Abstract
While mechanisms of resistance to major antifungal agents have been characterized in Candida albicans, little is known about the evolutionary trajectories during the emergence of drug resistance. Here, we examined the evolutionary dynamics of C. albicans that evolved in vitro in the presence or absence of fluconazole using the visualizing evolution in real-time (VERT) method, a novel experimental approach that facilitates the systematic isolation of adaptive mutants that arise in the population. We found an increase in the frequency of adaptive events in the presence of fluconazole compared to the no-drug controls. Analysis of the evolutionary dynamics revealed that mutations that led to increased drug resistance appeared frequently and that mutants with increased levels of resistance arose in independent lineages. Interestingly, most adaptive mutants with increased fitness in the presence of the drug did not exhibit a significant fitness decrease in the absence of the drug, supporting the idea that rapid resistance can arise from mutations in strains maintained in the population prior to exposure to the drug.
INTRODUCTION
The emergence of antimicrobial drug resistance in pathogens is a process of adaptive evolution, generally as a result of genetic mutations. Depending on the size of the population, the rate of mutation, and the relative fitness coefficients, the population may be heterogeneous, consisting of multiple resistant genotypes competing for expansion in a process called “clonal interference” (10, 12). Determining the evolutionary dynamics during adaptation is important for understanding the fundamental principles underlying how eukaryotic microbes evolve resistance to antimicrobial agents, a process that differs from that in bacteria because it is unlikely to involve horizontal gene transfer (1, 6).
It is important to understand the frequencies with which adaptive mutants arise and expand, the evolutionary trajectories (the order of occurrence of adaptive mutations), and the potential convergence or divergence in the adaptive mechanisms between parallel populations in order to better appreciate how drug resistance can emerge in pathogens growing within the host. For example, knowledge of the frequency and order in which drug-resistant mutants arise in the population and whether the early-arising resistance mechanisms play a role in the level of drug resistance ultimately reached in the population can be used to predict the likely trajectory of a clinical infection and to develop appropriate therapeutic strategies.
Candida albicans is the fourth most common cause of nosocomial infections, resulting in rates of mortality in U.S. hospitals that approach 50% (23, 24, 44). Emergence of resistance to almost all major antifungal agents used in treating C. albicans infections has been reported (15, 22, 29). Among the existing classes of antifungal drugs, azoles are the most commonly used, due to their low toxicity and their oral availability. The fact that they are fungistatic rather than fungicidal provides a clear opportunity for the evolution of drug resistance during long periods of prophylactic treatment (25).
The mechanisms of resistance to azole drugs have been studied extensively (18, 19, 21, 27, 28, 32, 36, 37, 40, 42, 43). However, a large percentage (50%) of drug-resistant clinical isolates from a reported collection have unknown mechanisms of resistance (40). In C. albicans, resistance has arisen both in vitro and in vivo and includes point mutations, loss of heterozygosity, and/or gross chromosomal-rearrangement mechanisms, (32, 34, 35). Studies of drug-resistant isolates from patients during the course of treatment, as well as from long-term laboratory evolution experiments, suggest that evolving populations are heterogeneous, include multiple resistance mechanisms (9, 35, 38, 39), and likely involve complex population dynamics. For example, in a series of 9 C. albicans isolates from a bone marrow transplant patient undergoing antifungal drug treatments, multiple resistance mechanisms were present and competing (34), further highlighting the need to understand the dynamics during the emergence of drug resistance.
Here, we report the first study to monitor the rise and expansion of adaptive events in C. albicans during in vitro evolution in continuous cultures under increasing concentrations of fluconazole. We used the visualizing evolution in real-time (VERT) approach, which was originally developed for analyzing evolution in the model eukaryote Saccharomyces cerevisiae (14). In VERT, a set of isogenic strains differing only in their expression of different fluorescent proteins (FP) is used to visualize when adaptive events occur (the rise and expansion of adaptive mutants), to determine how frequently they occur, and to facilitate the identification and isolation of adaptive mutants from the population. The evolutionary dynamics indicate that adaptive events occur more frequently in the presence of the drug than in the control populations that evolved in the absence of the drug. Adaptive clones isolated from each of the observed adaptive events and relative fitness coefficient measurements were used to estimate the fitness costs associated with increases in the level of drug resistance. Importantly, increases in drug resistance were not necessarily accompanied by significant fitness costs and in some cases were accompanied by fitness increases.
MATERIALS AND METHODS
Strains.
All strains used in this work are isogenic derivatives of the C. albicans strain SC5314. Three different fluorescent cassettes, each containing a NAT1 marker and a fluorescent protein (either red fluorescent protein [RFP], green fluorescent protein [GFP], or yellow fluorescent protein [YFP]), were amplified by PCR from plasmids pMG2261, pMG2120, and pMG2263, respectively, using two different sets of primers. The primers for PCR amplifying GFP and YFP cassettes were 5′-GAG AAT CGA AGA AGA ATT AGG TTC TGA AGC TAT CTA CGC TGG TAA AGA TTT CCA AAA GGC TTC TCA ATT GGG TGG TGG TTC TAA AGG TGA AGA ATT ATT-3′ and 5′-TTT AAT TAG TTC ATA TAT TCA AGA TGT TCC TAT AAA AGA AAA AAA AAG CAC CAG CTT TTT TTT ATT TAA TCG TAA AAC GAC GGC CAG TGA ATT C-3′. The primers for amplifying the RFP cassette were 5′-GAG AAT CGA AGA AGA ATT AGG TTC TGA AGC TAT CTA CGC TGG TAA AGA TTT CCA AAA GGC TTC TCA ATT GGG TGG TGG TGA TAA CAC TGA AGA TGT TAT T-3′ and 5′-TTT AAT TAG TTC ATA TAT TCA AGA TGT TCC TAT AAA AGA AAA AAA AAG CAC CAG CTT TTT TTT ATT TAA TCG TAA AAC GAC GGC CAG TGA ATT C-3′. The resulting PCR products were then transformed into strain SC5314 to form a translational fusion of fluorescent cassettes to the 3′ end of the ENO1 gene in the chromosome by homologous recombination. Transformants were selected on yeast extract-peptone-dextrose plus adenine (YPAD) agar plates with nourseothricin (400 μg/ml), and the fluorescence of each strain was confirmed by microscopy.
The strains were grown in yeast nitrogen base (YNB) without amino acids supplemented with 0.5% (wt/vol) ammonium sulfate and 2% (wt/vol) d-glucose unless otherwise specified.
Laboratory evolution experiments.
Cells expressing each fluorescent protein (GFP, YFP, or RFP) were mixed together in approximately equal numbers (as confirmed by flow cytometry) at the start of parallel chemostat (30 ml each) evolution experiments in YNB supplemented with 0.5% (wt/vol) ammonium sulfate and 0.2% dextrose (the limiting nutrient) with or without fluconazole. The populations evolved at steady state at 30°C at a dilution rate (D) of ∼0.2 h−1. In chemostat experiments, the generation time is determined by the dilution rate. Using a dilution rate of 0.2 h−1, the generation time is calculated as ln(2)/0.2, or 3.5 h/generation. Two parallel series of evolution experiments were conducted; each series contained one control population (no fluconazole) and one population exposed to stepwise increases of fluconazole, starting with 0.25 μg per ml and increasing the concentration by 4-fold every 50 to 70 generations. Samples were taken every 7 to 14 generations, and a portion (1.5 ml) of the cells were frozen in 17% glycerol for future analysis. Cell concentrations were measured using a hemocytometer, and the relative proportions of the three subpopulations (GFP, YFP, and RFP) were measured using a FACScan flow cytometer (BD Biosciences, San Jose, CA).
Isolation of adaptive mutants.
At the conclusion of the evolution experiment, we identified the time point samples likely to have a substantial fraction of an adaptive clone in one of the fluorescently marked subpopulations, based on the dynamics of the relative subpopulation sizes. Specifically, we identified the time point sample in which an expanding colored FP subpopulation appeared to reach its maximum proportion of the population. Population samples from the generation at which the expanding subpopulation reached a maximum proportion were plated on YNB plates. Using a Dark Reader (Clare Chemical), which can distinguish GFP-, YFP-, and RFP-expressing cells, at least seven isolates expressing the colored fluorescent protein of the expanding subpopulation were picked for fitness measurements. For all isolates from the same adaptive event, we performed pairwise competition experiments by growing each isolate in continuous culture together with the previous adaptive mutant (for the first observed adaptive event, the isolates were individually competed against the parental strain of a different FP). Figure S1 in the supplemental material shows the flow of the process. The experiments were carried out in the presence of the same concentration of fluconazole used during the evolution of those isolates. Of the seven or more clones tested from each observed adaptive event, the clone with the highest fitness coefficient was chosen as the adaptive clone for that adaptive event, except in cases where multiple isolates exhibited similar fitness coefficients. In those cases, an arbitrary isolate was chosen from the fittest group.
Pairwise competition experiments.
Chemostat experiments in YNB with 0.2% dextrose and 0.5% (wt/vol) ammonium sulfate and in the presence or absence of fluconazole were initiated with approximately equal numbers of the two clones to be competed (each carrying a different FP marker). The pairwise competition experiments were conducted at 30°C with a dilution rate of ∼0.2 h−1. Samples were taken every 1 to 5 generations, and the relative proportions of each of the two clones were measured using flow cytometry. The data were plotted on a linear scale with ln[x/(1 − x)] versus t, where x is the fraction of the sample clone and t is generations. The relative fitness coefficient was determined from the slope of the linear region of the curve using linear regression analysis. For statistical analysis, the relative fitness coefficients between pairs of the fluorescently labeled parental strains were measured either in the absence of fluconazole or in the presence of 0.25 μg/ml of fluconazole. A minimum of three biological replicates were performed for each pairwise competition experiment. A Student t test was used to determine whether the relative fitness coefficients measured for the adaptive mutants were statistically significant (P value cutoff, 0.05).
MIC measurements.
The fluconazole MIC for each isolated adaptive mutant was determined with an Etest strip according to the manufacturer's instructions (bioMérieux SA, France). The highest MIC value from 3 biological replicates was reported.
Stability analysis of adaptive mutants.
One of the parent strains (KKCa-14) and all isolated adaptive mutants from the DP-1 and DP-2 populations underwent stability tests in the absence of fluconazole. The strains were first plated on YNB plates supplemented with 2% dextrose, and a single colony was picked up and transferred into fresh YNB broth. Five microliters of cell culture for each strain was inoculated into 5 ml fresh YNB broth to make a 1,000-fold dilution and then was incubated at 30°C for 24 h. The transfer was repeated for two more days (for a total of ∼30 generations). The optical density of the cell culture was measured at 600 nm before each transfer. MIC measurements were performed for each strain to test the stability of the drug resistance after long-term incubation in serial batch transfer in the absence of the drug.
CHEF karyotype analysis.
Preparation of cells for contour-clamped homogeneous electric field (CHEF) karyotype analysis was performed as described previously (31). Whole-chromosome separation was performed on a Bio-Rad CHEF-DR III with the following program: 60- to 120-s switch, 6 V/cm, 120° angle for 36 h, followed by a 120- to 300-s switch, 4.5 V/cm, 120° angle for 12 h. SfiI digestion of CHEF gel plugs was performed using the following procedure. A portion of whole-chromosome CHEF plugs were washed twice for 10 min each time in 1 ml of 10 mM Tris (pH 8.0), 1 mM EDTA (TE buffer) and then incubated in 1 ml of TE buffer containing 0.1 mM phenylmethylsulfonyl fluoride for 10 min. The plugs were washed twice as before and incubated in 500 μl of SfiI restriction enzyme buffer containing bovine serum albumin (BSA) at 100 μg/ml for 15 min, and 40 units of SfiI enzyme was added. The SfiI-treated plugs were then incubated overnight at 50°C, washed in 100 mM EDTA (pH 8.0), added to a 1% Megabase agarose gel (Bio-Rad, Hercules, CA) in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA), and separated as previously described (35).
Ploidy analysis.
Strain ploidy was examined by flow cytometry. C. albicans strains were grown to log phase, and ∼4 × 106 cells were collected in a round-bottom microtiter plate by centrifugation. The pellets were resuspended in 20 μl of 50 mM Tris, pH 8, 50 mM EDTA (50/50 TE); 180 μl of 95% ethanol was added dropwise, and the suspensions were stored overnight at −20°C. The ethanol-fixed cell pellets were washed twice with 200 μl of 50/50 TE, resuspended in 200 μl of RNase A at 500 μg/ml in 50/50 TE, and incubated for 2 h at 37°C. The cells were centrifuged again, and the pellets were incubated in 200 μl of 1 mg/ml proteinase K in 50/50 TE for 2 h at 37°C. The cells were centrifuged again and washed in 50/50 TE, and the pellets were resuspended in 50 μl of Sybr green I (1:85 dilution; Invitrogen, Carlsbad, CA) in 50/50 TE and incubated overnight in the dark. Finally, the cells were centrifuged, the pellets were resuspended in 700 μl 50/50 TE, and ploidy was detected and analyzed on a FACScaliber flow cytometer (BD Biosciences, San Jose, CA).
RESULTS
Adaptive evolution in the presence of fluconazole.
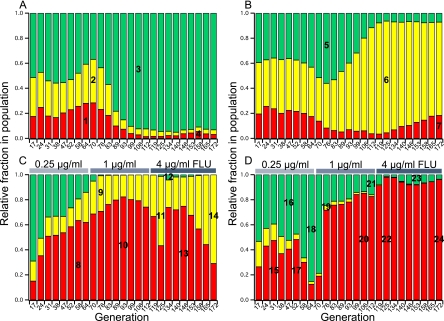
Two sets of parallel populations of C. albicans were allowed to evolve either in the absence (control populations, CP-1 and CP-2) or in the presence (DP-1 and DP-2) of increasing concentrations of fluconazole. The VERT system was used to follow the evolutionary dynamics of three subpopulations within each parallel population. At the start of the VERT experiment, the populations were initiated with approximately equal numbers of each FP cell type. The relative proportions of each FP subpopulation were measured using flow cytometry. For the fluconazole-exposed populations (DP-1 and DP-2), the concentration of fluconazole at the start of the experiment was 0.25 μg/ml, which is equal to twice the MIC of the parental strains. At generation 64, the fluconazole concentration had increased to 1 μg/ml, and at generation 112, it had further increased to 4 μg/ml. The evolutionary dynamics for all 4 populations over approximately 170 generations are shown in Fig. 1.
Fig. 1.
Evolutionary dynamics of experimental populations evolved either in the absence (CP-1 population [A] and CP-2 population [B]) or presence (DP-1 population [C] and DP-2 population [D]) of fluconazole. The colored bars represent the relative fractions of each colored subpopulation, RFP (red), GFP (green), and YFP (yellow), as determined using fluorescence-activated cell sorting (FACS). FLU, concentration of fluconazole in the feed. The subpopulations from which the adaptive mutants were isolated are numbered (details are in Table 1).
An observed expansion of a particular FP subpopulation is indicative of an adaptive event within that subpopulation, and clones that were likely to be responsible for the observed expansions were isolated and named according to the population from which they were isolated. For example, clone CP-1-M1 was the adaptive mutant isolated from the first observed expansion (M1) from the CP-1 population. Based on the evolutionary dynamics observed, an adaptive event occurred every ∼20 to 50 generations in the controls and every ∼10 to 40 generations in the fluconazole-exposed populations. Thus, the number of adaptive events observed over ∼170 generations was significantly higher in the populations exposed to drug than in those not exposed to drug, with approximately 7 and 10 adaptive events observed in the two fluconazole-exposed populations and 3 and 4 adaptive events observed in the control populations.
Dynamics of evolution in the presence of fluconazole.
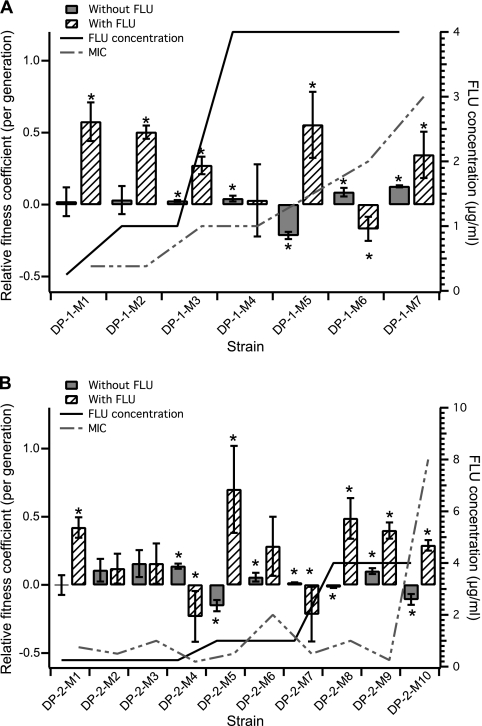
We expected that exposure of an expanding subpopulation to increased fluconazole concentrations would result in continued expansion of that FP subpopulation, as the most competitive clone is most likely present within the subpopulation that is expanding. Indeed, this expected trend was observed in the DP-1 population. Upon the first increase in the fluconazole concentration, the YFP subpopulation, which started expanding at approximately generation 58, continued to expand following the increase in the drug concentration at generation 64. In addition, the RFP subpopulation began to expand at this time point, suggesting that an adaptive clone arose in this subpopulation, as well. Indeed, the adaptive mutant isolated after the first increase in the fluconazole concentration (DP-1-M2) showed increased fitness compared to both the previous adaptive mutant (DP-1-M1) and the parental strain (KKCa-20) in the presence of the drug (Fig. 2 and 3 and Tables 1 and 2), despite the fact that the MIC for fluconazole was 0.38 μg/ml for both DP-1-M1 and DP-1-M2. Upon the second increase in the fluconazole concentration, at generation 112, the expanding YFP subpopulation again continued to expand as expected, suggesting that the expanding subpopulations prior to each increase in the drug concentration were composed of the most drug-resistant mutants. However, the isolated adaptive mutant after the second increase in the drug concentration (DP-1-M4) was not more fit than the previous adaptive mutant (DP-1-M3) in the presence of fluconazole but was slightly more fit in the absence of the drug.
Fig. 2.
Relative fitness coefficients of adaptive mutants isolated from the fluconazole-exposed populations compared with the previous adaptive mutant, DP-1 (A) and DP-2 (B), in the presence of fluconazole (hatched bars), measured at the drug concentration indicated by the solid line, or without fluconazole (solid bars). The data shown are the averages and standard deviations (SD) of a minimum of 3 biological replicates. The asterisks indicate that the relative fitness difference measured in the adaptive strains is statistically significant (P < 0.05, using an unpaired Student t test with unequal variance). The dashed line indicates the measured MIC of each adaptive mutant.
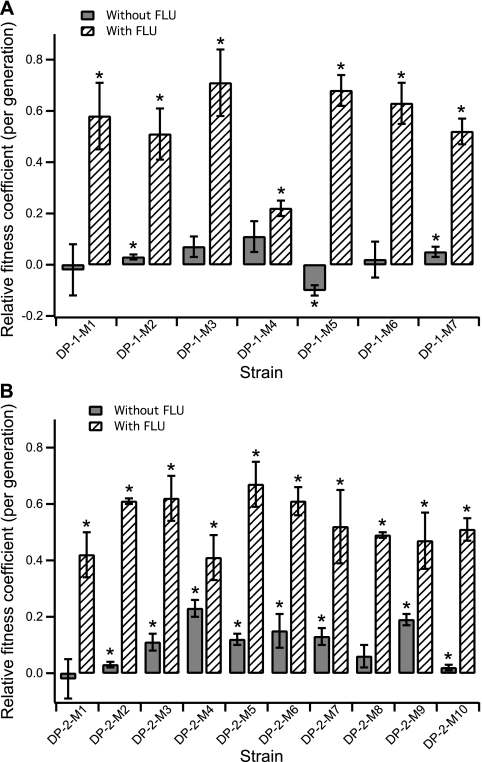
Fig. 3.
Relative fitness coefficients of adaptive mutants isolated from fluconazole-exposed populations compared with the parental strain (different FP), DP-1 (A) and DP-2 (B), in the presence of 0.25 μg/ml of fluconazole (hatched bars) or without fluconazole (solid bars). The data shown are the averages and SD of a minimum of 3 biological replicates. The asterisks indicate that the relative fitness difference measured in the adaptive strains is statistically significant (P < 0.05, using an unpaired Student t test with unequal variance).
Table 1.
Adaptive mutants isolated from both fluconazole-exposed (FLU) and control populations
| Adaptive event | Strain | Colora | FLU (μg/ml)b | No. of generationsc | No drug |
FLU |
MIC (μg/ml) | Ploidy | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Relative fitnessd/generation | P valued | Relative fitnessd/generation | P valued | |||||||
| Parental | KKCa-14 | RFP | 0.125 | 2N | ||||||
| Parental | KKCa-17 | GFP | 0.125 | 2N | ||||||
| Parental | KKCa-20 | YFP | 0.125 | 2N | ||||||
| 1 | CP-1-M1 | RFP | 0 | 64 | 0.16 ± 0.1 | 0.05 | NAe | NA | ∼2N | |
| 2 | CP-1-M2 | YFP | 0 | 70 | 0.05 ± 0.01 | 0.00 | NA | NA | ∼2N | |
| 3 | CP-1-M3 | GFP | 0 | 106 | 0.15 ± 0.02 | 0.00 | NA | NA | ∼2N | |
| 4 | CP-1-M4 | RFP | 0 | 158 | 0.24 ± 0.14 | 0.02 | NA | NA | ∼2N | |
| 5 | CP-2-M1 | GFP | 0 | 76 | 0.3 ± 0.02 | 0.00 | NA | NA | ∼2N | |
| 6 | CP-2-M2 | YFP | 0 | 125 | 0.11 ± 0.09 | 0.09 | NA | NA | ∼4N | |
| 7 | CP-2-M3 | RFP | 0 | 172 | 0.09 ± 0.11 | 0.12 | NA | NA | ∼2N | |
| 8 | DP-1-M1 | RFP | 0.25 | 58 | 0.02 ± 0.1 | 0.74 | 0.58 ± 0.13 | 0.01 | 0.38 | ∼2N |
| 9 | DP-1-M2 | YFP | 1 | 76 | 0.03 ± 0.1 | 0.53 | 0.5 ± 0.05 | 0.00 | 0.38 | ∼2N |
| 10 | DP-1-M3 | RFP | 1 | 93 | 0.02 ± 0.01 | 0.00 | 0.27 ± 0.06 | 0.01 | 1 | ∼2N |
| 11 | DP-1-M4 | YFP | 4 | 125 | 0.04 ± 0.02 | 0.02 | 0.03 ± 0.25 | 0.88 | 1 | ∼2N |
| 12 | DP-1-M5 | GFP | 4 | 134 | −0.21 ± 0.02 | 0.00 | 0.55 ± 0.23 | 0.05 | 1.5 | ∼2N |
| 13 | DP-1-M6 | RFP | 4 | 146 | 0.09 ± 0.03 | 0.03 | −0.17 ± 0.08 | 0.02 | 2 | ∼3N |
| 14 | DP-1-M7 | YFP | 4 | 172 | 0.13 ± 0.01 | 0.00 | 0.35 ± 0.16 | 0.06 | 3 | ∼2N |
| 15 | DP-2-M1 | RFP | 0.25 | 31 | 0 ± 0.07 | 0.95 | 0.42 ± 0.08 | 0.00 | 0.75 | ∼2N |
| 16 | DP-2-M2 | GFP | 0.25 | 47 | 0.11 ± 0.08 | 0.08 | 0.12 ± 0.11 | 0.20 | 0.5 | ∼2N |
| 17 | DP-2-M3 | RFP | 0.25 | 52 | 0.16 ± 0.1 | 0.11 | 0.16 ± 0.15 | 0.13 | 1 | ∼2N |
| 18 | DP-2-M4 | GFP | 0.25 | 64 | 0.14 ± 0.02 | 0.00 | −0.32 ± 0.03 | 0.00 | 0.19 | ∼2N |
| 19 | DP-2-M5 | YFP | 1 | 76 | −0.15 ± 0.04 | 0.02 | 0.68 ± 0.34 | 0.03 | 0.5 | ∼2N |
| 20 | DP-2-M6 | RFP | 1 | 106 | 0.06 ± 0.03 | 0.04 | 0.31 ± 0.20 | 0.12 | 2 | ∼2N |
| 21 | DP-2-M7 | GFP | 1 | 112 | 0.02 ± 0 | 0.01 | −0.26 ± 0.15 | 0.03 | 0.5 | ∼2N |
| 22 | DP-2-M8 | RFP | 4 | 125 | −0.02 ± 0.01 | 0.01 | 0.49 ± 0.15 | 0.03 | 1 | ∼2N |
| 23 | DP-2-M9 | GFP | 4 | 153 | 0.1 ± 0.02 | 0.01 | 0.4 ± 0.06 | 0.00 | 0.25 | ∼2N |
| 24 | DP-2-M10 | RFP | 4 | 172 | −0.11 ± 0.04 | 0.04 | 0.29 ± 0.04 | 0.00 | 8 | ∼2N |
Fluorescent marker.
Fluconazole concentration in the feed.
Generation from which the strain was isolated.
Relative to previous adaptive mutant.
NA, not applicable.
Table 2.
Relative fitness coefficients for isolated adaptive mutants compared to the parental isolate used to initiate the culture
| Adaptive event | Strain | No drug |
FLUb |
||
|---|---|---|---|---|---|
| Relative fitnessa/ generation | P valuea | Relative fitnessa/ generation | P valuea | ||
| 8 | DP-1-M1 | −0.02 ± 0.1 | 0.74 | 0.58 ± 0.13 | 0.01 |
| 9 | DP-1-M2 | 0.03 ± 0.01 | 0.05 | 0.51 ± 0.1 | 0.01 |
| 10 | DP-1-M3 | 0.07 ± 0.04 | 0.08 | 0.71 ± 0.13 | 0.01 |
| 11 | DP-1-M4 | 0.11 ± 0.06 | 0.07 | 0.22 ± 0.03 | 0.00 |
| 12 | DP-1-M5 | −0.1 ± 0.02 | 0.01 | 0.68 ± 0.06 | 0.00 |
| 13 | DP-1-M6 | 0.02 ± 0.07 | 0.50 | 0.63 ± 0.08 | 0.00 |
| 14 | DP-1-M7 | 0.05 ± 0.02 | 0.04 | 0.52 ± 0.05 | 0.00 |
| 15 | DP-2-M1 | −0.02 ± 0.07 | 0.63 | 0.42 ± 0.08 | 0.00 |
| 16 | DP-2-M2 | 0.03 ± 0.01 | 0.00 | 0.61 ± 0.01 | 0.00 |
| 17 | DP-2-M3 | 0.11 ± 0.03 | 0.03 | 0.62 ± 0.08 | 0.00 |
| 18 | DP-2-M4 | 0.23 ± 0.03 | 0.01 | 0.41 ± 0.08 | 0.01 |
| 19 | DP-2-M5 | 0.12 ± 0.02 | 0.01 | 0.67 ± 0.08 | 0.03 |
| 20 | DP-2-M6 | 0.15 ± 0.06 | 0.05 | 0.61 ± 0.05 | 0.00 |
| 21 | DP-2-M7 | 0.13 ± 0.03 | 0.01 | 0.52 ± 0.13 | 0.02 |
| 22 | DP-2-M8 | 0.06 ± 0.04 | 0.08 | 0.49 ± 0.01 | 0.00 |
| 23 | DP-2-M9 | 0.19 ± 0.02 | 0.00 | 0.47 ± 0.1 | 0.01 |
| 24 | DP-2-M10 | 0.02 ± 0.01 | 0.03 | 0.51 ± 0.04 | 0.00 |
Relative to the parental strain of a different color.
FLU, fluconazole.
Interestingly, we did not observe the same trend in the DP-2 population. After each of the stepwise increases in the fluconazole concentration, a different FP subpopulation started to expand. One possibility to account for this observation is that a more drug-resistant mutant was already present in the subpopulation that started expanding after the increase in the drug concentration, but it was present at a low frequency and thus was more difficult to observe (see below for further discussion).
Relative fitness coefficients in the presence and absence of drug.
The observed adaptive events and the colored FP corresponding to each adaptive expanding subpopulation at the generation in which it reached its maximum proportion of the population are illustrated in Fig. 1. Adaptive mutants (listed in Table 1) were isolated from each of the observed adaptive events (see Materials and Methods for details). The fitness coefficient of each isolated adaptive clone was determined by competing the isolate against the previous adaptive clone (for all isolated adaptive mutants) and the parental strain of a different color (for adaptive mutants isolated from the DP-1 and DP-2 populations), either in the presence (for the fluconazole-exposed populations) or in the absence (for both fluconazole-exposed and no-drug control populations) of drug (Fig. 2 and 3 and Tables 1 and 2). While not all isolated adaptive mutants from the fluconazole-exposed populations exhibited an increase in fitness compared to the previous adaptive mutants, all of them showed increases in fitness compared to the parental strain in the presence of fluconazole (Fig. 3 and Table 2).
Interestingly, for most of the adaptive clones isolated from the fluconazole-exposed populations, increases in the relative fitness coefficient in the presence of fluconazole were not accompanied by significant decreases in fitness in the absence of the drug. This was especially evident in the isolated adaptive mutants from the DP-2 population, where most mutants exhibited an increase in the relative fitness coefficient compared to the parental strain in the absence of the drug, suggesting that there was no fitness cost associated with the increase in drug resistance (Fig. 3 and Table 2). Furthermore, for most of the adaptive clones, there was no fitness cost associated with increases in the MIC (Fig. 2 and Table 1).
Chromosomal rearrangements.
Because aneuploidy has been associated with the acquisition of fluconazole resistance in a subset of C. albicans isolates (32, 34), we next asked if any of the adaptive mutants exhibited obvious signs of altered ploidy, using flow cytometry to measure the DNA content (Table 1). The majority of the adaptive mutants appeared to be diploids (2N). However, there were two exceptions: strain CP-2-M2, isolated from generation 125 of the control population, had an ∼4N genome content, suggesting that it is a tetraploid; strain DP-1-M6, isolated from generation 146 of the fluconazole-exposed population, had an ∼3N genome content, suggesting that it may be triploid or multiply aneuploid.
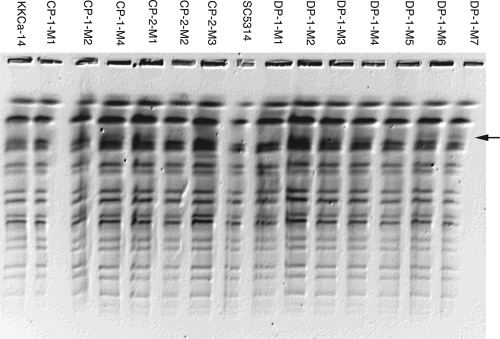
To examine if there were any obvious gross chromosomal rearrangements in the isolated adaptive mutants, we performed CHEF gel analysis (see Fig. S2 in the supplemental material). Alterations in the mobility of chromosome 6 (Chr6) and -7 are often detected and are likely due to alterations in the lengths of the major repeat sequence tracts (16). No alterations in the larger bands were observed, and no ectopic bands were evident. Analysis of genomic DNA digested with SfiI, which digests the genome into ∼2 or 3 resolvable bands per chromosome, detected new bands in two independent lineages: strains DP-1-M6 (RFP) and DP-1-M7 (GFP) (Fig. 4). To identify the DNA present in each of these bands, they were excised from the gel and hybridized to microarrays of C. albicans open reading frame (ORFs) (11, 35). In both cases, hybridization detected a region of ChrR spanning the approximate coordinates 1,353 to 1,907 kb. This SfiI fragment begins at a major repeat sequence (MRS), includes the centromere (CENR), and ends at an SfiI site just past the rDNA repeats. The most likely explanation is that the presence of the new band was due to an alteration in the lengths of the rDNA repeats. Thus, we did not detect any major genome rearrangements spanning nonrepetitive regions of the genome (33). In addition, we did not detect the appearance of chromosome 5L isochromosome [i(5L)] (M. McClellan and J. Berman, data not shown).
Fig. 4.
Detection of genome rearrangements using SfiI-digested chromosomes separated by CHEF gel electrophoresis. The arrow indicates a novel band migrating in strains DP-1-M6 and DP-1-M7. Subsequent analysis indicated that this band corresponds to ChrR and is hypothesized to include altered numbers of rDNA repeats (McClellan and Berman, data not shown).
Stability of increased drug resistance.
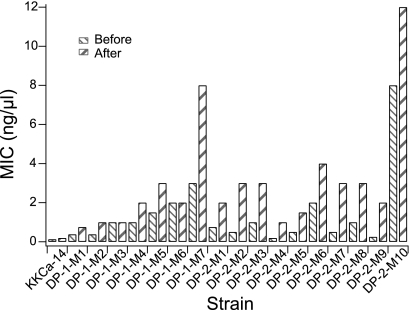
Increased fluconazole resistance in the adaptive mutants could arise from transient physiological changes or more stable genetic changes. To ask if the increased drug resistance was stable, adaptive mutants isolated from the DP-1 and DP-2 populations were subjected to serial transfer for approximately 30 generations in the absence of fluconazole, and then the MICs of the transferred isolates were compared to the MICs of the original mutants. Importantly, the MIC values of all of the adaptive mutants tested did not decrease following the nonselective serial transfer and always remained higher than the MIC values of the parental strain, suggesting that the drug resistance is stable for a period of at least 30 generations (Fig. 5) and is likely due to a genetic change.
Fig. 5.
Stability of drug resistance. Shown are the MIC values for one of the parental strains and all of the isolated adaptive mutants from the fluconazole-exposed populations before and after serial passage in the absence of the drug for more than 30 generations.
DISCUSSION
Fitness cost of drug resistance.
The fitness effect associated with a resistance mutation plays a key role in determining whether the resistant genotype can be maintained and whether it has the possibility of becoming dominant in the population (3, 4). In most bacteria, a drug-resistant phenotype is acquired via horizontal gene transfer and is usually accompanied by a fitness cost in the absence of the drug. Thus, it is expected that, in the absence of drug, the proportion of the population represented by the drug-resistant subpopulation will decrease or be eliminated from the population. Alternatively, further evolution in the absence of drug can give rise to compensatory mutations, resulting in improved fitness of drug-resistant mutants (4, 17, 20, 30). In fungal cells, where resistance is acquired via changes in the genome rather than via horizontal gene transfer, the situation appears to be more complex.
Most of the adaptive mutants that evolved in the presence of drug showed no significant decrease in relative fitness in the absence of the drug. This is similar to other studies in C. albicans, where the acquisition of antifungal drug resistance was accompanied by no immediate fitness cost (2, 7). In fact, several adaptive mutants isolated from the DP-2 population exhibited increases in relative fitness compared to the parental strain in the absence of drug (Fig. 3 and Table 2). In one study, the initial fitness costs that were likely associated with antifungal drug resistance in one population decreased upon further evolution (8). In another study, the drug resistance acquired by C. albicans after evolution in fluconazole was unstable and reverted to the drug-susceptible phenotype after serial passage in the absence of the drug (5). The reversion to a drug-susceptible phenotype in the absence of the drug suggests that some mechanisms of drug resistance are associated with a fitness cost.
The different degrees of reversion of this resistance are likely due to the specific mechanism that confers resistance, as well as to the genetic changes that gave rise to it. For example, increased levels of efflux pump activity likely incur a fitness cost because the cell expends energy in removing not only drug, but also nutrients, from the cell. Alternatively, a minor change in the conformation of an enzyme involved in sterol biosynthesis may not have much effect on the efficiency of the process in the absence of drug. Similarly, an aneuploidy that confers drug resistance, such as i(5L), may be acquired (and lost) rapidly via mitotic nondisjunction, irrespective of whether it incurs a large fitness cost. In contrast, once point mutations are acquired (which is a lower-frequency event), reversion of these mutations may require precise changes that also arise less frequently.
The appearance of adaptive mutants that had large increases in ploidy is of particular interest, as is the fact that these ploidy differences did not appear to persist in the populations. DP-1-M6, an isolate from a fluconazole-exposed population, has an ∼3N ploidy and did not exhibit a decrease in fitness compared with the parental strain but did show a decrease in relative fitness in the presence of the drug compared to the previous adaptive mutant from the same population. The other event appeared in the no-drug control population, where it is important to note that cells were maintained under limiting glucose, which is considered a stress condition (11). Importantly, we did not detect the appearance of i(5L) (McClellan and Berman, data not shown), a rearrangement that causes fluconazole resistance (34). The only detectable genome rearrangements were most likely due to alterations in the lengths of the rDNA repeat tracts on ChrR and alterations in the lengths of MRS tracts on Chr6 and -7. The lack of observed gross chromosomal rearrangements that are generally associated with increased fluconazole resistance and the stability of the increased drug resistance in the absence of the drug suggest that the resistance mechanisms are due to single nucleotide polymorphisms (SNPs) and other stable genome rearrangements that are not detectable by CHEF analyses (33). It is possible that some of the beneficial mutations were already present in the initial colony or during the batch growth phase prior to initiation of the continuous-culture experiments (13) and are thus not a result of the selection process in the chemostats. However, with the use of VERT, we were able to identify multiple adaptive events that occurred in different fluorescent subpopulations during the course of the laboratory evolution, and thus, it is unlikely that all the beneficial mutations were already present prior to the start of evolution. Future work will use whole-genome transcriptional profiling and resequencing to identify all genome changes in the isolated adaptive mutants and the parental strains, allowing us to address the possibility of the issue mentioned above and to shed light on their resistance mechanisms.
Relationship between the evolutionary dynamics and relative fitness of adaptive mutants.
Theoretically, each adaptive mutant isolated from an observed adaptive event should be fitter than the adaptive mutant isolated from the previous expanding subpopulation (e.g., M3 should be fitter than M2 from the same population). Consistent with this, most of the isolated adaptive mutants exhibit increased fitness compared with the previous adaptive mutant. Nonetheless, some exceptions appeared: adaptive mutants DP-1-M6 (generation 146), DP-2-M4 (generation 64), and DP-2-M7 (generation 112) exhibited negative relative fitness coefficients in the presence of the drug, suggesting that they were less drug resistant than the previous adaptive mutant. MIC measurements confirmed this for DP-2-M4 and DP-2-M7 but not for DP-1-M6. Here, we offer some potential explanations for these exceptions. The observed expansion leading to the isolation of the DP-2-M7 was likely due to noise in the measurements rather than to a bona fide adaptive event, since the expansion occurred during a quasi-steady state for the relative proportions of the three FP subpopulations from generation ∼100 to generation ∼112 and only one time point exhibited that increase in the GFP subpopulation at generation 112. The relative fitness coefficient measurements further supported this idea: this GFP isolate was less fit than the previous adaptive mutant (DP-2-M6 isolated from the RFP subpopulation) in the presence of drug. Alternatively, complex interactions in the population that cannot be detected with the 3-color VERT analysis could contribute to the transient increase in the GFP population. In contrast, the green expansion that led to the isolation of DP-2-M4 was clearly an adaptive event, based on the population dynamics. However, none of the 8 clones isolated from the green subpopulation at generation 64 from the DP-2 population showed an increase in fitness relative to DP-2-M3. Based on the rate of expansion of the green subpopulation, it is unlikely that the expanding adaptive mutant was in a small proportion of the population. Thus, either the actual adaptive mutant responsible for the observed expansion was not present among the 8 colonies picked or the condition used for the relative fitness coefficient measurements was not able to recapitulate the relative fitness benefit of DP-2-M4 during the evolution experiment. Even though the DP-2-M4 and DP-2-M7 clones may not have been responsible for the expansion of the colored subpopulations, both are adaptive mutants, as they had increased fitness compared to the parental strain in the presence of fluconazole.
We expected that the expanding subpopulation prior to each stepwise increase in the fluconazole concentration would continue expanding, as it likely contained the fittest mutant in the population at that time point. However, this trend was not observed in the DP-2 population; rather, after each stepwise increase in the fluconazole concentration, a different FP subpopulation began expanding. For example, prior to generation 64, the GFP subpopulation was expanding, but after the increase in the drug concentration, the RFP and YFP subpopulations started to expand, and the YFP adaptive mutant isolated (DP-2-M5) was indeed more drug resistant than the GFP adaptive mutant isolated prior to the drug increase (DP-2-M4). One explanation for this observation is that the adaptive mutant, DP-2-M5, may have been present in the YFP subpopulation prior to generation 64 but was at a low enough frequency or was not expanding fast enough to result in an observed expansion of the YFP subpopulation prior to the increase in the fluconazole concentration. This further demonstrates the effects of clonal interference on the evolving population structure and highlights the complex dynamics present in an evolving population.
Since evolving populations are likely to be heterogeneous, it is also possible that there are interactions between different mutants within the population similar to mutual and/or commensal interactions that occur between communities of organisms growing within the host. These types of interactions have been observed in vitro in a long-term adaptive-evolution experiment with Escherichia coli under glucose limitation (26), where adaptive mutants that exhibit cross-feeding of different metabolites establish a stable coexistence. In this scenario, the increase in fitness of the adaptive mutant in the evolving environment would not be detected in the pairwise competition experiments used to study fitness relationships. Thus, we cannot rule out the possibility that complex interactions also contributed to some of the observed expansions in the populations that evolved here.
Relationship between the fluconazole MIC and relative fitness coefficients.
The MIC of an antifungal agent is an important indicator of drug susceptibility (reviewed by White et al. [41]). However, the relationship between the MIC value of a pathogen and its ability to adapt to the environment is not linear either in vivo or in vitro (7, 25). The short-term adaptive-evolution experiments reported here for a continuous-culture system support the idea that the MIC alone does not predict the relative fitness of a strain in the presence of drug. Furthermore, for the majority of the adaptive clones isolated, there was no fitness cost associated with increases in the fluconazole MIC, which is consistent with other studies of in vitro-evolved isolates (35). Similarly, no correlation between relative fitness coefficients and MIC values was detected in the adaptive clones. For example, DP-2-M9 showed a significant increase in fitness in the presence of drug relative to DP-2-M8, yet its MIC was reduced relative to DP-2-M8 (Table 1). The different growth conditions and methods of measurement most likely explain these differences: the MIC was determined using Etest strip assays, where cells were grown on RPMI 1640 agar plates; fitness was determined by using pairwise competition experiments in chemostats with YNB liquid medium containing fluconazole. Thus, our data support the idea that the MIC does not provide a direct measure of competitive fitness (7).
Conclusions.
The emergence of drug resistance in fungal pathogens is dynamic. Time course samples collected from patients undergoing antifungal treatments provide data on the evolution of pathogens in vivo. However, due to the limited number of clones isolated, the population dynamics during the emergence of drug resistance in vivo has been difficult to assess. In vitro evolution of multiple populations in parallel provides a more controlled environment for the study of population dynamics. However, traditional adaptive-evolution experiments do not follow evolutionary dynamics due to the lack of real-time indications of when adaptive mutants arise and expand in the population. We addressed this challenge using VERT with C. albicans isolates evolving in the presence of fluconazole to isolate adaptive mutants in a more systematic manner. Importantly, we detected an increase in the frequency of adaptive events in the presence of the drug. Furthermore, isolates from one of the fluconazole-exposed populations evolved a general increase in fitness compared to the parental strain in the absence of the drug, suggesting that drug resistance mechanisms do not always come with an obvious fitness cost. In addition, these resistance mechanisms are stably maintained for at least 30 generations in the absence of the drug.
Supplementary Material
ACKNOWLEDGMENTS
We thank Maryam Gerami-Nejad for expert technical assistance with strain construction.
Partial financial support was provided by U.S. NSF grant MCB-1054276 and the Texas Engineering Experimental Station (TEES) to K.C.K. and NIH R01 AI0624273 to J.B.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Anderson J. B. 2005. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nat. Rev. Microbiol. 3:547–556 [DOI] [PubMed] [Google Scholar]
- 2. Anderson J. B., et al. 2003. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 163:1287–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andersson D. I. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452–456 [DOI] [PubMed] [Google Scholar]
- 4. Andersson D. I., Hughes D. 2010. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8:260–271 [DOI] [PubMed] [Google Scholar]
- 5. Calvet H. M., Yeaman M. R., Filler S. G. 1997. Reversible fluconazole resistance in Candida albicans: a potential in vitro model. Antimicrob. Agents Chemother. 41:535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cowen L. E., Anderson J. B., Kohn L. M. 2002. Evolution of drug resistance in Candida albicans. Annu. Rev. Microbiol. 56:139–165 [DOI] [PubMed] [Google Scholar]
- 7. Cowen L. E., Kohn L. M., Anderson J. B. 2001. Divergence in fitness and evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 183:2971–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cowen L. E., et al. 2002. Population genomics of drug resistance in Candida albicans. Proc. Natl. Acad. Sci. U. S. A. 99:9284–9289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cowen L. E., et al. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desai M. M., Fisher D. S. 2007. Beneficial mutation selection balance and the effect of linkage on positive selection. Genetics 176:1759–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunham M. J., et al. 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 99:16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gerrish P. J., Lenski R. E. 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102-103:127–144 [PubMed] [Google Scholar]
- 13. Gresham D., et al. 2008. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 4:e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kao K. C., Sherlock G. 2008. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat. Genet. 40:1499–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Katiyar S., Pfaller M., Edlind T. 2006. Candida albicans and Candida glabrata clinical isolates exhibiting reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 50:2892–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lephart P. R., Chibana H., Magee P. T. 2005. Effect of the major repeat sequence on chromosome loss in Candida albicans. Eukaryot. Cell 4:733–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levin B. R., Perrot V., Walker N. 2000. Compensatory mutations, antibiotic resistance and the population genetics of adaptive evolution in bacteria. Genetics 154:985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Löffler J., et al. 2000. Phospholipid and sterol analysis of plasma membranes of azole-resistant Candida albicans strains. FEMS Microbiol. Lett. 185:59–63 [DOI] [PubMed] [Google Scholar]
- 19. Lyons C. N., White T. C. 2000. Transcriptional analyses of antifungal drug resistance in Candida albicans. Antimicrob. Agents Chemother. 44:2296–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maisnier-Patin S., Andersson D. I. 2004. Adaptation to the deleterious effects of antimicrobial drug resistance mutations by compensatory evolution. Res. Microbiol. 155:360–369 [DOI] [PubMed] [Google Scholar]
- 21. Mansfield B. E., et al. 2010. Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 6:e1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marr K. A., Lyons C. N., Rustad T. R., Bowden R. A., White T. C. 1998. Rapid, transient fluconazole resistance in Candida albicans is associated with increased mRNA levels of CDR. Antimicrob. Agents Chemother. 42:2584–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McNeil M. M., et al. 2001. Trends in mortality due to invasive mycotic diseases in the United States, 1980-1997. Clin. Infect. Dis. 33:641–647 [DOI] [PubMed] [Google Scholar]
- 24. Pfaller M. A., Diekema D. J. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rex J. H., Rinaldi M. G., Pfaller M. A. 1995. Resistance of Candida species to fluconazole. Antimicrob. Agents Chemother. 39:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rosenzweig R. F., Sharp R. R., Treves D. S., Adams J. 1994. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics 137:903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sanglard D., Ischer F., Monod M., Bille J. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143:405–416 [DOI] [PubMed] [Google Scholar]
- 28. Sanglard D., et al. 1995. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 39:2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sanglard D., Odds F. C. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73–85 [DOI] [PubMed] [Google Scholar]
- 30. Schoustra S. E., Debets A. J., Slakhorst M., Hoekstra R. F. 2006. Reducing the cost of resistance; experimental evolution in the filamentous fungus Aspergillus nidulans. J. Evol. Biol. 19:1115–1127 [DOI] [PubMed] [Google Scholar]
- 31. Selmecki A., Bergmann S., Berman J. 2005. Comparative genome hybridization reveals widespread aneuploidy in Candida albicans laboratory strains. Mol. Microbiol. 55:1553–1565 [DOI] [PubMed] [Google Scholar]
- 32. Selmecki A., Forche A., Berman J. 2006. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313:367–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Selmecki A., Forche A., Berman J. 2010. Genomic plasticity of the human fungal pathogen Candida albicans. Eukaryot. Cell 9:991–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Selmecki A., Gerami-Nejad M., Paulson C., Forche A., Berman J. 2008. An isochromosome confers drug resistance in vivo by amplification of two genes, ERG11 and TAC1. Mol. Microbiol. 68:624–641 [DOI] [PubMed] [Google Scholar]
- 35. Selmecki A. M., Dulmage K., Cowen L. E., Anderson J. B., Berman J. 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5:e1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Singh S. D., et al. 2009. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5:e1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vanden Bossche H., et al. 1998. Antifungal drug resistance in pathogenic fungi. Med. Mycol. 3 6(Suppl. 1):119–128 [PubMed] [Google Scholar]
- 38. White T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. White T. C. 1997. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14alpha demethylase in Candida albicans. Antimicrob. Agents Chemother. 41:1488–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. White T. C., Holleman S., Dy F., Mirels L. F., Stevens D. A. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. White T. C., Marr K. A., Bowden R. A. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wirsching S., Michel S., Morschhauser J. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856–865 [DOI] [PubMed] [Google Scholar]
- 43. Wirsching S., Moran G. P., Sullivan D. J., Coleman D. C., Morschhauser J. 2001. MDR1-mediated drug resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 45:3416–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zaoutis T. E., et al. 2005. The epidemiology and attributable outcomes of candidemia in adults and children hospitalized in the United States: a propensity analysis. Clin. Infect. Dis. 41:1232–1239 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.