Abstract
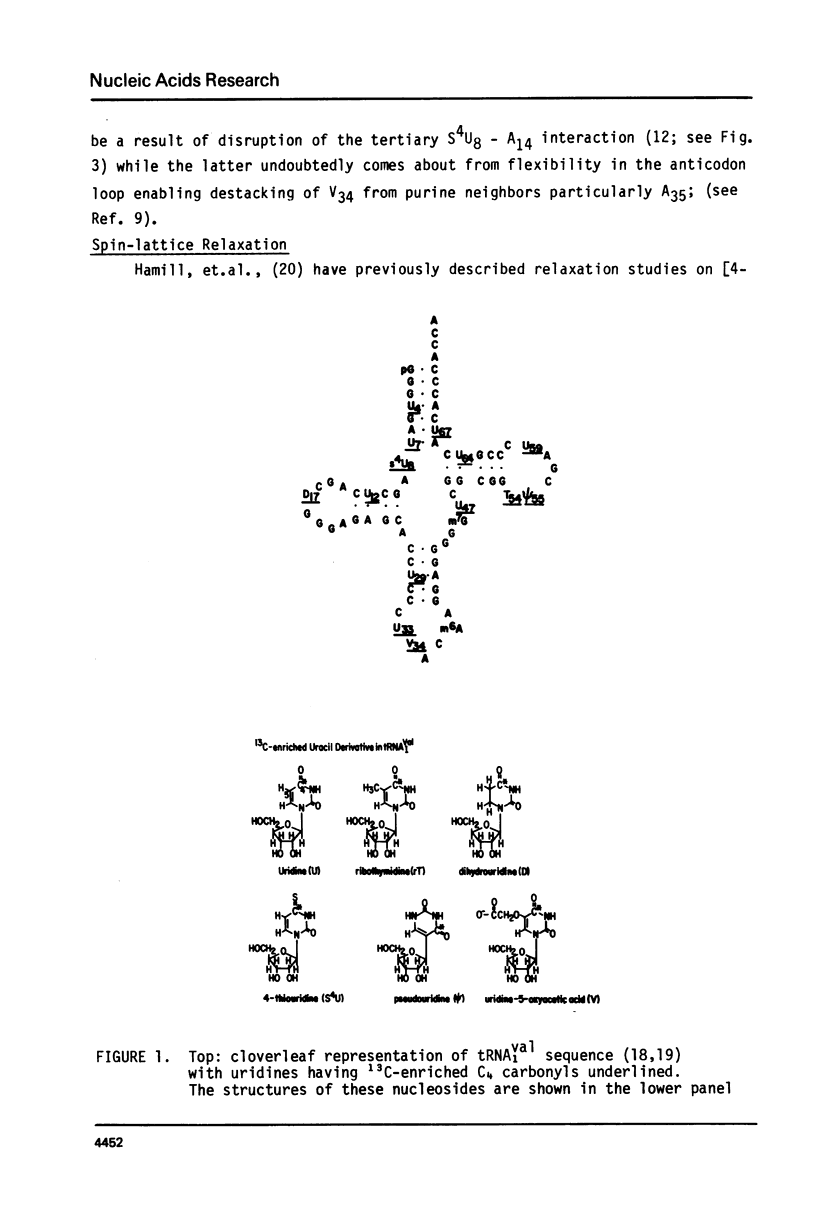
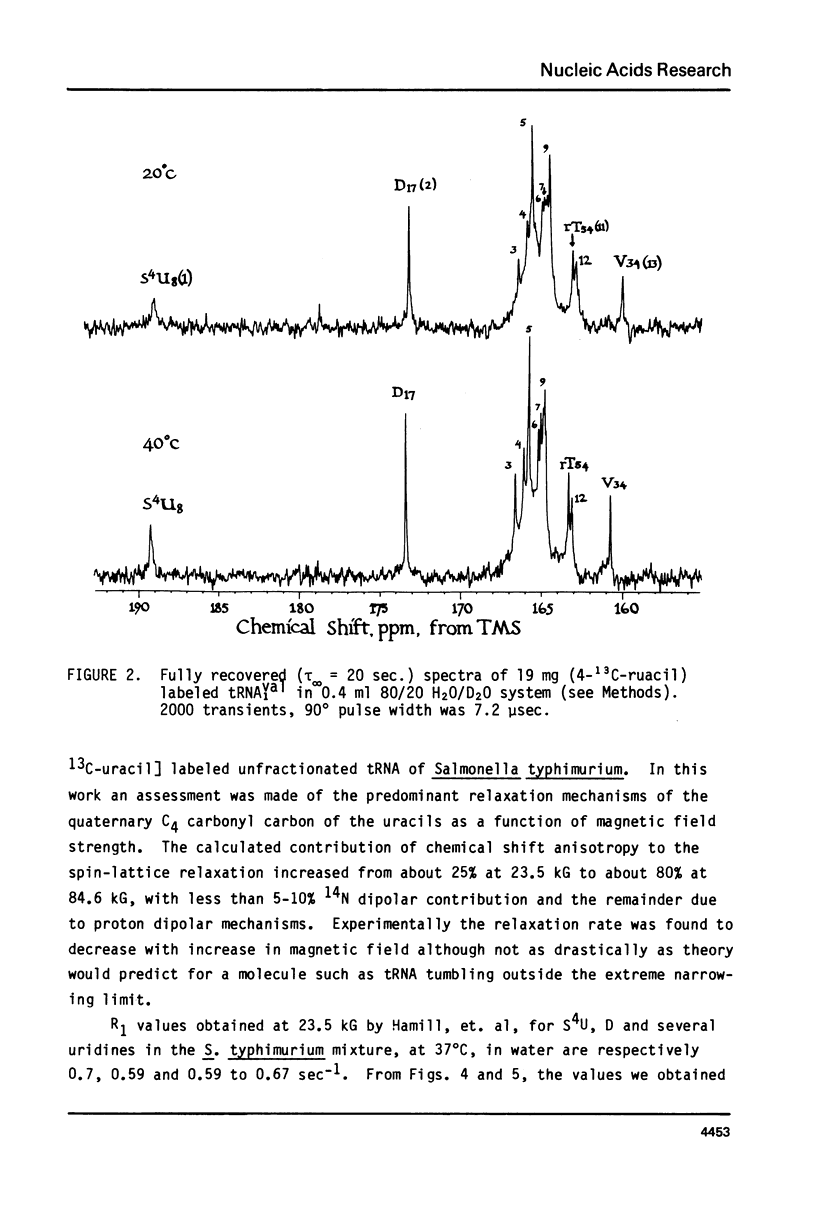
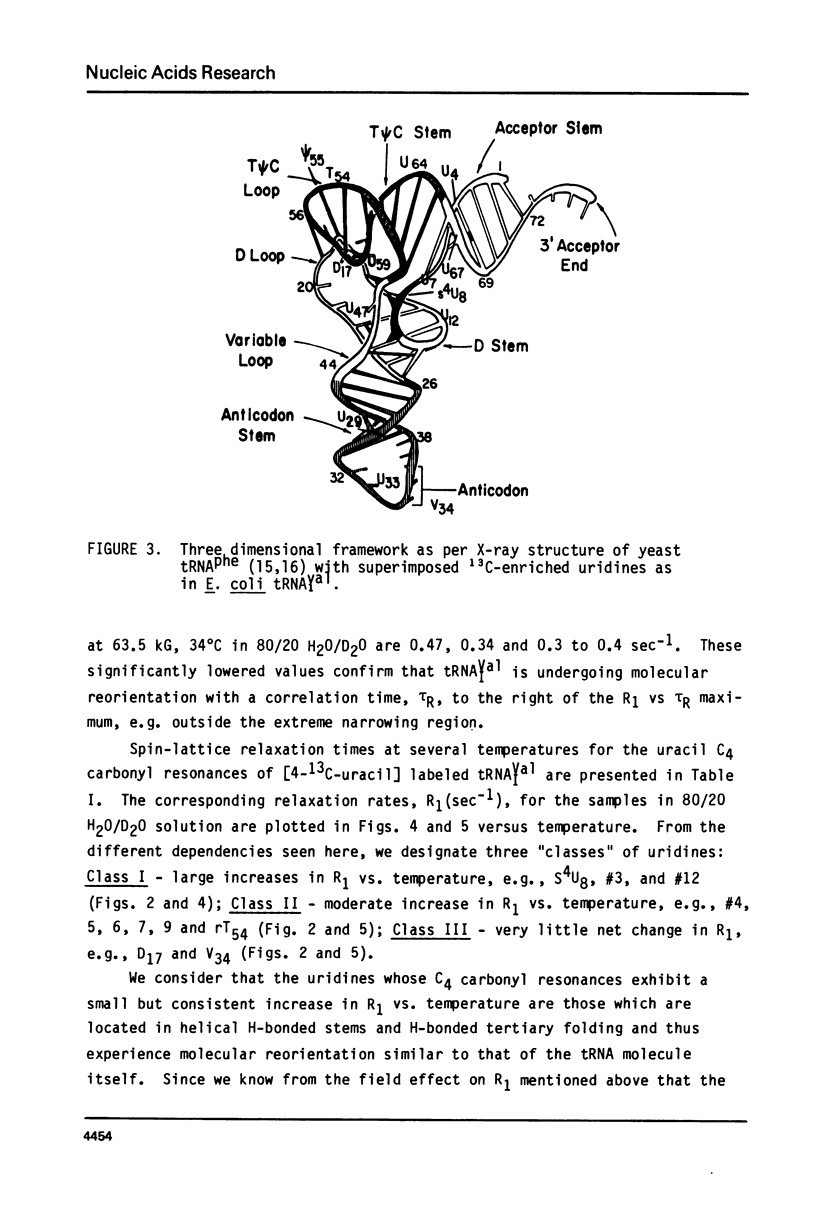
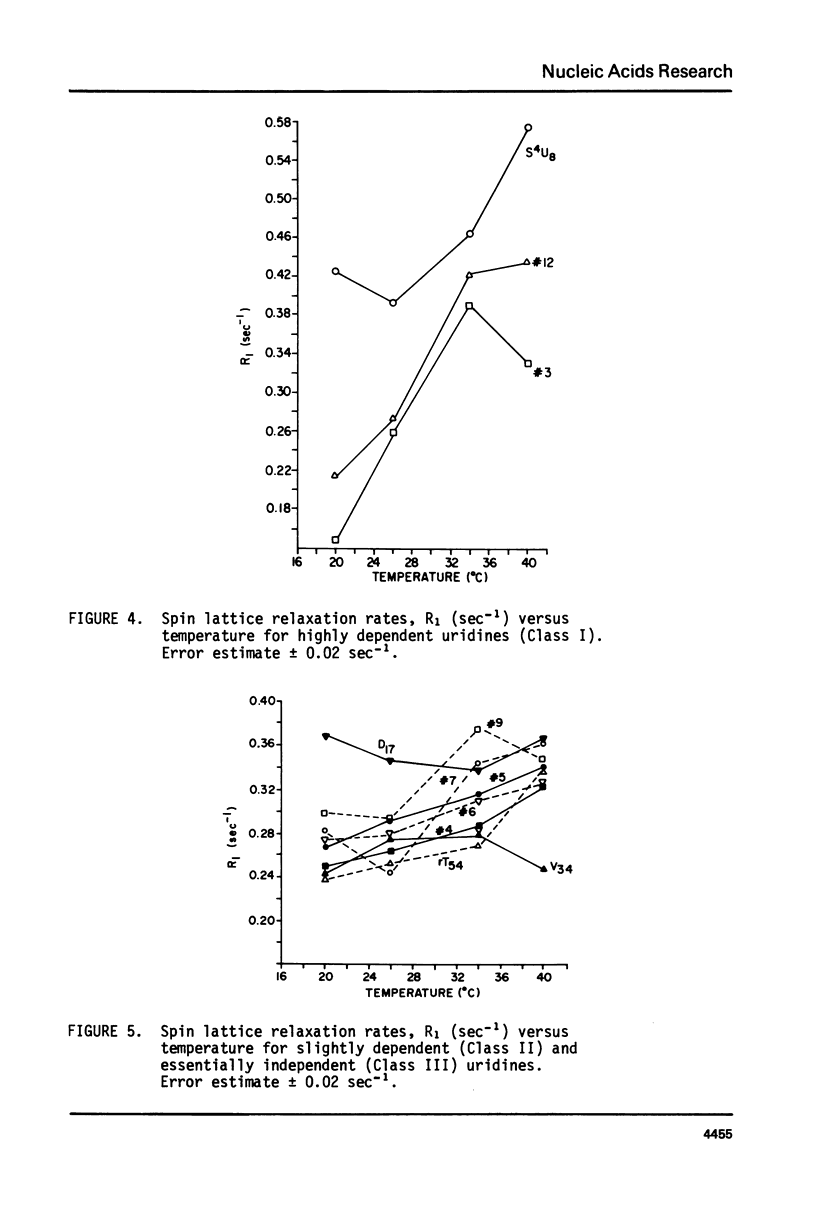
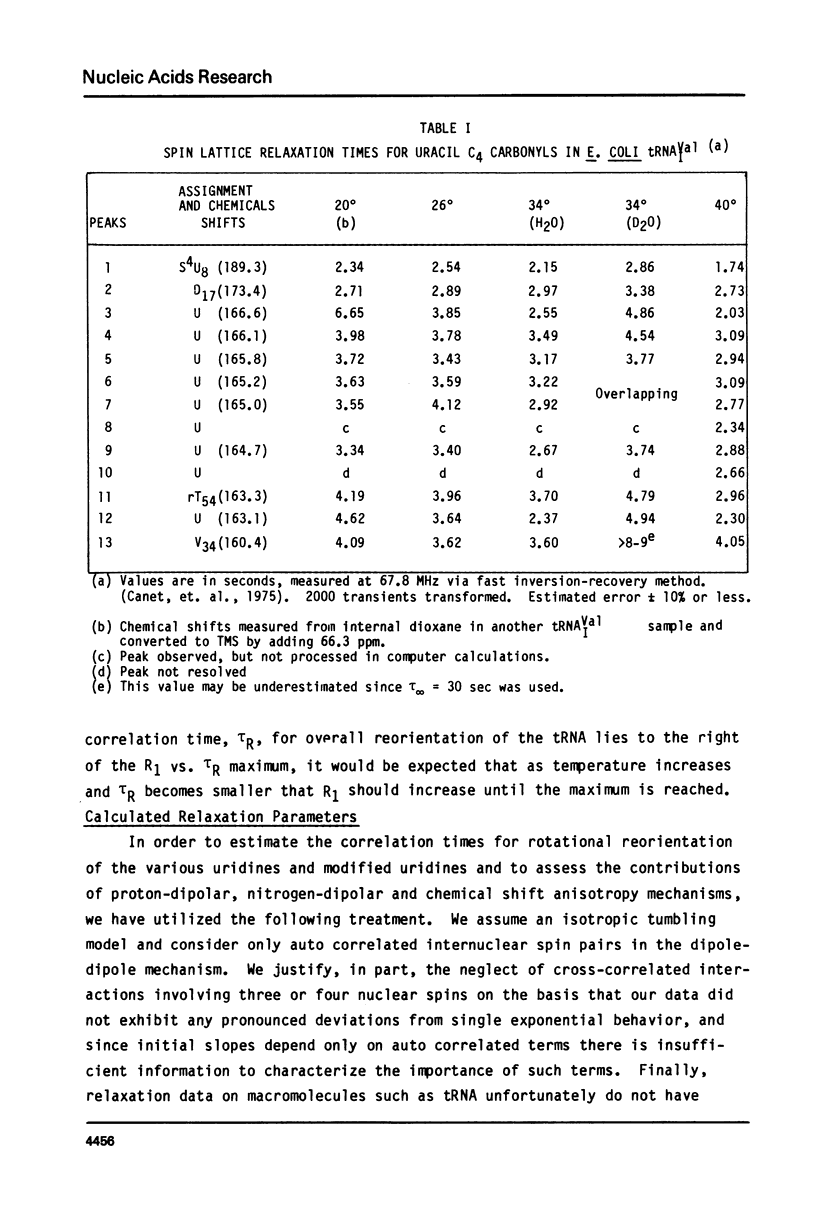
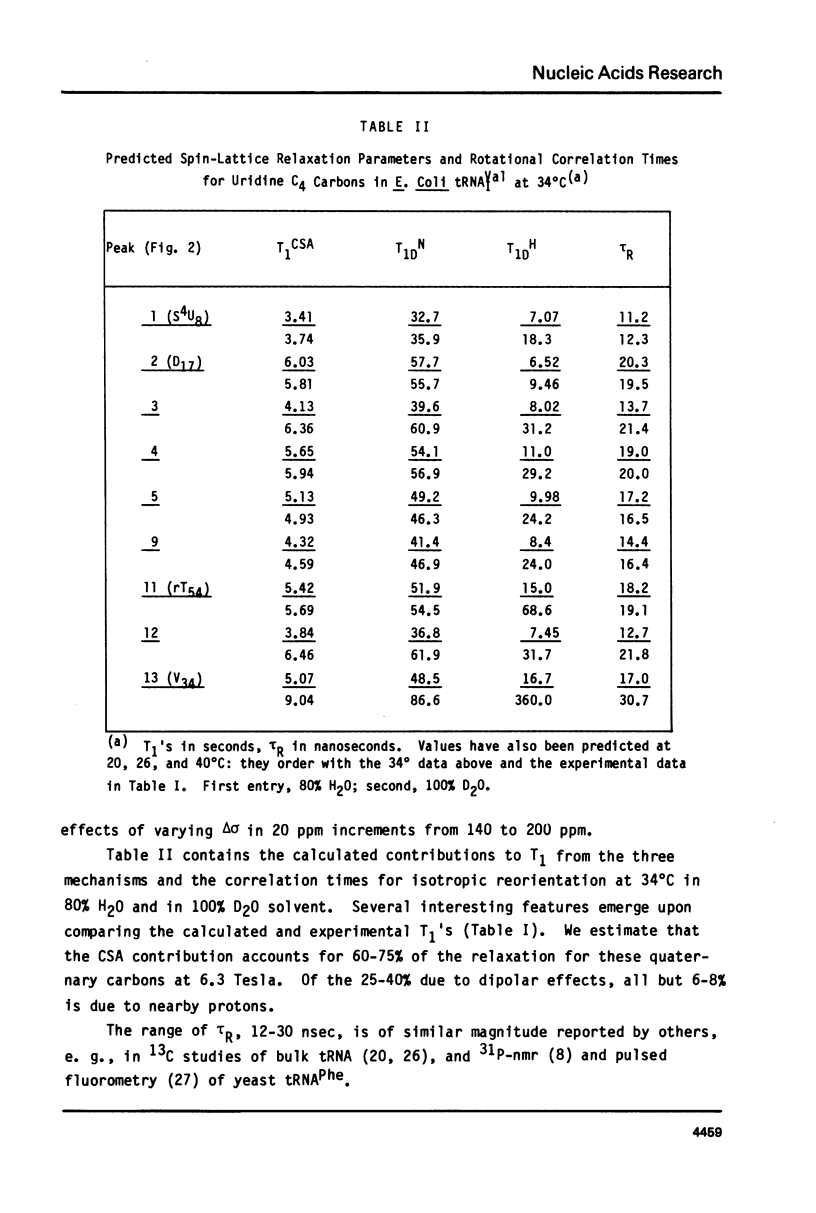
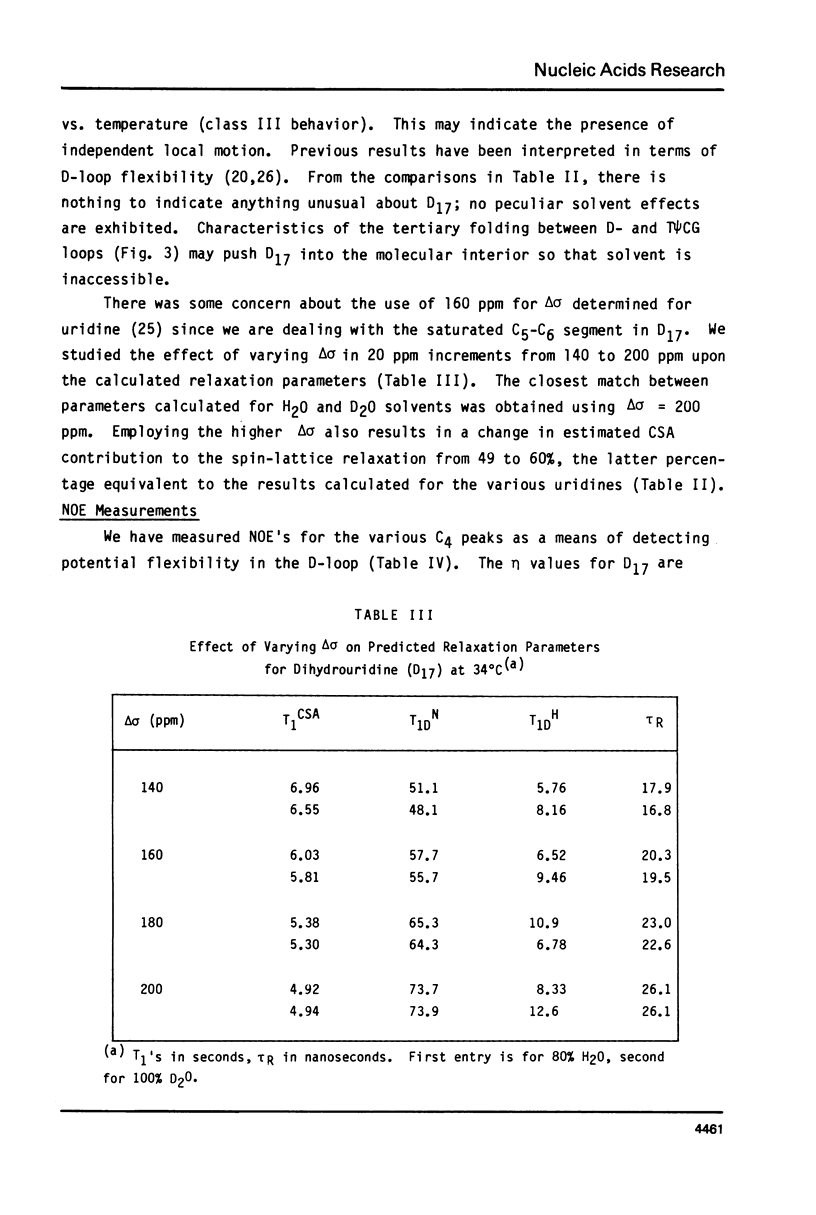
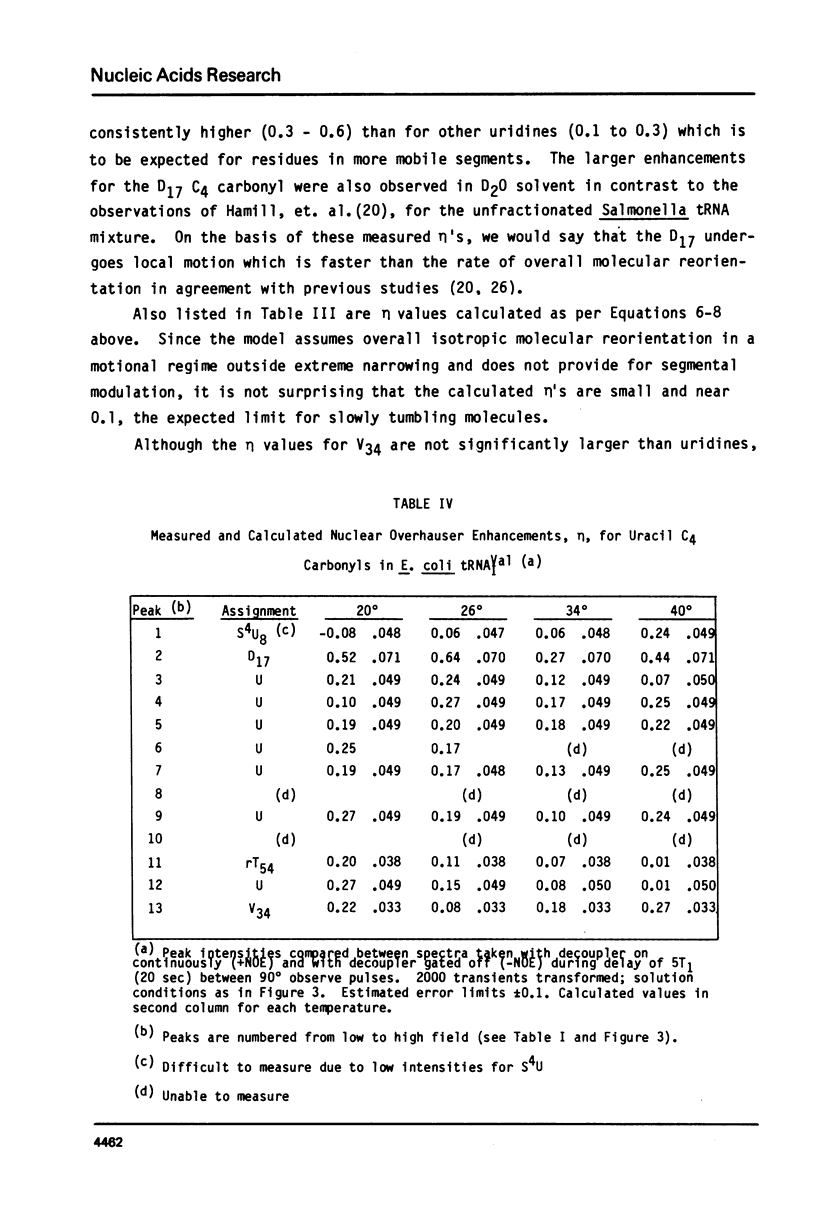
We report 67.8 MHz carbon-13 spin-lattice relaxation studies on [4-13C-uracil] labeled tRNAIVal purified from E. coli SO-187. Following 13C-enriched C4 carbonyl resonances from modified and unsubstituted uridines scattered throughout the polymer backbone enables us to determine dynamical features in both loop and helical stem regions. The experimental results have been analyzed in terms of a model of isotropic overall molecular reorientation. "Anomalous" residues for which the experimental data cannot be accounted for in terms of the model provide an assessment of local and regional properties. Thus, "native" tRNAIVal under physiological conditions of magnesium (10 mM) and temperature (20 degrees - 40 degrees C), exhibits the following characteristics: 1) uridines held rigidly in helical stems and tertiary interactions display correlation times for rotational reorientation of 15-20 nsecs, typical for overall tRNA motion; 2) uridines in loops such as the wobble residue uridine-5-oxyacetic acid (V34) are quite accessible to solvent; moreover V34 and another loop residue, D17, exhibit local mobility; 3) the tertiary interactions involving 4-thio uridine (s4U8) and A14 and ribothymidine (rT54) and A58 are weakened as temperature increases.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agris P. F., Schmidt P. G. Structure of transfer RNA by carbon NMR: resolution of single carbon resonances from 13C-enriched, purified species. Nucleic Acids Res. 1980 May 10;8(9):2085–2091. doi: 10.1093/nar/8.9.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenberg M., Rigler R., Wintermeyer W. On the structure and conformational dynamics of yeast phenylalanine-accepting transfer ribonucleic acid in solution. Biochemistry. 1979 Oct 16;18(21):4588–4599. doi: 10.1021/bi00588a020. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Luxon B. A. High-resolution phosphorus nuclear magnetic resonance spectra of yeast phenylalanine transfer ribonucleic acid. Melting curves and relaxation effects. Biochemistry. 1979 Aug 21;18(17):3796–3804. doi: 10.1021/bi00584a024. [DOI] [PubMed] [Google Scholar]
- Harada F., Kimura F., Nishimura S. Primary sequence of tRNA val from Escherichia coli B. I. Oligonucleotide sequences of digests of Escherichia coli tRNA val with RNase T and pancreatic RNase. Biochemistry. 1971 Aug 17;10(17):3269–3277. doi: 10.1021/bi00793a017. [DOI] [PubMed] [Google Scholar]
- Hurd R. E., Reid B. R. Nuclear magnetic resonance studies on transfer ribonucleic acid: assignment of AU tertiary resonances. Biochemistry. 1979 Sep 4;18(18):4005–4011. doi: 10.1021/bi00585a025. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., RICH A. The general structure of transfer RNA molecules. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4970–4974. doi: 10.1073/pnas.71.12.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura F., Harada F., Nishimura S. Primary sequence of tRNA-Val-1 from Escherichia coli B. II. Isolation of large fragments by limited digestion with RNases, and overlapping of fragments to reduce the total primary sequence. Biochemistry. 1971 Aug 17;10(17):3277–3283. doi: 10.1021/bi00793a018. [DOI] [PubMed] [Google Scholar]
- Komoroski R. A., Allerhand A. Natural-abundance carbon-13 Fourier-transform nuclear magnetic resonance spectra and spin lattice relaxation times of unfractionated yeast transfer-FNA. Proc Natl Acad Sci U S A. 1972 Jul;69(7):1804–1808. doi: 10.1073/pnas.69.7.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid B. R., McCollum L., Ribeiro N. S., Abbate J., Hurd R. E. Identification of tertiary base pair resonances in the nuclear magnetic resonance spectra of transfer ribonucleic acid. Biochemistry. 1979 Sep 4;18(18):3996–4005. doi: 10.1021/bi00585a024. [DOI] [PubMed] [Google Scholar]
- Reid B. R. NMR studies on RNA structure and dynamics. Annu Rev Biochem. 1981;50:969–996. doi: 10.1146/annurev.bi.50.070181.004541. [DOI] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Rohrer D. C., Sundaralingam M. Stereochemistry of nucleic acids and their constituents. VI. The crystal structure and conformation of dihydrouracil: a minor base of transfer-ribonucleic acid. Acta Crystallogr B. 1970 May 15;26(5):546–553. doi: 10.1107/s0567740870002789. [DOI] [PubMed] [Google Scholar]
- Salemink P. J., Swarthof T., Hilbers C. W. Studies of yeast phenylalanine-accepting transfer ribonucleic acid backbone structure in solution by phosphorus-31 nuclear magnetic resonance spectroscopy. Biochemistry. 1979 Aug 7;18(16):3477–3485. doi: 10.1021/bi00583a007. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Redfield A. G. Transfer RNA in solution: selected topics. Annu Rev Biophys Bioeng. 1980;9:181–221. doi: 10.1146/annurev.bb.09.060180.001145. [DOI] [PubMed] [Google Scholar]
- Schweizer M. P., Hamill W. D., Jr, Walkiw I. J., Horton W. J., Grant D. M. Carbon-13 NMR studies on [4-13C] uracil labelled E. coli transfer RNA1(Val1). Nucleic Acids Res. 1980 May 10;8(9):2075–2083. doi: 10.1093/nar/8.9.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler P. B. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4(00):477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Usuki K. M., Yamaizumi Z., Nishimura S., Miyazawa T. Tertiary structures of Escherichia coli tRNA as studied by NMR spectroscopy with 13C-labeling method. FEBS Lett. 1980 Sep 22;119(1):77–80. doi: 10.1016/0014-5793(80)81001-5. [DOI] [PubMed] [Google Scholar]


