Abstract
Motivation: High-throughput sequencing (HTS) has revolutionized gene regulation studies and is now fundamental for the detection of protein–DNA and protein–RNA binding, as well as for measuring RNA expression. With increasing variety and sequencing depth of HTS datasets, the need for more flexible and memory-efficient tools to analyse them is growing.
Results: We describe Pyicos, a powerful toolkit for the analysis of mapped reads from diverse HTS experiments: ChIP-Seq, either punctuated or broad signals, CLIP-Seq and RNA-Seq. We prove the effectiveness of Pyicos to select for significant signals and show that its accuracy is comparable and sometimes superior to that of methods specifically designed for each particular type of experiment. Pyicos facilitates the analysis of a variety of HTS datatypes through its flexibility and memory efficiency, providing a useful framework for data integration into models of regulatory genomics.
Availability: Open-source software, with tutorials and protocol files, is available at http://regulatorygenomics.upf.edu/pyicos or as a Galaxy server at http://regulatorygenomics.upf.edu/galaxy
Contact: eduardo.eyras@upf.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Gene expression is regulated through a complex network of protein interactions with RNA and DNA. Recent advances in high-throughput sequencing (HTS) technologies provide a very effective way to obtain information about these interactions at genome-wide level at reasonable cost. ChIP-Seq (chromatin immunoprecipitation followed by HTS) has become the preferred method for current analysis of transcriptional regulation in vivo, since it provides genome-wide coverage at a high sensitivity and signal-to-noise ratio (Johnson et al., 2007; Robertson et al., 2007). However, to unravel the complete gene expression network, it is also necessary to systematically identify protein–RNA interactions in cells. HITS-CLIP or CLIP-Seq (cross-linking immunoprecipitation followed by HTS) provides a powerful mean to obtain global information about direct protein–RNA interactions in cells (Licatalosi et al., 2008; Xue et al., 2009). Additionally, the RNA-Seq experiments provide a way to measure RNA levels at a higher sensitivity and coverage than microarray experiments. Despite the inherent biases, RNA-Seq can overcome some of the array limitations and provides the possibility to measure alternative splicing events (Pan et al., 2008; Wang et al., 2008a) or expression levels (Pepke et al., 2009; Trapnell et al., 2010), to detect single nucleotide polymorphisms (Wang et al., 2008b) and to discover novel genes (Khalil et al., 2009). Short-read sequencing platforms such as Illumina are very suitable for the study of protein–DNA and protein–RNA interactions, since they can produce enough data to perform accurate quantitative analysis. Furthermore, the resulting reads are of sufficient length to be mapped accurately to a reference sequence, but short enough to define the interaction sites with enough precision. However, not all interactions can be detected at the same coverage. For instance, transcription factors produce generally a clearly localized, or punctuated, signal (Park, 2009), whereas histone marks or RNA polymerase II (RNAPII) produce broad signals, covering more extended regions (Park, 2009). Most of the tools published for the analysis of ChIP-Seq data focus on punctuated signals (Fejes et al., 2008; Nix et al., 2008; Park, 2009; Pepke et al., 2009; Zhang et al., 2008), with only few methods devoted to analyse broad signals (Shin et al., 2009; Zang et al., 2009). However, these methods are not readily applicable to other HTS datatypes, like CLIP-Seq or RNA-Seq, making it necessary to incorporate additional tools to perform gene regulation studies, which generally involve multiple HTS experiments. The increase in variety and sequencing depth of such experiments, as well as the growing need for memory-efficient tools that can be applied to various datatypes, motivated us to develop Pyicos.
In this article, we describe Pyicos, a versatile toolkit operating on short HTS reads that have already been mapped to a reference coordinate system, like a genome or a transcriptome. We demonstrate its accuracy on punctuated ChIP-Seq data comparing it to MACS (Zhang et al., 2008), FindPeaks (Fejes et al., 2008) and USeq (Nix et al., 2008), using published ChIP-Seq datasets. We also describe the accuracy of Pyicos enrichment analysis (EA) to detect differentially expressed (DE) genes from RNA-Seq data by comparing it with the Bioconductor packages DEGseq (Wang et al., 2010), DESeq (Anders and Huber, 2010) and edgeR (Robinson et al., 2010) using published RNA-Seq data and microarray experiments on the same samples. Furthermore, we illustrate the flexibility of Pyicos by showing its effectiveness on other HTS datatypes: broad ChIP-Seq signals and CLIP-Seq data. Finally, we also discuss the performance of Pyicos in terms of memory usage and CPU time.
Pyicos provides different protocols to define significant signals from ChIP-Seq (both punctuated and broad), CLIP-Seq and RNA-Seq data; and all operations can be applied independently or combined in a protocol file, which makes Pyicos useful for the integrative analysis of a wide variety of HTS data. Moreover, the software has a modular architecture that allows re-usability in other Python applications. Pyicos thus offers a useful framework to facilitate the analysis of a variety of HTS datatypes, providing the basis for data integration into gene regulation studies.
2 METHODS
Pyicos is written in Python and is available under the GNU General Public License. It can operate from the command line or as a software library within Python programs (tested on GNU/Linux Fedora 12 and MAC OSX). All operations can be used independently and the parameters can be set by the user; thus providing as much flexibility as possible. Additionally, Pyicos can be run using protocols files (Supplementary Fig. S1 and Supplementary Material), which allows the user to apply them directly for standard analyses or to design customized workflows.
Pyicos has its own compressed format for peaks as explained in the Supplementary Material. This is also used as an internal representation for read clusters, and can be used as input and output formats. Furthermore, Pyicos can handle other various formats, providing converter capabilities. It can read eland, SAM, BED and wiggle (fixed and variable step); and write to SAM, BED and wiggle (fixed and variable step). Some formats are obligatory for some of the operations, which are specified in the corresponding command help.
2.1 Callpeaks protocol
The callpeaks protocol is applied to punctuated ChIP-Seq data and starts by optionally removing duplicated reads, i.e. redundant reads in terms of position and strand in a genome. Next, all reads that fall into a set of regions specified by the user, such as satellites and pericentromeric regions, are removed, as they generally lead to false positives (FPs). Reads are extended to the expected initial fragment size, given as input. Alternatively, Pyicos can also estimate this extension using a shift correlation analysis (Zhang et al., 2008). Next, reads are grouped into read clusters according to genomic overlap. To follow standard nomenclature, for punctuated data we will refer to read clusters as peaks. When a control experiment is available, callpeaks normalizes the sample with respect to control and subtracts the profile of the control from that of the sample with nucleotide resolution (Supplementary Material). At this stage, callpeaks removes two possible artefacts: peaks that are shorter than a certain length (100 nt by default), which may be produced by the subtraction step; and block-like peaks, which may occur when duplicates are kept. This removal is optional to the user. Pyicos also includes a split operation, which allows exploring the possibility that some peaks may describe multiple binding sites. Finally, callpeaks calculates the significance for a peak to correspond to a real binding site using Poisson analysis applied to three different parameters: the peak height, the number of reads per peak and the number of reads per nucleotide. For each chromosome, a P-value is calculated independently for each of these parameters. We also define a peak score as a combination of the Poisson P-value and the peak parameter P: peak score=[P − log10(P-value)]/2, for P equal to either the peak height or the peak read count.
2.2 Benchmarking of callpeaks
We compared Pyicos with MACS (Zhang et al., 2008) version 1.3.7.1, FindPeaks (Fejes et al., 2008) version 3.3 and USeq (Nix et al., 2008) version 7.2, using ChIP-Seq datasets for four different factors: the insulator (CTCF) in K562 cells (ENCODE Consortium, 2011), the CCAAT/enhancer-binding protein alpha (CEBPA) in liver cells (Schmidt et al., 2010), the neuron-restrictive silencer factor (NRSF) in Jurkat T cells (Johnson, Mortazavi, et al., 2007) and the progesterone receptor (PR) in T47D cells (Vicent et al., 2011). We have used various measures to evaluate the quality of peak definition on the punctuated ChIP-Seq data. Two of the descriptors of the peak definition used are the peak length and the distance between the peak centre and summit, i.e. the position with the highest read pileup. We further compared the rankings of the peaks provided by each method: the peak score (Pyicos), the peak height (FindPeaks), the value of −10·log10(P-value) (MACS) and the ranking provided by USeq.
For the accuracy evaluation, we also used the 83 positive and 30 negative regions validated by ChIP-qPCR for NRSF (Mortazavi et al., 2006) as done in Johnson et al. (2007). Since peaks differ a lot in their extensions (Supplementary Fig. S2), we took a region of 100 nt centred on the summit as the predicted binding region. We classified the peaks from each method as true positives (TPs) and FPs depending on whether the 100 nt long region centred on the summit overlapped a positive or negative validated region, respectively. The true positive rate (TPR) was calculated as TP over the total number of positive regions and the false positive rate (FPR) was calculated as FP over the total number of negative regions. Receiver operating characteristic (ROC) curves were calculated using incremental subsets of peaks along the ranked peaks from each method, with increasing subset sizes of 500 peaks.
An additional measure of the quality of the predictions is the fraction of peaks associated to the expected motif (Ji et al., 2008; Zhang et al., 2008). We thus measured the fraction of peaks with motif occurrence in the sequence underlying the summit, considering 100 nt for NRSF and PR peaks and 200 nt for CEBPA and CTCF peaks. For the motif analysis, we scanned the selected sequences from the peaks using the matrices for NRSF, CTCF, CEBPA and the three available matrices for PR from TRANSFAC (Knüppel et al., 1994) (accession numbers: M00256, M01200, M00116, M00954, M00957, M00960) using a custom script. We used as core similarity cut-off 0.99 and as total similarity cut-off 0.85. As the summit of a peak is associated more strongly with the binding motif than the peak centre (Supplementary Fig. S3), we considered it as a suitable reference position. Additionally, we used as fourth descriptor the spatial resolution, i.e. the distance from the summit position to the centre of the detected motif.
Finally, for all four ChIP-Seq datasets, we considered the Pyicos rankings using both the peak height and read count scores. For further analyses, we selected the score that gives the best motif content in top-scoring peaks: the peak height score for PR and CEBPA, and the read count score for CTCF and NRSF.
2.3 Enrichment protocol
Pyicos incorporates a method to calculate the significance of the enrichment of the signal between two samples based on the comparison with the distribution of enrichment values on the same regions for experimental or theoretical replicas, similarly to the methods MATR and MARS from DEGseq (Wang et al., 2010). Using subsets of ∼5% of neighbouring data points along the axis of average densities A, the enrichment values M calculated for genes between RNA-Seq samples for two liver replicas from Marioni et al. (2008) follow a normal distribution (Supplementary Fig. S4). Accordingly, Pyicos uses a sliding window along the A axis and calculates a Z-score for the enrichment M of each region from the comparison of the replicas (details in Supplementary Material). The region is assigned to a background window according to proximity: the region is compared with the values of the window which centre is closest to the region in terms of the value of A. If no replicas are provided, theoretical replicas are created by randomly rearranging the reads into two subsets, taking into account the relative sizes of the original samples. We integrated the TMM normalization (Robinson and Oshlack, 2010) in Pyicos EA. The calculation of the TMM factor on our read count lists yielded a value of 0.68 for liver versus kidney and a value of 1.01 for the comparison of the two liver replicas, which agrees with the results reported in Robinson and Oshlack (2010).
Pyicos EA can have read counts or reads per kilobase per million reads (RPKM) (Mortazavi et al., 2008) as input; or alternatively, directly the BED files with regions and read coordinates, overcoming the need of an additional tool to count reads. Additionally, Pyicos can calculate enriched regions de novo by scanning the samples provided, using two configurable parameters, the proximity between two reads to be taken as part of the same region, and the minimum number of reads in the region to be considered.
2.4 Benchmarking of EA
To assess the accuracy of Pyicos in the prediction of DE genes, we compared it to three other methods, DEGseq (Wang et al., 2010), DESeq (Anders and Huber, 2010) and edgeR (Robinson et al., 2010); using published datasets for RNA-Seq and equivalent microarray experiments for liver and kidney samples (Marioni et al., 2008). First, we mapped the microarray results to the Ensembl annotation (Flicek et al., 2011). Then, using the microarray data, we defined a benchmarking set composed of DE and non-DE genes. DE genes were defined to have False Discovery Rate (FDR) <0.001 and an absolute log2-fold change >0.5 (Marioni et al., 2008), regardless of whether they are up- or down-regulated. Non-DE genes were defined to have an FDR >0.01 and an absolute log2-fold change of at most 0.5. This benchmarking set is composed of 6700 DE genes and 7060 non-DE genes. To predict DE genes, we considered for each Ensembl locus the mean of the values for read counts or RPKM for the corresponding Ensembl transcripts in the locus. The RPKM and read count per transcript were calculated with reads falling within the exons of each transcript. A pseudocount of one read per transcript was added to be able to operate with transcripts whose exons did not have any reads in any of the samples. We estimated the TPs and FPs as the number of predicted DE (enriched or depleted) genes that were annotated as DE or non-DE genes in the benchmarking set, respectively. ROC curves were calculated by considering increasing absolute values of the Z-score or −log10(P-value) for the predictions for each method. We also calculated precision–recall curves along Z-score thresholds, where precision is the ratio of TP over the number of predicted cases and recall is the same as the TPR.
2.5 Enrichment on ChIP-Seq data
We performed EA on broad ChIP-Seq signals using ENCODE data for H3K36me3 and RNAPII, and correlated the results to the enrichment of RNA-Seq data, for the K562 and NHEK cells (ENCODE Consortium, 2011). The calculation of significantly enriched or depleted regions was based on RPKM values. Replicas 1 and 2 from K562 and replica 1 from NHEK were used to calculate enrichment on three different types of regions from the Ensembl annotation (Flicek et al., 2011): for H3K36me3 we used the transcript-body, spanning from the transcription start site (TSS) to the transcription termination site (TTS); for RNAPII we used a window of 4 kb around the TSS; and the transcript exons for RNA-Seq, as before. Significant regions were defined as those with an absolute Z-score >10. We calculated the Pearson's correlation coefficient of Z-scores from significant regions between H3K36me3 and RNA-Seq, as well as between RNAPII and RNA-Seq.
2.6 Memory usage and running time benchmarking
To test the memory usage and running time of the callpeaks protocol, we used ChIP-Seq data for the human transcription factor CEBPA (Schmidt et al., 2010). To simulate files of different sizes, we first pooled together all available reads and took random subsets with an increasing size step of 3 million reads, up to 30 million reads, separately for sample and control. In each run, we subsampled two random sets of equal size from the ChIP-Seq reads and from control, using steps of 3 million reads.
In order to test the performance of Pyicos EA, we compared it to DEGSeq and to a combination of BEDTools (Quinlan and Hall, 2010) with edgeR and DESeq, as these two programs do not accept as input BED files with the regions of interest and the positions of the mapped reads from the samples to be compared. In this way, all four calculations start with the same input: BED files for the samples and the region file. We used the same CEBPA data mentioned before and we calculated the enrichment in the region of 2 kb upstream of the TSS for RefSeq annotated genes. This time we sampled twice per run on the experiment, in order to get a simulated replica, and once for the control. We sampled reads increasing by 3 million reads in each run.
3 RESULTS
3.1 Analysis of punctuated ChIP-Seq data
The problem of peak detection entails two challenges: first, to determine significant peaks by distinguishing between real and false binding signal; and secondly, once we have selected the candidate peaks, we need to properly determine the site of interaction. In what follows, we use various methods to establish the accuracy and quality of peak prediction for Pyicos and three other published methods for punctuated ChIP-Seq analysis. But first, we assess the effectiveness of Pyicos operations for peak definition using ChIP-Seq datasets for NRSF, PR, CTCF and CEBPA.
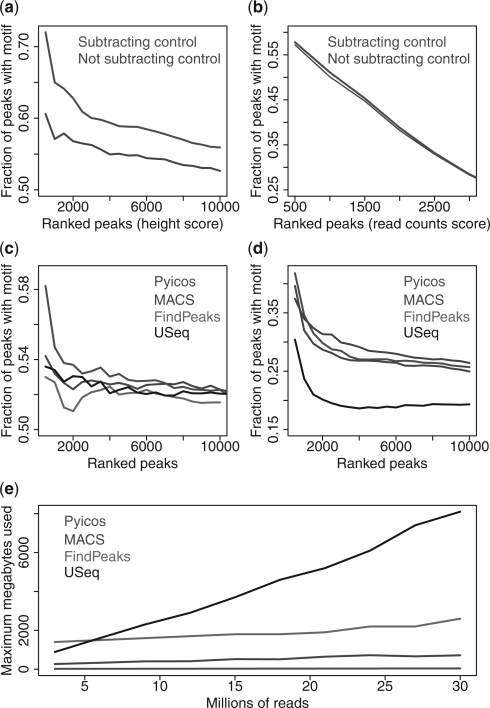
ChIP-Seq analysis often starts by removing duplicated reads, i.e. redundant reads in terms of position and strand in a genome. This has been suggested to eliminate amplification biases (Zhang et al., 2008). We observe that keeping duplicates does not produce any improvement on the four sets and that, in fact, the highest fraction of peaks with motifs is achieved when all duplicates are removed (Supplementary Fig. S5). An additional way to remove experimental biases, which may introduce FPs, is the use of a control. When this is available, callpeaks subtracts the control from the sample after normalization (Supplementary Material). To test whether Pyicos subtraction improves peak detection, we generated peaks with and without the subtraction of the control. We observe a great improvement in peak detection for CEBPA (Fig. 1a), CTCF and PR (Supplementary Fig. S6), while we only saw a slight increase for the top peaks for NRSF (Fig. 1b). Interestingly, the subtraction of the control results into almost no difference in the spatial resolution (Supplementary Fig. S7).
Fig. 1.
Properties of candidate peaks. Cumulative plots of the fraction of Pyicos peaks with a motif along the ranking selected by Poisson P-value cut-offs for peaks with and without subtraction for (a) CEBPA and (b) NRSF. Cumulative plot of the fraction of peaks with motifs along the ranking for the top 3000 peaks predicted by Pyicos, MACS, FindPeaks and USeq, for (c) PR and (d) CTCF. (e) Memory performance of the same four methods on the CEBPA ChIP-Seq data.
To explore the possibility that some regions of ChIP-Seq signal may describe multiple binding sites, Pyicos includes the option to split peaks (Supplementary Material). We found that splitting peaks showed improvement only on the CTCF dataset, while spatial resolution was maintained (Supplementary Fig. S8). The protocol callpeaks incorporates a Poisson test on the retained peaks to select those that are more likely to be functional (Section 2). Although the Poisson distribution seems not to fit well the background biases of ChIP-Seq experiments (Zhang et al., 2008), our analyses indicate that by subtracting the control we remove these biases and Poisson analysis can be applied. Each predicted peak is given a peak-score and a P-value (Section 2). The peak score gives a ranking for all peaks, whereas the P-value is used to select a subset of candidate peaks with certain significance. In order to show that the peak score produces an appropriate ranking, we calculated the fraction of peaks containing the expected motif along ranked peaks. We observe that for increasing cut-offs of the P-value, the fraction of peaks with motifs decreases with the peak score (Supplementary Fig. 9). The selection of candidate peaks by P-value is therefore consistent with the expected association of significant peaks to functional motifs. The peak score can be calculated using the peak height or read count. On the one hand, we found no differences in peak quality on NRSF and slight differences on CTCF and CEBPA, using peak height or read count. On the other hand, the peak height score for PR results into an increased fraction of peaks with motifs relative to the read count score (Supplementary Fig. S10).
3.2 Comparing peak detection with other methods
In order to further assess the quality of Pyicos peak calling, we compared Pyicos with three other methods: MACS, FindPeaks and USeq. We evaluated the peak definition of the four methods in terms of motif content (Section 2). They all show a similar trend along the ranking, with Pyicos and MACS showing higher densities for all ranking positions (Fig. 1c and d and Supplementary Fig. S11). Moreover, calculating the spatial resolution (Section 2), the four methods show the largest agreement for the PR dataset, whereas USeq shows the lowest spatial resolution on NRSF, CEBPA and CTCF (Supplementary Fig. S12).
We also evaluated the accuracy of peak definition measuring the overlap of the selected peaks with a benchmarking set of positive and negative NRSF binding regions validated by ChIP-qPCR (Section 2). All four methods show high agreement for their top 3000 NRSF peaks in terms of overlap (Supplementary Table S1) and in terms of the pairwise correlations (Supplementary Table S2); hence, we expect they would achieve similar accuracies. Indeed, the four methods perform similarly well with an area under the ROC curve (AUC) between 0.90 and 0.91 (Table 1). We also compared the accuracy of peak definition of the selected peaks by calculating the distances between summit and peak centre as well as the lengths of the peaks. We found that USeq, which produces the shortest peaks (Supplementary Fig. S2), achieves the shortest distance between summit and peak centre, closely followed by Pyicos (Supplementary Fig. S13).
Table 1.
AUC for peaks predicted by each tested method, using the ChIP-qPCR-validated NRSF regions for benchmarking
| Pyicos | MACS | FindPeaks | USeq | |
|---|---|---|---|---|
| AUC | 0.9 | 0.9 | 0.91 | 0.9 |
Finally, in order to test our software performance, we used ChIP-Seq data for the human transcription factor CEBPA (Schmidt et al., 2010) (Section 2). Running all four methods with conditions as similar as possible (Supplementary Material), we observed that Pyicos is more efficient in memory usage than the other three methods (Fig. 1e) and stays competitive in running time (Supplementary Fig. S14).
3.3 EA on RNA-Seq data
Methods to measure differential gene expression from RNA-Seq are generally based on EA between samples (Anders and Huber, 2010; Bullard et al., 2010; Oshlack et al., 2010; Robinson and Oshlack, 2010; Wang et al., 2010). Pyicos incorporates a method to detect regions of significant enrichment between two samples based on the comparison of the observed enrichment values with those measured between two replicas (Section 2). In order to establish the accuracy of Pyicos for detecting DE genes, we compared it first to the methods MATR (with replicas) and MARS (without replicas) from DEGseq (Wang et al., 2010), using RNA-Seq data from liver and kidney (Marioni et al., 2008). Both methods, DEGseq and Pyicos, can accept as input read counts or RPKM values for each gene; and they also can estimate a theoretical replica when an experimental one is not provided. We measured the significance of the enrichment of liver over kidney in all genes using four different combinations of input data: replicated count, non-replicated count, replicated RPKM and non-replicated RPKM. Comparing the Z-scores calculated by Pyicos and DEGseq, we observe a high correlation between both methods for all the inputs (Supplementary Table 3).
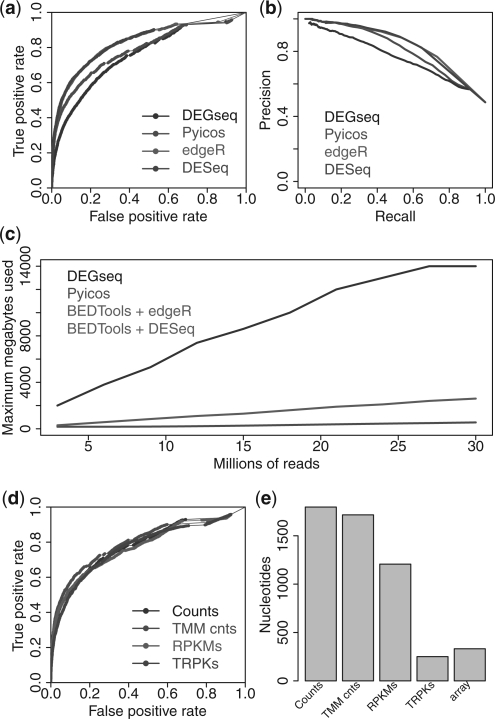
To further estimate the accuracy of Pyicos EA, we also ran DESeq (Anders and Huber, 2010) and edgeR (Robinson et al., 2010), on the same benchmarking sets, using again the four different input types. Using the microarray experiments from Marioni et al. (2008), we created a benchmarking set (Section 2). We found an overall good performance for Pyicos, with AUCs between 0.791 and 0.813 and a behaviour similar to DEGseq. All four methods agree similarly well with the benchmarking set when replicated read count data is used (Fig. 2a). However, we find greater disagreements between methods and lower accuracy for the other three input combinations (Supplementary Fig. S15 a–c). Moreover, the precision–recall curves show a similar level of agreement for replicated read count data (Fig. 2b) and confirm the good performance of Pyicos for all combination of inputs (Supplementary Fig. S15 d–f). For DESeq, it was not possible to make calculations on non-integer values, so we restricted the calculations to counts, which resulted in a high AUC when replicas were used. Although edgeR was developed for replicated count data, it was also run with non-replicated count data and performed similarly well.
Fig. 2.
Prediction of DE genes. (a) ROC curves for the benchmarking against the microarray data (Marioni et al., 2008) for DESeq, Pyicos, edgeR and DEGseq using read counts and replicated data. (b) Precision–recall curves for the benchmarking against microarray data for the same four methods using read counts and replicated data. (c) Memory performance of the same four methods on the EA of CEBPA ChIP-Seq dataset on the promoter region (Section 2). DESeq and edgeR are run in combination with BEDTools. (d) ROC curves of the different normalization methods: read counts (Counts), TMM-normalized counts (TMM counts), RPKMs and TRPKs, for the microarray benchmarking. (e) Absolute differences of the medians from the length distributions of DE and non-DE genes calculate with Pyicos using counts, TMM-normalized counts, RPKMs, TRPKs, and the corresponding value from the microarray data.
In order to test the memory usage and processing time of Pyicos EA calculation, we applied EA from all four methods on the CEBPA dataset to compare the ChIP-Seq sample to the control sample. While Pyicos and DEGseq can be run directly on BED files, edgeR and DESeq require count data as input. Hence, we combined these last two methods with BEDTools for the comparison. Since the most time consuming and memory intensive operation is the counting of reads per region, which is done with BEDTools, edgeR and DESeq show the same behaviour. The performance of DEGseq suffers in terms CPU time and especially memory usage. Although Pyicos is not as fast as the combination of BEDTools with edgeR or DESeq, it still can handle very large datasets at much lower memory usage (Supplementary Fig. S16 and Fig. 2c). BEDTools algorithm compromises memory usage in exchange of execution time, whereas Pyicos algorithm focuses on memory efficiency.
3.4 Normalization of the RNA-Seq data
As genes have different lengths and RNA-Seq samples may vary considerably in size, proper normalization of the input data is essential. The trimmed mean of M values (TMM) normalization (Robinson and Oshlack, 2010) aims to correct biases due to differences in samples sizes and expression patterns. TMM normalization on count data correctly places the M median on zero on our gene set (Supplementary Figs S17a and S17b). We considered the combination of TMM normalization with RPKM densities, which we hypothesize that it would improve results as it takes into account both gene lengths and sample sizes. We thus define a TRPK density as the TMM-normalized Read Per Kilobase density (Supplementary Material). First, as expected, using TRPK we can achieve a correction of the M-median for RPKM (Supplementary Figs S17c and S17d). Next, we assessed whether the TMM normalization also results in an improvement of the accuracy calculation using the array results. However, for this particular benchmarking set, we could not observe such an improvement (Fig. 2d).
Intriguingly, read counts seem to perform slightly better in our benchmarking analysis, as shown above. However, they have been observed to produce length biases in the determination of DE genes (Oshlack and Wakefield, 2009). To explore the correction effect of the various normalizations on the length biases, we calculated the lengths for all DE and non-DE genes predicted by Pyicos EA using counts, TMM normalized counts, RPKM and TRPK; and compared them to the lengths of the DE and non-DE genes from the microarray results (Supplementary Fig. S18). We observe a significant difference between the absolute length medians of DE and non-DE genes when normalized counts are not used. However, this difference decreases when we use the RPKM and TMM-normalized read counts, achieving the smallest difference and reaching a similar value to the one found for the microarray when TRPK is used (Fig. 2e).
3.5 EA on broad ChIP-Seq data
We next provide evidence that Pyicos EA approach is also suitable for the analysis of broad ChIP-Seq data when comparing two conditions. Broad ChIP-Seq data does not generally produce clearly delimited regions; hence, one cannot speak of peaks. However, it is interesting to be able to measure how the signal changes between two conditions or cell types in specific regions. Pyicos allows the calculation of significant enrichment in predefined regions, e.g. gene-body, promoter regions, etc., which are given as input in a BED file. To show the applicability of Pyicos on broad ChIP-Seq data, we tried to reproduce the relation between RNAPII activity and transcription (Sultan et al., 2008) and between the H3K36me3 chromatin signal and transcription (Joshi and Struhl, 2005; Pokholok et al., 2005). For this, we calculated the correlation of the enrichment of H3K36me3 and RNAPII, with that of RNA-Seq using ENCODE datasets (ENCODE Consortium, 2011). For each datatype, we calculated the EA values of K562 over NHEK, considering the variation between two K562 replicas (Section 2). We selected a cut-off of |Z-score|>10, obtaining 1238 genes with significant changes in RNAPII and RNA-Seq, and 377 genes with significant changes in H3K36me3 and RNA-Seq. For these gene sets, we measured a Pearson's correlation between the enrichment Z-scores of 0.87 and 0.90, for RNAPII versus RNA-Seq and H3K36me3 versus RNA-Seq, respectively.
3.6 CLIP-Seq data analysis
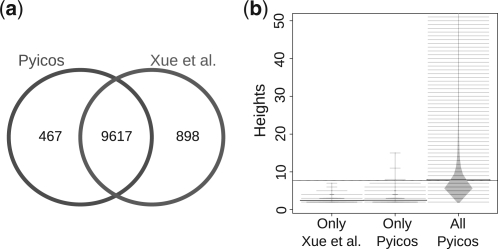
CLIP-Seq reads can be mapped to the genome or to the transcriptome in order to detect RNA binding activity. CLIP experiments normally use a control with an unspecific antibody to check the presence of unspecific binding. However, such a control is not always sequenced; hence, a method to calculate enrichment without a control was proposed (Xue et al., 2009; Yeo et al., 2009). This method, called modified false discovery rate (modFDR), is included in Pyicos (Supplementary Material). In order to show Pyicos applicability to CLIP-Seq experiments, we reproduced the results from Xue et al. (2009), where the interacting RNA sites for the polypyrimidine tract binding protein were mapped to the reference human genome. We used all the mapped reads from this experiment and ran Pyicos modFDR operation considering the gene-body of the RefSeq genes as regions to calculate the significant read clusters. Pyicos retrieved 92% of the 32 298 regions defined in Xue et al. (2009) on the same gene set, which means 91.5% of the reported 10 515 genes having at least one significant cluster (Fig. 3a). Moreover, the genes missed by Pyicos modFDR were those with a low number of reads: from the 467 genes missed by Pyicos, 89.7% contained clusters with heights of at most three reads (Fig. 3b), suggesting that they are likely FPs. Moreover, there were 898 genes selected by Pyicos that were not in the predictions from Xue et al. (2009), from which 11.1% had clusters that are >3, indicating that Pyicos may still recover real sites that are borderline. Nonetheless, the clusters missed by either case had P-values closely above the used 0.001 thresholds, indicating that the observed variation was mostly due to the random nature of the background model calculation.
Fig. 3.
Detecting significant clusters in CLIP-Seq. (a) Genes with at least one significant cluster using Pyicos CLIP-Seq protocol (red) and the results published in Xue et al. (2009) (blue). (b) Beanplots (Kampstra, 2008) showing the distribution of heights for three subsets of read clusters: the significant clusters exclusively detected in Xue et al. (2009) and not by Pyicos (Only Xue et al.), the significant clusters exclusively found by Pyicos (Only Pyicos) and all the significant clusters found by Pyicos (All Pyicos).
4 DISCUSSION
We have described Pyicos, a powerful tool for the analysis of mapped HTS reads. Pyicos framework facilitates the analysis of different HTS datatypes. We have described its application to ChIP-Seq, RNA-Seq and CLIP-Seq data using the three corresponding protocols: callpeaks, enrichment and clipseq. In callpeaks, we define a peak score in terms of a Poisson P-value, which is calculated independently for each chromosome, and in terms of one peak property, either the peak height or read count. The peak score therefore takes into account the differences across chromosomes and the fact that peaks with the same height or read count may have different P-values depending on the chromosome in which they are located. Using ChIP-Seq data for PR, CTCF, CEBPA and NRSF, we have shown that the peak score provides an appropriate ranking for peaks. Interestingly, using either the peak height or read count can make a difference depending on the dataset, as we found an increase of ∼7% in the fraction of the top 500 PR peaks with motifs, leading to a better peak definition compared with the other methods. We have further shown that the subtraction of the control is effective to increase the fraction of peaks with motif, indicating that it eliminates potential FPs.
Pyicos has the advantage that all operations described are configurable by the user to keep as much flexibility as possible, since not all of them may be applicable to a dataset. For instance, splitting peaks seems to result in improvements only for CTCF. Similarly, although Pyicos allows the user to choose the number of tolerated duplicated reads, we found that in all datasets the best peak definition is achieved by removing all duplicates.
Furthermore, using various measures, we have compared callpeaks with three other methods specifically developed for punctuated ChIP-Seq data: MACS, USeq and FindPeaks. Regarding peak definition, we have found that FindPeaks, Pyicos and MACS show very similar spatial resolutions, which are also higher than those for USeq peaks. Furthermore, we observed that peaks ranked by Pyicos and MACS show a slightly higher fraction of motif-containing peaks than those ranked by FindPeaks and USeq.
Peak detection has been assessed using ChIP-qPCR validated positive and negative regions for NRSF. We found that all methods perform similarly, probably due to high agreement between their generated peaks. The methods with the highest pairwise correlations are Pyicos, MACS and USeq. FindPeaks showed a lower correlation, probably due to a different handling of the control sample: whereas, FindPeaks compares a peak height with the distribution of peak heights from the control sample, the other three methods compare signal to control locally, using windows (MACS and USeq) or at base pair resolution (Pyicos). In summary, our results lead us to conclude that callpeaks provides an accurate protocol for peak detection for punctuated ChIP-Seq data. Moreover, due to the more flexible usage of the various operations compared with other methods, it allows to design a customized analysis of the ChIP-Seq data.
We have further illustrated Pyicos flexibility by applying it to other datatypes. Using data from RNA-Seq and microarray experiments on liver and kidney samples, we have shown that Pyicos can recover DE genes with high accuracy and that is in fact comparable to methods specifically designed for differential expression analysis from RNA-Seq. Moreover, Pyicos performs well using different inputs: replicated or non-replicated, read counts or RPKM. DEGseq, which can also work with RPKM, shows similar accuracy as Pyicos and both correlate well in terms of the predicted Z-scores. The other two methods, DESeq and edgeR, were not designed to work with RPKM, hence could not be included in the comparison. The fact that Pyicos performs well with simulated replicated data presents the advantage of making possible to analyse many of the published datasets that have been produced without replica.
It is surprising that the highest accuracy is achieved on count data, since it is known that using counts leads to a length bias in DE gene detection. This is probably because the genes selected from the microarray for benchmarking are not much affected by the length bias, since the accuracy hardly changes when using RPKM. This also suggests that RPKM alone does not provide the optimal normalization method. However, using a new density definition, TRPK, which combines RPKM with the TMM normalization, the length differences between DE and non-DE genes are reduced to a level close to that of the microarray. Nonetheless, the TMM normalization is based on the assumption that the majority of regions tested do not change significantly, which might not hold true for some pairs of samples or for certain sets of regions. Accordingly, Pyicos implements this as an option to the user.
The flexibility of Pyicos is further demonstrated by applying the EA protocol to broad ChIP-Seq data. In particular, using ENCODE ChIP-Seq data for H3K36me3 and RNAPII from the cell lines K562 and NHEK, we obtain a high correlation for the Z-scores of these signals when compared with the enrichment Z-scores for RNA-Seq between the same cell lines. Thus, the EA of Pyicos using RPKM provides a tool to analyse broad ChIP-Seq data of various sorts, which would otherwise require a combination of approaches to be analysed (Young et al., 2011). A further advantage is that Pyicos can directly accept BED files with mapped reads and regions of interest, unlike DESeq and edgeR. Pyicos can also calculate enriched regions de novo genome wide, using the reads from two experiments that are overlapping or sufficiently close in position. This is particularly useful for the analysis of signals for which the user does not know where to expect the enrichment relative to genome annotations, like enhancer elements. We have further shown that Pyicos can also be used to process CLIP-Seq data without a control. We expect that many of the basic Pyicos operations could be applicable to other datatypes and could possibly be combined to generate new analysis protocols.
Some of the operations described can become impracticable for some analysis tools due to the amount of reads produced by a single HTS experiment nowadays. The bottleneck of HTS data analysis mostly lies in the memory usage of the software and in the storage and retrieval of data. Indeed, HTS data generation seems to be outpacing the improvements in CPU and disk storage (Kahn, 2011). Moreover, HTS data has become ubiquitous in genomic research; hence, we should aim to provide software that can be adapted to the available computing resources and, in particular, that can run on the average desktop computer. For these reasons, we developed Pyicos minimizing RAM usage and maintaining reasonable CPU time usage. Pyicos callpeaks protocol for ChIP-Seq data outperforms the other tested methods concerning memory usage while it stays competitive in running time. FindPeaks and USeq algorithms load entire files in memory, allowing for better time performance. However, this is only practical on a computer with enough RAM. For example, an experiment with 100 million reads using as input a BED file, would require >4 GB available of RAM. For EA, the main bottleneck lies in the calculation of read counts or RPKM on the input regions. Although Pyicos is not as fast as BEDTools combined with DESeq or edgeR, its memory performance is far superior, allowing the handling of very large datasets. As made patent in the last few years, read files are increasing enormously in size with the development of the technology, making memory usage a critical feature in HTS analysis software.
We conclude that the added value of having a modular tool is not at the cost of accuracy or performance; hence, Pyicos provides a useful framework for the analysis and integration of heterogeneous HTS data. Finally, as Pyicos is open source, we encourage the addition of new operations in order to combine them with already existing ones and possibly to create new analysis protocols.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Christian Perez-Llamas, David González, Julien Lagarde and Robert Castelo for useful discussions, and Alba Jene for discussions and comments on the manuscript. We also acknowledge Mark Kon, Manolis Kellis and Ana Rojas for providing us the opportunity to present Pyicos in their institutions.
Funding: Generalitat de Catalunya by FI grant (to S.A.); Spanish Ministry of Science (MICINN) by FPI grant (to J.G.V.); MICINN grant BIO2008-01091 (to E.E.); European Commission grant EURASNET-(LSHG-CT-2005-518238) (to E.E.).
Conflict of Interest: none declared.
Footnotes
†The authors wish it to be known that, in their opinion, the last two authors should be regarded as joint First Authors.
REFERENCES
- Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J.H., et al. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Consortium. A User's Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes A.P., et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.S., et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kahn S.D. On the future of genomic data. Science. 2011;331:728–729. doi: 10.1126/science.1197891. [DOI] [PubMed] [Google Scholar]
- Khalil A.M., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knüppel R., et al. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol. 1994;1:191–198. doi: 10.1089/cmb.1994.1.191. [DOI] [PubMed] [Google Scholar]
- Licatalosi D.D., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni J.C., et al. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., et al. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nix D.A., et al. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A., Wakefield M.J. Transcript length bias in RNA-seq data confounds systems biology. Biol. Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A., et al. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S., et al. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D.K., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D., et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., et al. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Sultan M., et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- Trapnell C., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent G.P., et al. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev. 2011;25:845–862. doi: 10.1101/gad.621811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.T., et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008a;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS One. 2008b;3:e3839. doi: 10.1371/journal.pone.0003839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- Xue Y., et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., et al. ChIP-seq analysis reveals distinct H3K27me3 profiles that correlate with transcriptional activity. Nucleic Acids Res. 2011;39:7415–7427. doi: 10.1093/nar/gkr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C., et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.