Abstract
The discovery of mitochondrial-type genes in organisms thought to lack mitochondria led to the demonstration that hydrogenosomes share a common ancestry with mitochondria, as well as the discovery of mitosomes in multiple eukaryotic lineages. No examples of examined eukaryotes lacking a mitochondrion-related organelle exist, implying that the endosymbiont that gave rise to the mitochondrion was present in the first eukaryote. These organelles, known as hydrogenosomes, mitosomes or mitochondrion-like organelles (MLO), are typically reduced, both structurally and biochemically, relative to classical mitochondria. However, despite diversification and adaptation to different niches, all appear to play a role in Fe-S cluster assembly, as observed for mitochondria. Although evidence supports the use of common protein targeting mechanisms in the biogenesis of these diverse organelles, divergent features are also apparent. The metabolism and biogenesis of these organelles in parasitic protists is discussed here.
Keywords: hydrogenosome, mitosome, evolution, microaerophilic protists, biogenesis
Introduction
Deciphering the origins of eukaryotic cells is one of the more challenging problems in evolutionary biology today. The seminal work of Woese and colleagues led to the grouping of all living organisms into three domains of life: eubacteria, archaea (or archaebacteria), and eukaryotes (122). Over the past 15 years, extensive sequencing of the genomes of organisms from all 3 domains has revealed that eukaryotic genomes contain both eubacterial and archaeal contributions (35, 96). Information genes encoding proteins involved in processes such as translation, transcription, and replication appear to be homologous to archaeal genes. Eubacterial contributions to the eukaryotic genome are seen primarily in the so-called operational genes, which encode proteins such as metabolic enzymes, structural proteins, and membrane components (124).
While many core metabolic properties of eukaryotes have archaeal and eubacterial features, eukaryotes are more complex and exhibit several distinctive physical characteristics absent in archaea and eubacteria. These include an extensive endomembrane system and the compartmentalization of metabolic pathways into discrete membrane-bound organelles; one such organelle is the mitochondrion. How eukaryotic cells evolved these distinctive features and how these features have diversified during the radiation of evolutionarily distinct lineages remains the focus of much research (8). The acquisition of the mitochondrion during eukaryotic evolution may have been a central catalyst allowing further development of the many unique features of eukaryotic cells.
The mitochondrion was originally hypothesized to be the result of endosymbiosis by Margulis and colleagues (75). The ancient eukaryotic forerunner was proposed to have engulfed a bacterium, which was retained and eventually degenerated to the point of being dependent upon the host cell. Much evidence now supports the basic tenets of this theory, including the presence of a double membrane containing a unique bacterial lipid called cardiolipin that is present only on the inner membrane of the organelle. In addition, many mitochondria contain a genome. Sequencing of several mitochondrial genomes and subsequent phylogenetic analyses led to the conclusion that the mitochondrial endosymbiont was closely related to modern α-proteobacteria (123), with its closest existing relative being Rickettsia (4). This answered the question of who donated the endosymbiont; but what about the host cell?
Multiple arguments have been presented and continue to be discussed regarding the biological nature of the cell that engulfed the proto-mitochondrion (77). Two competing views posit that the proto-eukaryote was a member of the archea, and that acquisition of the mitochondrial endosymbiont led to rapid development of the accompanying features that now define eukaryotes. The second theory states that many of these eukaryotic features were already in place, i.e. that such subcellular structures as the endoplasmic reticulum (ER) and nucleus were likely already present in the proto-eukaryote, making alteration of the endosymbiont into a bona fide organelle a more easily explainable process (33). Key to distinguishing between these competing hypotheses is when the mitochondrion was acquired. Various theories have been proposed to explain the circumstances surrounding both the acquisition and retention of the mitochondrion. Many of these expound in particular upon the nature of the host, and invoke one or more endosymbiotic events to account for specific traits present in the protoeukaryotic cell. Several of these hypotheses have been described in detail (28, 72, 78, 81) and are reviewed extensively in Martin et al. 2001 (77).
Early constructions of the eukaryotic tree of life led to the formation of the Archezoan hypothesis (16, 103). These phylogenies, which were based on both rRNA and elongation factor (ef) protein sequences, placed three highly divergent branches containing Trichomonas vaginalis, Giardia lamblia, and microsporidia at the base of the tree (103). These organisms all appeared to have arisen prior to the acquisition of mitochondria, and it was speculated that they represented primitively amitochondriate eukaryotic cells. This would imply that the characteristic features of a eukaryotic cell such as a nucleus and flagella predated the mitochondrial endosymbiosis. Study such organisms would consequently aid in resolving the conundrum of protoeukaryotic identity.
The first indication that these organisms were not true archezoans came from studies of the microsporidia. Analysis of additional genes led to their grouping with fungi (52). Their original position in the tree was likely due to long branch attraction (LBA), an artifact that occurs when highly divergent genes that have evolved at different rates are compared, and appear as long branches in the tree. Additional analyses that eliminate LBA continue to support T. vaginalis and G. lamblia being basal eukaryotes, although it remains difficult to define the root of the eukaryotic tree (8, 20, 60, 124). What has been challenged is the claim that these organisms are amitochondriate. In fact, many genes of mitochondrial origin have since been identified in the ‘amitochondriates’. Moreover, these organisms contain highly modified, or even relic, organelles that are thought to be derived from the proto-mitochondrion, hydrogenosomes and mitosomes (Figure 1).
Figure 1.
Mitochondrion-related organelles are found in all eukaryotes. Eukaryotes can be divided into several major groups, all of which contain mitochondria (red), mitosomes or MLO (blue), or hydrogenosomes (green). Three of these groups have lineages with organisms that contain combinations of these organelles. This tree is adapted from Keeling et al. 2005.
T. vaginalis and G. lamblia have served as model organisms for the study of hydrogenosomes and mitosomes, respectively (27, 107). Hydrogenosomes produce ATP through substrate-level phosphorylation, creating hydrogen as a by-product (69). Mitosomes do not produce ATP, and until recently their potential metabolic role in the cell was somewhat of a mystery (reviewed in 114). Despite differences in metabolic functions, the majority of the data currently available support a single endosymbiosis that gave rise to mitochondria and also resulted in the presence of hydrogenosomes or mitosomes in a variety of protists. The vast differences observed in these organelles would then be the result of divergent evolution in highly specialized ecological niches. Although descent from a single, common, protomitochondrial endosymbiont has been the prevailing view for several years, distinguished scientists continue to raise important arguments contrary to this view (22, 76).
Hydrogenosomes
Hydrogenosomes were first described in trichomonads by Muller and colleagues following futile attempts to detect mitochondrial and peroxisomal activities in trichomonad cellular extracts (69). These organelles, which are 0.5–1 μm in diameter, were originally thought to be bound by a single membrane due to the close apposition of two membranes. This characteristic and the apparent lack of peroxisomes originally fostered a popular theory that the T. vaginalis hydrogenosome was a microbody related to, but distinct from, peroxisomes. Biochemical studies on crude hydrogenosomal fractions provided clues to the contrary. Muller and colleagues showed that hydrogenosomes are the site of fermentative metabolism of pyruvate leading to the production of ATP and molecular hydrogen. Complementary analyses by the groups of Cerkasov and Metenier indicated a role in cyanide-insensitive respiration, dependence on ADP, and the presence of a mitochondrial-like decarboxylating malate dehydrogenase (85). The demonstration of these metabolic properties of the organelle together with freeze fracture electron microscopy that revealed the presence of two membranes argued against a peroxisomal nature and instead suggested similarities with mitochondria (53).
The T. vaginalis genome sequence (15), partial hydrogenosomal proteomes (49), and various biochemical studies (31, 55, 83–84) have revealed the presence of other diverse activities compartmentalized within the T. vaginalis hydrogenosome. These include enzymes involved in amino acid metabolism in mitochondria (83, 84, unpublished data), oxidoreductases similar to components of respiratory complex I in mitochondria (31, 55), as well as novel putative peroxidises likely involved in oxygen stress responses (91). Our unpublished proteomic analyses of hydrogenosomes reveal the presence of over 500 proteins and indicate the presence of yet additional metabolic pathways within the organelle.
Organelles capable of producing molecular hydrogen, hence likewise categorized as hydrogenosomes, have also been described in the ciliate Nyctotherus ovalis, the rumen-dwelling chytrid fungus Neocallimastix frontalis, and the heterolobosean flagellate Psalteriomonas lanterna (23, 39, 44–45, 47). These organelles contain hydrogenase activity and are found exclusively in aerotolerant protists that lack mitochondria. Genetic evidence also supports the possible presence of hydrogenosomes or mitochondrion-related organelles in the free-living amoeba, Mastigamoeba balamuthi (39) and the preaxostyla flagellate Trimastix pyriformis (47). The phylogenetic distribution of hydrogenosome-bearing organisms scattered in various lineages of the eukaryotic tree, however, make it evident that they arose independently of one another (Figure 1; 32), albeit all hydrogenosomes appear to have arisen from either a protomitochondrion or, in the case of the ciliate hydrogenosome, from bonafide mitochondria.
Although most hydrogenosomes lack a genome, a hydrogenosomal genome has been identified and sequenced from the ciliate N. ovalis (9). While the metabolic activity of this hydrogenosome was unlike that of mitochondria, it was abundantly clear that this genome arose from a bona fide mitochondrion. Unlike the hydrogenosome of T. vaginalis, those of N. ovalis contain cardiolipin (a distinctively mitochondrial lipid) in their membranes and have cristae. The descent of the N. ovalis hydrogenosome from modern-day mitochondria and analyses of both the ciliate and chytrid hydrogenosome, have been recently reviewed (44–45).
The origin of the T. vaginalis hydrogenosome could not be determined by organelle genome sequencing, as one does not exist (19). The use of several alternative approaches aimed at determining the evolutionary history of T. vaginalis hydrogenosomes has fostered much research and controversy in recent years (41, 109). Such intense interest in this topic has served to broaden the community of researchers interested in the evolution of eukaryotic organelles and has accelerated our understanding of this fundamental process.
Phylogenetic Evidence of Primitive Mitochondrial Ancestry – the Chaperonin Story
Mitochondria maintain part of the original endosymbiont genome, though massive gene transfer to the host nucleus occurred during the transition to organelle (50). The T. vaginalis hydrogenosome, as well as mitosomes found in other ‘amitochondriate’ protists, have entirely dispensed with a genome. Thus mitochondrial-like genes present in their nuclear genomes were used in phylogenetic analyses in search of the origin of these organelles. One class of mitochondrial genes that are highly conserved, and therefore good candidates for phylogenetic analyses, are the chaperonin genes (42–43). Proteins are imported into organelles in a disordered state, and once inside are folded into their correct tertiary structure with the assistance of chaperonins. Three of these, Cpn60, Cpn10, and Hsp70, are conserved in most eukaryotes. Hsp70 aids in the import of proteins across the double membrane of mitochondria as part of the import motor of the inner membrane import complex (119). Cpn10 and Cpn60 function as a complex together to refold imported matrix proteins. The mechanism by which Cpn10 and Cpn60 fold proteins has been well-studied in the prokaryotic protein homologs GroES and GroEL (63, 119).
Genes for Cpn10, Cpn60, and Hsp70 are present in the nuclear genome of T. vaginalis and phylogenetic analyses indicated that all are of mitochondrial origin (12, 36, 54, 98). The mitochondrial-like Hsp70 sequences also contain two sequence motifs confined to Hsp70s of mitochondrial origin (12, 36). These data were the first to strongly support that T. vaginalis once harbored the mitochondrial endosymbiont.
Evidence of mitochondrial genes has now been identified in all examined ‘amitochondriate’ organisms, casting serious doubt on the archezoan hypothesis. Early support for a Cpn60-like protein in the parasite G. lamblia came from western blots and immunfluorescence microscopy that indicated a protein that cross-reacted with anti-Cpn60 antibodies from other organisms (104). The gene encoding Cpn60 was later cloned and subjected to phylogenetic analyses that strongly supported the descent of glCpn60 from its mitochondrial counterpart (99). A single Hsp70 gene has also been analyzed from G. lamblia, and it appears to be monophyletic with mitochondrial Hsp70 genes (82). Mitochondrial-type Cpn 60, Hsp70 and Cpn10 are also present and expressed in several species of Entamoeba (5–7, 18, 116). The apicomplexan parasite Cryptosporidium parvum was thought to be distinct from other members of its class, in that it apparently contains neither a plastid nor a mitochondrion. However, a Cpn60 and Hsp 70 gene are found in Cryptosporidium; both are expressed and share a common ancestry with genes from α-proteobacterial homologs(95, 101).
Microsporidial genomes also bear witness to a relic mitochondrion. Hsp70 sequences have been identified and characterized from several species, including Antonospora locustae, Trachipleistophora hominis, Vairimorpha necatrix, Glugea plecoglossi, Encephalitozoon cuniculi, and E. hellem (5, 37, 51, 89). Despite sequencing of the E. cuniculi genome, no trace of either Cpn10 or Cpn60 could be found, although a total of four Hsp70 genes were present, along with additional genes that appeared to be related to α-proteobacteria (58).
A full complement of mitochondrial chaperonins is encoded by T. vaginalis and Entamoeba. Unusually, both G. lamblia and C. parvum encode Cpn60, but appear to lack its partner Cpn10; Hsp70 is present in both. The microsporidia represent a more extreme state, as they have retained only mtHsp70. The absence of Cpn10 or both it and Cpn60 in certain mitosomes raises the question of how proteins are refolded upon import; and specifically how this differs from protein folding in mitochondria and hydrogenosomes that contain a full complement of chaperonins. In some mitosomes, Hsp70 may be able to refold imported proteins alone without the additional GroES/GroEL-like activity of Cpn10/Cpn60. Alternatively, the cpn60 in these organelles may function without its usual partner in crime, cpn10, or imported proteins are capable of folding correctly without chaperonin assistance, as has been described for some mitochondrial matrix proteins (100). These losses likely reflect the continual reductive evolution that has shaped the mitosomes (14).
The localization of mitochondrial-like chaperonins to relic organelles found in previously described ‘amitochondriate’ protists were crucial for the discovery of mitosomes. These organelles are double-membrane bound, like mitochondria and hydrogenosomes, but considerably smaller. In E. histolytica, immunofluroescence microscopy was used to localize Cpn60 to this previously unidentified organelle (73, 111). In G. lamblia, both Hsp70 and Cpn60 localize to the mitosome, though they lack discernable N-terminal presequences (94). In the apicomplexan C. parvum both Cpn60 and Hsp70 localize to a double-membrane bound cytosolic organelle (90, 101). Finally, antibodies against Hsp70 from the microsporidian T. hominis were localized by both light and electron microscopy to double-membrane organelles (121). Therefore not only did these previously ‘amitochondriate’ organisms once harbor the mitochondrial endosymbiont, but they retain more than genes as evidence. They also maintained organelles –but to what purpose?
Biochemical Activities – It’s not just about ATP
The analysis of chaperonin genes as well as additional metabolic genes has demonstrated that the mitochondrial endosymbiosis occurred extremely early in eukaryotic evolution. Mitochondria, hydrogenosomes, or mitosomes, are found in every eukaryotic domain, including the previously ‘amitochondriate’ Excavata lineage (Figure 1; 8, 46, 59). It is unlikely that any extant eukaryotic cells that predate the mitochondrial endosymbiosis exist – in fact, the acquisition of the mitochondrion may have been the defining event in eukaryotic evolution (77). This critical event in eukaryotic history appears to have happened only once. But since that event, the cells carrying the endosymbiont have become highly adapted to diverse environmental niches, and the endosymbiont they carried evolved accordingly.
Why have these remnant organelles been retained? The original proteobacterial endosymbiont was hypothesized to have been retained because the host cell obtained necessary metabolites from it (75, 78, 81). While hydrogenosomes lack many of the metabolic pathways of mitochondria, they do produce ATP through substrate-level phosphorylation. But no evidence of ATP production has been obtained for the mitosomes of G. lamblia, E. histolytica, C. parvum, or the microsporidia. ATP is produced either in the cytosol or is gleaned from the host cells, thus in these organisms the mitochondrial remnant must serve another purpose. Thus far the only unifying biochemical pathway for mitochondria, mitosomes, and hydrogenosomes appears to be Fe-S cluster formation, a process proposed to be the only essential function of the mitochondrion (67). Fe-S cluster formation in the mitochondrion is critical for formation of all Fe-S containing proteins in the cell, which are involved in various vital processes (66, 68).
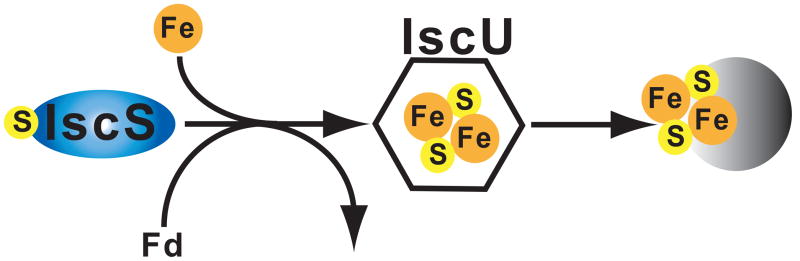
Proteins known to be required for Fe-S cluster assembly in yeast include IscS (NifS), which produces sulphur from cysteine for incorporation into the Fe-S scaffold, and IscU (NifU), which binds the Fe substrate for incorporation (Figure 2; 68). In addition to these, ferredoxin is known to interact, possibly in reducing the Fe or S, or as an intermediate in Fe-S formation. Hsp70 also participates, probably by binding the apoproteins and maintaining their structure prior to transfer of the Fe-S cluster. Frataxin often acts as an iron donor. Additional steps must occur, but the proteins that are involved and their mechanism are undefined (66). Several genes involved in Fe-S cluster assembly have been identified in recently published genomes from mitosome-carrying organisms, and a few of these proteins have even been demonstrated to localize to the T. vaginalis hydrogenosome and some mitosomes (Table 1).
Figure 2.
Fe-S cluster formation. The biogenesis of Fe-S clusters within mitochondria is similar to the bacterial ISC process. IscS is a cysteine desulferase that donates the S, and Fe is often donated by frataxin in mitochondria. Ferredoxin acts in electron transfer, perhaps to reduce the donated S. IscU is a scaffold protein on which the initial FeS cluster is formed before transfer to the apoprotein.
TABLE 1.
Core Proteins of Anaerobic Protists and Their Localization Where Known
| Organism | PFO | FeFe Hydrogenase | Ferredoxin | IscU | IscS | mtHsp70 |
|---|---|---|---|---|---|---|
| T. vaginalis | + (H) | + (H) | + (H) | + | + (H) | + (H) |
| G. lamblia | + (C) | + (C) | + (M) | + (M) | + (M) | + (M) |
| E. histolytica | + (C) | + (C) | + (C) | NifUc | NifSc | + (M) |
| C. parvum | +a | +b | + | + | + | + (M) |
| Blastocystis sp. | + (M) | + (M) | + | + | + | |
| microsporidia | + | + | + | + (M) |
Key: H=hydrogenosome M=mitosome C=cytosol
Cells left blank indicate that no homologs have been identified.
+ indicates that the gene has been sequenced. Protein localization is given when known.
C. parvum PFO is fused to an NADPH-cytochrome P450 reducatase domain, as also seen in Euglena gracilis.
C. parvum FeFe hydrogenase is more similar to those of eukaryotes than to other protist hydrogenases.
The NifU and NifS genes of E. histolytica are likely the result of LGT with an epsilon-proteobacterium and appear to be localized to both the mitosome and the cytosol.
IscS was the first Fe-S cluster assembly gene to be identified in T. vaginalis (106). It was shown to be expressed and localized to the hydrogenosome. Following publication of the genome in 2007, IscU was identified (15). While it has not been localized to the hydrogenosome, it does have a predicted presequence that would target it to this organelle (15). Frataxin, a likely Fe donor, has also been localized in hydrogenosomes, and is found in the same clade as other mitochondrial-like frataxin genes in phylogenies (24). It appears therefore that in addition to ATP production, the hydrogenosome, like mitochondria, also acts as a center for Fe-S cluster formation.
The case for mitosomal Fe-S cluster assembly is very strong for G. lamblia. Both IscS and IscU giardial genes are phylogenetically related to those of mitochondria, lending additional support to the mitosome’s heritage as an endosymbiotic relic (106, 112). It has been demonstrated that IscU, IscS, ferredoxin, and mtHsp70 all localize to the G. lamblia mitosome (94). Additionally a monothiol glutaredoxin containing an unusually long N-terminal presequence has been shown to co-localize with IscU (92). Phylogenies also support a mitochondrial ancestry for this gene, which is involved in transfer of Fe-S clusters from the IscU scaffold to apoproteins.
A similar story has unfolded for the mitosome of C. parvum. Mitochondrial-type IscS, IscU, a frataxin-like protein, and ferredoxin have been discovered in the genome of C. parvum (1, 64). The presence of a NifS-like gene further supports a role for the mitosome in Fe-S cluster assembly (90, 95). Interestingly, additional metabolic processes may occur in the C. parvum mitosome. Recently, an alternative oxidase gene related to those found in Trypanosoma brucei was identified–this protein can substitute for Complexes II and IV from mitochondria (90, 97). A gene encoding a subunit of Complex V (ATP synthase subunit β) was also identified, as well as superoxide dismutase (90). These proteins support the idea that the mitosome of C. parvum may be capable of modified aerobic metabolism in addition to Fe-S cluster assembly, but further biochemical analyses would be required before this conclusion is given a great deal of weight (90).
Fractionation of E. histolytica indicates that pyruvate metabolism occurs in the cytosol, or at least is not confined to any identified compartment (93). Other anerobic metabolic processes are also predicted to occur in the cytosol. An Fe-Fe hydrogenase lacking an N-terminal presequence is present in the E. histolytica genome and is found in the cytosol (38). Genes encoding E. histolytica proteins involved in Fe-S cluster assembly, namely NifU and NifS, were identified in the genome, however unlike other mitosomal genes, phylogenetic analysis indicates that these are not mitochondrial in origin but were likely acquired via lateral gene transfer from a member of the epsilon-proteobacteria (2–3, 115). This is also the case in the related amoeba M. balamuthi, indicating that acquisition of these genes occurred prior to the split between these lineages (39). When their distribution was examined, both NifU and NifS were found in the cytosol as well as in mitosomes (74). Ferredoxin is also found in the cytosol (73), while Hsp70 is known to localize to the mitosomes (110). This implies that at least some Fe-S cluster assembly may occur in this mitosome, similar to the requirement for mitochondrial assembly in cytosolic Fe-S protein maturation (68, 74). Recently it was demonstrated that the E. histolytica mitosome may also harbor a sulphate activation pathway, as three enzymes involved in this process (ATP sulfurylase, APS kinase, and inorganic pyrophosphatase) also localize to this organelle (80).
In microsporidia, where reductive evolution has affected every aspect of their biology, the purpose of the remnant mitosome becomes much more difficult to discern. The genome sequence of the microsporidium E. cuniculi revealed twenty-two genes homologous to yeast mitochondrial genes, six of which group with α-proteobacterial sequences (58). Of these six genes, ISU1/ISU2 (Nif-U like), NFS1 (similar to IscS and NifS), YAH1 (ferredoxin), and PDB1 (pyruvate dehydrogenase complex E1) are all involved in Fe-S cluster assembly. However, when the Fe-S cluster assembly pathway was localized, differing results were obtained in different species (40). In E. cuniculi, frataxin, Nfs1, and Isu1 all colocalized with Hsp70 in the mitosome. However, in T. hominis only Nfs1 and Hsp70 were found in the mitosome, while both Isu1 and frataxin appeared to be predominantly cytosolic (40). Further work remains to decipher conclusively the Fe-S cluster assembly pathway in microsporidia, although at least parts of this pathway appear in their mitosomes.
During the early evolution of the mitochondrion, the change from endosymbiont to organelle may have been triggered by the host cell’s reliance on metabolic products. Mitosomes have not retained ATP production, unlike the hydrogenosome, but are likely critical for Fe-S cluster assembly. The need to retain an organelle for this purpose may reflect a reliance on a membrane potential for maturation of Fe-S cluster proteins (67). Thus far, membrane potentials have been demonstrated for mitochondrion-related organelles in T. vaginalis and G. lamblia, as well as for C. parvum (10, 70, 97) but not for E. histolytica or microsporidia (73, 121). In eukaryotic cells retaining a canonical aerobic mitochondrion, it is known that cytosolic Fe-S proteins are reliant on Fe-S cluster formation within the mitochondrion (68). Possibly the same constraints exist in hydrogenosome and mitosome-bearing organisms (74).
An intermediate organelle in Blastocystis sp
Another exception to the “no DNA” rule for mitochondrion-related organelles is found in the mitochondrion-like organelle (MLO) of Blastocystis hominis, which has a genome of ~27–28 kb (88, 120). Blastocystis MLOs were first described as “cytochrome-free mitochondria” in 1986, and various enzymes associated with mitochondria were shown to be absent in these vestigal organelles (125–126). It was noted, however, that staining with the dye Rhodamine 123 indicated the presence of a weak membrane potential, a finding confirmed following the isolation of the organelles (86, 126).
Later, fractions enriched for Blastocytsis MLOs were examined for enzymes found in either mitochondria or hydrogenosomes and activities for malic enzyme, pyruvate:ferredoxin oxidoreductase (PFO), acetyl-CoA hydrolase, STK, alpha-ketoglutarate dehydrogenase, isocitrate dehydrogenase, and aconitase were detected (65, 86). Many of these enzymes are considered hallmarks of the T. vaginalis hydrogenosome (85). An incomplete Krebs cycle was also detected in these studies, likely because this work was performed anaerobically unlike previous studies.
Sequencing of the MLO genome of three strains of Blastocystis (88, 105, 120) unveiled the presence of 45 genes, 27 of which are ORFs and the remainder are structural RNA genes. All genes encoding cytochrome and ATPase subunits are lacking (88), as would be expected as no pathways utilizing these proteins have been detected (65, 126). Genes encoding an Fe-Fe hydrogenase and PFO, both of which group with eukaryotic sequences from anaerobes and green algae are present (Table 1, 105). Genes for subunits of complex I, complex II, NADH dehydrogenase, several carrier proteins have also been retained. Phylogenetic analyses of the complex I and NADH dehydrogenase genes indicate they are most closely related to α-proteobacteria, and as such the MLO of Blastocystis can be said to be a highly divergent mitochondrion with little to no controversy (88, 105). In addition, genes are present for frataxin, ferredoxin, IscS, glutaredoxin, and Isca2, all of which are involved in Fe-S cluster assembly (105). This organelle will be a good model for examining partial degeneration from mitochondrion to mitosome, as processes found in both hydrogenosomes and mitochondria are present.
Protein Targeting in Hydrogenosomes and Mitosomes
An early step in conversion from endosymbiont to organelle would have been the development of systems that allow exchange with the host cell cytosol (27, 57). Once gene transfer to the nucleus occurred, any proteins that were necessary for processes in the organelle would have to be imported utilizing translocases to cross both membranes. The imported proteins must be targeted, recognized by the organelle, imported across membranes, localized within the organelle, and properly folded for activity. Additional molecules must also be imported and exported and their carrier proteins therefore must be present (27). Bioinformatic and proteomic comparisons of mitochondrion-related organelles have revealed that some of these processes are likely to have been conserved, however much divergence is also evident. Biochemical analyses of these processes are sparse and are needed to clearly define conserved and unique proteins involved in the biogenesis of these divergent organelles.
N-terminal presequences
Most proteins destined for the mitochondria contain N-terminal presequences that exhibit a consistent pattern; these presequences are usually hydrophobic and are capable of forming an amphipathic α-helix (87). Proteins targeted to the hydrogenosome and several bound for mitosomes also contain presequences. Hydrogenosomal presequences are generally shortened, but retain characteristics similar to those of mitochondria (10, 28). Not all mitosomal proteins contain N-terminal presequences, and when present they are typically shorter and less defined than those of hydrogenosomes or mitochondria (13), with the exception of those for C. parvum mitosomes (64).
In hydrogenosomes, the first protein shown to contain a targeting presequence was ferredoxin (Fd) (56). Since then presequences have been identified on several proteins bound for the hydrogenosome. Many of these have also been shown to function in heterologous systems, by targeting various reporter sequences to yeast mitochondria (29, 48). However recent work has demonstrated that at least some hydrogenosomal proteins are localized correctly even when their presequence is deleted (79). This may indicate that these signals are in the process of being evolutionarily lost, or that additional unknown sequences are also involved in targeting proteins to the hydrogenosome. This could also imply that the hydrogenosomal protein translocases may differ substantially from those of mitochondria, an observation with some merit, as discussed below. Although unexpected, the lack of targeting sequences at the N-termini of a subset of T. vaginalis hydrogenosomal proteins is congruent with that observed for numerous mitosomal proteins.
In E. histolytica Cpn60 contains an N-terminal targeting sequence of approximately 22 amino acids that is cleaved in vivo (73). Removal of the first 15 of these amino acids results in a cytosolic distribution for Cpn60, and replacement of the Cpn60 presequence with a mitochondrial targeting sequence from T. cruzi Hsp70 restored targeting (73, 111). The protein targeting machinery of E. histolytica and T. cruzi appears to be conserved (111). Later experiments indicated that in addition to the N-terminal presequence, other targeting information is likely contained in the Cpn60 gene (2). Pyridine Nucleotide Transhydrogenase (PNT) also contains a putative N-terminal targeting sequence, but experimental work indicates it is not localized to the mitosome (2).
Thus far, all genes thought to target the relic mitochondrion of C. parvum contain predicted N-terminal presequences. Cpn60, the first mitochondrial gene described in C. parvum, contains an N-terminal extension of 38 amino acids, that when fused to GFP and expressed in yeast it targets GFP to the mitochondrion (95). CpHsp70 also has a presequence of 34 amino acids, that has been shown to be functional by targeting of a GFP fusion protein to the mitochondrion in yeast as well as in the related apicomplexan, Toxoplasma gondii (101). The Fe-S cluster assembly genes IscS and IscU contain predicted presequences of 37 amino acids and 27 amino acids respectively, and CpFd has a predicted targeting presequence of 35 amino acids (64). As noted above, C. parvum mitosomal presequences are longer than those typically seen on other mitosomal or T. vaginalis hydrogenosomal proteins. Indeed they are more in line with that observed for proteins targeted to the matrix of yeast mitochondria. However it has yet to be tested whether the full presequence, as defined, is required for translocation of protein into the C. parvum mitosome and thus shorter ones may be functional.
In mitosomal proteins of Giardia, it is less clear whether presequences are required for import. N-terminal presequences are not present on Cpn60, nor are any obvious on IscS (99, 106), both of which localize to the mitosome. Hsp70 does appear to have a very short hydrophobic N-terminus that may function as a targeting peptide, but this has not been verified by sequencing of the mature protein (94). Only IscU and ferredoxin retain presequences that are necessary for import into the mitosome and also are cleaved upon import (102, 112).
The role of presequences in mitosomal import becomes even less clear when microsporidian mitosomal proteins are examined. N-terminal presequences are rarely predicted by bioinformatic software in these organisms, and they have no consistent characteristics (14). When full-length mitosomal proteins from A. locustae or E. cuniculi were fused with GFP and expressed in yeast cells, only 6 out of 16 total proteins were targeted to the mitochondrion. Of those that were sent to the mitochondrion, there was no correlation with a distinguishable leader (14). In fact, only a single protein with a leader was sent to the mitochondrion in these experiments.
Presequence processing peptidases
Once imported into the matrix of the mitochondrion, the targeting presequence is cleaved by a mitochondrial processing peptidase (MPP) composed of an α and β subunit. In yeast mitochondria, these subunits must form a heterodimer to be functional (108). Initial studies in T. vaginalis were able to identify a β-subunit, but no evidence of a corresponding α subunit was forthcoming (11). Because presequences have been demonstrated to be cleaved in T. vaginalis it was initially proposed that the β-HPP functioned independently, and that the α-subunit was lost when the endosymbiont-containing T. vaginalis lineage split. Later work demonstrated that a glycine-rich loop protein was in fact a functional α-HPP subunit and that the HPP model looked very much like that of MPP (102). Further evidence supporting a common origin for mitochondria and hydrogenosomes was also borne out by these studies, as the mitochondrial MPP and hydrogenosomal HPP were clearly shown to reside within the same phylogenetic clade (11, 102).
With regard to the presequence processing enzyme in Giardia, it was shown that there is no α subunit, and that a single β-GPP is capable of cleavage of short leaders, similar to that described for a related proteinase from Rickettsia (61, 102). Interestingly, this monomeric giardial mitosomal processing peptidase is incapable of cleaving longer mitochondrial presequences (102). A single gene encoding a β-subunit of a processing peptidase has been identified in C. parvum but no biochemical analysis to determine whether it functions as a monomer as well has been reported (90). Blast searches of the recently published E. histolytica genome returned hits for sequences similar to a β-subunit, but none for an α-subunit (unpublished data). However, further biochemical analyses and careful in silico searches may allow the identification of α-subunits in these organisms. Thus far, no processing peptidase has been found in the Blastocystis sp., but it was hypothesized that a metalloprotease I protein that is present may serve this function (105). Likewise, no processing peptidases have yet been identified in any of the microsporidia species and it is thought that these proteins may no longer be necessary, as targeting sequences appear to have been disposed of in these organisms (13–14, 58).
TOM/TIM-type Protein Translocases
Similar to mitochondrial matrix proteins, hydrogenosomal, mitosomal, and MLO matrix proteins must traverse the double membranes that enclose these organelles. With the exception of Blastocystis and Nyctotherus, none of the “degenerate” organelles have genomes, therefore all proteins destined for processes within the organelles must be imported from the cytosol. In mitochondria this task is achieved by specific protein complexes called Translocase of the Outer Membrane (TOM) and Translocase of the Inner Membrane (TIM). TOM and TIM are multiple heterooligomeric complexes that include pore proteins, receptor proteins, motor proteins, chaperonins, and associated accessory proteins. In addition to the TIM/TOM pathways, the Sorting and Assembly Machinery (SAM) proteins assist in localization of β-barrel and integral outer membrane proteins and the Mia40 pathway moves proteins into the intermembrane space. These complexes have been best defined in yeast mitochondria but homologues of these proteins have been found in multiple organisms (for recent reviews, see 34, 87).
In yeast, the Tom40 complex recognizes proteins with N- terminal presequences, acts as the pore, and assists the protein across the outer membrane where it is then bound to the Tiny Tims in the intermembrane space. These “Tiny Tims” deliver unfolded matrix proteins to the Tim23 translocon, which is assisted by the Pam proteins and Hsp70 acting as a motor complex. In the case of integral inner membrane proteins, Tom40 again acts as pore but it is Tom70 that binds the incoming protein and guides it. A different set of Tiny Tims deliver inner membrane proteins to the Tim22 translocon, which is required for their insertion into the membrane. Less is known about how proteins are directed to the Sam pathway; although presequences are not required for proper targeting of membrane proteins, they are sometimes found on inner membrane proteins. For membrane proteins that do not have them their overall charge is likely involved both in their recognition and orientation into the membrane (87). The Mia40 pathway, which is involved in targeting intermembrane space proteins, was only recently discovered, and further work remains to fully understand it (34, 87).
Many import proteins have been identified by bioinformatic screening in organisms containing hydrogenosomes and mitosomes, usually by searching completed genomes and in some cases proteomes (Table 2). Those that have been found share very little sequence similarity with homologs in yeast or other organisms with canonical aerobic mitochondria. Using Hidden Markov Models (HMM), genes have been identified for Sam50, the Tim 17/22/23 family, Tim44, Hsp70, and Pam18 in T. vaginalis (25). Whether any of these proteins act as translocases in this organelle is unknown, though Hsp70 and Pam18 have been localized to the hydrogenosome (25–26). G. lamblia contains both Hsp70 and Pam18, both have presequences, and both localize to the mitosome (26, 94). Recent work has identified and localized a Tom40 homolog to the G. lamblia mitosome outer membrane, and it was also demonstrated to be part of a high-molecular weight complex (21). Genes for a putative Tom70, Tim9, Tim 17, Tim21, and Tim50 have been identified in the B. hominis genome, but no biochemical confirmation that they function as translocases exists yet (105).
TABLE 2.
Predicted Presence of Mitochondrial Translocase Homologs in Anaerobic Parasites
| Translocase | T. vaginalis | G. lamblia | E. histolytica | B. hominis | C. parvum | microsporidia |
|---|---|---|---|---|---|---|
| Sam50 | + | + | + | |||
| Tom40 | ? | + | + | + | + | |
| Tom70 | + | + | ||||
| Tim17/22/23 | + | + | + | + | ||
| Tim21 | ||||||
| Tim44 | + | ? | ||||
| Tim50 | + | |||||
| Pam16/18 | + | + | + | |||
| mtHsp70 | + | + | + | + | + | + |
? indicates minimal sequence similarity.
Cells left blank indicate that no homologs have been identified either via in silico methods or biochemically.
In searches of the E. histolytica genome, only Hsp70 and a putative Tom40 homologue have been identified (2, 71, 80, 116). In C. parvum a very limited analysis has identified Hsp70, a Sam50, a potential Tim44, and three genes related to the Tim17/22/23 family (25, 90). Genomic searches of the microspordian E. cuniculi revealed the presence of homologues for a shortened Tom70, a Sam50, and a highly divergent form of Tom40 (25, 58, 118). The Tom70 was shown to partially complement a Tom70 yeast mutant, indicating that it does appear to retain some function as a receptor; whether it functions in mitosomes is unknown (118). As far as inner membrane complexes in microsporidian mitosomes are concerned, a Tim17/22/23 family member, Tim50, and Pam16 were discovered using HMM analyses (118). None of these have been tested for localization.
A compilation of available bioinformatic studies indicates that the most highly conserved translocases in these parasitic protists are Sam50, Tom40, and members of the Tim17/22/23 family of inner pore complexes (Table 2). It is important to note that the presence of these proteins is usually predicted by in silico studies. As most components of the mitochondrial translocase machinery (with the exception of Sam50 and Hsp70) have no homologs in α-proteobacteria, this machinery likely evolved after the mitochondrial endosymbiotic event (25). Therefore much of the machinery need not be conserved. Furthermore restriction of import studies to primarily yeast mitochondria makes the current model even more limiting. It can safely be predicted that additional biochemical and functional analysis of import proteins will yield considerably more information about diversity in these organelles. Notably, the only member of the Tom40 complex that has been identified in these divergent organisms is Tom40 itself. Since Tom20 and 22 act as receptors that recognize the N-terminal presequence of yeast matrix proteins, it is possible that the loss or alteration of these proteins in protists has led to the reduced reliance on N-terminal presequences as discussed above.
Mitochondrial carrier family proteins
The mitochondrial carrier family (MCF) is composed of proteins that transport a variety of molecules such as NADH, ADP, ATP, and other metabolically important substrates (62). Several homologues of these family members have been identified in organisms harboring hydrogenosomes and mitosomes.
The ATP produced in hydrogenosomes needs to be transported into the cytosol for use by the cell and hence it makes sense that mitochondrial-like ADP/ATP carriers may be conserved in hydrogensome-containing organisms. An MCF member, Hmp31, was identified in T. vaginalis hydrogenosomes and is predicted to be localized to the inner membrane based on trypsin digest assays in intact organisms (29). This protein, which is one of the more abundant hydrogenosomal membrane proteins, has a structure composed of four helices, similar to the structure of other MCF proteins. Biochemical studies indicate that Hmp31 forms a homo-oligomer (29). Interestingly, an ADP/ATP carrier with similar properties has also been identified in the hydrogenosomes of Neocallimastix (117).
Although ATP production has not been reported in any mitosomes, proteins involved in Fe-S cluster assembly, as well as carriers required to transport Fe-S clusters into the cytosol are predicted to be present in these organelles. These are as yet poorly defined. A single MCF protein has been described in E. histolytica, which functions when expressed in bacteria (17), and several ESTs from B. hominis MLO indicate the presence of MCF proteins (105). C. parvum also has genes coding for carrier proteins (1). Thus far these have not been localized to the organelle, nor has their range of substrate affinities been confirmed.
Within the microspordian E. cuniculi four MCF proteins have been identified based on sequence analysis of the recently published genome, only one of which localizes to the mitosome (113). It is thought that this carrier actually supplies ATP from the cytosol to the mitosome, presumably for Fe-S cluster formation (113). This MCF protein is phylogenetically related to NAD/NADP+ MCF members, but biochemical characterization demonstrated that it actually carried ADP/ATP. This finding illustrates how important biochemical studies are in highly divergent organisms- initial bioinformatic analyses can prove misleading as to the actual function of proteins that have had many years to diverge from their original purpose within the cell.
Unclassified membrane proteins
Hydrogenosomes, mitosomes, and MLOs differ markedly from mitochondria in spite of their multiple similarities. It is therefore to be expected that many translocases that may have been shared by the progenitor organelle may have been lost or become so divergent as to be unrecognizable by bioinformatic analyses alone. In addition novel proteins that play key roles in the biogenesis of these organelles also may have arisen. For example, in the hydrogenosome of T. vaginalis an integral membrane protein of 35 kD has been identified that has a predicted secondary structure similar to pore-forming proteins (30). Called Hmp35, this protein lacks any strong similarity to any other proteins, and its origin and exact function remain enigmatic. Interestingly Hmp35 can be crosslinked to exogenous Fd as the latter is imported into the hydrogenosome (30), suggesting it is a novel transport protein. Experiments on other cryptic organelles will likely identify additional unique transport proteins or carriers that do not have ancestors found in mitochondria. Such studies will shed light on the evolutionary paths taken by these organelles as well as provide further clues to decipher the metabolic roles played by these organelles.
Conclusions and remaining questions
The overwhelming evidence points to a single endosymbiotic event producing both the mitochondrion and related organelles known as hydrogenosomes, mitosomes, or mitochondrion-like organelles. However, the exact timing of this event remains unknown. It is unlikely that mitosomes and the T. vaginalis hydrogenosomes developed from a full-fledged mitochondrion. Possibly the lineages containing these organelles diverged from other eukaryotes while the endosymbiont was still in the process of transitioning to an organelle. It is still generally accepted that T. vaginalis and G. lamblia are deeply divergent eukaryotes based on several phylogenies. Thus, their organelles are likely to have acquired or retained unique properties, some of which may be essential for their biogenesis and metabolic functions. It may also be appropriate to view these organelles as degenerate mitochondria, as is often argued. However, current data indicates their differences relative to mitochondria are likely greater than their similarities. Future studies aimed at better definition of the proteins present in these unusual organelles and the metabolic properties attributed to them are likely to unlock additional novel biological secrets.
One can also speculate as to why T. vaginalis hydrogenosomes either retained or developed novel metabolic pathways, while in mitosome-containing organisms most metabolic processes appear to have been transferred to the cytosol or lost altogether. These adaptations occurred subsequent to the transfer of endosymbiotic genes to the nuclear genome of all of these organisms, with the consequent development of N-terminal targeting sequences, at least for some proteins targeted to these organelles. Whether these sequences are typically sufficient to direct proteins to any of these organelles is now questionable. Clearly, more in depth studies are needed to sort out mechanisms involved in protein sorting per se.
The unifying features of mitochondria, mitosomes, MLOs, and hydrogenosomes are sparse. Thus far, Fe-S cluster assembly is the single metabolic process that links almost all these organelles, with the presence of cpn60 also a unifying feature. Limited conservation of the translocase machinery, presequence processing peptidase complex, and the chaperonins required for refolding translocated proteins is also evident. The retention of cpn60 in all these organelles is intriguing, particularly in light of the general trend for the organelles to undergo reduction and divergence. This would point to an indispensable role for cpn60. However, cpn10, which is essential for cpn60 function in mitochondria (63, 119) has not been found in any mitosome except E. histolytica, and the genes for both cpn60 and cpn10 are absent in microsporidia (58).
Energy production was postulated to be the driving force behind retention of the endosymbiotic eubacterium, but research on these mitochondrion-related organelles in the last 15 years clearly preclude energy metabolism as the sole factor in selection for organelle retention. ATP appears to be no longer (if ever) produced in all described mitosomes, and in the case of E. cuniculi and potentially others, ATP may actually be supplied to the mitosomes (113). Why then do these remnant organelles continue to sequester at least part of the pathway involved in Fe-S cluster production? Is it the requirement for a membrane potential in maturation of Fe-S proteins which can be maintained within the double-membranes of these organelles? It is unknown why Fe-S cluster formation appears to be tied to an organelle, or why this remains localized in mitosomes that lack an apparent membrane potential. Are there additional, unknown requirements for Fe-S assembly? Furthermore, for hydrogenosomes and mitosomes that have a membrane potential, virtually nothing is known about how this potential is generated. The elaborate inner membrane complexes that generate membrane potential in classical mitochondria are absent, thus other mechanisms must be at play. What are these, and are they conserved across these unusual organelles or have different organelles acquired different mechanisms?
Recent research on mitochondrion-related organelles in unicellular protists has greatly expanded our understanding of the evolution and function of eukaryotic organelles. As answers emerge, even more questions arise, laying a foundation for future studies that promises to reveal further unexpected, biological puzzles.
Literature Cited
- 1.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, et al. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–5. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 2.Aguilera P, Barry T, Tovar J. Entamoeba histolytica mitosomes: organelles in search of a function. Exp Parasitol. 2008;118:10–6. doi: 10.1016/j.exppara.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Ali V, Shigeta Y, Tokumoto U, Takahashi Y, Nozaki T. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J Biol Chem. 2004;279:16863–74. doi: 10.1074/jbc.M313314200. [DOI] [PubMed] [Google Scholar]
- 4.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–40. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 5.Arisue N, Sanchez LB, Weiss LM, Muller M, Hashimoto T. Mitochondrial-type hsp70 genes of the amitochondriate protists, Giardia intestinalis, Entamoeba histolytica and two microsporidians. Parasitol Int. 2002;51:9–16. doi: 10.1016/s1383-5769(01)00093-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakatselou C, Beste D, Kadri AO, Somanath S, Clark CG. Analysis of genes of mitochondrial origin in the genus Entamoeba. J Eukaryot Microbiol. 2003;50:210–4. doi: 10.1111/j.1550-7408.2003.tb00119.x. [DOI] [PubMed] [Google Scholar]
- 7.Bakatselou C, Clark CG. A mitochondrial-type hsp70 gene of Entamoeba histolytica. Arch Med Res. 2000;31:S176–7. doi: 10.1016/s0188-4409(00)00211-3. [DOI] [PubMed] [Google Scholar]
- 8.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–6. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 9.Boxma B, de Graaf RM, van der Staay GW, van Alen TA, Ricard G, et al. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–9. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- 10.Bradley PJ, Lahti CJ, Plumper E, Johnson PJ. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 1997;16:3484–93. doi: 10.1093/emboj/16.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown MT, Goldstone HM, Bastida-Corcuera F, Delgadillo-Correa MG, McArthur AG, Johnson PJ. A functionally divergent hydrogenosomal peptidase with protomitochondrial ancestry. Mol Microbiol. 2007;64:1154–63. doi: 10.1111/j.1365-2958.2007.05719.x. [DOI] [PubMed] [Google Scholar]
- 12.Bui ET, Bradley PJ, Johnson PJ. A common evolutionary origin for mitochondria and hydrogenosomes. Proc Natl Acad Sci U S A. 1996;93:9651–6. doi: 10.1073/pnas.93.18.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burri L, Keeling PJ. Protein targeting in parasites with cryptic mitochondria. Int J Parasitol. 2007;37:265–72. doi: 10.1016/j.ijpara.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Burri L, Williams BA, Bursac D, Lithgow T, Keeling PJ. Microsporidian mitosomes retain elements of the general mitochondrial targeting system. Proc Natl Acad Sci U S A. 2006;103:15916–20. doi: 10.1073/pnas.0604109103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–12. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalier-Smith T. The origin of eukaryotic and archaebacterial cells. Ann N Y Acad Sci. 1987;503:17–54. doi: 10.1111/j.1749-6632.1987.tb40596.x. [DOI] [PubMed] [Google Scholar]
- 17.Chan KW, Slotboom DJ, Cox S, Embley TM, Fabre O, et al. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr Biol. 2005;15:737–42. doi: 10.1016/j.cub.2005.02.068. [DOI] [PubMed] [Google Scholar]
- 18.Clark CG, Roger AJ. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc Natl Acad Sci U S A. 1995;92:6518–21. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clemens DL, Johnson PJ. Failure to detect DNA in hydrogenosomes of Trichomonas vaginalis by nick translation and immunomicroscopy. Mol Biochem Parasitol. 2000;106:307–13. doi: 10.1016/s0166-6851(99)00220-0. [DOI] [PubMed] [Google Scholar]
- 20.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci U S A. 2008;105:20356–61. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dagley MJ, Dolezal P, Likic VA, Smid O, Purcell AW, et al. The protein import channel in the outer mitosomal membrane of Giardia intestinalis. Mol Biol Evol. 2009;26:1941–7. doi: 10.1093/molbev/msp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Duve C. The origin of eukaryotes: a reappraisal. Nat Rev Genet. 2007;8:395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 23.de Graaf RM, Duarte I, van Alen TA, Kuiper JW, Schotanus K, et al. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol Biol. 2009;9:287. doi: 10.1186/1471-2148-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dolezal P, Dancis A, Lesuisse E, Sutak R, Hrdy I, et al. Frataxin, a conserved mitochondrial protein, in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2007;6:1431–8. doi: 10.1128/EC.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313:314–8. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 26.Dolezal P, Smid O, Rada P, Zubacova Z, Bursac D, et al. Giardia mitosomes and trichomonad hydrogenosomes share a common mode of protein targeting. Proc Natl Acad Sci U S A. 2005;102:10924–9. doi: 10.1073/pnas.0500349102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to organelles. Science. 2004;304:253–7. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 28.Dyall SD, Johnson PJ. Origins of hydrogenosomes and mitochondria: evolution and organelle biogenesis. Curr Opin Microbiol. 2000;3:404–11. doi: 10.1016/s1369-5274(00)00112-0. [DOI] [PubMed] [Google Scholar]
- 29.Dyall SD, Koehler CM, Delgadillo-Correa MG, Bradley PJ, Plumper E, et al. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol Cell Biol. 2000;20:2488–97. doi: 10.1128/mcb.20.7.2488-2497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyall SD, Lester DC, Schneider RE, Delgadillo-Correa MG, Plumper E, et al. Trichomonas vaginalis Hmp35, a putative pore-forming hydrogenosomal membrane protein, can form a complex in yeast mitochondria. J Biol Chem. 2003;278:30548–61. doi: 10.1074/jbc.M304032200. [DOI] [PubMed] [Google Scholar]
- 31.Dyall SD, Yan W, Delgadillo-Correa MG, Lunceford A, Loo JA, et al. Non-mitochondrial complex I proteins in a hydrogenosomal oxidoreductase complex. Nature. 2004;431:1103–7. doi: 10.1038/nature02990. [DOI] [PubMed] [Google Scholar]
- 32.Embley TM, Hirt RP. Early branching eukaryotes? Curr Opin Genet Dev. 1998;8:624–9. doi: 10.1016/s0959-437x(98)80029-4. [DOI] [PubMed] [Google Scholar]
- 33.Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–30. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- 34.Endo T, Yamano K. Multiple pathways for mitochondrial protein traffic. Biol Chem. 2009;390:723–30. doi: 10.1515/BC.2009.087. [DOI] [PubMed] [Google Scholar]
- 35.Esser C, Ahmadinejad N, Wiegand C, Rotte C, Sebastiani F, et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–60. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 36.Germot A, Philippe H, Le Guyader H. Presence of a mitochondrial-type 70-kDa heat shock protein in Trichomonas vaginalis suggests a very early mitochondrial endosymbiosis in eukaryotes. Proc Natl Acad Sci U S A. 1996;93:14614–7. doi: 10.1073/pnas.93.25.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germot A, Philippe H, Le Guyader H. Evidence for loss of mitochondria in Microsporidia from a mitochondrial-type HSP70 in Nosema locustae. Mol Biochem Parasitol. 1997;87:159–68. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 38.Ghosh S, Field J, Rogers R, Hickman M, Samuelson J. The Entamoeba histolytica mitochondrion-derived organelle (crypton) contains double-stranded DNA and appears to be bound by a double membrane. Infect Immun. 2000;68:4319–22. doi: 10.1128/iai.68.7.4319-4322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gill EE, Diaz-Trivino S, Barbera MJ, Silberman JD, Stechmann A, et al. Novel mitochondrion-related organelles in the anaerobic amoeba Mastigamoeba balamuthi. Mol Microbiol. 2007;66:1306–20. doi: 10.1111/j.1365-2958.2007.05979.x. [DOI] [PubMed] [Google Scholar]
- 40.Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, et al. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–8. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 41.Gray MW. Evolutionary biology: the hydrogenosome’s murky past. Nature. 2005;434:29–31. doi: 10.1038/434029a. [DOI] [PubMed] [Google Scholar]
- 42.Gupta RS. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol Microbiol. 1995;15:1–11. doi: 10.1111/j.1365-2958.1995.tb02216.x. [DOI] [PubMed] [Google Scholar]
- 43.Gupta RS, Aitken K, Falah M, Singh B. Cloning of Giardia lamblia heat shock protein HSP70 homologs: implications regarding origin of eukaryotic cells and of endoplasmic reticulum. Proc Natl Acad Sci U S A. 1994;91:2895–9. doi: 10.1073/pnas.91.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hackstein JHP, Baker SE, Hellemond JJv, Tielens AGM. Hydrogenosomes of Anaerobic Chytrids: An Alternative Way to Adapt to Anaerobic Environments. In: Tachezy J, editor. Hydrogenosomes and Mitosomes: Mitochodria of Anaerobic Eukaryotes. New York: Springer; 2008. pp. 147–62. [Google Scholar]
- 45.Hackstein JHP, Graaf RMd, Hellemond JJv, Tielens AGM. Hydrogenosomes of Anaerobic Ciliates. In: Tachezy J, editor. Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes. New York: Springer; 2008. pp. 97–112. [Google Scholar]
- 46.Hampl V, Hug L, Leigh JW, Dacks JB, Lang BF, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106:3859–64. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hampl V, Silberman JD, Stechmann A, Diaz-Trivino S, Johnson PJ, Roger AJ. Genetic evidence for a mitochondriate ancestry in the ‘amitochondriate’ flagellate Trimastix pyriformis. PLoS One. 2008;3:e1383. doi: 10.1371/journal.pone.0001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hausler T, Stierhof YD, Blattner J, Clayton C. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur J Cell Biol. 1997;73:240–51. [PubMed] [Google Scholar]
- 49.Henze K. The Proteome of T. vaginalis Hydrogenosomes. In: Tachezy J, editor. Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes. New York: Springer; 2008. pp. 163–78. [Google Scholar]
- 50.Herrmann JM. Converting bacteria to organelles: evolution of mitochondrial protein sorting. Trends Microbiol. 2003;11:74–9. doi: 10.1016/s0966-842x(02)00033-1. [DOI] [PubMed] [Google Scholar]
- 51.Hirt RP, Healy B, Vossbrinck CR, Canning EU, Embley TM. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: molecular evidence that microsporidia once contained mitochondria. Curr Biol. 1997;7:995–8. doi: 10.1016/s0960-9822(06)00420-9. [DOI] [PubMed] [Google Scholar]
- 52.Hirt RP, Logsdon JM, Jr, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci U S A. 1999;96:580–5. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honigberg BM, Volkmann D, Entzeroth R, Scholtyseck E. A freeze-fracture electron microscope study of Trichomonas vaginalis Donne and Tritrichomonas foetus (Riedmuller) J Protozool. 1984;31:116–31. doi: 10.1111/j.1550-7408.1984.tb04300.x. [DOI] [PubMed] [Google Scholar]
- 54.Horner DS, Hirt RP, Kilvington S, Lloyd D, Embley TM. Molecular data suggest an early acquisition of the mitochondrion endosymbiont. Proc Biol Sci. 1996;263:1053–9. doi: 10.1098/rspb.1996.0155. [DOI] [PubMed] [Google Scholar]
- 55.Hrdy I, Hirt RP, Dolezal P, Bardonova L, Foster PG, et al. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–22. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 56.Johnson PJ, d’Oliveira CE, Gorrell TE, Muller M. Molecular analysis of the hydrogenosomal ferredoxin of the anaerobic protist Trichomonas vaginalis. Proc Natl Acad Sci U S A. 1990;87:6097–101. doi: 10.1073/pnas.87.16.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson PJ, Lahti CJ, Bradley PJ. Biogenesis of the hydrogenosome in the anaerobic protist Trichomonas vaginalis. J Parasitol. 1993;79:664–70. [PubMed] [Google Scholar]
- 58.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–3. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 59.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–6. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 60.Keeling PJ, Palmer JD. Parabasalian flagellates are ancient eukaryotes. Nature. 2000;405:635–7. doi: 10.1038/35015167. [DOI] [PubMed] [Google Scholar]
- 61.Kitada S, Uchiyama T, Funatsu T, Kitada Y, Ogishima T, Ito A. A protein from a parasitic microorganism, Rickettsia prowazekii, can cleave the signal sequences of proteins targeting mitochondria. J Bacteriol. 2007;189:844–50. doi: 10.1128/JB.01261-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kunji ER. The role and structure of mitochondrial carriers. FEBS Lett. 2004;564:239–44. doi: 10.1016/S0014-5793(04)00242-X. [DOI] [PubMed] [Google Scholar]
- 63.Kusmierczyk AR, Martin J. Assembly of chaperonin complexes. Mol Biotechnol. 2001;19:141–52. doi: 10.1385/MB:19:2:141. [DOI] [PubMed] [Google Scholar]
- 64.LaGier MJ, Tachezy J, Stejskal F, Kutisova K, Keithly JS. Mitochondrial-type iron-sulfur cluster biosynthesis genes (IscS and IscU) in the apicomplexan Cryptosporidium parvum. Microbiology. 2003;149:3519–30. doi: 10.1099/mic.0.26365-0. [DOI] [PubMed] [Google Scholar]
- 65.Lantsman Y, Tan KS, Morada M, Yarlett N. Biochemical characterization of a mitochondrial-like organelle from Blastocystis sp. subtype 7. Microbiology. 2008;154:2757–66. doi: 10.1099/mic.0.2008/017897-0. [DOI] [PubMed] [Google Scholar]
- 66.Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–8. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- 67.Lill R, Kispal G. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci. 2000;25:352–6. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- 68.Lill R, Muhlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 69.Lindmark DG, Muller M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem. 1973;248:7724–8. [PubMed] [Google Scholar]
- 70.Lloyd D, Harris JC, Maroulis S, Wadley R, Ralphs JR, et al. The “primitive” microaerophile Giardia intestinalis (syn. lamblia, duodenalis) has specialized membranes with electron transport and membrane-potential-generating functions. Microbiology. 2002;148:1349–54. doi: 10.1099/00221287-148-5-1349. [DOI] [PubMed] [Google Scholar]
- 71.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–8. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Garcia P, Moreira D. Metabolic symbiosis at the origin of eukaryotes. Trends Biochem Sci. 1999;24:88–93. doi: 10.1016/s0968-0004(98)01342-5. [DOI] [PubMed] [Google Scholar]
- 73.Mai Z, Ghosh S, Frisardi M, Rosenthal B, Rogers R, Samuelson J. Hsp60 is targeted to a cryptic mitochondrion-derived organelle (“crypton”) in the microaerophilic protozoan parasite Entamoeba histolytica. Mol Cell Biol. 1999;19:2198–205. doi: 10.1128/mcb.19.3.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, et al. Bacterial-type oxygen detoxification and iron-sulphur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2009 doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- 75.Margulis L. Origin of eukaryotic cells; evidence and research implications for a theory of the origin and evolution of microbial, plant, and animal cells on the Precambrian earth. xxii. New Haven: Yale University Press; 1970. p. 349. [Google Scholar]
- 76.Margulis L, Chapman M, Guerrero R, Hall J. The last eukaryotic common ancestor (LECA): acquisition of cytoskeletal motility from aerotolerant spirochetes in the Proterozoic Eon. Proc Natl Acad Sci U S A. 2006;103:13080–5. doi: 10.1073/pnas.0604985103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382:1521–39. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- 78.Martin W, Muller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392:37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 79.Mentel M, Zimorski V, Haferkamp P, Martin W, Henze K. Protein import into hydrogenosomes of Trichomonas vaginalis involves both N-terminal and internal targeting signals: a case study of thioredoxin reductases. Eukaryot Cell. 2008;7:1750–7. doi: 10.1128/EC.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mi-Ichi F, Yousuf MA, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moreira D, Lopez-Garcia P. Symbiosis between methanogenic archaea and delta-proteobacteria as the origin of eukaryotes: the syntrophic hypothesis. J Mol Evol. 1998;47:517–30. doi: 10.1007/pl00006408. [DOI] [PubMed] [Google Scholar]
- 82.Morrison HG, Roger AJ, Nystul TG, Gillin FD, Sogin ML. Giardia lamblia expresses a proteobacterial-like DnaK homolog. Mol Biol Evol. 2001;18:530–41. doi: 10.1093/oxfordjournals.molbev.a003832. [DOI] [PubMed] [Google Scholar]
- 83.Mukherjee M, Brown MT, McArthur AG, Johnson PJ. Proteins of the glycine decarboxylase complex in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2006;5:2062–71. doi: 10.1128/EC.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mukherjee M, Sievers SA, Brown MT, Johnson PJ. Identification and biochemical characterization of serine hydroxymethyl transferase in the hydrogenosome of Trichomonas vaginalis. Eukaryot Cell. 2006;5:2072–8. doi: 10.1128/EC.00249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muller M. The hydrogenosome. J Gen Microbiol. 1993;139:2879–89. doi: 10.1099/00221287-139-12-2879. [DOI] [PubMed] [Google Scholar]
- 86.Nasirudeen AM, Tan KS. Isolation and characterization of the mitochondrion-like organelle from Blastocystis hominis. J Microbiol Methods. 2004;58:101–9. doi: 10.1016/j.mimet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 87.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–49. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 88.Perez-Brocal V, Clark CG. Analysis of two genomes from the mitochondrion-like organelle of the intestinal parasite Blastocystis: complete sequences, gene content, and genome organization. Mol Biol Evol. 2008;25:2475–82. doi: 10.1093/molbev/msn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peyretaillade E, Broussolle V, Peyret P, Metenier G, Gouy M, Vivares CP. Microsporidia, amitochondrial protists, possess a 70-kDa heat shock protein gene of mitochondrial evolutionary origin. Mol Biol Evol. 1998;15:683–9. doi: 10.1093/oxfordjournals.molbev.a025971. [DOI] [PubMed] [Google Scholar]
- 90.Putignani L, Tait A, Smith HV, Horner D, Tovar J, et al. Characterization of a mitochondrion-like organelle in Cryptosporidium parvum. Parasitology. 2004;129:1–18. doi: 10.1017/s003118200400527x. [DOI] [PubMed] [Google Scholar]
- 91.Putz S, Gelius-Dietrich G, Piotrowski M, Henze K. Rubrerythrin and peroxiredoxin: two novel putative peroxidases in the hydrogenosomes of the microaerophilic protozoon Trichomonas vaginalis. Mol Biochem Parasitol. 2005;142:212–23. doi: 10.1016/j.molbiopara.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 92.Rada P, Smid O, Sutak R, Dolezal P, Pyrih J, et al. The monothiol single-domain glutaredoxin is conserved in the highly reduced mitochondria of Giardia intestinalis. Eukaryot Cell. 2009;8:1584–91. doi: 10.1128/EC.00181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reeves RE, Warren LG, Susskind B, Lo HS. An energy-conserving pyruvate-to-acetate pathway in Entamoeba histolytica. Pyruvate synthase and a new acetate thiokinase. J Biol Chem. 1977;252:726–31. [PubMed] [Google Scholar]
- 94.Regoes A, Zourmpanou D, Leon-Avila G, van der Giezen M, Tovar J, Hehl AB. Protein import, replication, and inheritance of a vestigial mitochondrion. J Biol Chem. 2005;280:30557–63. doi: 10.1074/jbc.M500787200. [DOI] [PubMed] [Google Scholar]
- 95.Riordan CE, Ault JG, Langreth SG, Keithly JS. Cryptosporidium parvum Cpn60 targets a relict organelle. Curr Genet. 2003;44:138–47. doi: 10.1007/s00294-003-0432-1. [DOI] [PubMed] [Google Scholar]
- 96.Rivera MC, Jain R, Moore JE, Lake JA. Genomic evidence for two functionally distinct gene classes. Proc Natl Acad Sci U S A. 1998;95:6239–44. doi: 10.1073/pnas.95.11.6239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roberts CW, Roberts F, Henriquez FL, Akiyoshi D, Samuel BU, et al. Evidence for mitochondrial-derived alternative oxidase in the apicomplexan parasite Cryptosporidium parvum: a potential anti-microbial agent target. Int J Parasitol. 2004;34:297–308. doi: 10.1016/j.ijpara.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 98.Roger AJ, Clark CG, Doolittle WF. A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis. Proc Natl Acad Sci U S A. 1996;93:14618–22. doi: 10.1073/pnas.93.25.14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Roger AJ, Svard SG, Tovar J, Clark CG, Smith MW, et al. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc Natl Acad Sci U S A. 1998;95:229–34. doi: 10.1073/pnas.95.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rospert S, Looser R, Dubaquie Y, Matouschek A, Glick BS, Schatz G. Hsp60-independent protein folding in the matrix of yeast mitochondria. EMBO J. 1996;15:764–74. [PMC free article] [PubMed] [Google Scholar]
- 101.Slapeta J, Keithly JS. Cryptosporidium parvum mitochondrial-type HSP70 targets homologous and heterologous mitochondria. Eukaryot Cell. 2004;3:483–94. doi: 10.1128/EC.3.2.483-494.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smid O, Matuskova A, Harris SR, Kucera T, Novotny M, et al. Reductive evolution of the mitochondrial processing peptidases of the unicellular parasites trichomonas vaginalis and giardia intestinalis. PLoS Pathog. 2008;4:e1000243. doi: 10.1371/journal.ppat.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sogin ML. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991;1:457–63. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 104.Soltys BJ, Gupta RS. Presence and cellular distribution of a 60-kDa protein related to mitochondrial hsp60 in Giardia lamblia. J Parasitol. 1994;80:580–90. [PubMed] [Google Scholar]
- 105.Stechmann A, Hamblin K, Perez-Brocal V, Gaston D, Richmond GS, et al. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–5. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tachezy J, Sanchez LB, Muller M. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol Biol Evol. 2001;18:1919–28. doi: 10.1093/oxfordjournals.molbev.a003732. [DOI] [PubMed] [Google Scholar]
- 107.Tachezy J, Smid O. Mitosomes in Parasitic Protists. In: Tachezy J, editor. Hydrogenosomes and Mitosomes: Mitochondria of Anaerobic Eukaryotes. New York: Springer; 2008. pp. 201–30. [Google Scholar]
- 108.Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, et al. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9:615–25. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- 109.Tielens AG, Rotte C, van Hellemond JJ, Martin W. Mitochondria as we don’t know them. Trends Biochem Sci. 2002;27:564–72. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- 110.Tovar J, Cox SS, van der Giezen M. A mitosome purification protocol based on percoll density gradients and its use in validating the mitosomal nature of Entamoeba histolytica mitochondrial Hsp70. Methods Mol Biol. 2007;390:167–77. doi: 10.1007/978-1-59745-466-7_11. [DOI] [PubMed] [Google Scholar]
- 111.Tovar J, Fischer A, Clark CG. The mitosome, a novel organelle related to mitochondria in the amitochondrial parasite Entamoeba histolytica. Mol Microbiol. 1999;32:1013–21. doi: 10.1046/j.1365-2958.1999.01414.x. [DOI] [PubMed] [Google Scholar]
- 112.Tovar J, Leon-Avila G, Sanchez LB, Sutak R, Tachezy J, et al. Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature. 2003;426:172–6. doi: 10.1038/nature01945. [DOI] [PubMed] [Google Scholar]
- 113.Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–6. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 114.van der Giezen M. Hydrogenosomes and mitosomes: conservation and evolution of functions. J Eukaryot Microbiol. 2009;56:221–31. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 115.van der Giezen M, Cox S, Tovar J. The iron-sulfur cluster assembly genes iscS and iscU of Entamoeba histolytica were acquired by horizontal gene transfer. BMC Evol Biol. 2004;4:7. doi: 10.1186/1471-2148-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van der Giezen M, Leon-Avila G, Tovar J. Characterization of chaperonin 10 (Cpn10) from the intestinal human pathogen Entamoeba histolytica. Microbiology. 2005;151:3107–15. doi: 10.1099/mic.0.28068-0. [DOI] [PubMed] [Google Scholar]
- 117.van der Giezen M, Slotboom DJ, Horner DS, Dyal PL, Harding M, et al. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 2002;21:572–9. doi: 10.1093/emboj/21.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Waller RF, Jabbour C, Chan NC, Celik N, Likic VA, et al. Evidence of a reduced and modified mitochondrial protein import apparatus in microsporidian mitosomes. Eukaryot Cell. 2009;8:19–26. doi: 10.1128/EC.00313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Walter S, Buchner J. Molecular chaperones--cellular machines for protein folding. Angew Chem Int Ed Engl. 2002;41:1098–113. doi: 10.1002/1521-3773(20020402)41:7<1098::aid-anie1098>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 120.Wawrzyniak I, Roussel M, Diogon M, Couloux A, Texier C, et al. Complete circular DNA in the mitochondria-like organelles of Blastocystis hominis. Int J Parasitol. 2008;38:1377–82. doi: 10.1016/j.ijpara.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 121.Williams BA, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–9. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 122.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985;82:4443–7. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yutin N, Makarova KS, Mekhedov SL, Wolf YI, Koonin EV. The deep archaeal roots of eukaryotes. Mol Biol Evol. 2008;25:1619–30. doi: 10.1093/molbev/msn108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zierdt CH. Cytochrome-free mitochondria of an anaerobic protozoan--Blastocystis hominis. J Protozool. 1986;33:67–9. doi: 10.1111/j.1550-7408.1986.tb05559.x. [DOI] [PubMed] [Google Scholar]
- 126.Zierdt CH, Donnolley CT, Muller J, Constantopoulos G. Biochemical and ultrastructural study of Blastocystis hominis. J Clin Microbiol. 1988;26:965–70. doi: 10.1128/jcm.26.5.965-970.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]