Abstract
Helicobacter pylori has been associated with the development of two malignant diseases: gastric adenocarcinoma and gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Although the cag pathogenicity island, especially the cagA gene, has been linked with adenocarcinoma, few data concerning H. pylori pathogenic factors involved in low-grade gastric MALT lymphoma are available. The goal of this study was to analyze the prevalence of and correlation between genes coding for seven H. pylori virulence factors (cagA, cagE, vacA, iceA, babA, hopQ, and oipA) and two novel adhesins (sabA and hopZ) by comparing a collection of 43 H. pylori strains isolated from patients with low-grade gastric MALT lymphoma to 39 strains isolated from age-matched patients with gastritis only. Our results show that taken individually, none of the nine genes tested can be considered associated with MALT strains and allow us to conclude that MALT pathogenesis is not linked with more proinflammatory H. pylori strains. We demonstrated that in patients infected with strains harboring the iceA1 allele, sabA functional status, and hopZ “off” status, the odds of developing a MALT lymphoma were 10 times higher. However, the low prevalence of such strains (10 of 43 MALT strains) renders this triple association a low-sensitivity marker for MALT strains. Our data confirmed that H. pylori virulence factors are correlated with one another. If the involvement of H. pylori in MALT lymphoma is well established, the pathomechanism by which gastric lymphoma occurs remains to be identified.
Helicobacter pylori infection is the essential etiological factor of type B chronic gastritis and peptic ulcer disease (29, 35). It is also the first bacterium discovered to be involved in the development of malignant diseases: i.e., gastric carcinoma and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (14, 21, 40). Following H. pylori colonization, inflammation occurs. Untreated gastritis may persist for years with different grades of severity. In some individuals, immunological stimulation induces lymphoid follicles in the gastric mucosa and provides the background for MALT lymphoma development (11), the polyclonal lymphoid hyperplasia evolving further toward a monoclonal lymphoid population (36). The reasons for this evolution, which is accompanied by genetic abnormalities, remain to be elucidated (33, 37). MALT lymphoma induction and growth are probably antigen driven, and it has been shown that H. pylori could be the trigger (18, 23, 24, 26). Moreover, there is a demonstrated causal association between H. pylori infection and MALT-type lymphoma development, because it is possible to cure this cancer by eradicating H. pylori, suggesting that bacterial virulence factors exist and are associated with the disease development. H. pylori is the cause of approximately 80% of MALT lymphomas (8, 57).
Among the factors that may be linked to the outcome of infection, bacterial virulence factors such as cytotoxins and the cag pathogenicity island (PAI) have been extensively investigated but have not been clearly associated with MALT lymphoma strains (19, 39). The goal of this study was to analyze the prevalence of and correlation between genes coding for seven H. pylori virulence factors (cagA, cagE, vacA, iceA, babA, hopQ, and oipA) and two novel adhesins (sabA [sialic acid-binding adhesin] and hopZ [H. pylori outer membrane protein]) by comparing a collection of 43 H. pylori strains isolated from patients with low-grade gastric MALT lymphoma to strains isolated from age-matched patients with gastritis only (4, 10, 20, 22, 25, 44, 46, 55, 58, 60, 61). However, because MALT lymphoma is a rare disease, few studies have evaluated these new putative virulence markers in strains isolated from gastric MALT lymphoma.
Because disease-associated bacterial virulence factors may be correlated with one another, the aim of our study was to analyze the prevalence of and correlation between all of these genes by using a collection of 43 H. pylori strains isolated from patients with low-grade gastric MALT lymphoma and comparing them with 39 strains isolated from age-matched patients with gastritis only.
MATERIALS AND METHODS
Patients and H. pylori strains.
We examined 82 H. pylori strains: 43 were isolated from patients with gastric low-grade MALT lymphoma (27 men and 16 women; mean age, 48.2 ± 13.4 years), and 39 were isolated from age-matched patients with histological gastritis only (29 men and 10 women; mean age, 48.3 ± 14.7 years) suffering from dyspepsia. Patients with gastric MALT lymphoma were enrolled from different areas of France in prospective multicenter studies carried out by the Groupe d'Etude Français des Lymphomes Digestifs of the Fédération Française de Cancérologie Digestive (GELD) (48) and the Groupe d'Etude des Lymphomes de l'Adulte (GELA) (32).
Briefly, once a lesion suspected to be a MALT lymphoma was observed during endoscopy, the patient was invited to come back for a complete exploration, including H. pylori detection within 3 weeks.
Isolation of H. pylori was carried out from two gastric biopsy specimens (one from the lesion and one from macroscopically normal mucosa) by standard culture methods as previously described (31).
All strains were cloned, and genomic DNA was extracted from culture originating from simple colonies by using a commercial kit (Qiagen SA, Courtaboeuf, France).
In addition, the two H. pylori strains for which the genome has been sequenced, 26695 and J99, were used as reference strains (3, 52, 53).
Detection of genes encoding pathogenic factors.
The genes explored (Table 1) were detected by PCR, except for the determination of the functional status of the oipA, hopZ, and sabA genes, for which sequencing was also performed. All of the primers used in this study were desalted and provided by Q-BIOgene (Strasbourg, France).
TABLE 1.
Comparison of genes from H. pylori strains isolated from patients suffering from gastric MALT lymphoma and from patients with gastritis only
| Gene characteristic | Strain type
|
Pa | |||
|---|---|---|---|---|---|
| MALT
|
Gastritis
|
||||
| n | % | n | % | ||
| cagA | |||||
| Absent | 21 | 48.8 | 19 | 48.7 | 1.000 |
| Present | 22 | 51.2 | 20 | 51.3 | |
| cagE | |||||
| Absent | 22 | 51.2 | 17 | 43.6 | 0.515 |
| Present | 21 | 48.8 | 22 | 56.4 | |
| vacA | |||||
| s1m1 | 13 | 30.2 | 22 | 56.4 | 0.064 |
| s1m2 | 12 | 27.9 | 6 | 15.4 | |
| s2m2 | 18 | 41.9 | 11 | 28.2 | |
| iceA | |||||
| A1 | 24 | 55.8 | 15 | 38.5 | 0.128 |
| A2 | 19 | 44.2 | 24 | 61.5 | |
| babA | |||||
| A1 | 24 | 55.8 | 18 | 46.1 | 0.507 |
| A2 | 19 | 44.2 | 21 | 53.9 | |
| hopQ | |||||
| I | 21 | 48.8 | 24 | 61.5 | 0.274 |
| II | 22 | 51.2 | 15 | 38.5 | |
| oipA | |||||
| Off | 23 | 53.5 | 18 | 46.1 | 0.659 |
| On | 20 | 46.5 | 21 | 53.9 | |
| sabAb | |||||
| Off | 16 | 39.0 | 20 | 57.1 | 0.167 |
| On | 25 | 61.0 | 15 | 42.9 | |
| hopZc | |||||
| Off | 23 | 56.1 | 14 | 40.0 | 0.176 |
| On | 18 | 43.9 | 21 | 60.0 | |
P value by Fisher's exact test.
sabA gene was not amplified for two MALT strains and four gastritis strains.
hopZ gene was not amplified for two MALT strains and three gastritis strains. The hopZ gene was absent in one gastritis strain.
cag PAI status was evaluated by PCR for two loci: cagA (with two sets of primers) (54) and cagE (59). A cagA-positive status was defined when the cagA gene was detected by at least one of the two primer pairs. Specific primers for the cag empty site were also used to confirm the absence of the cag PAI (1, 27). The vacA (s [signal] and m [middle] regions), babA, and iceA alleles were detected by PCR as previously described [5, 6, 13, 22; R. M. Peek, S. A. Thompson, J. C. Atherton, M. J. Blaser, and G. G. Miller, abstract from the Ninth International Workshop on the Gastroduodenal Pathology of Helicobacter pylori 1996, Gut 39(Suppl. 2):A71, 1996]. The hopQ type I and II alleles were determined by allele-specific primers (Table 2). The functional status of the oipA (HP638/jhp581), sabA (HP725/jhp662), and hopZ genes is regulated by a slipped strand repair based on the number of CT dinucleotide repeats in the signal coding region. The signal sequences of the oipA and sabA genes including the CT repeats were amplified by PCR with the primers listed in Table 2 with sense and antisense primers designed on the flanking gene and behind the CT dinucleotide repeats, respectively. The primer sense was also used for sequencing. For the amplification of the hopZ gene, we used the previously described primers (41), and sequencing was done with the R2-hopZ primer (Table 2).
TABLE 2.
Designation and sequences of the primers designed for this study
| Region amplified | Primer designation | Sequences (5′ to 3′) |
|---|---|---|
| Type I hopQ | F2-hopQ-T1 | GCGGTGGGAGCACAAATAG (sense) |
| R3-hopQ-T1 | GATGTGGTTACATGCGCTTC (antisense) | |
| Type II hopQ | F1-hopQ-T2 | CATATGCGGAGGCTATACGG (sense) |
| R3-hopQ-T2 | GCGTTGGTTGCTGTTTTAATG (antisense) | |
| oipA flanking gene and oipA | F1-HP0637-jhp580 | CCCCACAAGCGCTTAACAG (sense) |
| F1-HP0638-jhp581 | GAGAGTGCCTAAACCCTATAATCC (antisense) | |
| sabA flanking gene and sabA | F1-HP726-jhp663 | TTTTTGTCAGCTACGCGTTC (sense) |
| R1-HP725-jhp662 | ACCGAAGTGATAACGGCTTG (antisense) | |
| hopZ flanking genea | R2-hopZ | GCGTGTTBGCTTTTAAATTATCG (antisense) |
This primer was only used for sequencing the region containing the CT repeats of the hopZ gene.
PCR amplifications for cagA, cagE, the cag PAI empty site, vacA, iceA, babA2, hopQ, oipA, and sabA were carried out in a 25-μl volume containing 2.5 μl of 10× PCR buffer (Eurobio, Les Ulis, France), 1.5 mM MgCl2 (Eurobio), 200 μM (each) deoxynucleoside triphosphates (dNTPs) (Eurobio), 2 U of Taq DNA polymerase (Eurobio), 1 μM (each) primers, and 10 ng of H. pylori DNA. After 4 min of denaturation at 94°C, each reaction mixture was amplified for 35 cycles as follows: 30 s at 94°C, 30 s of annealing at 60 (for cagA, iceA, hopQ, and oipA), 58 (for cagE and cag PAI empty site), 55 (for babA2 and sabA), or 53°C (for vacA); and 30 s at 72°C. After the last cycle, extension was continued for another 7 min (except for vacA [2 min]).
The hopZ gene was amplified in a 25-μl volume containing 2.5 μl of 10× PCR buffer (Invitrogen, Cergy Pontoise, France), 1.5 mM MgCl2 (Invitrogen), 200 μM (each) dNTPs (Eurobio), 1 U of Platinum Taq polymerase (Invitrogen), 1.25 μM (each) primers, and 10 ng of H. pylori DNA. After 4 min of denaturation at 94°C, each reaction mixture was amplified for 40 cycles as follows: 30 s at 94°C then 30 s of annealing at 58°C and 2 min 30 s at 72°C. After the last cycle, extension was continued for another 7 min.
All PCR products were analyzed on a 1.5% agarose gel (except for iceA [3% agarose]) stained with ethidium bromide.
To determine the oipA, hopZ, and sabA status, amplified DNA fragments were purified with Microspin S-400 HR columns (Amersham Pharmacia Biotech, Inc., Uppsala, Sweden), and direct sequencing was performed with the ABI PRISM BigDye Terminators v3.0 cycle sequencing kit (PE Applied Biosystems, Foster City, Calif.) using an ABI 3700 analyzer DNA sequencer (PE Applied Biosystems).
Statistical analysis.
Statistical analysis was performed with STATA 7.0 statistical software (Stata Corporation, Texas). A first comparison between MALT and gastritis strains was performed by univariate analysis and Fisher's exact test. Spearman rank coefficients (r) were also determined to study the association between the different characteristics of the strains.
In order to create a variable describing how the different strain characteristics were associated, a multiple correspondence analysis (MCA) was performed (47). To confirm the results, a cluster analysis was used to draw a dendrogram based on the same variables, by using the average aggregation method and Euclidian distance. Both analyses were performed with the Statbox 2.5 program (Grimmer Logiciels, Paris, France). These analyses were performed for all of the subjects included in this study for whom information on all genotypes considered was available as well as for the groups of patients with MALT lymphoma and with gastritis, separately.
The odds for association with MALT lymphoma and the 95% confidence interval (CI) were compared within groups of strains and between the different strata of the summary variables obtained by the MCA.
RESULTS
Relationship between each of the nine H. pylori virulence factors tested in strains of patients suffering from gastric MALT lymphoma and gastritis (Table 1). (i) cag PAI.
Twenty-two MALT strains (51.2%) and 20 gastritis strains (51.3%) were cagA positive. Twenty-one MALT strains (48.8%) and 22 gastritis strains (56.4%) were cagE positive. Among the 43 MALT strains, 41 (95.3%) were either cagA positive, cagE positive, and cag PAI empty site-negative or cagA negative, cagE negative, and empty site positive, suggesting that the cag PAI is usually either complete or totally deleted. However, one MALT strain was cagA negative, cagE negative, and cag PAI empty site negative, and another one was cagA positive, cagE negative, and cag PAI empty site negative. Among the 39 gastritis strains, 34 (87.2%) were either cagA or cagE positive and cag empty site negative or cagA or cagE negative and empty site positive (data not shown). Two of these strains were cagA or cagE negative, but the cag PAI empty site PCR revealed an amplification fragment of 1,630 and 2,036 bp, respectively, suggesting that in these two strains, the cag PAI had partial deletions that allowed PCR amplification under our PCR conditions. Three other gastritis strains were cagA negative, cagE positive, and cag PAI empty site negative. At least one cagA or cagE-negative strain and one cagA-positive, cagE-negative strain were empty site negative. The distribution of cagA and cagE genes was not significantly different in H. pylori isolates from MALT patients compared to isolates from gastritis patients (P = 1.0 and 0.515, respectively).
(ii) vacA alleles.
All 82 strains were classified according to the vacA gene signal region (either s1 or s2) or middle region (m1 or m2). The distributions of the different alleles s1m1, s1m2, and s2m2 among the MALT strains were 13 (30.2%), 12 (27.9%), and 18 (41.9%), respectively, and those for the gastritis strains were 22 (56.4%), 6 (15.4%), and 11 (28.2%), respectively. The type s2m1 was never detected in either group of strains. The distribution of vacA genotypes was not significantly different (P = 0.064). However, when considering the m genotype only, a slight association was found between m2 genotype and MALT strains (P = 0.025).
(iii) iceA status.
The primers used allowed us to determine iceA genotypes in all strains. Twenty-four MALT strains (55.8%) were iceA1, and 19 (44.2%) were iceA2 versus 15 (38.5%) iceA1 and 24 (61.5%) iceA2 among the gastritis strains. With iceA2-specific primers, PCR products for 10 gastritis and 6 MALT strains were longer than the expected size (344 versus 230 bp), suggesting the presence of iceA2 variants in these strains. The prevalence of the two genotypes was not different in the two groups (P = 0.128).
(iv) babA2 status.
Nineteen MALT strains (44.2%) and 21 gastritis strains (53.9%) were babA2 positive. The prevalence of babA2 was not significantly different between the two groups (P = 0.507).
(v) hopQ status.
The primers used for this new PCR allowed us to determine hopQ alleles in all strains. Type I hopQ alleles were detected in 21 MALT strains (48.8%) and 24 gastritis strains (61.5%), and type II hopQ alleles were detected in 22 MALT strains (51.2%) and 15 gastritis strains (38.5%). The distribution of type I and II hopQ alleles was not different in both groups of strains (P = 0.274).
(vi) oipA functional status.
The primers used for this new PCR amplified the oipA gene in all strains. Based on the CT dinucleotide repeats present in the signal sequence coding region of the oipA gene, 20 MALT strains (46.5%) and 21 gastritis strains (53.9%) were considered to be oipA “on,” and 23 (53.5%) and 18 (46.2%) strains, respectively, were considered oipA “off.” The number of CT dinucleotide repeats in MALT strains ranged from 5 to 11, and the number in gastritis strains ranged from 5 to 12, in agreement with Yamaoka et al. (58). CT repeats began at position +23 in 72.1% of the MALT strains and 74.4% of the gastritis strains. There was no difference in the distributions of oipA functional status between these two groups of strains (P = 0.659).
(vii) sabA functional status.
The primers used for this new PCR failed to obtain sabA gene amplification for two MALT strains and four gastritis strains. For the 76 remaining strains, 25 (61.0%) MALT strains and 15 (42.9%) gastritis strains contained a functional sabA gene. The number of CT dinucleotide repeats, which began at position +18, ranged from 3 to 12 for both MALT and gastritis strains (Table 3). When three CT repeats were observed, a thymidine-thymidine was always present in the middle of the repetition in frame with the rest of the gene. Interestingly, for strains harboring seven CT repeats, the sabA gene can be in or out of frame depending on the nucleotide sequence after the CT repeats (Table 3). There was no difference in sabA functional statuses between the two groups of strains (P = 0.167).
TABLE 3.
Signal sequence coding region of the sabA gene and deduced amino acid sequences of 18 different Helicobacter pylori strainsa
| Strain | Sequence of the signal peptide coding region | No. of CT repeats | Deduced amino acid sequence | Gene status |
|---|---|---|---|---|
| G1 | ATGAAAAAACGATTTTTACTTTCTCTATCCC--------------TTGCATCGTCATTACTTTATGCTGAAGACAACGGCTTTTTTGTGA | 3 | MKKRFLLSLSL----ASSLLYAEDNGFFVSAGYQIGEAVQMVKNTGEL | On |
| M30 | ATGAAAAAACGATTTTTACTTTCTCTATCCC--------------TTGCAGCGTCATTACTTTATGCTGAAGACAACGGCTTTTTTGTGA | 3 | MKKRFLLSLSL----AASLLYAEDNGFFVSAGYQIGEAVQMVKNTGEL | On |
| M32 | ATGAAAAAGACAATTCTGCTCTCTCTC--------------------GCTTCATCGCTCTTGCACGCTGAAGACAACGGCTTTTTTGTGA | 4 | MKKTILLSL------ASSLLHAEDNGFFVSAGYQIGEAVQMVKNTGEL | On |
| M45 | ATGAAAAAACGATTTTTACTCTCTCTCTC--------------GCTTGCGGTATCATCGCTCCATGCTGAAGACAACGGCTTTTTTGTGG | 5 | MKKRFLLSLSL----AVSSLHAEDNGFFVGVGYQIGEAVQMVKNTGEL | On |
| G15 | ATGAAAAAACGATTTTTACTCTCTCTCTCTC------------GTTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTATCG | 6 | MKKRFLLSLS----RLRYHRSTLKTTAFLSAWAIKSAKRCKWSKTPVN | Off |
| M59 | ATGAAAAAGACAATTCTACTCTCTCTCTCTC----------------GCTTCATCGCTCTTGCACGCTGAAGACAACGGCTTTTTTGTGG | 6 | MKKTILLSLS----RFIALAR* | Off |
| G5 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTC----------GCTTGCGGTACCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGG | 7 | MKKRFLLSLSL-ACGTIAPR* | Off |
| M61 | ATGAAAAAGACAATTTTACTCTCTCTCTCTCTC--------------GCTTCATCGCTCTTGCATGCTGAAGACAACGGCTTTTTTGTAG | 7 | MKKTILLSLSL----ASSLLHAEDNGFFVGVGYQIGEAVQMVKNTGEL | On |
| G21 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTC--------GTTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGA | 8 | MKKRFLLSLSLSF--AVSSLHAEDNGFFVSAGYQIGEAVQMVKNTGEL | On |
| M23 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTC--------GCTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGA | 8 | MKKRFLLSLSLSL--AVSSLHAEDNGFFVSAGYQIGEAVQMVKNTGEL | On |
| G16 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTC------GCTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTATCG | 9 | MKKRFLLSLSLS--RLRYHRSTLKTTAFLSVWAIKSAKRCKWSKTPVN | Off |
| M25 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTC------GTTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTATCG | 9 | MKKRFLLSLSLS--RLRYHRSTLKTTAFLSVWAIKSAKRCKWSKTPVN | Off |
| G13 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTCTC----GCTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGG | 10 | MKKRFLLSLSLSLACGIIAPR* | Off |
| M31 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTCTC----GCTTGCGGCATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGA | 10 | MKKRFLLSLSLSLACGIIAPR* | Off |
| G26 | ATGAAAAAACGAATTTTACTCTCTCTCTCTCTCTCTCTCTC--GCTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTAA | 11 | MKKRILLSLSLSLSLAVSSLHAEDNGFFVXVGYQIGEAVQMVKNTGEL | On |
| M65 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTCTCTC--GCTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGA | 11 | MKKRFLLSLSLSLSLAVSSLHAEDNGFFVSAGYQIGEAVQMVKNTGEL | On |
| G6 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTCTCTCTCGCTTGCGGCGCCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTGG | 12 | MKKRFLLSLSLSLSRLRRHRSTLKTTAFLWARAIKSVKRCKWSKTPAN | Off |
| M38 | ATGAAAAAACGATTTTTACTCTCTCTCTCTCTCTCTCTCTCTCGTTTGCGGTATCATCGCTCCACGCTGAAGACAACGGCTTTTTTGTAG | 12 | MKKRFLLSLSLSLSRLRYHRSTLKTTAFL* | Off |
G, gastritis strain; M, MALT strain. The stop codon is boxed for strains having the off status (except for strains G15, G16, M25, and G6), and the end of the protein is indicated by an asterisk. Concerning these strains, the stop codon is at positions 133 and 144 for the G15 and G6 strains, respectively, and position 139 for the G16 and M25 strains. All 86 strains tested in this study having the on status presented the italicized conserved motif. CT repeats are underlined, and the thymidine-thymidine sequences in the middles of the repetitions are in boldface. The sabA gene sequences from G1 to M38 strains were submitted to GenBank and assigned accession no. AY299975 to AY29992, respectively.
(viii) hopZ functional status.
We failed to obtain PCR hopZ gene amplification for two MALT strains and three gastritis strains. In addition, for one gastritis strain, an 800-bp PCR product, was obtained (versus products of approximately 3,000 bp according to the 26695 and J99 reference strains), and direct sequencing revealed the lack of the hopZ gene. For the remaining strains, 18 (43.9%) MALT strains and 21 (60.0%) gastritis strains contained a functional hopZ gene. Analysis of the gene sequences revealed that CT repeats began at position +18, and the number of repeats ranged from 6 to 12 as previously described (41). Interestingly, two MALT strains contained four dinucleotide repeats in frame with the residual gene (GenBank accession no. AY299993 and AY299994). Moreover, 11 (26.8%) MALT strains and 4 (11.4%) gastritis strains contained eight dinucleotide repeats out of frame (GenBank accession no. AY299995 to AY300009). There was no difference in the distributions of hopZ functional status between the two groups of strains (P = 0.176).
Relationship between the nine H. pylori virulence factors tested in strains of patients suffering from gastric MALT lymphoma and gastritis.
In MALT strains, we found a positive correlation between cagA and cagE (r = 0.9545, P < 10−4) and between vacA s1m1 genotype (r = 0.6431, P < 10−4) and babA2 (r = 0.4009, P = 0.008), and oipA on (r = 0.7245, P < 10−4) and hopQ type I (r = 0.7684, P < 10−4), but there was a negative correlation between cagA and vacA s2m2 (r = 0.7742, P < 10−4). The babA2 gene was correlated with cagE (r = 0.4423, P = 0.003) and hopQ type I (r = 0.3486, P = 0.0022). Among the gastritis strains, cagA was associated with the same markers as in MALT strains, with the exception of babA2, which was correlated with vacA s1m1 alleles (r = 0.5346, P = 0.0005) and with hopQ type I (r = 0.4310, P = 0.0062).
Results of the MCA for construction of the summary variable.
First, the MCA was performed on 70 strains for which complete data were available for all variables, yielding the F1/F2 plane, which accounted for 82.4% of the total information (73.9% for the horizontal axis [F1] and 8.6% for the vertical axis [F2]). Two main clusters were identified on each side of the horizontal axis: the first included strains positive for cagA, cagE, vacA s1m1 type, babA2, oipA on, and hopQ type I (group A), and the second included strains negative for cagA, cagE, vacA s2m2 type, babA1, oipA off, and hopQ type II (group B). On the vertical axis, iceA1 and sabA on strains were on one side, and iceA2 and sabA off strains were on the other. The hopZ strains were found on the third axis (F3) (data not shown).
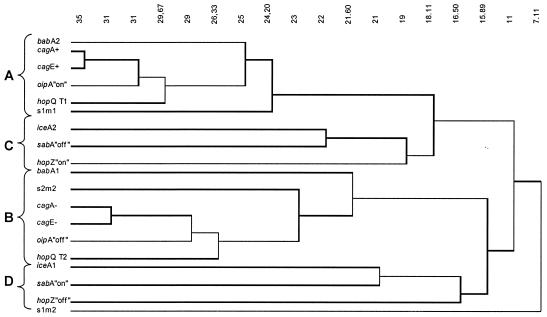
The dendrogram drawn from the results of the cluster analysis of both populations considered together confirmed the presence of four groups: the A and B groups, which were formally identified; and two others. Group C contained iceA2, sabA off, and hopZ on strains, and group D contained iceA1 and sabA on strains (Fig. 1).
FIG. 1.
Dendrogram of the distribution of 70 H. pylori strains from both strain populations for which complete data were available for all variables. Strains have been grouped according to cagA, cagE, vacA, babA, iceA, hopQ, oipA, hopZ, and sabA status.
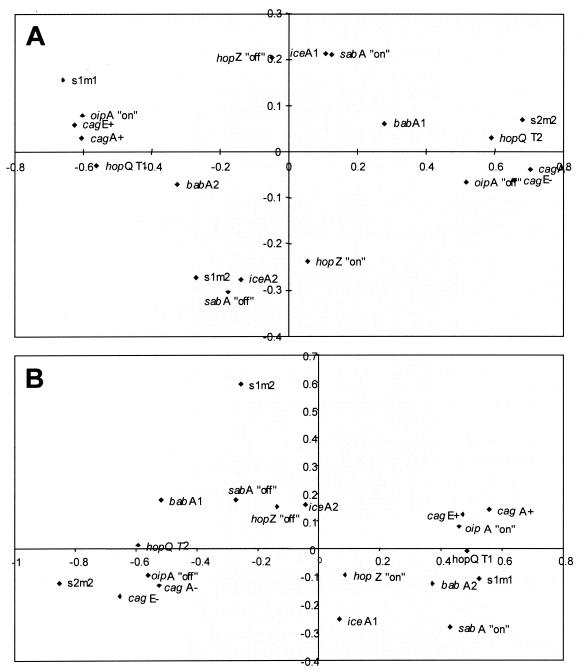
The same approach was then performed for each population: i.e., MALT patients (39 strains) or gastritis patients (31 strains) from whom complete data were available for each variable. The analysis of the MCAs revealed that groups A and B were similar and that the main difference between MALT and gastritis strains was based on the distribution of iceA genotype, sabA status, and hopZ status (Fig. 2A and B). In MALT strains, a cluster absent in gastritis strains and containing iceA1, sabA on, and hopZ off was identified (Fig. 2B).
FIG. 2.
MCA plot of 39 and 31 H. pylori strains isolated from patients suffering from gastric MALT lymphoma (A) and from gastritis (B), respectively, for which complete data were available according to cagA, cagE, vacA, babA, iceA, hopQ, oipA, hopZ, and sabA status.
Relationship between the genotype of H. pylori strains and MALT diagnosis.
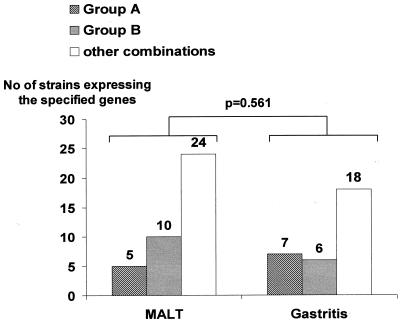
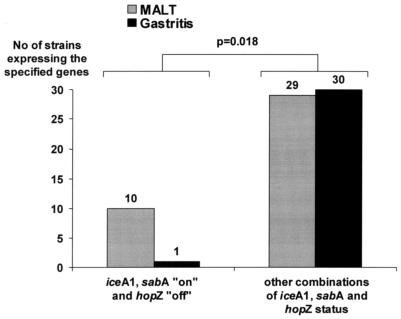
The possible association between the presence of MALT and the characteristics of strains cultured from these patients was tested. As shown in Table 4, the distribution of the dendrogram deduced A and B groups (based on cagA, cagE, and vacA alleles; babA and oipA status; and hopQ genotype) revealed no difference between MALT and gastritis strains (P = 0.561) (Table 4 and Fig. 3). The proportion of MALT patients was not different in those harboring strains with the sabA gene on, with iceA1, or with the hopZ gene on (62.5, 61.5, and 75.7%, respectively). The odds ratios (ORs) were in line with these results. In contrast, when analyzing the summary variables according to the MCA results, it was remarkable that the odds of having MALT lymphoma among patients harboring iceA1, sabA on, and hopZ off strains (OR, 10.3; 95% CI, 1.2 to 86.0) were 10 times higher than for the other defined groups; indeed among a total of 11 strains having these three markers, 10 (90.9%) corresponded to MALT strains (P = 0.018) (Table 4 and Fig. 4).
TABLE 4.
Relationship between the genotypes of H. pylori strains and MALT diagnosisa
| Gene characteristic | Population with gene | Population with MALT | % MALT (95% CI)b | OR (95% CI) |
|---|---|---|---|---|
| sabA | 76 | |||
| Off | 36 | 16 | 44.4 (27.9-61.9) | Baseline |
| On | 40 | 25 | 62.5 (45.8-77.3) | 2.1 (0.8-5.2) |
| iceA | 82 | |||
| iceA2 | 43 | 19 | 44.2 (29.1-60.1) | Baseline |
| iceA1 | 39 | 24 | 61.5 (44.6-76.6) | 2.0 (0.8-4.9) |
| hopZ | 76 | |||
| Off | 39 | 18 | 46.2 (30.1-62.8) | Baseline |
| On | 37 | 28 | 75.7 (58.8-88.2) | 1.9 (0.8-4.8) |
| MCA summary variables | 70 | |||
| ice and sabA genes | ||||
| Others | 49 | 24 | 49.0 (34.4-63.7) | Baseline |
| iceA1 and sabA on | 21 | 15 | 71.4 (47.8-88.7) | 2.6 (0.9-7.8) |
| iceA, sabA, and hopZ genes | ||||
| Others | 59 | 29 | 49.2 (35.9-62.5) | Baseline |
| iceA1, sabA on, and hopZ off | 11 | 10 | 99.9 (58.7-99.8) | 10.3 (1.2-86.0) |
| 3rd MCA variable | ||||
| Group Ac | 12 | 5 | 41.7 (15.2-72.3) | Baseline |
| Group Bd | 16 | 10 | 62.5 (35.4-84.8) | 2.3 (0.5-10.8) |
| Others | 42 | 24 | 57.1 (41.0-72.3) | 1.9 (0.5-6.9) |
ORs are presented as univariate results.
CI, exact binomial 95% CI.
Group A, isolates cagA positive, cagE positive, vacA s1m1 babA2 hopQ T1 oipA on.
Group B, isolates cagA negative, cagE negative, vacA s2m2 babA1 hopQ T2 oipA off.
FIG. 3.
Distribution of the strains harboring the group A and group B combinations among Helicobacter pylori strains isolated from patients with gastric MALT lymphoma or gastritis only.
FIG. 4.
Distribution of strains harboring the triple combination iceA1, sabA on, and hopZ off among H. pylori strains isolated from patients with gastric MALT lymphoma or gastritis only.
DISCUSSION
The present study investigated the influence of the cagA, cagE, vacA, babA, iceA, hopQ, oipA, hopZ, and sabA genes on the outcome of H. pylori-associated disease by comparing two populations of H. pylori strains involved in gastric MALT lymphoma and in gastritis only. In comparison with previous studies concerning MALT lymphoma, the present study evaluated the prevalence and correlation of most H. pylori virulence factors and included the largest collection of strains.
We found that among the various putative virulence factors tested considered individually, none can be associated with MALT strains; however, the MCA allowed us to define two main clusters (groups A, cagA positive, cagE positive, vacA s1m1 babA2 hopQ T1 oipA on; and group B, cagA negative, cagE negative, vacA s2m2 babA1 hopQ T2 oipA off). The cagA gene, the first to be found differentially present in H. pylori strains, is considered a marker for the presence of the cag PAI (15). This region includes a number of other genes forming a type IV secretion system and is associated with increased virulence and severe clinical outcome, such as severe gastritis, duodenal ulceration, and gastric adenocarcinoma (2, 12). However, the region has not been clearly associated with gastric MALT lymphoma. In 1997, Witherell et al. (56) reviewed all of the publications that examined the role of cagA-positive strains in gastric MALT lymphoma and concluded that CagA-positive strains did not contribute to the disease outcome. Since then, several controversial results essentially based on serological data (detection of CagA antibodies) have been published (17, 30, 45, 50, 51). However, a consensus seems to have been found, indicating that H. pylori strains expressing CagA protein are not associated with gastric MALT lymphoma but are associated with diffuse large B-cell lymphoma. Our results based on a large collection of well-characterized MALT strains are consistent with this observation and allowed us to conclude that the CagA pathogenicity marker is not associated with gastric MALT lymphoma. We also tested the cagE gene of the cag PAI, which was found associated with the development of precancerous lesions in the Mongolian gerbil model (38). We observed a strong correlation between cagA and cagE, which is in favor of the accuracy of our results. Furthermore, it may happen that the cag PAI even when present is not functional (i.e., is not capable of transferring CagA into the cells), and this possibility has not been taken into account in most of the studies. Approximately 50% of the H. pylori strains produced the VacA cytotoxin (16). Specific vacA genotypes have been associated with different levels of cytotoxin production as well as with different clinical outcomes (5, 7). While the distributions of vacA alleles were not different between MALT and gastritis strains, we found as expected a good correlation between the vacA s1m1 genotype (corresponding to a high level of cytotoxin production) and cagA and cagE, suggesting that these virulence genes are closely associated: however, this does not explain the evolution toward MALT lymphoma.
Our results concerning iceA are in line with previous published data (20, 43) that showed that a heterogeneity of the iceA2 gene did exist, due to the presence of a varying number of cassettes.
Increased interleukin-8 (IL-8) production has been found to be linked to the presence of a functional oipA gene. This IL-8 production is enhanced when the cag PAI is present (61). The oipA gene has also been used to discriminate duodenal ulcer from gastritis (58). Our results are in agreement with those of the previous study, which showed a good correlation between oipA on status and cagA, cagE, and vacA s1m1 genotypes. The oipA on status was significantly related to H. pylori density and neutrophil infiltration (58). Because we found no difference between MALT and gastritis strains in oipA functional status as well as for cag PAI, we can hypothesize that the MALT pathogenesis is not linked to proinflammatory properties of H. pylori strains.
The babA2 and hopQ genes encoding outer membrane proteins were reported to be related to the cag PAI and might contribute to virulence (49). For example, babA2, which plays a central role in H. pylori adherence (9, 25), has been associated with duodenal ulcer and gastric cancer (22), and the type I hopQ allele has been associated with cagA-positive s1 vacA-type strains from patients with ulcer disease (10). Our results confirm this correlation for both groups of strains (MALT and gastritis). However, Gerhard et al. found an inverse correlation of triple-positive strains (babA2, cagA positive, and vacA s1m1 genotype) with the presence of MALT strains (22). But, in their study, 29 MALT strains isolated in Germany were compared to a group of strains isolated mainly from gastric adenocarcima and duodenal ulcer patients, which were essentially cagA positive (92.6 and 100%, respectively), and were not compared to gastritis strains only, in contrast to our study. This can explain the different results. The distribution of the A and B groups (based on cagA, cagE, vacA alleles, babA, oipA status, and hopQ genotypes) did not allow identification of any difference, but the association of these genes in a cluster probably indicates that such strains have a selective advantage.
Nevertheless, MCA results also show that for patients harboring iceA1, sabA on, and hopZ off strains, the odds of developing a gastric MALT lymphoma are 10 times higher. The iceA gene, which has a significant homology to a type II restriction endonuclease, has been described as a pathogenic factor. The iceA1 allelic variant is upregulated upon contact of H. pylori with human gastric epithelial cells and has been associated with peptic ulcer disease and gastric adenocarcinoma (28, 42, 44, 55). The sabA and hopZ genes code for two adhesins that have been identified, but no association with a clinical outcome of H. pylori infection has yet been reported (34, 41). For the first time, we have demonstrated in this study an association between the presence of a sabA functional status with an iceA1 allele and hopZ off status of H. pylori strains and MALT lymphoma. The association between a gene that is activated by contact with the gastric epithelium and an adhesin seems to be in line with their primary roles. The analysis of the sabA functional status confirms the heterogeneity of this gene among H. pylori strains and shows for the first time that this marker can be associated with an iceA1 allele and, to a lesser extent, with a clinical outcome. However, the low prevalence of such strains (10 of 43 MALT strains) makes this triple association as a marker poorly sensitive while specific for MALT strains. It has been shown that sabA inactivation does not affect adherence mediated by the babA adhesin, implying that these two adhesins are organized and expressed as independent units (34). Our results support this observation, because we found no correlation between them, and moreover the sabA functional status and babA2 are not present in the same clusters.
Since a very low proportion of H. pylori-infected patients will develop a gastric MALT lymphoma, our results show that current pathogenicity markers cannot identify the strains associated with gastric MALT lymphoma. Although the involvement of H. pylori in this disease is now well established, the pathomechanism by which a clonal expansion of B cells occurs in a background of chronic gastritis remains to be identified. However, our current data identified a polymorphism in the recently described sabA gene that supports the hypothesis that a small proportion of MALT strains may use the SabA adhesin to adhere to sialylated glycoconjugates expressed during chronic inflammation and thus may be able to contribute to the chronicity of H. pylori infection.
Acknowledgments
We want to thank the Fédération Française de Cancérologie Digestive, the Assistance Publique-Hôpitaux de Paris (AP-HP), and the Groupe d'Etude des Lymphomes de l'Adulte, for supplying H. pylori strains; and the Conseil Régional d'Aquitaine and the Fond de Recherche of the Societé Nationale Française de Gastroentérologie for financially supporting the project.
Editor: V. J. DiRita
REFERENCES
- 1.Achtman, M., T. Azuma, D. E. Berg, Y. Ito, G. Morelli, Z. J. Pan, S. Suerbaum, S. A. Thompson, A. van der Ende, and L. J. van Doorn. 1999. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 32:459-470. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. L. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Arents, N. L., A. A. van Zwet, J. C. Thijs, A. M. Kooistra-Smid, K. R. van Slochteren, J. E. Degener, J. H. Kleibeuker, and L. J. van Doorn. 2001. The importance of vacA, cagA, and iceA genotypes of Helicobacter pylori infection in peptic ulcer disease and gastroesophageal reflux disease. Am. J. Gastroenterol. 96:2603-2608. [DOI] [PubMed] [Google Scholar]
- 5.Atherton, J. C., P. Cao, R. M. Peek, M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori—association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 6.Atherton, J. C., T. L. Cover, R. J. Twells, M. R. Morales, C. J. Hawkey, and M. J. Blaser. 1999. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J. Clin. Microbiol. 37:2979-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atherton, J. C., R. M. Peek, K. T. Tham, T. L. Cover, and M. J. Blaser. 1997. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology 112:92-99. [DOI] [PubMed] [Google Scholar]
- 8.Bayerdorffer, E., A. Neubauer, B. Rudolph, C. Thiede, N. Lehn, S. Eidt, M. Stolte et al. 1995. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. Lancet. 345:1591-1594. [DOI] [PubMed] [Google Scholar]
- 9.Boren, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 10.Cao, P., and T. L. Cover. 2002. Two different families of hopQ alleles in Helicobacter pylori. J. Clin. Microbiol. 40:4504-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, S. J., H. Yokoo, and A. Vanagunas. 1996. Progression of gastritis to monoclonal B-cell lymphoma with resolution and recurrence following eradication of Helicobacter pylori. JAMA 275:937-939. [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA 93:14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chisholm, S. A., E. L. Teare, B. Patel, and R. J. Owen. 2002. Determination of Helicobacter pylori vacA allelic types by single-step multiplex PCR. Lett. Appl. Microbiol. 35:42-46. [DOI] [PubMed] [Google Scholar]
- 14.Correa, P., J. Fox, E. Fontham, B. Ruiz, Y. Lin, D. Zavala, N. Taylor, D. Mackinley, E. De Lima, H. Portilla, and G. Zarama. 1990. Helicobacter pylori and gastric carcinoma. Cancer 66:2569-2574. [DOI] [PubMed] [Google Scholar]
- 15.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover, T. L. 1996. The vacuolating cytotoxin of Helicobacter pylori. Mol. Microbiol. 20:241-246. [DOI] [PubMed] [Google Scholar]
- 17.Delchier, J. C., D. Lamarque, M. Levy, E. M. Tkoub, C. Copie Bergman, L. Deforges, M. T. Chaumette, and C. Haioun. 2001. Helicobacter pylori and gastric lymphoma: high seroprevalence of CagA in diffuse large B-cell lymphoma but not in low-grade lymphoma of mucosa-associated lymphoid tissue type. Am. J. Gastroenterol. 96:2324-2328. [DOI] [PubMed] [Google Scholar]
- 18.D'Elios, M. M., A. Amedei, M. Manghetti, F. Costa, C. T. Baldari, A. S. Quazi, J. L. Telford, S. Romagnani, and G. Del Prete. 1999. Impaired T-cell regulation of B-cell growth in Helicobacter pylori-related gastric low-grade MALT lymphoma. Gastroenterology 117:1105-1112. [DOI] [PubMed] [Google Scholar]
- 19.Eck, M., B. Schmausser, R. Haas, A. Greiner, S. Czub, and H. K. Muller Hermelink. 1997. MALT-type lymphoma of the stomach is associated with Helicobacter pylori strains expressing the CagA protein. Gastroenterology 112:1482-1486. [DOI] [PubMed] [Google Scholar]
- 20.Figueiredo, C., W. G. V. Quint, R. Sanna, E. Sablon, J. P. Donahue, Q. Xu, G. G. Miller, R. M. Peek, M. J. Blaser, and L. J. van Doorn. 2000. Genetic organization and heterogeneity of the iceA locus of Helicobacter pylori. Gene 246:59-68. [DOI] [PubMed] [Google Scholar]
- 21.Forman, D., P. Webb, and J. Parsonnet. 1994. Helicobacter pylori and gastric cancer. Lancet 34:243-244. [PubMed] [Google Scholar]
- 22.Gerhard, M., N. Lehn, N. Neumayer, T. Boren, R. Rad, W. Schepp, S. Miehlke, M. Classen, and C. Prinz. 1999. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc. Natl. Acad. Sci. USA 96:12778-12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussell, T., P. G. Isaacson, J. E. Crabtree, and J. Spencer. 1996. Helicobacter pylori-specific tumour-infiltrating T cells provide contact dependent help for the growth of malignant B cells in low-grade gastric lymphoma of mucosa-associated lymphoid tissue. J. Pathol. 178:122-127. [DOI] [PubMed] [Google Scholar]
- 24.Hussell, T., P. G. Issacson, J. E. Crabtree, and J. Spencer. 1993. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 342:571-574. [DOI] [PubMed] [Google Scholar]
- 25.Ilver, D., A. Arnqvist, J. Ogren, I. M. Frick, D. Kersulyte, E. T. Incecik, D. E. Berg, A. Covacci, L. Engstrand, and T. Boren. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 26.Isaacson, P. G., and J. Spencer. 1995. The biology of low grade MALT lymphoma. J. Clin. Pathol. 48:395-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kersulyte, D., H. Chalkauskas, and D. E. Berg. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31:31-43. [DOI] [PubMed] [Google Scholar]
- 28.Kidd, M., R. M. Peek, A. J. Lastovica, D. A. Israel, A. F. Kummer, and J. A. Louw. 2001. Analysis of iceA genotypes in South African Helicobacter pylori strains and relationship to clinically significant disease. Gut 49:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuipers, E. J., A. Lee, E. C. Klinkenbergknol, and S. G. M. Meuwissen. 1995. The development of atrophic gastritis—Helicobacter pylori and the effects of acid suppressive therapy. Aliment. Pharmacol. Ther. 9:331-340. [DOI] [PubMed] [Google Scholar]
- 30.Lamarque, D., T. Gilbert, F. Roudot-Thoraval, L. Deforges, M. T. Chaumette, and J. C. Delchier. 1999. Seroprevalence of eight Helicobacter pylori antigens among 182 patients with peptic ulcer, MALT gastric lymphoma or non-ulcer dyspepsia. Higher rate of seroreactivity against CagA and 35-kDa antigens in patients with peptic ulcer originating from Europe and Africa. Eur. J. Gastroenterol Hepatol. 11:721-726. [DOI] [PubMed] [Google Scholar]
- 31.Lehours, P., A. Ruskone Fourmestraux, A. Lavergne, F. Cantet, and F. Mégraud. 2003. Which test to use to detect Helicobacter pylori infection in patients with low-grade gastric mucosa-associated lymphoid, tissue lymphoma? Am. J. Gastroenterol. 98:291-295. [DOI] [PubMed] [Google Scholar]
- 32.Levy, M., C. Copie Bergman, C. Traulle, A. Lavergne Slove, N. Brousse, J. F. Flejou, A. de Mascarel, F. Hemery, P. Gaulard, and J. C. Delchier. 2002. Conservative treatment of primary gastric low-grade B-cell lymphoma of mucosa-associated lymphoid tissue: predictive factors of response and outcome. Am. J. Gastroenterol. 97:292-297. [DOI] [PubMed] [Google Scholar]
- 33.Liu, H. X., A. Ruskone-Fourmestraux, A. Lavergne Slove, H. T. Ye, T. Molina, Y. Bouhnik, R. A. Hamoudi, T. C. Diss, A. Dogan, F. Megraud, J. C. Rambaud, M. Q. Du, and P. G. Isaacson. 2001. Resistance of t(11;18) positive gastric mucosa-associated lymphoid tissue lymphoma to Helicobacter pylori eradication therapy. Lancet 357:39-40. [DOI] [PubMed] [Google Scholar]
- 34.Mahdavi, J., B. Sonden, M. Hurtig, F. O. Olfat, L. Forsberg, N. Roche, J. Angstrom, T. Larsson, S. Teneberg, K. A. Karlsson, S. Altraja, T. Wadstrom, D. Kersulyte, D. E. Berg, A. Dubois, C. Petersson, K. E. Magnusson, T. Norberg, F. Lindh, B. B. Lundskog, A. Arnqvist, L. Hammarstrom, and T. Boren. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mégraud, F., and H. Lamouliatte. 1992. Helicobacter pylori and duodenal ulcer—evidence suggesting causation. Digest. Dis. Sci. 37:769-772. [DOI] [PubMed] [Google Scholar]
- 36.Miyamoto, M., K. Haruma, T. Hiyama, T. Kamada, H. Masuda, F. Shimamoto, K. Inoue, and K. Chayama. 2002. High incidence of B-cell monoclonality in follicular gastritis: a possible association between follicular gastritis and MALT lymphoma. Virchows Arch. 440:376-380. [DOI] [PubMed] [Google Scholar]
- 37.Morgner, A., E. Bayerdorffer, A. Neubauer, and M. Stolte. 2001. Helicobacter pylori associated gastric B cell MALT lymphoma: predictive factors for regression. Gut 48:290-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ogura, K., S. Maeda, M. Nakao, T. Watanabe, M. Tada, T. Kyutoku, H. Yoshida, Y. Shiratori, and M. Omata. 2000. Virulence factors of Helicobacter pylori responsible for gastric diseases in Mongolian gerbil. J. Exp. Med. 192:1601-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsonnet, J., G. D. Friedman, N. Orentreich, and H. Vogelman. 1997. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 40:297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parsonnet, J., S. Hansen, L. Rodriguez, A. B. Gelb, R. A. Warnke, E. Jellum, N. Orentreich, J. H. Vogelman, and G. D. Friedman. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 41.Peck, B., M. Ortkamp, K. D. Diehl, E. Hundt, and B. Knapp. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peek, R. M., M. J. Blaser, D. J. Mays, M. H. Forsyth, T. L. Cover, S. Y. Song, U. Krishna, and J. A. Pietenpol. 1999. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 59:6124-6131. [PubMed] [Google Scholar]
- 43.Peek, R. M., Jr., L. J. van Doorn, J. P. Donahue, K. T. Tham, C. Figueiredo, M. J. Blaser, and G. G. Miller. 2000. Quantitative detection of Helicobacter pylori gene expression in vivo and relationship to gastric pathology. Infect. Immun. 68:5488-5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peek, R. M., Jr., S. A. Thompson, J. P. Donahue, K. T. Tham, J. C. Atherton, M. J. Blaser, and G. G. Miller. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Am. Soc. Physicians 110:531-544. [PubMed] [Google Scholar]
- 45.Peng, S. Y., S. P. Chou, and H. C. Hsu. 1998. Association of downregulation of cyclin D1 and of overexpression of cyclin E with p53 mutation, high tumor grade and poor prognosis in hepatocellular carcinoma. J. Hepatol. 29:281-289. [DOI] [PubMed] [Google Scholar]
- 46.Rad, R., M. Gerhard, R. Lang, M. Schoniger, T. Rosch, W. Schepp, I. Becker, H. Wagner, and C. Prinz. 2002. The Helicobacter pylori blood group antigen-binding adhesin facilitates bacterial colonization and augments a nonspecific immune response. J. Immunol. 168:3033-3041. [DOI] [PubMed] [Google Scholar]
- 47.Rouanet, H., and B. Le Roux. 1993. Analyse des données multidimensionnelles, vol. 1. Dunod, Paris, France.
- 48.Ruskoné Fourmestraux, A., A. Lavergne, P. H. Aegerter, F. Mégraud, L. Palazzo, A. de Mascarel, T. Molina, and J. C. L. Rambaud. 2001. Predictive factors for regression of gastric MALT lymphoma after anti-Helicobacter pylori treatment. Gut 48:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmausser, B., M. Eck, A. Greiner, M. Kraus, and H. K. Müller-Hermelink. 2000. Mucosal humoral immune response to CagA shows a high prevalence in patients with gastric MALT-type lymphoma. Virchows Arch. 436:115-118. [DOI] [PubMed] [Google Scholar]
- 51.Taupin, A., A. Occhialini, A. Ruskoné Fourmestraux, J. C. Delchier, J. C. Rambaud, and F. Mégraud. 1999. Serum antibody responses to Helicobacter pylori and the cagA marker in patients with mucosa-associated lymphoid tissue lymphoma. Clin. Diagn. Lab. Immunol. 6:633-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomb, J. F. 1999. The complete genome sequence of the gastric pathogen Helicobacter pylori. Comment. Scientist 13:17. [Google Scholar]
- 53.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. Smith, O., C. M. Fraser, J. C. Venter et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. (Erratum, 389:412.) [DOI] [PubMed] [Google Scholar]
- 54.Tummuru, M. K. R., T. L. Cover, and M. J. Blaser. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun. 61:1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Doorn, L.-J., C. Figueiredo, R. Sanna, A. Plaisier, P. Schneeberger, W. De Boer, and W. Quint. 1998. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 115:58-66. [DOI] [PubMed] [Google Scholar]
- 56.Witherell, H. L., S. Hansen, E. Jellum, N. Orentreich, J. H. Vogelman, and J. Parsonnet. 1997. Risk for gastric lymphoma in persons with CagA+ and CagA− Helicobacter pylori infection. J. Infect. Dis. 176:1641-1644. [DOI] [PubMed] [Google Scholar]
- 57.Wotherspoon, A. C., C. Doglioni, T. C. Diss, L. Pan, A. Moschini, M. de Boni, and P. G. Isaacson. 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342:575-577. [DOI] [PubMed] [Google Scholar]
- 58.Yamaoka, Y., S. Kikuchi, H. M. T. El-Zimaity, O. Gutierrez, M. S. Osato, and D. Y. Graham. 2002. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology 123:414-424. [DOI] [PubMed] [Google Scholar]
- 59.Yamaoka, Y., T. Kodama, M. Kita, J. Imanishi, K. Kashima, and D. Y. Graham. 1999. Relation between clinical presentation, Helicobacter pylori density, interleukin 1β and 8 production, and cagA status. Gut 45:804-811. (Erratum, 46:584, 2000.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaoka, Y., T. Kodama, O. Gutierrez, J. G. Kim, K. Kashima, and D. Y. Graham. 1999. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J. Clin. Microbiol. 37:2274-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaoka, Y., D. H. Kwon, and D. Y. Graham. 2000. A M(r) 34,000 proinflammatory outer membrane protein (oipA) of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 97:7533-7538. (Erratum, 97:11133.) [DOI] [PMC free article] [PubMed] [Google Scholar]