Abstract
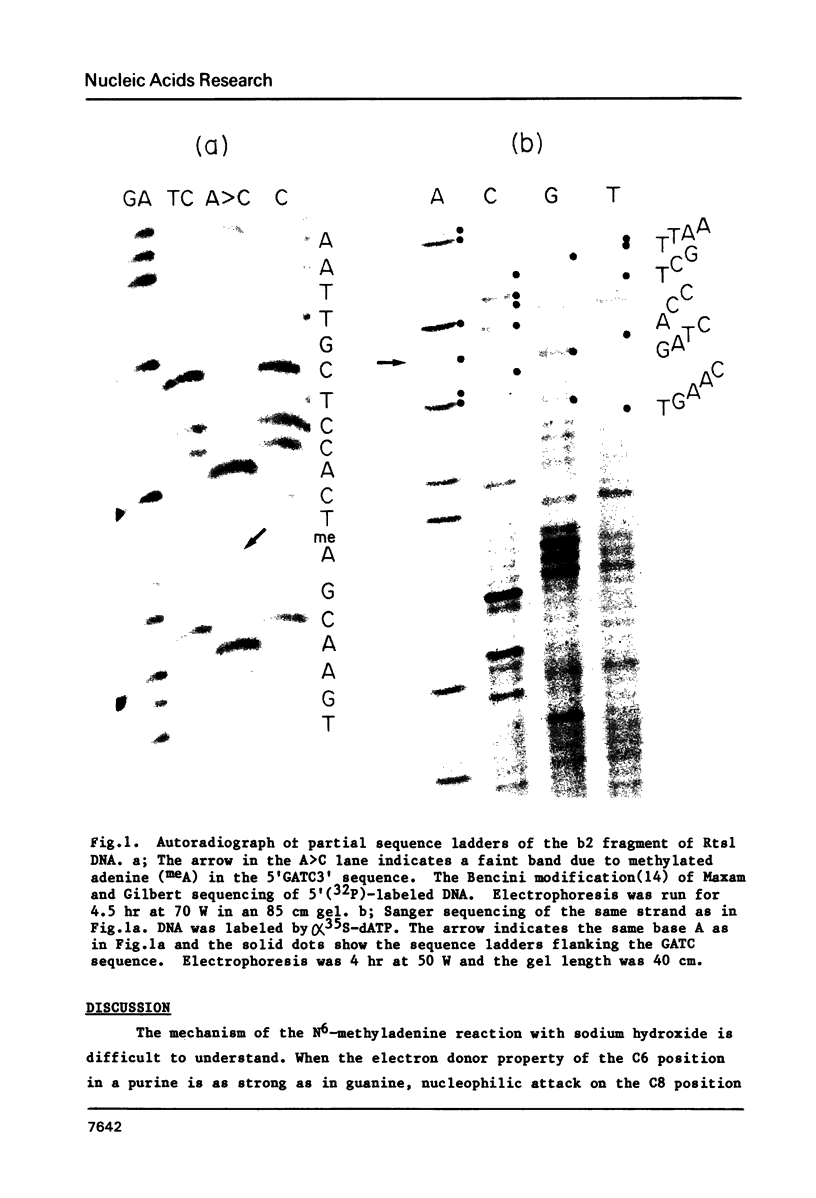
When the Maxam and Gilbert DNA sequencing method which is modified by Bencini et al. (Biotechniques Jan/Feb pp4-5, 1984) is applied to DNA containing methylated adenine in a GATC sequence, the cleavage reaction by sodium hydroxide is found to be greatly reduced in comparison to that of non-methylated adenine. Thus, a faint band in A greater than C reaction suggests a methyl adenine and can be used for its detection. That the faint band corresponds to a methyladenine was confirmed by Sanger sequencing of the same fragment and further by Maxam and Gilbert sequencing of the complementary strand of DNA, which was replicated in an E. coli strain either having or lacking methylation enzymes.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K. A cautionary note on the use of certain restriction endonucleases with methylated substrates. Gene. 1980 Oct;11(1-2):169–171. doi: 10.1016/0378-1119(80)90097-9. [DOI] [PubMed] [Google Scholar]
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garoff H., Ansorge W. Improvements of DNA sequencing gels. Anal Biochem. 1981 Aug;115(2):450–457. doi: 10.1016/0003-2697(81)90031-2. [DOI] [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Lu A. L., Welsh K., Clark S., Su S. S., Modrich P. Repair of DNA base-pair mismatches in extracts of Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:589–596. doi: 10.1101/sqb.1984.049.01.066. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa N., Yoshimoto H., Kaji A. Identification of an Rts1 DNA fragment conferring temperature-dependent instability to vector plasmids. Plasmid. 1985 Mar;13(2):88–98. doi: 10.1016/0147-619x(85)90061-7. [DOI] [PubMed] [Google Scholar]
- Razin A., Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki D. L., Erlanger B. F., Beiser S. M. Immunochemical detection of minor bases in nucleic acids. Science. 1971 Oct 1;174(4004):70–72. doi: 10.1126/science.174.4004.70. [DOI] [PubMed] [Google Scholar]
- Störl H. J., Simon H., Barthelmes H. Immunochemical detection of N6-methyladenine in DNA. Biochim Biophys Acta. 1979 Aug 29;564(1):23–30. doi: 10.1016/0005-2787(79)90184-9. [DOI] [PubMed] [Google Scholar]
- Terawaki Y., Takayasu H., Akiba T. Thermosensitive replication of a kanamycin resistance factor. J Bacteriol. 1967 Sep;94(3):687–690. doi: 10.1128/jb.94.3.687-690.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Dohet C., Jones M., Doutriaux M. P., Hutchinson F., Radman M. Involvement of Escherichia coli mismatch repair in DNA replication and recombination. Cold Spring Harb Symp Quant Biol. 1984;49:611–615. doi: 10.1101/sqb.1984.049.01.069. [DOI] [PubMed] [Google Scholar]
- Yousaf S. I., Carroll A. R., Clarke B. E. A new and improved method for 3'-end labelling DNA using [alpha-32P]ddATP. Gene. 1984 Mar;27(3):309–313. doi: 10.1016/0378-1119(84)90075-1. [DOI] [PubMed] [Google Scholar]