Abstract
Mammalian cells have evolved sophisticated DNA repair systems to correct mispaired or damaged bases and extrahelical loops. Emerging evidence suggests that, in some cases, the normal DNA repair machinery is “highjacked” to become a causative factor in mutation and disease, rather than act as a safeguard of genomic integrity. In this review, we consider two cases in which active MMR leads to mutation or to cell death. There may be similar mechanisms by which uncoupling of normal MMR recognition from downstream repair allows triplet expansions underlying human neurodegenerative disease, or cell death in response to chemical lesion.
Keywords: Mismatch repair, MSH2, MSH3, MSH6, DNA repair, microsatellite instability, trinucleotide repeats, CAG repeats, CTG repeats, base excision repair, break repair, OGG1, Huntington’s Disease, Myotonic dystrophy, cisplatin lesions, DNA lesions, chemical lesions
INTRODUCTION
The mismatch repair [MMR] system protects genomic integrity by removing mismatched, chemically modified, or extrahelical DNA in an ATP dependent reaction. The first step of MMR is carried out in both bacteria, yeast and in mammalian cells by MutSα-homologue [MSH] recognition complexes, which couple lesion binding with ATP hydrolysis, and initiate an ordered series of events leading to repair of the lesion and to mutation avoidance [1–6]. Compromise of the MMR system in cells and tissues leads to an increase in spontaneous mutation rate, which is typically referred to as a mutator phenotype [1–3]. The mutational spectrum in these mismatch repair-defective tumors reflects the inability to correct post-replicative errors throughout the genome [1–3], and result in both base substitution mutations and microsatellite instability. For example, about 15% of patients with hereditary non-polyposis colorectal cancer [HNPCC] have wide-spread genome instability, characterized by changes in copy number at repetitive tracts throughout the genome [1–6].
There is a second, less-well studied class of MMR defects, which does not involve mutant forms of MMR machinery [Figure 1][4–6]. Instead, lesion binding itself induces deleterious changes in the MMR initiation complexes that lead to defects in downstream signaling. Defective repair arises from a deleterious conformational change typically during recognition of complex lesions such as looped-out CAG tracts [4] or bulky lesions such as platinum (Pt) [5, 6] or O6-methylguanine [4–6]. In these cases, the MMR system not only fails to act as a guardian of the genome, but become causative factors, leading, for example, to CAG expansion [7–9], the lethal mutation underlying Huntington’s Disease [HD], or to cell death in the case of Pt or O6-methylguanine [6]. This review focuses on mechanisms by which the native MMR proteins are co-opted to cause mutation [Figure 1]. We refer to this process as “highjacking”.
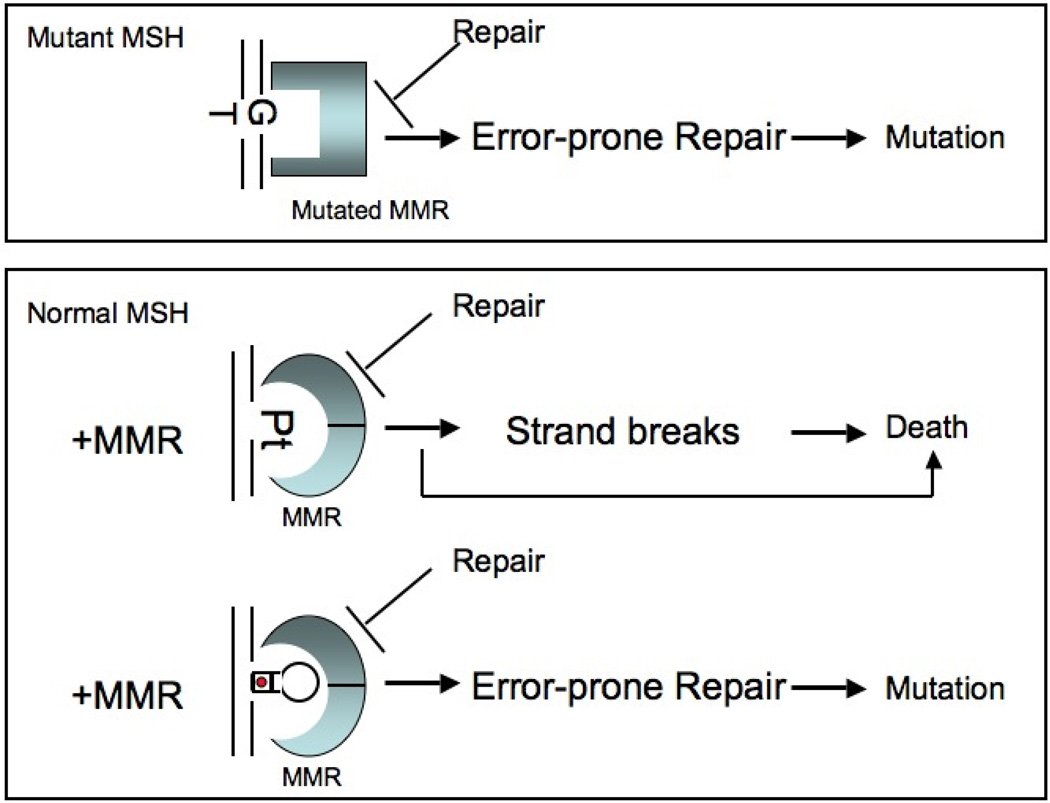
Figure 1. Defects of MMR leading to mutation and cell death. [top].
Mutant form of MMR machinery [mMSH] recognizes a single base G/T lesion. Blue square indicates a mutated MSH2 recognition protein. Faulty processing of the lesion leads to missense mutations. [bottom] Normal MSH protein [blue rings] recognizes bulky lesions. Pt signifies platinum inter-strand crosslinking. Interaction of the MSH protein with the lesion blocks repair. Faulty repair induces toxic strand breaks due to futile cycling, which leads to death. Alternatively, cell death arises from direct damage signaling [arrow]. At the hairpin site, MSH2 binds to the lesion, undergoes conformational change that inhibits faithful loop repair. Faulty repair leaves behind unrepaired loops, which are incorporated into the DNA to cause expansion mutations. The hairpin represents a CAG fold-back structure with an A/A mispaired base every third nucleotide. Red dots in hairpin represent A/A mismatched base in the stem.
A. A FUNCTIONAL MISMATCH REPAIR COMPLEX CAUSES TRINUCLEOTIDE EXPANSION IN VIVO
I. A native MSH2/MSH3 complex causes CAG/CTG expansion in Huntington’s Disease and Myotonic Dystrophy
HD is one of several progressive neurodegenerative disorders in which the underlying mutation is an expansion in a simple trinucleotide sequence [7–9]. Expansion causes disease when a particular base sequence is repeated beyond the normal range, interfering with the expression or properties of a gene product [7–9]. Disease severity and onset depend on the number of CAG repeats. As the length of the repeat grows, so also do the size of the successive expansions and the likelihood of another unstable event [Figure 2]. In Huntington’s disease, for example, instability and pathogenesis are not observed at 28 repeats, occur frequently at 36 repeats and are certain above 60 repeats [Figure 2]. The CAG repeat codes for the amino acid glutamine. As the CAG repeat number grows, the growing polyglutamine tract produces an HD gene product (called mutant huntingtin (mhtt)) with increasingly aberrant properties that cause death of brain cells controlling movement [red, Figure 2].
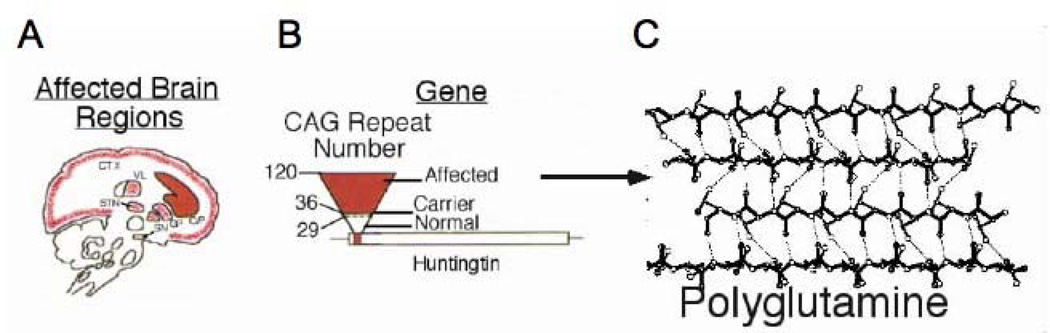
Figure 2. CAG expansion and toxicity in Huntington’s Disease.
[A] Huntington’s Disease is a severe neurodegenerative disorder, which is characterized by regional cell death. mhtt targets cells in the enkephalinergic, medium spiny neurons in the striatum and in the cortex for death. Red regions indicate the major areas of neuronal loss in HD patients with 36–120 CAG repeats. These brain regions control movement. C/P is the caudate/putamen; CTX is the cortex; GP is globus pallidus; STN is subthalamic nucleus; VL is ventrolateral thalamic nucleus; SN is substantia nigra. [B] The relationship between CAG repeat number and HD pathophysiology. [right] Schematic representation of the HD gene: the open bar represents the coding region of the Huntington’s gene [called huntingtin]; the lines indicate the non-coding portions of the gene; the small red bar indicates the position of the CAG repeat stretch located within the N-terminal portion of the coding sequence. Upside down triangle represents increasing number of CAG repeats. Base of triangle in white represents normal unaffected individuals with 6–29 CAG repeats. Dotted lines indicate unaffected carriers for disease with 29–35 CAG repeats. Red part of the triangle indicates affected individuals with 36–120 CAG repeats. [C] Schematic representation of β-sheet conformation of the huntingtin protein harboring a long polyglutamine tract. Dotted lines indicate hydrogen bonding.
Unexpectedly, in crosses of HD transgenic mice with mice lacking MSH2, expansion of triplet repeats was abolished in somatic tissue of their hHD/MSH2[‒/‒] progeny [10,11] and their germ cells [11] [Figure 3]. This unexpected finding revealed that The MMR recognition complex is causative for the mutation. The MMR system typically corrects biosynthetic errors, recombination errors, and is involved as an accessory factor in a number of other DNA repair pathways [1–3]. In eukaryotes, MMR is initiated by one of two heterodimeric MutS homolog [MSH] complexes, MSH2/MSH6 and MSH2/MSH3, with different but overlapping lesions specificities [12–24][Figure 4]. The ideal substrates for repair by MSH2/MSH3 are small, extra-helical loops [IDL], while MSH2/MSH6 prefers single base mismatches. MSH2/MSH3 and MSH2/MSH6 can repair some single base mismatches [17] and single base bulges [18], but MSH2/MSH3 is highly specific for repair looped DNA lesions beyond one base [12–16, 20–24][Figure 4]. CAG hairpin loops form with mispaired bases every third nucleotide [25], and therefore exhibit features recognized by both MMR recognition complexes. However, in vivo, expansion of long disease-length alleles strongly depends on MSH2/MSH3 complex rather than an MSH2/MSH6 complex [26, 27][Figure 4]. Loss of MSH3 abrogates expansion in transgenic mice harboring the 5’CTG3’ repeat in the 3’ non-coding region of the DM transgene [26] as well as 5-’CAG-3’ tract occurring within the coding sequence of the hHD transgene [27]. In both systems, loss of MSHS6 had marginal, tissue specific effects, which did not block expansion, but modulated the size of the expanded tracts. The mechanism by which MSH2/MSH3 causes expansion of TNR tracts is not understood. However, data from HD patients, bacteria, yeast, and animals have provided hints.
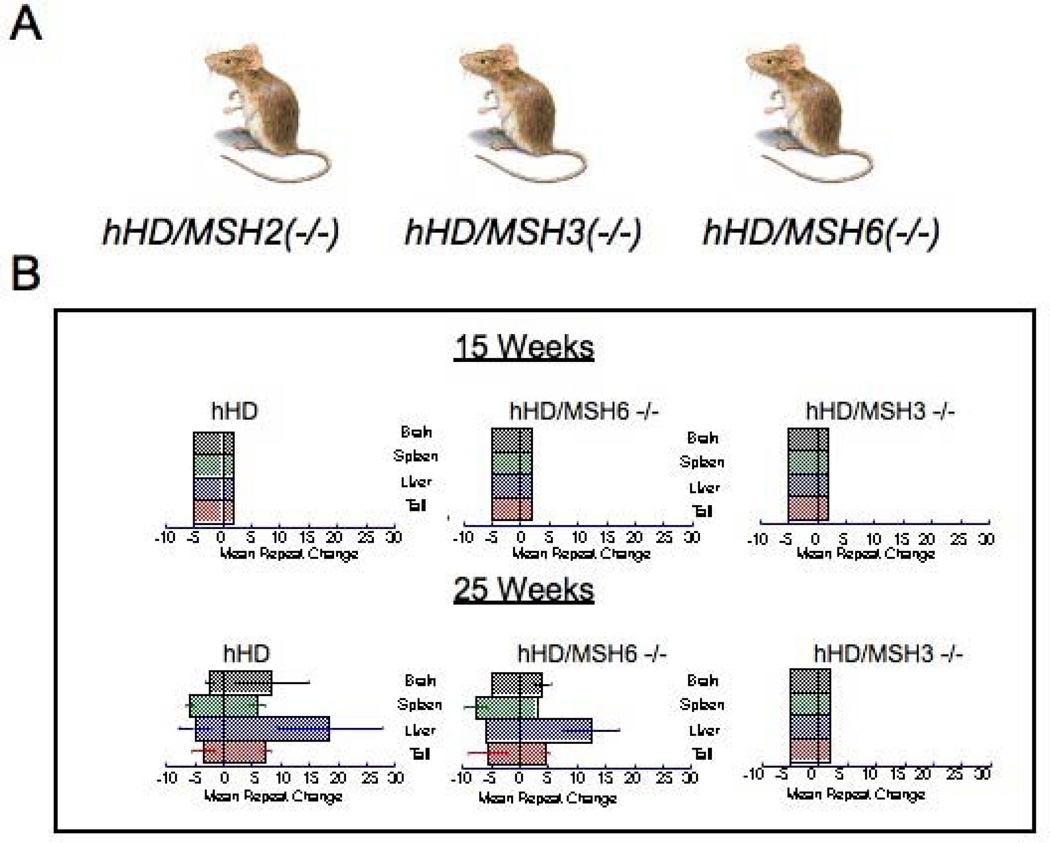
Figure 3. MSH2/MSH3 but not MSH2/MSH6 play a causative role in CAG expansion.
[A] The CAG repeat sizes of the indicated mouse crosses used in the analysis. hHD is the parental R6/1 animals expressing exon 1 of the human Huntington’s Disease. Crosses with hHD lacking MSH3 or MSH6 are indicated. [B] The changes in CAG repeats sizes [mean repeat change] for hHD/MSH2[‒/‒], hHD/MSH3[‒/‒], and hHD/MSH6[‒/‒] animals at 15 weeks and 25 weeks, as indicated. No expansion is observed in any line at 15 weeks of age. However, expansion is prevalent in hHD and hHD/MSH6[‒/‒] animals, while no expansion is observed in hHD/MSH2[‒/‒] and hHD/MSH3[‒/‒] animals. Colored bars represent the indicated tissues.
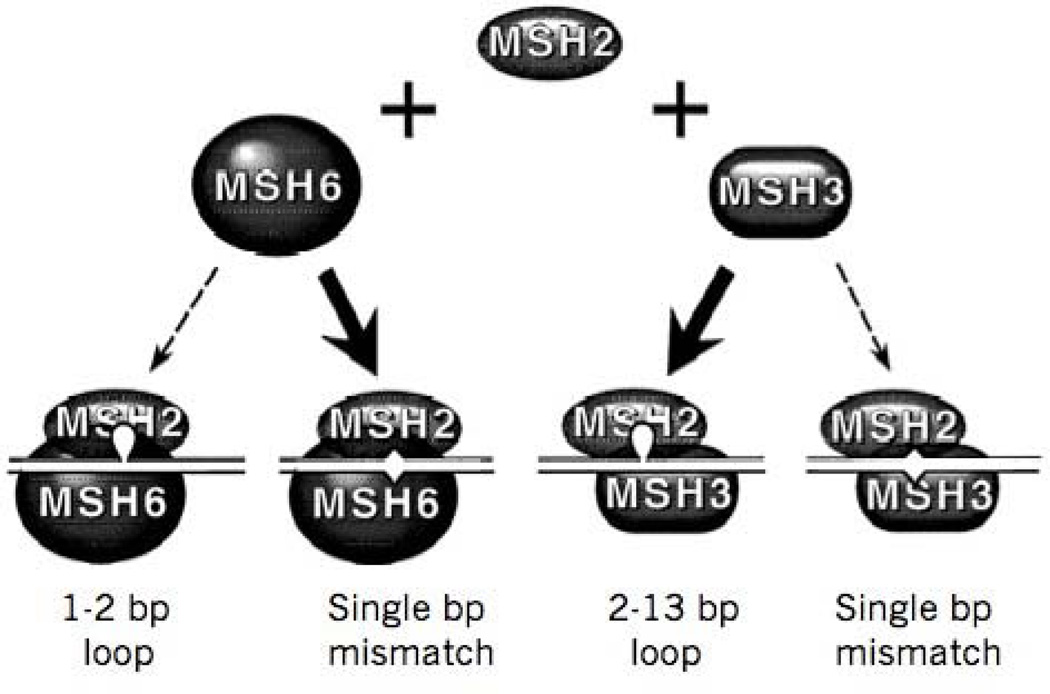
Figure 4. Eukaryotic mismatch repair complexes.
Two eukaryotic heterodimeric proteins are homologous to the bacterial MutS, and both share a common MSH2 subunit. MSH2/MSH3 and MSH2/MSH6 are responsible for recognition of lesions in DNA, but with different specificities. The MSH2/MSH6 complex has a strong preference for repairing single base mismatches, but has very little ability to repair small loops beyond 2 nucleotides. Both MSH2/MSH3 and MSH2/MSH6 can recognize single base bulges. However, MSH2/MSH3 has strong specificity for small heteroduplex loops between 2–13 nucleotides. MSH2/MSH3 can also repair some single base mismatches.
II. Triplet repeat disorders are “single-site” mutations
In HD patients, expansion is largely limited to the long disease-length alleles, which are rare elsewhere in the genomes of normal individuals. The “single-site” pattern of mutation in HD patients is distinctly different from that found in patients with hereditary non-polyposis colon cancer [HNPCC][28]. Therefore, expansion in HD patients could not be readily explained by mutations in the MMR machinery. In HNPCC patients, mutations in MMR that compromise MMR integrity result in widespread microsatellite instability [mutator phenotype] because the mutated MMR proteins do not function properly at most post-replicative lesions in the genome [28]. In contrast, instability in HD patients occurs only in the disease allele and not elsewhere in the genome. The same is true for FRDA and other triplet diseases. Based on these data, the repair defect in trinucleotide diseases is unlikely to reflect faulty function of the mutated repair machinery. More likely, expansion arises from the inability of the normal repair machinery to work at specific sites. Specifically, in such a model, mismatch repair proteins would be expected to operate normally at most sites in the genome, but act improperly at specific sites, i.e., the long disease-length triplet allele. Such a model is consistent with the fact that HD patients develop striatal pathology and neurodegeneration due to the expanded allele, but are not prone to cancer relative to the unaffected population.
III. Stable hydrogen bonding blocks repair of hairpin loops in vivo
While the animal data unambiguously pointed to MMR as a causative factor in the expansion mutation, the involvement of MMR was counterintuitive. Expansion, by definition, is a process in which “extra” DNA is incorporated into the DNA template, yet the cellular role for MSH2/MSH3 is to excise heteroduplex DNA [1–6]. These apparently conflicting data raised the issue that MMR might recognize heteroduplex loops, but fail to excise the lesion and carry out repair. Several lines of evidence suggested that such a model was possible in vivo. Moore and co-workers [29] forced recombination in yeast at the His locus to produce stable heteroduplex loops from at least 10 different triplets [including CAG, CTG, GTT and ATT]. Heteroduplex loops generated from triplets capable of forming stable hairpin structures were maintained as loops and escaped repair, while loops formed from non-structure forming triplets [e.g. GTT] were efficiently removed from the genome [29]. By making the appropriate point mutations, TNR tracts with base substitutions designed to destabilize hairpin structures were also repaired efficiently. These data suggested that repair of hairpins is blocked through stable hydrogen bonding and active formation of secondary structure, not just the trinucleotide sequence [29][Figure 5]. The latter analysis was performed in cells undergoing meiosis. However, in another line of investigation, the same conclusion was reached by experiments in proliferating yeast cells [30–33]. Using a 5-fluororotic acid selection method, LaHue and co-workers quantified the frequency of expansions in vivo for a number of triplet repeat tracts CAG, CTG [30, 31], GTT, GAC [32], and CGG [33]. Their analysis revealed that triplet repeats capable of forming secondary structures increased the frequency of expansion in yeast anywhere from 5–1000-fold [30–33]. Equivalent repeat tracts lacking the capacity to form secondary structure did not expand in vivo and the mutation frequency could not be distinguished from background.
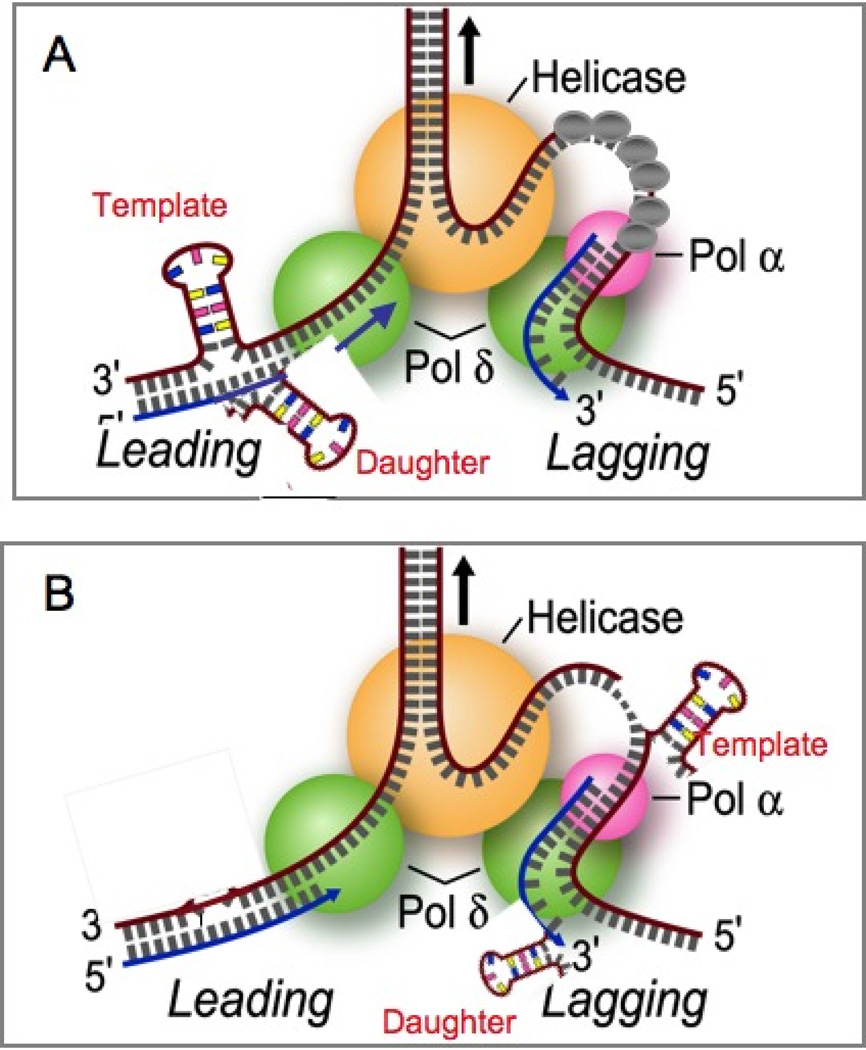
Figure 5. Polymerase slippage model for expansion based on E. coli.
During replication, the TNR units can misalign resulting in an extrahelical DNA loop that increases TNR length if it occurs on the daughter strand (daughter in red), and a decrease in TNR length if it occurs on the template strand (template in red). [A] Model for hairpin formation during leading strand synthesis. [B] Model for hairpin formation during lagging strand synthesis. In either model, hairpins can theoretically form on either the template or the daughter strand (shown in red). Grey bars are random DNA bases. Green balls are polymerase, orange ball is the helicase at the replication fork, grey balls are the single strands binding proteins. Pink ball is primase.
These data indicated that not only were heteroduplex structures able to form in vivo, but they constituted the key intermediates in the expansion mechanism [34]. Physical analyses over the years have confirmed that structure-forming ability is a common feature among expansion capable sequences. To name a few, CAG or CTG [25] and CGG [35] formed stable stem-loop structures comprising a repeat unit of two GC pairs and a mismatched pair. The mutation underlying Friedreich’s Ataxia [FRDA] is a GAA repeat which can form a pyrimidine-purine-pyrimidine (YRY) or purine-purine-pyrimidine (RRY) triple helix containing non Watson-Crick pairs [33, 35–38]. Similar to hairpins, the triplex structures mediate intergenerational instability in 96% of transmissions in FRDA patients [35]. Several structures have been reported for CGG and CCG and more complex minisatellites [such as telomeres] including i-motif [39], e-motif [40] and quadruplex DNA [41].
IV. Potential mechanisms by which MMR might cause CAG expansion
A role for MMR in mediating expansion, however, was far from clear. It made sense that if heteroduplex looped DNA could form in vivo, it could serve as a substrate for MMR. What mechanisms might allow structures to form in duplex DNA, however, and the role of MMR in this process were not obvious. MMR cooperates in many cellular repair processes in addition to its functions in post-replicative MMR [1–6]. At CAG repeats, for example, hairpins have been reported to form at the site of single-strand breaks [27, 42, 43], at origins of replication [44], during recombination [45–48], or by polymerase slippage during proliferation [49–54, 43]. Pearson and colleagues, in this issue, discuss some of these mechanisms. Although initially an overwhelming list of possibilities, these mechanisms can be broadly grouped into two functional classes, those that depend on cell proliferation and those that depend on break-dependent replication. However, the requirement for MMR has, at least in part, been successful in pointing to those mechanisms most likely to operate in mammalian cells [7–9, 54].
MMR is designed to correct post-replicative base mismatches and loops [1–6]. Thus, slippage during cell proliferation was one of the earliest models for expansion [7–9, 49, 50, 52–58]. In fact, a defect in canonical processing of hydrogen-bonded loops is an attractive model for expansion. During replication, the TNR units can misalign resulting in an extrahelical DNA loop that increases TNR length if it occurs on the daughter strand, and a decrease in TNR length if it occurs on the template strand [7–9, 54] [Figure 5]. In both prokaryotic and eukaryotic models, it is well established that microsatellite instability is sensitive to the length of the repeat [7–9, 51–55], to its structure forming potential [25, 29–37, 54], and to its proximity to the origin of replication [44]. DNA loops are the preferred substrate for MSH2/MSH3, and are the intermediates for expansion [7–9]. Thus, a role for MSH2/MSH3 in faulty processing during post-replicative repair was consistent with both the lesion specificity of MSH2/MSH3 [1–3, 14–24] and its requirement for expansion in vivo. However, several observations indicated that inhibition of post-replicative MMR repair per se was not the source of the expansions. Yeast and bacteria have been used to examine the effect of proliferation on TNR expansion [7–9, 54]. Theoretically, expansion should occur if slippage is on the daughter strand. [7–9]. However, deletions of TNR tracts are favored over insertions during proliferation in yeast and bacteria by a factor of 10–1000 in wild type cells [30–32, 55, 56]. Moreover, the deletion bias is relatively independent of the MMR [55–57].
Another issue was the size of the slipped structures. Loop size formed by replication slippage is small since the energy requirements for unpairing more than a few bases is larger than for duplex formation [58]. Since the size of any one slip is inconsistent with the tract increases observed in human disease, it has been suggested that expansion could arise if multiple slips occur along the CAG template [59]. Indeed, it has been widely reported in primer extension assays in vitro that triplet repeats inhibit polymerase progression and pausing along the tract [49, 50]. In vitro, both 5’CAG and 5’CTG inhibit polymerase progression [49, 50], and slippage should occur equally on the daughter and template strand. Yet, expansions are infrequent and, in bacteria and in yeast, there is a strong deletion bias during cell proliferation. Constraints during proliferation, at least in a simple model, do not favor slippage on the daughter strand (7–9, 51, 53). Moreover, slippage should occur whether polymerase passage occurs on the leading or lagging orientation, yet there is an orientation-dependence to expansion in vivo. 5’-CTG or 5’CGG repeats have greater susceptibility to deletions when they serve as a template for lagging strand DNA synthesis. However, as the leading strand template, 5’-CTG or 5’ CGG repeats are relatively stable [31, 49–54, 60–65]. Thus, it is not obvious why slippage should be limited to only one orientation if the same sequence is being copied [66]. If some aspect of the lagging strand configuration promotes slippage, there is no compelling explanation for why it should be limited to the template strand. Recent in vitro experiments have provided, however, at least one explanation [60].
In those experiments, both 5’CAG and 5’CTG repeats inhibits polymerase progression, but 5’-CAG is the most severe [60]. The impediment occurs only on the leading strand because SSB protein eliminates the impediment on both sequences [60]. This outcome indicates that the instability would be driven on the leading strand, consistent with the observation that instability is greater when 5’CTG is in the lagging strand. Thus, the DNA synthesis rate becomes slower than the helicase unwinding rate when 5’CAG is the leading template and threatens functional coupling. To avoid uncoupling, the DNA polymerase pushes away and passes over a small tract of 5’-CAG DNA in the leading strand template, saving synthesis time, and the unreplicated 5’-CAG hairpin on the leading strand template is trapped after polymerase passage, The trapping of loops may explain why MSH2/MSH3 has only modest effects on deletion, and why deletion is favored over insertions during replication. This study supports previous measurements in which leading strand can produce deletions [52], and 5’CTG can inhibit polymerase progression [49, 50]. However, such a model argues against a mechanism in which MSH2/MSH3 causes expansion during post-replicative repair.
Although there are notable exceptions [69–70], most CAG tracts in proliferating HD patient fibroblasts are also stable or delete trinucleotide repeats even in the presence of an intact MMR system [71–73]. Thus, a simple replication slippage model is not entirely consistent with the requirement of MSH2/MSH3 in causing expansion. MSH2/MSH3 is needed for somatic age-dependent expansion in HD and DM animals [43]. However, at least in HD animals, expansion occurs in post-mitotic neurons well after replication has ceased [43, 74]. Simple slippage during proliferation, although it occurs, does not mediate the expansion observed in human disease, nor does it predict the deletion bias observed in rapidly replicating bacteria and yeast [54].
Replication could, however, cause expansion in the context of strand break repair [Figures 6 and 7]. Indeed, several lines of evidence supported this hypothesis. CTG tracts appear to be fragile sites. In one assay, a system was developed in which breakage within the CAG/CTG tracts could be corrected to restore expression of a selectable marker that promoted survival in the presence of 5-fluoroorotic acid. Indeed, breakage occurred at CAG tracts of 130 but not at smaller tracts, and the frequency of recombination increased with the length of the repeat [45]. Moreover, RAD1 and FEN-1 had the most impact on the insertion events. Given the fact that FEN-1 has functions in both replication and recombination, these data suggested that expansion might occur during gap-filling synthesis in recombination by single strand annealing to repair breaks in DNA [45]. Other recombination assays have yielded similar conclusions. For example, in yeast, a chromosomal site was engineered to harbor a single HO endonuclease site. After cleavage, the HO-induced chromosomal break can be corrected by gene conversion from a plasmid containing homologous sequences and a CAG/CTG tract [46]. Surviving cells repaired the site using the plasmid as the sequence donor. Both expansion and contraction were observed at the chromosomal break. However, in the same cell, only deletions were found within CAG or CTG repeats of the replicating donor plasmid [46]. Thus, increases in tract length expansion were associated exclusively at the chromosomal break site.
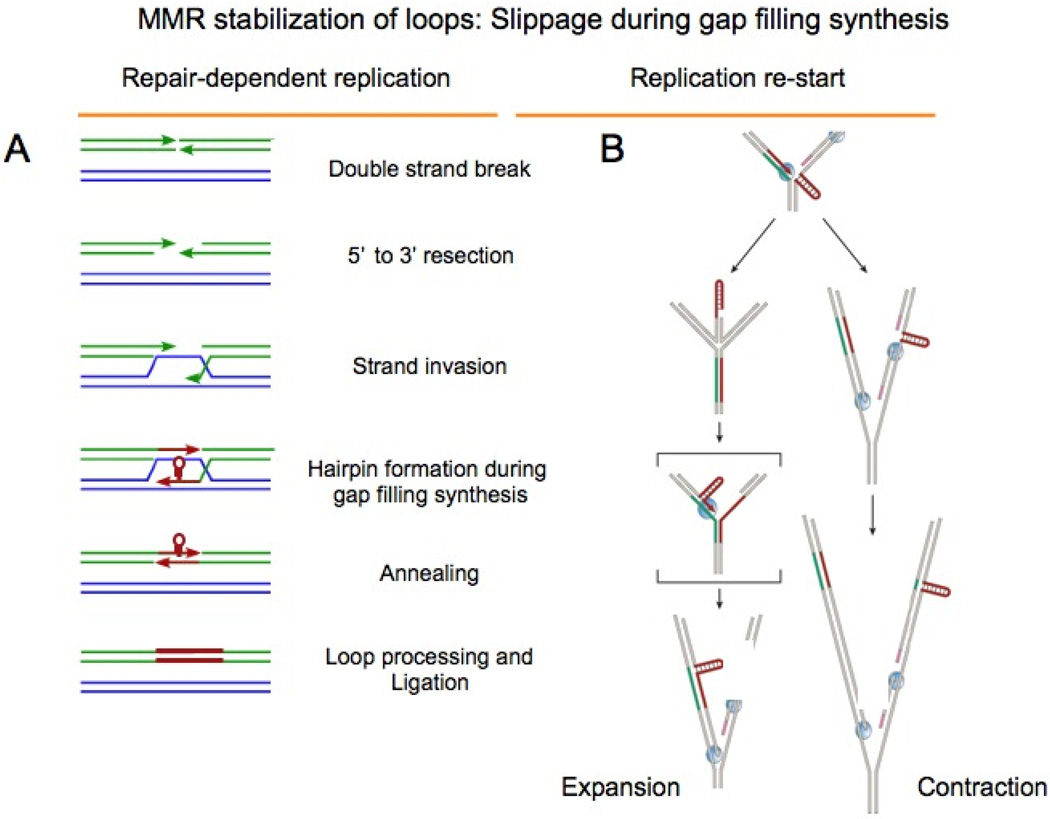
Figure 6. Break-dependent replication; expansion during gap filling synthesis.
Recombination models for repeat expansions. [A] Double strand breaks are repaired on the homologous chromosomes (shown in green and blue) and repetitive DNA strands are shown in red. The two ends are resected to leave unpaired overhangs that can invade the homologous chromosome or a sister chromatid. Slippage during gap filling synthesis can generate hairpins. The strands containing the hairpin reanneal, and expansion occurs after loop processing (in red). (B) A model for repeat instability based on fork stalling and replication restart within the repetitive region [taken from [7] Repeat contractions (upper pathway) occur when the machinery for the lagging-strand synthesis skips the repetitive hairpin on the lagging-strand template. Repeat expansions (lower pathway) can occur during replication fork reversal and restart, leading to the formation of a repetitive hairpin on the nascent leading strand. The structure-prone strand of the repetitive run is shown in red, its complementary strand in green, and flanking DNA in beige. DNA polymerases are shown in blue, primers for Okazaki fragments in pink, and single-stranded-DNA-binding proteins as grey circles. The bracketed intermediate contains a hairpin on the nascent strand, which can be stabilized by MSH2/MSH3.
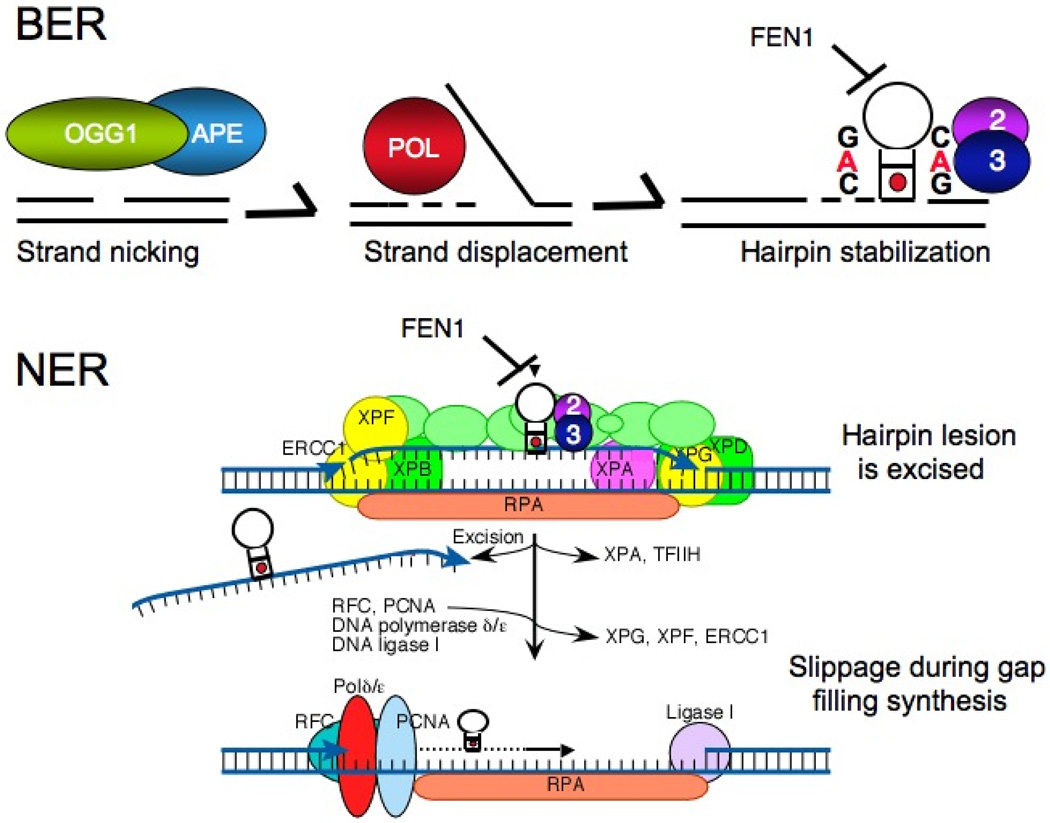
Figure 7. Excision repair models for expansion.
[A] The base excision repair model for expansion. Expansion can be directly induced under conditions of oxidation in mammalian cells. The 7,8-dihydro-8-oxo-guanine-DNA glycosylase [OGG1](green ball), a DNA glycosylase responsible for removal of oxidized guanines creates a single strand nick in the DNA. Together with the apo-endonucelase (APE)(blue ball) process the end to allow polymerase (Pol in red) to undertake gap-filling synthesis (dotted lines). The hairpins form by strand displacement together with any slippage that might occur during synthesis. The loading of FEN1 is inhibited on the hairpin since its 5’ end is hidden. The binding of MSH2/MSH3 stabilizes the repetitive hairpin, preventing flap removal. The slipped-stranded intermediate is converted into an expansion by an error-prone repair pathway. [B] The nucleotide excision repair model for expansion. There has been no direct data to support such an NER mechanism, to date. However, the hairpin can act as a complex lesion. After lesion recognition, a 10–20 nucleotide bubble is opened by TFIIH containing the XPB an XPD helicases followed by XPA binding. Strand cleavage occurs on the 5’ or 3’ side of the lesion by ERCC1/XPF and XPG, respectively. The hairpins lesion is removed, but small slipped structures form during gap filling synthesis by the replication machinery including RFC, PCNA processivity factor, and DNA polymerases δ/ε. The loops are sealed into the DNA with DNA ligase I, and processed to generate an expansion.
Taken together, the results from simple organisms suggested that replication could contribute to expansion in the context of break repair [45, 46, 74–77]. Stalling of polymerase during replication has long been associated with strand breaks. Recently, two-dimensional gel analysis of replication intermediates has provided compelling evidence that polymerase indeed stalls on tracts of CAG repeats [74], CGG repeats [75], and GAA repeats [76] in vivo, and, under these conditions, the rate of expansion increases. These key experiments provide an integrated model for expansion, which accounts for aspects of replication, fragility, and break repair. Explicit break-dependent mechanisms for expansion might be envisioned to include recombination [7–9][Figure 6], replication re-start [77] [Figure 6], or single strand break repair [42, 43][Figure 7].
To what degree mechanisms deduced from model organisms operate in mammalian cells remains an unresolved question. Patient fibroblasts containing long repeats do not lack repair proteins. Thus, their role in causing expansion is difficult to test. Further, tests of expansion among cell types have not always painted a consistent picture due to the fact that the effects of strand breaks cannot be separated from the effects of replication in proliferating cells [67–72, 78–80]. However, the use of transgenic animals has been particularly informative [70, 80–86]. Loss of MSH2 stops expansion in germ cells and in somatic cells during development in animals [10, 11, 26, 70, 80, 81], and, as widely observed, leads to deletion during intergenerational transmission in mouse models of HD [70, 80, 81, 84]. Interpretation of these effects is debatable, but it appears that, in the absence of MSH2, recombination in the germ cells, or cell proliferation early in embryogenesis is the source of the deletion events. Results from transgenic animals argue against the former [11, 81–87]. While loss of MSH2 blocked expansion in HD or DM animals, loss of RAD 52, RAD 54, and Ku in DM mice has no effect on expansion of the CTG repeat [81]. Thus, TNR expansion requires MMR, but does not require enzymes generally needed for homologous recombination or non-homologous end-joining in animals. Since expansion occurs in post-mitotic neurons of HD animals and in DRPLA patients, these results suggest that replication is also not required for expansion and, most often, results in deletion [43, 73]. Exclusion of these mechanisms indicates that expansion in animals may arise in the context of single strand break repair, as a consequence of base excision repair [BER], or potentially from nucleotide excision repair [NER][Figure 7]. To date, there is little evidence to support a NER mechanism, but removal of oxidative DNA damage by BER appears to be a relevant model for expansion, at least in somatic cells [43].
In HD mice, age-dependent expansion at the HD locus occurs concomitantly with the accumulation of oxidative DNA damage [43], and expansion can be directly induced under conditions of oxidation in mammalian cells that, otherwise, would not expand their repeats in culture [43]. Importantly, loss of 7,8-dihydro-8-oxo-guanine-DNA glycosylase [OGG1], a DNA glycosylase responsible for removal of oxidized guanines, suppresses expansion in HD mice [43] [Figure 7]. Suppression in HD animals is not observed for other glycosylases tested. Single strand breaks are intermediates of BER. Thus, age-dependent somatic mutation associated with HD implicate repair of single strand breaks as part of the mechanism. Since oxidation occurs throughout life, progressive somatic expansion could be envisioned to occur by a toxic oxidation cycle [43], and provides a plausible mechanism by which expansion might occur in post-mitotic neurons. The role for MSH2/MSH3 in removal of oxidative DNA damage is not entirely understood. However, MSH2 null animals suppress expansion in the presence of OGG1 [11], OGG1 null animals suppress expansion of their repeats in the presence of MSH2 [43], and loss of MSH2 results in tract deletion [70, 80, 81]. Thus, MSH2 and OGG1 are likely to operate in the same mechanistic pathway. There is a large body of literature, which implicates the MMR system in the removal of oxidative DNA damage [87]. Thus, it is possible that physical or functional interactions between the MSH2/MSH3 and OGG1 can cooperate to cause expansion in somatic cells [Figure 7]. Further testing will address whether such a mechanism is correct or whether putative interactions between the two repair proteins would be relevant during intergenerational transmission.
V. A role for MMR in Loop stabilization?
While the exact mechanism that generates looped intermediates is not well understood, expansion, by definition arises from incorporation of DNA loop intermediates into genomic DNA. In that regard, MSH2/MSH3 might cause expansion by either failing to correct post-replicative slippage errors, or by stabilizing a looped intermediate at a break. Although far from clear, current in vivo and in vitro data suggest that the latter is more likely. In one report, circular plasmids were hybridized to single stranded complementary DNA to create long circular templates in which a primer tail existed on one site of the plasmid [88]. Single strand breaks were placed on either side of a preformed hairpin trapped between the primer and the break. These plasmid templates were incubated with repair-competent cell extracts, and the fate of the preformed CAG and CTG loops was followed in vitro during polymerase passage. For trapped loops, expansion in vitro depended on the position of the hairpin with respect to the break [88]. In contrast to mouse models, however, expansion in vitro was independent of MSH2, as cell extracts lacking MSH2 had no effect on loop processing [88]. Since the loops were preformed and trapped in the substrates in the in vitro system, a role for MSH2/MSH3 forming the loop could not be tested directly. However, the fact that changes in the CAG tract length occurred in the absence of MSH2 argues that MMR is not required in vitro once the hairpins are formed. Overall, the requirement for MSH2 in vivo and its dispensable nature in vitro tend to support the notion that MSH2/MSH3 is involved in generating the loop intermediate. A loop stabilization model is attractive since it is consistent with in vivo results that expansion depends on MSH2 in animals, and in its absence, failure to stabilize loops results in CAG tract deletion as it is transmitted in subsequent generations.
VI. Hijacking biochemical functions of Mismatch Repair
Whatever the precise mechanisms for loop formation, MSH2/MSH3 fails to process them faithfully. If MSH2/MSH3 causes expansion by stabilizing looped DNA, then such a model implies that the MSH2/MSH3-hairpin complex is dysfunctional in signaling repair. Indeed, recent analysis of the MSH2/MSH3-CAG hairpin complex reveals that CAG/CTG hairpin binding suppresses most of the biochemical functions required for lesion recognition and initiation of MMR [27]. To carry out repair, all MSH complexes must bind DNA, bind ATP, hydrolyze ATP, and properly couple DNA binding and ATP hydrolysis to signal downstream components of the MMR machinery [1–6]. The structural features of the MSH proteins are exquisitely designed to carry out these functions [18, 89–91][Figure 8]. MSH proteins contain two conserved Walker-type nucleotide binding domains in their carboxy-termini [18, 89–91]. Walker A and Walker B motifs have been shown to both bind and hydrolyze ATP [89–96], which is essential for recognition and repair of lesions in DNA. Nucleotide binding is structurally linked to DNA binding located in two domains at the N-terminus [18, 89–91]. Although the subunits of bacterial MutS are identical, DNA binding creates an asymmetry between them, in which one subunit is in direct contact with a mispaired base and the other making few non-specific or no contacts with the phosphodiester backbone [18, 89–91]. In MutS and in MSH2/MSH6, the subunit that contacts DNA contains a conserved phenylalanine [Phe] residue that stacks onto a single mispaired DNA base, and is essential for its efficient repair [97]. How lesion recognition is coupled to ATP binding is not well understood. However, crosslinking experiments have recently demonstrated that MSH2 binds ADP with high affinity, while MSH6 binds ATP with high affinity [98], and each nucleotide binds with half-site occupancy [93–96, 98, 99]. There is general agreement that transient double occupancy in MutS and MSH2/MSH6 is possible. However, there is no general agreement as to the role of double occupancy states in mediating MMR [ADP/ADP, ATP/ADP, or ATP/ATP], and as to what liganded state correlates with specific functions in MMR [98, 99]. Crosslinking experiments have led to the suggestion that the ATP/ATP bound state in MSH2/MSH6 forms an ATP hydrolysis-independent clamp, which allows sliding and movement away from the lesion to signal repair [20, 98]. Far less is known about MSH2/MSH3. However, MSH3 that is unable to bind or hydrolyze ATP results in a mutator phenotype [1–6]. Thus, nucleotide binding within the subunits of MSH2/MSH3 will also be key in regulating mismatch recognition and downstream signaling during MMR [1– 6].
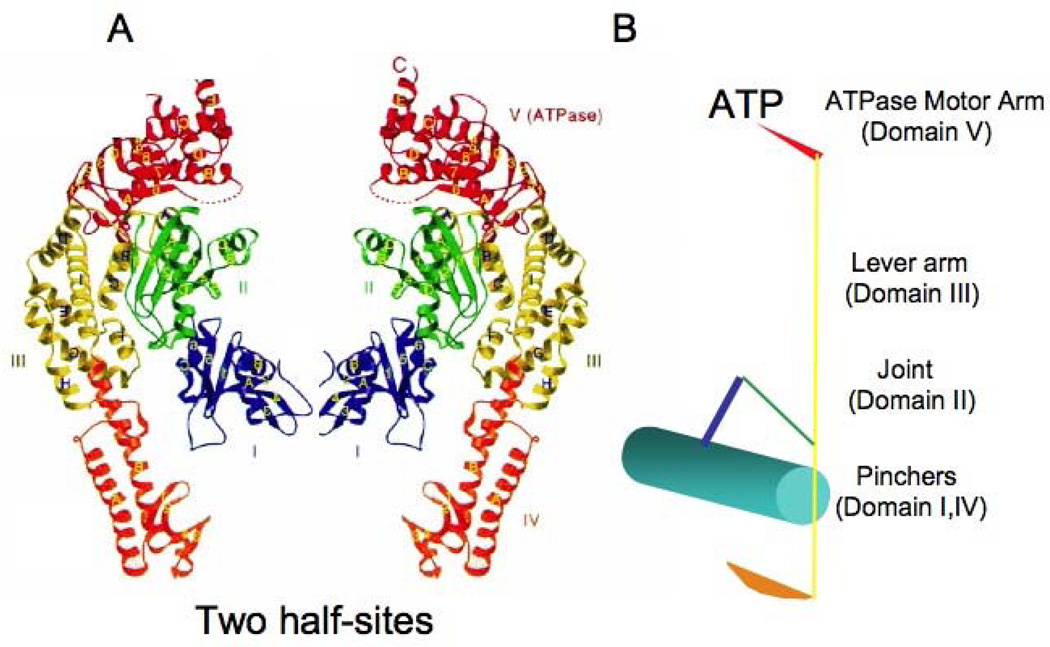
Figure 8. The Structure of MutS.
[A] Crystal structure of the TAQ MutS-DNA complex. The two protein half-sites are mirror images and form a homodimer represented by ribbon diagrams. Ribbon diagram of subunit A. The five domains are colored blue (DNA binding, domain I), green (connector, domain II), yellow (lever arm, domain III), orange (DNA binding, domain IV) and red (ATPase activity, domain V) from the N to the C terminus. MSH proteins contain a conserved Walker-type nucleotide binding domain in their carboxy-termini. Walker A and Walker B motifs have been shown to both bind and hydrolyze ATP, which is essential for recognition and repair of lesions in DNA. Nucleotide binding is structurally linked to DNA binding located within the N-terminus. The linker regions connect the C- and N-termini, and connector regions connect the DNA binding domain to the linker. Secondary structures are named in alphabetic or numeric order for [α]-helices and [β]-strands, respectively, following their order in the primary sequence in each domain. [B] Each functional unit is schematically represented in stick figure as a clamp. The colors represent each domain of MutS in the half site in [A].
Since in all MSH recognition complexes, disruption of nucleotide binding and hydrolysis prevents faithful repair, it is significant that binding of MSH2/MSH3 to the CAG hairpin inhibits its ATPase activity, alters its binding stoichiometry to DNA, and changes its nucleotide affinity [27]. All of these changes in MSH2/MSH3 require the A/A mispaired bases in the stem of the hairpin [27]. Placement of A/A mismatches within a duplex DNA does not inhibit the ATPase activity of MSH2/MSH3 indicating that the structure of the hairpin not just its sequence appears to be required for the inhibitory effects. Surprisingly, inhibition of ATPase activity occurs without significant alterations in DNA binding affinity relative to loop templates typically corrected by MSH2/MSH3 [27]. Thus, the DNA-induced repair defect in MSH2/MSH3 appears to be an “uncoupling” of DNA binding from ATP hydrolysis at the CAG hairpin site. The detail or extent of the conformational changes induced in MSH2/MSH3 by hairpin binding is unknown. However, candidate defects include loss of bending and sliding away from the lesion [20–23, 98]. For MSH2/MH6, binding to single base mismatches introduce a 60° DNA bend at the site of the mismatch [18, 89–91], which is facilitated by intercalation of a conserved phenylalanine residue in the DNA binding site. The bending appears to be a critical step in recognition, and bent to unbent transitions have been reported during recognition of DNA lesions by MSH2/MSH6 [100]. In contrast, inhibition of ATP hydrolysis in the MSH2/MSH3-hairpin complex is independent of the length of the template, suggesting that the hairpin prevents sliding off the lesion [27]. MSH2/MSH3, once bound, appears to be “stuck” to the hairpin, which may prevent bending and flexibility in the DNA needed for regulation and coupling. An uncoupling model predicts that MSH2/MSH3 recognizes the mispaired bases in the hairpins stem, but, once bound, fails to correct them. In such a model, the mismatch repair system would operate normally at most sites in the genome, but its normal lesion binding function is “hijacked” to cause expansion at the site of CAG hairpin formation.
Although 5’-CAG and 5’-CTG have the same dependence on MSH2/MSH3, it has been debatable as to whether the same mechanism of expansion applies to all triplet repeats. A confounding issue is the size of repeat tract changes, which differ depending on whether the triplet repeat exists in a coding or a non-coding region of a gene. 5’-CAG is the only triplet found in the coding sequence of genes. For 5’CAG tracts, the size of the repeat expansions is small (3–15 repeats). In contrast, triplet tracts located in non-coding regions (5’CTG and 5’CGG) can expand up to thousands. The basis for these size differences is not entirely clear. However, in the coding region of genes, very long 5’CAG tracts serve as a template for an increasingly toxic gene product, which may not support life. Thus, it is generally suspected that the smaller 5’CAG expansions are the result of “selection” rather than a different expansion mechanism relative to non-coding repeats. In other words, repeat tracts in both coding and non-coding regions may undergo large expansions, but the larger expansion of coding tracts would not be detected since cells expressing the expanded gene products would not survive. Beyond size differences, there are striking similarities among expanding triplets including a common dependence on MSH2/MSH3, a comparable expansion threshold, and the frequent and progressive growth of the repeat. Thus, the mechanism postulated to cause CAG expansion in HD is likely to play roles in the other triplet repeats, although that hypothesis will bear further testing.
VII. Lesion specificity: MSH2/MSH3 but not MSH/MSH6 causes expansion
Both MSH2/MSH3 and MSH2/MSH6 bind and hydrolize ATP to initiate MMR, the hijacking model does not readily explain why the CAG expansion displays an unexpected specificity for MSH2/MSH3, while MSH2/MSH6 has little effect. MSH2/MSH3 has a higher affinity and specificity for insertion/deletion DNA loops [IDL] composed of 2–13 bases [15, 22] and branched DNAs [101] [Figure 4]. However, the hairpin lesion also contains multiple mispaired bases within its stem, offering a number of preferred substrates for MSH2/MSH6. Thus, the structural basis for the specificity of expansion on MSH2/MSH3 is unclear. The DNA binding domain of MSH2/MSH3 differs significantly from its MSH2/MSH6 counterpart [18, 89–91, 102]. While the Phe residue of MSH6 is required for stacking onto the mispaired base, MSH3 has a lysine in the analogous position. Thus, repair by MSH2/MSH3 and MSH2/MSH6, even for the same lesion, is unlikely to occur by the same mechanism.
Although MSH2/MSH3 and MSH2/MSH6 share a common MSH2 subunit, lesion specificity for the expanded CAG site may also be influenced by differences in the interactions with the MSH2 subunit with MSH3 and MSH6. For human MSH2/MSH6, the MSH2 subunit makes no contact with DNA even within the phosphodiester backbone [92], and, in yeast, loss of DNA binding Domain I of MSH2 has little effect on repair activity of MSH2/MSH6 [102]. However, loss of the same domain in MSH2 inactivates MMR and recombination activities of MSH2/MSH3 in yeast [102]. Furthermore, when the DNA-binding domain of MSH3 is switched for the DNA-binding domain of MSH6, the resulting chimera complements the mutator phenotype of MSH3[‒/‒] strains in vivo [16]. However, when the MSH6 DNA-binding domain is switched for the MSH3 DNA-binding domain, neither MSH3 nor MSH6 deletion mutants were competent in repair [16]. These data suggest that the MSH3 and MSH6 subunits are both structurally and functionally different. Thus, the dependence of expansion on MSH2/MSH3 and not MSH2/MSH6 may reflect aspects of the normal lesion specificity of MSH2/MSH3 for looped DNA. A DNA-induced conformational change in MSH2/MSH3 may uncouple MSH2/MSH3 from signaling canonical MMR. Instead, the protein-DNA complex may divert signaling to an alternative pathway, which repairs the CAG/CTG site inefficiently. Understanding the role of MSH2/MSH3 will be aided by mapping occupancy of nucleotide within its subunits, as well as detailed structural analysis of the MSH2/MSH3-hairpin complex.
B. CHEMICAL LESIONS
Repair deficits such as the one mediated by MSH2/MSH3 in HD are reminiscent of mismatch repair defects in which the normal MSH2/MSH6 complex dissociates normal coupling and signals cell death at some lesions. MSH2/MSH6 recognizes a variety of damaged DNA substrates and complex lesions including the common alkylation lesion mismatches O6-methylguanine-C and O6-methylguanine–T [4–6, 103, 104]. Agents that methylated bases in DNA by an SN1 mechanism, such as N-methyl-N’-nitro-N’nitrosoguanididne [MMMG] and N-methyl-N-nitrosourea [MMU] are mutagenic and cytotoxic [4–6, 103, 104]. The O6-methyl-G [O6-meG] DNA adducts induce cell cycle arrest and apoptosis in a manner requiring the DNA mismatch repair [MMR] proteins MutSα and MutLβ [1–6]. MSH2/MSH6 also recognizes mismatches containing 8-oxoguanine [87] and cis-Platinum [Pt][105–107]. The most abundant lesion generated by cisplatin is 1,2-d[GpG] intrastrand cross-link formed by the cross-linking of a platinum residue with the N-7 atom of two adjacent guanine residues [105]. This lesion can give rise to a compound lesion, namely an intrastrand adduct in one strand and a mismatch in the other [106]. Such compound lesions arise from the misincorporation of a base opposite the platinated guanine of the 1,2-intrastrand cross-link during replication bypass [106]. Pt treatment is widely used for treatment of ovarian cancer, and in this case, the cytotoxic properties are an advantage [4–6, 104].
As with the CAG expansion mutation, MMR-dependent signaling at chemical lesions is thought to occur through molecular mechanisms that differ from those required for normal post-replicative mismatch repair [4–6]. For example, the ATP-dependent biochemical properties of Escherichia coli MutSα differs when the protein interacts with a DNA oligonucleotide containing a GT mismatch versus a more complex lesion [108]. MutS exhibits substantial affinity for cisplatin 1,2-d[GpG] intrastrand cross-link with a mispaired thymine opposite the 3′ platinated guanine, whether or not it binds ATP [108]. Similar to the effects of CAG hairpin binding on MSH2/MSH3 function [27], Pt binding inhibits MutSα ATPase activity to a rate equivalent to that induced by nonspecific homoduplex DNA [108]. Loss or mutations of MSH2, MSH3, and MSH6 typically result in a 2- to 4-fold increase in lesion tolerance [109], which contributes significantly to the failure of cancer therapy [109, 110]. Thus, biochemical properties induced by Pt are remarkably similar to those imparted by the CAG hairpin on MSH2/MSH3.
Although the specific type of lesion likely contributes to the choice of repair or cell death, the recent crystal structure of MSH2/MSH6 reveals that its conformation is unaltered by binding to either a cytotoxic Pt lesion or to a repair competent GT mispaired base [91]. Furthermore, the nucleotide binding properties of MSH2/MSH6 remain unchanged when bound to G/T mismatch base pairs [98]. Thus, the structural basis for uncoupling of lesion binding and ATP hydrolysis by MSH2/MSH6 at Pt lesions is unknown. However, molecular modeling and mutational analysis has been informative in identifying residues that affect the repair and cell death responses independently [106, 111–113]. The conserved glutamate in the Phe-X-Glu DNA binding domain appears to be a key contact with cisplatin-DNA, and is instrumental in the resulting cytotoxicity [111–113]. Mutation of this residue increases non-specific DNA binding, and reduces cisplatin toxicity [111– 113]. In contrast, the conserved phenylalanine that is indispensable for mismatch recognition during post-replicative repair is not required for cisplatin cytotoxicity [111– 113]. These differences in protein–DNA interactions appear to be translated into localized conformational changes that affect nucleotide requirements and inter-subunit interactions. Specifically, coupling of ATP binding and hydrolysis is necessary for repair of G/T mismatches, but has little consequence for the MMR-dependent damage response. In the former, the MutS conformation is sufficient to support interaction with downstream proteins and to carry out post-replicative repair. However, in the latter, an aberrant conformational change in the MSH-related proteins either directly aborts coupling with MLH1/PMS2 and to the downstream MMR machinery, or diverts the MMR-lesion complex to an alternative pathway, which is not competent for repair.
Thus, there are distinct functional similarities in the effects of MSH2/MSH6 at Pt lesions with those of MSH2/MSH3 at CAG hairpins, despite the fact that one is cytotoxic and one is mutagenic. Two models have been mainly proposed to explain the role of MMR proteins in DNA-damage response [1–6]. The futile cycling model proposes that repetitive repair attempts are triggered at a mispaired base opposite a persistent lesion, resulting in the accumulation of strand breaks, which further activate cell death [114– 116]. The direct signaling model is based on the concept that MMR proteins have two separate roles, one in the repair of mismatches and the other in the DNA damage response [4–6, 111–116]. Indeed, MMR proteins interact with an intricate network of proteins involved in cell cycle checkpoint and apoptotic pathways, among which are p73, ATM, CHK2, BRCA1 and PCNA [1–6]. Checkpoint signaling in response to DNA methylation occurs during S phase and requires DNA replication that gives rise to O6-meG/T mispairs. DNA binding studies reveal that MutSα specifically recognizes O6-meG/T mispairs, but not O6-meG/C. In an in vitro assay, ATR-ATRIP, but not RPA, is preferentially recruited to O6-meG/T mismatches in a MutSα- and MutLα-dependent manner [103]. Furthermore, ATR kinase is activated to phosphorylate Chk1 in the presence of O6-meG/T mispairs and MMR proteins. These results suggest that MMR proteins can act as direct sensors of methylation damage and help recruit ATR-ATRIP to sites of cytotoxic O6-meG adducts to initiate ATR checkpoint signaling [103]. Thus, MutSα may directly signal cell death under the correct conformational setting [115, 116].
In conclusion, there are many differences between toxicity induced by chemical lesions and those associated with disease states such as HD. Chemical exposures can directly signal apoptoic pathways, as well as induce strand breaks in futile attempts at repair [4– 6, 115]. HD, on the other hand, is a single site mutation in which toxicity involves both synthesis of a toxic protein and elevation of oxidative DNA damage in cells. However, both conditions initiate from a DNA-induced conformational change in a normal mismatch repair complex that is functionally and structurally related. The similarities raise interesting questions as to whether there were common and measurable states by which a DNA-induced conformation change, in an otherwise normal MMR complex, might uncouple lesion binding from ATP hydrolysis to cause cell death.
ACKNOWLEDGMENT
This work was supported by the Mayo Foundation, the National Institutes of Health grants NS40738 [CTM], NS060115 [CTM] and GM 066359 [CTM].
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Modrich P. Mechanisms in eukaryotic mismatch repair. J. Biol. Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 3.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 4.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 5.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol. Med. 2006;12:440–50. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 8.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 9.Kovtun IV, McMurray CT. Models for DNA repair underlying triplet repeat expansion. In: Wells RD, Wells R, Ashizawa T, editors. Genetic Instabilities and Hereditary Neurological Diseases. Volume 2. Academic Press; 2006. pp. 679–690. [Google Scholar]
- 10.Manley K, Shirley TL, Flaherty L, Messer A. MSH2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat. Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 11.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nature Genetics. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 12.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMsh3 and hMsh6. Proc.Natl. Acad.Sci.USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast Msh3 and Msh6 genes for Msh2-dependent genomic stability. J.Biol.Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 14.Marsischky GT, Filosi N, Kane MF, Kolodner RD. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 15.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSb, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 1996:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 16.Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc Natl. Acad. Sci. USA. 2007;104:10956–10961. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrington JM, D Kolodner R. Saccharomyces cerevisiae Msh2- Msh3 acts in repair of base-base mispairs. Mol. Cell Biol. 2007;27:6546–6554. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 19.Sacho EJ, Kadyrov FA, Modrich P, Kunkel TA, Erie DA. Direct visualization of asymmetric adenine nucleotide-induced conformational changes in MutL alphα. Mol Cell. 2008;29:112–121. doi: 10.1016/j.molcel.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 Forms a Hydrolysis-Independent Sliding Clamp on Mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 21.Gradia S, Acharya S, Fishel R. The role of mismatched oligonucleotides in activating the hMSH2-MSH6 molecular switch. J. Biol. Chem. 2000;275:3922–3930. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- 22.Wilson T, Guerrette S, Fishel R. Dissociation of mismatch recognition and ATPase activity by hMSH2-MSH3. J. Biol. Chem. 1999;274:21659–21664. doi: 10.1074/jbc.274.31.21659. [DOI] [PubMed] [Google Scholar]
- 23.Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol. Cell. Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alani E, Lee S, Kane MF, Griffith J, Kolodner RD. Saccharomyces cerevi\siae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J.Mol.Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 25.Gacy AM, Goellner G, Juranic N, Macura S, T C. McMurray, Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 26.van den Broek WJ, Nelen MR, Wansink DG, Coerwinkel MM, Riele H, Groenen PJ, Wieringa B. Somatic expansion behavior of the [CTG]n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–199. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 27.Owen BA, Yang Z, Lai M, Gajek M, Badger 2nd JD, Hayes JJ, Edelmann W, Kucherlapati R, Wilson TM, McMurray CT. [CAG][n]-hairpin DNA binds to Msh2-Msh3 and changes properties of mismatch recognition. Nat. Struct. Mol. Biol. 2005;12:663–70. doi: 10.1038/nsmb965. [DOI] [PubMed] [Google Scholar]
- 28.Goellner GM, Tester D, Thibodeau S, Almqvist E, Goldberg YP, Hayden MR, T C. McMurray, Different mechanisms underlie DNA instability in Huntington Disease and Colorectal Cancer. Am. J. Hum. Genet. 1997;60:879–890. [PMC free article] [PubMed] [Google Scholar]
- 29.Moore H, Greewell PW, Liu CP, Arnheim N, Petes TD. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl. Acad. Sci. USA. 1999;96:1504–1509. doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miret JJ, Pessoa-Brandão L, S Lahue R. Instability of CAG and CTG trinucleotide repeats in Saccharomyces cerevesiae. Mol. Cell Biol. 1997;17:3382–3387. doi: 10.1128/mcb.17.6.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miret JJ, Pessoa-Brandao L, Lahue RS. Orientation-dependent and sequence-specific expansion of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Acad. Sci. USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiro C, et al. Inhibition of FEN-1 processing by DNA secondary structure at trinucleotide repeats. Mol. Cell. 1999;4:1079–1085. doi: 10.1016/s1097-2765(00)80236-1. [DOI] [PubMed] [Google Scholar]
- 33.Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication an expansion of trinucleotide repeats in yeast. Mol. Cell Biol. 2003;23:1349–57. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMurray CT. DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. USA. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Mariappan SV, Catasti P, Ratliff R, Moyzis RK, Laayoun A, Smith SS, Bradbury EM, Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc. Natl. Acad.Sci. [USA] 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A. Friedreich’s Ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 37.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury E, Dyer RB, Mikesell MJ, Yao JZ, Johnson AJ, et al. GAA instability in Friedriech’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 38.Sakamoto N, Chastain PD, Parniewski P, Ohshima K, Pandolfo M, Griffith JD, Wells RD. Sticky DNA: self-association properties of long GAA.TTC repeats in R.R.Y triplex structures from Friedreich’s ataxia. Mol. Cell. 1999;4:465–75. doi: 10.1016/s1097-2765(00)80474-8. [DOI] [PubMed] [Google Scholar]
- 39.Catasti P, Chen X, Deaven LL, Moyzis RK, Bradbury EM, Gupta G. Cystosine-rich strands of the insulin minisatellite adopt hairpins with intercalated cytosine+.cytosine pairs. J. Mol. Biol. 1997;272:369–382. doi: 10.1006/jmbi.1997.1248. [DOI] [PubMed] [Google Scholar]
- 40.Zheng M, Huang X, Smith GK, Yang X, Gao X. Genetically unstable CXG repeats are structurallydynamic and have a high propensity for foldingt. An NMR and UV spectroscopic study. J. Mol. Biol. 1996;264:323–336. doi: 10.1006/jmbi.1996.0643. [DOI] [PubMed] [Google Scholar]
- 41.Shafer RH. Stability and structure of model DNA triplexes and quadruplexes and their interactions with small ligands. Prog. Nuc. Acids Res. 1998;59:55–94. doi: 10.1016/s0079-6603(08)61029-6. [DOI] [PubMed] [Google Scholar]
- 42.Lyons-Darden T, Topal MD. Abasic sites induce triplet-repeat expansion during DNA replication in vitro. J. Biol. Chem. 1999;274:25975–25978. doi: 10.1074/jbc.274.37.25975. [DOI] [PubMed] [Google Scholar]
- 43.Kovtun IV, Li Y, Bjoras M, Klungland A, Wilson SH, McMurray CT. OGG1 initiates age-dependent CAG expansion in somatic cells during base excision repair of oxidized bases in vitro and in vivo. Nature. doi: 10.1038/nature05778. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cleary JD, Nichol K, Wang YH, Pearson CE. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 2002;31:37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 45.Freudenreich CH, Kantrow SM, Zakian VA. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 46.Richard GF, Goellner GM, McMurray CT, Haber JE. Recombination-induced CAG trinucleotide repeat expansions in yeast involve the MRE11/RAD50/XRS2 complex. EMBO J. 2000;19:2381–2390. doi: 10.1093/emboj/19.10.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jakupciak JP, Wells RD. Genetic instabilities in [CTG CAG] repeats occur by recombination. J. Biol. Chem. 1999;274:23468–23479. doi: 10.1074/jbc.274.33.23468. [DOI] [PubMed] [Google Scholar]
- 48.Cemal CK, Huxley C, Chamberlain S. Insertion of expanded CAG trinucleotide repeatmotifs into a yeast artificial chromosome containing the human Machado- Joseph disease gene. Gene. 1999;236:53–61. doi: 10.1016/s0378-1119(99)00273-5. [DOI] [PubMed] [Google Scholar]
- 49.Kang S, Ohshima K, Shimizu M, Amirhaeri S, Wells RD. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 50.Petruska J, Hartenstine MJ, Goodman MF. Analysis of strand slippage in DNA polymerase expansions of CAG/CTG triplet repeats associated with neurodegenerative disease. J Biol. Chem. 1998;273:5204–10. doi: 10.1074/jbc.273.9.5204. [DOI] [PubMed] [Google Scholar]
- 51.Kang S, Jaworski A, Ohshima K, Wells RD. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E.coli. Nature Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 52.Iyer RR, Wells RD. Expansion and deletion of triplet repeat sequences in Escherichia coli occur on the leading strand of DNA replication. J.Biol. Chem. 1999;274:3865–3877. doi: 10.1074/jbc.274.6.3865. [DOI] [PubMed] [Google Scholar]
- 53.Freudenreich CH, Stavenhagen JB, Zakian VA. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol. Cell. Biol. 1997;17:2090–2097. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 55.Schweitzer JK, Livingston DM. The effect of DNA replication mutations on CAG tract stability in yeast. Genetics. 1999;152:953–963. doi: 10.1093/genetics/152.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sia EA, Dominska M, Stefanovic L, Petes TD. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast, Mol. Cell Biol. 2001;21:8157–8167. doi: 10.1128/MCB.21.23.8157-8167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sia EA, Kokoska RJ, Dominska M, Greenwell P, Petes TD. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell Biol. 1997;17:2851–2858. doi: 10.1128/mcb.17.5.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McMurray CT. Mechanisms of DNA expansion. Chromosoma. 1995;104:2–13. doi: 10.1007/BF00352220. [DOI] [PubMed] [Google Scholar]
- 59.Monckton DG, Coolbaugh MI, T Ashizawa K, Siciliano MJ, Caskey CT. Hypermutable myotonic dystrophy CTG repeats in transgenic mice, Nat. Genet. 1997;15:193–196. doi: 10.1038/ng0297-193. [DOI] [PubMed] [Google Scholar]
- 60.Delagoutte E, Goellner GM, Guo J, Baldacci G, McMurray CT. Single stranded DNA binding protein in vitro eliminates the orientation dependent impediment to polymerase passage on CAG/CTG repeats. J Biol Chem. 2008 doi: 10.1074/jbc.M800153200. online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinder DJ, Blake CE, Lindsey JC, Leach DR. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 1998;28:719–727. doi: 10.1046/j.1365-2958.1998.00831.x. [DOI] [PubMed] [Google Scholar]
- 62.Hashem VI, Rosche WA, Sinden RR. Genetic assays for measuring rates of [CAG].[CTG] repeat instability in Escherichia coli. Mutat. Res. 2002;502:25–37. doi: 10.1016/s0027-5107(02)00026-x. [DOI] [PubMed] [Google Scholar]
- 63.Pollard LM, Sharma R, Gomez M, Shah S, Delatycki MB, Pianese L, Monticelli A, Keats BJ, Bidichandani SI. Replication-mediated instability of the GAA triplet repeat mutation in Friedreich ataxia. Nucleic Acids Res. 2004;3:5962–5971. doi: 10.1093/nar/gkh933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hirst MC, White PJ. Cloned human FMR1 trinucleotide repeats exhibit a length- and orientation-dependent instability suggestive of in vivo lagging strand secondary structure. Nucleic Acids Res. 1998;26:2353–2358. doi: 10.1093/nar/26.10.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Balakumaran BS, Freudenreich CH, Zakian VA. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 2000;9:93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- 66.Kroutil LC, Kunkel TA. Deletion errors generated during replication of CAG repeats. Nucleic Acids Res. 1999;27:3481–3486. doi: 10.1093/nar/27.17.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z, Lau R, Marcadier JL, Chitayat D, Pearson CE. Replication inhibitors modulate instability of an expanded trinucleotide repeat at the myotonic dystrophy type 1 disease locus in human cells. Am. J. Hum. Genet. 2003;73:1092–1010. doi: 10.1086/379523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomes-Pereira M, Fortune MT, Monckton DG. Mouse tissue culture models of unstable triplet repeats: in vitro selection for larger alleles, mutational expansion bias and tissue specificity, but no association with cell division rates. Hum. Mol. Genet. 2001;10:845–854. doi: 10.1093/hmg/10.8.845. [DOI] [PubMed] [Google Scholar]
- 69.Manley K, Pugh J, Messer A. Instability of the CAG repeat in immortalized fibroblast cell cultures from Huntington’s disease transgenic mice. Brain Res. 1999;835:74–79. doi: 10.1016/s0006-8993(99)01451-1. [DOI] [PubMed] [Google Scholar]
- 70.Kovtun IV, Thornhill AR, McMurray CT. Somatic deletion events occur during early embryonic development and modify the extent of CAG expansion in subsequent generations. Hum. Mol. Genet. 2004;13:3057–3068. doi: 10.1093/hmg/ddh325. [DOI] [PubMed] [Google Scholar]
- 71.Spiegel R, La Spada AR, Kress W, Fischbeck KH, Schmid W. Somatic stability of the expanded CAG trinucleotide repeat in X-linked spinal and bulbar muscular atrophy. Hum. Mutat. 1996;8:32–37. doi: 10.1002/(SICI)1098-1004(1996)8:1<32::AID-HUMU4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 72.Wohrle D, Hennig I, Vogel W, Steinbach P. Mitotic stability of fragile X mutations in differentiated cells indicates early post-conceptional trinucleotide repeat expansion. Nat. Genet. 1993;4:140–142. doi: 10.1038/ng0693-140. [DOI] [PubMed] [Google Scholar]
- 73.Wantanabe H, Tanaka F, Doyu M, Riku S, Yoshida M, Hashizume Y. Differential somatic CAG repeat instability in variable brain lineage in dentatorubral pallidoluysian atrophy [DRPLA]: a laser capture microdissection [LCM]-based analysis. Hum. Genet. 2000;107:452–457. doi: 10.1007/s004390000400. [DOI] [PubMed] [Google Scholar]
- 74.Samadashwily GM, Raca G, Mirkin SM. Trinucleotide repeats affect DNA replication in vivo. Nature Genet. 1997;17:298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 75.Pelletier R, Krasilnikova MM, Samadashwily GM, Lahue R, Mirkin SM. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krasilnikova MM, Mirkin SM. Replication stalling at Friedreich’s ataxia [GAA]n repeats in vivo. Mol. Cell Biol. 2004;24:2286–2295. doi: 10.1128/MCB.24.6.2286-2295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim S-H, Pytlos MJ, Sinden RR. Replication restart: A pathway for [CTG].[CAG] repeat deletion in Escherichia coli. Mutat. Res. 2006;595:5–22. doi: 10.1016/j.mrfmmm.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 78.Yoon SR, Dubeau L, de Young M, Wexler NS, Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed, Proc. Natl. Acad. Sci. USA. 2003;100:8834–8838. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leeflang EP, Tavare S, Marjoram P, Neal COS, Srinidhi J, MacDonald ME, de Young M, Wexler NS, Gusella JF, Arnheim N. Analysis of germline mutation spectra at the Huntington’s disease locus supports a mitotic mutation mechanism. Hum. Mol. Genet. 1999;8:173–183. doi: 10.1093/hmg/8.2.173. [DOI] [PubMed] [Google Scholar]
- 80.Wheeler VC, Lebel LA, Vrbanac V, Teed A, te Riele H, MacDonald ME. Mismatch repair gene Msh2 modifies the timing of early disease in Hdh(Q111) striatum. Hum. Mol. Genet. 2003;12:273–281. doi: 10.1093/hmg/ddg056. [DOI] [PubMed] [Google Scholar]
- 81.Savouret C, Brisson E, Essers J, Kanaar R, Pastink A, te Riele H, Junien C, Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seznec H, Lia-Baldini A-S, Duros C, Fouquet C, Lacroix C, Hofmann-Radvanyi H, Junien C, Gourdon G. Transgenic mice carrying large human genomic sequences with expanded CTG repeat mimic closely the DM CTG repeat intergenerational and somatic instability. Hum. Mol. Genet. 2000;9:1185–1194. doi: 10.1093/hmg/9.8.1185. [DOI] [PubMed] [Google Scholar]
- 83.Wohrle D, Kennerknecht I, Wolf M, Enders H, Schwemmle S, Steinbach P. Heterogeneity of DM kinase repeat expansion in different fetal tissues and further expansion during cell proliferation in vitro: evidence for a casual involvement of methyl-directed DNA mismatch repair in triplet repeat stability. Hum. Mol. Genet. 1995;4:1147–1153. doi: 10.1093/hmg/4.7.1147. [DOI] [PubMed] [Google Scholar]
- 84.Wheeler VC, et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum. Mol. Genet. 1999;8:115–122. doi: 10.1093/hmg/8.1.115. [DOI] [PubMed] [Google Scholar]
- 85.Mangiarini L, et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- 86.Sato T, Oyake M, Nakamura K, Nakao K, Fukusima Y, Onodera O, Igarashi S, Takano H, Kikugawa K, Ishida Y, et al. Transgenic mice harboring a full- length human mutant DRPLA gene exhibit age-dependent intergenerational and somatic instabilities of CAG repeats comparable with those in DRPLA patients. Hum. Mol. Genet. 1999;8:99–106. doi: 10.1093/hmg/8.1.99. [DOI] [PubMed] [Google Scholar]
- 87.Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair DNA Repair. 2006 doi: 10.1016/j.dnarep.2006.10.015. online. [DOI] [PubMed] [Google Scholar]
- 88.Panigrahi GB, Lau R, Montgomery SE, Leonard MR, Pearson CE. Slipped [CTG]*[CAG] repeats can be correctly repaired, escape repair or undergo error prone repair. Nat Struct Mol. Biol. 2005;12:654–62. doi: 10.1038/nsmb959. [DOI] [PubMed] [Google Scholar]
- 89.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 90.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol. Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 91.Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579–592. doi: 10.1016/j.molcel.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 92.Lamers MH, Winterwerp HH, Sixma TK. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J. 2003;22:746–756. doi: 10.1093/emboj/cdg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Antony E, Khubchandani S, Chen M.M S. Hingorani, Contribution of Msh2 and Msh6 subunits to the asymmetric ATPase and DNA mismatch binding activities of Saccharomyces cerevisiae Msh2-Msh6 mismatch repair protein, DNA Repair. Amst. 2005;3:153–162. doi: 10.1016/j.dnarep.2005.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Blackwell LJ, Bjornson KP, Allen DJ, Modrich P. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem. 2001;276:34339–34347. doi: 10.1074/jbc.M104256200. [DOI] [PubMed] [Google Scholar]
- 95.Blackwell LJ, Bjornson KP, Allen DJ, Modrich P. Distinct MutS DNA-binding modes that are differentially modulated by ATP binding and hydrolysis. J. Biol. Chem. 2001;276:34339–34347. doi: 10.1074/jbc.M104256200. [DOI] [PubMed] [Google Scholar]
- 96.Drotschmann K, Yang W, Kunkel TA. Evidence for sequential action of two ATPase active sites in yeast MSH2-MSH6. DNA Repair. 2002;1:742–753. doi: 10.1016/s1568-7864(02)00081-2. [DOI] [PubMed] [Google Scholar]
- 97.Yamamoto A, Schofield MJ, Biswas I, Hsieh P. Requirement for Phe36 for DNA binding and mismatch repair by Escherichia coli MutS protein. Nucleic Acids Res. 2000;28:3564–3569. doi: 10.1093/nar/28.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mazur DJ, Mendillo ML, Kolodner RD. Inhibition of Msh6 ATPase activity by mispaired DNA induces a Msh2[ATP]-Msh6[ATP] state capable of hydrolysis-independent movement along DNA. Mol. Cell. 2006;22:39–49. doi: 10.1016/j.molcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 99.Martik D, Baitinger C, Modrich P. Differential specificities and simultaneous occupancy of human MutSα nucleotide binding. J. Biol.Chem. 2004;279:28402–28410. doi: 10.1074/jbc.M312108200. [DOI] [PubMed] [Google Scholar]
- 100.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, Erie DA. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc Natl. Acad. Sci. U S A. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Surtees JA, Alani E. Mismatch repair factor MSH2-MSH3 binds and alters the conformation of branched DNA structures predicted to form during genetic recombination. J. Mol. Biol. 2006;360:523–536. doi: 10.1016/j.jmb.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 102.Lee SD, Surtees JA, Alani E. Saccharomyces cerevisiae MSH2-MSH3 and MSH2-MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J. Mol. Biol. 2007;366:53–66. doi: 10.1016/j.jmb.2006.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts, Mol. Cell. 2006;22:501. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sedletska Y, Giraud-Panis MJ, Malinge JM. Cisplatin is a DNA-damaging antitumour compound triggering multifactorial biochemical responses in cancer cells: importance of apoptotic pathways. Current Medicinal Chemistry - Anti-Cancer Agents. 2005;5:251–265. doi: 10.2174/1568011053765967. [DOI] [PubMed] [Google Scholar]
- 105.Chaney SG, Campbell SL, Temple B, Bassett E, Wu Y, Faldu M. Protein interactions with platinum-DNA adducts: from structure to function. Journal of Inorganic Biochemistry. 2004;98:1551–1559. doi: 10.1016/j.jinorgbio.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 106.Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chemistry & Biology. 1996;3:579–89. doi: 10.1016/s1074-5521(96)90149-0. [DOI] [PubMed] [Google Scholar]
- 107.Sharma S, Gong P, Temple B, Bhattacharyya D, Dokholyan NV, Chaney SG. Molecular dynamic simulations of cisplatin- and oxaliplatin-d[GG] intrastand cross-links reveal differences in their conformational dynamics. Journal of Molecular Biology. 2007;373:1123–1140. doi: 10.1016/j.jmb.2007.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sedletska Y, Fourrier L, Malinge JM. Modulation of MutS ATP-dependent functional activities by DNA containing a cisplatin compound lesion [base damage and mismatch] J. Mol. Biol. 2007;369:27–40. doi: 10.1016/j.jmb.2007.02.048. 2007. [DOI] [PubMed] [Google Scholar]
- 109.Aebi S, Kurdi-Haidar B, Gordon R, Cenni B, Zheng H, Fink D, Christen RD, Boland CR, Koi M, Fishel R, B Howell S. Loss of DNA MMR in acquired resistance to cisplatin. Cancer Res. 1996;56:3087–3090. [PubMed] [Google Scholar]
- 110.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin, Critical Reviews in Oncology-Hematology. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 111.Salsbury FR, Jr, Clodfelter JE, Gentry MB, Hollis T, Scarpinato KD. The molecular mechanism of DNA damage recognition by MutS homologs and its consequences for cell death response. Nucleic Acids Res. 2006;34:2173–85. doi: 10.1093/nar/gkl238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Clodfelter JE, Gentry MB, Drotschmann K. MSH2 missense mutations alter cisplatin cytotoxicity and promote cisplatin-induced genome instability. Nucleic Acids Res. 2005;33:3323–3330. doi: 10.1093/nar/gki646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Drotschmann K, Topping RP, Clodfelter JE, Salsbury FR. Mutationsin the nucleotide-binding domain of MutS homologs uncouple cell death from cell survival. DNA Repair, Amst. 2004;3:729–742. doi: 10.1016/j.dnarep.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 114.Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–1937. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- 115.Li GM. The role of mismatch repair in DNA damage-induced apoptosis. Oncol, Res. 1999;11:393–400. [PubMed] [Google Scholar]