deletion of msrB |
| msrB2 | GTC CGC TCC TGT GGA ATA ATT TGC TGA ATC GTT TTT TCA GCC TAT GAA TAT CCT CCT TAG | Deletion of msrB |
| msrB test 1 | GATTCGTGAGCCGCCATTTC | Confirmation of msrB deletion |
| msrB test 2 | GATACACTTCCGGCGTCATG | Confirmation of msrB deletion |
| metC_For | CAT GCT AGT TTA GAC ATC CAG ACG GTTAAA ATC AGG AAA CGC AAC GTA GGC TGG AGC TGC TTC | Deletion of metC |
| metC_Rev | CAG ACT TTT CCA CGG AAA TTG TCT GCA TAT ATG TCC ATC CCC GCC TAT GAA TAT CCT CCT TAG | Deletion of metC |
| k metC_For | CGT CGC CAG GGT GCA GAT GG | Confirmation of metC deletion |
| k metC_Rev | GCC TTG ATC CCG GAC GCA AC | Confirmation of metC deletion |
| msrC_For | GAC CAC GAA GTG ATT ATA TAA TGA GCA AAA CAG AAC TAT ACG CGG GTA GGC TGG AGC TGC TTC | Deletion msrC |
| msrC_Rev | CAA ACG CCG TGC TAT CTA TAT CCA GCA CGC CGA TAA TCC GTT CGC TAT GAA TAT CCT CCT TAG | Deletion msrC |
| k msrC_For | GAG CAA GAA CGC ATT TAA TGC | Confirmation of msrC deletion |
| k msrC_Rev | GCG TAC GCA GGC CGT GT TC | Confirmation of msrC deletion |
| msrA1_HindIII | ATA TAT AAG CTT CTC AGG CAC AGT AAG CTA AC | Complementation with msrA |
| msrA2_HindIII | ATA TAT AAG CTT TTC AAC TCC GAC AAG TTC CC | Complementation with msrA |
| msrC_ HindIII | ATA TAT AAG CTT CTG CGT CAT TTG CCA GAT GC | Complementation with msrC |
| msrC_ HindIII | ATA TAT AAG CTT TCG GCC AGA AAC GCG ATA AC | Complementation with msrC |
| msrA_F_NheI | ATA TAT GCT AGC ATG AGT TTA TTT GAT AAA AAA CAT CTG | Overexpression MsrA |
| msrA_R_BamHI | ATA TAT GGA TCC TCA CGC GTC AGG CGG CAG G | Overexpression MsrA |
| msrB_F_NheI | ATA TAT GCT AGC GTG AGC ACG TTT AAA GTG AGA TG | Overexpression MsrB |
| msrB_R_BamHI | ATA TAT GGA TCC TCA GCC TTT CAG TTG ATC GCC G | Overexpression MsrB |
Table 7. Plasmids used in this study.
| plasmid | selection marker | source |
| pKD3 | chloramphenicol, ampicillin | [8] |
| pKD4 | kanamycin, ampicillin | [8] |
| pKD46 | ampicillin | [8] |
| pCP20 | chloramphenicol, ampicillin | [8] |
| pBAD30 | ampicillin | [16] |
| pBAD30::msrA | ampicillin | this study |
| pBAD30::msrC | ampicillin | this study |
| pET28c | kanamycin | Novagen |
| pET28c::msrA | kanamycin | this study |
| pET28c::msrB | kanamycin | this study |
Briefly, primers were designed that consist of a 42 – 45 bp region homologous to the flanking regions of the gene to be deleted, and an 18 bp region homologous to the resistance cassette of pKD3 or pKD4, respectively (Table 6). PCR products contain a FRT-flanked chloramphenicol (pKD3) or kanamycin (pKD4) resistance cassette flanked by 42 – 45 bp regions homologous to the adjacent regions of the gene to be deleted.
The helper plasmid pKD46 (Table 7) carrying the Red Recombinase under the control of an arabinose inducible ParaB promotor was transformed into an electrocompetent S. Typhimurium receiver strain. After electroporation transformants were selected at 30°C on LB agar plates with ampicillin. Next step the pKD46 carrying clone was cultivated in 50 ml LB broth with ampicillin and 20 mM L-arabinose for induction of the Red Recombinase to OD600 0.3 - 0.4. Cells were prepared for electroporation and purified PCR products specific for deletion of msrA, msrB, metC or msrC, respectively, were transformed into the receiver strain. Site-specific recombination mediated by the Red recombinase was carried out during 3 h of incubation after electroporation without shaking. 100 µl of the bacteria suspension were plated on LB agar with the appropriate antibiotic and incubated over night at 37°C. Chloramphenicol or kanamycin resistant clones were selected and subcultured twice in 5 ml LB at 42°C to get rid off the temperature sensitive helper plasmid pKD46. Obtained mutants were confirmed by PCR using primers that bind in the flanking regions of the disrupted gene (Table 6).
When necessary, antibiotic resistance genes were eliminated by the temperature-sensitive plasmid pCP20 (Table 7) carrying a FLP recombinase. Mutants containing FRT-flanked antibiotic resistance cassettes were transformed with pCP20 and ampicillin-resistant transformants were selected at 30°C. To get rid off the temperature sensitive helper plasmid pCP20 the mutants were cultivated at 42°C.
The construction of the double ΔmsrAΔmsrB deletion mutant was performed by using the single msrA mutant as background strain. Mutations were transferred between different strains by transduction with the bacteriophage P22int [37] to construct the multiple deletion mutants ΔmetCΔmsrB and ΔmetCΔmsrC, ΔmetCΔmsrBΔmsrC and ΔmsrBΔmsrC .
For complementation of the deletion mutants with wild type genes from S. Typhimurium 14028 the open reading frames msrA and msrC including upstream regions between 250bp – 400 bp were amplified by PCR using Phusion™ High Fidelity DNA Polymerase (NEB). Primers (Table 6) included restriction sites for subsequent cloning into the vector pBAD30 [38]. All genes were under control of their own promoter.
Growth of S. Typhimurium in LB medium under aerobic conditions
Over night cultures of S. Typhimurium were diluted to an OD600 of 0.02 and were then incubated aerobically under shaking conditions in LB medium (37°C, 130 rpm). The OD600 was measured every 45 min. Experiments were done in duplicates.
Susceptibility of S. Typhimurium to oxidative stress in vitro
Hydrogen peroxide was purchased from Sigma. Over night cultures of S. Typhimurium were diluted to an OD600 of 0.02 and 100 µl diluted culture were mixed with 100 µl of hydrogen peroxide (4 mM) at a final concentration of 2 mM. The assay was carried out in LB broth and the 96-well-plates were incubated at 37°C without shaking. Killing of bacteria was determined by plating for colony forming units on LB agar after 3, 6 and 9 h of exposure. Experiments were done in duplicates.
For experiments using the OxyBlot™ protein oxidation detection kit (Millipore), S. Typhimurium strains were grown in 25 ml LB to OD600 1. Cultures of S. Typhimurium were exposed to H2O2 at a final concentration of 0.75 mM and incubated for 3 hours at 37°C without shaking. Cells were centrifuged at 9273×g, dissolved in 500 µl 10 mM Tris, 25 mM NaCl and 50 mM DTT pH 7.5 and transferred to Lysing Matrix B tubes (MP Biomedicals). Cells were disrupted mechanically using the Hybaid Ribolyser for 40 seconds at speed 6.0. After cooling down the samples on ice cell debris were removed 3 min at 16000×g, the supernatant was transferred into a new reaction tube.
Detection of whole cell protein profiles using the OxyBlot™ protein oxidation detection kit (Millipore) was performed according to the manual provided by the manufacturer. Briefly, 10 µg protein (5 µl) from each culture supernatant was mixed with 5 µl of 12% SDS, subsequently mixed with 10 µl of the derivatizing agent 1×DNPH (dinitrophenyl hydrazine) and incubated at room temperature for 15 min. Reaction was stopped by adding 7.5 µl neutralization solution to all tubes, and samples were loaded on a 12.5% SDS-PAGE.
The proteins on the SDS-PAGE were electrotransferred onto a nitrocellulose membrane (Whatman Protran nitrocellulose), and subsequently blocked for 1 h in blocking buffer containing PBS, 1% BSA and 0.05% Tween 20. The membrane was incubated in rabbit anti-DNP-antibody (1∶150) diluted in TBS with 1% BSA over night at 4°C, and subsequently in goat anti-rabbit IgG horseradish peroxidase conjugate (1∶300) diluted in TBS with 1% BSA for 1 h. Oxidized proteins were visualized using the ECL substrate (SuperSignal West Pico Chemiluminescent Substrate, Thermo Scientific) according to manufacturer's directions.
Infection of the macrophage cell line RAW 264.7
RAW 264.7 macrophages (ECACC) were grown in RPMI (Gibco) supplemented with 10% heat inactivated FCS (Sigma), 4 mM Glutamine (Gibco) and 20 mM Hepes (Gibco). Infection experiments were performed as described earlier [39]. Briefly, cells were seeded in 24-well plates at a density of 5×104 cells per well. Over night cultures of S. Typhimurium were diluted to an OD600 of 0.2 and 1.5 ml of bacteria were harvested, washed twice with PBS, and resuspended in 1 ml RPMI medium without any supplements. The bacterial suspension was further diluted 1∶100 in RPMI and 100 µl were used for infection of RAW 264.7 cells in 900 µl (MOI of 10∶1) RPMI medium without any supplements. Plates were centrifuged at 220×g for 5 min and incubated 30 min at 37°C and 5% CO2. Extracellular bacteria were removed by washing three times with PBS and incubated 60 min with RPMI containing 100 µg ml−1 gentamicin. Medium was replaced by RPMI with 10 µg ml−1 gentamicin for the rest of the experiment. To assess the bacterial load in the macrophages 2 h and 16 h post infection, cells were washed three times with PBS and lysed with 1 ml sterile bidistilled water. Bacteria were plated on LB agar for colony forming units (cfu). To calculate bacterial growth, cfu count 16 h post infection was divided by the cfu count 2 h post infection. For activation, macrophages were stimulated with murine IFN-γ (Sigma) 18–24 h prior infection. All experiments were performed in triplicates.
Competitive infection of Balb/cJ mice with S. Typhimurium
Fitness of mutant strains was compared to wild type by a competition experiment in mice. Strains were taken from an over night LB agar plate, resuspended in PBS and the number of cells was adjusted to 5×103 bacteria for each strain. For infection a 1∶1 mixture of two strains with a total of 1×104 bacteria in 100 µl sterile PBS was used for infection. For each time point and mutant groups of 5 female BALB/cJ mice at the age of 6 to 8 weeks were infected intraperitoneally. Mice were sacrificed on day 1 and day 3 post infection and the livers and spleens were removed and homogenized in PBS. Bacteria were plated for cfu on LB agar and on LB agar containing the appropriate antibiotic (chloramphenicol for ΔmsrA, ΔmsrAΔmsrB, ΔmsrC, ΔmsrBΔmsrC, kanamycin for ΔmsrB) to distinguish between wild type and mutant. Competitive indices (CI) were calculated as described previously [26]. Briefly, the CI is the ratio between the mutant and the wild type at day 1 or 3 divided by the ratio of mutant and wild type at day 0. The competitive index measures the attenuation of the tested mutant strain.
Culture of Salmonella on methionine and methionine sulfoxide
Overnight cultures of S. Typhimurium in LB were harvested by centrifugation, washed three times in M9 minimal medium, and inoculated in M9 minimal medium at an OD600 0.05. 13 µg ml−1 of methionine or Met-R-SO, respectively, were added as methionine source. The cells were incubated aerobically under shaking conditions (37°C, 130 rpm). Growth was measured by OD600 every hour for 9 h of cultivation. Experiments were done in triplicates.
Overexpression and purification of S. Typhimurium His-tagged MsrA and MsrB
The open reading frames of msrA (STM4408) and msrB (STM1291) from S. Typhimurium were amplified from genomic DNA by PCR. Primers (Table 6) included restriction sites for subsequent cloning into the T7 overexpression vector pET28c (Novagen, Table 7) according to manufacturer's instructions. Plasmids, designated pET28c::msrA and pET28c::msrB (Table 7), respectively, were transformed into E. coli T7 lysY Express (NEB) for expression of the recombinant His-tagged proteins. Expression of MsrA and MsrB was induced by addition of 0.5 mM IPTG at an OD600 ∼0,8, and cells were grown for another 5 h. Bacteria were harvested by centrifugation (3000×g, 25 min, 4°C) and resuspended in binding buffer containing 50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole (pH 8.0 for MsrA, pH 8.5 for MsrB). Cells were lysed by gently shaking with lysozyme (0.3 mg ml−1, Roth) for 1 h at 4°C, and by sonication in a SONOPLUS sonicator (Bandelin). The supernatant was purified using a Ni-NTA column (QIAGEN) and the Bio Logic Duo Flow System (Biorad). Buffer used for pre-equilibration of the Ni-NTA column and washing steps (binding buffer, described above) contained 20 mM imidazole, buffer used for elution contained 500 mM imidazol. The His-tagged proteins were eluted by applying a linear gradient from 0 to 100% elution buffer to the column. Fractions with MsrA or MsrB were pooled and desalted using size-exclusion chromatography into 10 mM Tris and 1 mM EDTA (pH 8.0 for MsrA, pH 8.5 for MsrB). Proteins were concentrated by centrifugation (2500×g, 4°C) using VIVASPIN 20 concentrators (Sartorius Stedim Biotech). Protein concentrations of MsrA and MsrB were determined by the Quick Start Bradford Protein Assay (Bio-Rad).
NADPH linked reductase activity assay
MsrA and MsrB activities were measured by monitoring NADPH oxidation at 340 nm by means of a coupled test system. Thioredoxin B (TrxB) and thioredoxin reductase (TrxR) from M. tuberculosis were used to regenerate MsrA and MsrB to its reduced form. All enzyme assays were performed at 25°C in a final volume of 500 µl, using an UVmc2 spectrophotometer (Safas S.A.). The reaction mixture contained 10 µM purified MsrA or MsrB, 10 µM TrxR, 10 µM TrxB, 450 µM NADPH, and 5 µM to 500 µM of (R) or (S) methyl p-tolyl sulfoxide (Sigma) or (R) or (S) N-acetyl methionine sulfoxide (MOLISA), respectively. After 10 min pre-incubation reactions were started by the addition of the substrate.
Acetylation of Met-S-SO or Met-R-SO
Met-S-SO or Met-R-SO (102 mg, 0.62 mmol) and NaHCO3 (130 mg, 1.54 mmol; 2.5 eq) were dissolved in ice-cold water (1.5 ml). The solution was treated with acetic anhydride (88 µl, 0.93 mmol; 1.5 eq) and stirred vigorously for 3 hours at room temperature. It was then adjusted to pH ∼3 with 1 M KHSO4-solution, and diluted with MeOH, transferred to a 100 ml-flask, and evaporated to dryness in vacuo. Silica gel and CH2Cl2/MeOH (1∶1) were added, and the mixture was again evaporated in vacuo to yield a dry powder which was poured onto a pre-filled chromatography column. Regular flash chromatography (CH2Cl2/MeOH = 3∶1+1% HOAc) and co-evaporation with toluene in vacuo yielded the product as colorless glass (58–75 mg; 45–58%).
We used 1H-NMR spectrum to confirm the structure of N-acetyl Met-S-SO and of N-acetyl Met-R-SO, respectively:
N-acetyl Met-S-SO: 1H-NMR (600 MHz, MeOH-d4), δ = 4.52 (dd, 8.4 Hz, 5.3 Hz, 1 H; C2-H), 2.94-2.83 (m, 2 H; C4-H2), 2.65 (s, 3 H; C6-H3), 2.36–2.28 (m, 1 H, C3-H'), 2.14–2.06 (m, 1 H, C3-H″), 2.00 (s, 3 H; N-CO-CH3) ppm
N-acetyl Met-R-SO: 1H-NMR (600 MHz, MeOH-d4), δ = 4.50 Δ(dd, 8.7 Hz, 4.8 Hz, 1 H; C2-H), 2.94 (ddd, 13.2 Hz, 10.0 Hz, 6.4 Hz, 1 H; C4-H'), 2.80 (ddd, 13.2 Hz, 10.0 Hz, 4.9 Hz, 1 H; C4-H″), 2.65 (s, 3 H; C6-H3), 2.35–2.28 (m, 1 H, C3-H'), 2.14–2.07 (m, 1 H, C3-H″), 2.00 (s, 3 H; N-CO-CH3) ppm.
Statistical analysis
The t-test was applied for statistical analyses and P-values <0.05 were considered to be significant. Significance of the competitive indices was calculated using the Wilcoxon Signed Rank Test.
Supporting Information
Growth curve of S . Typhimurium wild type and mutants in LB medium. Over night cultures of S. Typhimurium were diluted to an OD600 of 0.02 and growth of all strains was measured every 45 min.
(TIF)
Growth curve of S . Typhimurium wild type and mutants in LB medium. Over night cultures of S. Typhimurium were diluted to an OD600 of 0.02 and growth of all strains was measured every 45 min.
(TIF)
Acknowledgments
We thank S. Suerbaum for his support. We thank Kristin Heller for the help with the protein expression, purification and the enzymatic assay.
Footnotes
Competing Interests: Timo Jäger was employed by MOLISA GmbH at the time when the experiments for this manuscript were being performed. Timo Jäger has neither financial nor non-financial, nor professional, nor personal conflicts of interest with respect to his position in the company and his author on the present manuscript. The authors' adherence to all the PLoS ONE policies on sharing data and materials is not affected. LAD, SAH, SFR, VK, MR, FCB have declared that no competing interests exist.
Funding: This work was supported by the International Research Training Group 1273 funded by the German Research Foundation to SAH, LAD, and SFR. MR was supported by the Swedish Medical Research Council. Publication fee for this study was funded by the German Research Foundation (Deutsche Forschungsgemeinschaft). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nat Rev Microbiol. 2008;6:53–66. doi: 10.1038/nrmicro1788. [DOI] [PubMed] [Google Scholar]
- 2.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med. 2000;192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hebrard M, Viala JP, Meresse S, Barras F, Aussel L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009;191:4605–4614. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horst SA, Jaeger T, Denkel LA, Rouf SF, Rhen M, et al. Thiol peroxidase protects Salmonella enterica from hydrogen peroxide stress in vitro and facilitates intracellular growth. J Bacteriol. 2010;192:2929–2932. doi: 10.1128/JB.01652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aussel L, Zhao W, Hebrard M, Guilhon AA, Viala JP, et al. Salmonella detoxifying enzymes are sufficient to cope with the host oxidative burst. Mol Microbiol. 2011;80:628–640. doi: 10.1111/j.1365-2958.2011.07611.x. [DOI] [PubMed] [Google Scholar]
- 7.Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, et al. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch Biochem Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [DOI] [PubMed] [Google Scholar]
- 8.Levine RL, Mosoni L, Berlett BS, Stadtman ER. Methionine residues as endogenous antioxidants in proteins. Proc Natl Acad Sci U S A. 1996;93:15036–15040. doi: 10.1073/pnas.93.26.15036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boschi-Muller S, Olry A, Antoine M, Branlant G. The enzymology and biochemistry of methionine sulfoxide reductases. Biochim Biophys Acta. 2005;1703:231–238. doi: 10.1016/j.bbapap.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Lin Z, Johnson LC, Weissbach H, Brot N, Lively MO, et al. Free methionine-(R)-sulfoxide reductase from Escherichia coli reveals a new GAF domain function. Proc Natl Acad Sci U S A. 2007;104:9597–9602. doi: 10.1073/pnas.0703774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le DT, Lee BC, Marino SM, Zhang Y, Fomenko DE, et al. Functional analysis of free methionine-R-sulfoxide reductase from Saccharomyces cerevisiae. J Biol Chem. 2009;284:4354–4364. doi: 10.1074/jbc.M805891200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee BC, Le DT, Gladyshev VN. Mammals reduce methionine-S-sulfoxide with MsrA and are unable to reduce methionine-R-sulfoxide, and this function can be restored with a yeast reductase. J Biol Chem. 2008;283:28361–28369. doi: 10.1074/jbc.M805059200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas T, Daniel DS, Parida BK, Jagannath C, Dhandayuthapani S. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J Bacteriol. 2004;186:3590–3598. doi: 10.1128/JB.186.11.3590-3598.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh VK, Moskovitz J. Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology. 2003;149:2739–2747. doi: 10.1099/mic.0.26442-0. [DOI] [PubMed] [Google Scholar]
- 15.St John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc Natl Acad Sci U S A. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Atack JM, Kelly DJ. Oxidative stress in Campylobacter jejuni: responses, resistance and regulation. Future Microbiol. 2009;4:677–690. doi: 10.2217/fmb.09.44. [DOI] [PubMed] [Google Scholar]
- 17.Zhao C, Hartke A, La Sorda M, Posteraro B, Laplace JM, et al. Role of methionine sulfoxide reductases A and B of Enterococcus faecalis in oxidative stress and virulence. Infect Immun. 2010;78:3889–3897. doi: 10.1128/IAI.00165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alamuri P, Maier RJ. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol Microbiol. 2004;53:1397–1406. doi: 10.1111/j.1365-2958.2004.04190.x. [DOI] [PubMed] [Google Scholar]
- 19.Skaar EP, Tobiason DM, Quick J, Judd RC, Weissbach H, et al. The outer membrane localization of the Neisseria gonorrhoeae MsrA/B is involved in survival against reactive oxygen species. Proc Natl Acad Sci U S A. 2002;99:10108–10113. doi: 10.1073/pnas.152334799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee WL, Gold B, Darby C, Brot N, Jiang X, et al. Mycobacterium tuberculosis expresses methionine sulphoxide reductases A and B that protect from killing by nitrite and hypochlorite. Mol Microbiol. 2009;71:583–593. doi: 10.1111/j.1365-2958.2008.06548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bong SM, Kwak GH, Moon JH, Lee KS, Kim HS, et al. Structural and kinetic analysis of free methionine-R-sulfoxide reductase from Staphylococcus aureus: conformational changes during catalysis and implications for the catalytic and inhibitory mechanisms. J Biol Chem. 2010;285:25044–25052. doi: 10.1074/jbc.M110.103119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruez A, Libiad M, Boschi-Muller S, Branlant G. Structural and biochemical characterization of free methionine-R-sulfoxide reductase from Neisseria meningitidis. J Biol Chem. 2010;285:25033–25043. doi: 10.1074/jbc.M110.134528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 24.Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, et al. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 25.Mintz KP, Moskovitz J, Wu H, Fives-Taylor PM. Peptide methionine sulfoxide reductase (MsrA) is not a major virulence determinant for the oral pathogen Actinobacillus actinomycetemcomitans. Microbiology. 2002;148:3695–3703. doi: 10.1099/00221287-148-11-3695. [DOI] [PubMed] [Google Scholar]
- 26.Beuzon CR, Holden DW. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 2001;3:1345–1352. doi: 10.1016/s1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
- 27.Ejiri SI, Weissbach H, Brot N. Reduction of methionine sulfoxide to methionine by Escherichia coli. J Bacteriol. 1979;139:161–164. doi: 10.1128/jb.139.1.161-164.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 29.Sasindran SJ, Saikolappan S, Dhandayuthapani S. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2007;2:619–630. doi: 10.2217/17460913.2.6.619. [DOI] [PubMed] [Google Scholar]
- 30.Ezraty B, Bos J, Barras F, Aussel L. Methionine sulfoxide reduction and assimilation in Escherichia coli: new role for the biotin sulfoxide reductase BisC. J Bacteriol. 2005;187:231–237. doi: 10.1128/JB.187.1.231-237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhandayuthapani S, Jagannath C, Nino C, Saikolappan S, Sasindran SJ. Methionine sulfoxide reductase B (MsrB) of Mycobacterium smegmatis plays a limited role in resisting oxidative stress. Tuberculosis (Edinb) 2009;89(Suppl 1):S26–S32. doi: 10.1016/S1472-9792(09)70008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, et al. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh VK, Moskovitz J, Wilkinson BJ, Jayaswal RK. Molecular characterization of a chromosomal locus in Staphylococcus aureus that contributes to oxidative defence and is highly induced by the cell-wall-active antibiotic oxacillin. Microbiology. 2001;147:3037–3045. doi: 10.1099/00221287-147-11-3037. [DOI] [PubMed] [Google Scholar]
- 34.Lee BC, Gladyshev VN. The biological significance of methionine sulfoxide stereochemistry. Free Radic Biol Med. 2011;50:221–227. doi: 10.1016/j.freeradbiomed.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moskovitz J, Singh VK, Requena J, Wilkinson BJ, Jayaswal RK, et al. Purification and characterization of methionine sulfoxide reductases from mouse and Staphylococcus aureus and their substrate stereospecificity. Biochem Biophys Res Commun. 2002;290:62–65. doi: 10.1006/bbrc.2001.6171. [DOI] [PubMed] [Google Scholar]
- 36.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 38.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravortty D, Hansen-Wester I, Hensel M. Salmonella pathogenicity island 2 mediates protection of intracellular Salmonella from reactive nitrogen intermediates. J Exp Med. 2002;195:1155–1166. doi: 10.1084/jem.20011547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curve of S . Typhimurium wild type and mutants in LB medium. Over night cultures of S. Typhimurium were diluted to an OD600 of 0.02 and growth of all strains was measured every 45 min.
(TIF)
Growth curve of S . Typhimurium wild type and mutants in LB medium. Over night cultures of S. Typhimurium were diluted to an OD600 of 0.02 and growth of all strains was measured every 45 min.
(TIF)