This second part of a four-part series deals with pharmacogenetic variability in drug transport and anticancer phase I drug metabolism, and emphasizes opportunities for patient-tailored pharmacotherapy based on the current knowledge in the field of pharmacogenetics in oncology.
Keywords: Pharmacogenetics, Drug transport, Phase I metabolism, Personalized medicine, Oncology, Anticancer drugs
Learning Objectives
After completing this course, the reader will be able to:
List currently identified candidate genes involved in phase I metabolism that are potential pharmacogenetic markers in anticancer therapy.
Describe the general effect on standard treatment of allelic variants of the candidate genes and the implications for individualized treatment.
This article is available for continuing medical education credit at CME.TheOncologist.com
Abstract
Equivalent drug doses in anticancer chemotherapy may lead to wide interpatient variability in drug response reflected by differences in treatment response or in severity of adverse drug reactions. Differences in the pharmacokinetic (PK) and pharmacodynamic (PD) behavior of a drug contribute to variation in treatment outcome among patients. An important factor responsible for this variability is genetic polymorphism in genes that are involved in PK/PD processes, including drug transporters, phase I and II metabolizing enzymes, and drug targets, and other genes that interfere with drug response. In order to achieve personalized pharmacotherapy, drug dosing and treatment selection based on genotype might help to increase treatment efficacy while reducing unnecessary toxicity.
We present a series of four reviews about pharmacogenetic variability in anticancer drug treatment. This is the second review in the series and is focused on genetic variability in genes encoding drug transporters (ABCB1 and ABCG2) and phase I drug-metabolizing enzymes (CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, DPYD, CDA and BLMH) and their associations with anticancer drug treatment outcome. Based on the literature reviewed, opportunities for patient-tailored anticancer therapy are presented.
Introduction to the Series
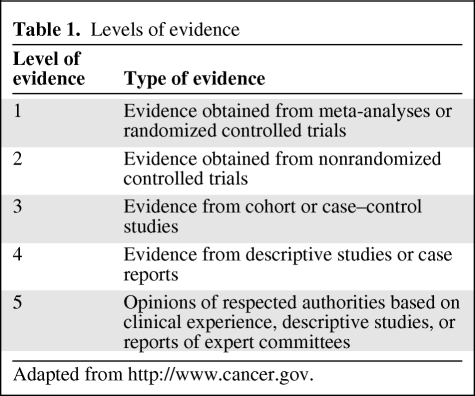
We present a series of four reviews about pharmacogenetic variability in anticancer phase I and II drug metabolism, drug transport, and pharmacodynamic drug effects. The first review focused on the molecular biological background and methodologies and technologies in pharmacogenetic research. This second part in the series deals with pharmacogenetic variability in drug transport and anticancer phase I drug metabolism, and emphasizes opportunities for patient-tailored pharmacotherapy based on the current knowledge in the field of pharmacogenetics in oncology. The level of evidence of the reviewed studies was graded according to the levels reported in Table 1.
Table 1.
Levels of evidence
Adapted from http://www.cancer.gov.
Drug Transport by ATP-Binding Cassette Transporters
The ATP-binding cassette (ABC) transporters are a family of transmembrane proteins that use ATP-derived energy to actively transport a variety of substrates across cell membranes. Thereby, they are heavily involved in the absorption and disposition of many clinically used drugs, including anticancer drugs. Based on the sequence homology of ABC transporters, seven subfamilies (ABCA to ABCG) are distinguished, two of which—ABCB1 (P-glycoprotein [P-gp]) and ABCG2 (breast cancer resistance protein [BCRP])—are discussed.
P-gp (ABCB1)
P-gp (ABCB1) is expressed in the intestine, liver, kidney, brain, and placenta, with highly varying expression levels among individuals [1–3]. The substrate affinity of P-gp is broad, and many anticancer drugs are transported by P-gp, including etoposide, teniposide, doxorubicin, vinblastine, vincristine, daunorubicin, irinotecan, paclitaxel, and docetaxel [4].
The gene encoding P-gp is ABCB1, which contains various functional polymorphisms that range in allele frequency among various ethnicities [4–8]. A widely investigated single nucleotide polymorphism (SNP) in ABCB1 is 3435C>T (Ile1145Ile; ABCB1*6), which is in strong linkage disequilibrium with another silent SNP, 1236C>T (Gly412Gly; ABCB1*8) and the triallelic variant 2677G>T/A (Ala893Ser/Thr) [7, 9]. The combination of these three SNPs (i.e., haplotype) is also designated as P-gp*2 [7]. There is debate about the functional effect of 3435C>T. Some studies reported that this SNP affects mRNA stability and results in lower mRNA expression and thereby lower protein levels [5, 10–12], whereas others reported higher expression levels and enhanced activity of P-gp [7, 13, 14].
With regard to ABCB1 polymorphism and irinotecan treatment outcome, the homozygous P-gp*2 variant haplotype was shown to be associated with lower renal clearance of irinotecan and its active metabolite SN-38 [8] and showed a lower area under the plasma concentration–time curve (AUC) of SN-38 glucuronide in 2677TT/3435TT individuals than in wild-type patients [15]. Furthermore, 3435TT was significantly associated with grade 3 diarrhea in 107 patients with non-small cell lung cancer (NSCLC) given irinotecan and cisplatin [15].
Besides irinotecan, taxanes are also substrates for P-gp. In 62 patients with NSCLC treated with docetaxel and cisplatin, 3435TT allele carriers also more frequently (33%) experienced grade ≥2 diarrhea than heterozygous (4%) and wild-type (11%) patients [16]. The pharmacogenetic analysis from the Scottish Randomised Trial in Ovarian Cancer 1 (SCOTROC1) trial, however, did not demonstrate a relationship between genetic polymorphism in ABCB1 and toxicity or treatment outcome in 914 patients with ovarian cancer who had received either docetaxel or paclitaxel combined with carboplatin [17].
Polymorphism in ABCB1 has also been investigated in patients with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL). In the treatment of childhood ALL according to Berlin-Frankfurt-Münster protocols, a matched case–control study in white patients showed a lower rate of central nervous system relapse for 3435C>T variant allele carriers than for wild-type patients [18]. Similarly, in 405 white AML patients receiving etoposide, mitoxantrone, or daunorubicin, a significantly shorter overall survival duration and higher probability of relapse were observed in 3435C>T wild-type patients than in hetero- or homozygous patients [14]. In contrast, a smaller study in Asian patients with AML reported a higher response rate and 3-year event-free survival rate for patients with the wild-type genotype [19].
In conclusion, polymorphisms in ABCB1 have been shown to possibly affect treatment outcome with chemotherapy, especially irinotecan. However, some of the observed associations with clinical outcome for other anticancer drugs were not always consistent. This might result from differences in ethnicity, population size, and type of treatment regimen in the various populations that have been studied. For this reason, genetic polymorphism in ABCB1 is currently not suitable yet for patient-tailored anticancer therapy. The study results obtained, however, should encourage the conduction of additional pharmacogenetic studies. Given the highly polymorphic character of ABCB1 in differing among ethnicities, a haplotype analysis that includes additional genetic variants in ABCB1 besides the above-mentioned SNPs might help to better predict treatment outcome with P-gp (anticancer) drug substrates.
BCRP (ABCG2)
One of the most important ABC transporters of the ABCG family is ABCG2, also known as BCRP. ABCG2 is highly expressed in the gastrointestinal tract, liver, kidney, brain, heart, and placenta [20]. Anticancer drugs that are known substrates for ABCG2 include, among others, mitoxantrone, methotrexate, SN-38, topotecan, imatinib, and gefitinib, but as for P-gp, substrate affinity of ABCG2 is very broad and it transports many other drugs as well [21].
Multiple polymorphisms in ABCG2 have been identified that may modulate the functional activity of ABCG2 [22–24]. Particularly relevant SNPs in ABCG2 appear to be 421C>A (Gln141Lys) and the nonsense SNP 376C>T (Gln126stop). Until now, the nonsense SNP 376C>T has only been identified in Japanese individuals [25–27]. The allele frequency of 421C>A is also higher in Japanese than in white subjects (30% versus 10%). 421C>A has been reported to affect the translation efficiency of ABCG2 and to result in lower ABCG2 (placental) protein expression [25, 26]. Indeed, additional in vitro research showed greater drug accumulation and less drug resistance for patients with the 421C>A polymorphism [27–29]. However, in white [30] and Asian [31] patients treated with irinotecan, 421C>A did not significantly affect the pharmacokinetics of irinotecan or its metabolites, although one of two homozygous mutated allele carriers showed extensive accumulation of SN-38 and SN-38 glucuronide [30].
The clinical effect of 421C>A has also been investigated in patients treated with the tyrosine kinase inhibitors imatinib and gefitinib. One study in 82 patients with gastrointestinal tumors treated with imatinib showed no significant pharmacokinetic effect [32], whereas another study in 67 patients did show a 22% lower clearance of imatinib in 421C>A heterozygous patients [33]. Likewise, in gefitinib-treated patients, 421C>A was associated with a higher accumulation of gefitinib [34] and with grade 1 or 2 diarrhea [35]. However, in that study, the majority of heterozygous patients did not develop any diarrhea, and the single homozygous patient had no noticeable toxicity. Moreover, this association was not confirmed in a similar, but Asian, study population [36].
Overall, despite preclinical evidence that 421C>A functionally impairs ABCG2 activity, a significant association with toxicity was only observed in white patients treated with gefitinib. With other anticancer drugs, the clinical relevance of 421C>A in ABCG2 appears to be thus far of limited importance. Additional trials among various geographic populations are awaited to evaluate the exact clinical relevance of polymorphisms and haplotypes of ABCG2, especially in patients treated with gefitinib.
Phase I Anticancer Drug Metabolism
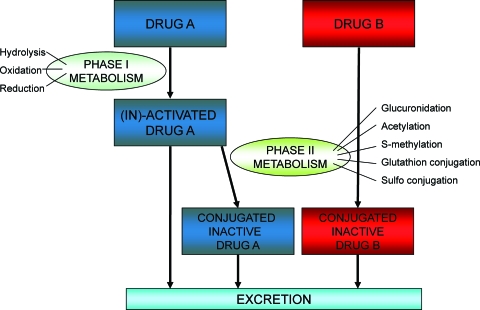
Phase I drug-metabolizing enzymes mediate drug oxidation, reduction, and hydrolysis reactions, by which drugs may be activated or inactivated (Fig. 1). In addition, phase I metabolism generally increases the polarity of a drug, and thereby facilitates excretion from the body. Phase I reactions may be followed by phase II reactions to further increase solubility; however, preceding phase I reactions are not a prerequisite. Typical phase II reactions are glucuronidation, acetylation, S-methylation, and glutathione- or sulfo-conjugation of drugs. Genetic polymorphism in phase I metabolism may modulate the pharmacokinetics and disposition of drugs and thereby affect the toxicity and efficacy of treatment, and is discussed in the following sections.
Figure 1.
Phase I and phase II drug metabolism. Phase I drug-metabolizing enzymes mediate drug oxidation, reduction, or hydrolysis reactions, by which drugs may be activated or inactivated. This may be followed by phase II reactions to further increase solubility and thereby facilitate excretion from the body. Preceding phase I reactions are not a prerequisite.
Oxidizing Phase I Metabolizing Enzymes
The cytochrome P450 (CYP450) system is involved in oxidation reactions. The CYP450 genes particularly involved in anticancer therapy are CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5.
CYP2B6
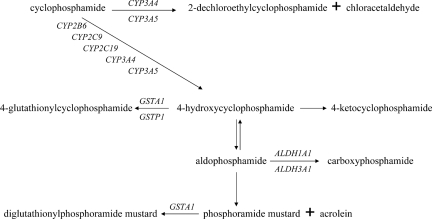
Cyclophosphamide and ifosfamide undergo extensive metabolism by CYP450. CYP2A6, CYP2B6, CYP2C8, CYP2C9, CYP2C19, and CYP3A4 are involved in the activation as well as inactivation of cyclophosphamide and ifosfamide (Fig. 2). CYP2B6 activates cyclophosphamide to 4-hydroxycyclophosphamide, whereas CYP2B6 inactivates the CYP3A4-derived hydroxylated active form of ifosfamide, 4-hydroxy-ifosfamide [37, 38]. In addition, thiotepa is a minor substrate for, but acts also as an inhibitor of, CYP2B6 [39, 40].
Figure 2.
Biotransformation of cyclophosphamide. The biotransformation of cyclophosphamide involves multiple drug-metabolizing enzymes that are subject to genetic polymorphism, which in turn may affect the disposition of cyclophosphamide and its metabolites. However, because of the fact that its metabolism is regulated by several phase I and phase II enzymes, a genetic defect in a single gene might go unnoticed because other metabolic enzymes may serve as escape metabolic routes.
Multiple functional polymorphisms in CYP2B6 exist [41–45]. A commonly occurring genetic variant is CYP2B6*6, which is compromised of two SNPs, 516G>T (Q172H) and 785A>G (K262R). In vitro investigations on the functional effect of CYP2B6*6 showed inconsistent findings—on the one hand greater enzyme activities were reported [46, 47], but on the other hand, lower enzyme activities was reported as well [41, 48, 49]. This inconsistency in study results is possibly an effect of other (still unknown) mutations linked to these SNPs, creating various haplotypes with different enzyme activities.
The relationship between CYP2B6*6 and the pharmacokinetics of cyclophosphamide was investigated in several studies. Greater CYP2B6-mediated activation of cyclophosphamide to 4-hydroxycyclophosphamide for 516G>T variant allele carriers has been observed [50, 51], as well as a higher clearance and shorter half-life of cyclophosphamide for CYP2B6*6 homozygous mutant patients than for wild-type patients [52]. These findings, however, could not be confirmed by others in a cohort of 124 patients with solid tumors [53]. Despite the fact that a few studies reported a significant pharmacokinetic effect of cyclophosphamide by CYP2B6*6, no significant associations with adverse events [54, 55], disease-free survival or overall survival [56] were observed in cancer patients treated with cyclophosphamide combination chemotherapy. Therefore, the clinical relevance of CYP2B6*6 appears to be limited in cyclophosphamide treatment. However, because cyclophosphamide is a substrate for several subfamilies of the CYP450 system (Fig. 2), an effect of a genetic defect in a single gene might go unnoticed because other metabolic enzymes may serve as escape metabolic routes. A combined analysis that would include multiple genes involved in the pharmacological pathway could possibly help to clarify the broad range in drug response for compounds that are substrates for multiple metabolizing enzymes.
CYP2C8
CYP2C8 is an important inactivating enzyme of the taxane paclitaxel [57]. Several polymorphisms have been identified, such as CYP2C8*2 (805A>T, Ile269Phe), CYP2C8*3 (416G>A, Arg139Lys and 1196A>G, Lys399Arg), and CYP2C8*4 (792C>G, Ile264Met) [58–60].
Although in vitro results showed a lower metabolism of paclitaxel by up to 15% for CYP2C8*3 [59, 60] carriers, no effect of CYP2C8 genetic polymorphism on the clearance of unbound paclitaxel was observed in patients treated with paclitaxel [61, 62]. Moreover, a study in 914 patients receiving either docetaxel or paclitaxel combined with carboplatin showed that CYP2C8 polymorphisms were not associated with toxicity or efficacy of treatment [17].
In conclusion, polymorphisms in CYP2C8 have thus far not been demonstrated to affect paclitaxel treatment outcome and are therefore not yet suitable for patient-tailored therapy with paclitaxel.
CYP2C9
CYP2C9 metabolizes, among others, the anticancer agents cyclophosphamide, etoposide, ifosfamide, and tamoxifen, and the experimental anticancer drug indisulam (E7070). CYP2C9 harbors many allelic variants, of which at least two SNPs, CYP2C9*2 (430C>T, Arg144Cys) and CYP2C9*3 (1075A>C, Ile359Leu), are known to decrease CYP2C9 enzyme activity [63–65]. Despite these significant in vitro observations, four recent studies in patients with cancer did not demonstrate a significant effect of CYP2C9 polymorphism on the pharmacokinetics of cyclophosphamide [51, 53, 66] or tamoxifen [67]. In addition, no relationship between CYP2C9 genotype and survival was observed in patients with breast cancer treated with tamoxifen [68, 69]. However, a study in 67 patients treated with the experimental anticancer drug indisulam revealed a lower elimination rate of 27% and a significantly higher risk for severe neutropenia in heterozygous CYP2C9*3 carriers [70].
To conclude, allelic variants of CYP2C9 do not appear to affect treatment outcome with cyclophosphamide or tamoxifen, but possibly do affect indisulam treatment outcome. This suggests that a substrate-specific pharmacogenetic effect might be present. Further studies are awaited to draw definite conclusions.
CYP2C19
Besides cyclophosphamide, ifosfamide, and tamoxifen, thalidomide is also a substrate for CYP2C19 and is activated by CYP2C19-mediated hydroxylation [71]. There are two SNPs in CYP2C19 that lead to the poor metabolizer phenotype. These are 681G>A (CYP2C19*2), which results in a splicing defect, and 636G>A (CYP2C19*3), which introduces a premature stop codon. Both allelic variants have no residual activity left, and approximately 99% of the CYP2C19 poor metabolizer phenotype is explained by these two SNPs [72]. Thus far, only one study investigated CYP2C19 polymorphism in relationship to response to treatment with thalidomide. In 92 patients with multiple myeloma treated with thalidomide, extensive metabolizers experienced a significantly higher response rate (63%) than CYP2C19*2-induced poor metabolizers (33%) [73]. Further studies are awaited.
With regard to cyclophosphamide and CYP2C19 activity, poor metabolizers are theoretically expected to have a poor response and low toxicity probability upon therapy with cyclophosphamide, because its CYP2C19-mediated activation is eliminated. Indeed, one study in 60 white cancer patients showed a CYP2C19*2-dependent lower clearance of cyclophosphamide at doses <1,000 mg/m2 [66]; however, no effect on the pharmacokinetics of cyclophosphamide for CYP2C19*2 and CYP2C19*3 was observed in two larger trials conducted in Japanese [52] and European [53] patients, and no relationship with clinical outcome was reported [54].
In summary, CYP2C19*2 and CYP2C19*3 result in a CYP2C19 poor metabolizer phenotype. Their clinical relevance appears limited in cyclophosphamide treatment, but not in thalidomide treatment. Additional investigation is required before definitive conclusions can be drawn.
CYP2D6
The enzyme CYP2D6 is particularly important in the treatment of breast cancer patients with tamoxifen. CYP2D6 oxidizes tamoxifen to 4-hydroxytamoxifen, the antiestrogen potency of which is 50 times higher than that of tamoxifen itself [74]. Furthermore, the conversion of N-desmethyltamoxifen to endoxifen is primarily mediated by CYP2D6. The potency of endoxifen is also higher than that of tamoxifen, and comparable with the binding affinity and suppression of estradiol-stimulated cell proliferation of 4-hydroxy-tamoxifen [75]. Thus, theoretically, CYP2D6 poor metabolizers are expected to benefit less from therapy with tamoxifen because of a lower rate of formation of the active substrate.
CYP2D6 is highly polymorphic. Multiple allelic variants have been described, of which, some result in lower, or even absent, enzyme activity [76]. Furthermore, copy number variants of CYP2D6 exist with either two, three, four, five, or 13 gene copies, which consequently lead to the ultrarapid metabolizer phenotype. The most abundant and functionally important SNPs are CYP2D6*4 (1846G>A), resulting in a splicing defect, CYP2D6*5, characterized by complete CYP2D6 gene deletion, CYP2D6*6 (1707delT), resulting in a frameshift at amino acid 118, and CYP2D6*10 (100C>T), which markedly reduces enzyme activity [77–80].
A few studies in patients with breast cancer treated with tamoxifen showed that plasma levels of endoxifen are lower in CYP2D6 poor metabolizers than in extensive metabolizers. Besides genetic variants, potent inhibitors of CYP2D6, such as paroxetine or fluoxetine, also led to lower levels of endoxifen [67, 81, 82]. Moreover, several retrospective clinical trials demonstrated a shorter time to recurrence or shorter survival time for women with the poor metabolizer phenotype [68, 83–89]; however, this could not be confirmed in other retrospective studies [90–93]. Prospective evaluations are currently lacking. Plausible explanations for inconsistent findings among the various studies are, among other things, the retrospective study design; incomplete CYP2D6 genotyping; a lack of stratification for coadministration of no, weak, or strong CYP2D6 inhibitors; the inability to account for drug compliance; and differences in patient selection, duration of treatment, and dose of tamoxifen. In addition, some studies analyzed tumor DNA whereas others used germline DNA. Notwithstanding, the concordance rate between tumor and germline DNA for CYP2D6 appears to be 100% [88, 89].
In conclusion, poor metabolizers as a result of genetic defects appear to benefit less from treatment with tamoxifen, though inconsistent findings have been reported. Treatment with tamoxifen is also negatively affected by simultaneous use of potent CYP2D6 inhibitors. Well-defined prospective trials are needed, with complete CYP2D6 genotyping, that are supported by pharmacokinetic analyses. These trials should additionally differentiate the strengths of coadministered inhibitors of CYP2D6 for tamoxifen [94], to establish the exact role of CYP2D6 polymorphism in tamoxifen treatment.
CYP3A4 and CYP3A5
The CYP3A subfamily is highly expressed in the liver and small intestine, and metabolizes >50% of clinically used drugs, including several anticancer drugs such as etoposide, teniposide, docetaxel, paclitaxel, irinotecan, toremifene, vinblastine, vincristine, vinorelbine, cyclophosphamide, ifosfamide, thiotepa, gefitinib, and erlotinib [95–97]. Enzyme activity of CYP3A ranges widely among subjects, and besides genetic polymorphism, its activity is largely affected by nongenetic factors such as age, endogenous hormone levels, transcription factor activity, health status, and environmental stimuli [98, 99].
To date, approximately 40 allelic variants have been described for CYP3A4, of which some reduce its activity, such as CYP3A4*6, CYP3A4*8, and CYP3A4*17 [100]. In addition, a common SNP, −392A>G (CYP3A4*1B, CYP3A4-V), appears to influence CYP3A4 expression as a result of altered nuclear protein binding affinity to the polymorphic element [101]. In CYP3A5, the main SNP of interest is 6986G>A (CYP3A5*3), which leads to a splicing defect that results in severely lower enzyme activity. Most white people are homozygous for this genetic defect and consequently live with a CYP3A5 deficiency [102, 103].
Docetaxel is metabolized by CYP3A4 and CYP3A5 up to 93% [104]. Therefore, variability in CYP3A enzyme activity is hypothesized to affect the metabolism of docetaxel and hence its toxicity and possibly efficacy.
Although two studies showed a higher clearance of docetaxel for the CYP3A4*1B variant allele in patients treated with docetaxel [105, 106], this was not observed by others [107]. For paclitaxel, a taxane as well, no associations were observed with CYP3A genotype and treatment outcome [17, 61, 108].
In treatment with cyclophosphamide-based chemotherapy, controversial results have been reported with regard to CYP3A4*1B and treatment outcome. Two studies reported a shorter (disease-free) survival time for variant allele carriers [56, 109], whereas this could not be confirmed by others [53, 54].
With regard to other anticancer drugs, in one study in 42 patients with advanced NSCLC treated with irinotecan and carboplatin, CYP3A4*1B was not associated with toxicity [110]; however, a nonsignificant association with skin rash grade ≥2 for CYP3A4*1B and CYP3A5*3 was observed in a prospective study in 80 cancer patients receiving erlotinib monotherapy [111].
Obviously, further research is warranted. It can be concluded though that genetic variability in CYP3A alone is insufficient to explain its widely ranging enzyme activity [112]. Possibly, CYP3A4 phenotypic approaches, although often more costly, might serve as better predictors of treatment outcome.
Additional Oxidizing Phase I Metabolizing Enzymes
Other typical phase I oxidation enzymes are monoamine oxidase (MAO), cyclooxygenase (COX), alcohol dehydrogenase (ADH), and aldehyde dehydrogenase (ALDH). The enzymes MAO and COX are not involved in the biotransformation of anticancer drugs, but there is an increasing interest in COX inhibitors in the prevention and treatment of cancer [113–115]. In addition, polymorphisms in ADH have been associated with a higher risk for developing cancer, especially in high alcohol consumers [116, 117].
ALDH oxidizes acetaldehyde (a metabolite of alcohol) and also oxidizes cyclophosphamide and ifosfamide. A study in 124 white patients treated with high-dose chemotherapy showed that two polymorphisms in ALDH (ALDH1A1*2 and ALDH3A1*2) did not affect the pharmacokinetics of cyclophosphamide; however, a significantly higher risk for liver toxicity and hemorrhagic cystitis was observed [53, 54]. Notwithstanding, this association was not observed in Asian patients [55]. Besides differences in ethnicity, this discrepancy might also be a result of differences in patient selection or treatment regimen, and therefore additional studies are warranted before definitive conclusions can be drawn.
Reducing, Hydrolyzing, and Deaminating Phase I Metabolizing Enzymes
Dihydropyrimidine dehydrogenase (DPD) is a phase I reduction enzyme and a key detoxification enzyme of fluoropyrimidines. Other inactivating enzymes of anticancer drugs are cytidine deaminase (CDA) for gemcitabine and cytarabine and bleomycin hydrolase (BLMH) for bleomycin.
DPD
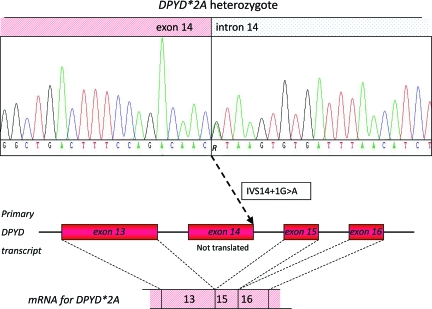
The primary step in the 5-fluorouracil (5-FU) degradation pathway is mediated by DPD [118, 119]. Furthermore, DPD also inactivates the 5-FU oral prodrugs capecitabine and tegafur. About 3%–5% of the population has a (partial) DPD deficiency, which increases the risk for 5-FU–induced severe toxicity in these individuals [120]. Currently, >50 polymorphisms in DPYD, the gene encoding DPD, have been identified [121]. The most predominant polymorphism associated with DPD deficiency is IVS14+1G>A (DPYD*2A). This SNP results in complete skipping of exon 14 during pre-mRNA splicing, and consequently creates a truncated protein that has no residual activity left (Fig. 3) [122–124]. Another polymorphism in DPYD that negatively affects DPD enzyme activity, mainly by interfering with cofactor binding, is 2846A>T (Asp949Val) [125–127].
Figure 3.
Functional effect of DPYD*2A (IVS14+1G>A). The polymorphism DPYD IVS14+1G>A is a single nucleotide polymorphism that is located at the first position of intron 14. This polymorphism results in complete skipping of exon 14 during the process of pre-mRNA splicing, which thereby creates a truncated protein with absent dihydropyrimidine dehydrogenase activity.
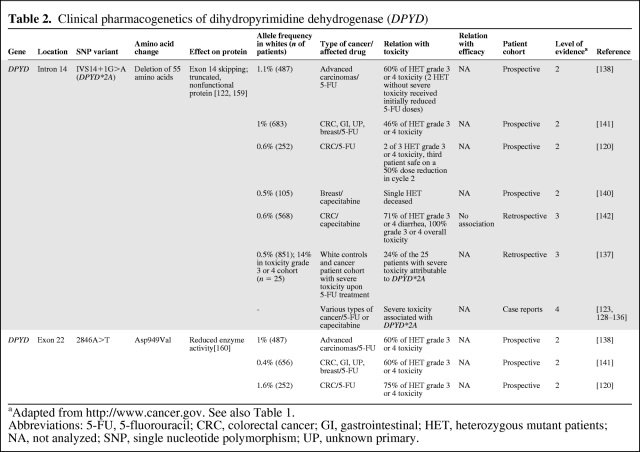
Table 2 provides an overview of various clinical studies that investigated the effect of DPYD*2A and 2846A>T on treatment outcome with fluoropyrimidines. Multiple case reports have been described, reporting on patients with severe, even lethal, toxicity following 5-FU–based chemotherapy who proved to be polymorphic for DPYD*2A [123, 128–136]. In addition, others showed that DPYD*2A was present in approximately 25% of patients presenting with severe toxicity following treatment with fluoropyrimidines [135, 137]. Moreover, several retro- and prospective population trials consisting of hundreds of patients per trial showed that, on average, >70% of all patients polymorphic for DPYD*2A developed severe, including lethal, toxicity following treatment with 5-FU or capecitabine [120, 138–143].
Table 2.
Clinical pharmacogenetics of dihydropyrimidine dehydrogenase (DPYD)
aAdapted from http://www.cancer.gov. See also Table 1.
Abbreviations: 5-FU, 5-fluorouracil; CRC, colorectal cancer; GI, gastrointestinal; HET, heterozygous mutant patients; NA, not analyzed; SNP, single nucleotide polymorphism; UP, unknown primary.
Similarly, the polymorphism 2846A>T in DPYD is also associated with severe toxicity to fluoropyrimidines, as demonstrated by multiple cohort studies [120, 138, 141, 142]. Though DPYD 2846A>T is slightly less predictive of severe toxicity than DPYD*2A, the majority of patients polymorphic for 2846A>T still develop severe toxicity following 5-FU–based treatment. Moreover, the simultaneous presence of both variant alleles (DPYD*2A and 2846A>T) in an individual, a rarely (<1 in 1,000 patients) occurring phenomenon, however, was shown to be lethal in multiple cases shortly after the start of fluoropyrimidine treatment [120, 129].
In conclusion, these data demonstrate the clinical significance of DPYD*2A and 2846A>T in fluoropyrimidine treatment, suggesting prospective screening prior to the start of therapy to avoid severe toxicity in patients with the variant genotype. Possibly, initial fluoropyrimidine dose reductions of 50% in DPYD*2A and 25% in 2846A>T heterozygous patients followed by further dose titration upon clinical tolerability could be a safe and effective strategy [142] that needs to be assessed in additional, prospective clinical trials.
CDA
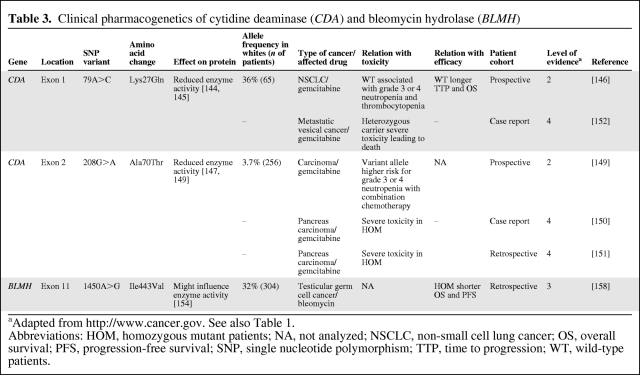
The enzyme CDA inactivates gemcitabine to 2′,2′-difluorodeoxyuridine and inactivates cytarabine as well. Two nonsynonymous SNPs in CDA, 79A>C (Lys27Gln) and 208G>A (Ala70Thr) were shown to reduce CDA enzyme activity [144–147]. The 208G>A SNP, however, is likely to occur only in Japanese and Korean subjects, and has not yet been detected in African, white, and Chinese Americans [148]. A few studies evaluated the predictive value of these SNPs in gemcitabine treatment (Table 3). A study in 256 Japanese patients treated with gemcitabine-based chemotherapy showed a higher AUC and maximum concentration of gemcitabine in patients heterozygous polymorphic for 208G>A. In addition, 208G>A was associated with grade ≥3 neutropenia in patients who were coadministered 5-FU and a platinum analog [149]. Furthermore, homozygosity for this SNP in Japanese patients has been associated with severe toxicity to gemcitabine [150, 151].
Table 3.
Clinical pharmacogenetics of cytidine deaminase (CDA) and bleomycin hydrolase (BLMH)
aAdapted from http://www.cancer.gov. See also Table 1.
Abbreviations: HOM, homozygous mutant patients; NA, not analyzed; NSCLC, non-small cell lung cancer; OS, overall survival; PFS, progression-free survival; SNP, single nucleotide polymorphism; TTP, time to progression; WT, wild-type patients.
For 79A>C, one case report of a patient with lethal toxicity following treatment with gemcitabine was described, who proved to be heterozygous polymorphic for 79A>C but wild-type for 208G>A. Additional phenotyping in that patient showed a 75% lower CDA enzyme activity than in nontoxic controls [152]. However, it appears unlikely that 79A>C alone caused CDA deficiency in that patient because no effect on the pharmacokinetics of gemcitabine for 79A>C has been observed in Japanese [149] and white [153] patients. Moreover, a study in 65 chemotherapy-naïve NSCLC patients treated with gemcitabine and cisplatin showed that wild-type 79A>C patients more frequently experienced grade ≥3 neutropenia and thrombocytopenia and had a longer time to progression and overall survival time as well [146].
In summary, inconsistent findings have been reported for 79A>C in CDA, showing positive and negative associations with clinical outcome with gemcitabine. This might be partly a result of differences in patient selection, treatment regimen, and ethnicity, but as yet undetected polymorphisms might also possibly play a role. However, for CDA 208G>A, clear associations with severe toxicity from gemcitabine have been shown in Japanese patients. Caution and possibly initial dose reductions of gemcitabine for at least homozygous 208G>A carriers appear indicated. CDA 208G>A has the potential to become a predictive marker in gemcitabine treatment in Japanese patients, and this requires additional studies for independent confirmation.
BLMH
BLMH is the primary enzyme in the inactivation of bleomycin. The enzymatic activity of BLMH is, among other things, regulated by its C-terminal region [154–156]. A SNP that is located in this C-terminal region, 1450A>G, was shown in vitro to affect bleomycin-induced chromatid breaks per cell [157]. Moreover, a retrospective study in patients with testicular germ cell cancer treated with bleomycin showed shorter progression-free and overall survival times for homozygous 1450A>G variant allele carriers [158]. To determine whether this SNP is of clinical relevance, further (pre-)clinical studies on the functional effect of BLMH 1450A>G and on its effect on the clinical pharmacokinetics, toxicity, and efficacy of bleomycin are required.
Conclusion: Implications for Clinical Practice—Opportunities for Patient-Tailored Anticancer Therapy
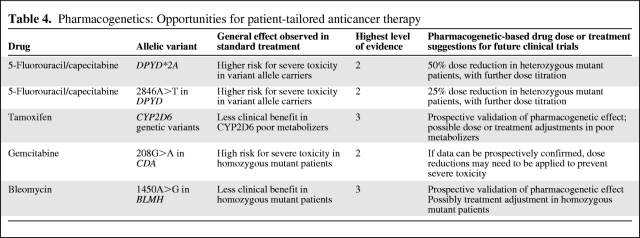
Based on the literature reviewed, genetic polymorphisms in at least four candidate genes involved in phase I metabolism could potentially serve as pharmacogenetic markers in anticancer therapy to enable more safe, and possibly more effective, anticancer pharmacotherapy. These are DPYD*2A and 2846A>T in DPYD in fluoropyrimidine treatment, CYP2D6 polymorphism in breast cancer patients receiving tamoxifen, CDA 208G>A (which, however, appears to only occur in Asians) in gemcitabine treatment, and possibly 1450A>G in BLMH in patients treated with bleomycin (Table 4).
Table 4.
Pharmacogenetics: Opportunities for patient-tailored anticancer therapy
As determined in several studies, DPYD*2A and 2846A>T in DPYD consistently showed significant relationships with severe, possibly lethal, toxicity following treatment with standard-dose fluoropyrimidines, with a level of evidence of 2. Initial dose reductions ≥50% in DPYD*2A and ≥25% in 2846A>T heterozygous polymorphic patients, both followed by further dose titration upon clinical tolerability, are recommended.
Despite some inconsistent findings, genetic polymorphism in CYP2D6 appears to negatively affect survival in the treatment of breast cancer with tamoxifen, because of a lower rate of formation of active metabolites of tamoxifen in CYP2D6 poor metabolizers. Whether this genetic subgroup of patients should be given higher doses of tamoxifen or another type of treatment, such as, for example, aromatase inhibitors, is currently unknown. Additional, prospective studies, preferentially supported by pharmacokinetic analyses, will help to address these important questions.
With gemcitabine treatment, CDA 208G>A homozygous patients, in particular, but also CDA 208G>A heterozygous patients, appear to be predisposed for severe gemcitabine toxicity. If this finding can be independently confirmed by additional, prospective studies, the question arises of whether or not severe gemcitabine toxicity is preventable by initial dose reductions in at least CDA 208G>A homozygous variant allele carriers without negatively affecting treatment response.
For bleomycin, a single retrospective study in patients with testicular germ cell cancer treated with bleomycin-based chemotherapy reported that patients homozygous polymorphic for 1450A>G in BLMH experienced shorter overall and progression-free survival times (level of evidence, 3). Prospective studies should evaluate whether these findings can be confirmed. If so, the question evolves of whether or not this genetically defined subgroup of patients would benefit more from another type of chemotherapeutic regimen that does not include bleomycin.
Overall, genetic polymorphism in candidate genes involved in phase I metabolism has been shown to potentially affect the pharmacokinetics of anticancer drugs, and the toxicity and efficacy of treatment. A few selected candidate polymorphisms are, or at least have the potential to become, predictive markers for anticancer treatment outcome. These results should encourage the continuation of pharmacogenetic research in anticancer therapy, in an effort to implement personalized medicine in daily clinical practice.
Author Contributions
Conception/Design: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
Collection and/or assembly of data: Maarten J. Deenen
Data analysis and interpretation: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
Manuscript writing: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
Final approval of manuscript: Maarten J. Deenen, Annemieke Cats, Jos H. Beijnen, Jan H.M. Schellens
References
- 1.Thiebaut F, Tsuruo T, Hamada H, et al. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci U S A. 1987;84:7735–7738. doi: 10.1073/pnas.84.21.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cordon-Cardo C, O'Brien JP, Casals D, et al. Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proc Natl Acad Sci U S A. 1989;86:695–698. doi: 10.1073/pnas.86.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuetz EG, Furuya KN, Schuetz JD. Interindividual variation in expression of P-glycoprotein in normal human liver and secondary hepatic neoplasms. J Pharmacol Exp Ther. 1995;275:1011–1018. [PubMed] [Google Scholar]
- 4.Schwab M, Eichelbaum M, Fromm MF. Genetic polymorphisms of the human MDR1 drug transporter. Annu Rev Pharmacol Toxicol. 2003;43:285–307. doi: 10.1146/annurev.pharmtox.43.100901.140233. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascorbi I, Gerloff T, Johne A, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69:169–174. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 7.Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70:189–199. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- 8.Sai K, Kaniwa N, Itoda M, et al. Haplotype analysis of ABCB1/MDR1 blocks in a Japanese population reveals genotype-dependent renal clearance of irinotecan. Pharmacogenetics. 2003;13:741–757. doi: 10.1097/00008571-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Kroetz DL, Pauli-Magnus C, Hodges LM, et al. Sequence diversity and haplotype structure in the human ABCB1 (MDR1, multidrug resistance transporter) gene. Pharmacogenetics. 2003;13:481–494. doi: 10.1097/00008571-200308000-00006. [DOI] [PubMed] [Google Scholar]
- 10.Siegsmund M, Brinkmann U, Schaffeler E, et al. Association of the P-glycoprotein transporter MDR1(C3435T) polymorphism with the susceptibility to renal epithelial tumors. J Am Soc Nephrol. 2002;13:1847–1854. doi: 10.1097/01.asn.0000019412.87412.bc. [DOI] [PubMed] [Google Scholar]
- 11.Hitzl M, Drescher S, van der Kuip H, et al. The C3435T mutation in the human MDR1 gene is associated with altered efflux of the P-glycoprotein substrate rhodamine 123 from CD56+ natural killer cells. Pharmacogenetics. 2001;11:293–298. doi: 10.1097/00008571-200106000-00003. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Johnson AD, Papp AC, et al. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 13.Nakamura T, Sakaeda T, Horinouchi M, et al. Effect of the mutation (C3435T) at exon 26 of the MDR1 gene on expression level of MDR1 messenger ribonucleic acid in duodenal enterocytes of healthy Japanese subjects. Clin Pharmacol Ther. 2002;71:297–303. doi: 10.1067/mcp.2002.122055. [DOI] [PubMed] [Google Scholar]
- 14.Illmer T, Schuler US, Thiede C, et al. MDR1 gene polymorphisms affect therapy outcome in acute myeloid leukemia patients. Cancer Res. 2002;62:4955–4962. [PubMed] [Google Scholar]
- 15.Han JY, Lim HS, Yoo YK, et al. Associations of ABCB1, ABCC2, and ABCG2 polymorphisms with irinotecan-pharmacokinetics and clinical outcome in patients with advanced non-small cell lung cancer. Cancer. 2007;110:138–147. doi: 10.1002/cncr.22760. [DOI] [PubMed] [Google Scholar]
- 16.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxel-cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15:1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 17.Marsh S, Paul J, King CR, et al. Pharmacogenetic assessment of toxicity and outcome after platinum plus taxane chemotherapy in ovarian cancer: The Scottish Randomised Trial in Ovarian Cancer. J Clin Oncol. 2007;25:4528–4535. doi: 10.1200/JCO.2006.10.4752. [DOI] [PubMed] [Google Scholar]
- 18.Stanulla M, Schaffeler E, Arens S, et al. GSTP1 and MDR1 genotypes and central nervous system relapse in childhood acute lymphoblastic leukemia. Int J Hematol. 2005;81:39–44. doi: 10.1532/ijh97.e0418. [DOI] [PubMed] [Google Scholar]
- 19.Kim DH, Park JY, Sohn SK, et al. Multidrug resistance-1 gene polymorphisms associated with treatment outcomes in de novo acute myeloid leukemia. Int J Cancer. 2006;118:2195–2201. doi: 10.1002/ijc.21666. [DOI] [PubMed] [Google Scholar]
- 20.Maliepaard M, Scheffer GL, Faneyte IF, et al. Subcellular localization and distribution of the breast cancer resistance protein transporter in normal human tissues. Cancer Res. 2001;61:3458–3464. [PubMed] [Google Scholar]
- 21.Cusatis G, Sparreboom A. Pharmacogenomic importance of ABCG2. Pharmacogenomics. 2008;9:1005–1009. doi: 10.2217/14622416.9.8.1005. [DOI] [PubMed] [Google Scholar]
- 22.Bäckström G, Taipalensuu J, Melhus H, et al. Genetic variation in the ATP-binding cassette transporter gene ABCG2 (BCRP) in a Swedish population. Eur J Pharm Sci. 2003;18:359–364. doi: 10.1016/s0928-0987(03)00038-1. [DOI] [PubMed] [Google Scholar]
- 23.Bosch TM, Kjellberg LM, Bouwers A, et al. Detection of single nucleotide polymorphisms in the ABCG2 gene in a Dutch population. Am J Pharmacogenomics. 2005;5:123–131. doi: 10.2165/00129785-200505020-00005. [DOI] [PubMed] [Google Scholar]
- 24.Zamber CP, Lamba JK, Yasuda K, et al. Natural allelic variants of breast cancer resistance protein (BCRP) and their relationship to BCRP expression in human intestine. Pharmacogenetics. 2003;13:19–28. doi: 10.1097/00008571-200301000-00004. [DOI] [PubMed] [Google Scholar]
- 25.Imai Y, Nakane M, Kage K, et al. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol Cancer Ther. 2002;1:611–616. [PubMed] [Google Scholar]
- 26.Kobayashi D, Ieiri I, Hirota T, et al. Functional assessment of ABCG2 (BCRP) gene polymorphisms to protein expression in human placenta. Drug Metab Dispos. 2005;33:94–101. doi: 10.1124/dmd.104.001628. [DOI] [PubMed] [Google Scholar]
- 27.Mizuarai S, Aozasa N, Kotani H. Single nucleotide polymorphisms result in impaired membrane localization and reduced ATPase activity in multidrug transporter ABCG2. Int J Cancer. 2004;109:238–246. doi: 10.1002/ijc.11669. [DOI] [PubMed] [Google Scholar]
- 28.Tamura A, Wakabayashi K, Onishi Y, et al. Re-evaluation and functional classification of non-synonymous single nucleotide polymorphisms of the human ATP-binding cassette transporter ABCG2. Cancer Sci. 2007;98:231–239. doi: 10.1111/j.1349-7006.2006.00371.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparreboom A, Loos WJ, Burger H, et al. Effect of ABCG2 genotype on the oral bioavailability of topotecan. Cancer Biol Ther. 2005;4:650–658. doi: 10.4161/cbt.4.6.1731. [DOI] [PubMed] [Google Scholar]
- 30.de Jong FA, Marsh S, Mathijssen RH, et al. ABCG2 pharmacogenetics: Ethnic differences in allele frequency and assessment of influence on irinotecan disposition. Clin Cancer Res. 2004;10:5889–5894. doi: 10.1158/1078-0432.CCR-04-0144. [DOI] [PubMed] [Google Scholar]
- 31.Jada SR, Lim R, Wong CI, et al. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C>A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci. 2007;98:1461–1467. doi: 10.1111/j.1349-7006.2007.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardner ER, Burger H, van Schaik RH, et al. Association of enzyme and transporter genotypes with the pharmacokinetics of imatinib. Clin Pharmacol Ther. 2006;80:192–201. doi: 10.1016/j.clpt.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Petain A, Kattygnarath D, Azard J, et al. Population pharmacokinetics and pharmacogenetics of imatinib in children and adults. Clin Cancer Res. 2008;14:7102–7109. doi: 10.1158/1078-0432.CCR-08-0950. [DOI] [PubMed] [Google Scholar]
- 34.Li J, Cusatis G, Brahmer J, et al. Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients. Cancer Biol Ther. 2007;6:432–438. doi: 10.4161/cbt.6.3.3763. [DOI] [PubMed] [Google Scholar]
- 35.Cusatis G, Gregorc V, Li J, et al. Pharmacogenetics of ABCG2 and adverse reactions to gefitinib. J Natl Cancer Inst. 2006;98:1739–1742. doi: 10.1093/jnci/djj469. [DOI] [PubMed] [Google Scholar]
- 36.Akasaka K, Kaburagi T, Yasuda S, et al. Impact of functional ABCG2 polymorphisms on the adverse effects of gefitinib in Japanese patients with non-small-cell lung cancer. Cancer Chemother Pharmacol. 2010;66:691–698. doi: 10.1007/s00280-009-1211-6. [DOI] [PubMed] [Google Scholar]
- 37.Granvil CP, Madan A, Sharkawi M, et al. Role of CYP2B6 and CYP3A4 in the in vitro N-dechloroethylation of (R)- and (S)-ifosfamide in human liver microsomes. Drug Metab Dispos. 1999;27:533–541. [PubMed] [Google Scholar]
- 38.Boddy AV, Yule SM. Metabolism and pharmacokinetics of oxazaphosphorines. Clin Pharmacokinet. 2000;38:291–304. doi: 10.2165/00003088-200038040-00001. [DOI] [PubMed] [Google Scholar]
- 39.Rae JM, Soukhova NV, Flockhart DA, et al. Triethylenethiophosphoramide is a specific inhibitor of cytochrome P450 2B6: Implications for cyclophosphamide metabolism. Drug Metab Dispos. 2002;30:525–530. doi: 10.1124/dmd.30.5.525. [DOI] [PubMed] [Google Scholar]
- 40.Jacobson PA, Green K, Birnbaum A, et al. Cytochrome P450 isozymes 3A4 and 2B6 are involved in the in vitro human metabolism of thiotepa to TEPA. Cancer Chemother Pharmacol. 2002;49:461–467. doi: 10.1007/s00280-002-0453-3. [DOI] [PubMed] [Google Scholar]
- 41.Lang T, Klein K, Fischer J, et al. Extensive genetic polymorphism in the human CYP2B6 gene with impact on expression and function in human liver. Pharmacogenetics. 2001;11:399–415. doi: 10.1097/00008571-200107000-00004. [DOI] [PubMed] [Google Scholar]
- 42.Hiratsuka M, Takekuma Y, Endo N, et al. Allele and genotype frequencies of CYP2B6 and CYP3A5 in the Japanese population. Eur J Clin Pharmacol. 2002;58:417–421. doi: 10.1007/s00228-002-0499-5. [DOI] [PubMed] [Google Scholar]
- 43.Lamba V, Lamba J, Yasuda K, et al. Hepatic CYP2B6 expression: Gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther. 2003;307:906–922. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 44.Rotger M, Tegude H, Colombo S, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007;81:557–566. doi: 10.1038/sj.clpt.6100072. [DOI] [PubMed] [Google Scholar]
- 45.Zukunft J, Lang T, Richter T, et al. A natural CYP2B6 TATA box polymorphism (−82T-> C) leading to enhanced transcription and relocation of the transcriptional start site. Mol Pharmacol. 2005;67:1772–1782. doi: 10.1124/mol.104.008086. [DOI] [PubMed] [Google Scholar]
- 46.Ariyoshi N, Miyazaki M, Toide K, et al. A single nucleotide polymorphism of CYP2B6 found in Japanese enhances catalytic activity by autoactivation. Biochem Biophys Res Commun. 2001;281:1256–1260. doi: 10.1006/bbrc.2001.4524. [DOI] [PubMed] [Google Scholar]
- 47.Jinno H, Tanaka-Kagawa T, Ohno A, et al. Functional characterization of cytochrome P450 2B6 allelic variants. Drug Metab Dispos. 2003;31:398–403. doi: 10.1124/dmd.31.4.398. [DOI] [PubMed] [Google Scholar]
- 48.Hofmann MH, Blievernicht JK, Klein K, et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325:284–292. doi: 10.1124/jpet.107.133306. [DOI] [PubMed] [Google Scholar]
- 49.Hesse LM, He P, Krishnaswamy S, et al. Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics. 2004;14:225–238. doi: 10.1097/00008571-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 50.Xie HJ, Yasar U, Lundgren S, et al. Role of polymorphic human CYP2B6 in cyclophosphamide bioactivation. Pharmacogenomics J. 2003;3:53–61. doi: 10.1038/sj.tpj.6500157. [DOI] [PubMed] [Google Scholar]
- 51.Xie H, Griskevicius L, Ståhle L, et al. Pharmacogenetics of cyclophosphamide in patients with hematological malignancies. Eur J Pharm Sci. 2006;27:54–61. doi: 10.1016/j.ejps.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima M, Komagata S, Fujiki Y, et al. Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenet Genomics. 2007;17:431–445. doi: 10.1097/FPC.0b013e328045c4fb. [DOI] [PubMed] [Google Scholar]
- 53.Ekhart C, Doodeman VD, Rodenhuis S, et al. Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4-hydroxycyclophosphamide. Pharmacogenet Genomics. 2008;18:515–523. doi: 10.1097/FPC.0b013e3282fc9766. [DOI] [PubMed] [Google Scholar]
- 54.Ekhart C, Rodenhuis S, Smits PH, et al. Relations between polymorphisms in drug-metabolising enzymes and toxicity of chemotherapy with cyclophosphamide, thiotepa and carboplatin. Pharmacogenet Genomics. 2008;18:1009–1015. doi: 10.1097/FPC.0b013e328313aaa4. [DOI] [PubMed] [Google Scholar]
- 55.Low SK, Kiyotani K, Mushiroda T, et al. Association study of genetic polymorphism in ABCC4 with cyclophosphamide-induced adverse drug reactions in breast cancer patients. J Hum Genet. 2009;54:564–571. doi: 10.1038/jhg.2009.79. [DOI] [PubMed] [Google Scholar]
- 56.Gor PP, Su HI, Gray RJ, et al. Cyclophosphamide-metabolizing enzyme polymorphisms and survival outcomes after adjuvant chemotherapy for node-positive breast cancer: A retrospective cohort study. Breast Cancer Res. 2010;12:R26. doi: 10.1186/bcr2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar GN, Oatis JE, Jr, Thornburg KR, et al. 6 alpha-hydroxytaxol: Isolation and identification of the major metabolite of taxol in human liver microsomes. Drug Metab Dispos. 1994;22:177–179. [PubMed] [Google Scholar]
- 58.Bahadur N, Leathart JB, Mutch E, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64:1579–1589. doi: 10.1016/s0006-2952(02)01354-0. [DOI] [PubMed] [Google Scholar]
- 59.Dai D, Zeldin DC, Blaisdell JA, et al. Polymorphisms in human CYP2C8 decrease metabolism of the anticancer drug paclitaxel and arachidonic acid. Pharmacogenetics. 2001;11:597–607. doi: 10.1097/00008571-200110000-00006. [DOI] [PubMed] [Google Scholar]
- 60.Soyama A, Saito Y, Hanioka N, et al. Non-synonymous single nucleotide alterations found in the CYP2C8 gene result in reduced in vitro paclitaxel metabolism. Biol Pharm Bull. 2001;24:1427–1430. doi: 10.1248/bpb.24.1427. [DOI] [PubMed] [Google Scholar]
- 61.Henningsson A, Marsh S, Loos WJ, et al. Association of CYP2C8, CYP3A4, CYP3A5, and ABCB1 polymorphisms with the pharmacokinetics of paclitaxel. Clin Cancer Res. 2005;11:8097–8104. doi: 10.1158/1078-0432.CCR-05-1152. [DOI] [PubMed] [Google Scholar]
- 62.Marsh S, Somlo G, Li X, et al. Pharmacogenetic analysis of paclitaxel transport and metabolism genes in breast cancer. Pharmacogenomics J. 2007;7:362–365. doi: 10.1038/sj.tpj.6500434. [DOI] [PubMed] [Google Scholar]
- 63.King BP, Khan TI, Aithal GP, et al. Upstream and coding region CYP2C9 polymorphisms: Correlation with warfarin dose and metabolism. Pharmacogenetics. 2004;14:813–822. doi: 10.1097/00008571-200412000-00004. [DOI] [PubMed] [Google Scholar]
- 64.Shintani M, Ieiri I, Inoue K, et al. Genetic polymorphisms and functional characterization of the 5′-flanking region of the human CYP2C9 gene: In vitro and in vivo studies. Clin Pharmacol Ther. 2001;70:175–182. doi: 10.1067/mcp.2001.117367. [DOI] [PubMed] [Google Scholar]
- 65.Takanashi K, Tainaka H, Kobayashi K, et al. CYP2C9 Ile359 and Leu359 variants: Enzyme kinetic study with seven substrates. Pharmacogenetics. 2000;10:95–104. doi: 10.1097/00008571-200003000-00001. [DOI] [PubMed] [Google Scholar]
- 66.Timm R, Kaiser R, Lötsch J, et al. Association of cyclophosphamide pharmacokinetics to polymorphic cytochrome P450 2C19. Pharmacogenomics J. 2005;5:365–373. doi: 10.1038/sj.tpj.6500330. [DOI] [PubMed] [Google Scholar]
- 67.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 68.Schroth W, Antoniadou L, Fritz P, et al. Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol. 2007;25:5187–5193. doi: 10.1200/JCO.2007.12.2705. [DOI] [PubMed] [Google Scholar]
- 69.Jernström H, Bågeman E, Rose C, et al. CYP2C8 and CYP2C9 polymorphisms in relation to tumour characteristics and early breast cancer related events among 652 breast cancer patients. Br J Cancer. 2009;101:1817–1823. doi: 10.1038/sj.bjc.6605428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zandvliet AS, Huitema AD, Copalu W, et al. CYP2C9 and CYP2C19 polymorphic forms are related to increased indisulam exposure and higher risk of severe hematologic toxicity. Clin Cancer Res. 2007;13:2970–2976. doi: 10.1158/1078-0432.CCR-06-2978. [DOI] [PubMed] [Google Scholar]
- 71.Ando Y, Price DK, Dahut WL, et al. Pharmacogenetic associations of CYP2C19 genotype with in vivo metabolisms and pharmacological effects of thalidomide. Cancer Biol Ther. 2002;1:669–673. doi: 10.4161/cbt.318. [DOI] [PubMed] [Google Scholar]
- 72.De Morais SM, Wilkinson GR, Blaisdell J, et al. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 73.Li Y, Hou J, Jiang H, et al. Polymorphisms of CYP2C19 gene are associated with the efficacy of thalidomide based regimens in multiple myeloma. Haematologica. 2007;92:1246–1249. doi: 10.3324/haematol.11319. [DOI] [PubMed] [Google Scholar]
- 74.Dehal SS, Kupfer D. CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res. 1997;57:3402–3406. [PubMed] [Google Scholar]
- 75.Johnson MD, Zuo H, Lee KH, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- 76.Human Cytochrome P450 (CYP) Allele Nomenclature Committee. CYP2D6 Allele Nomenclature. [accessed June 26, 2010]. Available at http://www.cypalleles.ki.se/cyp2d6.htm.
- 77.Aklillu E, Persson I, Bertilsson L, et al. Frequent distribution of ultrarapid metabolizers of debrisoquine in an Ethiopian population carrying duplicated and multiduplicated functional CYP2D6 alleles. J Pharmacol Exp Ther. 1996;278:441–446. [PubMed] [Google Scholar]
- 78.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): Clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 79.Johansson I, Lundqvist E, Bertilsson L, et al. Inherited amplification of an active gene in the cytochrome P450 CYP2D locus as a cause of ultrarapid metabolism of debrisoquine. Proc Natl Acad Sci U S A. 1993;90:11825–11829. doi: 10.1073/pnas.90.24.11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanioka N, Kimura S, Meyer UA, et al. The human CYP2D locus associated with a common genetic defect in drug oxidation: A G1934—A base change in intron 3 of a mutant CYP2D6 allele results in an aberrant 3′ splice recognition site. Am J Hum Genet. 1990;47:994–1001. [PMC free article] [PubMed] [Google Scholar]
- 81.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19:56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 82.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: Implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80:61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 83.Bijl MJ, van Schaik RH, Lammers LA, et al. The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat. 2009;118:125–130. doi: 10.1007/s10549-008-0272-2. [DOI] [PubMed] [Google Scholar]
- 84.Goetz MP, Knox SK, Suman VJ, et al. The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat. 2007;101:113–121. doi: 10.1007/s10549-006-9428-0. [DOI] [PubMed] [Google Scholar]
- 85.Kiyotani K, Mushiroda T, Sasa M, et al. Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci. 2008;99:995–999. doi: 10.1111/j.1349-7006.2008.00780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Newman WG, Hadfield KD, Latif A, et al. Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin Cancer Res. 2008;14:5913–5918. doi: 10.1158/1078-0432.CCR-07-5235. [DOI] [PubMed] [Google Scholar]
- 87.Schroth W, Goetz MP, Hamann U, et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu Y, Sun Y, Yao L, et al. Association between CYP2D6 *10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol. 2008;19:1423–1429. doi: 10.1093/annonc/mdn155. [DOI] [PubMed] [Google Scholar]
- 89.Goetz MP, Rae JM, Suman VJ, et al. Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol. 2005;23:9312–9318. doi: 10.1200/JCO.2005.03.3266. [DOI] [PubMed] [Google Scholar]
- 90.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat. 2005;91:249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 91.Wegman P, Vainikka L, Stål O, et al. Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res. 2005;7:R284–R290. doi: 10.1186/bcr993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wegman P, Elingarami S, Carstensen J, et al. Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res. 2007;9:R7. doi: 10.1186/bcr1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okishiro M, Taguchi T, Jin Kim S, et al. Genetic polymorphisms of CYP2D6 10 and CYP2C19 2, 3 are not associated with prognosis, endometrial thickness, or bone mineral density in Japanese breast cancer patients treated with adjuvant tamoxifen. Cancer. 2009;115:952–961. doi: 10.1002/cncr.24111. [DOI] [PubMed] [Google Scholar]
- 94.Sideras K, Ingle JN, Ames MM, et al. Coprescription of tamoxifen and medications that inhibit CYP2D6. J Clin Oncol. 2010;28:2768–2776. doi: 10.1200/JCO.2009.23.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25:1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 96.Li J, Zhao M, He P, et al. Differential metabolism of gefitinib and erlotinib by human cytochrome P450 enzymes. Clin Cancer Res. 2007;13:3731–3737. doi: 10.1158/1078-0432.CCR-07-0088. [DOI] [PubMed] [Google Scholar]
- 97.Guengerich FP. Cytochrome P-450 3A4: Regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 98.Ozdemir V, Kalow W, Tang BK, et al. Evaluation of the genetic component of variability in CYP3A4 activity: A repeated drug administration method. Pharmacogenetics. 2000;10:373–388. doi: 10.1097/00008571-200007000-00001. [DOI] [PubMed] [Google Scholar]
- 99.Wojnowski L, Kamdem LK. Clinical implications of CYP3A polymorphisms. Expert Opin Drug Metab Toxicol. 2006;2:171–182. doi: 10.1517/17425255.2.2.171. [DOI] [PubMed] [Google Scholar]
- 100.Human Cytochrome P450 (CYP) Allele Nomenclature Committee. CYP3A4 Allele Nomenclature. [accessed June 28, 2010]. Available at http://www.cypalleles.ki.se/cyp3a4.htm.
- 101.Rodriguez-Antona C, Sayi JG, Gustafsson LL, et al. Phenotype-genotype variability in the human CYP3A locus as assessed by the probe drug quinine and analyses of variant CYP3A4 alleles. Biochem Biophys Res Commun. 2005;338:299–305. doi: 10.1016/j.bbrc.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 102.Hustert E, Haberl M, Burk O, et al. The genetic determinants of the CYP3A5 polymorphism. Pharmacogenetics. 2001;11:773–779. doi: 10.1097/00008571-200112000-00005. [DOI] [PubMed] [Google Scholar]
- 103.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 104.Shou M, Martinet M, Korzekwa KR, et al. Role of human cytochrome P450 3A4 and 3A5 in the metabolism of taxotere and its derivatives: Enzyme specificity, interindividual distribution and metabolic contribution in human liver. Pharmacogenetics. 1998;8:391–401. doi: 10.1097/00008571-199810000-00004. [DOI] [PubMed] [Google Scholar]
- 105.Tran A, Jullien V, Alexandre J, et al. Pharmacokinetics and toxicity of docetaxel: Role of CYP3A, MDR1, and GST polymorphisms. Clin Pharmacol Ther. 2006;79:570–580. doi: 10.1016/j.clpt.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 106.Baker SD, Verweij J, Cusatis GA, et al. Pharmacogenetic pathway analysis of docetaxel elimination. Clin Pharmacol Ther. 2009;85:155–163. doi: 10.1038/clpt.2008.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lewis LD, Miller AA, Rosner GL, et al. A comparison of the pharmacokinetics and pharmacodynamics of docetaxel between African-American and Caucasian cancer patients: CALGB 9871. Clin Cancer Res. 2007;13:3302–3311. doi: 10.1158/1078-0432.CCR-06-2345. [DOI] [PubMed] [Google Scholar]
- 108.Gréen H, Söderkvist P, Rosenberg P, et al. Pharmacogenetic studies of paclitaxel in the treatment of ovarian cancer. Basic Clin Pharmacol Toxicol. 2009;104:130–137. doi: 10.1111/j.1742-7843.2008.00351.x. [DOI] [PubMed] [Google Scholar]
- 109.Petros WP, Hopkins PJ, Spruill S, et al. Associations between drug metabolism genotype, chemotherapy pharmacokinetics, and overall survival in patients with breast cancer. J Clin Oncol. 2005;23:6117–6125. doi: 10.1200/JCO.2005.06.075. [DOI] [PubMed] [Google Scholar]
- 110.Pillot GA, Read WL, Hennenfent KL, et al. A phase II study of irinotecan and carboplatin in advanced non-small cell lung cancer with pharmacogenomic analysis: Final report. J Thorac Oncol. 2006;1:972–978. [PubMed] [Google Scholar]
- 111.Rudin CM, Liu W, Desai A, et al. Pharmacogenomic and pharmacokinetic determinants of erlotinib toxicity. J Clin Oncol. 2008;26:1119–1127. doi: 10.1200/JCO.2007.13.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ingelman-Sundberg M, Sim SC, Gomez A, et al. Influence of cytochrome P450 polymorphisms on drug therapies: Pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 113.Sahin M, Sahin E, Gümüslü S. Cyclooxygenase-2 in cancer and angiogenesis. Angiology. 2009;60:242–253. doi: 10.1177/0003319708318378. [DOI] [PubMed] [Google Scholar]
- 114.Clevers H. Colon cancer—understanding how NSAIDs work. N Engl J Med. 2006;354:761–763. doi: 10.1056/NEJMcibr055457. [DOI] [PubMed] [Google Scholar]
- 115.Aparicio Gallego G, Diaz Prado S, Jiménez Fonseca P, et al. Cyclooxygenase-2 (COX-2): A molecular target in prostate cancer. Clin Transl Oncol. 2007;9:694–702. doi: 10.1007/s12094-007-0126-0. [DOI] [PubMed] [Google Scholar]
- 116.Seitz HK, Becker P. Alcohol metabolism and cancer risk. Alcohol Res Health. 2007;30:38–41. 44–47. [PMC free article] [PubMed] [Google Scholar]
- 117.Yokoyama A, Omori T. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and risk for esophageal and head and neck cancers. Alcohol. 2005;35:175–185. doi: 10.1016/j.alcohol.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 118.Diasio RB, Harris BE. Clinical pharmacology of 5-fluorouracil. Clin Pharmacokinet. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 119.Heggie GD, Sommadossi JP, Cross DS, et al. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987;47:2203–2206. [PubMed] [Google Scholar]
- 120.Boisdron-Celle M, Remaud G, Traore S, et al. 5-Fluorouracil-related severe toxicity: A comparison of different methods for the pretherapeutic detection of dihydropyrimidine dehydrogenase deficiency. Cancer Lett. 2007;249:271–282. doi: 10.1016/j.canlet.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 121.Maekawa K, Saeki M, Saito Y, et al. Genetic variations and haplotype structures of the DPYD gene encoding dihydropyrimidine dehydrogenase in Japanese and their ethnic differences. J Hum Genet. 2007;52:804–819. doi: 10.1007/s10038-007-0186-6. [DOI] [PubMed] [Google Scholar]
- 122.Meinsma R, Fernandez-Salguero P, Van Kuilenburg AB, et al. Human polymorphism in drug metabolism: Mutation in the dihydropyrimidine dehydrogenase gene results in exon skipping and thymine uracilurea. DNA Cell Biol. 1995;14:1–6. doi: 10.1089/dna.1995.14.1. [DOI] [PubMed] [Google Scholar]
- 123.Wei X, McLeod HL, McMurrough J, et al. Molecular basis of the human dihydropyrimidine dehydrogenase deficiency and 5-fluorouracil toxicity. J Clin Invest. 1996;98:610–615. doi: 10.1172/JCI118830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vreken P, Van Kuilenburg AB, Meinsma R, et al. A point mutation in an invariant splice donor site leads to exon skipping in two unrelated Dutch patients with dihydropyrimidine dehydrogenase deficiency. J Inherit Metab Dis. 1996;19:645–654. doi: 10.1007/BF01799841. [DOI] [PubMed] [Google Scholar]
- 125.Mattison LK, Johnson MR, Diasio RB. A comparative analysis of translated dihydropyrimidine dehydrogenase cDNA: Conservation of functional domains and relevance to genetic polymorphisms. Pharmacogenetics. 2002;12:133–144. doi: 10.1097/00008571-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 126.Van Kuilenburg AB, Haasjes J, Richel DJ, et al. Clinical implications of dihydropyrimidine dehydrogenase (DPD) deficiency in patients with severe 5-fluorouracil-associated toxicity: Identification of new mutations in the DPD gene. Clin Cancer Res. 2000;6:4705–4712. [PubMed] [Google Scholar]
- 127.Dobritzsch D, Schneider G, Schnackerz KD, et al. Crystal structure of dihydropyrimidine dehydrogenase, a major determinant of the pharmacokinetics of the anti-cancer drug 5-fluorouracil. EMBO J. 2001;20:650–660. doi: 10.1093/emboj/20.4.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Blasco H, Boisdron-Celle M, Bougnoux P, et al. A well-tolerated 5-FU-based treatment subsequent to severe capecitabine-induced toxicity in a DPD-deficient patient. Br J Clin Pharmacol. 2008;65:966–970. doi: 10.1111/j.1365-2125.2008.03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ezzeldin H, Johnson MR, Okamoto Y, et al. Denaturing high performance liquid chromatography analysis of the DPYD gene in patients with lethal 5-fluorouracil toxicity. Clin Cancer Res. 2003;9:3021–3028. [PubMed] [Google Scholar]
- 130.Johnson MR, Hageboutros A, Wang K, et al. Life-threatening toxicity in a dihydropyrimidine dehydrogenase-deficient patient after treatment with topical 5-fluorouracil. Clin Cancer Res. 1999;5:2006–2011. [PubMed] [Google Scholar]
- 131.Saif MW, Ezzeldin H, Vance K, et al. DPYD*2A mutation: The most common mutation associated with DPD deficiency. Cancer Chemother Pharmacol. 2007;60:503–507. doi: 10.1007/s00280-006-0392-5. [DOI] [PubMed] [Google Scholar]
- 132.Steiner M, Seule M, Steiner B, et al. 5-Fluorouracil/irinotecan induced lethal toxicity as a result of a combined pharmacogenetic syndrome: Report of a case. J Clin Pathol. 2005;58:553–555. doi: 10.1136/jcp.2004.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Kuilenburg AB, Vreken P, Beex LV, et al. Heterozygosity for a point mutation in an invariant splice donor site of dihydropyrimidine dehydrogenase and severe 5-fluorouracil related toxicity. Eur J Cancer. 1997;33:2258–2264. doi: 10.1016/s0959-8049(97)00261-x. [DOI] [PubMed] [Google Scholar]
- 134.Van Kuilenburg AB, Muller EW, Haasjes J, et al. Lethal outcome of a patient with a complete dihydropyrimidine dehydrogenase (DPD) deficiency after administration of 5-fluorouracil: Frequency of the common IVS14+1G>A mutation causing DPD deficiency. Clin Cancer Res. 2001;7:1149–1153. [PubMed] [Google Scholar]
- 135.Van Kuilenburg AB, Meinsma R, Zoetekouw L, et al. High prevalence of the IVS14 + 1G>A mutation in the dihydropyrimidine dehydrogenase gene of patients with severe 5-fluorouracil-associated toxicity. Pharmacogenetics. 2002;12:555–558. doi: 10.1097/00008571-200210000-00007. [DOI] [PubMed] [Google Scholar]
- 136.Maring JG, Van Kuilenburg AB, Haasjes J, et al. Reduced 5-FU clearance in a patient with low DPD activity due to heterozygosity for a mutant allele of the DPYD gene. Br J Cancer. 2002;86:1028–1033. doi: 10.1038/sj.bjc.6600208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Raida M, Schwabe W, Haüsler P, et al. Prevalence of a common point mutation in the dihydropyrimidine dehydrogenase (DPD) gene within the 5′-splice donor site of intron 14 in patients with severe 5-fluorouracil (5-FU)- related toxicity compared with controls. Clin Cancer Res. 2001;7:2832–2839. [PubMed] [Google Scholar]
- 138.Morel A, Boisdron-Celle M, Fey L, et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther. 2006;5:2895–2904. doi: 10.1158/1535-7163.MCT-06-0327. [DOI] [PubMed] [Google Scholar]
- 139.Braun MS, Richman SD, Thompson L, et al. Association of molecular markers with toxicity outcomes in a randomized trial of chemotherapy for advanced colorectal cancer: The FOCUS trial. J Clin Oncol. 2009;27:5519–5528. doi: 10.1200/JCO.2008.21.6283. [DOI] [PubMed] [Google Scholar]
- 140.Largillier R, Etienne-Grimaldi MC, Formento JL, et al. Pharmacogenetics of capecitabine in advanced breast cancer patients. Clin Cancer Res. 2006;12:5496–5502. doi: 10.1158/1078-0432.CCR-06-0320. [DOI] [PubMed] [Google Scholar]
- 141.Schwab M, Zanger UM, Marx C, et al. Role of genetic and nongenetic factors for fluorouracil treatment-related severe toxicity: A prospective clinical trial by the German 5-FU Toxicity Study Group. J Clin Oncol. 2008;26:2131–2138. doi: 10.1200/JCO.2006.10.4182. [DOI] [PubMed] [Google Scholar]
- 142.Deenen MJ, Tol J, Burylo AM, et al. Relationship between single nucleotide polymorphisms and haplotypes in DPYD and toxicity and efficacy of capecitabine in advanced colorectal cancer. Clin Cancer Res. 2011 doi: 10.1158/1078-0432.CCR-10-2209. in press. [DOI] [PubMed] [Google Scholar]
- 143.Boige V, Mendiboure J, Pignon JP, et al. Pharmacogenetic assessment of toxicity and outcome in patients with metastatic colorectal cancer treated with LV5FU2, FOLFOX, and FOLFIRI: FFCD 2000–05. J Clin Oncol. 2010;28:2556–2564. doi: 10.1200/JCO.2009.25.2106. [DOI] [PubMed] [Google Scholar]
- 144.Gilbert JA, Salavaggione OE, Ji Y, et al. Gemcitabine pharmacogenomics: Cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin Cancer Res. 2006;12:1794–1803. doi: 10.1158/1078-0432.CCR-05-1969. [DOI] [PubMed] [Google Scholar]
- 145.Kirch HC, Schröder J, Hoppe H, et al. Recombinant gene products of two natural variants of the human cytidine deaminase gene confer different deamination rates of cytarabine in vitro. Exp Hematol. 1998;26:421–425. [PubMed] [Google Scholar]
- 146.Tibaldi C, Giovannetti E, Vasile E, et al. Correlation of CDA, ERCC1, and XPD polymorphisms with response and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2008;14:1797–1803. doi: 10.1158/1078-0432.CCR-07-1364. [DOI] [PubMed] [Google Scholar]
- 147.Yue L, Saikawa Y, Ota K, et al. A functional single-nucleotide polymorphism in the human cytidine deaminase gene contributing to ara-C sensitivity. Pharmacogenetics. 2003;13:29–38. doi: 10.1097/00008571-200301000-00005. [DOI] [PubMed] [Google Scholar]
- 148.Sugiyama E, Lee SJ, Lee SS, et al. Ethnic differences of two non-synonymous single nucleotide polymorphisms in CDA gene. Drug Metab Pharmacokinet. 2009;24:553–556. doi: 10.2133/dmpk.24.553. [DOI] [PubMed] [Google Scholar]
- 149.Sugiyama E, Kaniwa N, Kim SR, et al. Pharmacokinetics of gemcitabine in Japanese cancer patients: The impact of a cytidine deaminase polymorphism. J Clin Oncol. 2007;25:32–42. doi: 10.1200/JCO.2006.06.7405. [DOI] [PubMed] [Google Scholar]
- 150.Yonemori K, Ueno H, Okusaka T, et al. Severe drug toxicity associated with a single-nucleotide polymorphism of the cytidine deaminase gene in a Japanese cancer patient treated with gemcitabine plus cisplatin. Clin Cancer Res. 2005;11:2620–2624. doi: 10.1158/1078-0432.CCR-04-1497. [DOI] [PubMed] [Google Scholar]
- 151.Ueno H, Kaniwa N, Okusaka T, et al. Homozygous CDA*3 is a major cause of life-threatening toxicities in gemcitabine-treated Japanese cancer patients. Br J Cancer. 2009;100:870–873. doi: 10.1038/sj.bjc.6604971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mercier C, Raynal C, Dahan L, et al. Toxic death case in a patient undergoing gemcitabine-based chemotherapy in relation with cytidine deaminase downregulation. Pharmacogenet Genomics. 2007;17:841–844. doi: 10.1097/FPC.0b013e32825ea6e3. [DOI] [PubMed] [Google Scholar]
- 153.Maring JG, Wachters FM, Slijfer M, et al. Pharmacokinetics of gemcitabine in non-small-cell lung cancer patients: Impact of the 79A>C cytidine deaminase polymorphism. Eur J Clin Pharmacol. 2010;66:611–617. doi: 10.1007/s00228-010-0799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Koldamova RP, Lefterov IM, Gadjeva VG, et al. Essential binding and functional domains of human bleomycin hydrolase. Biochemistry. 1998;37:2282–2290. doi: 10.1021/bi9722204. [DOI] [PubMed] [Google Scholar]
- 155.Morris G, Mistry JS, Jani JP, et al. Neutralization of bleomycin hydrolase by an epitope-specific antibody. Mol Pharmacol. 1992;42:57–62. [PubMed] [Google Scholar]
- 156.Brömme D, Rossi AB, Smeekens SP, et al. Human bleomycin hydrolase: Molecular cloning, sequencing, functional expression, and enzymatic characterization. Biochemistry. 1996;35:6706–6714. doi: 10.1021/bi960092y. [DOI] [PubMed] [Google Scholar]
- 157.Tuimala J, Szekely G, Gundy S, et al. Genetic polymorphisms of DNA repair and xenobiotic-metabolizing enzymes: Role in mutagen sensitivity. Carcinogenesis. 2002;23:1003–1008. doi: 10.1093/carcin/23.6.1003. [DOI] [PubMed] [Google Scholar]
- 158.de Haas EC, Zwart N, Meijer C, et al. Variation in bleomycin hydrolase gene is associated with reduced survival after chemotherapy for testicular germ cell cancer. J Clin Oncol. 2008;26:1817–1823. doi: 10.1200/JCO.2007.14.1606. [DOI] [PubMed] [Google Scholar]
- 159.Nauck M, Gierens H, März W, et al. Rapid detection of a common dihydropyrimidine dehydrogenase mutation associated with 5-fluorouracil toxicity and congenital thymine uraciluria using fluorogenic hybridization probes. Clin Biochem. 2001;34:103–105. doi: 10.1016/s0009-9120(01)00188-6. [DOI] [PubMed] [Google Scholar]
- 160.Seck K, Riemer S, Kates R, et al. Analysis of the DPYD gene implicated in 5-fluorouracil catabolism in a cohort of Caucasian individuals. Clin Cancer Res. 2005;11:5886–5892. doi: 10.1158/1078-0432.CCR-04-1784. [DOI] [PubMed] [Google Scholar]