Abstract
Most epigenetic studies assess methylation of 5′-CpG-3′ sites but recent evidence indicates that non-CpG cytosine methylation occurs at high levels in humans and other species. This is most prevalent at 5′-CHG-3′, where H = A, C or T, and it preferentially occurs at 5′-CpA-3′ and 5′-CpT-3′ sites. With the goal of facilitating the detection of non-CpG methylation, the restriction endonucleases ApeKI, BbvI, EcoP15I, Fnu4HI, MwoI and TseI were assessed for their sensitivity to 5-methylcytosine at GpCpA, GpCpT, GpCpC or GpCpG sites, where methylation is catalyzed by the DNA 5-cytosine 5′-GpC-3′ methyltransferase M.CviPI. We tested a variety of sequences including various plasmid-based sites, a cloned disease-associated (CAG)83•(CTG)83 repeat and in vitro synthesized tracts of only (CAG)500•(CTG)500 or (CAG)800•(CTG)800. The repeat tracts are enriched for the preferred CpA and CpT motifs. We found that none of the tested enzymes can cleave their recognition sequences when they are 5′-GpC-3′ methylated. A genomic site known to convert its non-CpG methylation levels upon C2C12 differentiation was confirmed through the use of these enzymes. These enzymes can be useful in rapidly and easily determining the most common non-CpG methylation status in various sequence contexts, as well as at expansions of (CAG)n•(CTG)n repeat tracts associated with diseases like myotonic dystrophy and Huntington disease.
Key words: non-CpG methylation, CpG methylation, 5-methylcytosine, trinucleotide repeats, ApeKI, BbvI, EcoP151, Fnu4HI, MwoI and TseI
Introduction
DNA methylation has important roles in genetic regulation, development and disease etiology.1 In several neurological, neurodegenerative and neuromuscular diseases, characterized by the high instability of gene-specific repeat tracts,2,3 the presence of aberrant 5′-CpG-3′ methylation flanking the unstable repeat has been reported. For example, aberrant CpG methylation within the expanded CGG repeats and flanking sequences is observed in several rare fragile site diseases like fragile X mental retardation FRAXA, FRAXE, FRAXF, FRAX10A, FRAX11B, FRA12A and FRAXA16A and associated with gene silencing.2,4 For myotonic dystrophy (DM1), Friedreich's ataxia (FRDA) and Huntington disease (HD) involving the non-CpG repeat motifs (CTG)n, (GAA)n and (CAG)n, respectively, acquisition of CpG methylation profiles has been observed adjacent to the mutated repeat alleles.2,5–11
Most studies of DNA methylation assess 5′-CpG-3′ but recent evidence indicates that non-CpG cytosine methylation occurs at high levels in humans and other species,12 this being most prevalent at 5′-CpA-3′ and 5′-CpT-3′, which are present in the disease-associated repeats (CAG)n and (CTG)n. Human cells show abundant DNA methylation in non-CG contexts (meCHG and meCHH, where H = A, C or T), which comprises ∼25% of all methylated cytosines in the genome.12 Notably, a genome-wide study found that methylation levels were higher for CHG than CHH, and preferentially found more often at CAG than at CTG.12 This preference supports previous gene-specific observations.13–21 Interestingly, the CAG/CTG site preference of non-CpG methylation matches the sequence preference of some mammalian DNA methyltransferases.22,23 Non-CpG methylation has typically been assessed using bisulfite sequencing, a method with which most labs are not familiar.
The presence of non-CpG methylation deserves further attention. It is noteworthy that non-CpG methylation has been reported in genes containing CNG trinucleotide repeat tracts, although only in non-expanded samples.24 An easy assay to rapidly assess non-CpG methylation at any site, including trinucleotide repeats, is not currently available.
Here, we assessed the influence of non-CpG methylation (5′-GpC-3′ context) on the activity of six restriction endonucleases: Fnu4HI (5′-GC↓NGC-3′), TseI (5′-G↓CWGC-3′), MwoI (5′-GCNNNNN↓NNGC-3′), EcoP15I (5′-CAGCAG(N)25↓-3′), BbvI (5′-GCAGC(N)8↓-3′) and ApeKI (5′-G↓CWGC-3′). Each of these enzymes recognizes sites that can harbor or overlap with the preferred non-CpG methylatable sites CpA and CpT, including the repeat (CAG)n•(CTG)n recognition motifs. Cytosine methylation at GpC sites in the CAGCAG and CTGCTG tracts conforms to the preferred non-CpG sites 5′-CpApG-3′ and 5′-CpTpG-3′.12 (CTG)n and (CAG)n tracts are the most common repeat sequences associated with a class of neurodegenerative and neuromuscular diseases2,3 ApeKI, BbvI and EcoP15I have shown no sensitivity to CpG methylation.25 In contrast, Fnu4HI has been reported to be sensitive to overlapping CpG methylation,25–28 while MwoI and TseI are sensitive to some combinations of overlapping 5-CpG methylation.28 However, nothing is known about the 5-methylcytosine sensitivity of any of these enzymes outside the CpG context. Two approaches were followed in order to analyze differential GpC methylation status in vitro: (1) digestion of the pKH11 DNA, a plasmid containing various forms of each of the restriction sites as well as a cloned (CAG)83•(CTG)83 repeat and DM1 flanking sequences (Fig. 1); (2) digestion of in vitro synthesized DNA tracts containing only (CAG)500•(CTG)500 or (CAG)800•(CTG)800 repeat units. All 5′-GpC-3′ sites were methylated on the cytosine residues using the methylase M.CviPI (a DNA 5-cytosine methyltransferase), which has specificity for 5′-GpC-3′.29
Figure 1.
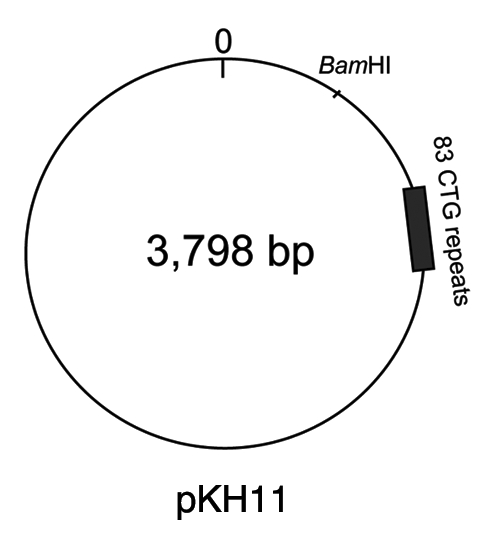
Plasmid pKH11, harboring numerous plasmid-based restriction sites and a DM1 sequence with (CTG)83 repeats. For restriction site details see Supplemental Figure S1. For cloning details see Materials and Methods.
Results
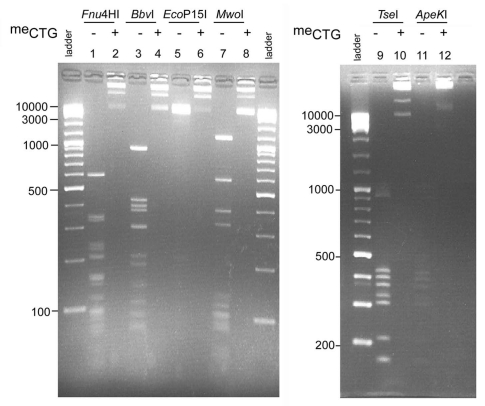
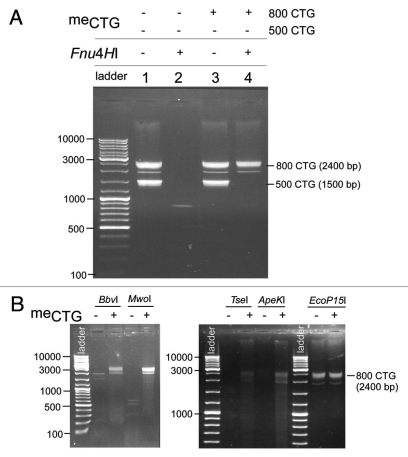
All enzymes tested were sensitive to pre-methylation at 5′-GpC-3′ sites. Figure 2 shows the results from plasmid pKH11-(CTG)83 digestions, before and after methylase treatment. Lanes 1, 3, 5, 7, 9 and 11 reflect the DNA restriction fragment profiles in absence of methylation. Patterns of digestion products are representative of the expected complete digestion for Fnu4HI, TseI, MwoI, ApeKI and BbvI as shown by the predicted cleavage profiles (left parts in Sup. Fig. S1). EcoP15I requires the presence of two inversely oriented 5′-CAGCAG recognition sites for efficient DNA cleavage, where sites can be separated by as much as 3.5 kb.33 EcoP15I digestion of the pKH11 plasmid containing a (CAG)83 tract yielded a smear of products (Fig. 2 and lane 5) cutting at overlapping sites within the repeat track and the unique plasmid-based site (Sup. Fig. S1). Lanes 2, 4, 6, 8, 10 and 12 show the total cleavage inhibition under presence of 5′-GpC-3′ methylation, including through the (CTG)83 repeat tract, for all six restriction endonucleases tested. Digestion with BamHI (unique cleavage site in the pKH11 plasmid) prior to or following EcoP15I digestion, showed the expected linearized 3,798 bp fragment, indicating that resistance to EcoP15I was in fact due to GpC methylation (data not shown). For all enzymes except EcoP15I, the cleavage of DNAs containing only long CTG/CAG tracks was sensitive to GpC methylation (Fig. 3). In the absence of GpC methylation, complete cleavage of the DNA repeat fragment is observed (Fnu4HI, in part A, lanes 2 and 4; MwoI, BbvI, TseI and ApekI in B). In contrast, the presence of GpC methylation on the 800 repeat DNA fragment completely inhibited cleavage by these five enzymes (Fig. 3 and lane 4 in A; + lanes in B). The exception was EcoP15I which, in the absence of a CTGCTG sequence in the opposite orientation on these pure-CAGCAG repeat DNAs, could not cleave prior nor after GpC methylation (Fig. 3B).
Figure 2.
Restriction of plasmid pKH11 prior to (odd lanes) and following (even lanes) methylation of 5′-GpC-3′ sites by the DNA 5-cytosine methyl-transferase M.CviPI (Materials and Methods). pKH11 was digested with Fnu4HI, BbvI, EcoP15I, MwoI, TseI or ApeKI and products resolved by electrophoresis on 2% agarose gels. The marker was the 100 bp ladder.
Figure 3.
(A) As a negative internal control, in vitro synthesized (CAG)800•(CTG)800 DNA (MATERIALS & METHODS) were completely methylated at all 5′-GpC-3′ sites by DNA 5-cytosine methyl-transferase M.CviPI (indicated by +) and mixed at equimolar amounts with in vitro synthesized (CAG)500•(CTG)500 DNA that were mock-treated with M.CviPI GpC methylase (indicated by −). These DNA mixtures were restriction digested with Fnu4HI and products resolved by electrophoresis on 1% agarose gels. The 100 bp ladder was loaded in the leftmost lane. (B) In vitro synthesized (CAG)800•(CTG)800 DNA was mock-treated with GpC methylase (−) or methylated (+) at all GpC sites. The DNA was restriction digested with BbvI, MwoI, TseI, ApeKI or EcoP15I and products resolved by electrophoresis on 1% agarose gels. DNA marker has been loaded in the leftmost lanes.
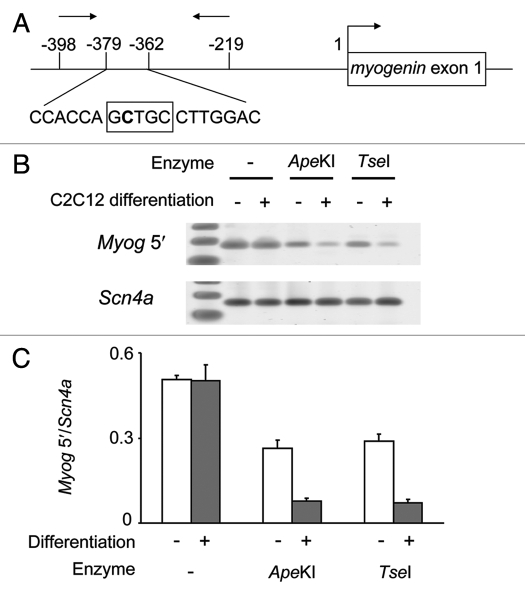
We then tested the utility of two of these enzymes to detect non-CpG methylation at a genomic locus known to display non-CpG methylation. We used the C2C12 mouse muscle cell line to examine non-CpG methylation at a mammalian endogenous locus. Previous studies have shown that methylation of non-CpG cytosine in the myogenin (Myog) 5′-flanking region is dependent on the differentiation state of these cells.34 Genomic DNA samples from differentiated and undifferentiated C2C12 cells were subjected to restriction digestion by ApeKI or TseI. After PCR amplification, the relative amount of undigested Myog 5′-flanking region was higher in undifferentiated C2C12 cells than in differentiated cells (Fig. 4). This result suggests that these enzymes are capable of assessing non-CpG methylation status in a genomic DNA.
Figure 4.
(A) Diagram showing the site of non-CpG methylation in myogenin promoter region. The target non-CpG site is shown in bold. The box indicates the recognition site for ApeKI and TseI. Arrows indicate primer positions. (B) PCR products of Myog 5′-flanking and Scn4a exon 6 regions after digestion with ApeKI or TseI. (C) Relative amount of PCR products of Myog 5′-flanking region from undifferentiated (white bar) or differentiated (gray bar) C2C12 DNA. DNA digestion and PCR were performed in triplicate.
Discussion
The cytosine methylation sensitivities in non-CpG contexts determined by this method can be applied directly to epigenetic studies of various sequences, including those involved in human diseases. The use of any of the six restriction endonucleases (Fnu4HI, TseI, MwoI, BbvI, ApeKI and EcoP15I) tested here provides an excellent method to determine if high levels of methylation may exist in various sequence contexts, including the CTG•CAG tracts of various diseases or other loci. Such non-CpG methylation may correlate with the already detected presence of CpG methylation close to expanded repeat tracts.11,35–38 The use of EcoP15I would be helpful only for those CTG/CAG loci that have adjacent CTG sites in the opposite orientations within 3.5 kb, as previously noted for the HD35 or DM1 loci (www.ncbi.nlm.nih.gov/pubmed, DMPK GeneID: 1760). The approach may be particularly useful for methylation detection through long expanded repeat tracts, as observed in DM1 (>80–6,550 repeats), since methylation analysis by bisulfite and sequencing will only be applicable to those short repeat tracts that can be PCR amplified across (≤300–500 bp). Assessing GpC methylation in genomic samples by the restriction endonuclease(s) approach presented here can also be applied to specific restriction sites outside of repetitive tracts.
We report the sensitivity of various enzymes to non-CpG methylation, specifically assessing the effect of 5-methylcytosine at GpCpA, GpCpT, GpCpC or GpCpG sites, where methylation is executed by the GpC 5-cytosine methylase. In either the (CTG)n or (CAG)n repeats, a GpC site appears and the C that is methylated is followed by a T or an A. This is the case in the most frequent non-CpG sites recently reported to be methylated, CpT and CpA. In addition, these sites are located in the preferred CHG context.12 We also show that the methylation status of many plasmid-based non-CpG sites that are outside of the (CAG)n or (CTG)n repeat context can also be assessed (Fig. S1). The method we present reveals new properties of a series of restriction enzymes that now permits the analysis of a relatively newly recognized form of epigenetic DNA modification, non-CpG DNA methylation. This method is timely, as the biological significance of non-CpG methylation has only just become apparent.12 The enzymatic sensitivities that we reveal match the predominant non-CpG methylation marks reported by Lister et al. for the human genome.12 Specifically meCpA and meCpT, which are part of the trinucleotides (CAG)n and (CTG)n and the other non-repeat sites we assessed. A wide variety of sequences recognized by the series of restriction enzymes are sensitive to GpC methylation. This supports the potential utility of this method for the wide community of epigenetic researchers. We modestly suggest that commercial providers of these restriction enzymes include a comment on their sensitivity to non-CpG sites, as this information would be very valuable to many researchers studying these sequences. Now all labs, even those that have not set-up bisulfite sequencing, can rapidly and easily assess non-CpG methylation at a variety of sites using any of the enzymes that we have showed to be methyl-sensitive.
Materials and Methods
Plasmid pKH11 was constructed by cloning a 615 bp fragment of the DM1 locus containing 83 CTG repeats (plus 142 bp human flanking sequence upstream and 224 bp downstream), including the flanking CTCF binding sites from patient fetal fibroblasts in pCRScript-Amp (Stratagene). A 219 nt fragment containing the SV40 sequence (viral position 5210/5211) was cloned as a blunted XbaI fragment into the downstream blunted SapI site.
The (CAG)500•(CTG)500 or (CAG)800•(CTG)800 DNAs were created by in vitro synthesis using ϕ29 DNA polymerase-mediated rolling circle amplification, as previously described in reference 30.
All enzymes if not otherwise specified, are from New England Biolabs Inc., and reactions performed as described. Briefly, methylation DNAs at all 5′-GpC-3′ sites by the DNA 5-cytosine methyl-transferase M.CviPI, in 1x GC Reaction Buffer (50 mM Tris-HCl, 50 mM NaCl, 10 mM Dithiothreitol, pH 8.5 at 25°C, supplemented with 160 µM S-adenosylmethionine), as described in reference 29. As per Xu et al. we adopted the use of HaeIII to detect completion of GpmeC methylation; they and others have found that HaeIII cleaves GGCC but not GGmeCC sequences.31,32 Restriction digestions were performed as described. Briefly, ApeKI digests were in NEBuffer #3 at 75°C, BbvI digests were in NEBuffer #2 at 37°C, EcoP15I digests were in NEBuffer #3 at 37°C, Fnu4HI digests were in NEBuffer #4 at 37°C, MwoI digests were in NEBuffer #3 at 60°C and TseI digests were in NEBuffer #4 at 65°C.
Restriction digested DNAs were resolved on 1–2% agarose gels, in 1x TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA, pH 8.5) and electrophoresed under 75 V for ∼1 hour. Products were visualized with ethidium bromide staining and UV light.
C2C12 mouse muscle cell line was cultured in DMEM (Gibco) supplemented with 10% FBS, 100 units/ml penicillin and 100 mg/ml streptomycin. For differentiation, C2C12 cells were cultured for five days in DMEM (Gibco) supplemented with 1% FBS and the antibiotics. Genomic DNA was extracted from undifferentiated or differentiated C2C12 cells using the Gentra Puregene Cell Kit (Qiagen), and then digested by ApeKI or TseI. After heat inactivation or removal of enzymes, the myogenin (Myog) 5′ flanking region was amplified for 33 cycles using primers 5′-TGT TCC CTT CCT GCC CTG TC-3′ and 5′-AGG CCG TCG GCT GTA ATT TG-3′. As a standard for semi-quantitative analysis, Scn4a exon 6 region, which possesses no recognition sites for these enzymes, was also amplified for 30 cycles using primers 5′-CCA TGA ATG ACA CCA ACA CCA C-3′ and 5′-TCC CTT CGT CAT TGA TGT AGG C-3′. PCR products were analyzed on agarose gels followed by SYBR Green I (Invitrogen) staining using ImageQuant software (Molecular Dynamics).
Acknowledgements
This work was supported by the Canadian Institutes of Health Research (C.E.P.), the Muscular Dystrophy Association Canada (C.E.P.), the University of Rochester Paul Wellstone Muscular Dystrophy Cooperative Research Center with support from the NIH (grant U54NS48843, to C.A.T. & C.E.P.), a post-doctoral fellowship from The Hospital for Sick Children Research Training Centre (A.L.C.) and the Muscular Dystrophy Association and postdoctoral fellowships from the Cell Science Research Foundation and the Uehara Memorial Foundation (M.N.).
Supplementary Material
References
- 1.Gopalakrishnan S, Van Emburgh BO, Robertson KD. DNA methylation in development and human disease. Mutat Res. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 3.López Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 4.Winnepenninckx B, Debacker K, Ramsay J, Smeets D, Smits A, Fitz-Patrick DR, et al. CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13. Am J Hum Genet. 2007;80:221–231. doi: 10.1086/510800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumari D, Usdin K. Chromatin remodeling in the noncoding repeat expansion disease. J Biol Chem. 2009;284:7413–7417. doi: 10.1074/jbc.R800026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park CW, Chen Z, Kren BT, Steer CJ. Double-stranded siRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem Biophys Res Commun. 2004;323:275–280. doi: 10.1016/j.bbrc.2004.08.096. [DOI] [PubMed] [Google Scholar]
- 8.Theilmann JL, Robbins CA, Hayden MR. Methylation at the D4S95 locus and predictive testing. Am J Hum Genet. 1989;45:477–479. [PMC free article] [PubMed] [Google Scholar]
- 9.Pritchard CA, Cox DR, Myers RM. Methylation at the Huntington disease-linked D4S95 locus. Am J Hum Genet. 1989;45:335–336. [PMC free article] [PubMed] [Google Scholar]
- 10.Reik W, Maher ER, Morrison PJ, Harding AE, Simpson SA. Age at onset in Huntington's disease and methylation at D4S95. J Med Genet. 1993;30:185–188. doi: 10.1136/jmg.30.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López Castel A, Nakamori M, Tomé S, Chitayat D, Gourdon G, Thornton CA, et al. Expanded CTG repeat demarcates a boundary for abnormal CpG methylation in myotonic dystrophy patient tissues. Hum Mol Genet. 2010;20:1–15. doi: 10.1093/hmg/ddq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandjean V, Yaman R, Cuzin F, Rassoulzadegan M. Inheritance of an epigenetic mark: the CpG DNA methyltransferase 1 is required for de novo establishment of a complex pattern of non-CpG methylation. PLoS One. 2007;2:1136. doi: 10.1371/journal.pone.0001136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burden AF, Manley NC, Clark AD, Gartler SM, Laird CD, Hansen RS. Hemimethylation and non-CpG methylation levels in a promoter region of human LINE-1 (L1) repeated elements. J Biol Chem. 2005;280:14413–14419. doi: 10.1074/jbc.M413836200. [DOI] [PubMed] [Google Scholar]
- 15.Imamura T, Kerjean A, Heams T, Kupiec JJ, Thenevin C, Pàldi A. Dynamic CpG and non-CpG methylation of the Peg1/Mest gene in the mouse oocyte and preimplantation embryo. J Biol Chem. 2005;280:20171–20175. doi: 10.1074/jbc.M501749200. [DOI] [PubMed] [Google Scholar]
- 16.Inoue S, Oishi M. Effects of methylation of non-CpG sequence in the promoter region on the expression of human synaptotagmin XI (syt11) Gene. 2005;348:123–134. doi: 10.1016/j.gene.2004.12.044. [DOI] [PubMed] [Google Scholar]
- 17.Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation. Gene. 2002;289:41–48. doi: 10.1016/s0378-1119(02)00469-9. [DOI] [PubMed] [Google Scholar]
- 18.White GP, Watt PM, Holt BJ, Holt PG. Differential patterns of methylation of the IFN gamma promoter at CpG and non-CpG sites underlie differences in IFN gamma gene expression between human neonatal and adult CD45RO-T cells. J Immunol. 2002;168:2820–2827. doi: 10.4049/jimmunol.168.6.2820. [DOI] [PubMed] [Google Scholar]
- 19.Haines TR, Rodenhiser DI, Ainsworth PJ. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–598. doi: 10.1006/dbio.2001.0504. [DOI] [PubMed] [Google Scholar]
- 20.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodcock DM, Crowther PJ, Diver WP. The majority of methylated deoxycytidines in human DNA are not in the CpG dinucleotide. Biochem Biophys Res Commun. 1987;145:888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]
- 22.Aoki A, Suetake I, Miyagawa J, Fujio T, Chijiwa T, Sasaki H, et al. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 2001;29:3506–3512. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. J Mol Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Jang SJ, Benoit N, Hoque MO, Califano JA, Trink B, et al. Presence of 5-methylcytosine in CpNpG trinucleotides in the human genome. Genomics. 2010;96:67–72. doi: 10.1016/j.ygeno.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:234–236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramsahoye BH, Burnett AK, Taylor C. Restriction endonuclease isoschizomers ItaI, BsoFI and Fsp4HI are characterized by differences in their sensitivities to CpG methylation. Nucleic Acids Res. 1997;25:3196–3198. doi: 10.1093/nar/25.16.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen RS, Gartler SM, Scott CR, Chen SH, Laird CD. Methylation analysis of CGG sites in the CpG island of the human FMR1 gene. Hum Mol Genet. 1992;1:571–578. doi: 10.1093/hmg/1.8.571. [DOI] [PubMed] [Google Scholar]
- 28.Pietrobono R, Pomponi MG, Tabolacci E, Oostra B, Chiurazzi P, Neri G. Quantitative analysis of DNA demethylation and transcriptional reactivation of the FMR1 gene in fragile X cells treated with 5-azadeoxycytidine. Nucleic Acids Res. 2002;30:3278–3285. doi: 10.1093/nar/gkf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu M, Kladde MP, Van Etten JL, Simpson RT. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 1998;26:3961–3966. doi: 10.1093/nar/26.17.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osborne RJ, Thornton CA. Cell-free cloning of highly expanded CTG repeats by amplification of dimerized expanded repeats. Nucleic Acids Res. 2008;36:24. doi: 10.1093/nar/gkn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann MB, Smith HO. Specificity of HpaII and HaeIII DNA methylases. Nucleic Acids Res. 1977;4:4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Backman K. A cautionary note on the use of certain restriction endonucleases with methylated substrates. Gene. 1980;11:169–171. doi: 10.1016/0378-1119(80)90097-9. [DOI] [PubMed] [Google Scholar]
- 34.Fuso A, Ferraguti G, Grandoni F, Ruggeri R, Scarpa S, Strom R, et al. Early demethylation of non-CpG, CpC-rich, elements in the myogenin 5′-flanking region: a priming effect on the spreading of active demethylation. Cell Cycle. 2010;9:3965–3976. doi: 10.4161/cc.9.19.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Möncke-Buchner E, Reich S, Mücke M, Reuter M, Messer W, Wanker EE, et al. Counting CAG repeats in the Huntington's disease gene by restriction endonuclease EcoP15I cleavage. Nucleic Acids Res. 2002;30:83. doi: 10.1093/nar/gnf082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucleic Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 38.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.