Abstract
Domesticated maize and its wild ancestor (teosinte) differ strikingly in morphology and afford an opportunity to examine the connection between strong selection and diversity in a major crop species. The tb1 gene largely controls the increase in apical dominance in maize relative to teosinte, and a region of the tb1 locus 5′ to the transcript sequence was a target of selection during maize domestication. To better characterize the impact of selection at a major “domestication” locus, we have sequenced the upstream tb1 genomic region and systematically sampled nucleotide diversity for sites located as far as 163 kb upstream to tb1. Our analyses define a selective sweep of ≈60–90 kb 5′ to the tb1 transcribed sequence. The selected region harbors a mixture of unique sequences and large repetitive elements, but it contains no predicted genes. Diversity at the nearest 5′ gene to tb1 is typical of that for neutral maize loci, indicating that selection at tb1 has had a minimal impact on the surrounding chromosomal region. Our data also show low intergenic linkage disequilibrium in the region and suggest that selection has had a minor role in shaping the pattern of linkage disequilibrium that is observed. Finally, our data raise the possibility that maize-like tb1 haplotypes are present in extant teosinte populations, and our findings also suggest a model of tb1 gene regulation that differs from traditional views of how plant gene expression is controlled.
Domestication has had far-reaching effects on crop genomes that are only now being understood. A common feature of domestication has been reduction of genetic diversity in crops relative to wild progenitors (1). This reduction has resulted from two major forces. First, most domestication events are thought to have involved initial populations of small size (relative to wild ancestors) that constrained genome-wide levels of genetic diversity (hereafter referred to as “bottleneck effects”). The severity of genetic loss ascribed to bottleneck effects varies greatly among crop species (1, 2). The second factor to have an impact on crop genomes is selection for the agronomic traits that distinguish crops from their ancestors. Selection is expected both (i) to reduce diversity at selected loci as favorable alleles are driven to high frequency and (ii) to reduce diversity at linked loci through the effect of genetic hitchhiking. Selection is also predicted to elevate linkage disequilibrium (LD) near selected sites relative to that observed at neutral (not selected) genomic locations.
Historically, the effect of selection on genomic diversity in crops has been difficult to assess because the identity of selected loci has remained largely unknown. Recently, a few genes that were targets of domestication or subsequent crop improvement have been identified. Examples include BoCAL for inflorescence morphology in cauliflower (Brassica oleracea subsp. botrytis) (3), fw2.2 for fruit size in tomato (Lycopersicon esculentum var. esculentum) (4), Rht-B1 and Rht-D1 for plant stature and yield in wheat (Triticum species) (5), and Waxy for kernel biochemistry in rice (Oryza sativa) (6). Where it has been examined, genetic diversity at selected loci is typically reduced relative to the diversity expected from bottleneck effects alone (6, 7).
In maize (Zea mays subsp. mays), the teosinte branched1 (tb1) gene affords the opportunity to examine the connection between selection and diversity in a crop species. The tb1 gene is largely responsible for a major-effect quantitative trait locus on chromosome 1 that controls differences in plant architecture between maize and its wild relative, teosinte (Z. mays subsp. mexicana and subsp. parviglumis) (8–11). The tb1 gene has been cloned (12) and is a member of the TCP family of transcriptional regulators (12, 13). In an initial study, Wang et al. (7) sampled single-nucleotide polymorphism (SNP) diversity at tb1 and showed that the 5′ untranslated region was selected during maize domestication, even though the pattern of diversity in the coding and 3′ region of tb1 was consistent with neutral evolution. The observation that selection acted on the 5′ region suggests that tb1 regulation was the target of early agriculturalists and is consistent with expression studies that have shown a difference in tb1 message levels between maize and teosinte (12, 14, 15). However, the 5′ boundary of the selective sweep upstream to tb1 has not been determined, and the critical question of whether the selective sweep affects genes in addition to tb1 has remained unanswered. Here we characterize SNP diversity in the tb1 genomic region to define the extent of the selective sweep at the tb1 locus and to understand the impact of selection at tb1 on diversity and LD at nearby genomic regions.
Materials and Methods
Bacterial Artificial Chromosome (BAC) Selection, Sequencing, and Analysis. Probes for the tb1 gene were used to screen a maize B73 HindIII BAC library (16), and a clone (ZMMBBb0178A11) containing the tb1 gene was sequenced. This clone was used to identify a DNA fingerprinting contig by webfpc (www.genome.arizona.edu/fpc/maize, contig 615 with release on Dec. 16, 2002), and a second clone (ZMMBBb0013I05) that extended farthest 5′ to tb1 was also sequenced. For sequencing, two libraries of an average insert size of 2 and 5 kb were prepared by using the PUC119 vector with BAC insert DNA sheared with a HydroShear device (Genemachines, San Carlos, CA). These libraries were sequenced by using M13 universal primers from both directions on an ABI3700 capillary sequencer with BigDye terminator chemistry. Base calling and quality assessment were performed by using phred (17, 18), assembly was performed by using phrap, and editing was done by using consed (19). Finishing of BAC sequences was performed as described by Ramakrishna et al. (20).
For sequence annotation, we first identified repetitive sequences by using the repeatmasker program (A. F. A. Smit and P. Green, repeatmasker at http://ftp.genome.washington.edu/RM/RepeatMasker.html) with speed/sensitivity set to “default” and DNA source set to “Grasses.” Nonrepetitive DNA was analyzed for genes by using the fgenesh gene prediction software (www.softberry.com/berry.phtml/) with the monocot training set and by homology searches to all sequences in GenBank by using the blast algorithm (21). To examine gene structure and extent of sequence conservation at tb1, we compared maize tb1 sequences with those from homologous loci in other grasses by using the main vista (mvista) program (22–24) with window length set to 100 bp. For these comparisons, maize sequence was aligned to genomic regions from sorghum (Sorghum bicolor, 8,821–29,960 bp of AF466204) and rice (7,859–29,025 bp of AC091775).
Sampling Strategy and Plant Material. We surveyed nucleotide diversity in the tb1 region by PCR amplification and sequencing of ≈500- to 1,000-bp segments of genomic DNA from a collection of maize and teosinte individuals (Table 3, which is published as supporting information on the PNAS web site). For maize, we used a set of 24 individuals, including 16 landraces, 2 tropical inbreds, and 6 U.S. inbreds, that is nearly identical with that used by Tenaillon et al. (25) to survey maize diversity but is less biased toward U.S. inbred material. Samples of landrace DNA are from haploid plants as described (25). Alleles of tb1 were also amplified from Z. mays subsp. parviglumis and subsp. mexicana and from a more distant relative (Zea diploperennis). We attempted to recover products for approximately eight subsp. parviglumis and four subsp. mexicana alleles and for one Z. diploperennis allele.
PCR and Sequencing. We PCR-amplified products from genomic DNA by using SuperMix High Fidelity Enzyme mixture (Invitrogen) for 35 cycles with primers designed to match maize genomic sequence (primer sequences are available on request). Where DNA templates were known to harbor a single allele for the tb1 region (maize inbred and haploid material), PCR products were purified (QIAquick PCR Purification Kit, Qiagen, Valencia, CA) and sequenced on capillary machines (Applied Biosystems) at the University of Wisconsin Biotechnology Center. For heterozygous teosinte samples, products were cloned (pCR 2.1-TOPO kit, Invitrogen), and at least five clones per allele were sequenced from each PCR to control for PCR errors, or, preferably, three products were sequenced that had each been cloned from independent PCR amplifications. To ensure correct sequence calls, we sequenced amplicons in both orientations. Sequence reads were assembled and edited with sequencher software (Gene Codes, Ann Arbor, MI).
In addition, we have included in our analyses sequences for the tb1 coding and immediate 5′ region that have been reported (see Results and refs. 7 and 25). We evaluated singleton changes for one study (25) by resequencing and have submitted new sequences to GenBank as appropriate (accession nos. AY428407–AY428435).
Sequence Analysis. Sequences were aligned by using se-al Version 2.0a11 (A. Rambaut, 1996. Se-Al: Sequence Alignment Editor, http://evolve.zoo.ox.ac.uk). Molecular population genetic statistics were estimated by using the software dnasp, Version 3.51 (26), unless otherwise noted. For the Hudson–Kreitman–Aguade (HKA) test, the Z. diploperennis sequence was used to calculate divergence, and adh1 (25, 27), adh2 (28), te1 (29), and glb1 (25, 27) were used as neutral control genes. For calculation of r2, a pairwise measure of LD, insertion/deletion (indel) changes were included in the analysis unless they were associated with simple sequence repeats. Fay and Wu's H test (30) to detect hitchhiking after a selective sweep was performed with 10,000 simulations at http://crimp.lbl.gov/htest.html. Z. diploperennis was used to infer ancestral character states. For this test, an estimate of population recombination (R) was calculated from the data, and divergence at synonymous and noncoding sites was multiplied by 3/8 to estimate the rate of back mutation as described (30). Neighbor-joining phylogenies were generated by using the paup Version 4.0b10 software package (31). All phylogenies were midpoint rooted, and robustness of trees was assessed with 1,000 bootstrap replicates by using the “Fast” stepwise-addition method (trees were constructed with distance measure set to uncorrected “p”).
Results
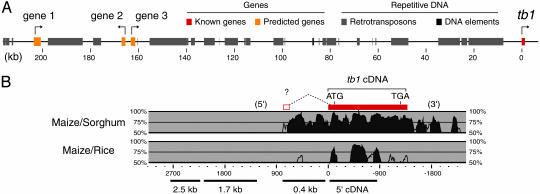
Genomic Sequence of the tb1 Region. To determine the full impact of the selective episode in the tb1 region, we isolated and sequenced two BAC clones that together form a contig of 220.9 kb (Fig. 1A). The sequence from this contig extends 214.3 kb 5′ and 5.4 kb 3′ to the tb1-coding sequence. The contig sequence was annotated by using a combination of gene identification programs and homology searches (see Materials and Methods) that predicted three genes 5′ to tb1 (here referred to as genes 1, 2, and 3; Fig. 1 A). Gene 2 is predicted to encode a 227-aa product with homology to metal-transporting ATPases. Genes 1 and 3 have homology to predicted genes of unknown function in several plant genomes, including those of rice and Arabidopsis thaliana, and contain ORFs of 2,304 and 1,272 bp, respectively. Gene 3 is the nearest gene 5′ to tb1, and the length of the intergenic region between tb1 and gene 3 is 161.4 kb. The tb1 region also harbors a large number of repetitive elements that belong primarily to retrotransposon families, and the program repeatmasker (see Materials and Methods) identified 45% of the sequence contig as highly repetitive. The tb1 upstream region thus appears relatively gene-poor (compare with ref. 32), and the large size of the intergenic segment between gene 3 and tb1 is particularly striking.
Fig. 1.
Genomic region and gene structure. (A) A schematic representation of the tb1 region indicating predicted genes and repetitive sequence features. (B)vista plots compare identity of maize genomic sequence for tb1 with that of sorghum and rice (similarity indicated by shading). The position of the previously identified cDNA (7) is shown with putative start and termination sites for translation. The location of a possible 5′ exon is as indicated (open box, “?”; ref. 7). The locations of sites surveyed for diversity in the current study (2.5-, 1.7-, and 0.4-kb and 5′ cDNA sites) are shown at the bottom.
tb1 Gene Structure. Relatively little is known about tb1 gene organization. The longest identified cDNA clone from the tb1 locus is 1,306 bp and contains the highly conserved coding sequence (7, 33). The sequence from this clone is contiguous with genomic DNA (Fig. 1B and ref. 33), and the length of the clone is consistent with the size of tb1 message detected by Northern blot analysis (33). In addition, a small exon located 5′ to the cDNA sequence has also been identified from a single RT-PCR product (Fig. 1B and ref. 33), although it is unclear that the message containing this exon can initiate translation inframe to make the conserved TB1 protein. Moreover, we have been unable to repeat the amplification of the 5′ maize exon by RT-PCR (suggesting that inclusion of the exon is a rare event); and using the 5′ RACE technique, we have never recovered spliced products from the tb1 locus (data not shown).
To gain additional insight into tb1 gene structure, we have compared genomic tb1 sequences from maize with corresponding genomic sequences from a sorghum tb1-like gene and with the rice ortholog, OsTB1 (34). Sorghum is a close relative to maize within the tribe Andropogoneae (35), whereas rice is a distant relative to maize within the family Poaceae (36). Pairwise identity plots to the maize sequence are shown in Fig. 1B. A large region conserved between maize and sorghum extends for ≈1kb both 5′ and 3′ to the known maize cDNA sequence. As expected, less conservation is observed when maize is compared with the more distantly related rice OsTB1, although a small region of high identity is apparent between maize and rice in the region 5′ to the coding sequence (Fig. 1B). It is possible that sequences conserved with either sorghum or rice that are located 5′ or 3′ to the known maize cDNA sequence are required for conserved aspects of tb1 regulation. Although conservation of the putative 5′ exon sequence with sorghum is difficult to interpret because of the extensive similarity between maize and sorghum sequences, the comparison with rice does not provide evidence for conservation of this exon. Collectively, our data suggest that the functional tb1 message may be transcribed from the single large exon that contains the conserved coding sequence.
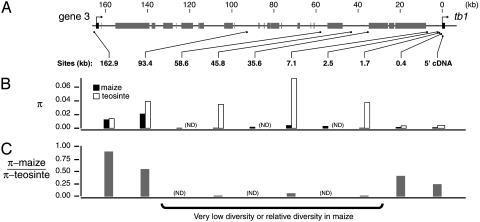
Genetic Diversity in the tb1 Region. To assess the impact of selection at tb1, we have examined levels of DNA polymorphism for ten sites ranging from 467 to 1,024 bp in length located in the region from tb1 to gene 3 (Table 1 and Figs. 1B and 2A). The 5′ cDNA site (Fig. 1B) corresponds to the 5′ most 839 bp of the previously published tb1 cDNA sequence (33). Of this, 783 bp are coding if the first in-frame ATG in the tb1 ORF is defined as the translational start site. The remaining nine regions are noncoding and are referred to by distance (in kilobases) from the 5′ cDNA site. The 162.9-kb site is located immediately 5′ to the gene 3 ORF (Fig. 2 A).
Table 1. Diversity statistics.
| Maize
|
parviglumus + mexicana
|
parviglumus
|
mexicana
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus | Length, bp | n | θ × 103 | π × 103 | n | θ × 103 | π × 103 | n | θ × 103 | π × 103 | n | θ × 103 | π × 103 |
| 162.9-kb | 467 | 18 | 10.7 | 12.3 | 10 | 16.2 | 13.9 | 5 | 12.1 | 11.9 | 5 | 19.0 | 19.3 |
| 93.4-kb | 485 | 14 | 27.1 | 20.8 | 13 | 41.8 | 38.9 | 8 | 38.6 | 37.5 | 5 | 40.3 | 43.1 |
| 58.6-kb | 520 | 23 | 0.5 | 0.2 | — | — | — | — | — | — | — | — | — |
| 45.8-kb | 1,003 | 24 | 1.1 | 0.3 | 13 | 33.8 | 34.7 | 9 | 31.1 | 32.9 | 4 | 32.8 | 36.7 |
| 35.6-kb | 1,024 | 24 | 3.1 | 1.7 | — | — | — | — | — | — | — | — | — |
| 7.1-kb | 842 | 24 | 6.7 | 4.0 | 12 | 59.8 | 72.2 | 8 | 17.6 | 12.7 | 4 | 11.6 | 10.8 |
| 2.5-kb | 534 | 24 | 3.5 | 2.8 | — | — | — | — | — | — | — | — | — |
| 1.7-kb | 935 | 24 | 0.6 | 0.3 | 14 | 35.7 | 37.3 | 8 | 34.1 | 34.9 | 6 | 38.8 | 38.8 |
| 0.4-kb | 761 | 32 | 3.4 | 1.4 | 10 | 5.0 | 3.6 | 7 | 4.6 | 3.6 | 3 | 3.7 | 3.7 |
| 5′ cDNA | 839 | 32 | 1.8 | 1.0 | 10 | 6.3 | 4.2 | 7 | 6.8 | 5.2 | 3 | 0.8 | 0.8 |
Length of sampled regions (bp) is relative to B73 maize inbred genomic sequence, and diversity was estimated for maize, for subsp. parviglumis and subsp. mexicana, and for each teosinte subspecies individually. Sites for which we did not obtain teosinte data are indicated by dashes. For the 0.4-kb and 5′ cDNA sites, we analyzed sequence data reported in two studies (7, 25).
Fig. 2.
Diversity survey. (A) The location of sampled sites relative to sequence features in the gene 3 to tb1 region. (B) The levels of nucleotide diversity (π) for maize and teosinte. (C) The relative ratio of π in maize to teosinte. Values of π for teosinte were calculated from combined subsp. parviglumis and subsp. mexicana sequences. ND, regions for which teosinte data are not determined.
We calculated two estimates of diversity from our data. Nucleotide polymorphism (θ) is calculated from the total number of segregating sites with correction for sample size (37), and nucleotide diversity (π) is the probability that two randomly selected sequences will possess different nucleotides at a site. In maize, θ and π are very low for all sites extending from the tb1 transcript to the 58.6-kb site, with diversity at the 1.7-, 45.8-, and 58.6-kb sites (Table 1 and Fig. 2B) among the lowest reported for any maize loci (25, 29, 38, 39). At the 93.4- and 162.9-kb sites diversity is markedly higher in maize.
Although low diversity is consistent with selection, absolute levels of diversity are also expected to vary according to levels of functional constraints on sequence evolution (e.g., coding vs. noncoding). To correct for this, we examined nucleotide diversity in maize (πM) relative to that in teosinte (πT) with the expectation that a reduction in the πM/πT ratio relative to that observed for neutral genes should reflect past selection specific to the maize lineage (Fig. 2C). Although the ratio for neutrally evolving maize loci is ≈0.75 (39, 40), a striking reduction in relative diversity is observed at the 1.7-, 7.1-, and 45.8-kb sites (πM/πT < 0.009 for the 1.7- and 45.8-kb sites; Fig. 2C). For the 5′ cDNA, 0.4- and 93.4-kb sites, relative diversity is somewhat higher (each >0.23), and at the 162.9-kb site the value of the πM/πT ratio is typical of that expected solely from the effect of the domestication bottleneck. The region of strong selection identified by Wang et al. (7) at tb1 begins 0.9 kb 5′ to the tb1 coding sequence, and our data show that the region of selection extends to between the 58.6- and 93.4-kb sites. Our data provide no evidence that SNP diversity at the closest gene 5′ to tb1 was reduced by selection.
Tests for Selection. The HKA selection test (41) examines whether selection has reduced diversity at a candidate locus relative to that observed at neutral loci. We applied the HKA test to our data by using four loci sampled in previous studies (see Materials and Methods) as neutral controls and Z. diploperennis as an outgroup (Table 2). For the 1.7- and 7.1-kb sites, HKA test results are significant for each comparison, whereas for the 35.6-kb site results are highly significant for three of the four tests. In each case, at least three tests remain significant if the conservative Bonferroni procedure is used to correct for multiple tests within each site. For the 5′ cDNA site, test results are marginally significant for two of the four tests, although not after a Bonferroni correction is applied. No tests were significant for any comparisons for other sites in the region. Collectively, the HKA test results suggest selection within the region of low maize diversity 5′ to the tb1 coding sequence and indicate that the 5′ boundary of the selective sweep at tb1 does not extend as far upstream as the 93.4-kb site.
Table 2. Tests for selection and hitchhiking.
| Maize
|
Teosinte statistics
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| HKA tests, P values
|
Statistics
|
|||||||||
| Region | Sites, bp | adh1 | adh2 | te1 | glb1 | Tajima's D | Fu and Li's D* | H | Tajima's D | Fu and Li's D* |
| 162.9-kb | 446 | 0.854 | 0.665 | 0.547 | 0.697 | 0.563 | 1.540† | -3.059† | -0.652 | -0.623 |
| 93.4-kb | 431 | 0.950 | 0.827 | 0.393 | 0.896 | -1.014 | -0.972 | -2.418 | -0.314 | -0.321 |
| 58.6-kb | — | — | — | — | — | -1.161 | -1.591 | — | — | — |
| 45.8-kb | — | — | — | — | — | -1.884† | -2.796† | -4.022† | 0.122 | 0.279 |
| 35.6-kb | 464 | 0.009‡ | 0.004‡ | 0.129 | 0.006‡ | -1.589 | -1.593 | — | — | — |
| 7.1-kb | 841 | <0.001‡ | <0.001‡ | 0.042† | <0.001‡ | -1.480 | -0.861 | -10.283† | 0.974 | 0.517 |
| 2.5-kb | — | — | — | — | — | -0.600 | 0.629 | — | — | — |
| 1.7-kb | 601 | <0.001‡ | <0.001‡ | <0.001‡ | <0.001‡ | -0.920 | -0.661 | — | 0.195 | 0.456 |
| 0.4-kb | 730 | 0.219 | 0.167 | 0.818 | 0.148 | -1.814† | -2.980† | -0.540 | -1.284 | -1.509 |
| 5′ cDNA | 833 | 0.051 | 0.039† | 0.389 | 0.030† | -1.273 | -1.150 | -0.923 | -1.595 | -1.577 |
We were unable to apply the HKA test to the 2.5-, 45.8-, and 58.6-kb sites for which we were not able to recover the Z. diploperennis outgroup sequence. Data from subsp. parviglumis and subsp. mexicana were combined to calculate Tajima's D and Fu and Li's D* statistics for teosinte. For statistical tests, †, P < 0.05; ‡, P < 0.01.
We have also calculated Tajima's D statistic (42) to test for nonneutral evolution. For this statistic, negative values are consistent with directional selection. Tajima's D statistic is negative for all sites in maize with the exception of the 162.9-kb site (Table 2), although no significant deviations remain after application of a Bonferroni correction. For teosinte, values of D are typically closer to zero for most sites (Table 2), and no significant deviations from neutrality were detected. Fu and Li's D* (43), an additional statistic that has an interpretation similar to Tajima's D statistic, gives similar results (Table 2). Although the D test statistics provide little statistical support for selection in the tb1 region, these statistics are known to have low power to detect selection and can be influenced by additional population genetic and demographic factors (44). As descriptive statistics, however, the negative values in maize relative to teosinte for the region of low maize diversity are consistent with the expectation of a selective sweep.
Finally, we calculated the H statistic (30) to test for genetic hitchhiking. We observed significant results (P < 0.05) with this test at three of six sites (7.1, 45.8, and 162.9 kb), whereas only 1 of 20 significant tests (5%) would be expected by chance. This finding suggests that diversity at some of the sites was affected by hitchhiking. In theory, H values should be nonsignificant at the selected site itself, significant at sites in the region of hitchhiking, and not significant at more distant locations for which the hitchhiking effect has been diminished by recombination (30). The pattern we observe is roughly consistent with selection at a site upstream to tb1 and hitchhiking in the 7.1- to 162.9-kb region. The nonsignificant result at 93.4 kb may be a function of the small sample size at this site (n = 14).
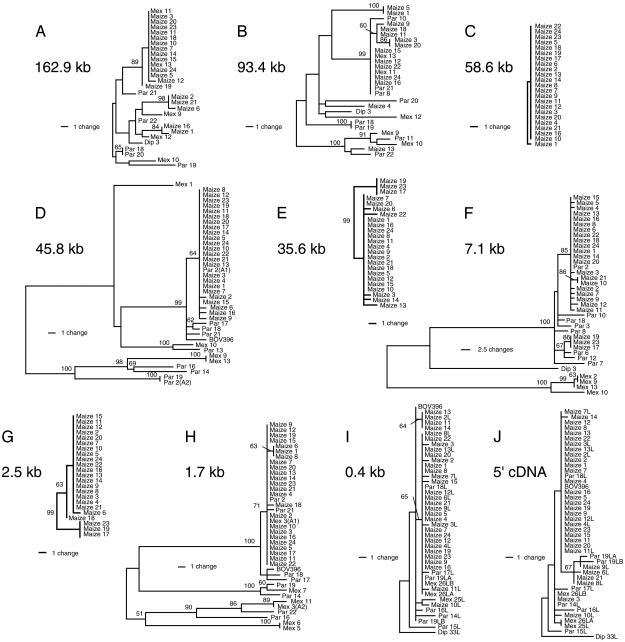
Phylogenetic Analysis. In phylogenetic studies of neutral loci, maize sequences are typically dispersed among different clades that include closely related teosinte sequences (28, 38, 45). The observed trees can be explained by incomplete lineage sorting among the closely related Z. mays subspecies. In the simplest model of selection, in which a single favored haplotype at a locus became fixed during domestication, maize sequences would be expected to form a single clade with only minor differences (7).
We have generated phylogenies from our data to test this expectation and to investigate the relationships between maize and teosinte haplotypes for the tb1 region (Fig. 3). For both the 93.4- and 162.9-kb sites (Fig. 3 A and B), maize alleles are dispersed among well supported clades that include teosinte samples. Wang et al. (7) reported a similar neutral topology for sequences that include the 0.4-kb and 5′ cDNA sites (Fig. 1B). The phylogenies we constructed for the 0.4-kb and 5′ cDNA sites (Fig. 3 I and J) include the sequences of Wang et al. (7) but also an additional 21 maize sequences (see Materials and Methods and ref. 25). The combined data sets continue to give neutral topologies for the 0.4-kb to 5′ cDNA region, although the neutral topology is less evident for the 5′ cDNA site. In contrast, trees for regions located from the 1.7- to the 58.6-kb sites have topologies that are consistent with the expectation of a selective sweep (Fig. 3 C–H). In particular, for the 1.7-, 45.8-, and 58.6-kb sites, all maize samples that we included in our diversity survey are nearly identical (Fig. 3 C, D, and H). For the 2.5-, 7.1-, and 35.6-kb sites, the majority of maize samples also fall within clades of nearly identical sequences, whereas three maize sequences (samples 17, 19, and 23) fall into a second distinct clade that is close to the major maize clade at each site (Fig. 3 E–G).
Fig. 3.
Phylogenies. Neighbor-joining trees are shown for 10 sites in the tb1 region with percent bootstrap support indicated at nodes when >50%. In the absence of an outgroup for all sites, we used midpoint rooting to generate tree outputs that facilitate comparisons of topologies. Teosinte samples are indicated as either Par (subsp. parviglumis) or Mex (subsp. mexicana), and Z. diploperennis samples (Dip) are shown for sites where full-length sequence was obtained. Multiple alleles recovered from heterozygous teosinte plants are indicated (sample name plus A1 or A2). For the 0.4-kb and 5′ cDNA sites, sample names that end with L, LA, or LB are from Wang et al. (7). However, sample maize 1L from the previous study (7) was changed to BOV396 to facilitate discussion (see text).
Wang et al. (7) observed that, whereas most maize sequences in the region immediately 5′ to tb1 fall into a single clade, a haplotype for one maize individual (BOV396) was slightly different. To examine how the sequence from this sample relates to the major maize types at additional intergenic sites, we sampled an allele from this individual for the 1.7- and 45.8-kb sites, where all maize sequences we used to sample diversity form single clades. The BOV396 sample remains distinct from other maize sequences at these sites (Fig. 3 D and H), although it falls near the major maize clade in each case. We also note that a teosinte allele from one subsp. parviglumis individual (Par-2) is identical with the major maize haplotype for all sites in the selected region for which we sampled teosinte sequences (Fig. 3 D, F, and H).
Finally, we searched for polymorphisms that are fixed in maize but are absent from all teosinte sequences with the expectation that such changes might underlie phenotypic differences between maize and teosinte. We found no such fixed changes in the regions we surveyed.
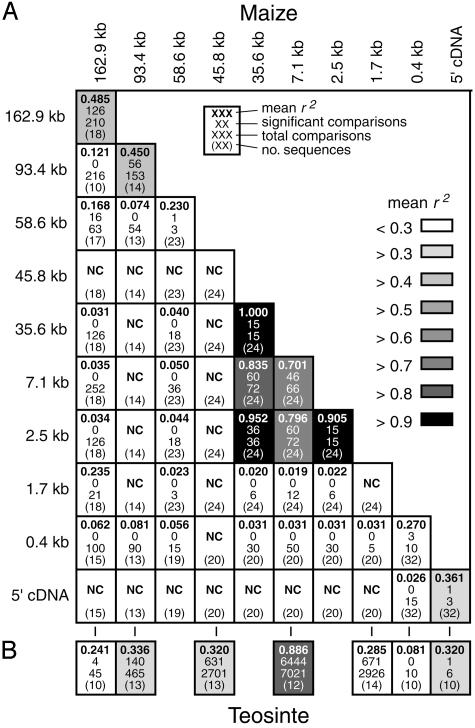
LD. We have examined the pattern of LD in the tb1 region with the expectation that LD should be higher near a selected site (or sites). We caution, however, that our sampling strategy was not designed specifically to assess LD (our sample sizes are small and vary between sites), and conclusions should be interpreted with caution.
To determine within- and between-region LD for maize we calculated mean r2 values and determined the number of significant pairwise comparisons by using Fisher's exact test (Fig. 4A). At the 93.4- and 162.9-kb sites, for which we have not detected obvious selective signatures, mean r2 values are between 0.4 and 0.5, and 60% and 36% of comparisons are significant, respectively. For between-site comparisons that include the 93.4- and 162.9-kb sites LD is very low and virtually no significant comparisons are observed. Although the low level of polymorphism in maize for regions 3′ to the 93.4-kb site limits our ability to assess LD, we do observe strong within- and between-site LD for the 2.5-, 7.1-, and 35.6-kb sites (0.7 < r2 < 1.0; significant comparisons >69% of total comparisons for each analysis).
Fig. 4.
LD. Within- and between-region pairwise LD comparisons for maize (A) and within-region pairwise LD comparisons for teosinte (B). Magnitude of mean r2 is indicated by shading (see scale, A). No pairwise comparisons (NC) could be calculated where insufficient polymorphism was present.
To better understand the pattern of LD in maize, we also assessed within-region LD in teosinte (Fig. 4B). For the most part, the pattern of LD in teosinte is similar to that observed for maize, and LD is sharply elevated at the 7.1-kb site (corresponding to the region of elevated LD in maize). This finding suggests that factors in addition to selection during domestication may explain (or partially explain) the observed pattern of LD for maize (see Discussion). We note, however, that the high divergence of subsp. parviglumis and subsp. mexicana sequences at the 7.1-kb site (Table 1 and Fig. 3F) may bias the estimate of LD for teosinte at this site.
Discussion
Selection and Genetic Diversity. The extent to which selection at a locus constrains diversity at neighboring loci is a key factor for understanding overall patterns of genomic diversity. We have systematically sampled SNP diversity at and nearby tb1, a major domestication locus, to ascertain the genomic impact of selection. We detected clear evidence for a selective sweep that extends at least 58.6 kb 5′ to the tb1 coding region, but the 5′ boundary of the selective sweep resides no farther than 93.4 kb 5′ to tb1. Current statistical approaches detect no nonneutral drop in SNP diversity in flanking regions.
The region between tb1 and gene 3 is characterized by a mixture of nongenic unique sequences separated by intervening tracts of repetitive elements, including retrotransposons. This type of genomic organization resembles that reported for other regions of the maize genome (32, 46), although gene density in the tb1 region is low. The core selective sweep 5′ to tb1 is entirely contained within the region between tb1 and gene 3. This region does not appear to harbor additional genic sequences, although we cannot rule out the possibility that a gene or genes may exist in this region that are difficult to detect by current methods (e.g., genes producing small products or functional RNAs). We also note that several recent studies have shown that haplotypes can differ significantly for the presence or absence of sequences between inbred maize strains (47, 48). Therefore, because our genomic sequence data come from a single maize strain, our description of the genomic organization for the tb1 region may not be representative of all Z. mays germ plasm. Nevertheless, the simplest interpretation of our analyses is that selection at tb1 has not appreciably affected genomic diversity at other genes. If genes in addition to tb1 contribute to the chromosome 1 quantitative trait locus effect for which tb1 is a candidate (8–11), our data suggest that they are not included as part of the selective sweep at tb1 itself.
The limited impact of selection at tb1 on genomic diversity is striking given that tb1 was under strong selection during domestication (7). In contrast, selected regions in other species have typically been found to include multiple genes. In Drosophila melanogaster, Sáez et al. (49) defined a selective sweep of ≈41–54 kb near the Sod locus that is similar in chronological age to the sweep at tb1 (but probably far older in generation number). The estimated selection coefficient for this sweep (49) is similar to that estimated for tb1 (7). Although the physical size of the selective sweep near Sod is smaller than we observe at tb1, at least five transcripts are within the core region of low diversity in the Drosophila sweep (49). Thus, the impact of selection in the SOD region can be viewed as more extensive than that observed at tb1. In the malaria parasite Plasmodium falciparum, selective sweeps of >200 kb (50) and ≈100 kb (51) have also been characterized for resistance to antimalarial drugs. In each case, multiple genes are included in the region affected by strong selection (50, 51).
Species differences for the impact of selection on diversity include factors that have been well characterized from theory, such as recombination rate, population size, population structure, and breeding systems. The observation that selection has minimally affected diversity at regions adjacent to tb1 is probably explained in part by a large population size during maize domestication (2) and by a bottleneck of short generation number (39). Furthermore, maize is a wind-pollinated, outcrossing species and therefore has a high effective recombination rate that is also expected to reduce the genomic effect of selection.
LD Pattern in the tb1 Region. The level of LD in genomes has attracted recent interest because it controls the resolution and practicability of association-mapping studies (52). LD is governed largely by recombination and decays with genetic distance. However, many historical, demographic, and population genetic factors also influence LD (53), and increased LD has been used to pinpoint selective sweeps even in the absence of selective signatures detectable by traditional selection tests (54). Tenaillon et al. (25) examined interlocus LD among 21 loci distributed on chromosome 1 (including the tb1 locus) and detected no evidence for substantial LD between sites located at centimorgan distances. To our knowledge our study is the first to examine the pattern of LD among adjacent sites in a large maize sequence contig, and we detected little evidence for significant LD between sites outside the region of the selective sweep upstream to tb1. Our results are consistent with the observation that LD typically decays rapidly within individual maize loci (25, 52), although loci in regions of low recombination (i.e., near centromeres) may have more extensive LD (52).
Although lack of polymorphism limited our analysis of LD for much of the selected region in maize, the high level of LD that we did observe within this region is consistent with the expectation of selection. However, in the maize genome, recombination is thought to occur largely at “hotspots” (55, 56), and recombination in intergenic regions may be several orders of magnitude lower than that observed in transcribed regions (56). Thus, it may be that LD is elevated within the selected region 5′ to tb1 transcribed sequences, in part, because recombination is rare in this portion of the intergenic region between tb1 and gene 3 (this would elevate LD regardless of selection). Consistent with this possibility, LD is high in teosinte for one site within the selected region for which LD is also high in maize. This observation raises the possibility that differences in the frequency of recombination across the tb1 to gene 3 region may have significantly contributed to the observed pattern of LD in maize. If recombination is low within the region of the selective sweep, selection for a single or a small number of causative polymorphisms in maize could account for the extensive region of extremely low diversity 5′ to tb1, even though diversity and LD patterns at sites that immediately flank the selected sweep are consistent with neutral patterns.
Location and Origin of Selected Polymorphisms. Conservation of the 0.4-kb site with sorghum and rice is consistent with a regulatory role for sequences immediately 5′ to the tb1 transcript. However, we detected no evidence for selection at this site (see also ref. 7), even though tb1 regulation appears to have been a domestication target (12, 15). Instead, the core region of selection at tb1 extends 5′ from the 0.4-kb site for as much as ≈90 kb. Although traditional views of gene regulation in plants have posited that regulatory sequences are located within several kilobases of transcript sequences, our data suggest that sequences more distant from the presumed promoter region may also influence tb1 expression. Precedent for this view is provided from studies of the maize b1 gene, where an enhancer required for b1 regulation was localized ≈100 kb 5′ to transcribed sequences (57).
We identified a teosinte haplotype (Par-2) that was identical with the major maize haplotype at each site that we sequenced in the selected region. The extent to which variation at tb1 alleles controls plant architecture in teosinte is not known, and it is possible that haplotypes that confer maize-like tb1 phenotypes predated maize domestication. This view differs from one in which major mutations arising during domestication contributed to cultivation. “Domestication” alleles at BoCAL in cauliflower and fw2.2 in tomato are also present in wild-progenitor populations (3, 58), although the possibility of hybridization between the cultivated varieties and wild populations remains a caution for these studies (3, 58) and ours. It is nevertheless intriguing that the Par-2 sequence is from the subsp. parviglumis population of teosinte that is likely the direct progenitor to maize (59) and that subsp. parviglumis and maize are thought to hybridize infrequently (60).
We also identified a single maize sequence (BOV396) that differs from the major maize clades for at least two sites in the selected region. This finding suggests that several tb1 allele types can confer maize-like morphology. We do caution, however, that because the causative polymorphisms at tb1 are not known, recombination or gene-conversion events could complicate inferences for haplotype genealogies.
Domestication Impact and Crop Improvement. The extent to which genetic diversity has been lost during domestication has profound implications for the management of conventional breeding programs for crop improvement (1) and necessitates a broader understanding of the effect of selection on diversity in crop genomes. The genetic bottleneck during maize domestication is among the mildest observed in a major crop species (2), and our results show that it is possible for selection at a major domestication locus in maize to have little effect on genomic diversity at neighboring genes. Although it is likely that stronger effects of selection on diversity will be observed in maize where selected loci are in regions of low recombination (e.g., centromeric regions; see also ref. 52), it may be that the major effect of selection on the maize genome is limited largely by the number of loci that were directly targeted during domestication. In the next few years, advances in technologies and resources for genome science in maize will enable a systematic assessment of this hypothesis.
Supplementary Material
Acknowledgments
We thank S. Kavchok, S. Young, A. Bronzino, and G. Keizer for technical assistance, Y. Vigouroux and Q. Zhao for advice on experiments, and B. Gaut, A. Rafalski, and K. Bomblies for critical reading of the manuscript. This work was supported by National Science Foundation Grant 9975618 (to J.M.), National Institutes of Health Grant GM-58816 (to J.F.D.), and National Institutes of Health Award F32 GM-65008 (to R.M.C.).
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
Abbreviations: LD, linkage disequilibrium; SNP, single-nucleotide polymorphism; HKA, Hudson–Kreitman–Aguade; BAC, bacterial artificial chromosome.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF464738, AY325816, and AY428163–AY428435).
See accompanying Biography on page 697.
References
- 1.Tanksley, S. D. & McCouch, S. R. (1997) Science 277, 1063–1066. [DOI] [PubMed] [Google Scholar]
- 2.Buckler, E. S., IV, Thornsberry, J. M. & Kresovich, S. (2001) Genet. Res. 77, 213–218. [DOI] [PubMed] [Google Scholar]
- 3.Purugganan, M. D., Boyles, A. L. & Suddith, J. I. (2000) Genetics 155, 855–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frary, A., Nesbitt, T. C., Grandillo, S., Knaap, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K. B. & Tanksley, S. D. (2000) Science 289, 85–88. [DOI] [PubMed] [Google Scholar]
- 5.Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., Beales, J., Fish, L. J., Worland, A. J., Pelica, F., et al. (1999) Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- 6.Olsen, K. M. & Purugganan, M. D. (2002) Genetics 162, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang, R. L., Stec, A., Hey, J., Lukens, L. & Doebley, J. (1999) Nature 398, 236–239. [DOI] [PubMed] [Google Scholar]
- 8.Doebley, J., Stec, A., Wendel, J. & Edwards, M. (1990) Proc. Natl. Acad. Sci. USA 87, 9888–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doebley, J. & Stec, A. (1991) Genetics 129, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doebley, J. & Stec, A. (1993) Genetics 134, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doebley, J., Stec, A. & Gustus, C. (1995) Genetics 141, 333–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doebley, J., Stec, A. & Hubbard, L. (1997) Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- 13.Cubas, P., Lauter, N., Doebley, J. & Coen, E. (1999) Plant J. 18, 215–222. [DOI] [PubMed] [Google Scholar]
- 14.Lukens, L. & Doebley, J. (1999) Genet. Res. 74, 291–302. [Google Scholar]
- 15.Hubbard, L., McSteen, P., Doebley, J. & Hake, S. (2002) Genetics 162, 1927–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yim, Y. S., Davis, G. L., Duru, N. A., Musket, T. A., Linton, E. W., Messing, J. W., McMullen, M. D., Soderlund, C. A., Polacco, M. L., Gardiner, J. M. & Coe, E. H., Jr. (2002) Plant Physiol. 130, 1686–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ewing, B. & Green, P. (1998) Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- 18.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- 19.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195–202. [DOI] [PubMed] [Google Scholar]
- 20.Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P. & Bennetzen, J. L. (2002) Plant Cell 14, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 22.Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., Pachter, L. S. & Dubchak, I. (2000) Bioinformatics 16, 1046–1047. [DOI] [PubMed] [Google Scholar]
- 23.Bray, N., Dubchak, I. & Pachter, L. (2003) Genome Res. 13, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dubchak, I., Brudno, M., Loots, G. G., Pachter, L., Mayor, C., Rubin, E. M. & Frazer, K. A. (2000) Genome Res. 10, 1304–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenaillon, M. I., Sawkins, M. C., Long, A. D., Gaut, R. L., Doebley, J. F. & Gaut, B. S. (2001) Proc. Natl. Acad. Sci. USA 98, 9161–9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- 27.Tiffin, P. & Gaut, B. S. (2001) Genetics 158, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goloubinoff, P., Pääbo, S. & Wilson, A. C. (1993) Proc. Natl. Acad. Sci. USA 90, 1997–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White, S. E. & Doebley, J. F. (1999) Genetics 153, 1455–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fay, J. C. & Wu, C. I. (2000) Genetics 155, 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swofford, D. L. (2003) paup*, Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.
- 32.Fu, H., Park, W., Yan, X., Zheng, Z., Shen, B. & Dooner, H. K. (2001) Proc. Natl. Acad. Sci. USA 98, 8903–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doebley, J. & Stec, A. (1998) Maize Genet. Coop. News Lett. 73, 82–83. [Google Scholar]
- 34.Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., Matsuoka, M. & Ueguchi, C. (2003) Plant J. 33, 513–520. [DOI] [PubMed] [Google Scholar]
- 35.Mathews, S., Spangler, R. E., Mason-Gamer, R. J. & Kellogg, E. A. (2002) Int. J. Plant Sci. 163, 441–450. [Google Scholar]
- 36.Kellogg, E. A. (2001) Plant Physiol. 125, 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watterson, G. A. (1975) Theor. Popul. Biol. 7, 188–193. [DOI] [PubMed] [Google Scholar]
- 38.Hilton, H. & Gaut, B. S. (1998) Genetics 150, 863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eyre-Walker, A., Gaut, R. L., Hilton, H., Feldman, D. L. & Gaut, B. S. (1998) Proc. Natl. Acad. Sci. USA 95, 4441–4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, L., Peek, A. S., Dunams, D. & Gaut, B. S. (2002) Genetics 162, 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson, R. R., Kreitman, M. & Aguade, M. (1987) Genetics 116, 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tajima, F. (1989) Genetics 123, 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu, Y. X. & Li, W. H. (1993) Genetics 133, 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreitman, M. (2000) Annu. Rev. Genomics Hum. Genet. 1, 539–559. [DOI] [PubMed] [Google Scholar]
- 45.Hanson, M. A., Gaut, B. S., Stec, A. O., Fuerstenberg, S. I., Goodman, M. M., Coe, E. H. & Doebley, J. F. (1996) Genetics 143, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song, R., Llaca, V. & Messing, J. (2002) Genome Res. 12, 1549–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song, R. & Messing, J. (2003) Proc. Natl. Acad. Sci. USA 100, 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu, H. & Dooner, H. K. (2002) Proc. Natl. Acad. Sci. USA 99, 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saez, A. G., Tatarenkov, A., Barrio, E., Becerra, N. H. & Ayala, F. J. (2003) Proc. Natl. Acad. Sci. USA 100, 1793–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wootton, J. C., Feng, X., Ferdig, M. T., Cooper, R. A., Mu, J., Baruch, D. I., Magill, A. J. & Su, X. Z. (2002) Nature 418, 320–323. [DOI] [PubMed] [Google Scholar]
- 51.Nair, S., Williams, J. T., Brockman, A., Paiphun, L., Mayxay, M., Newton, P. N., Guthmann, J. P., Smithuis, F. M., Hien, T. T., White, N. J., et al. (2003) Mol. Biol. Evol. 20, 1526–1536. [DOI] [PubMed] [Google Scholar]
- 52.Remington, D. L., Thornsberry, J. M., Matsuoka, Y., Wilson, L. M., Whitt, S. R., Doebley, J., Kresovich, S., Goodman, M. M. & Buckler, E. S., IV (2001) Proc. Natl. Acad. Sci. USA 98, 11479–11484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nordborg, M. & Tavare, S. (2002) Trends Genet. 18, 83–90. [DOI] [PubMed] [Google Scholar]
- 54.Saunders, M. A., Hammer, M. F. & Nachman, M. W. (2002) Genetics 162, 1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, X., Hsia, A. P., Zhang, L., Nikolau, B. J. & Schnable, P. S. (1995) Plant Cell 7, 2151–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fu, H., Zheng, Z. & Dooner, H. K. (2002) Proc. Natl. Acad. Sci. USA 99, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stam, M., Belele, C., Ramakrishna, W., Dorweiler, J. E., Bennetzen, J. L. & Chandler, V. L. (2002) Genetics 162, 917–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nesbitt, T. C. & Tanksley, S. D. (2002) Genetics 162, 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuoka, Y., Vigouroux, Y., Goodman, M. M., Sanchez, G. J., Buckler, E. & Doebley, J. (2002) Proc. Natl. Acad. Sci. USA 99, 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkes, H. G. (1977) Econ. Bot. 31, 254–293. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.