Background: A dedicated chaperone is required to target the secretin PulD to the outer membrane.
Results: The unstructured C-terminal 28 residues of PulD fold upon interaction with its chaperone to form a high affinity complex.
Conclusion: An unstructured chaperone-binding domain seems to provide a balance between efficient targeting and proteolysis.
Significance: Recognition of intrinsically disordered regions facilitates key interactions for protein targeting and assembly of trans-envelope machineries.
Keywords: Chaperone Chaperonin, Escherichia coli, Intrinsically Disordered Proteins, Membrane Proteins, Secretion, Outer Membrane, Periplasm
Abstract
Interaction of bacterial outer membrane secretin PulD with its dedicated lipoprotein chaperone PulS relies on a disorder-to-order transition of the chaperone binding (S) domain near the PulD C terminus. PulS interacts with purified S domain to form a 1:1 complex. Circular dichroism, one-dimensional NMR, and hydrodynamic measurements indicate that the S domain is elongated and intrinsically disordered but gains secondary structure upon binding to PulS. Limited proteolysis and mass spectrometry identified the 28 C-terminal residues of the S domain as a minimal binding site with low nanomolar affinity for PulS in vitro that is sufficient for outer membrane targeting of PulD in vivo. The region upstream of this binding site is not required for targeting or multimerization and does not interact with PulS, but it is required for secretin function in type II secretion. Although other secretin chaperones differ substantially from PulS in sequence and secondary structure, they have all adopted at least superficially similar mechanisms of interaction with their cognate secretins, suggesting that intrinsically disordered regions facilitate rapid interaction between secretins and their chaperones.
Introduction
Secretins form large homomultimeric protein complexes in the outer membrane (OM)3 of Gram-negative bacteria to provide the gateway to the extracellular milieu for many trans-envelope secretion machineries such as type II secretion, type IV pilus, type III secretion needle assembly, and filamentous bacteriophage secretion systems (1). Secretin complexes are highly stable, SDS-resistant oligomers of 12–14 subunits that form gated channels of ∼6 nm diameter (2–6). They all have a similar domain organization comprising a relatively divergent N-terminal periplasmic region of varying length and repeat sequences that associates with other components of the secretion machinery and/or confers substrate specificity (1). Adjacent to this region is the more conserved C domain (PRAM secretin, PF00263) that is involved in multimerization and membrane insertion (1). The C domain is extended by short (30–70 amino acids) divergent sequences that, in some cases, interact with dedicated chaperones (7–11). PulD, an archetypical member of the secretin family, is part of the Klebsiella oxytoca type II secretion system that secretes the amylolytic enzyme pullulanase (PulA) (12). PulD belongs to a subclass of secretins that are strictly dependent on chaperones called pilotins (PulS in the case of PulD (13)) for correct targeting to the OM.
Most secretins require dedicated chaperones or accessory proteins for stabilization, oligomerization, targeting, and membrane association (14). Although many of these chaperones perform at least superficially similar functions, they are surprisingly unrelated in sequence and structure. Most of them are small OM lipoproteins (120–150 residues). MxiM, the Shigella chaperone for the type III secretion system secretin MxiD, is composed of eight antiparallel β-strands that form a pseudo-barrel that is “cracked” on one side by a 310 helix and an α-helix that form a deep hydrophobic cavity (15). In contrast, the PilQ chaperones PilF from Pseudomonas aeruginosa and PilW from Neisseria meningitidis comprise 13 antiparallel α-helices that fold into six tetratricopeptide motifs (16–18), known to be involved in protein-protein interactions. MxiM, PilF, and PilW differ substantially from PulS (<15% sequence identity). Furthermore, although many accessory proteins like PulS, OutS (the PulS homolog from Erwinia chrysanthemi), and MxiM bind the C-terminal region of secretins (10, 11), other chaperones, for example YscW from Yersinia enterocolitica, bind elsewhere (19). Comparisons of secretin chaperone sequences and functions allow their classification into different families; the PulS family, called pilotins, is found in only a limited number of type II secretion systems that share high sequence similarity and appears to have a different mode of function from other secretin chaperones such as MxiM and PilF/W, leading us to predict that its structure would also be different.
PulS is a triacylated lipoprotein that localizes to the OM of Escherichia coli and interacts specifically with the S domain (Sdom) of PulD to protect PulD monomers from degradation and to target them to the OM via the conserved OM lipoprotein-sorting pathway (7, 9, 20, 21). In vitro studies demonstrated that PulD does not require PulS for multimerization or membrane insertion (22). In the absence of PulS, or when Sdom is deleted, PulD mislocalizes to the inner membrane, where it induces the phage shock response (7, 9). The fusion of PulD Sdom to the C terminus of the f1 bacteriophage phage secretin pIV increased the efficiency of its targeting to the OM in the presence of PulS, but the absence of PulS rendered the chimera susceptible to proteolysis (20). Likewise, fusion of Sdom to the C terminus of periplasmic maltose-binding protein (MalE) rendered it more susceptible to proteolysis (9). Furthermore, a soluble periplasmic version of PulS (MalE-PulS) stabilized PulD but did not target it to the OM, thereby uncoupling protection and targeting functions of PulS (9). PulD completely lacking Sdom was degraded even in the presence of PulS, but removal of only the last 25 residues of Sdom abolished targeting but not protection by PulS (20, 21, 23).
To understand better how PulS protects and targets PulD to the OM, we studied the interaction between PulS and a soluble polypeptide corresponding to Sdom in vitro. We determined their stoichiometry and affinity, as well as their secondary structures before and after association, and we identified a minimal PulS binding region in Sdom. We discuss the data in relation to the structures of other secretin chaperones and their interactions with their cognate secretins.
EXPERIMENTAL PROCEDURES
Materials
Unless otherwise indicated, chemicals were of molecular biology grade and were purchased from Sigma. Thermolysin (Sigma), thrombin (Novagen), and Factor Xa (New England Biolabs) were used according to the manufacturers' recommendations. DNA polymerases, restriction endonucleases, and DNA modification enzymes were from New England Biolabs, Invitrogen, or Roche Applied Science. Vectors pMal-c2 and pMal-p2X were purchased from New England Biolabs, and pQE30 was obtained from Qiagen. Plasmid purification, PCR purification, and gel extraction kits were purchased from Qiagen and used according to the manufacturers' recommendations.
Plasmid Construction
DNA encoding PulS without its signal peptide was cloned into the pMal-p2X vector, previously modified to include an additional His6 tag at the MalE N terminus (17). pulS was amplified using forward primer PNN19 (see supplemental Table 1 for primer sequences) with a SnaBI restriction site to replace Cys18+ with Val18+ after Factor Xa cleavage, and reverse primer PNN12 with a HindIII site, and cloned into XmnI-HindIII digested pMal-p2X. DNA encoding the C-terminal 71 amino acids of PulD (Sdom) was amplified with primers PNN28 and PNN29 and cloned into the BamHI-HindIII sites of pQE30. Alternatively, DNA encoding Sdom, SdomA, and SdomB were constructed as MalE fusions with primers PNN55 and PNN56, PNN55 and PNN58, and PNN57 and PNN56, respectively, in pMal-c2 digested with EcoRI and HindIII. Primers were designed to encode an N-terminal thrombin cleavage site to allow removal of MalE. The region corresponding to SdomA (Glu599–Leu628) in full-length pulD, from vector pCHAP3635 (21), was either deleted using primer pair PNN67 and PNN68 or replaced with the linker region (ENSSSNNNNNNNNNNLGIEGRISEFGSSRV) of the pMal-c2 vector using primer pair PNN69 and PNN70 by two-step inverse PCR, and the resulting plasmids were used for in vivo complementation studies. All constructs were verified by DNA sequencing.
Protein Preparation
E. coli producing MalE-PulS and His6-Sdom were grown in Terrific Broth (24, 25) to A600 of 0.5 and induced with 0.5 mm isopropylthiogalactoside for 4 h at either 30 or 37 °C. Cells were harvested, resuspended in lysis buffer (20 mm sodium phosphate buffer, pH 7.5, 500 mm NaCl, 10 mm imidazole, Complete EDTA-free protease inhibitor mixture (Roche Applied Science) and 20 μg/ml of DNase and RNase), lysed either by sonication or with a French press and centrifuged at 27,000 × g for 20 min to remove the debris. Recombinant proteins were purified from the supernatant fraction by affinity chromatography using a 5-ml HiTrap chelating column (GE Healthcare) charged with nickel and equilibrated in 20 mm sodium phosphate buffer, pH 7.5, with 500 mm NaCl and 10 mm imidazole. Proteins were eluted with a linear 10–500 mm imidazole gradient. MalE-PulS was dialyzed against 50 mm HEPES, pH 7.5, 20 mm NaCl before overnight digestion with either Factor Xa (1:50) or thermolysin (1:1000), and PulS was purified by cation exchange using a HiTrap 1-ml SP-Sepharose column (GE Healthcare). The sample was further purified, and the buffer was exchanged to 20 mm Tris-Cl, pH 8.0, 150 mm NaCl by size exclusion chromatography using a HiLoad 16/60 Superdex 200 column (GE Healthcare). Factor Xa cleaved specifically between the MalE fusion and the PulS protein to yield full-length PulS with Cys18+ changed to Val18+. Alternatively, limited proteolysis trials revealed that PulS could also be cleaved from the MalE fusion by thermolysin treatment, cleaving specifically at Val30+ of PulS. All data obtained with thermolysin-treated PulS was indistinguishable from those obtained with full-length PulS; therefore, only the former are shown.
After affinity purification, His6-Sdom was dialyzed against 20 mm Tris-Cl, pH 8.0, and further purified on a 1-ml Resource Q column (GE Healthcare). MalE fused to Sdom, SdomA, or SdomB (see text for description of these polypeptide fragments) was purified from the soluble fraction of cell lysates in 20 mm Tris-Cl, pH 8.0, 200 mm NaCl, and 1 mm EDTA on amylose-agarose resin (New England BioLabs) and eluted with 20 mm maltose. Pooled fractions of the chimeras with or without digestion by thrombin (1:1000 molar ratio of protease to fusion protein) to remove MalE were dialyzed against 20 mm Tris-Cl, pH 8.0, and purified on a HiTrap Capto Q 1-ml column (GE Healthcare) with a linear NaCl gradient. All batch purifications were performed on an ÄKTA Purifier FPLC system (GE Healthcare), and analytical studies were performed on an Ettan LC HPLC system. Analytical size exclusion chromatography of proteins and complexes was performed with a prepacked Superdex 75 PC 3.2/30 column with a bed volume of 2.4 ml (GE Healthcare). A280 was measured to estimate the protein content in the samples, and the concentration was calculated from extinction coefficients obtained from the ExPASy ProtParam tool. All purified proteins and processing events were confirmed by Edman N-terminal sequencing and mass spectrometry, and the absence of aggregates was confirmed by dynamic light scattering measurements (DynaPro Plate reader, Wyatt Technology).
Strains, Growth Conditions, and in Vivo Complementation Studies
E. coli strain PAP105 (Δ(lac-pro) F′ (lacIq1 ΔlacZM15 proAB+ Tn10)) was used for plasmid manipulations and complementation studies. Strains were grown under aeration conditions at 30 °C in LB broth or LB with 10% M63 minimal media, with 0.4% maltose for induction of the secretion machinery and antibiotics where appropriate (chloramphenicol, 25 μg·ml−1; ampicillin, 100 μg·ml−1). For secretion assays, pCHAP8243 (a derivative of pCHAP231 carrying the full pul operon with a pulD deletion and encoding soluble nonacylated variant of PulA (26)) complemented with pulD (pCHAP3635) and pulD variants was grown to an A600 of ∼1.5 and centrifuged to separate cells and supernatant. To observe PulS protection and targeting of PulD, PAP105 carrying wild type PulD or the PulD variants was grown with or without the presence of PulS (pCHAP585 (20)). Samples were separated by 10% SDS-PAGE, transferred to nitrocellulose membrane, probed with protein-specific antibodies, and detected using enhanced chemiluminescence (GE Healthcare).
Limited Proteolysis and SDS-PAGE
50 μm PulS, 50 μm Sdom, or an equimolar mixture of the two were digested with 30, 15, or 7.5 nm thermolysin for 4 h at room temperature, and the reaction was stopped by adding 2 mm 1,10-phenanthroline. Samples were heated to 100 °C in SDS-PAGE loading buffer with 5 mm DTT and separated by electrophoresis in 12% polyacrylamide gels (SDS-PAGE) using Tris-glycine-HCl buffers. Protein markers PageRuler Plus and Spectra Multicolor low range protein ladders were from Fermentas.
Circular Dichroism (Far-UV) Spectroscopy
CD spectra were recorded on an Aviv 215 spectropolarimeter (Aviv Biomedical) with protein samples at 0.4–0.8 mg·ml−1 in 20 mm Tris-Cl, pH 8.0, 150 mm NaF. Far-UV CD spectra were recorded between 190 and 260 nm using a cylindrical cell with a 0.01-cm path length and an averaging time of 1 s per step. Three consecutive scans from each sample were merged to produce an averaged spectrum and corrected using buffer base lines measured under the same conditions. Data were normalized to the molar peptide bond concentration and path length and expressed as mean residue ellipticity ([θ] degree·cm2·dmol−1). The relative secondary structure content was estimated from the far-UV CD spectra using the CDSSTR routine (27) of the DICHROWEB server (28, 29) run on the SP175 reference dataset (30) containing 72 proteins representing a large panel of secondary structures. Similar results were obtained from different datasets (31) and using the CONTIN/LL routine (32). Thermal denaturation of the proteins was measured by monitoring the change in ellipticity at 206 or 222 nm over the range of 20–100 °C, in increments of 1 °C and an averaging time of 4 s per step. The experimental denaturation profiles were analyzed by a nonlinear least squares fit assuming a two-state transition and used to calculate the melting temperature (Tm) and enthalpy of unfolding (ΔH) (33, 34). Dual cell cuvettes containing two adjacent chambers of 0.44 cm each were used for mixing experiments to determine the gain in secondary structure of the complex. CD spectra from 203 to 260 nm were recorded for 50 μm PulS or 50 μm Sdom, with the second compartment filled with buffer as a control. Subsequently, PulS and Sdom were placed in either one of the two chambers, and their combined spectra were acquired before mixing and after mixing. The change in spectra was attributed to a gain or loss of secondary structure upon complex formation. Raw data are presented as ellipticity in millidegrees.
Fluorescence Spectroscopy
Fluorescence studies were performed on a PTI Quanta-Master QM4CW spectrofluorometer (Photon Technology International) at 25 °C using a 10-mm wide quartz cell. The excitation wavelength was 295 nm, and intrinsic fluorescence emission of the single tryptophan residue of PulS was recorded from 310 to 400 nm. Bandwidths of excitation and emission monochromators were set at 1 and 10 nm, respectively. Cells were charged with 1 μm of PulS in a final volume of 1 ml, and Sdom was added in 0.1 μm increments.
Nuclear Magnetic Resonance Spectroscopy
The 1H NMR spectrum of 0.3 mm Sdom (30 mm Tris-Cl, pH 8.0, 200 mm NaCl, 10% D2O, total volume of 240 μl) was acquired on an INOVA spectrometer (Varian) operating at 600-MHz 1H frequency and equipped with a cryogenic probe, with 64 scans and 1024 points at 25 °C. The water signal was suppressed by excitation sculpting with gradients.
Analytical Ultracentrifugation
Equilibrium and velocity experiments were performed at 20 °C in 20 mm Tris-Cl, pH 8.0, 150 mm NaCl on a Proteomelab XL-I analytical ultracentrifuge (Beckman Coulter) equipped with an AN60-Ti rotor. The protein concentration as a function of radial position and time was measured by absorbance at 280 nm. The buffer viscosity η = 1.021 centipoise and density ρ = 1.00499 g·ml−1 were calculated with Sednterp 1.09, as well as the partial specific volume of PulS, v̄ = 0.723 ml·g−1. The partial specific volume of Sdom, v̄ = 0.667 ml·g−1 was measured by sedimentation equilibrium experiments using the computed molecular mass. The partial specific volume of the PulS-Sdom mixture was calculated by weight average of PulS and Sdom partial specific volume values, v̄ = 0.699 ml·g−1. For sedimentation equilibrium experiments, Sdom samples (120 μl at 25 μm, 50 μm, 100 μm) were loaded in a 1.2-mm thick double sector aluminum centerpiece and centrifuged successively for 27 h at rotor speeds of 26,000 rpm, 23 h at 32,000 rpm, and 16 h at 45,000 rpm. Data were recorded for each speed after controlling that the sedimentation/diffusion equilibrium had been reached. The base line was measured at 60,000 rpm after 6 h. Radial distributions were analyzed by global fitting of the three speeds using the one species model of the UltraScan 9.9 software (35). For sedimentation velocity experiments, the protein samples (400 μl each) of 100 μm PulS, 100 μm Sdom, and of the mixture of PulS-Sdom at a ratio of 50:100, 75:75, and 100:50 μm, were loaded in 1.2-mm thick double sector aluminum centerpiece and spun at 60,000 rpm. Sedimentation velocity profiles were monitored at 4-min intervals. Data were analyzed with the Sedfit 12.0 software using a continuous size distribution c(s) model (36).
Intrinsic Viscosity, Molecular Mass, and Viscosity Increment Measurements
200-μl samples of 200 μm Sdom or 130 μm 1:1 molar ratio PulS-Sdom complex were separated according to their hydrodynamic size on a Superdex-75 column (GE Healthcare), in 20 mm Tris-Cl, pH 8.0, 150 mm NaCl at 20 °C, connected on line to a model 302 triple detector array (Viscotek, Malvern Instruments Ltd.). The triple detector array contains four in-line detectors as follows: (i) a static light-scattering cell with two photodiode detectors at 7° for low angle light scattering and at 90° for right angle laser light scattering; (ii) a deflection refractometer; (iii) a photometer, and (iv) a 4-capillary differential viscometer that measures pressure imbalances of the eluted sample by differential pressure transducers. Protein concentration was determined using both the photometer and the deflection refractometer, and right angle laser light scattering and low angle light scattering data coupled to the concentration provided molecular mass determination and differential viscometer measurements combined with concentration gave the intrinsic viscosity [η]. General procedures and conditions have been described previously (37). BSA was used for molecular mass calibration, and polyethylene oxide standards (Malvern Instruments Ltd.) were used for intrinsic viscosity calibration. All data were acquired and processed using the Omnisec software. The viscosity increment (ν) and the hydration (δ) were calculated as described previously (37, 38). Briefly, the viscosity increment ν (also called the Simha-Einstein hydrodynamic function) was calculated from the Einstein viscosity relation using the following equation (39): M[η] = νVHNA, where VH is the hydrodynamic volume defined by VH = 4πRH/3, and NA is Avogadro's number.
Surface Plasmon Resonance
Assays were performed on the Biacore 2000 instrument (GE Healthcare) equilibrated at 25 °C in 20 mm Tris-Cl, pH 8.0, 150 mm NaCl supplemented with 0.005% Tween 20. The monoclonal α-MalE mAb565 antibody (40) was immobilized on three flow cells of a CM5 sensor chip (GE Healthcare) to a level of 12,000–15,000 resonance units (resonance units ∼pg/mm2). For separate experiments, 1,200–2,000 resonance units of ligand (MalE-PulS, MalE-Sdom, or MalE-SdomB) were captured on the surface, while maintaining one free flow cell as an α-MalE blank. Preliminary studies determined that it was not possible to measure directly the true binding affinity using MalE-PulS captured on the surface and Sdom flowed over the surface as the analyte due to mass transport limitations. As an alternative, competition experiments designed to measure binding constants in solution provided a reliable estimation of the true binding affinity (41). For competition experiments, 10 or 30 nm PulS alone or pre-equilibrated with competitor (Sdom, SdomA, or SdomB; 0.2 nm to 1 μm) was injected over the surface loaded with either MalE-Sdom or MalE-SdomB for 4 min at 50 μl/min to allow steady-state equilibrium to be reached. The appropriate conditions and protein concentrations were used to allow accurate determination of Sdom and SdomB affinities. All profiles were double referenced by subtracting the signals from the reference surface and the blank injections of running buffer using the Scrubber 2.0 software (BioLogic Software). Data analysis and affinity calculations were carried out using the BIAEvaluation software (GE Healthcare).
RESULTS
PulS Chaperone Interacts Specifically with the C-terminal 71 Residues of PulD
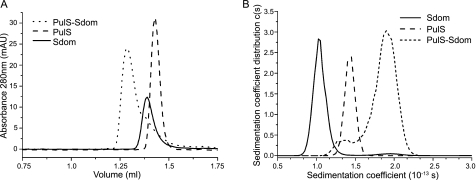
Genetic and in vivo studies on PulD demonstrated that the PulS-binding site is within the C-terminal 71 amino acids (20, 23). To understand better the interaction between PulD and PulS, we constructed soluble variants of both proteins. PulS has a conserved intramolecular disulfide bond that is required for function (42). To preserve the disulfide bond, PulS was exported to the E. coli periplasm as a C-terminal MalE chimera, from which MalE was subsequently removed by thermolysin digestion. PulD spontaneously forms multimers and inserts into membranes, and it can only be purified in ZW3-14 detergent, making it difficult to study the interaction with its pilotin in vitro. Therefore, DNA encoding the periplasmically exposed C-terminal 71 amino acids of PulD (Sdom) was cloned into the pQE30 vector system incorporating DNA for an N-terminal His6 tag. Both polypeptides were soluble and were purified to homogeneity, as determined by SDS-PAGE, N-terminal sequencing, mass spectrometry, and dynamic light scattering (see under “Experimental Procedures”). Size exclusion chromatography showed that the purified proteins migrated as single species that formed a stable complex when mixed in a 1:1 ratio (Fig. 1A), in line with previously determined stoichiometry of PulD and PulS in complexes purified in ZW3-14 (3, 4).
FIGURE 1.
PulS and Sdom form a stable 1:1 complex by size exclusion chromatography and analytical ultracentrifugation. A, size exclusion chromatography elution profiles of 40 μm PulS, 40 μm Sdom, and an equimolar ratio of PulS and Sdom, preformed at 20 °C. B, continuous sedimentation coefficient distribution analysis of 100 μm Sdom, 100 μm PulS, and PulS/Sdom mixture (75 μm Sdom, 75 μm PulS). Sedimentation coefficients are expressed in Svedberg units (1 S = 10−13 s).
Because Sdom does not have any tryptophan residues and PulS has only one, steady-state tryptophan fluorescence was measured as an indicator of Sdom binding to PulS. PulS had a maximum fluorescence emission (λmax) of 347 nm and underwent a 3-nm blue shift and concurrent decrease in intensity upon complex formation. Titration of PulS with Sdom showed that the complex was in a 1:1 stoichiometry with an affinity in the low nanomolar range (supplemental Fig. S1).
PulS and Sdom Form a 1:1 Complex and Sdom Is Elongated
Ultracentrifugation sedimentation velocity experiments were performed to characterize the behavior of PulS, Sdom, and the complex in solution (Fig. 1B and Table 1). PulS and Sdom had sedimentation characteristics of monomers in solution (Fig. 1B). An equimolar mixture or mixtures with an excess of either protein sedimented as expected for a 1:1 stoichiometry complex with corresponding absorbance and interference signal (Fig. 1B and data not shown). The hydrodynamic radii (RH) calculated from the sedimentation coefficient (Table 1) corresponded to the values determined from dynamic light scattering analysis. The frictional ratio (f/f0) of PulS, calculated from the molecular mass and the sedimentation coefficient, was 1.2, typical of a globular protein, whereas that of Sdom was 1.9, much higher than the theoretical 1.4 value for a globular protein of the same molecular mass and amino acid sequence. An equimolar mixture of PulS and Sdom contained a new main species with a sedimentation coefficient of 1.9 ± 0.1 S and a frictional ratio of 1.5.
TABLE 1.
Hydrodynamic properties of PulS, Sdom, and the PulS-Sdom complex
| PulS | Sdom | PulS-Sdom | |
|---|---|---|---|
| Molecular massa | NDb | 9.7 ± 0.1 kDa | 22.1 ± 0.7 kDa |
| Sedimentation coefficient (10−13 s)c | 1.4 ± 0.1 | 1.1 ± 0.1 | 1.9 ± 0.1 |
| RH (nm)c | 1.7 | 2.5 | 2.7 |
| Frictional ratio (f/f0)d | 1.2 | 1.9 | 1.5 |
| Intrinsic viscosity (ml·g−1)a | NDb | 16.9 ± 0.7 | 5.9 ± 0.1 |
| Viscosity increment, νe | NDb | 4.4 | 2.5 |
a Data were determined by static light scattering and viscometry experiments using size exclusion chromatography-coupled triple detector array.
b ND means not determined.
c Data were determined by analytical ultracentrifugation experiments.
d Data were calculated from the experimental hydrodynamic radius and the hydrodynamic radius of an anhydrous and spherical molecule with equivalent molecular mass and partial specific volume.
e Data were calculated using Einstein's viscosity relation, see under “Experimental Procedures.”
In conclusion, soluble Sdom behaves as either an elongated protein or a coil-like, intrinsically disordered protein with an unusually large frictional ratio value for a protein of its size, whereas PulS behaves like a typical globular protein, as determined from distributions of the frictional ratios of characterized proteins as a function of their molar mass (43, 44). The complex behaves like an intermediate state protein with characteristics similar to a premolten globule-like protein. Thus, PulS and Sdom interact to form a stable 1:1 complex, and Sdom behaves as an elongated and/or unfolded protein.
Circular Dichroism and NMR Analysis Confirm That Sdom Is Intrinsically Disordered
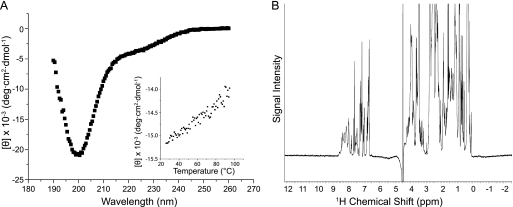
Sdom elutes as an oligomer or elongated monomer during size exclusion chromatography, migrates aberrantly upon SDS-PAGE, and has a higher frictional ratio than would be expected for a globular protein of its molecular mass, indicative of an unfolded or extended conformation. To characterize Sdom further, far-UV circular dichroism (CD) was used to analyze its secondary structure. Sdom had a CD spectrum typical of a predominantly unfolded or intrinsically disordered protein (43), as indicated by the strong negative band around 200 nm and a weak negative shoulder near 220 nm (Fig. 2A). Consistent with these results, Sdom did not show a clear cooperative unfolding transition during thermal denaturation, confirming the absence of secondary structure (Fig. 2A, inset). One-dimensional 1H NMR analysis of Sdom revealed a spectral dispersion of the amide and aromatic protons in the range of 7–9 ppm. This range is more restricted than the well separated signal dispersion (6–11 ppm) typically observed in folded proteins, consistent with the predicted unfolded state of Sdom (Fig. 2B).
FIGURE 2.
Sdom is an intrinsically disordered polypeptide. A, far-UV CD spectra of 100 μm Sdom from 190 to 260 nm. The inset shows the thermal denaturation profile of Sdom from 20 to 100 °C monitored by CD at 206 nm, in increments of 1 °C, and an averaging time of 4 s per step. [θ] means mean residual ellipticity. B, NMR one-dimensional proton spectrum of 300 μm Sdom recorded at 25 °C.
Increase in Secondary Structure upon Complex Formation
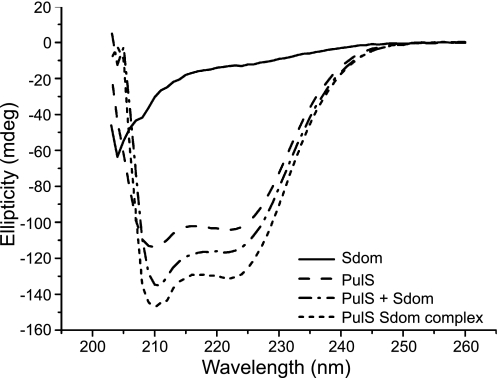
CD was used to track secondary structure changes upon interaction of Sdom with PulS. The far-UV spectrum of PulS was characteristic of an α-helical protein with minima at 208 and 222 nm (supplemental Fig. S2), and deconvolution of the secondary structure predicted a helical content of 84% (Table 2). The spectrum of an equimolar PulS-Sdom complex revealed mainly α-helices with some unordered regions. The sum of the individual spectra from PulS and Sdom did not add up to the combined α-helical signal obtained with the complex (Table 2), suggesting that Sdom and/or PulS gain additional secondary structure when they interact. This gain in secondary structure was quantified by dual cuvette analysis, which revealed a 10% gain in ellipticity at 222 nm, consistent with an overall gain of α-helical content upon complex formation (Fig. 3). Because PulS is almost fully structured, we hypothesize that the gain in α-helical content occurs predominantly in Sdom, indicating a disorder-to-order mode of interaction with PulS. Thermal denaturation analysis showed that the PulS-Sdom complex is more stable than PulS alone (Table 2). Therefore, Sdom and PulS form a complex that is more stable than either of the proteins alone.
TABLE 2.
Secondary structure of PulS, Sdom, and the PulS-Sdom complex
| Sample | Relative secondary structurea |
Tmb | ΔHb | |||
|---|---|---|---|---|---|---|
| α | β | Turn | Unordered | |||
| °C | kcal/mol | |||||
| PulS | 84 | 0 | 7 | 8 | 77.3 ± 0.1 | 60.8 ± 1.0 |
| Sdom | 17 | 19 | 19 | 46 | NAc | NA |
| PulS + Sdomd | 54 | 5 | 12 | 30 | NA | NA |
| Complex | 63 | 3 | 11 | 24 | 80.6 ± 0.1 | 70.2 ± 1.0 |
a Relative secondary structure was estimated from the far-UV CD spectra using the CDSSTR routine of the DICHROWEB server run on the SP175 reference dataset, see “Experimental Procedures.”
b The thermal denaturation profiles were analyzed by a nonlinear least squares fit assuming a two-state transition and were used to calculate the melting temperature (Tm) and enthalpy of unfolding (ΔH); see under “Experimental Procedures.”
c NA means not applicable.
d Theoretical spectrum of an equimolar mixture of PulS + Sdom was calculated as a mean of each individual spectrum weighted according to their masses and the number of each amino acid in each protein and deconvoluted to give the predicted secondary structure.
FIGURE 3.
Dual cuvette circular dichroism analysis shows an increase in secondary structure upon PulS-Sdom complex formation. Far-UV CD spectra of Sdom + buffer, PulS + buffer, and PulS + Sdom before mixing and PulS Sdom complex after mixing were recorded in a dual cuvette from 203 to 260 nm.
Sdom Undergoes Compaction upon Complex Formation
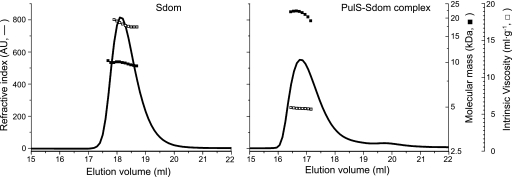
To characterize further the mechanism of complex formation, the hydrodynamic properties of Sdom and the PulS-Sdom complex were measured by size exclusion chromatography coupled with a triple detector array (Viscotek) (results summarized in Table 1), allowing measurement of molecular mass, hydrodynamic radius, and intrinsic viscosity of the eluted peak (Fig. 4). The results confirmed the molecular mass (9.7 ± 0.3 kDa), the sedimentation coefficients, and hydrodynamic radii of Sdom and the complex obtained by sedimentation analysis (described above). The intrinsic viscosity of Sdom, 16.9 ± 0.7 ml·g−1, was measured as a function of pressure imbalances generated using the differential pressure inducer. This high intrinsic viscosity value, which is compatible with the frictional ratio measured by sedimentation analysis, is typical of an elongated and/or highly hydrated molecule. The PulS-Sdom complex had a molecular mass of 22.1 ± 0.7 kDa and an intrinsic viscosity of 5.9 ± 0.1 ml·g−1, which is again similar to the data obtained by sedimentation analysis. Even though the intrinsic viscosity of the complex is smaller than that of Sdom alone, it is still somewhat higher than that of a globular protein such as BSA (4.1 ml·g−1), showing that the complex is slightly elongated and/or is highly hydrated. The viscosity increment (ν) or shape factor of Sdom (4.4) was between that of a sphere (ν = 2.5) and an elongated protein such as IgG (ν = 7.5) (45), whereas the viscosity increment of the PulS-Sdom complex was 2.5, showing that it is globular. Thus, the hydrodynamic parameters calculated from the experimental data support the idea that Sdom is intrinsically disordered and that it undergoes significant compaction upon complex formation.
FIGURE 4.
High intrinsic viscosity of Sdom decreases upon complex formation with PulS. Hydrodynamic properties of Sdom (left panel) and PulS-Sdom complex (right panel) analyzed by size exclusion chromatography connected to a triple detector array (“Experimental Procedures”). The concentration determined from the refractive index (solid line, left axis) was combined with the hydrodynamic radius from the light scattering detector to calculate the molecular mass (solid squares, inside right axis) or with the differential viscometer measurements to determine the intrinsic viscosity (open squares, outside right axis).
Limited Proteolysis Identifies a Minimal PulS-binding Region of 28 Residues in Sdom
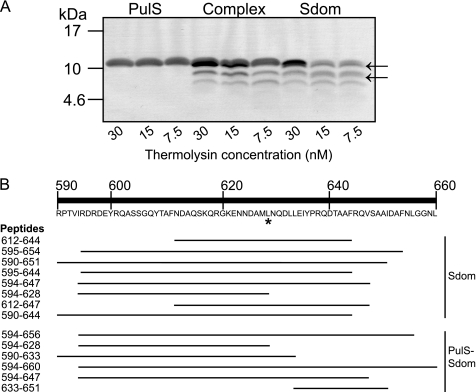
Limited proteolysis experiments coupled to mass spectrometry were performed with different proteases to determine the minimal PulS-binding region in Sdom. SDS-PAGE revealed that thermolysin did not cleave PulS and that binding of PulS to Sdom prevented the appearance of two Sdom cleavage products (arrows in Fig. 5). Mass spectrometry of digested samples (Fig. 5 and supplemental Table 2) indicated that thermolysin cleaved at the same site near the center of Sdom (Fig. 5, 628–629, asterisk) irrespective of the presence or absence of PulS and that changes in cleavage profile occurred on either side of this site, suggesting the presence of two binding sites. For further analysis, Sdom was considered to consist of two parts, SdomA(594–628) and SdomB(629–656), consistent with previous in vivo results showing that residues 590–660 of PulD are required for both protection and OM localization by PulS, whereas the absence of residues 636–660 of PulD caused loss of OM targeting but not protection by PulS (7, 21). To test the existence of separate binding sites in these two hypothetical subdomains of Sdom, SdomA and SdomB were produced in E. coli and purified as either MalE chimeras or peptides cleaved from MalE by thrombin and were analyzed for complex formation with PulS by size exclusion chromatography. The results indicated that PulS interacted only with SdomB (supplemental Fig. S3).
FIGURE 5.
Limited proteolysis and mass spectrometry identified the minimal binding site for PulS. A, 50 μm Sdom, 50 μm PulS, or an equimolar complex was digested with 30, 15, or 7.5 nm thermolysin and peptide fragments were separated by SDS-PAGE. Arrows indicate the additional bands found upon proteolysis of the Sdom sample but not detected after proteolysis of the complex. B, schematic representation of the most relevant peptide fragments identified by mass spectrometry (see supplemental Table S2 for list of total peptides identified), including amino acid coordinates and listed in descending order from the peptide with the highest relative intensity. Top, amino acid sequence and numbering of the Sdom (His tag not included), asterisk indicates the common cleavage event between both samples.
PulS Binds to Sdom with Nanomolar Affinity
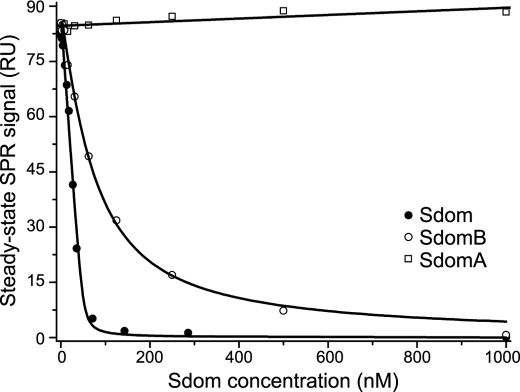
Surface plasmon resonance was used to study further the interaction between PulS and Sdom (Fig. 6 and see supplemental Fig. S4 for raw data). It was impractical to measure affinities accurately with tethered MalE-PulS chimera as the ligand and Sdom as the analyte, possibly due to mass transport limitations linked to the coupling of Sdom folding and binding. This problem was overcome by competition experiments in which MalE-Sdom or MalE-SdomB was captured on the surface as ligand, and PulS alone or in competition with Sdom, SdomA, or SdomB was flown over the surface as the analyte. Steady-state measurements indicated a binding affinity of 1.74 ± 0.13 nm with Sdom and 49.28 ± 7.04 nm with SdomB and confirmed that SdomA does not interact with PulS.
FIGURE 6.
PulS binds with nanomolar affinity to Sdom and SdomB. Experimentally determined steady-state SPR signals were plotted versus competitor (Sdom, SdomA, and SdomB) concentration for the determination of the Kd values, see text. RU, resonance unit. For experimental data see supplemental Fig. S4.
SdomA Is Required for Efficient Pullulanase Secretion
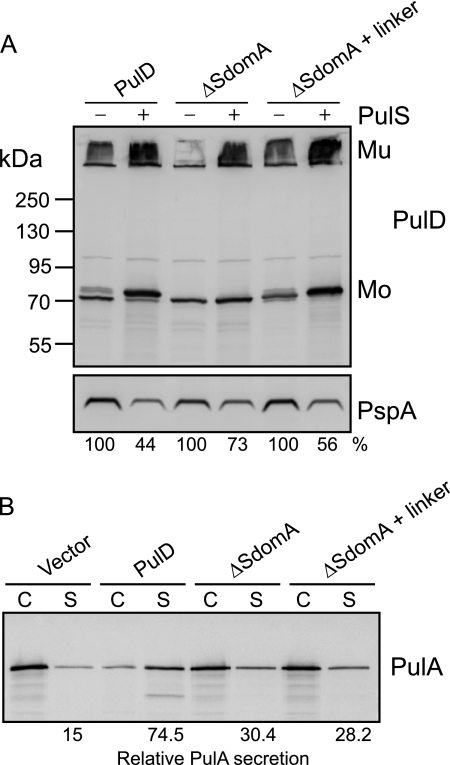
In vitro analysis defined the minimal binding region of PulS as 28 residues near the C terminus of the S domain (SdomB). To determine the role, if any, of SdomA, PulD variants were constructed that either lacked SdomA or in which SdomA was replaced by a random linker. PulD and PulD variants were produced in E. coli in the presence or absence of PulS to observe PulD multimerization, protection, and targeting to the outer membrane. All PulD variants were produced in similar amounts of PulD monomer and multimer, confirming that the mutations did not affect PulD stability or multimerization (Fig. 7A). Furthermore, PulS protected the PulD variants from proteolysis, as indicated by the higher yields of PulD and by the protection from proteolytic trimming, as observed with the wild type PulD. The Psp response, characterized by increased PspA production caused by mislocalization of PulD to the inner membrane, was also alleviated by the presence of PulS, suggesting that the PulD variants were targeted to the outer membrane (Fig. 7A), as was confirmed by membrane fractionation and sucrose gradient centrifugation experiments (supplemental Fig. S5).
FIGURE 7.
SdomA is not required for in vivo targeting by PulS but is necessary for efficient secretion of PulA. A, total cell extracts of E. coli producing PulD or PulD variants in the absence and presence of PulS were examined by SDS-PAGE and immunoblotting with PulD and PspA antiserum. Relative percent of PspA was measured with respect to its corresponding control without PulS (100%). Mu, PulD multimer; Mo, PulD monomer. B, total cell and culture supernatant fractions of E. coli PAP105(pCHAP8243) producing all the Pul factors except PulD and a soluble variant of PulA, complemented with wild type or SdomA mutant PulD. An equivalent of 0.015 absorbance units of culture were separated by SDS-PAGE and immunoblotted with PulA-specific antiserum. Secretion levels are the proportion (%) of the PulA detected in the supernatant fraction.
Although the PulD variants interacted with PulS and were thereby targeted to the outer membrane, they were only able to substitute partially for wild type PulD in PulA secretion (Fig. 7B). These results confirm that SdomB interacts specifically with PulS for PulD targeting to the outer membrane and provide the first evidence that SdomA is required for efficient PulD function.
DISCUSSION
PulS and the C-terminal region of PulD (Sdom) interact in vitro to form a stable 1:1 complex, consistent with the copurification of PulD and PulS in equal molar concentrations from the outer membrane in detergent (3, 4). Surface plasmon resonance studies indicated a tight interaction with an affinity of 1.74 nm. Limited proteolysis identified a high affinity binding site in the C-terminal 28 amino acid residues of Sdom, defined here as SdomB. In vivo data confirmed that SdomB is sufficient to target PulD to the outer membrane. Although the upstream part of Sdom (SdomA) is not required for localization or multimerization, it is necessary for efficient PulA secretion, indicating that Sdom plays at least two roles in PulD targeting and function. As the N-terminal regions of secretins extend into the periplasm and confer substrate specificity (46, 47), SdomA might function in late steps of PulA release or ejection. It is important to note, however, that we cannot rule out a possible influence of SdomA on PulS binding to SdomB, because the removal of the SdomA diminished the apparent affinity of Sdom for PulS. Furthermore, SdomA appears to retain some ability to interact with PulS when present in a variant of PulD that lacks SdomB, because PulS protects this construct from proteolysis, although it cannot target it to the outer membrane (21, 23).
Circular dichroism and one-dimensional nuclear magnetic resonance experiments demonstrating that purified Sdom is unfolded or intrinsically disordered were confirmed by other approaches indicating that Sdom is elongated with a high frictional coefficient. Upon binding to PulS, Sdom undergoes compaction and gains secondary structure, consistent with a coupled binding and folding model of interaction. We hypothesize that Sdom adopts the same unstructured, elongated, and highly hydrated state in full-length PulD, although we cannot rule out that regions N-terminal to Sdom in PulD affect Sdom organization and binding to PulS. However, sequence alignments show that Sdom is a separate domain that is not found in all secretins, and its fusion to the C terminus of other secretins both destabilizes these proteins in the absence of PulS and allows them to be correctly targeted to the E. coli OM and protected against proteolysis when PulS is present (20, 48), providing strong evidence that Sdom is a separate domain that can fold and function independently of PulD.
The unfolded nature of the PulD Sdom and the coupled binding and folding model of interaction are similar to the secretin-chaperone interaction of the type III secretion system of Shigella flexneri. MxiM is required for stabilization and multimerization of secretin MxiD, but it is unclear whether it plays a role in targeting to the OM (49). The C-terminal residues 553–570 of MxiD are intrinsically disordered but fold into an amphipathic α-helix upon binding its cognate chaperone MxiM, for which it has an affinity of 71 nm (10). However, the predicted α-helical structure of PulS is clearly different from that of MxiM, which is essentially composed of β-strands (15). Thus, although PulS and MxiM do not share any sequence or structural similarity, they share a common mechanism of interaction with their respective secretins.
Other OM proteins require nonspecific chaperones such as SurA, Skp, DegP, and FkpA for correct folding, localization, and insertion into the OM. Therefore, it is reasonable to compare the binding affinities of dedicated chaperones like PulS and MxiM to those of general chaperones (50). Skp, a 141-amino acid trimeric polypeptide that is important in the early stages of OM protein folding, forms 1:1 complexes with OmpA, OmpG, and NalP, with affinities of 12–50 nm (51). DegP, a protease with chaperone activity involved in the degradation of irreversibly misfolded proteins or unfolding of aggregated or misfolded proteins, has an affinity of 0.175 nm for the OM autotransporter EspP and 0.756 nm for OmpC (52). In comparison, SurA has an affinity of 3.25 nm for EspP, whereas FkpA, which forms a homodimer, has an affinity of 64.2 nm for unfolded EspP (53). Thus, periplasmic chaperones of all classes have high affinities for their targets, suggesting that high affinity interactions are necessary in the periplasm for correct assembly of OM proteins.
The studies reported here and work by Okon et al. (10) indicate that the intrinsically disordered C-terminal region of secretins is probably important for their targeting to the OM or their assembly. Coupled binding and folding interactions frequently involve intrinsically disordered proteins or regions. For example, the N-terminal 30 residues of bacterial type III secretion system effectors SseJ, SptP, SopD-2, and GtgE and YopH of Yersinia pestis are all intrinsically disordered in solution but adopt helical structures in the presence of stabilizing agents (54). Buchko et al. (54) suggest that structural disorder might be a universal signal for effector secretion in this system. In the type I secretion system of Bordetella pertussis, secretion and function of the adenylate cyclase toxin (CyaA) are dependent on a disorder-to-order transition of the repeat in toxin motif. Repeat in toxin motifs are tandem repeats of calcium-binding nonapeptide sequences that are present in proteins secreted by this system that undergo drastic compaction, dehydration, and folding in the presence of calcium (37). Chenal et al. (37) speculated that the intrinsically disordered nature of CyaA facilitates secretion through the secretion machinery and that the high levels of calcium present in the external medium could trigger folding as CyaA emerges from the machinery. Another important example of a disorder-to-order transition is the well characterized flagellin protein of bacterial flagella. The 65 N-terminal and 45 C-terminal residues are disordered but become α-helical upon polymerization, and their interaction is responsible for the proper folding of the filament core (55–57). Therefore, intrinsically disordered proteins or regions play important roles in the assembly of several bacterial trans-envelope machineries and in protein secretion.
Intrinsically disordered proteins or regions lack stable three-dimensional structures in solution and exist instead as dynamic ensembles of conformations (43, 58). As described above, intrinsically disordered proteins can acquire structure upon binding to other proteins, and these disorder-to-order transitions are specifically linked to function (58, 59). Flexibility in disordered regions enables binding of or to numerous structurally distinct targets, overcomes steric restrictions by enabling larger surfaces of interaction in protein complexes than are possible with rigid partners, or allows protein interactions to occur with both high specificity and low affinity (43). Intrinsically disordered regions facilitate binding to their partners by increasing the capture radius for its target, the so-called “fly-casting” mechanism (60), and reduce the free energy barrier for coupled folding and binding interactions, providing an additional kinetic advantage (61). Thus, fewer encounters are required before the final complex is formed. However, intrinsically disordered or unfolded regions are more susceptible to proteolysis, and structural disorder provides a signal for protein degradation (62). PulD and unrelated proteins fused to Sdom are highly susceptible to proteolytic trimming (within Sdom) and degradation in the absence of PulS, and in vitro assays showed that Sdom is more susceptible to cleavage by thermolysin and trypsin in the absence of PulS (Fig. 5 and data not shown). Therefore, we suspect that the intrinsically disordered S domain of PulD maintains a sensitive balance between fast association with PulS for correct targeting to the OM and degradation by periplasmic proteases.
PulS is a lipoprotein that transits through an intermediate phase during which it is anchored to the inner membrane with the polypeptide chain exposed in the periplasm while diacylglyceride is added, the signal peptide is removed, the N-terminal cysteine is fatty acylated, and the mature lipoprotein interacts with LolCDE for release into the periplasm (63). Release is coordinated with capture by LolA, which is followed by routing to the outer membrane, where PulS interacts with LolB to release LolA (7). The precise moment at which PulS interacts with PulD is unknown, but we speculate that the interaction must occur quickly to prevent trimming of Sdom and potentially detrimental misrouting (64). LolA is a low abundance protein (200–400 copies per cell) (65) that transports over 105 lipoproteins to the outer membrane per generation (<60 min under laboratory conditions). Thus, LolA must recycle more than four times every minute to ensure that OM lipoproteins do not accumulate in the inner membrane, which would likely be detrimental to the cell (66). The data presented here suggest that the intrinsic disorder of the PulS binding domain in PulD ensures that all of the PulD exported to the periplasm interacts quickly with PulS, either before PulS is released from the membrane or in complex with LolA. The production of large amounts of PulS through constitutive expression of its gene (in contrast to pulD, which is part of the maltose regulon and thus only induced when the substrate for the secreted enzyme pullulanase is present in the environment (67)) is a second mechanism that ensures that PulD finds PulS quickly, even though this means that a large proportion of PulS produced reaches the OM without encountering PulD.
Supplementary Material
Acknowledgments
We are grateful to the following: Ingrid Guilvout and Séverine Collin for their help and support; Nicolas Bayan and members of the Molecular Genetics Unit for their helpful discussions; Olivera Francetic for the generous gift of plasmid pCHAP8243; Sylviane Hoos (Plate-forme de Biophysique des Macromolécules et de leurs Interactions) for help with surface plasmon resonance experiments; Jacques d'Alayer (Plate-forme d'Analyse et de Microséquençage des Protéines, Institut Pasteur) for N-terminal sequencing; Jean Claude Rousselle and Abdelkader Namane (Plate-forme de Protéomique, Institut Pasteur) for mass spectrometry analysis; and Luca Mollica (Groupe Flexibilité et Dynamique des Protéines par RMN, Institut de Biologie Structurale) for one-dimensional NMR measurements.
This work was supported in part by French National Research Agency Grants ANR-05-0307-01 and ANR-09-BLAN-0291.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Tables 1 and 2, and an additional reference.
This article was selected as a Paper of the Week.
- OM
- outer membrane
- Sdom
- S domain.
REFERENCES
- 1. Korotkov K. V., Gonen T., Hol W. G. (2011) Trends Biochem. Sci. 36, 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chami M., Guilvout I., Gregorini M., Rémigy H. W., Müller S. A., Valerio M., Engel A., Pugsley A. P., Bayan N. (2005) J. Biol. Chem. 280, 37732–37741 [DOI] [PubMed] [Google Scholar]
- 3. Nouwen N., Ranson N., Saibil H., Wolpensinger B., Engel A., Ghazi A., Pugsley A. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8173–8177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nouwen N., Stahlberg H., Pugsley A. P., Engel A. (2000) EMBO J. 19, 2229–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Opalka N., Beckmann R., Boisset N., Simon M. N., Russel M., Darst S. A. (2003) J. Mol. Biol. 325, 461–470 [DOI] [PubMed] [Google Scholar]
- 6. Reichow S. L., Korotkov K. V., Hol W. G., Gonen T. (2010) Nat. Struct. Mol. Biol. 17, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Collin S., Guilvout I., Nickerson N. N., Pugsley A. P. (2011) Mol. Microbiol. 80, 655–665 [DOI] [PubMed] [Google Scholar]
- 8. Hardie K. R., Lory S., Pugsley A. P. (1996) EMBO J. 15, 978–988 [PMC free article] [PubMed] [Google Scholar]
- 9. Hardie K. R., Seydel A., Guilvout I., Pugsley A. P. (1996) Mol. Microbiol. 22, 967–976 [DOI] [PubMed] [Google Scholar]
- 10. Okon M., Moraes T. F., Lario P. I., Creagh A. L., Haynes C. A., Strynadka N. C., McIntosh L. P. (2008) Structure 16, 1544–1554 [DOI] [PubMed] [Google Scholar]
- 11. Shevchik V. E., Condemine G. (1998) Microbiology 144, 3219–3228 [DOI] [PubMed] [Google Scholar]
- 12. d'Enfert C., Reyss I., Wandersman C., Pugsley A. P. (1989) J. Biol. Chem. 264, 17462–17468 [PubMed] [Google Scholar]
- 13. D'Enfert C., Pugsley A. P. (1989) J. Bacteriol. 171, 3673–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bayan N., Guilvout I., Pugsley A. P. (2006) Mol. Microbiol. 60, 1–4 [DOI] [PubMed] [Google Scholar]
- 15. Lario P. I., Pfuetzner R. A., Frey E. A., Creagh L., Haynes C., Maurelli A. T., Strynadka N. C. (2005) EMBO J. 24, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Koo J., Tammam S., Ku S. Y., Sampaleanu L. M., Burrows L. L., Howell P. L. (2008) J. Bacteriol. 190, 6961–6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trindade M. B., Job V., Contreras-Martel C., Pelicic V., Dessen A. (2008) J. Mol. Biol. 378, 1031–1039 [DOI] [PubMed] [Google Scholar]
- 18. Kim K., Oh J., Han D., Kim E. E., Lee B., Kim Y. (2006) Biochem. Biophys. Res. Commun. 340, 1028–1038 [DOI] [PubMed] [Google Scholar]
- 19. Burghout P., Beckers F., de Wit E., van Boxtel R., Cornelis G. R., Tommassen J., Koster M. (2004) J. Bacteriol. 186, 5366–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Daefler S., Guilvout I., Hardie K. R., Pugsley A. P., Russel M. (1997) Mol. Microbiol. 24, 465–475 [DOI] [PubMed] [Google Scholar]
- 21. Guilvout I., Hardie K. R., Sauvonnet N., Pugsley A. P. (1999) J. Bacteriol. 181, 7212–7220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guilvout I., Chami M., Berrier C., Ghazi A., Engel A., Pugsley A. P., Bayan N. (2008) J. Mol. Biol. 382, 13–23 [DOI] [PubMed] [Google Scholar]
- 23. Guilvout I., Nickerson N. N., Chami M., Pugsley A. P. (2011) Res. Microbiol. 162, 180–190 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Appendix A.2, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Tartof K. D., Hobbs C. A. (1987) Focus 9, 12 [Google Scholar]
- 26. Franceti O., Pugsley A. P. (2005) J. Bacteriol. 187, 7045–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Johnson W. C. (1999) Proteins 35, 307–312 [PubMed] [Google Scholar]
- 28. Whitmore L., Wallace B. A. (2004) Nucleic Acids Res. 32, W668–W673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Whitmore L., Wallace B. A. (2008) Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 30. Lees J. G., Miles A. J., Wien F., Wallace B. A. (2006) Bioinformatics 22, 1955–1962 [DOI] [PubMed] [Google Scholar]
- 31. Sreerama N., Woody R. W. (2000) Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 32. Provencher S. W., Glöckner J. (1981) Biochemistry 20, 33–37 [DOI] [PubMed] [Google Scholar]
- 33. Santoro M. M., Bolen D. W. (1988) Biochemistry 27, 8063–8068 [DOI] [PubMed] [Google Scholar]
- 34. Swint L., Robertson A. D. (1993) Protein Sci. 2, 2037–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Demeler B. (2005) in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., Rowe A. J. eds) pp. 210–229, Royal Society of Chemistry, Cambridge, UK [Google Scholar]
- 36. Brown P. H., Schuck P. (2006) Biophys. J. 90, 4651–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chenal A., Guijarro J. I., Raynal B., Delepierre M., Ladant D. (2009) J. Biol. Chem. 284, 1781–1789 [DOI] [PubMed] [Google Scholar]
- 38. Karst J. C., Sotomayor Pérez A. C., Guijarro J. I., Raynal B., Chenal A., Ladant D. (2010) Biochemistry 49, 318–328 [DOI] [PubMed] [Google Scholar]
- 39. Harding S. E. (1995) Biophys. Chem. 55, 69–93 [DOI] [PubMed] [Google Scholar]
- 40. England P., Brégégère F., Bedouelle H. (1997) Biochemistry 36, 164–172 [DOI] [PubMed] [Google Scholar]
- 41. Nieba L., Krebber A., Plückthun A. (1996) Anal. Biochem. 234, 155–165 [DOI] [PubMed] [Google Scholar]
- 42. Pugsley A. P., Bayan N., Sauvonnet N. (2001) J. Bacteriol. 183, 1312–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Uversky V. N., Longhi S. (eds) (2010) Instrumental Analysis of Intrinsically Disordered Proteins, John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 44. Uversky V. N. (2002) Eur. J. Biochem. 269, 2–12 [DOI] [PubMed] [Google Scholar]
- 45. Lu Y., Longman E., Davis K. G., Ortega A., Grossmann J. G., Michaelsen T. E., de la Torre J. G., Harding S. E. (2006) Biophys. J. 91, 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Login F. H., Fries M., Wang X., Pickersgill R. W., Shevchik V. E. (2010) Mol. Microbiol. 76, 944–955 [DOI] [PubMed] [Google Scholar]
- 47. Shevchik V. E., Robert-Baudouy J., Condemine G. (1997) EMBO J. 16, 3007–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arts J., van Boxtel R., van Ulsen P., Tommassen J., Koster M. (2007) Assembly of the Pseudomonas aeruginosa Type II Secretion System. Ph.D. dissertation, Utrecht University, Utrecht, Germany [Google Scholar]
- 49. Schuch R., Maurelli A. T. (2001) J. Bacteriol. 183, 6991–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mogensen J. E., Otzen D. E. (2005) Mol. Microbiol. 57, 326–346 [DOI] [PubMed] [Google Scholar]
- 51. Qu J., Mayer C., Behrens S., Holst O., Kleinschmidt J. H. (2007) J. Mol. Biol. 374, 91–105 [DOI] [PubMed] [Google Scholar]
- 52. Ruiz-Perez F., Henderson I. R., Leyton D. L., Rossiter A. E., Zhang Y., Nataro J. P. (2009) J. Bacteriol. 191, 6571–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ruiz-Perez F., Henderson I. R., Nataro J. P. (2010) Gut Microbes 1, 339–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Buchko G. W., Niemann G., Baker E. S., Belov M. E., Smith R. D., Heffron F., Adkins J. N., McDermott J. E. (2010) Mol. Biosyst. 6, 2448–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aizawa S. I., Vonderviszt F., Ishima R., Akasaka K. (1990) J. Mol. Biol. 211, 673–677 [DOI] [PubMed] [Google Scholar]
- 56. Vonderviszt F., Kanto S., Aizawa S., Namba K. (1989) J. Mol. Biol. 209, 127–133 [DOI] [PubMed] [Google Scholar]
- 57. Kostyukova A. S., Pyatibratov M. G., Filimonov V. V., Fedorov O. V. (1988) FEBS Lett. 241, 141–144 [DOI] [PubMed] [Google Scholar]
- 58. Dyson H. J., Wright P. E. (2005) Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 59. Dunker A. K., Brown C. J., Lawson J. D., Iakoucheva L. M., Obradovi Z. (2002) Biochemistry 41, 6573–6582 [DOI] [PubMed] [Google Scholar]
- 60. Shoemaker B. A., Portman J. J., Wolynes P. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Huang Y., Liu Z. (2009) J. Mol. Biol. 393, 1143–1159 [DOI] [PubMed] [Google Scholar]
- 62. Tompa P., Prilusky J., Silman I., Sussman J. L. (2008) Proteins 71, 903–909 [DOI] [PubMed] [Google Scholar]
- 63. Tokuda H., Matsuyama S. (2004) Biochim. Biophys. Acta 1693, 5–13 [DOI] [PubMed] [Google Scholar]
- 64. Guilvout I., Chami M., Engel A., Pugsley A. P., Bayan N. (2006) EMBO J. 25, 5241–5249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Matsuyama S., Tajima T., Tokuda H. (1995) EMBO J. 14, 3365–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Robichon C., Vidal-Ingigliardi D., Pugsley A. P. (2005) J. Biol. Chem. 280, 974–983 [DOI] [PubMed] [Google Scholar]
- 67. d'Enfert C., Chapon C., Pugsley A. P. (1987) Mol. Microbiol. 1, 107–116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.