Abstract
The response regulator Spo0A governs multiple developmental processes in Bacillus subtilis, including most conspicuously sporulation. Spo0A is activated by phosphorylation via a multicomponent phosphorelay. Previous work has shown that the Spo0A protein is not rate limiting for sporulation. Rather, Spo0A is present at high levels in growing cells, rapidly rising to yet higher levels under sporulation-inducing conditions, suggesting that synthesis of the response regulator is subject to a just-in-time control mechanism. Transcription of spo0A is governed by a promoter switching mechanism, involving a vegetative, σA-recognized promoter, Pv, and a sporulation σH-recognized promoter, Ps, that is under phosphorylated Spo0A (Spo0A∼P) control. The spo0A regulatory region also contains four (including one identified in the present work) conserved elements that conform to the consensus binding site for Spo0A∼P binding sites. These are herein designated O1, O2, O3, and O4 in reverse order of their proximity to the coding sequence. Here we report that O1 is responsible for repressing Pv during the transition to stationary phase, that O2 is responsible for repressing Ps during growth, that O3 is responsible for activating Ps at the start of sporulation, and that O4 is dispensable for promoter switching. We also report that Spo0A synthesis is subject to a posttranscriptional control mechanism such that translation of mRNAs originating from Pv is impeded due to RNA secondary structure whereas mRNAs originating from Ps are fully competent for protein synthesis. We propose that the opposing actions of O2 and O3 and the enhanced translatability of mRNAs originating from Ps create a highly sensitive, self-reinforcing switch that is responsible for producing a burst of Spo0A synthesis at the start of sporulation.
INTRODUCTION
The Gram-positive bacterium Bacillus subtilis has the capacity to decide among a variety of cell fates at the end of the exponential phase of growth. At the heart of the decision-making process for several of these fates is the response regulatory protein Spo0A, which governs spore formation, biofilm formation, and cannibalism and is also required for competence (21). The activity of Spo0A is controlled by phosphorylation via a multicomponent phosphorelay, at the head of which are five histidine kinases (KinA to KinE) (4). Discrimination among alternative cell fates is determined in part by the cellular levels of phosphorylated Spo0A (Spo0A∼P), with the regulatory sites for genes with moderate to high affinity for Spo0A∼P (e.g., genes involved in cannibalism and biofilm formation) firing at low to intermediate levels of the phosphoprotein and those with low affinity being turned on only at high levels (14, 15).
Phosphorylation induces a conformational change in Spo0A, allowing it to dimerize and bind to target sequences (2). More than 100 genes are under the direct positive or negative control of Spo0A∼P (23). Genes activated by Spo0A∼P include those with promoters recognized by RNA polymerase containing the housekeeping sigma factor σA, as well as genes whose promoters are recognized by RNA polymerase containing the alternative sigma factor σH (including the Ps promoter for spo0A investigated here) (25). The target sites reveal a consensus binding sequence, TTTGTCRAA, which is known as the 0A box (23). The crystal structure of the C-terminal (DNA-binding) domain of the Bacillus stearothermophilus ortholog in a complex with 0A box sequences has been solved (32). This structure reveals contacts of the DNA-binding domain with bases in the 0A box and with the DNA backbone and has led to the suggestion that Spo0A∼P dimers are capable of forming tandem arrays in a head-to-tail arrangement along the DNA (32). It was suggested that such arrays may form in the regulatory region for genes such as spoIIA, spoIIE, and spoIIG that exhibit multiple 0A boxes in tandem extending far upstream from the start site of transcription. That said, the mechanism by which Spo0A∼P activates transcription remains largely unclear, with few examples that have been well characterized, two for promoters recognized by σA RNA polymerase (PspoIIG and Pskf) (10, 26) and one (Pspo0F) for promoters recognized by σH RNA polymerase (2).
Interestingly, Spo0A is maintained at relatively high levels (∼2,000 molecules/cell) during exponential phase, rapidly rising to even higher levels (∼20,000 molecules/cell) under sporulation-inducing conditions (9, 13). We have attributed the high maintenance level of Spo0A and the rapid increase to yet higher levels to a just-in-time regulatory system that ensures that Spo0A molecules do not become rate limiting at both low and high rates of flux of phosphoryl groups through the phosphorelay (9).
How are Spo0A protein levels regulated during the transition to stationary phase? The spo0A gene is transcribed from two promoters, whose start sites are located 204 and 45 bp upstream from the start codon for the open reading frame (Fig. 1A) (12, 29)). The more upstream promoter, Pv, is expressed during the exponential phase of growth under the control of σA-RNA polymerase. This transcription fluctuates strikingly and in a manner that correlates with small changes in the growth rate (22) but then shuts off during the transition to stationary phase (12). The downstream promoter, Ps, in contrast, is induced after the end of exponential-phase growth under the control of σH-RNA polymerase and Spo0A∼P. Thus, transcription of spo0A switches from Pv to Ps as cells exit exponential-phase growth (11, 12, 29). Transcription from Ps is required in order for Spo0A to reach the high levels needed for the activation of key sporulation genes, such as spoIIA, spoIIE, and spoIIG, but not for the low levels required for efficient entry into competence (14, 18). Insights into the mechanism of promoter switching has come from the work of Strauch and coworkers, who showed that it is mediated by Spo0A∼P and that the protein binds three 0A boxes in the regulatory region (29).
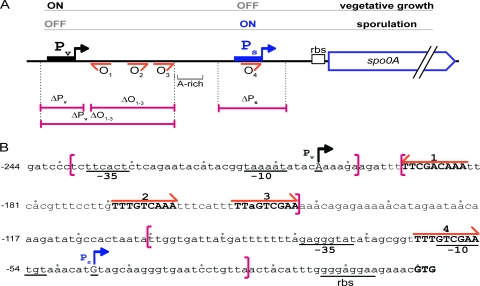
Fig. 1.
The spo0A regulatory region. (A) Cartoon of the regulatory region. Pv and Ps are indicated by black and blue boxes with arrows, respectively. Sequences matching the Spo0A consensus binding site (0A box), which are designated operator sites O1, O2, O3, and O4, are labeled with orange arrows and numbered in order from 5′ to 3′. The region extending from O1 to O3 (−192 to −143) is referred as O1–3 and the AT-rich region from position −142 to −99 (relative to the start codon) as “A-rich.” Regions deleted in mutants are indicated by pink bars with corresponding mutant names. (B) Sequence of the regulatory region. The start sites of transcription (+1) for the Pv and Ps promoters are underlined and highlighted by a vertical arrow. Positions are numbered relative to the initiation codon GTG (indicated in bold). Sequences matching 0A boxes are indicated with orange arrows and numbered as in panel A. Brackets indicate limits of the deletion in ΔPv (Abs894), ΔO1–3 (Abs883), ΔPv ΔO1–3 (Abs875), or ΔPs (Abs786).
Here we revisit the mechanism of promoter switching, which has not been investigated since the early work of Chibazakura (11) and Strauch (29). We report the discovery of a fourth 0A box that plays a key role in promoter switching and other features of the regulatory region that are conserved among multiple Bacillus spp. We refer to the four 0A boxes as O1, O2, O3, and O4 in the reverse order of their distance from the open reading frame (renaming them to accommodate the newly identified site O2). Our principal findings are that O1 is a negatively acting element that is responsible for repressing Pv at the end of exponential phase, that the newly identified O2 site is a negatively acting element that represses Ps during growth, that O3 is a positively acting element that activates Ps upon entry into stationary phase, and that O4 is dispensable. We also report the discovery of a translational control mechanism that impedes translation of mRNAs originating from Pv but not that originating from Ps. We also show that Pv provides a basal level of Spo0A that is required for efficient entry into the state of genetic competence and strong activation of Ps, and we suggest that Pv plays a pump-priming role in the activation of the Spo0A∼P-controlled promoter. An intricate regulatory region mediates a just-in-time regulatory system that helps to ensure an adequate supply of Spo0A molecules to meet the needs of the phosphorelay.
MATERIALS AND METHODS
Media and general methods.
Media, culture, cloning procedures, preparation of competent cells, β-galactosidase assay, immunoblot analysis, and sporulation assays were carried out as previously described (7, 9, 16, 28). RNA secondary structure prediction was performed with the RNA2 prediction software program using default settings and sequence alignments with ClustalV.
Plasmids and strain constructions.
All the Bacillus subtilis strains used in the present study were derivatives of the PY79 strain (31) and are listed in Table S1 in the supplemental material. Plasmids used are listed in Table S2 and oligonucleotides in Table S3. Markerless deletions of the vegetative promoter of spo0A (ΔPv), O1 to O3 (ΔO1–3), and both (ΔPv ΔO1–3) were constructed using pMAD (1) derivative plasmids: pAC328, pAC313, and pAC306. For this, two NcoI/EcoRI and EcoRI/BamHI DNA fragments corresponding to the spoIVB-spo0A locus were generated by PCR using AC-739/740 and AC-741/742 and cloned between NcoI/BamHI restriction sites of pMAD to give plasmid pAC328. pAC306 was generated likewise, except that AC-744 was used instead of AC-741, whereas in addition AC-743 was also used in place of AC-740 to generate pAC313. Markerless deletion of Ps was generated as previously described by Siranosian et al. (27) using pSK5, with the exception that PY79 was used as the recipient strain. All constructions in PY79 were checked for deletion by PCR amplification and sequencing.
Translational and transcriptional fusions with lacZ were obtained by cloning PCR-generated EcoRI/SalI or EcoRI/BamHI DNA fragments, respectively, into the corresponding sites of pDG1728 (see Tables S2 and S3 in the supplemental material for details). Fusions with mutated promoters were obtained either by using the QuikChange site-directed mutagenesis kit (Stratagene) using the manufacturer's protocol or by cloning fusion PCR products. Insertion mutants were obtained by adding a 5-bp (TATCT) or 11-bp (TATCTAGAGGC) sequence at position −136/−135. For construction of promoter fusions in which the SigH binding sequence of Ps was replaced by that of PspoVG, TAGAGGGTATATAGCGGTTTTGTCGAATGTA was replaced by AGCAGGATTTCAGAAAAAATCGTGGAATTGA (see Fig. S4 in the supplemental material).
Primer extension.
Primer extension was performed mainly as previously described (8), with the exception that 2 μg RNA was incubated with 0.1 pmol of the radiolabeled oligonucleotide AC-766.
Spo0A purification and DNase I footprinting.
Spo0A was purified by affinity chromatography, with a procedure derived from the work of Lewis et al. (20). In brief, Spo0A was extracted from an Escherichia coli strain harboring pAC571, cultured at 30°C. Cells resuspended in TEN buffer (50 mM Tris-HCl [pH 7.5], 10 mM EDTA, and 50 mM NaCl) were disrupted by sonication, and clarified lysate was loaded on an S column (Amersham-Pharmacia) preequilibrated with TEN.
For the footprinting assay, the radiolabeled oligonucleotide AC-470 was used with cold AC-781 to PCR amplify DNA fragments corresponding to a wild-type or mutant version of the spo0A promoter, using as template DNA pAC407 (wild type), pAC463 (m1), pAC458 (m2), or pAC507 (m3). Conditions of the assay were mainly as previously published (6), with the exception that after purification, resulting radiolabeled PCR products were incubated with freshly purified native Spo0A for 20 min at room temperature before DNase I treatment.
RESULTS
Both vegetative and sporulation promoters are required for rapid and high-level accumulation of Spo0A.
As a starting point for this investigation, we constructed a series of deletion mutations within the spo0A regulatory region and examined their consequences for spo0A transcription and for the accumulation of Spo0A. The spo0A gene is transcribed from an upstream, vegetative promoter, Pv, by σA-containing RNA polymerase and from a downstream sporulation promoter, Ps, by RNA polymerase containing the alternative sigma factor σH (12). Located between Pv and Ps are three 0A boxes, here referred to as O1, O2, and O3 (Fig. 1). Two of these elements (O1 and O3) had been identified previously and confirmed as Spo0A∼P binding sites by Strauch and coworkers (29). As discussed below, we detected a third 0A box located between O1 and O3, which we designate O2 in Fig. 1. Finally, the regulatory region contains a fourth binding site, O4, which overlaps with Ps (29). Based on these considerations, we created deletions that removed Pv (ΔPv), Ps (ΔPs) (27), O1 to O3 (ΔO1–3), and both Pv and O1-O3 (ΔPv ΔO1–3) (Fig. 1).
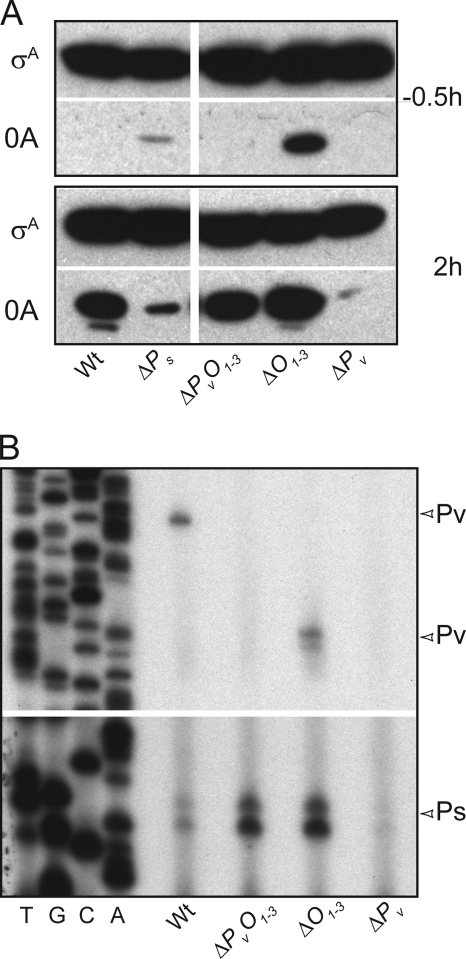
We used immunoblot analysis to investigate the effect of the deletion mutations on Spo0A accumulation. Because Spo0A accumulates to high levels by hour two of sporulation (9), we compared the levels of Spo0A at the mid-exponential phase of growth (T−0.5, representing 0.5 h prior to the end of exponential phase) with those at T2 (Fig. 2A; see also Fig. S1A in the supplemental material). Whereas Spo0A levels markedly increased by T2 in the wild type, little accumulation was observed at this time in the absence of either Pv or Ps. These findings are consistent with previous reports that the removal of either promoter impairs Spo0A-directed transcription (12, 27). (Note that little Spo0A could be seen at T−0.5 in the immunoblot of Fig. 2A but that Spo0A was readily detected during growth with longer exposure times and higher loading levels, as previously described [9] and as seen in the experiment of Fig. S1B.) Unexpectedly, however, we found that the removal of O1-O3 (ΔO1–3) boosted Spo0A to high levels as early as T−0.5 and bypassed the dependence of Spo0A accumulation on Pv; that is, in a mutant in which both Pv and O1-O3 had been removed (ΔPv ΔO1–3), Spo0A reached levels comparable to that seen for the wild type.
Fig. 2.
O1-O3 contains a regulatory element controlling Ps. (A) Immunoblot analysis of Spo0A (0A) accumulation during sporulation. Blotting was performed using samples taken at 0.5 h before sporulation induction by resuspension (upper panel) and 2 h postinduction (bottom panel). Equal amounts of protein, as quantified by the Bradford technique, were loaded as confirmed by the control immunoblot using anti-SigA (σA) antibodies. Samples were taken from shaking culture at 37°C of wild-type cells (Wt; PY79) or Ps (ΔPs; Abs786), O1–3 (ΔO1–3; Abs883), Pv (ΔPv; Abs894), or Pv plus O1–3 (ΔPvO1–3; Abs875) mutant cells. (B) Primer extension analysis of spo0A transcripts. Analysis was performed on RNA extracted from exponential-phase wild-type cells or mutant cells as described for panel A. Start sites (+1) are indicated on the right by arrowheads. Note the shift in the site of extension products originating from Pv caused by the shortening of the transcript in the ΔO1–3 deletion.
To determine which of the two promoters controlling spo0A was being affected by the deletion of O1-O3, we performed primer extension analysis using RNA from cells in the exponential phase of growth (Fig. 2B). As expected, Pv-directed transcripts were seen in the wild type but were absent from a mutant lacking the vegetative promoter (ΔPv) and from a mutant lacking both Pv and O1-O3 (ΔPv ΔO1–3). (Note that the absence of Pv also impaired transcription from Ps, a point to which we will return.) However, little or no effect was seen on transcription from Pv by the simple removal of the O1-O3 sequence (ΔO1–3). (Note the shorter length of the extension products due to the absence of the downstream O1-O3 sequence.) In sharp contrast, removal of O1-O3 markedly increased the level of transcription originating from the sporulation promoter Ps, both in the presence and the absence of Pv. We conclude that the accumulation of Spo0A to high levels in the absence of O1-O3 was due to enhanced transcription from Ps.
The results so far are consistent with a pump-priming model in which Pv allows enough Spo0A molecules to be produced to activate the Spo0A-dependent Ps promoter. Transcription from Ps, in turn, leads to the production of additional Spo0A molecules, thereby setting up a self-reinforcing cycle. Thus, in the absence of Pv, insufficient Spo0A is produced to activate Ps, as we had seen in Fig. 2B. We further surmise that the O1-O3 region contains a regulatory element that impedes transcription from Ps during the exponential phase of growth. In our model, removal of O1-O3 elevates transcription from Ps during growth, allowing enough Spo0A to accumulate to prime the pump even when Pv is absent.
The regulatory region for spo0A is conserved.
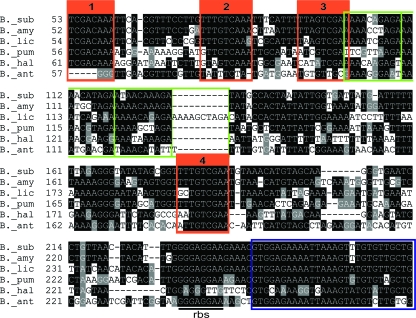
As noted above, the regulatory region for spo0A contains four 0A boxes, three of which have been described previously and a fourth, the newly identified O2 site, being a perfect match to the consensus sequence TTTGTCRAA (23). We wondered whether all four 0A boxes were conserved among Bacillus spp. Accordingly, we performed an alignment of the spo0A promoter region among six Bacillus species (Fig. 3). Strikingly, not only were all four 0A boxes conserved, but the entire regulatory region exhibited features that were conserved among all of the species examined with the exception of Bacillus anthracis. One such feature was the presence of an AT-rich region (80% in B. subtilis) located just downstream of O3. AT richness is known to enhance DNA flexibility and to cause intrinsic DNA bending, which can facilitate promoter activation (19, 24, 30), a point we consider further below. Another conserved feature is the 10-bp motif AAAWNNDAGA, which was directly repeated three times in several of the promoter regions and four times in that of Bacillus licheniformis.
Fig. 3.
The regulatory region of spo0A is conserved among Bacillus spp. Shown is a sequence alignment performed using spo0A promoter sequences from Bacillus subtilis (B._sub), Bacillus amyloliquefaciens (B._amy), Bacillus licheniformis (B._lic), Bacillus pumilis (B._pum), Bacillus halodurans (B._hal), and Bacillus anthracis (B._ant). Shown are alignments extending from the first 0A box to the 9th codon of the open reading frame. O1 to O4 are boxed in orange. The beginning of the open reading frames is boxed in blue. The conserved repeated motif AAAWNDAGA is highlighted in green.
O2 and O3 have opposing effects on Ps.
To further investigate the influence of the O1-O3 region on Ps, we created an in-frame fusion of lacZ to spo0A that extended from 23 codons downstream of the start codon to the 5′ end of the O1-O3 region (and hence lacked Pv). This fusion and mutant derivatives of it were introduced into the chromosome at amyE in cells that had a wild-type copy of spo0A at its normal location. (Thus, in contrast to the experiment of Fig. 2, the results with the gene fusions were carried out under conditions in which Spo0A was being produced from the wild-type copy of spo0A.) The results in Fig. 4A show that the full-length fusion (extending upstream through the O1-O3 region) was silent at T-0.5 and sharply induced between T0.5 and T1.5. As expected, induction was blocked in a mutant lacking the phosphorelay protein Spo0B (□), consistent with the idea that Ps is strongly dependent on Spo0A∼P.
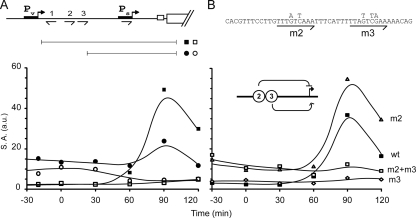
Fig. 4.
O2 and O3 have opposing effects on Ps expression. (A) Deletion of O1-O3 increased expression of a spo0A-lacZ translational fusion during exponential growth phase and reduced its expression during sporulation. The fusions were integrated at amyE. spo0A sequences in the fusions extended from position +72 to position −197 (▪, Abs988; □, Abs955) or position −145 (○, Abs1112; •, Abs1149) relative to the GTG start codon. Expression was compared between the wild type (▪ and •) and spo0B mutant cells (□ and ○). A typical experiment is presented, in which β-galactosidase specific activity (S.A.) is plotted as a function of time (a.u. [arbitrary units]). Samples were taken every 30 min starting 30 min before induction of sporulation by resuspension and up to 120 min postinduction. (B) Expression from spo0A-lacZ translational fusions integrated at amyE. The sequences in the fusions extended from position −197 to +72 and were wild type (wt) (▪; Abs988), O2 mutant (m2) (▵; Abs1055), mutant for O3 (m3) (⋄; Abs1205), or mutant for both (m2+m3) □; Abs1172). The nature of the mutation is described above the graphs. A typical experiment is presented in which β-galactosidase specific activity (S.A.) is plotted as a function of time (a.u. [arbitrary units]). Samples were taken every 30 min starting 30 min before induction of sporulation by resuspension and up to 120 min postinduction.
We then investigated the contribution of the O1-O3 region to this pattern of expression using a truncation mutant (Fig. 4A). The results show that the pattern of expression was markedly altered, with Ps exhibiting a high basal level of activity during growth and impaired induction during sporulation. A similar pattern of expression was observed with a truncation that removed both the O1-O3 region and the downstream AT-rich region (ΔO1–3 ΔAT-rich) (see Fig. S2 in the supplemental material). Thus, consistent with the results obtained by primer extension analysis, the O1-O3 region contains a regulatory element that inhibits Ps activity during growth.
Next, we investigated the contributions of O1, O2, and O3 individually to the activity of Ps by separately mutating each 0A box. Among the nine base pairs in the consensus Spo0A-binding sequence, the most critical seem to be the G and C at positions 4 and 6 (TTTGACRAA), with which Spo0A∼P form hydrogen bonds (32). Less highly conserved is position 7, which is most often a G. Accordingly, we created nucleotide substitutions at positions 4 and 6 in O2 (which has an A at position 7) and at 4, 6, and 7 in O3 (see Materials and Methods). Mutating O1 had little effect on expression of the fusion (see Fig. S2 in the supplemental material) and hence is not considered further. As seen in Fig. 4B, inactivation of O2 was sufficient to allow premature expression from Ps. In other words, mutating O2 alone was sufficient to mimic the effect of deleting the entire O1-O3 region in derepressing Ps during growth. On the other hand, and unlike the deletion of O1-O3 region, mutating O2 mildly but reproducibly enhanced the post-exponential-phase induction of transcription from Ps.
Conversely, mutation of O3 almost completely abolished the expression of the reporter, an unexpected result considering that deletion of the O1-O3 region was not blocked in Ps activity. To investigate this observation further, we built a double mutant in which both O2 and O3 were mutated. The pattern of expression from the double mutant resembled that of the O1-O3 deletion mutant.
We interpret these results as follows. O2 is a negative element that inhibits Ps, whereas O3 is a positively acting element that is required for activation of Ps during sporulation. When both O2 and O3 are mutated, a high level of transcription from Ps is seen during growth and sporulation, while induction of Ps during sporulation is prevented.
Evidence for DNA looping.
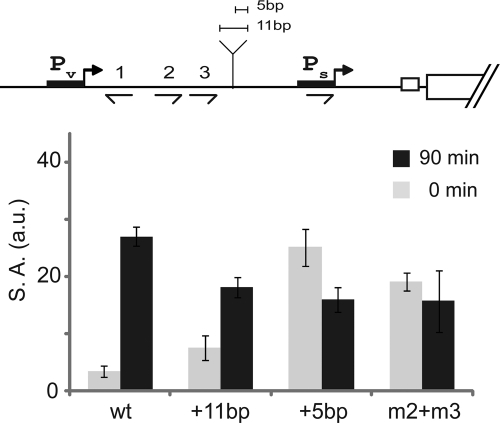
The large gap between O2 and O3 and the promoter (70 and 90 bp, respectively) raised the question of how Spo0A molecules bound so far upstream are able to control the activity of Ps. Two models, which are not mutually exclusive, are that the intervening sequence forms a loop and/or that Spo0A bound at O2 or O3 spreads to the promoter region along the intervening DNA. In an effort to distinguish between these models, we introduced insertions of 5 and 11 bp in the intervening DNA with the expectation that increasing the spacing by about half a turn of the helix but not by approximately a full turn would impair function to a greater extent if the intervening DNA forms a loop. Indeed, as seen in Fig. 5, an 11-bp insertion had little effect on the pattern of expression whereas the 5-bp insertion had a marked effect, creating a phenocopy of a double mutant of O2 and O3. The simplest interpretation of these results is that Spo0A∼P bound at O2 or O3 interacts with the promoter region via the formation of a loop. Reinforcing this model is the observation that, as noted above, the intervening DNA is rich in A·T base pairs, which is known to enhance the flexibility of DNA (30). At the same time, our results do not exclude the possibility that spreading also occurs. Indeed, DNase I footprinting experiments revealed a region of protection that extended downstream of O3 (see Fig. S3 in the supplemental material), a point we will discuss further.
Fig. 5.
Efficient regulation by O2 and O3 depends on their position relative to the face of the helix. Shown is expression from spo0A-lacZ translational fusions integrated at amyE. Bars represent the average values of three independent experiments. Sequences in the fusions extended from position −197 to +72 and were wild type (wt) (Abs988) or O2 and O3 mutant (m2+m3) (Abs1172) or contained a 5-bp insert (+5bp) (Abs1034) or an 11-bp insert (+11bp) (Abs1035). The inserts were at position −64. Samples were taken just before induction of sporulation by resuspension (0 min; gray bars) and at 1.5 h postinduction (90 min; black bars).
O4 is dispensable.
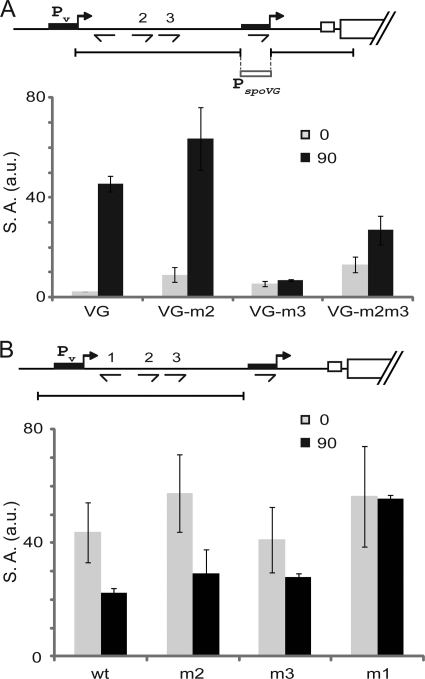
Next, we investigated the function of the fourth 0A box, O4, which is embedded in Ps. Because O4 (TTTGTCGAA) overlaps the −10 sequence of Ps (GTCGAATGT, in which the bold letters indicate bases shared with O4), 0A box positions 4 and 7 are the R and G at positions 1 and 4 of the −10 sequence, which are conserved in σH-recognized promoters (RnnGAATww) (3). Consequently, it was not possible to replace the conserved bases at positions 4, 6, and 7 of the 0A box without mutating the σH-recognized, −10 sequence of Ps. Thus, we decided to make a chimeric regulatory region in which a 31-bp stretch of sequence that included the −35 and −10 sequences of Ps was replaced with the corresponding region of the spoVG promoter (PspoVG) (see Fig. S4 in the supplemental material). PspoVG is a σH-controlled promoter that is not directly activated by Spo0A (Fig. 6A) (33).
Fig. 6.
Functional replacement of Ps with a σH-controlled promoter lacking an 0A box and repression of Pv by O1. (A) Expression from spo0A-lacZ translational fusions integrated at amyE. The fusions contained sequences extending from position −197 to +72 relative to the initiating codon (and hence excluding Pv) except that the 31-bp sequence from −6 to −36 containing Ps was replaced with a 31-bp sequence containing the spoVG promoter extending from −7 to −37 (relative to their start sites). In addition, the fusions were either wild type (VG) (Abs1223) or O2 mutant (VG-m2) (Abs1224), O3 mutant (VG-m3) (Abs1226), or both (VG-m2m3) (Abs1227). Samples were taken just before induction of sporulation by resuspension (0 min; gray bars) and 1.5 h postinduction (90 min; black bars). Values are the averages of data from two independent experiments. (B) Expression from spo0A-lacZ translational fusions containing sequences extending from position −305 to −78 (and hence excluding Ps). In addition, the fusions either wild type (wt) (Abs1229), O2 mutant (m2) (Abs1230), O3 mutant (m3) (Abs1231), or O1 mutant (m1) (Abs1234) are analyzed. Samples were taken just before induction of sporulation by resuspension (0 min; gray bars) and 1.5 h postinduction (90 min; black bars). Values are the averages of data from three independent experiments.
Remarkably, the pattern of expression with the chimeric regulatory region was strikingly similar to that of the parental regulatory region. That is, PspoVG was silent during growth and markedly induced by T1.5 (Fig. 6A). Next, we further modified the chimeric regulatory region by introducing mutations in O2, O3, or both. As we observed with Ps, induction of the chimera was strongly dependent on O3 when O2 was intact (but not when both O2 and O3 were mutants), and the basal level of expression was increased 5-fold during exponential-phase growth in the absence of O2. We infer that neither the function of O2 nor that of O3 requires the presence of O4. We further conclude that O2 and O3 are sufficient to confer a Spo0A-dependent mode of expression on a promoter that lacks a 0A box.
O1 is responsible for repression of Pv during sporulation.
Finally, we returned to the question of the function of O1, a mutant of which had little detectable effect on Ps expression (see Fig. S2 in the supplemental material). It was previously reported that promoter switching occurs during the transition to stationary phase, with Pv being turned off and Ps turned on (12). Because O1 is proximal to Pv, we postulated that it may be responsible for repression of Pv. To test this hypothesis, transcriptional fusions between Pv and lacZ (this time excluding Ps) were constructed. Again, we compared the wild-type sequence or sequences mutant for O1, O2, or O3 (Fig. 6B). Whereas the wild-type fusion and mutants of O2 or O3 exhibited decreased levels of β-galactosidase during sporulation, the enzyme level in cells containing a fusion with a mutant of O1 were maintained at a high level. The results are consistent with the idea that O1 contributes to the repression of Pv at the start of sporulation.
Translational control of Spo0A synthesis.
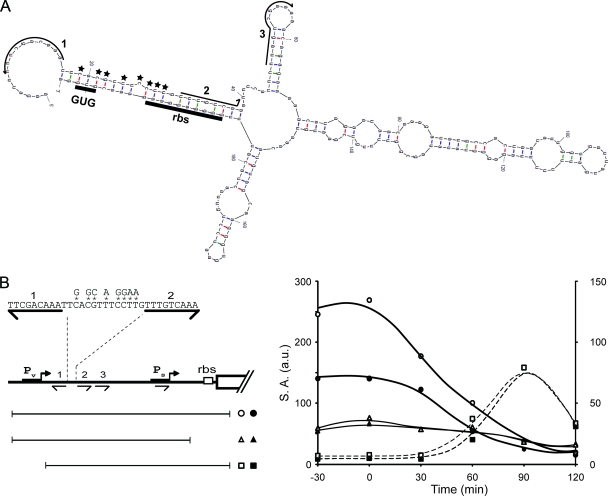
Finally, we noticed that transcripts originating from Pv exhibited an extensive secondary structure, including in particular complementarity between the region containing the ribosome binding site and start codon for the spo0A open reading frame and the region downstream of the O1 sequence and including the O2 sequence (Fig. 7A). We postulated that this secondary structure would be responsible for reducing the translational efficiency of mRNAs originating from Pv but not mRNAs originating from Ps. To test this possibility, we mutated 8 bases in the sequence between sites 1 and 2 (Fig. 7B, left panel) so as to weaken its interaction with the ribosome binding site and the start codon GUG without perturbing either 0A box. As shown in Fig. 7B, the presence of the mutations (open symbols) significantly increased the expression level of a lacZ translational fusion (circles) but had little effect on a transcriptional fusion (triangles). (The above-described experiment was carried out in the presence of a sigH mutation to limit transcription to Pv.) As a control, little or no effect on expression levels was observed when the same mutations were introduced into a translational fusion that lacked Pv (squares). (In this case the cells were wild type for sigH so as to allow transcription from Ps.) We conclude that the switch from Pv to Ps is accompanied by an increase in mRNA translation efficiency, thereby enhancing the rate of Spo0A synthesis at the start of sporulation. (Note that this also suggests that the high level of Spo0A observed in the ΔO1–3 strain at the mid-exponential phase [Fig. 2A] reflects both derepression of Ps due to the absence of O2 and enhanced translation of transcripts originating from Pv, given that the translation-inhibiting sequences are absent in the deletion mutant.)
Fig. 7.
RNA secondary structure impedes translation mRNAs originating from Pv. (A) Secondary structure prediction for the 5′ region of transcripts originating from Pv. RNA sequences corresponding to O1, O2, and O3 are indicated with arrows. The ribosome-binding site (RBS) and initiation codon (GUG) are underlined. Positions of nucleotide substitution mutations are indicated with stars. (B) Expression of spo0A-lacZ fusions integrated at amyE. A typical experiment is presented. The fusions were either translational, containing the full regulatory region (from −305 to +72) (circles) (Abs1019 and Abs1021) or lacking Pv (from −197 to +72) (squares) (Abs988 and Abs957) or were transcriptional, containing the full regulatory region (from −305 to −16) (triangles) (Abs1049 and Abs1050). The fusions either contained the wild-type sequence (black symbols) or were mutants, with the eight nucleotide substitutions indicated in the left panel. Expression of fusions containing the full regulatory region (circles and triangles) were determined in mutant cells lacking σH (sigH) to prevent expression from Ps. Dotted lines correspond to the right axis and continuous lines to the left axis.
DISCUSSION
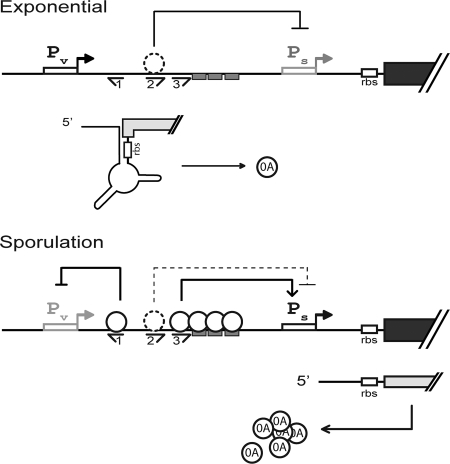
Building on the early work of Chibazakura et al. (11) and Strauch et al. (29) and our recent findings on the accumulation of Spo0A (9, 15), we propose a pump-priming model for the regulation of spo0A that involves transcriptional, translational, and posttranslational mechanisms. At the heart of the model is promoter switching from the vegetative promoter Pv to the σH-controlled, Spo0A∼P-dependent promoter Ps during the transition to stationary phase and the 0A boxes O1, O2, and O3. We propose (Fig. 8) that early during the exponential phase of growth, synthesis of Spo0A is principally if not exclusively driven by Pv. This synthesis is maintained at a relatively low level by a translational control mechanism that sequesters the start codon and ribosome-binding site for spo0A in the secondary structure of transcripts originating from the upstream promoter. Meanwhile, the downstream promoter, Ps, is silent due to the absence of both Spo0A∼P and σH. Then, at the mid-exponential phase of growth, the gene (sigH) for σH is derepressed (9) and Spo0A∼P begins to accumulate due to activation of kinases at the head of the phosphorelay other than KinA (5, 9). Nonetheless, Spo0A synthesis continues to be maintained at a basal (although significant) level due to repression of Ps via O2. Finally, during the transition to stationary phase, increasing levels of KinA lead to a surge in Spo0A∼P levels (13), overpowering O2-mediated repression of Ps and activating Ps via the binding of Spo0A∼P to O3. Meanwhile, the binding of Spo0A∼P to O1 represses Pv, effecting the switch from the vegetative to the sporulation promoter. The switch results in yet higher rates of Spo0A synthesis as a consequence of unimpeded translation from mRNAs originating from the downstream promoter. Thus, Pv primes the pump by maintaining a pool of Spo0A molecules in growing cells, which upon phosphorylation activates Ps, leading to yet higher levels of Spo0A and downregulation of Pv.
Fig. 8.
Model for the control of spo0A expression. The upper panel shows that during the exponential phase of growth, O2 represses transcription from Ps. Thus, transcripts originating from Pv are the chief source of mRNA, which due to secondary structure is impaired in translation. The dotted circle conveys that the identity of the molecule bound at O2 has not been confirmed as being Spo0A∼P biochemically. The lower panel shows that during the transition to stationary phase in sporulation-inducing medium, Pv is repressed by Spo0A∼P bound at O1, and Spo0A∼P molecules bound at O3 and downstream activate Ps, overriding repression by O2. Cooperative binding of Spo0A∼P molecules renders the switch from Pv to Ps highly sensitive to small changes in Spo0A∼P levels. The rate of Spo0A synthesis is enhanced both by the switch to the strong Ps promoter and the enhanced translatability of mRNA originating from Ps.
Our model raises several questions about the mechanism of action of the upstream 0A boxes as we now consider the following.
O1.
Because O1 is close to and downstream of the start site for Pv, the binding of Spo0A∼P to O1 likely represses Pv simply by interfering with the binding of RNA polymerase to the promoter.
O2.
We presume that O2, which is a perfect match to an 0A box, functions via the binding of Spo0A∼P, but neither we (see Fig. S3 in the supplemental material) nor Strauch and collaborators observed protection of O2 in DNase I footprinting experiments (29). Assuming that Spo0A∼P does bind to O2 in vivo despite the failure to detect such an interaction in vitro, an appealing possibility would be that Spo0A∼P bound at O2 forms a loop with an unidentified downstream sequence that occludes the binding of RNA polymerase to Ps. An alternative possibility is that a regulatory protein other than Spo0A∼P that functions during the transition to stationary phase binds to O2. If so, evidence indicates that the protein is not SinR, CodY, or Hpr (ScoC) (data not shown).
O3.
O3 is a positively acting 0A box that governs the Spo0A∼P-dependent activation of Ps. Binding of Spo0A∼P to O3 has been confirmed biochemically. We presume that O3 works via the direct interaction of Spo0A∼P bound at this site with σH-RNA polymerase, facilitating the binding of the transcription enzyme to Ps. Evidence presented in Results is consistent with the idea that this interaction occurs via looping of the intervening sequence. We further suggest that loop formation is facilitated by the observed binding and oligomerization of Spo0A∼P toward Ps. Zhao et al. (32) propose that dimers of Spo0A∼P oligomerize in a head-to-tail manner. In this oligomeric state, only one Spo0A∼P molecule in each dimer can interact with the Spo0A-binding sequence (chiefly with Gs), the other molecule being tilted and making interactions with the phosphate backbone (32). Thus, it is tempting to speculate that in curved DNA both Spo0A∼P molecules of a dimer contact bases, possibly Gs, thereby stabilizing or changing the degree of curvature. In support of this idea, the AT richness of the intervening DNA would be expected to impart flexibility and/or natural bending to the intervening DNA, and the G in the repeated, 10-bp motifs (AAAWNNDAGA) downstream of O3 (see Results) could be a contact site for Spo0A∼P molecules. If so, cooperative binding of multiple Spo0A∼P molecules to the intervening DNA could render activation of Ps sensitive to small changes in Spo0A∼P levels, creating a high-sensitivity switch and counteracting the repressive effect of O2.
O4.
The promoter-proximal, Spo0A∼P binding site O4 evidently does not play an important role in promoter switching given the demonstration that we could functionally replace Ps with another σH-controlled promoter (PspoVG) that is Spo0A∼P independent without apparently impeding the function of O2 or O3. Thus, in our view Ps is intrinsically simply a σH-controlled promoter, and the function of an embedded 0A box in Ps remains mysterious.
What is the biological significance of such a complicated and intricate regulatory region? We suggest that the regulatory region allows the cell to produce extremely large amounts of Spo0A (rising from ∼2,000 to ∼20,000 molecules per cell) on demand as the phosphorelay is activated such that the Spo0A protein never becomes rate limiting for the generation of Spo0A∼P. In this just-in-time scenario, Pv generates a basal level of Spo0A (∼2,000 molecules/cell) that can prime the pump for rapidly producing Spo0A when the phosphorelay is activated. Thus, in its absence, Spo0A accumulation is greatly delayed and sporulation efficiency is slightly impaired (see Fig. S1 and S5 in the supplemental material) (12). We have also discovered that entry into the state of genetic competence is impaired in a mutant lacking Pv (see Fig. S5) (but not in a mutant lacking Ps [27]). It was known that genetic competence requires Spo0A, and our present results indicate that Pv is responsible for providing enough Spo0A molecules for entry into this state. This is in agreement with the view that Spo0A from Pv-directed transcription is limiting for development of competence (22). Nonetheless, we are left with the puzzling finding that Pv is dispensable when O1, O2, and O3 are removed. That is, the pattern of Spo0A accumulation when both Pv and O1-O3 were removed was similar to that seen when the regulatory region was left intact (Fig. 2A). Thus, a truncated regulatory region, simply representing a σH-controlled promoter (Ps), was sufficient to mimic, at least at a coarse level, the post-exponential-phase induction of Spo0A synthesis seen in the wild type. We presume that Ps alone is not as tightly regulated as the entire intact regulatory region in a manner that confers a fitness advantage to the cell. For example, the intact regulatory region may be needed to suppress noise and minimize cell-to-cell variation in Spo0A levels during growth. If so, additional detailed experiments will be required to uncover differences between the behavior of Ps and the intact regulatory region.
In summary, we propose that Pv and O2 maintain Spo0A at a high, basal level (∼2,000 molecules/cell) during growth. As a result, the cells are poised to respond rapidly to signals triggering activation of the phosphorelay. Phosphorylation of Spo0A as a result of flux through the relay sets up a self-reinforcing cycle that rapidly amplifies Spo0A production to extremely high levels (∼20,000 molecules/cell), preventing Spo0A from becoming limiting for the accumulation of Spo0A∼P. Whereas the basal level of Spo0A may be relatively constant from cell to cell (as we propose), Spo0A∼P levels vary considerably from cell to cell at the start of sporulation (9). This heterogeneity likely originates from noise in the phosphorelay and is the basis for the diversification of cell types during the transition to stationary phase, resulting in cannibals, biofilm formers, and spore formers (17, 21).
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Grossman and C. Lee for plasmid psk5.
This work was supported by NIH grant GM18568 to R.L.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Arnaud M., Chastanet A., Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asayama M., Yamamoto A., Kobayashi Y. 1995. Dimer form of phosphorylated Spo0A, a transcriptional regulator, stimulates the spo0F transcription at the initiation of sporulation in Bacillus subtilis. J. Mol. Biol. 250:11–23 [DOI] [PubMed] [Google Scholar]
- 3. Britton R. A., et al. 2002. Genome-wide analysis of the stationary-phase sigma factor (Sigma-H) regulon of Bacillus subtilis. J. Bacteriol. 184:4881–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burbulys D., Trach K. A., Hoch J. A. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 5. Castilla-Llorente V., Salas M., Meijer W. J. 2008. kinC/D-mediated heterogeneous expression of spo0A during logarithmical growth in Bacillus subtilis is responsible for partial suppression of phi 29 development. Mol. Microbiol. 68:1406–1417 [DOI] [PubMed] [Google Scholar]
- 6. Chastanet A., Fert J., Msadek T. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061–1073 [DOI] [PubMed] [Google Scholar]
- 7. Chastanet A., Losick R. 2007. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol. Microbiol. 64:139–152 [DOI] [PubMed] [Google Scholar]
- 8. Chastanet A., Prudhomme M., Claverys J. P., Msadek T. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chastanet A., et al. 2010. Broadly heterogeneous activation of the master regulator for sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 107:8486–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen G., Kumar A., Wyman T. H., Moran C. P., Jr 2006. Spo0A-dependent activation of an extended −10 region promoter in Bacillus subtilis. J. Bacteriol. 188:1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chibazakura T., Kawamura F., Asai K., Takahashi H. 1995. Effects of spo0 mutations on spo0A promoter switching at the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177:4520–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chibazakura T., Kawamura F., Takahashi H. 1991. Differential regulation of spo0A transcription in Bacillus subtilis: glucose represses promoter switching at the initiation of sporulation. J. Bacteriol. 173:2625–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eswaramoorthy P., Dinh J., Duan D., Igoshin O. A., Fujita M. 2010. Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells. Microbiology 156:2294–2304 [DOI] [PubMed] [Google Scholar]
- 14. Fujita M., Gonzalez-Pastor J. E., Losick R. 2005. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 187:1357–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujita M., Losick R. 2005. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 19:2236–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujita M., Losick R. 2002. An investigation into the compartmentalization of the sporulation transcription factor SigmaE in Bacillus subtilis. Mol. Microbiol. 43:27–38 [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez-Pastor J. E., Hobbs E. C., Losick R. 2003. Cannibalism by sporulating bacteria. Science 301:510–513 [DOI] [PubMed] [Google Scholar]
- 18. Grossman A. D. 1995. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu. Rev. Genet. 29:477–508 [DOI] [PubMed] [Google Scholar]
- 19. Haran T. E., Mohanty U. 2009. The unique structure of A-tracts and intrinsic DNA bending. Q. Rev. Biophys. 42:41–81 [DOI] [PubMed] [Google Scholar]
- 20. Lewis R. J., et al. 2000. Domain swapping in the sporulation response regulator Spo0A. J. Mol. Biol. 297:757–770 [DOI] [PubMed] [Google Scholar]
- 21. Lopez D., Vlamakis H., Kolter R. 2009. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33:152–163 [DOI] [PubMed] [Google Scholar]
- 22. Mirouze N., Prepiak P., Dubnau D. 2011. Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PLoS Genet. 7:e1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Molle V., et al. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 24. Pérez-Martin J., Rojo F., de Lorenzo V. 1994. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 58:268–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seredick S., Spiegelman G. B. 2001. Lessons and questions from the structure of the Spo0A activation domain. Trends Microbiol. 9:148–151 [DOI] [PubMed] [Google Scholar]
- 26. Seredick S. D., Spiegelman G. B. 2007. Bacillus subtilis RNA polymerase recruits the transcription factor Spo0A ∼P to stabilize a closed complex during transcription initiation. J. Mol. Biol. 366:19–35 [DOI] [PubMed] [Google Scholar]
- 27. Siranosian K. J., Grossman A. D. 1994. Activation of spo0A transcription by sigma H is necessary for sporulation but not for competence in Bacillus subtilis. J. Bacteriol. 176:3812–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sterlini J. M., Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Strauch M. A., Trach K. A., Day J., Hoch J. A. 1992. Spo0A activates and represses its own synthesis by binding at its dual promoters. Biochimie 74:619–626 [DOI] [PubMed] [Google Scholar]
- 30. Travers A. A. 2004. The structural basis of DNA flexibility. Philos. Transact. A Math. Phys. Eng. Sci. 362:1423–1438 [DOI] [PubMed] [Google Scholar]
- 31. Youngman P., Perkins J. B., Losick R. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1–9 [DOI] [PubMed] [Google Scholar]
- 32. Zhao H., Msadek T., Zapf J., Madhusudan, Hoch J. A., Varughese K. I. 2002. DNA complexed structure of the key transcription factor initiating development in sporulating bacteria. Structure 10:1041–1050 [DOI] [PubMed] [Google Scholar]
- 33. Zuber P., Losick R. 1987. Role of AbrB in Spo0A- and Spo0B-dependent utilization of a sporulation promoter in Bacillus subtilis. J. Bacteriol. 169:2223–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.