Abstract
Fap1, a serine-rich repeat glycoprotein (SRRP), is required for bacterial biofilm formation of Streptococcus parasanguinis. Fap1-like SRRPs are found in many Gram-positive bacteria and have been implicated in bacterial fitness and virulence. A conserved five-gene cluster, secY2-gap1-gap2-gap3-secA2, located immediately downstream of fap1, is required for Fap1 biogenesis. secA2, gap1, and gap3 encode three putative accessory Sec proteins. SecA2 mediates export of mature Fap1, and Gap1 and Gap3 are required for Fap1 biogenesis. Interestingly, gap1 and gap3 mutants exhibited the same phenotype as a secA2 mutant, implying that Gap1 and Gap3 may interact with SecA2 to mediate Fap1 biogenesis. Glutathione S-transferase pulldown experiments revealed a direct interaction between SecA2, Gap1, and Gap3 in vitro. Coimmunoprecipitation analysis demonstrated the formation of a SecA2-Gap1-Gap3 complex. Homologues of SecA2, Gap1, and Gap3 are conserved in many streptococci and staphylococci. The corresponding homologues from Streptococcus agalactiae also interacted with each other and formed a protein complex. Furthermore, the Gap1 homologues from S. agalactiae and Streptococcus sanguinis rescued the Fap1 defect in the Gap1 mutant, indicating the functional conservation of the accessory Sec complex. Importantly, canonical SecA interacted with the accessory Sec protein complex, suggesting that the biogenesis of SRRPs mediated by the accessory Sec system is linked to the canonical Sec system.
INTRODUCTION
Binding of oral streptococci to the tooth surface is the first step in the formation of dental plaque (5, 9, 16). Streptococcus parasanguinis is a primary colonizer of the dental plaque (14) and also plays an important role in subacute bacterial endocarditis (4). Adhesion of S. parasanguinis is mediated by its long, peritrichous fimbriae (13). Fap1, a serine-rich repeat glycoprotein (SRRP), is the major subunit of the long fimbriae and is required for bacterial adhesion and biofilm formation (31, 32). SRRPs have been identified in many Gram-positive bacteria and implicated in bacterial fitness and virulence (37).
Fap1 biogenesis is mediated by a five-gene cluster, secY2-gap1-gap2-gap3-secA2, located immediately downstream of fap1 (30, 33). The gene cluster encodes five putative accessory secretory proteins, SecY2, Gap1, Gap2, Gap3, and SecA2, and its genetic organization is highly conserved in every genome that contains genes for SRRPs (37), implying the importance of this locus in the biogenesis of SRRPs. Mutation of secA2 results in defects in the export of a number of SRRPs, including Fap1 (1, 7, 19, 24). Mature Fap1 is not detected in the membrane and cytoplasm fractions of a secA2 mutant (7). Gap1 and Gap3 interact with each other, and mutagenesis of either one blocks Fap1 maturation (18, 20). Study of Gap1 and Gap3 homologues in Streptococcus gordonii has also indicated that Asp2 and Asp3 interact with SecA2 to modulate export of GspB (22). However, it is unknown whether Gap1 and Gap3 directly interact with the accessory Sec system in S. parasanguinis.
Sequence analysis has revealed similarity between SecA2 and canonical SecA. Biochemical analysis demonstrates that SecA2 is an ATPase that shares several structural features with SecA (2), suggesting that SecA2 may have a similar role in export of SRRP preproteins. Mutagenesis of SecA2 not only abolished the export of Fap1, GspB, and other SRRPs (1, 7, 19, 24) but also inhibited secretion of a major virulent lipoprotein, FimA, of S. parasanguinis (4, 7). As the lipoprotein FimA is exported via canonical SecA, these data imply that there is cross talk between SecA2 and SecA. Indeed, only glycosylated Fap1 and GspB are exported via the SecA2-mediated pathway, while unglycosylated Fap1 and GspB can be exported through the SecA system (3, 6), suggesting that there is an unknown mechanism regulating involvement of SecA2 and SecA in secretion of SRRPs. It is not clear whether the canonical SecA interacts directly with the accessory Sec components to facilitate this process.
In this study, we examined the protein-protein interactions between SecA2 and the Gap1-Gap3 complex and between the accessory Sec complex and canonical SecA. Our findings provide new insights into SecA2-mediated accessory secretion mechanisms in Gram-positive bacteria.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Streptococci were cultivated statically in 5% CO2 in Todd-Hewitt (TH) broth or on TH agar plates at 37°C. The Escherichia coli strain was grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C with shaking.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli Top10 | For propagation of the recombinant plasmids | Invitrogen |
| S. parasanguinis FW213 | Wild type | 10 |
| S. parasanguinis fap1 mutant | Wild type; fap1::aphA3; Kanr | 32 |
| S. parasanguinis gap1 mutant | Wild type; gap1::aphA3; Kanr | 18 |
| S. parasanguinis gap3 mutant | Wild type; gap3::aphA3; Kanr | 18 |
| S. parasanguinis secA2 mutant | Wild type; secA2::Tn5; Kanr | 18 |
| S. agalactiae J48 | Wild type | 23 |
| S. agalactiae 2603V/R | Wild type | 27 |
| S. sanguinis SK36 | Wild type | 34 |
| S. pneumoniae TIGR4 | Wild type | 28 |
| Plasmids | ||
| pGEX-5X-1 | GST fusion protein expression vector; Ampr | Amersham |
| pGEX-Gap1 | gap1 from FW213 cloned in pGEX-5X-1; Ampr | This study |
| pGEX-Asp1 (J48) | asp1 from J48 cloned in pGEX-5X-1; Ampr | This study |
| pGEX-Gap3 | gap3 from FW213 cloned in pGEX-5X-1; Ampr | 18 |
| pGEX-Asp3 (J48) | asp3 from J48 cloned in pGEX-5X-1; Ampr | This study |
| pGEX-SecA2 | secA2 from FW213 cloned in pGEX-5X-1; Ampr | This study |
| pGEX-SecA2 (J48) | secA2 from J48 cloned in pGEX-5X-1; Ampr | This study |
| pGEX-SecA | secA from FW213 cloned in pGEX-6P-1; Ampr | This study |
| pGBKT7 | In vitro translation vector; Kanr | Clontech |
| pBD-Gap1 | gap1 from FW213 cloned in pGBKT7; Kanr | 18 |
| pBD-Asp1 (J48) | asp1 from J48 cloned in pGBKT7; Kanr | This study |
| pBD-Gap3 | gap3 from FW213 cloned in pGBKT7; Kanr | 18 |
| pBD-Asp3 (J48) | asp3 from J48 cloned in pGBKT7; Kanr | This study |
| pBD-SecA2 | secA2 from FW213 cloned in pGBKT7; Kanr | This study |
| pBD-SecA2 (J48) | secA2 from J48 cloned in pGBKT7; Kanr | This study |
| pBD-SecA | secA from FW213 cloned in pGBKT7; Kanr | This study |
| pVT1666 | E. coli-Streptococcus shuttle vector; Ermr | 8 |
| pVPT-Gap1 | gap1 from FW213 cloned in pVT1666; Ermr | 36 |
| pVPT-Asp1 (J48) | asp1 from J48 cloned in pVT1666; Ermr | This study |
| pVPT-Asp1 (2603) | asp1 from 2603V/R cloned in pVT1666; Ermr | This study |
| pVPT-Asp1 (SK36) | asp1 from SK36 cloned in pVT1666; Ermr | This study |
| pVPT-Asp1 (TIGR4) | asp1 from TIGR4 cloned in pVT1666; Ermr | This study |
| pVPT-Gap3 | gap3 from FW213 cloned in pVT1666; Ermr | 20 |
Production of Fap1 in gap1, gap3, and secA2 mutants.
To examine Fap1 production in gap1, gap3, and secA2 mutants, 1 ml of exponentially grown (A470 ≈ 0.6) mutant cells was harvested and lysed as previously described (18). The cell lysates were mixed with 5× sodium dodecyl sulfate (SDS) loading buffer and subjected to Western blotting using the Fap1-specific monoclonal antibodies E42, D10, and F51 (25).
Construction of expression plasmids for GST fusion proteins.
Plasmids pGEX-Gap1, pGEX-SecA2, and pGEX-SecA were constructed to express glutathione S-transferase (GST)-Gap1, GST-SecA2, and GST-SecA fusion proteins. In brief, the gap1, secA2, and secA genes were amplified from the genomic DNA of S. parasanguinis FW213 by PCR using the primer pairs Gap1-EcoRI-1F and Gap1-XhoI-1575R, SecA2-BamHI-F and SecA2-BamHI-R, and SecA-SmaI-F and SecA-XhoI-R, respectively (Table 2). The PCR products were digested with EcoRI/XhoI, BamHI, or SmaI/XhoI and then cloned into pGEX-5X-1 to generate the desired fusion constructs.
Table 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Gap1-EcoRI-1F | GATCAGAATTCATGTTTTATTTTGTACCTTC |
| Gap1-XhoI-1575R | GATCTCTCGAGTTTCTTTTTTAGCATACCTTTC |
| SecA2-BamHI-F | CGGGATCCATATGGAAAAAATAAAACAAAAG |
| SecA2-BamHI-R | CGGGATCCTACAAAATAAATTGATAAACC |
| SecA2-PstI-R | AACTGCAGTACAAAATAAATTGATAAACC |
| SecA-SmaI-F | GATCACCCGGGTATGGCAAATTTATTAAAAAC |
| SecA-XhoI-R | GATCACTCGAGTCAAAATTTACGTCCGTGAC |
| SecA-NdeI-F | GATCACATATGGCAAATTTATTAAAAAC |
| SecA-PstI-R | GATCACTGCAGTCAAAATTTACGTCCGTGAC |
| Asp1-J48-BamHI-F | GATCAGGATCCCCATGTTTTATTTTATTCCTTCG |
| Asp1-J48-BamHI-R | GATCAGGATCCTTATTCTTTTTCTAATAATTTTCG |
| Asp3-J48-BamHI-F | GATCAGGATCCCCATGATTTTGGGAGAGTGTTTAG |
| Asp3-J48-BamHI-R | GATCAGGATCCTCATTTTTTATCCTTAGAAAATG |
| SecA2-J48-BamHI-F | GATCAGGATCCCCATGAAAAAAGTTGTTGTTAAACC |
| SecA2-J48-BamHI-R | GATCAGGATCCTTATACATAATATATTGAGTAACTTC |
| Asp1-J48-SalI-F | GATCAGTCGACATGTTTTATTTTATTCCTTCG |
| Asp1-2603-SalI-F | GATCAGTCGACATGTTTCATTTTATTCCATC |
| Asp1-2603-KpnI-R | GATCAGGTACCATAACTATTTATAATTCTTTC |
| Asp1-SK36-SalI-F | GATCAGTCGACATGTATTACTTTATTCCTTC |
| Asp1-SK36-BamHI-R | GATCAGGATCCTTATTTTGCATTCTCAGC |
| Asp1-TIGR4-SalI-F | GATCAGTCGACATGTATTATTTTATTCCAG |
| Asp1-TIGR4-BamHI-R | GATCAGGATCCTTAAGTTCCTTTGACATTTTTAAC |
Plasmids pGEX-Asp1 (J48), pGEX-Asp3 (J48), and pGEX-SecA2 (J48) were constructed and used to express GST-Asp1, GST-Asp3, and GST-SecA2 fusion proteins. The asp1, asp3, and secA2 genes were amplified from the genomic DNA of Streptococcus agalactiae J48 by PCR using the primer pairs Asp1-J48-BamHI-F and Asp1-J48-BamHI-R, Asp3-J48-BamHI-F and Asp3-J48-BamHI-R, and SecA2-J48-BamHI-F and SecA2-J48-BamHI-R, respectively (Table 2). The resulting PCR products were digested by BamHI and ligated into pGEX-5X-1 to generate the corresponding GST fusion constructs.
Construction of plasmids for in vitro translation of c-Myc-tagged proteins.
Plasmids pBD-SecA2 and pBD-SecA were constructed and used to generate in vitro-translated SecA2 and SecA proteins. In brief, the secA2 and secA genes were amplified from the genomic DNA of S. parasanguinis FW213 by PCR using the primer pairs SecA2-BamHI-F and SecA2-PstI-R and SecA-NdeI-F and SecA-PstI-R, respectively. The resulting PCR products were digested by BamHI and PstI for secA2 and by NdeI and PstI for secA and then ligated into pGBKT7 to generate pBD-SecA2 and pBD-SecA.
The plasmids pBD-Asp1 (J48), pBD-Asp3 (J48), and pBD-SecA2 (J48) were constructed and used to prepare in vitro-translated fusion proteins. The asp1, asp3, and secA2 genes were amplified from the genomic DNA of S. agalactiae J48 by PCR using the primer sets Asp1-J48-BamHI-F and Asp1-J48-BamHI-R, Asp3-J48-BamHI-F and Asp3-J48-BamHI-R, and SecA2-J48-BamHI-F and SecA2-J48-BamHI-R, respectively. The three PCR products were digested with BamHI and then ligated to pGBKT7 to create pBD fusion constructs.
GST pulldown assays to determine protein-protein interactions.
To examine in vitro protein interactions, the relevant GST fusion proteins and in vitro-translated products were prepared. The GST fusion proteins were bound to glutathione-Sepharose 4B beads (Amersham). The in vitro-translated products with the c-Myc tag were produced using a TnT quick coupled transcription-translation system (Promega) following the manufacturer's instructions. Five micrograms of the glutathione-Sepharose-GST fusion proteins was mixed with 5 μl of the c-Myc-tagged proteins in the presence of NETN buffer (20 mM Tris-HCl [pH 7.2], 100 mM NaCl, 1 mM EDTA, 0.2% NP-40) and incubated overnight on a rotary shaker at 4°C. The beads were washed three times with 600 μl NETN buffer, and the proteins were eluted with SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer, boiled for 10 min, and subjected to Western blotting with anti-c-Myc antibody.
Coimmunoprecipitation (co-IP) to determine in vivo protein-protein interactions.
An E. coli-streptococcus shuttle vector pVT1666 (8) with a green fluorescent protein (GFP) tag, a plasmid (pVPT-Gap1) with GFP-tagged Gap1, and a plasmid (pVPT-Gap3) with GFP-tagged Gap3 were transformed into the S. parasanguinis wild-type strain. In addition, plasmid pVT1666 and pVPT-Gap1 were also transformed into the gap3 mutant. All strains were grown to exponential growth phase (A470 ≈ 0.6). Cell lysates were prepared as described previously (18). Prepared cell lysates (400 μg) were incubated with anti-GFP monoclonal antibody or anti-SecA2 polyclonal antibody for 4 h at 4°C on a rotator prior to incubation with 50 μl of a 50% slurry of immobilized protein A/G agarose (Invitrogen) overnight at 4°C. Immunoprecipitates were collected by centrifugation, washed three times with NETN buffer, eluted with SDS sample buffer, and subjected to Western blot analyses using SecA2, Gap3, and SecA polyclonal antibodies.
Complementation of the S. parasanguinis gap1 mutant.
The Gap1 homologues were amplified from streptococcal genomic DNAs (23, 27, 28, 34) by PCR using primer pairs Asp1-J48-SalI-F and Asp1-J48-BamHI-R, Asp1-2603-SalI-F and Asp1-2603-KpnI-R, Asp1-TIGR4-SalI-F and Asp1-TIGR4-BamHI-R, and Asp1-Sk36-SalI-F and Asp1-Sk36-BamHI-R (Table 2). The PCR products were digested and cloned into the E. coli-streptococcus shuttle vector pVT1666 to generate the gap1 complementation plasmids pVPT-Asp1 (J48), pVPT-Asp1 (2603), pVPT-Asp1 (TIGR4), and pVPT-Asp1 (SK36). These plasmids were then transformed into the gap1 mutant via electroporation. The transformants were selected on TH agar plates containing kanamycin (125 μg ml−1) and erythromycin (10 μg ml−1). The ability of streptococcal Asp1 genes to restore mature Fap1 production was examined by Western blotting analyses using the Fap1 peptide-specific monoclonal antibody E42, the glycan-specific antibody D10, and the mature Fap1-specific antibody F51.
Production of SecA2, Gap1, and Gap3 in SecA2 and Gap3 mutants.
To examine production of SecA2, Gap1, and Gap3 in the SecA2 and Gap3 mutants, 1 ml of the mutant cells was grown to exponential growth phase (A470 ≈ 0.6), harvested by centrifugation, and lysed as previously described (18). The cell lysates were then subjected to Western blotting using SecA2, Gap1, or Gap3 polyclonal antibody. Production of FimA in S. parasanguinis was determined by anti-FimA antibody and used as a loading control.
RESULTS
The SecA2 mutant exhibited a phenotype similar to that of Gap1 and Gap3 mutants.
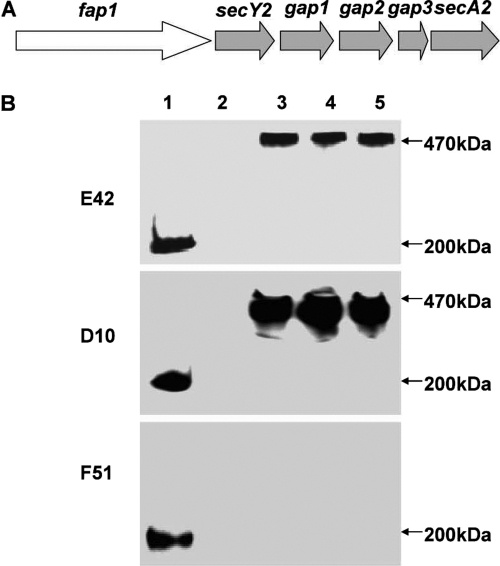
The Fap1 expression profile can be detected by three available Fap1-specific antibodies. mAbE42 is a peptide-specific antibody. mAbD10 is a glycan-specific antibody, and mAbF51 reacts with mature Fap1. Previous studies had revealed that the gap1 and gap3 mutants exhibited similar phenotypes in Fap1 biogenesis; both produce a 470-kDa high-molecular-weight (HMW) Fap1 precursor (18). A similar HMW Fap1 protein was also detected in the secA2 mutant by Western blot analyses using mAbE42 (Fig. 1B, top, lane 3) and mAbD10 (Fig. 1B, middle, lane 3). The mutants failed to react with mAbF51 (Fig. 1, bottom, lane 3), which recognizes a 200-kDa mature Fap1 produced by the wild-type bacterium (Fig. 1, bottom, lane 1), suggesting that SecA2, Gap1, and Gap3 are involved in the same process for the Fap1 biogenesis.
Fig. 1.
Expression of Fap1 in S. parasanguinis variants. (A) Organization of the fap1-secA2 locus. (B) Fap1 production in S. parasanguinis variants. Cell lysates of the wild type (lane 1) and the fap1 (lane 2), secA2 (lane 3), gap1 (lane 4), and gap3 (lane 5) mutants were subjected to Western blot analyses using Fap1-specific antibodies.
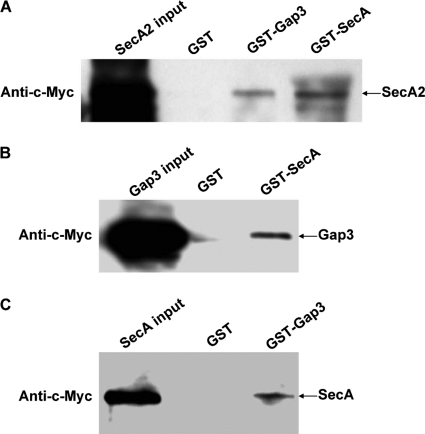
SecA2 interacted with the Gap1-Gap3 complex in vitro.
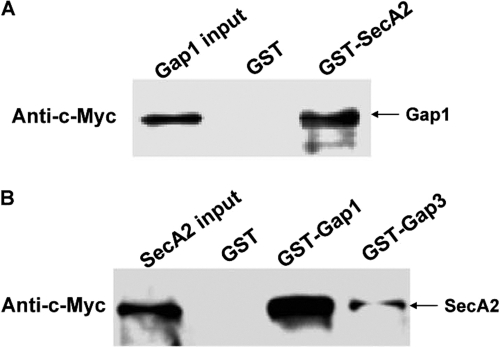
Mutants that are defective in different genes exhibit similar phenotypes; proteins encoded by such genes often form a complex (15, 29). We hypothesize that SecA2 interacts with the Gap1-Gap3 complex. GST pulldown experiments were performed to evaluate the potential interactions. Incubation of GST fusion proteins with in vitro-translated c-Myc-tagged proteins revealed a strong interaction between SecA2 and Gap1 (Fig. 2A and B) and a relatively weak interaction between SecA2 and Gap3 (Fig. 2B), whereas GST alone did not interact with c-Myc-tagged proteins (Fig. 2A and B), demonstrating the direct interactions between SecA2 and the Gap1-Gap3 complex.
Fig. 2.
In vitro GST pulldown assays to detect SecA2, Gap1, and Gap3 interactions. (A) Equal amounts of purified GST and GST-SecA2 bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-Gap1. (B) Equal amounts of purified GST, GST-Gap1, and GST-Gap3 bound to glutathione Sepharose 4B beads were incubated with in vitro-translated c-Myc-SecA2. The pulldown protein complexes were analyzed by Western blotting using c-Myc monoclonal antibody. Inputs are the in vitro-translated protein products.
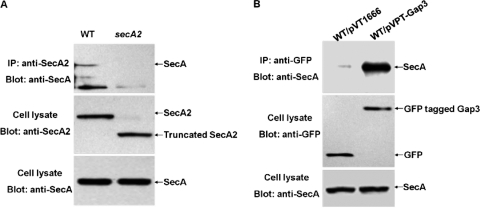
SecA2 associated with the Gap1-Gap3 complex in vivo.
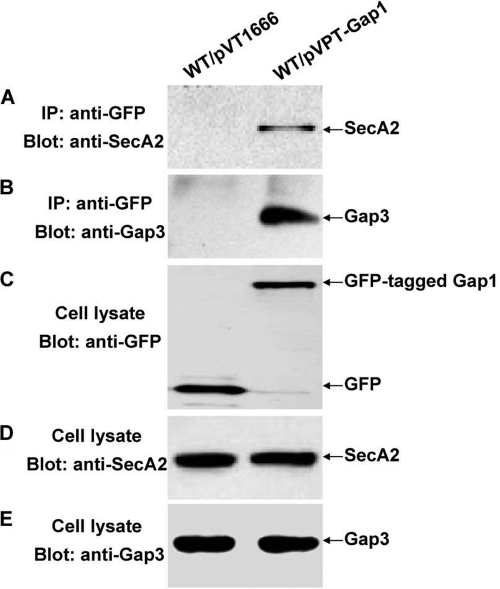
To determine whether SecA2 interacts with the Gap1-Gap3 complex in vivo, we performed immunoprecipitation assays. GFP-tagged Gap1 coprecipitated with native SecA2 and Gap3, whereas GFP itself did not (Fig. 3A and B). The same levels of SecA2, Gap3, GFP, and GFP-tagged Gap1 were detected in the recombinant strains (Fig. 3C to E). These data suggest that SecA2 interacts with the Gap1-Gap3 complex in vivo.
Fig. 3.
In vivo coimmunoprecipitation for SecA2, Gap1 and Gap3 interactions. Cell lysates of the wild-type strain with pVT1666 (WT/pVT1666) or pVPT-Gap1 (WT/pVPT-Gap1) were subjected to immunoprecipitation with monoclonal GFP antibody and then analyzed by Western blotting using polyclonal anti-SecA2 (A) and anti-Gap3 (B) antibodies, respectively. The expression of GFP fusion proteins, SecA2, and Gap3 was detected by Western blot analyses using anti-GFP (C), anti-SecA2 (D), and anti-Gap3 (E) antibodies, respectively.
Gap1 homologues from S. agalactiae and Streptococcus sanguinis complemented the corresponding S. parasanguinis mutants.
SecA2, Gap1, and Gap3 homologues are highly conserved at the primary amino acid sequence level. Phylogenetic analyses indicated that SecA2, Gap1, and Gap3 from S. parasanguinis are very similar to the corresponding homologues from S. agalactiae J48 and 2603V/R (data not shown). To examine whether they are functionally conserved, we determined if the Gap1 homologues from other streptococci can complement the corresponding mutant from S. parasanguinis (Fig. 4). The gap1 homologues from S. agalactiae J48 and S. sanguinis Sk36 (Fig. 4) partially restored the production of mature Fap1 by the S. parasanguinis gap1 mutant, suggesting that the Gap1 homologues are functionally conserved.
Fig. 4.
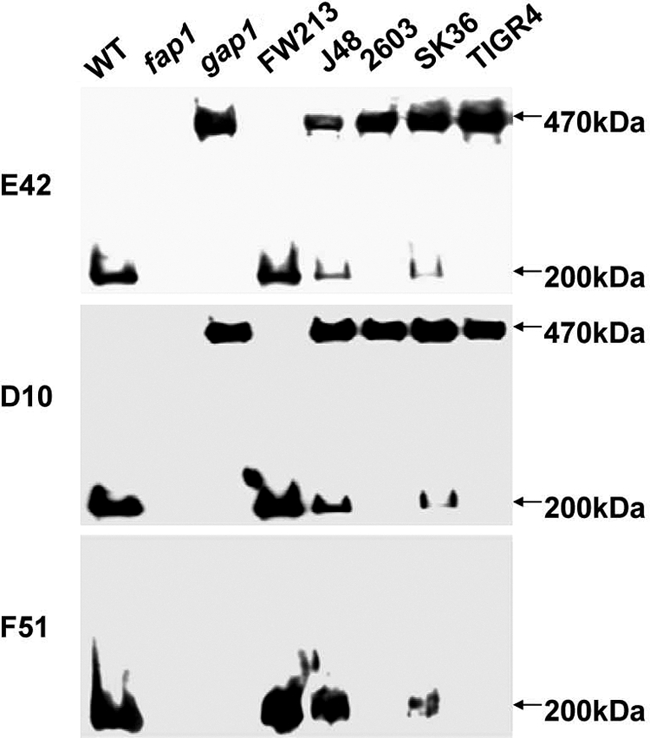
Complementation of the S. parasanguinis gap1 mutant with Gap1 homologues. Cell lysates from the gap1 mutant complemented with pVPT-Asp1 from a variety of streptococci were subjected to Western blot analyses using Fap1-specific antibodies.
Gap1, Gap3, and SecA2 homologues from S. agalactiae form a protein complex.
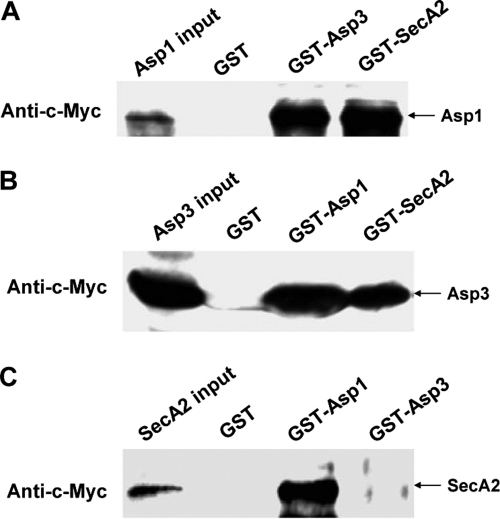
To determine whether Gap1, Gap3, and SecA2 homologues (Asp1, Asp3, and SecA2, respectively) of S. agalactiae form a protein complex as well, we performed GST pulldown experiments. GST-SecA2 indeed can pull down both Asp1 and Asp3 (Fig. 5A and B). GST-Asp1 could also pull down both Asp3 and SecA2 readily (Fig. 5B and C). In addition, GST-Asp3 could pull down Asp1 but not SecA2 (Fig. 5A and C). These results demonstrated that Asp1, Asp3, and SecA2 from S. agalactiae also formed a protein complex. Asp1 and Asp3 as well as Asp1 and SecA2 interacted with each other strongly in the complex.
Fig. 5.
In vitro GST pulldown assays for SecA2, Asp1, and Asp3 interactions by S. agalactiae. (A) Equal amounts of purified GST, GST-Asp3, and GST-SecA2 bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-Asp1. (B) Equal amounts of purified GST, GST-Asp1, and GST-SecA2 bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-Asp3. (C) Equal amounts of purified GST, GST-Asp1, and GST-Asp3 bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-SecA2. The pull-down protein complexes were analyzed by Western blotting using anti-c-Myc monoclonal antibody. Inputs represent the in vitro-translated protein products.
Canonical SecA associated with the SecA2-Gap1-Gap3 complex.
Canonical SecA, which has homology to SecA2, might interact with the SecA2-Gap1-Gap3 complex. To test this, we performed coimmunoprecipitation assays. SecA was found to be coimmunoprecipitated with SecA2 (Fig. 6A, top), and Gap3 (Fig. 6B, top). No SecA was coimmunoprecipitated in the secA2 mutant (Fig. 6A, top) or in the GFP control strain (Fig. 6B, top). All the proteins analyzed for immunoprecipitation were loaded equally (Fig. 6A, middle; Fig. 6A and B, bottom; Fig. 6B, middle). Interestingly, no interaction between Gap1 and SecA was detected by co-IP (data not shown). These results demonstrate that SecA is associated with the SecA2-Gap1-Gap3 complex.
Fig. 6.
In vivo coimmunoprecipitation for the interaction between SecA and the SecA2-Gap1-Gap3 complex. (A) Cell lysates of the wild-type strain (WT) and the secA2 mutant (secA2) were subjected to immunoprecipitation with polyclonal anti-SecA2 antibody and then analyzed by Western blotting using polyclonal anti-SecA antibody. Production of SecA2 and SecA in WT and secA2 strains was detected by Western blot analyses. (B) Cell lysates of the wild-type strain with pVT1666 (WT/pVT1666) or pVPT-Gap3 (WT/pVPT-Gap3) were subjected to immunoprecipitation with GFP monoclonal antibody and then analyzed by Western blotting using polyclonal anti-SecA antibody. Expression of GFP fusion proteins and SecA in WT/pVT1666 and WT/pVPT-Gap3 strains was detected by Western blot analyses.
To determine whether SecA directly interacts with the SecA2-Gap1-Gap3 complex, in vitro GST pulldown experiments were carried out. GST-SecA interacted with SecA2 (Fig. 7A) and Gap3 (Fig. 7B), and GST-Gap3 interacted with SecA2 (Fig. 7A) and SecA (Fig. 7C), while GST alone failed to interact (Fig. 7A to C). These data indicate that SecA directly interacts with the SecA2-Gap1-Gap3 complex.
Fig. 7.
In vitro GST pulldown assays for interactions between SecA and the SecA2-Gap1-Gap3 complex. (A) Equal amounts of purified GST, GST-Gap3, and GST-SecA bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-SecA2. (B) Equal amounts of purified GST and GST-SecA bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-Gap3. (C) Equal amounts of purified GST and GST-Gap3 bound to glutathione-Sepharose 4B beads were incubated with in vitro-translated c-Myc-SecA. The pulldown protein complexes were analyzed by Western blotting using anti-c-Myc monoclonal antibody. Inputs represent the in vitro-translated protein products.
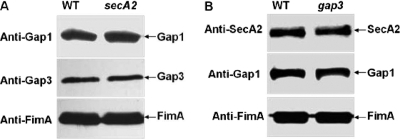
Mutation of SecA2 and Gap3 had no effect on production of their interacting partners.
The SecA2 deficiency had no effect on production of Gap1 (Fig. 8A, top) and Gap3 (Fig. 8A, top). The Gap3 deficiency also had no effect on production of SecA2 (Fig. 8B, top) or Gap1 (Fig. 8B, middle). No difference in production of a control protein, FimA, was observed among the S. parasanguinis strains (Fig. 8, bottom). These results suggest that mutation of the accessory Sec proteins does not affect the stability of their interacting partners.
Fig. 8.
Western blot analysis of production of SecA2, Gap1, and Gap3 in SecA2 and Gap3 mutants. (A) Cell lysates of the wild type (WT) and the secA2 mutant of S. parasanguinis were subjected to Western blot analysis using Gap1, Gap3, and FimA antibodies. (B) Cell lysates of the wild type (WT) and the gap3 mutant of S. parasanguinis were subjected to Western blot analysis using SecA2, Gap1, and FimA antibodies, respectively.
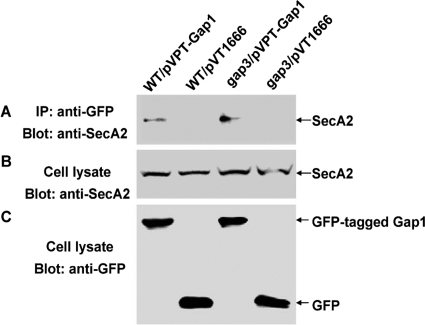
Lack of Gap3 did not affect the interaction between SecA2 and Gap1.
To determine whether SecA2 still interacts with Gap1 in the gap3 mutant in vivo, we performed immunoprecipitation assays. GFP-tagged Gap1 coprecipitated with SecA2 in both the wild type and the gap3 mutant, whereas GFP itself did not bind to SecA2 (Fig. 9A). The same levels of SecA2, GFP-tagged Gap1, and GFP were produced in the recombinant strains (Fig. 9B and C). These data suggest that the interaction between SecA2 and Gap1 is independent of Gap3.
Fig. 9.
In vivo coimmunoprecipitation for SecA2 and Gap1 from the gap3 mutant. Cell lysates of the wild-type strain with pVPT-Gap1 (WT/pVPT-Gap1) or pVT1666 (WT/pVT1666) and cell lysates of the gap3 mutant with pVPT-Gap1 (gap3/pVPT-Gap1) or pVT1666 (gap3/pVT1666) were subjected to immunoprecipitation with monoclonal GFP antibody and then analyzed by Western blotting using polyclonal anti-SecA2 (A). The expression of SecA2 and GFP fusion proteins was detected by Western blot analyses using anti-SecA2 (B) and anti-GFP (C) antibodies, respectively.
DISCUSSION
Genes encoding accessory Sec components are clustered with genes coding for SecA and SecY homologues in Gram-positive bacteria that possess SRRPs (21, 37), suggesting that a Sec-like protein secretion mechanism is adopted by these Gram-positive bacteria. A number of accessory Sec components have been identified and implicated in secretion of SRRPs (1, 7, 18, 19, 22, 24, 26, 30, 35, 36); however, the precise roles of each component have yet to be defined.
In this study, we explored protein function by examining their interactions and demonstrate that Gap1 and SecA2-Gap3 strongly interact with each other in vitro and in vivo. Since the locus secY2-gap1-gap2-gap3-secA2 is highly conserved in the Gram-positive bacteria that contain SRRPs, the SecA2-Gap1-Gap3 interaction is likely to be conserved. In fact, the homologues from S. agalactiae displayed a similar interaction profile. Furthermore, Asp1, a Gap1 homologue from S. agalactiae and S. sanguinis, rescued the Fap1 defect from the S. parasanguinis gap1 mutant. These findings are consistent with a recent report that described the interaction between accessory Sec components from S. gordonii (22), suggesting that the formation of the accessory Sec protein complex is common in Gram-positive bacteria that contain SRRPs.
There are two significant differences between the S. parasanguinis and S. gordonii accessory Sec systems: (i) unlike the strong interaction detected between Gap1 and SecA2 by S. parasanguinis, the interaction between homologous Asp1 and SecA2 was not detected in S. gordonii, and (ii) no in vivo interactions between Asp3 and SecA2 were identified in S. gordonii. The differences may manifest phenotypically, as inactivation of Gap1 diminishes but does not abolish export of Fap1, while inactivation of the Asp1 (Gap1 homologue in S. gordonii) completely inhibits export of glycosylated GspB (22).
Mature glycosylated Fap1 is exported via a SecA2-mediated accessory Sec pathway, whereas Fap1 that lacks glycosylation can be exported via a canonical SecA pathway (6). Export of GspB is conducted in a similar fashion in S. gordonii (3). These findings imply that there is a fine-tuning mechanism that directs glycosylated and unglycosylated substrates to their respective translocation apparatus, via either a SecA2-mediated accessory Sec system or a SecA-mediated canonical Sec system. What drives the selection of the secretion route is not yet clear. Since accessory SecA2 shares considerable homology with canonical SecA (7) and exhibits ATPase activity (2), SecA2 may interact with SecA. The interactions of canonical SecA with accessory SecA2, as well as with another accessory Sec component, Gap3, identified in this study, provide direct evidence that the accessory Sec system engages in cross talk with the canonical Sec system. The interaction may play a modulatory role in monitoring and directing the export of glycosylated and unglycosylated serine-rich proteins. Canonical SecA exists in both monomeric and dimeric forms (11, 17). The monomeric and dimeric SecA interact with SecYEG to mediate the function of the canonical Sec translocon. The ratio of SecA monomers to dimers bound to the SecYEG complex regulates the activity associated with the translocon (12). Therefore, it is possible that the ratio of homodimeric to heterodimeric SecA and SecA2 in bacterial cells may direct the affinity of the accessory Sec translocon to glycosylated or unglycosylated SRRPs, thereby modulating the selectivity for the accessory Sec apparatus.
Interestingly, Gap3 but not Gap1 can interact with both SecA and SecA2. The Gap3 homologue Asp3 from S. gordonii has been proposed to function as a scaffold protein to orchestrate the accessory Sec complex (Asp1-Asp2-Asp3-SecA2). Asp3 is also involved in binding to the serine-rich repeat (SRR) domains of GspB as a potential chaperone that facilitates the biogenesis of GspB (35). The ability to interact with both SecA and SecA2 indicates that Gap3 and its homologues are capable of bridging accessory and canonical Sec complexes, further supporting their role as a scaffold protein.
In summary, we have identified an accessory Sec protein complex and its association with a canonical Sec system, which provides new insights into the biogenesis of SRRPs.
ACKNOWLEDGMENTS
We thank Jeannine Brady from University of Florida and David Pritchard from UAB for providing us with the S. agalactiae clinical isolates.
This study was supported by NIH R01DE017954 (H. Wu) from the National Institute of Dental and Craniofacial Research and by grant 30900034 (M. Zhou) from the National Natural Science Foundation of China.
Footnotes
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Bensing B. A., Sullam P. M. 2002. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol. Microbiol. 44:1081–1094 [DOI] [PubMed] [Google Scholar]
- 2. Bensing B. A., Sullam P. M. 2009. Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J. Bacteriol. 191:3482–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bensing B. A., Takamatsu D., Sullam P. M. 2005. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol. Microbiol. 58:1468–1481 [DOI] [PubMed] [Google Scholar]
- 4. Burnette-Curley D., et al. 1995. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect. Immun. 63:4669–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Carlsson J., Grahnen H., Jonsson G., Wikner S. 1970. Establishment of Streptococcus sanguis in the mouths of infants. Arch. Oral Biol. 15:1143–1148 [DOI] [PubMed] [Google Scholar]
- 6. Chen Q., Sun B., Wu H., Peng Z., Fives-Taylor P. M. 2007. Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J. Bacteriol. 189:7610–7617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Q., Wu H., Fives-Taylor P. M. 2004. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis [sic] FW213. Mol. Microbiol. 53:843–856 [DOI] [PubMed] [Google Scholar]
- 8. Chen Q., Wu H., Kumar R., Peng Z., Fives-Taylor P. M. 2006. SecA2 is distinct from SecA in immunogenic specificity, subcellular distribution and requirement for membrane anchoring in Streptococcus parasanguis [sic]. FEMS Microbiol. Lett. 264:174–181 [DOI] [PubMed] [Google Scholar]
- 9. Cisar J., Brennan M., Sandberg A. 1985. Lectin-specific interaction of actinomyces fimbriae with oral streptococci. American Society for Microbiology, Washington, DC [Google Scholar]
- 10. Cole R. M., Calandra G. B., Huff E., Nugent K. M. 1976. Attributes of potential utility in differentiating among “group H” streptococci or Streptococcus sanguis. J. Dent. Res. 55:A142–A153 [DOI] [PubMed] [Google Scholar]
- 11. Ding H., Hunt J. F., Mukerji I., Oliver D. 2003. Bacillus subtilis SecA ATPase exists as an antiparallel dimer in solution. Biochemistry 42:8729–8738 [DOI] [PubMed] [Google Scholar]
- 12. Duong F. 2003. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 22:4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fives-Taylor P. M., Thompson D. W. 1985. Surface properties of Streptococcus sanguis FW213 mutants nonadherent to saliva-coated hydroxyapatite. Infect. Immun. 47:752–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Froeliger E. H., Fives-Taylor P. 2001. Streptococcus parasanguis [sic] fimbria-associated adhesin fap1 is required for biofilm formation. Infect. Immun. 69:2512–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goh K. I., et al. 2007. The human disease network. Proc. Natl. Acad. Sci. U. S. A. 104:8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jenkinson H. F., Lamont R. J. 1997. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 8:175–200 [DOI] [PubMed] [Google Scholar]
- 17. Jilaveanu L. B., Zito C. R., Oliver D. 2005. Dimeric SecA is essential for protein translocation. Proc. Natl. Acad. Sci. U. S. A. 102:7511–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., et al. 2008. A conserved domain of previously unknown function in Gap1 mediates protein-protein interaction and is required for biogenesis of a serine-rich streptococcal adhesin. Mol. Microbiol. 70:1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mistou M. Y., Dramsi S., Brega S., Poyart C., Trieu-Cuot P. 2009. Molecular dissection of the secA2 locus of group B Streptococcus reveals that glycosylation of the Srr1 LPXTG protein is required for full virulence. J. Bacteriol. 191:4195–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng Z., et al. 2008. Identification of critical residues in Gap3 of Streptococcus parasanguinis involved in Fap1 glycosylation, fimbrial formation and in vitro adhesion. BMC Microbiol. 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rigel N. W., Braunstein M. 2008. A new twist on an old pathway—accessory Sec [corrected] systems. Mol. Microbiol. 69:291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Seepersaud R., Bensing B. A., Yen Y. T., Sullam P. M. 2010. Asp3 mediates multiple protein-protein interactions within the accessory Sec system of Streptococcus gordonii. Mol. Microbiol. 78:490–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seifert K. N., et al. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology 152:1029–1040 [DOI] [PubMed] [Google Scholar]
- 24. Siboo I. R., Chaffin D. O., Rubens C. E., Sullam P. M. 2008. Characterization of the accessory Sec system of Staphylococcus aureus. J. Bacteriol. 190:6188–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stephenson A. E., et al. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis [sic] in an in vitro tooth model. Mol. Microbiol. 43:147–157 [DOI] [PubMed] [Google Scholar]
- 26. Takamatsu D., Bensing B. A., Sullam P. M. 2004. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol. Microbiol. 52:189–203 [DOI] [PubMed] [Google Scholar]
- 27. Tettelin H., et al. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tettelin H., et al. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506 [DOI] [PubMed] [Google Scholar]
- 29. van Driel M. A., Bruggeman J., Vriend G., Brunner H. G., Leunissen J. A. 2006. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 14:535–542 [DOI] [PubMed] [Google Scholar]
- 30. Wu H., Bu S., Newell P., Chen Q., Fives-Taylor P. 2007. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis [sic]. J. Bacteriol. 189:1390–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu H., Fives-Taylor P. M. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis [sic]. Mol. Microbiol. 34:1070–1081 [DOI] [PubMed] [Google Scholar]
- 32. Wu H., Mintz K. P., Ladha M., Fives-Taylor P. M. 1998. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis [sic] FW213. Mol. Microbiol. 28:487–500 [DOI] [PubMed] [Google Scholar]
- 33. Wu H., Zeng M., Fives-Taylor P. 2007. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect. Immun. 75:2181–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu P., et al. 2007. Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 189:3166–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yen Y. T., Seepersaud R., Bensing B. A., Sullam P. M. 2011. Asp2 and Asp3 interact directly with GspB, the export substrate of the Streptococcus gordonii accessory Sec system. J. Bacteriol. 193:3165–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou M., Peng Z., Fives-Taylor P., Wu H. 2008. A conserved C-terminal 13-amino-acid motif of Gap1 is required for Gap1 function and necessary for the biogenesis of a serine-rich glycoprotein of Streptococcus parasanguinis. Infect. Immun. 76:5624–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou M., Wu H. 2009. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology 155:317–327 [DOI] [PubMed] [Google Scholar]