Abstract
The coordinated expression of ribosomal protein genes (RPGs) has been well documented in many species. Previous analyses of RPG promoters focus only on Fungi and mammals. Recognizing this gap and using a comparative genomics approach, we utilize a motif-finding algorithm that incorporates cross-species conservation to identify several significant motifs in Drosophila RPG promoters. As a result, significant differences of the enriched motifs in RPG promoter are found among Drosophila, Fungi, and mammals, demonstrating the evolutionary dynamics of the ribosomal gene regulatory network. We also report a motif present in similar numbers of RPGs among Drosophila species which does not appear to be conserved at the individual RPG gene level. A module-wise stabilizing selection theory is proposed to explain this observation. Overall, our results provide significant insight into the fast-evolving nature of transcriptional regulation in the RPG module.
Keywords: Ribosomal protein, Evolution, Promoter, Motif
1. Introduction
Ribosomal proteins are the essential components of the translation machine and one of the most evolutionarily conserved gene groups (Zhang et al., 2002). For example, yeast and rat share all but one ribosomal protein, with an average of 60% protein sequence identity (Zhang et al., 2002), despite the divergence time between them exceeding 1 billion years (Nei et al., 2001). Between human and rat, the average protein sequence identity of ribosomal proteins is 99%, with 32 out of 72 known ribosomal proteins being identical (Wool, 1996). In addition, the coordinated expression of RPGs is a characteristic which leads to a roughly equimolecular accumulation of ribosomal proteins needed for ribosomal synthesis (Perry, 2005). Evidence suggests that similar promoter strength provides the basis for the equimolecular levels of ribosomal proteins through the coordinated transcription of RPGs (Meyuhas and Perry, 1980; Hariharan et al., 1989). In fact, the RPGs are significantly co-expressed at the mRNA level across many species (Stuart et al., 2003). Therefore, it is compelling to understand the mechanism governing the transcriptional regulation of RPGs and to investigate their evolution.
The change of transcriptional regulatory circuit has long been viewed as a major driving force of species evolution (Tautz, 2000; Ludwig, 2002; Wray et al., 2003), and has recently received much attention (Gasch et al., 2004; Ihmels et al., 2005). For example, the Brachyury gene is similarly expressed in the notochord precursor cells of two distantly related species, Halocynthia roretzi and Ciona intestinalis, but the regulatory modules and binding sites do not appear to be related (Takahashi et al., 1999). Consistent with these findings, the motifs found in Fungi RPG promoter regions (Tanay et al., 2005a) have no overlap with the motifs found in mammalian RPG promoter regions (Perry, 2005). In addition, a systematic study of the core promoter region in D. melanogaster showed that most DNA motifs identified are distinctive between Drosophila and human (FitzGerald et al., 2006). All these findings suggest that the cis-elements of gene expression regulatory networks evolve at a relatively high rate, which likely accounts for the phenotypic differences between species sharing a large fraction of genes (Li and Saunders, 2005). In order to further elucidate the evolution of transcriptional regulation network, it would be interesting to study the evolutionarily most conserved RPGs in well characterized species such as Drosophila.
In this paper, we study the evolution of RPG transcriptional regulation in Drosophila species, which lie between Fungi and mammal in evolution. We showed that there is only weak overlap of the RPG motifs among Fungi, Drosophila and mammal, suggesting rapid evolution of the cis-regulatory elements in the RPG gene group. Moreover, we found that RPG motifs evolve particularly fast even within the Drosophila species. We used a highly conserved palindrome motif DRE (FitzGerald et al., 2006) to further demonstrate this observation. The DRE motif appears to be frequently changing among individual Drosophila RPG promoters, while the total number of Drosophila RPGs containing instances of this motif is kept stable in all Drosophila species. This is an intriguing contradiction, as the conserved number of this motif implies a strong negative selection force, while frequent changes among individual RPG genes imply the absence of such a strong selection force. We resolved this inconsistency via postulating and validating a module-wise selection theory, which extends the traditional stabilizing selection theory to functional module.
2. Materials and methods
We downloaded RPG annotations of D. melanogaster from Ribosomal Protein Gene Database (RPGDB) (Nakao et al., 2004). However, RPGDB does not include the upstream data for other Drosophila species. Therefore, genomic data from the UCSC genome browser database (Kent et al., 2002) were also downloaded in addition to RPGDB. We manually extended the upstream sequence to a pre-specified distance (1 kb) from the annotated translation start site for the D. melanogaster RPGs. The aligned RPG genes of 5 other Drosophila species (D. yakuba, D. ananassae, D. pseudoobscura, D. mojavensis and D. virilis) were also downloaded from the UCSC genome browser. We manually inspected the alignment of these Drosophila species with D. melanogaster and corrected the apparent misannotation of translation start codon ATG, which results from the existence of short first exons (the upstream 1 kb sequences for each RPG of all 6 Drosophila species are provided as a supplementary file). The DNA sequence of the transcription factor DREF is also downloaded from the UCSC genome browser database.
Our objective is to find sequence motifs that are significantly conserved in RPG promoter regions across the Drosophila species. Conservation of a motif across species increases the likelihood of it being a functional site. We use a previously developed method, the cross-species conservation (CSC) method (Li et al., 2005), which jointly utilizes the motif over-representation and conservation property to find the motifs. This method differs from the other motif discovery methods in that it first identifies motifs by over-representation and then models the motifs in the context of the evolutionary divergence of neutral and functional sequences to evaluate their significance. We briefly describe the method below, and the details can be found in (Li et al., 2005).
2.1. Motif discovery by over-representation
For the upstream 1 kb sequences of RPGs obtained above in every species, RepeatMasker (Smit et al., 1996–2004) is used to mask the repeats in these sequences. We then use MEME (Baileyand Elkan,1994) to search for over-represented motifs. MEME uses the expectation maximization (EM) algorithm to detect motifs that have enriched instances in the input sequence sets compared with the genomic background. The current implementation of MEME enumerates all potential motif seeds, thus minimizing the drawback of missing motifs due to computational limitations. Parameters are set accordingly so that MEME will report motifs of 6 to 14 bp in length and an E-value statistic of <1E8. To overcome the effect of parameters, we tried several parameters (E-value 1E12, motif length 5 to 15, 8 to 16) and obtained similar motif results.
2.2. Assess the significance of motif conservation across species
Functional sequences tend to be more conserved than nonfunctional sequences. The key component of the CSC method (Li et al., 2005) is the calculation of the P value of the conservation of a motif. (1) It first models the evolutionarily neutral sequences by alignment. The background nucleotide distribution of the ancestral species and the speciation time can be estimated from the alignment. A 4×4 substitution matrix representing the neutral evolution is obtained subsequently. (2) The CSC method then identifies motifs that are significantly conserved in at least two species. An ancestral motif position specific weight matrix, as well as a threshold of this weight matrix, is constructed for each motif between two or more species. (3) The significance of conservation (a P value) of each conserved motif is then calculated as a function of the phylogenetic tree, the 4×4 substitution matrix, the ancestral motif position specific weight matrix and the corresponding threshold. A P value <1E19, the empirical cutoff, is considered to be significant, and the corresponding motif will be reported.
3. Results
3.1. Comparing RPG promoter motifs in Fungi, Drosophila and mammals
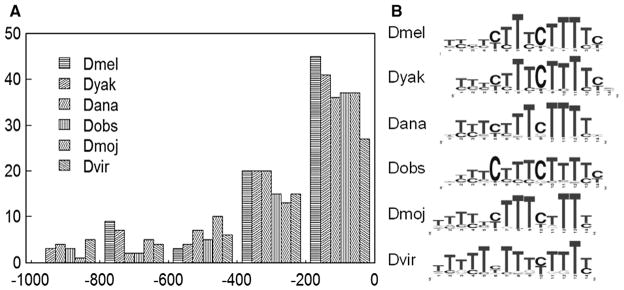
Using the RPG core promoter regions (1 kb upstream to translation start site) of Drosophila species, we identified 5 significant motifs that are conserved in at least four Drosophila species with the principle of over-representation and cross-species conservation (Li et al., 2005) (see Table 1; the position specific weight matrices are in a supplementary file). With the exception of the TOP (Terminal OligoPyrimidine) motif, the other 4 motifs we identified are also present in (Ohler et al., 2002), where a subset of the D. melanogaster genes is used. Moreover, the DRE motif has been identified via a systematic study of the D. melanogaster promoter sequence (FitzGerald et al., 2006). The TOP motif is known to be very close to the transcription start point of all mammalian RPGs (Perry, 2005), suggesting a shared transcription initiation mechanism of RPGs between Drosophila and mammals. We also find that the TOP sequence is rather close to the translation start codon ATG, suggesting a short five prime untranslated region (5′ UTR) (Fig. 1). It has been observed that an intron often separates the 5′ UTR region from the protein coding region of mammalian RPGs (Perry, 2005), also known as “intron appending.” In support of Perry’s theory that intron appending happened in early eukaryotes (Perry, 2005), this phenomenon is also found in Drosophila species (see Supplementary materials S1).
Table 1.
Motifs conserved in the RPG promoters across at least four Drosophila species found by the CSC method
| Motif | Name | CSC P value | Logo* | Fungi RPG? | Human RPG? |
|---|---|---|---|---|---|
| 1 | TOP | 1e-39 |

|
No [Tanay] | Yes [Perry] |
| 2 | DRE | 1e-137 |

|
No [Tanay] | Yes [Yamashita] |
| 3 | NFI | 1e-36 |

|
No [Tanay] | No [Perry] |
| 4 | Homol-D | 1e-36 |
|
Yes [Tanay] | No [Perry] |
| 5 | - | 1e-29 |

|
No [Perry] | No [Perry] |
Significant motifs are arranged according to the cross-species conservation score (CSC P value).
In columns 5 and 6, three papers (Perry, 2005; Tanay et al., 2005b; Yamashita et al., 2007) are abbreviated as “Perry”, “Tanay” and “Yamashita” respectively.
Sequence logo is based on the motif-finding result of MEME in D. melanogaster.
Fig. 1.
The TOP sequences. TOP sequences are close to the translation start codon of ribosomal protein genes across all Drosophila species (Panel A). Here y-axis represents the number of RPG genes having TOP motif at a given distance from the translation start codon as shown in x-axis. For each ribosomal protein gene, its TOP motif is defined as the best matching sequence in the upstream 1 kb region using the estimated position weight matrix in corresponding species, which is shown in panel B.
Once we have identified these five significant Drosophila motifs, we next hoped to find commonalities of the cis-regulatory motifs of RPG promoters among mammals, Fungi, and Drosophila, an intermediate species between the other two. As it turned out, the Drosophila RPG promoter motifs are not well conserved in Fungi and human (see Table 1). For example, the Homol-D motif is observed in the RPG promoters of both Fungi (Tanay et al., 2005a) and Drosophila species, while it is not observed in the RPG promoters of mammals (Perry, 2005). On the other hand, some motifs reported in mammals, including GABP, SP1, and YY1, are not observed in Drosophila species. This absence of conserved DNA motifs among Fungi, Drosophila and mammals demonstrates that the regulatory network of RPG modules evolves very fast and such a fast rewiring of the regulatory circuit may be the driving force of species evolution (FitzGerald et al., 2006).
DNA motifs often function together as a cis-element module to control the expression of the corresponding genes. We therefore wanted to test if there are any cis-element modules in Drosophila RPG promoters. Fisher’s exact test was used to detect dependency of occurrences between motif pairs among RPG promoters in each Drosophila species. However, we did not detect any significantly associated motif pairs between the highly conserved cis-elements across all Drosophila species (data not shown). It is also interesting to note that no dependency between motifs is found in mammals (Fisher’s exact test), as indicated by Fig. 6 in Perry, 2005. Thus, on the one hand, we are inclined to suggest that the independent appearance of regulatory motifs in RPGs may actually enable the expression of some RPGs such that they can be regulated differentially under specific genetic/environmental conditions. Such a differentially regulated expression could enable some RPGs to have functions other than protein biosynthesis, a subject which is reviewed by Wool, 1996. On the other hand, the coordinated expression of RPGs predicts a common transcriptional regulator of the DRE, NF-I and Homol-D genes. Since no obvious motif modules are detected in either Drosophila or mammals, we restrict our investigation to individual motifs at the gene level in the following.
3.2. The DRE motif
We next investigate the conservation of motifs at the individual gene level across Drosophila RPGs. There is, however, an obstacle associated with carrying out this type of investigation. Specifically, the study of motifs at the level of individual genes is hindered by the lack of experimentally verified binding data across Drosophila species for the same genes. Fortunately, we were able to identify the motif termed DNA-replication related element (DRE) (FitzGerald et al., 2006), which is so conserved that the consensus sequence TATCGATA is sufficient to accurately identify its position weight matrix. This enables us to conduct a detailed analysis of its evolution at the individual gene level. To accomplish this, we first demonstrate the likelihood that DRE is a regulatory motif of Drosophila RPG promoters by studying its positional conservation in RPG promoters and the conservation of its cognate transcription factor DREF. We then study the distribution of DRE motif across Drosophila RPG promoters and demonstrate a new selection scenario of the Drosophila RPGs.
3.3. Positional conservation of DRE motif in Drosophila RPG promoters
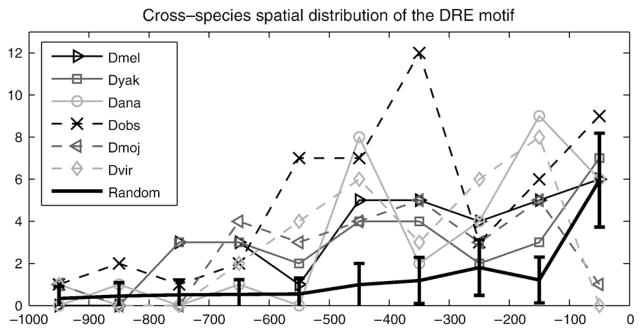
Strong positional conservation of a motif provides another level of evidence that it is functional. We therefore asked where the DRE motif localizes in the promoters of Drosophila RPGs. We scanned the promoter sequences using the derived position weight matrix. In Fig. 2, the numbers of RPG promoters harboring DRE in a given length of promoter region are depicted for all Drosophila species. In fact, Drosophila RPG promoters harbor more DRE motif than the D. melanogaster genomic background. To demonstrate this, we bootstrapped 1000 datasets, each consisting of 86 (the same number as Drosophila RPGs) D. melanogaster gene promoters, which are defined as the 1 kb upstream sequences (15,646 genes in total; obtained from UCSC genome browser database). We obtained the mean and the standard deviation of the numbers of promoters in these synthesized datasets harboring motif DRE as null distribution, which is illustrated by the error bar. It can be observed that there is a significant accumulation of RPG promoters harboring DRE in the upstream 600 base pair region. However, once the region extends upstream of 600 base pairs, this accumulation slows down dramatically. Although there is also an increase in the null distribution, it can be seen that the DRE is significantly more enriched in the upstream 1 kb regions of RPG promoters than in the null distribution. Together with the conclusion in (FitzGerald et al., 2006), this enrichment implies that the DRE motif we identified is likely a functional motif of Drosophila RPGs.
Fig. 2.
Cross-species spatial distribution of the DRE motif. DRE motifs are close to the translation start codon of ribosomal protein genes across all Drosophila species. Shown in y-axis is the number of RPGs harboring DRE in the promoter region with corresponding distance from translation start codon ATG shown in x-axis. The error bar indicated by “Random” corresponds to the null distribution of DRE in the promoter regions of all D. melanogaster genes (1000 bootstrap samples, each with 86 genes).
3.4. DREF is conserved across Drosophila species
The conservation of the DNA-binding domain of a transcription factor is a strong indicator that the corresponding binding motif is well conserved across species. Therefore, based on this finding, we next study how the DNA-binding domain of DREF, which binds DRE, is conserved across Drosophila species. DREF has been shown to bind to the palindrome TATCGATA for transcriptional regulation (Hirose et al., 1993). We retrieved the DREF protein sequences of all Drosophila species from the UCSC genome browser and aligned them (provided as a supplementary file). Pair-wise amino acid similarity ranges from 73% to 96%, with the lowest score of 73% between D. melanogaster and D. virilis, two of the most distantly related Drosophila species. It is also known that the DNA binding domain of DREF lies within its N-terminal (16–115 amino acids) (Kuge et al., 1997). We found that the DNA binding domain is almost exactly the same among all Drosophila species, as shown in Fig. 3. This suggests that the transcription factor DREF is subject to strong negative selection across Drosophila species and that the DRE motif we identified is, most likely, a potential regulatory motif of RPG promoters across Drosophila species.
Fig. 3.
DNA binding domain of the transcription factor DREF. The DREF DNA binding domain is conserved across Drosophila species. DNA binding domain is aligned in CLUSTALW (Higgins et al., 1994).
3.5. Contradiction of DRE motif appearance at individual gene level and module level
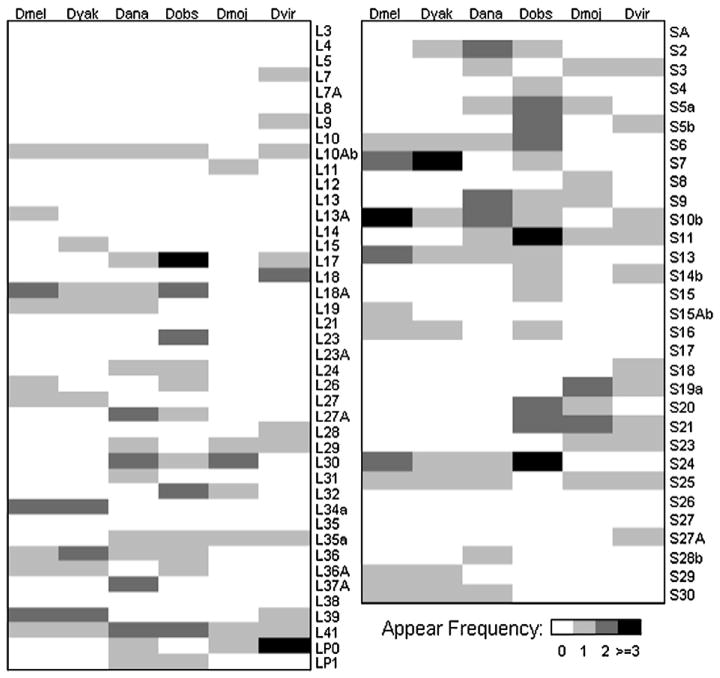
The positional conservation of DRE in Drosophila RPG promoters and the conservation of the DNA-binding domain of DREF across Drosophila species suggest that strong selection pressure is operating on the DRE binding site. This leads to the next logical step: studying the conservation of the DRE motif instances in every individual Drosophila RPG promoter. The Drosophila RPG promoters harboring DRE are depicted in Fig. 4, where, for illustrative purposes, large subunit proteins are shown in the left panel and small subunit proteins are shown in the right panel. Although the presence of the DRE motif is well conserved at the individual gene level among evolutionarily close species, frequent changes (gain and loss) of the DRE motifs at individual gene level are, conversely, observed among distant Drosophila species, e.g., D. melanogaster and D. pseudoobscura.
Fig. 4.
Occurrences of DRE in the Drosophila RPG core promoters (the upstream 1 kb region of translation start codon ATG). Proteins of the large subunit are in the left panel and those of small subunit are in the right panel. Color schemes are illustrated in bottom right.
A quantitative measure of this frequent change at the individual gene level is to check the dependency of DRE appearance in RPG promoters between Drosophila species, as follows. Specifically, we denote the number of RPGs with DRE in both species A and species B as a, the number of RPGs without DRE in species A and with DRE in species B as b, the number of RPGs with DRE in species A and without DRE in species B as c and the number of RPGs without DRE in both species A and species B as d. The dependency of DRE appearance in RPG promoters between Drosophila species is calculated using Fisher’s exact test on how numbers a, b, c, and d deviate from their expected values if species A and B are not related. A lack of significant dependency would imply frequent changes of the DRE motifs at individual gene level. As shown in Table 2, the dependency of appearance of DRE is not found for distant drosophila species, such as D. melanogaster and D. virilis. This result suggests that there are considerable turnover and gain events of DRE motif occurrence at the individual RPG gene level during the evolution of Drosophila species. Such a high rate of turnover and gain events of DRE motif implies a neutral role of DRE motif at individual RPG level during the evolution of Drosophila species.
Table 2.
Dependency of the appearance of DRE in RPGs between different Drosophila species
| Dyak | Dana | Dobs | Dmoj | Dvir | |
|---|---|---|---|---|---|
| Dmel | 4.13E-12 | 0.110033 | 0.114414 | 0.124786 | 0.57743 |
| Dyak | 0.016126 | 0.062177 | 0.127666 | 0.775589 | |
| Dana | 0.001174 | 0.045782 | 0.429024 | ||
| Dobs | 0.275559 | 1.00000 | |||
| Dmoj | 0.005883 |
P-values of Fisher’s exact test are listed.
Despite the seemingly neutral role of DRE motif at individual RPG gene level, it appears that the total numbers of promoters in the RPG module (i.e., the ribosomal protein genes as a whole) harboring DRE are roughly similar among Drosophila species as well as human (Yamashita et al., 2007), as can be seen from Table 3. We thus test the null hypothesis that the total numbers of RPG promoters with DRE motif are more similar to each other among observed Drosophila species than the expected numbers under the neutral evolutionary process. To test this hypothesis, we used the coefficient of variation (cv) as our statistic. Specifically, we denote the total number of RPG promoters with at least one DRE motif as Xi, with i =1, 2, …,6 for each Drosophila species. The coefficient of variation statistic is defined as cv (X)=std(X)/mean(X), where mean(X) and std(X) are the mean and standard deviation of vector X̄ = {Xi i = 1, 2, …, 6}. The null distribution of cv is obtained by simulating the neutral evolution process as follows. We first reconstructed (Yang, 1997) the common ancestral sequence of each RPG promoter sequence of these six Drosophila species, using Mosquito RPG promoter sequences as an out-group control. The evolutionary process was simulated 1000 times according to the established phylogenetic tree and evolutionary distance of these six Drosophila species (obtained from the RPG promoters) to estimate the null distribution of cv. As it turned out, the observed cv (=0.24) is significantly (P value=0.011) smaller than that under neutral process for DRE (defined simply as an 8-mer TATCGATA; see supplementary file for the null distribution of cv under neutral process). We note that the result is still significant (P value=0.033) when using the position weight matrix of DRE in (Ohler et al., 2002) with threshold 1200. Therefore, we conclude that the total numbers of promoters in RPG module harboring DRE in each species are kept similar among Drosophila species under strong selection pressure.
Table 3.
Numbers of RPG promoters with DRE are similar among Drosophila species
| Dmel | Dyak | Dana | Dobs | Dmoj | Dvir | Human* | |
|---|---|---|---|---|---|---|---|
| #RPGs annotated | 86 | 85 | 82 | 79 | 81 | 79 | 79 |
| Gene-level | 21 | 20 | 27 | 30 | 17 | 22 | 37 |
| Total DRE | 30 | 25 | 34 | 45 | 20 | 25 | 47 |
Total numbers of RPG promoters (upstream 1 kb) harboring DRE in each species are listed. For the sake of completeness, total numbers of DRE in the RPG promoters in each species (i.e., the DRE are counted more than once in some promoter sequences) are also listed as “Total DRE.”
: from (Yamashita et al., 2007).
Thus, our result suggests that the DRE motif is subject to strong selection pressure at the RPG module level, while it is not subject to selection pressure at the individual gene level. Such a surprising contradiction by looking the DRE motif at different level (i.e., individual gene and gene module) suggests the need to extend the traditional stabilizing selection theory at individual gene level (Ludwig, 2002) to gene module level, as detailed below.
3.6. DRE motif is under module-wise stabilizing selection
Frequent change of DRE motif appearance at the individual Drosophila RPG promoter regions along the evolutionary process would seem to contradict the strong selection pressure exerted on both DRE and DREF. However, we reached the conclusion that the total numbers of RPG promoters harboring DRE in each species are kept similar among Drosophila species. While this finding implies that the DRE element is under strong selection pressure, the selection force is actually operating on a functional module level where the RPGs are considered as a whole group. Since RPGs are functionally homogeneous, it would be reasonable to theorize that strong selection pressure is exerted on the functional module instead of individual RPGs. We will call this module-wise stabilizing selection, which would naturally result in roughly similar numbers of RPG promoters harboring DRE across Drosophila species (Table 3). On the other hand, stabilization of the total number of RPGs with a DRE-containing promoter may also be explained by rapid shuffling of promoter sequences during, or right after, speciation followed by extensive concerted evolution (Arnheim, 1983). However, no evidence suggests that extensive concerted evolution is present in RPGs. In fact, high variability of the length of the ribosomal protein genes in D. melanogaster eliminated the possibility of concerted evolution actively operating in their promoter sequences (data not shown). Therefore, module-wise stabilizing selection is most likely the driving force in shaping the DRE motif instance pattern among Drosophila RPGs.
4. Discussion
It has long been proposed that the evolution of regulatory interactions is a major driving force of species evolution. Results in this paper and others (Perry, 2005; Tanay et al., 2005a) consistently indicate that the regulatory circuit of RPGs evolves faster than the corresponding genes, which are highly conserved not only in amino acid sequences, but also in the coordinated expressions. The dramatic changes of the RPG regulatory circuit hint the rewiring of other more complex regulatory modules. Within Drosophila species, the appearance of DRE motif, which is subject to strong selection pressure at the RPG module, is also fast evolving. Overall, therefore, our results provide significant insight into the fast-evolving nature of transcriptional regulation in the RPG module.
One of our key results indicated that module-wise stabilizing selection is most likely the driving force in shaping the DRE motif instance pattern among Drosophila RPGs. Following this logic, the module-wise stabilizing selection would have significant impact on the traditional motif-finding strategy of comparative genomics, where promoters of homologous genes are analyzed to search for potential motifs. One such strategy, phylogenetic footprinting, assumes that the functional motifs are kept intact in the orthologous sequences during evolution. However, as we have shown in the Drosophila RPG module, the motif may only be conserved across species at the gene-module level, rather than at the orthologous individual gene level. Thus, a comparative genomics search over individual homologous genes may miss functional motifs. Based on our observation, we propose that a successful search should take both evolutionary conservation information and gene family information into consideration.
Acknowledgments
The authors would like to thank Chao Cheng and Matthew Lebo for their help and suggestions on the manuscript. X.M is supported by NIH/NSF Joint Mathematical Biology Initiative DMS-0241102 and by NIH P50 HG 002790 to Fengzhu Sun. K.Z. is supported by NIH grant 1R01GM074163 to X. Jasmine Zhou. X.L. is supported in part by the Showalter Award and the NIH grant R01HG004359.
Abbreviations
- ATG
adenosine thymidine guanosine
- CSC
cross-species conservation
- cv
coefficient of variation
- Dana
D. ananassae
- Dmel
D. melanogaster
- Dmoj
D. mojavensis
- Dobs
D. pseudoobscura
- Dvir
D.virilis
- Dyak
D. yakuba
- DRE
DNA-replication related element
- DREF
transcription factor related to DRE motif
- MEME
multiple EM for motif elicitation
- RPG
ribosomal protein genes
- RPGDB
ribosomal protein gene database
- TOP
Terminal OligoPyrimidine
- UTR
untranslated region
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.gene.2008.10.025.
References
- Arnheim N. Concerted evolution of multigene families. In: Nei M, Koehn R, editors. Evolution of Genes and Proteins. MA, Sunderland: 1983. pp. 38–61. [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- FitzGerald P, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7 (7):R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. Conservation and evolution of cis-regulatory systems in ascomycete fungi. PLoS Biol. 2004;2:e398. doi: 10.1371/journal.pbio.0020398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N, Kelley DE, Perry RP. Equipotent mouse ribosomal protein promoters have a similar architecture that includes internal sequence elements. Genes Dev. 1989;3:1789–1800. doi: 10.1101/gad.3.11.1789. [DOI] [PubMed] [Google Scholar]
- Higgins D, Thompson J, Gibson T, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose F, Yamaguchi M, Handa H, Inomata Y, Matsukage A. Novel 8-base pair sequence (Drosophila DNA replication-related element) and specific binding factor involved in the expression of Drosophila genes for DNA polymerase alpha and proliferating cell nuclear antigen. J Biol Chem. 1993;268:2092–2099. [PubMed] [Google Scholar]
- Ihmels J, et al. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge M, Fujii Y, Shimizu T, Hirose F, Matsukage A, Hakoshima T. Use of a fusion protein to obtain crystals suitable for X-ray analysis: crystallization of a GST-fused protein containing the DNA-binding domain of DNA replication-related element-binding factor, DREF. Protein Sci. 1997;6:1783–1786. doi: 10.1002/pro.5560060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Saunders MA. The chimpanzee and us. Nature. 2005;437:50–51. doi: 10.1038/437050a. [DOI] [PubMed] [Google Scholar]
- Li X, Zhong S, Wong WH. Reliable prediction of transcription factor binding sites by phylogenetic verification. Proc Natl Acad Sci U S A. 2005;102:16945–16950. doi: 10.1073/pnas.0504201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ. Functional evolution of noncoding DNA. Curr Opin Genet Dev. 2002;12:634–639. doi: 10.1016/s0959-437x(02)00355-6. [DOI] [PubMed] [Google Scholar]
- Meyuhas O, Perry RP. Construction and identification of cDNA clones for mouse ribosomal proteins: application for the study of r-protein gene expression. Gene. 1980;10:113–129. doi: 10.1016/0378-1119(80)90129-8. [DOI] [PubMed] [Google Scholar]
- Nakao A, Yoshihama M, Kenmochi N. RPG: the Ribosomal Protein Gene database. Nucleic Acids Res. 2004;32:168–170. doi: 10.1093/nar/gkh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Xu P, Glazko G. Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc Natl Acad Sci U S A. 2001;98:2497–2502. doi: 10.1073/pnas.051611498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Liao G-c, Niemann H, Rubin G. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3(12):research0087.0081–0087.0012. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. The architecture of mammalian ribosomal protein promoters. BMC Evol Biol. 2005;13(15) doi: 10.1186/1471-2148-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004. [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Mitani Y, Satoh G, Satoh N. Evolutionary alterations of the minimal promoter for notochord-specific Brachyury expression in ascidian embryos. Development. 1999;126:3725–3734. doi: 10.1242/dev.126.17.3725. [DOI] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci U S A. 2005a;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Regev A, Shamir R. Conservation and evolvability in regulatory networks: the evolution of ribosomal regulation in yeast. Proc Natl Acad Sci U S A. 2005b;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10:575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]
- Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21:164–5. [PubMed] [Google Scholar]
- Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Yamashita D, et al. hDREF regulates cell proliferation and expression of ribosomal protein genes. Mol Cell Biol. 2007;27 (6):2003–2013. doi: 10.1128/MCB.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl BioSciences. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Harrison P, Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–1482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]