Abstract
Cytochrome c maturation in many bacteria, archaea, and plant mitochondria involves the integral membrane protein CcmF, which is thought to function as a cytochrome c synthetase by facilitating the final covalent attachment of heme to the apocytochrome c. We previously reported that the E. coli CcmF protein contains a b-type heme that is stably and stoichiometrically associated with the protein and is not the heme attached to apocytochrome c. Here, we show that mutation of either of two conserved transmembrane histidines (His261 or His491) impairs stoichiometric b-heme binding in CcmF and results in spectral perturbations in the remaining heme. Exogeneous imidazole is able to correct cytochrome c maturation for His261 and His491 substitutions with small side chains (Ala or Gly), suggesting that a “cavity” is formed in these CcmF mutants in which imidazole binds and acts as a functional ligand to the b-heme. The results of resonance Raman spectroscopy on wild-type CcmF are consistent with a hexacoordinate low spin b-heme with at least one endogeneous axial His ligand. Analysis of purified recombinant CcmF proteins from diverse prokaryotes reveals that the b-heme in CcmF is widely conserved. We have also determined the reduction potential of the CcmF b-heme (Em,7 = -147 mV). We discuss these results in the context of CcmF structure and functions as a heme reductase and cytochrome c synthetase.
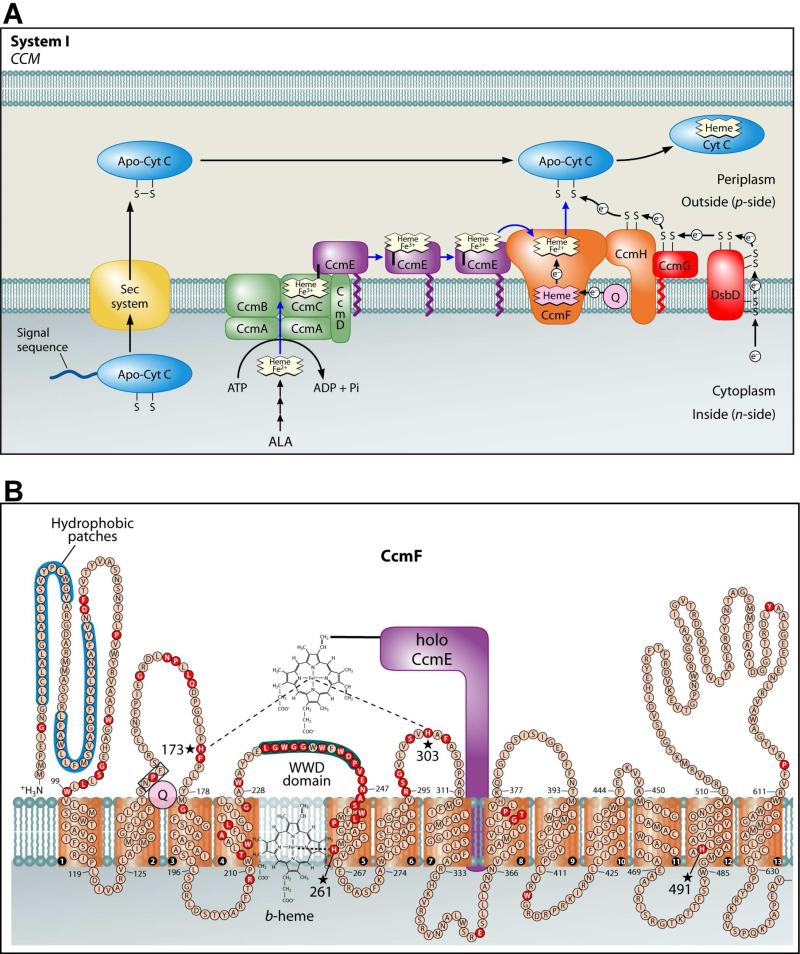
The c-type cytochromes are heme proteins present in nearly all organisms that participate in electron transfer, typically for the generation of an electrochemical proton gradient. C-type cytochromes function outside of the cytoplasmic membrane in prokaryotes, in the intermembrane space of the mitochondrion, and in the lumen of the chloroplast. Cytochromes c are commonly distinguished by covalent attachment of the two vinyl groups (α-carbons) of heme to the thiols of a conserved Cys-Xxx-Xxx-Cys-His motif in the apocytochrome. For thioether formation between heme and apocytochrome, the heme iron and the cysteine thiols must be in the reduced state (1, 2). In many prokaryotes, nearly all plant mitochondria and some protozoan mitochondria, cytochromes c are assembled by the cytochrome c maturation (ccm) pathway called system I (Figure 1A), which typically comprises eight or nine integral membrane proteins (In E. coli, CcmABCDEFGH) (3-6).
Figure 1.
Adapted from reference (5). (A) Current working model of the system I cytochrome c biogenesis pathway. Model includes trafficking and oxidation states of heme as well as the subpathways for apocytochrome translocation and reduction. (B) Topology of the CcmF integral membrane protein from E. coli. Possible histidine axial ligands to heme are starred. The highly conserved WWD domain is shaded as are the hydrophobic patches. Completely conserved amino acids (red) were identified by individual protein alignments using CcmF ORFs from the following organisms: the alpha proteobacteria, Agrobacterium tumefaciens C58, R. capsulatus, Caulobacter crescentus CB15, and Bradyrhizobium japonicum; the beta proteobacteria, Nitrospira mutliformis ATCC 25196 and Nitrosomonas europaea ATCC 19718; the gamma proteobacteria, E. coli K-12 MG1655, Pseudomonas fluorescens Pf01, Shewanella oneidensis MR-1 and Vibrio parahaemolyticus RIMD 2210633; the delta proteobacteria, Myxococcus xanthus and Desulfovibrio desulfuricans; and the deinococci, Deinococcus geothermalis and Thermus thermophilus.
In system I, heme is initially bound at a site in CcmC (Figure 1A) (7-9). Here, a unique covalent attachment is formed between His130 of CcmE and the β-carbon of the 2-vinyl of heme (10-12). CcmE with covalent heme is released from CcmC by the action of CcmAB (an ABC transporter) (13-15). CcmE is believed to traffic heme to the site of cytochrome c assembly, putatively the CcmF/H synthetase complex, where the heme from holoCcmE is attached to apocytochrome c (8, 16, 17). Although studies have shown that CcmF and CcmH form an integral membrane complex (8, 16, 18), the function of this complex, including its role as a synthetase, is poorly understood.
We have proposed that formation of the covalent bond between CcmE and heme (within the holoCcmCDE complex) is favored by heme oxidation (to Fe3+) (5, 7, 8). Consistent with this, the purified holoCcmCDE complex (7, 8) and purified holoCcmE (8, 11, 12) contain covalent heme in the oxidized (Fe3+) state. However, heme attachment to the apocytochrome requires that the heme from holoCcmE is reduced (Fe2+) prior to ligation (1, 2, 5). A possible mechanism for reducing heme in holoCcmE before heme attachment to apocytochrome c has recently emerged: we discovered that CcmF contains a b-type heme, independent of all other Ccm proteins, that exists in 1:1 stoichiometry with the protein (8). We proposed that reduction of the heme in holoCcmE by the CcmF b-heme would facilitate release of the CcmE His130 covalent bond and prepare the heme for attachment to the apocytochrome (5, 7).
It is important to establish the spectral and redox properties of the b-heme in CcmF to understand the function(s) of CcmF in the maturation of cytochromes c. Topological studies have revealed that CcmF contains 13 transmembrane domains (TMDs; Figure 1B) (8, 19). There are four completely conserved histidines that might serve as axial ligands to the b-heme: two are predicted to be in periplasmic loops (His173 and His303 in CcmF from E. coli) and two are predicted to reside in TMDs (His261 and His491) (see stars in Figure 1B). Alanine substitutions at each of these histidines show severe defects in cytochrome c assembly (16). To identify the axial ligands to the b-heme in CcmF, we use spectroscopic and site-directed mutagenesis approaches as well as chemical complementation with exogeneous imidazole. We show that the b-heme is present in recombinant CcmF proteins from diverse organisms and determine the reduction potential of the CcmF b-heme. Our findings address the location of the b-heme in CcmF and allow us to propose redox control mechanisms in the CcmF synthetase reactions.
EXPERIMENTAL PROCEDURES
Bacterial Growth Conditions
Escherichia coli strains (Table S1) were grown at 37°C by shaking at 240 rpm in Luria-Bertani broth (LB; Difco) supplemented with the appropriate antibiotics (Sigma-Aldrich) and other media additives at the following concentrations, unless otherwise noted: carbenicillin, 50 μg ml-1; chloramphenicol, 20 μg ml-1; gentamicin, 10 μg ml-1; kanamycin, 100 μg ml-1; isopropyl-β-D-thiogalactopyranoside (IPTG, Gold Biotechnology), 1 mM; arabinose (Gold Biotechnology), 0.1 % (wt/vol).
Protein Expression and Purification
For expression, E. coli strains RK103 (17) or RK113 (this work, see Table S1) were used. Starter cultures were initiated from a single colony and grown overnight at 37oC in 10 mL of LB with the appropriate antibiotics. 1 L of LB was inoculated to 1 % and was grown to an OD600 of 1.8, at which point the culture was induced with 1 mM IPTG for pGEX-based expression or 0.1 % arabinose (wt/vol) for pBAD-based expression for 14-16 hr. Cells were harvested at 5,000 × g for 12 min and frozen at -80°C. Cell pellets were thawed and resuspended in 10 mL/gram wet wt of a modified 1 × TALON (Clontech) buffer (20 mM Tris-HCl, pH 8; 100 mM NaCl) and treated with 4-(2-aminoethyl) bezenesulfonyl fluoride hydrochloride (AEBSF, Gold Biotechnology; 1 mM) and egg white lysozyme (Sigma-Aldrich; 100 μg ml-1) for 30 min while shaking on ice. Cells were disrupted by repeated sonication for 30 sec bursts on a Branson 250 sonicator (50 % duty, 60 % output) until clearing of the suspension was observed. Crude sonicate was centrifuged at 24,000 × g for 12 min to remove unbroken cells and cell debris, and membranes were isolated by centrifugation at 100,000 × g for 45 min. Membrane pellets were solubilized in 1 × modified TALON buffer with 1 % (wt/vol) Anatrace n-Dodecyl-β-D-Maltopyranoside (DDM, Affymetrix). Solubilized membranes (called “L” or load in Figures) were passed over TALON resin per the manufacturer's recommendations and bound protein was washed in 1 × modified TALON buffer with 0.1 % DDM with increasing concentrations of imidazole (in Figures, denoted as wash 1 “W1”, 0 mM imidazole; wash 2 “W2”, 2 mM imidazole; or wash 3 “W3”, 5 mM imidazole). Bound hexahistidine-tagged protein was eluted in 1 × modified TALON buffer containing 0.1 % DDM and 75 mM imidazole (called “E” or elution in Figures). The purified protein was concentrated and subjected to buffer exchange in an Amicon Ultra Centrifugal Filter 30,000 MWCO (Millipore) after purification (called “EC” or concentrated elution in Figures) to reduce imidazole to less than 100 μM, unless otherwise noted. To address the accumulation of DDM, purification of CcmF was alternately carried out using 0.02 % DDM instead of 0.1 %. For these purifications, imidazole was removed by successive dialysis using Pierce SnakeSkin Pleated Dialysis Tubing 7,000 MWCO (Thermo Scientific) against 20 mM Tris, pH 8, 100 mM NaCl, 0.02 % DDM, and the protein was concentrated in an Amicon Ultra Centrifugal Filter 100,000 MWCO (Millipore). Using this method, the concentration of DDM was maintained at 0.02 % throughout purification and concentration.
Resonance Raman spectroscopy
Resonance Raman (rR) spectra were obtained using 30 to 40 μM CcmF protein from E. coli, purified with the endogeneous b-heme. Sample solutions contained 20 mM Tris, pH 8.0, 100 mM NaCl, and dodecyl maltoside at 0.02 %, 0.48 %, or 3.2 %. Heme carbonyl complexes of CcmF were prepared in 5 mm NMR tubes by exhaustive equilibration of the CcmF solution with water-saturated N2 followed by titration of the sample with sodium dithionite to reduce the heme. The inert atmosphere in the tube was then displaced with either natural isotopic abundance CO or 13CO (95 atom % 13C). Raman scattering was excited using 413.1 nm emission from a Kr+ laser. Sample tubes were spun at ~ 20 Hz and spectra were recorded at ambient temperature. Scattered light was collected in the 135° backscattering geometry, passed through a holographic notch filter to remove Rayleigh scattered light and depolarized. The Raman spectrum was recorded using a 0.6 m spectrograph equipped with a 2400 g/mm holographic grating and a LN2 cooled CCD detector. Spectra were calibrated versus toluene, DMSO d6, and dibromomethane. Vibrational frequencies reported here are reproducible to ±1 cm−1.
Heme stains and other methods
Heme stains and immunoblots were performed as described in (17, 20). Proteins were separated by 12.5 % SDS-PAGE and transferred to Hybond C nitrocellulose membranes (GE Healthcare). Anti-His antibodies (Santa Cruz technologies) were used at a dilution of 1:5000. Protein A peroxidase (Sigma-Aldrich) was used as the secondary label. The chemiluminescent signal for heme stains and anti-His westerns was developed using the Pierce SuperSignal Femto kit (Thermo Scientific), and detected with an LAS-1000 Plus detection system (Fujifilm-GE Healthcare). Protein concentrations were determined using the Pierce BCA Protein Assay Kit (Thermo Scientific) using BSA as a standard. The concentration of heme in the CcmF protein preparations was determined by UV-visible absorption spectroscopy, pyridine extraction as described in (21) or heme stain as described in (22). Protein purity was assessed by Coomassie Blue staining of SDS-PAGE.
UV/Vis absorption spectroscopy
UV-visible absorption spectra were recorded with a Shimadzu UV-2101 PC UV-Vis scanning spectrophotometer at room temperature as described in (23). Chemically reduced spectra were generated by addition of sodium dithionite (sodium hydrosulfite) to the purified sample. Unless otherwise indicated, all spectra were recorded in 20 mM Tris-HCl (pH 8), 100 mM NaCl, 0.1 % DDM. To analyze the effect of imidazole on the electronic spectrum, small quantities of a concentrated imidazole solution (1 M, pH 7) were added to purified protein and spectra were recorded.
Cytochrome Reporter and Imidazole Complementation Assays
Cytochrome c4:His6 production was assayed by the following methods using two different strains. (Method 1) Strain RK111 (Δccm carrying the arabinose-inducible chromosomal integrate of the cyt c4:His6 gene) harboring pRGK406 (pGEX ΔccmF) and one of the following pBAD ccmF plasmids (pRGK408, 409, 410, 411, 412, 413, 414, 415, 416, 417, or 418; see Table S1 in Supporting Information) was grown aerobically in 5 mLs of LB with appropriate antibiotics for 3 hr and induced for an additional 3 hr with 0.8 % arabinose (wt/vol). (Method 2) Strain RK103 ((17); Δccm) harboring pRGK332 ((17); carrying the arabinose-inducible cyt c4:His6 gene) and one of the following pGEX system I-based plasmids (pRGK386 (17), pRGK403, pRGK404, pRGK405, or pRGK407; see Table S1 in Supporting Information) was grown aerobically in 5 mL LB with appropriate antibiotics for 3 hr and induced for an additional 3 hr with 0.2 % arabinose (wt/vol) and 1 mM IPTG. For both methods, cells were harvested by centrifugation at 10,000 × g for 5 min and the cell pellet was resuspended in 200 μL of Bacterial Protein Extraction Reagent (BPER, Thermo Scientific) to lyse cells and extract protein. Total protein concentration was determined using the Nanodrop 1000 spectrophotometer (Thermo Scientific) and 100 μg was analyzed by SDS-PAGE followed by heme stain. Imidazole complementation assays were performed in the same way with 10 mM imidazole (pH 7) added to the media prior to inoculation.
Determination of the Reduction Potential of CcmF
The reduction potential for the b-heme in CcmF was determined by reduction of a dye of known potential (24), using a modification of the Massey method (25). The assay contained 20 mM Tris-HCl, pH 7, 100 mM NaCl, DDM at 0.02 % or 1.2 %, 500 μM xanthine (Sigma-Aldrich), 20 μM CcmF and one of the following redox dyes: nile blue chloride (Em = -116 mV) (26), resorufin (Em = -50 mV) (26), or safranin O (Em = -280 mV) (26). 50 to 100 nM of xanthine oxidase enzyme (Sigma-Aldrich) was added to initiate the reaction. The reaction was performed at 25°C in a Coy Anaerobic Airlock Chamber after all solutions were allowed to equilibrate with N2 overnight. Visible spectra were recorded every 2 min using a Shimadzu UV-1800 spectrophotometer until reduction was complete (typically between 1 and 2 hrs). The absorbance change for the b-heme peak was measured at 426 nm, where the change in absorbance of the dye is negligible, and the absorbance change for the dye was measured at 632 nm, where there is little contribution from heme absorbance. The changes in absorbance were analyzed by the Nernst equation terms: [25 mV ln (b-hemered/b-hemeox)] for the one-electron reduction of heme and [12.5 mV ln (dyered/dyeox)] for the two-electron reduction of dye, where b-hemered/b-hemeox and dyered/dyeox represent ratios of the molar concentrations of the reduced and oxidized forms of the b-heme and the dye, respectively. Values corresponding to the Nernst equation terms for the b-heme and the dye at each time point during the titration were plotted against each other on the x- and y-axes, respectively, yielding a straight line with a slope of 1. The y-intercept of this line represents the difference in potential between the b-heme and the known potential of the reference dye. Using this method, the reduction potential for the b-heme was calculated to within ± 2 mV based on four independent titrations. To establish the accuracy of this method, two heme proteins of known potential (cytochrome c and myoglobin) were titrated using the above method and their respective reduction potentials were found to be within 2 mV of published values. Potentials are given versus the standard hydrogen electrode.
RESULTS
Resonance Raman Spectroscopy
Resonance Raman (rR) spectroscopy of heme proteins can yield considerable insight into structural, conformational and electronic properties of the heme cofactor. Specifically, it is possible to determine coordination number, oxidation number, and spin state of the iron, as well as heme conformation, axial ligand identity, the nature of axial ligand bonding, and nonbonded interactions with the protein (27-30). To gain some of this insight into the heme b that purifies with CcmF, we recorded Soret-excited rR spectra of ferric and ferrous WT CcmF. We also recorded spectra of the CcmF-CO complex to further probe the heme pocket.
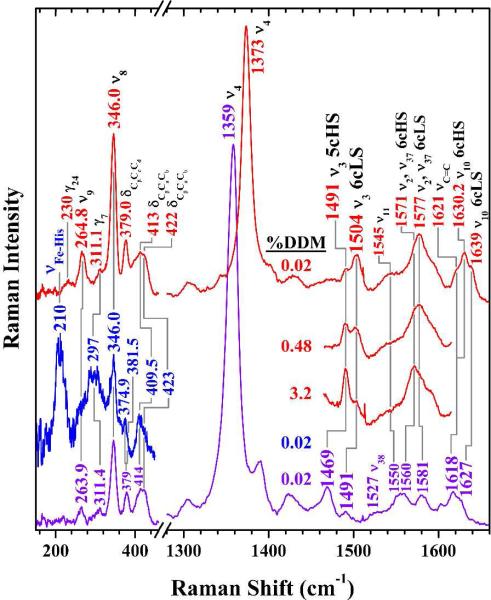
Figure 2 shows the high- and low-frequency Soret excited rR spectra of ferric (top, red) and ferrous (bottom, violet) CcmF in 0.02 % DDM. The bands in the high frequency regions of these spectra report oxidation and spin states of the heme iron as well as the coordination number (29, 30). The v3 regions of the these spectra exhibit two bands (1491 and 1504 cm-1 for ferric CcmF; 1469 and 1491 cm-1 for ferrous CcmF) typical of pentacoordinate high-spin (5cHS) and hexacoordinate low-spin (6cLS) heme complexes in both oxidation states. Thus, the heme b in ferric and ferrous CcmF exists as an equilibrium mixture of 5cHS and 6cLS states. Note that for ferrous CcmF, the 1469 cm−1 v3 band corresponding to the 5cHS fraction (Figure 2, bottom) is considerably more intense than its 6cLS counterpart. However, this intensity ratio does not reflect the relative populations of the two states of the ferrous heme because the relative rR scattering cross sections are not the same. In an effort to estimate the population ratio, the UV-visible spectrum was fit so that the Soret band intensities from the HS and LS populations could be compared (Figure S1). Based on this analysis, ferrous CcmF contains approximately 20 % 5cHS heme.
Figure 2.
Soret-excited resonance Raman spectra of the heme b in ferric (red) and ferrous (violet, blue) CcmF. Sample solutions were 88 μM in holoCcmF, 20 mM in Tris, pH 8, 100mM in NaCl, 0.02 % in dodecyl maltoside, ~ 2 nm in imidazole. HoloCcmF concentrations in the 3.2 % and 0.48 % DDM samples were 23 and 25 μM, respectively. The red and violet spectra were recorded using 10 mW of laser light at 413.1 nm (line focus of emission from Kr+ laser). The low-frequency blue spectrum was recorded with 2 mW of 441.6-nm emission from a HeCd laser to identify the Fe−His stretching band.
To assess whether the heme heterogeneity is intrinsic to CcmF or induced by solubilizing the protein in detergent, Soret-excited rR spectra of ferric and ferrous CcmF were recorded from solutions containing 0.02 % (Figure 2), 0.48 %, and 3.2 % DDM. The high-frequency spectra of ferric CcmF are compared in Figure 2 (partial traces, red). The rR spectra clearly show that higher DDM concentrations correspond to greater HS heme populations. Thus, although heterogeneity was apparent in all of the preparations, low DDM concentrations clearly favor the 6cLS heme population of ferric CcmF. Two possible explanations for this detergent dependence are envisioned at this time. First, the two states of the heme could be reporting conformational states of the protein that are accessed as part of its native function. For example, these could be states whose populations are modulated by in vivo interaction with CcmH, holoCcmE, or apocytochrome c. Alternatively, the population of two ferric heme states may arise from a non-physiological CcmF conformer whose formation is induced by solubilizing the protein in DDM detergent. It is important to note that the relative populations of 5cHS and 6cLS ferrous CcmF were only slightly sensitive to detergent concentration (data not shown).
Raman scattering by νFe-His modes of 5cHS ferrous hemes is typically well enhanced with blue excitation and usually occurs within the range of 200 to 245 cm-1 (32). Thus, if the axial ligand is an imidazole side chain of a histidine residue, we would expect to see a strong band in that frequency range. The second spectrum from the bottom (blue) in the low frequency range of Figure 2 shows the 441.6-nm excited rR spectrum of ferrous CcmF(WT). This spectrum exhibits an intense band at 210 cm−1, within the frequency range of Fe-His stretching modes. Such bands are typical for a Fe−His unit wherein the proximal histidine ligand is the donor to a weak H-bond within the proximal heme pocket (28, 31, 32). Such H-bonds can involve water in a H-bond network or O atoms from backbone carbonyl groups. Thus, in the 5cHS ferrous form, the proximal ligand is almost certainly the imidazole side chain of a histidine residue.
Vibrational frequencies of the FeCO group in heme—CO complexes are useful probes of the distal heme pocket. To further investigate the proximal ligand properties of CcmF with respect to the heme b, we recorded spectra of the CcmF—CO complex. By virtue of the π backbonding that typifies FeCO moieties, there is an inverse relationship between the sensitivity of the Fe—C and C—O bond strengths to the bonded and nonbonded interactions of the FeCO unit (34-36). This inverse relationship is clearly revealed on a heme FeCO backbonding correlation plot by the negative slope of the Fe—C stretching frequency versus the C—O stretching frequency for a large number of iron porphyrinates, including carbonyl complexes of heme proteins. Because the donor strength of the proximal ligand modulates the vertical position of a heme carbonyl complex on the correlation plot, the nature of the proximal ligand can be discerned from the position of the heme carbonyl complex relative to the positions of other heme carbonyls with known proximal ligands.
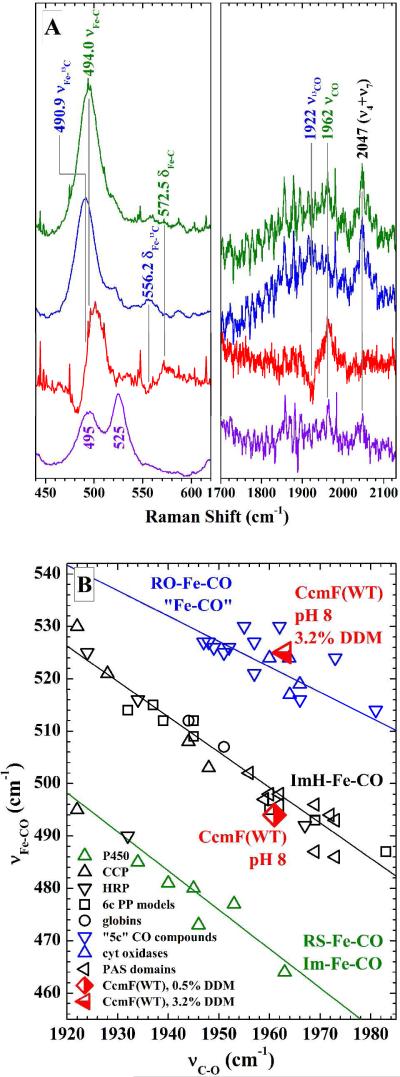
In order to place a particular complex on the correlation plot, the frequencies must be identified in the rR spectrum by isotope editing of the vibrational frequencies with 13CO. By this method, the frequencies of modes involving distortions of the FeCO unit can be unambiguously identified. Figure 3A shows the regions of the rR spectrum of the CcmF—CO (green) and CcmF—13CO (blue) complexes where bands arising from FeCO vibrations occur (33). Bands were assigned based on their 13C sensitivity, which is apparent from the difference spectra shown in red. The bands at 494 and 572 cm-1 are assigned to the vFe—C and the δFeCO modes, respectively. The band at 1962 cm-1 was assigned to the vC—O mode. Interestingly, the low frequency region of the CcmF—CO spectrum recorded in 3.2 % DDM (bottom, violet) reveals two Fe—C stretching bands: one at 495 cm−1 (the same Raman shift observed at low DDM concentrations) and the other at 525 cm−1. Thus, high DDM concentrations induce formation of a second CcmF—CO complex.
Figure 3.
(A) Soret(413.1 nm)-excited rR spectra of natural abundance CcmF−CO (green) and CcmF−13CO (blue) in the vFe-C and vC-O frequency regions. The red traces are the difference spectra, whose amplitudes have been multiplied by a factor of two for ease of viewing. The difference features reveal the bands whose frequencies are 13C dependent, thereby facilitating their assignments to FeCO modes, which are indicated in the labels above the bands. The violet trace shows the spectrum of natural abundance CcmF−CO in 3.2 % DDM. Note that this spectrum exhibits two Fe−C stretching bands. This heterogeneity is attributed to protein conformational changes driven by the high detergent concentration. (B) Heme FeCO backbonding correlation plot showing the positions of the two CcmF−CO conformers. The CcmF−CO complex that falls on the middle line (0.48 % DDM) most likely contains a proximal imidazole ligand from a His residue. Its position low and to the right on the imidazole line shows that the CO ligand interacts only weakly with the heme pocket. The CcmF−CO complex that falls on the top line (3.2 % DDM) is either pentacoordinate or has a proximal ligand that is bound through an oxygen atom.
Figure 3B shows the FeCO backbonding correlation plot described above, with three inverse correlations between vFe−C and vC−O frequencies for a wide selection of heme carbonyl complexes. The bottom (green) line in Figure 3B correlates Fe—C and C—O bond strengths for proximal ligands having anionic character, such as the strongly H-bond donating proximal imidazoles of the heme peroxidases and the proximal thiolates of the cytochromes P450. The middle (black) line correlates the analogous bond strengths in proteins that have charge neutral proximal ligands that are bound to the heme iron through an N atom. Nearly all of these complexes have proximal imidazole ligands from histidine residues. The top (blue) line correlates heme—CO complexes that are either pentacoordinate or have proximal O-atom donor ligands. The position of CcmF—CO in 0.48 % DDM is indicated by the half-filled red diamond in Figure 3B. This point falls on the proximal histidine correlation line, consistent with at least one endogenous axial histidine ligand for the heme b of CcmF. Note that this analysis does not facilitate identification of the specific coordinating histidine residue. The heme carbonyl that was only observed in 3.2 % DDM falls on the top line in Figure 3B, as shown by the half-filled red triangle. This position constitutes compelling evidence that the heme—CO complex in high DDM is either pentacoordinate or has a sixth axial ligand coordinated to iron through an oxygen atom. This result shows that in high DDM solutions, the second protein-based axial ligand that gives rise to 6cLS heme is unavailable to the heme in a significant fraction of these ferrous heme—CO complexes. This behavior is consistent with the correlation between the HS ferric heme population and DDM concentration illustrated in Figure 2.
Heme content of Ala substitutions
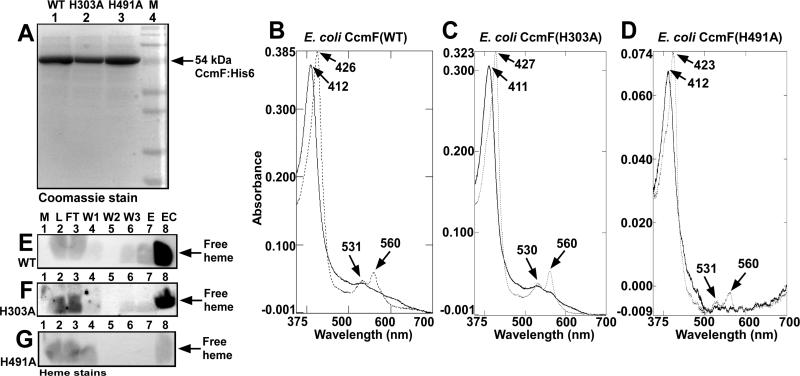
To identify the ligands to b-heme iron in CcmF, we focused on the four conserved histidines (His173 and His303 in periplasmic loops, and His261 and His491 in TMDs). Previously, we showed that purified CcmF with a His173Ala substitution contained wild-type (WT) levels of b-heme, while CcmF with a His261Ala substitution had approximately four-fold less heme (8). Here, we analyze heme levels in purified CcmF with alanine substitutions at His303 or His491. Figure 4A shows that CcmF His303Ala and CcmF His491Ala were each purified as full-length proteins with minimal degradation. Comparison of the Soret maxima in the electronic spectra indicates that CcmF His303Ala purified with heme levels similar to WT CcmF while CcmF His491Ala purified with less than 20 % of WT levels (note the differences in absorbance in Figures 4B, 4C, and 4D). This was confirmed by semi-quantitative heme stain to detect the b-heme, which runs at the dye front during SDS-PAGE (Figures 4E, 4F, and 4G). The purified concentrated preparations (lane 8 of Figures 4E, 4F, and 4G) show much less heme for CcmF His491Ala. The same concentration of purified CcmF was analyzed for each of the proteins; thus, the fractional heme loading in each protein can be directly compared. These data and previous findings in ref. (8), suggest that periplasmic His173 and His303 are not b-heme ligands, while transmembrane His261 and His491 are good candidates for ligands to the b-heme. In agreement with this, CcmF with glycine substitutions at His261 and His491 also contained approximately 20 % heme content relative to WT CcmF (data not shown).
Figure 4.
Heme levels in E. coli wild-type CcmF (WT), CcmF His303Ala (H303A), and CcmF His491Ala (H491A) proteins. Coomassie stain (A) of CcmF(WT), CcmF(H303A), and CcmF(H491A) showing purified full-length 54 kDa proteins (indicated by arrow). UV/Vis absorption spectra of CcmF(WT) (B), CcmF(H303A) (C), and CcmF(H491A) (D) as purified (oxidized; solid lines) and dithionite-reduced (dotted lines). Absorption maxima are indicated with arrows. Heme stains of CcmF(WT) (E), CcmF(H303A) (F), and CcmF(H491A) (G); free heme is indicated with arrows; M, molecular weight standards; L, load; FT, flow-through; W1-W3, washes 1-3; E, elution; EC, concentrated elution, as described in Experimental Procedures. 20 μM purified CcmF protein was analyzed by UV/Vis absorption spectroscopy (B, C, D) and 30 μg was analyzed by heme stain or Coomassie stain (A; E, F, G, lane 8).
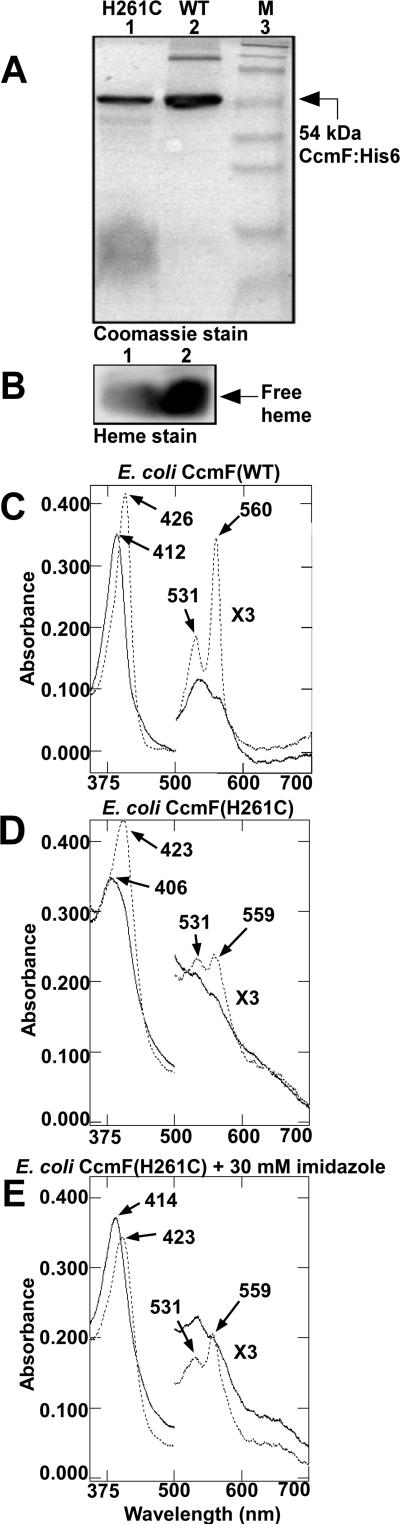
Spectral perturbations in His261Cys
In making substitutions to His261 and His491, we observed that a His261Cys substitution in CcmF retained a high level of function relative to WT (see below) and that purified CcmF His261Cys (Figure 5A) contained approximately 50 % of the b-heme as WT (Figure 5B). Cysteine has been shown to function as a ligand in place of the natural histidine ligand in many heme proteins, including myoglobin (37-39), cytochrome b5 (40), and flavocytochrome b2 (41). The UV/Vis absorption spectrum of CcmF His261Cys has several distinct features in comparison to the spectrum of WT CcmF (compare Figures 5C and 5D). Oxidized (“as purified”) WT CcmF exhibits a sharp Soret maximum at 412 nm, whereas the Soret band for CcmF His261Cys is broad with a maximum at 406 nm. Additionally, the α and β absorptions in the visible region are much less pronounced for CcmF His261Cys than for WT CcmF. The position of the Soret at 406 nm is indicative of high-spin heme (40), but the breadth of the Soret band and the α and β absorptions suggest that ferric CcmF His261Cys may also contain a population of 6cLS heme (42, 43). Thus, while the UV/Vis absorption spectrum of ferric WT CcmF is dominated by features characteristic of 6cLS heme (40), the spectrum of ferric CcmF His261Cys is consistent with a mixture of 5cHS and 6cLS heme. The dithionite-reduced spectrum for CcmF His261Cys shows a Soret of 423 nm and exhibits clear α and β absorptions, although the α:β peak ratio is different than for WT CcmF. Based on the position of the Soret band and the clearly resolvable α and β maxima, ferrous CcmF His261Cys appears to contain predominantly low-spin heme (40). The presence of 6cLS heme in CcmF His261Cys suggests possible coordination by His491 and Cys261. While low-spin hemes with cysteine ligands typically exhibit Soret bands near 450 nm (44), Soret absorptions near 425 nm have been observed when the sulfur-iron bond is weakened or displaced (39, 42, 43, 45). These spectral perturbations observed in ferric and ferrous CcmF His261Cys (relative to the spectrum of WT CcmF) suggest that in the WT protein, His261 is an axial ligand to the b-heme.
Figure 5.
Spectral analysis of CcmF His261Cys (H261C) and wild-type CcmF (WT) proteins. Coomassie stain (A) and heme stain (B) of CcmF(WT) and CcmF(H261C) proteins. UV/Vis absorption spectra of CcmF(WT) (C), CcmF(H261C) (D), and CcmF(H261C) in the presence of 30 mM imidazole (E), oxidized (solid lines) and dithionite-reduced (dotted lines). Absorption maxima are indicated with arrows. Absorbance values between 500 nm and 700 nm have been multiplied by a factor of 3. 30 μg of purified protein was analyzed by Coomassie and heme stain (A, B). For UV/Vis absorption spectra approximately 20 μM purified protein was analyzed for wild-type, and approximately 40 μM purified protein was analyzed for His261Cys in the presence and absence of 30 mM imidazole.
Imidazole, the side chain of histidine, has been used as a surrogate proximal ligand to probe heme binding sites in proteins in vitro (e.g., 40, 42, 46). To investigate the effect of imidazole on heme in the His261Cys CcmF protein, UV/Vis spectra of purified CcmF His261Cys were recorded in the presence of imidazole. At a concentration of 30 mM imidazole, the spectrum of the oxidized His261Cys mutant shifts to resemble that of WT CcmF (compare solid lines in Figures 5C and 5E). The Soret band occurs at 414 nm (compared to 406 nm in the absence of imidazole) and pronounced α and β absorptions similar to WT CcmF can be observed. Upon reduction with sodium dithionite, the His261Cys (+ 30 mM imidazole) exhibits a Soret band at 423 nm and an α to β peak ratio similar to that of reduced WT CcmF. Addition of 30 mM imidazole to WT CcmF preparations did not change the UV/Vis spectrum (not shown). These data suggest that in the presence of imidazole, heme in His261Cys is predominantly 6cLS in both the oxidized and reduced states. The similarities in the spectra of CcmF His261Cys in the presence of imidazole and WT CcmF suggest that, in the WT protein, the b-heme may be liganded by the imidazole side chains of two His residues.
Functional restoration of CcmF His261 and His491 mutants by imidazole
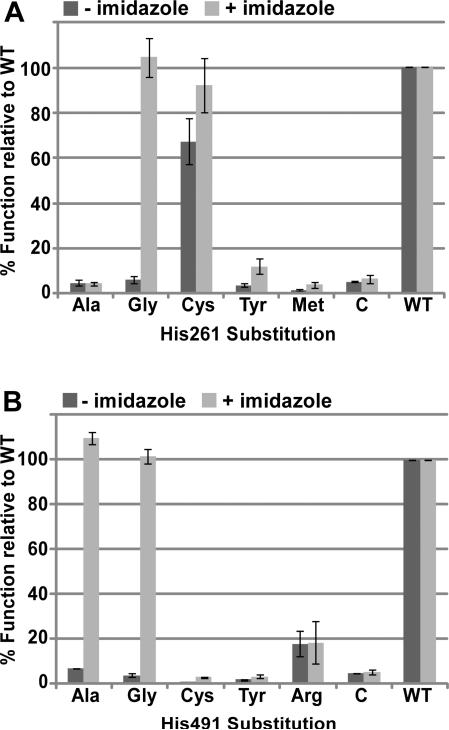
Alanine substitutions at each of the four conserved histidine residues (His173, His261, His303, and His491) abolished in vivo function (cytochrome c assembly) in E. coli under aerobic conditions (Figure S2). Initially, we analyzed whether these alanine substitutions could be corrected for in vivo function by adding imidazole to E. coli cultures (Figure S1). In vivo chemical complementation by exogeneous imidazole is conceptually similar to the correction of heme binding by imidazole in the recombinant myoglobin His93Gly “cavity” mutant, for which 10 mM imidazole was optimal (47). This approach has also been used to correct the function of histidine mutants in the hydrogenase cytochrome b of Azotobacter vinelandii (48) and the CcsBA system II cytochrome c synthetases from Helicobacter hepaticus (23) and Wolinella succinogenes (49). The His491Ala mutant of CcmF was corrected for cytochrome c4 assembly at 10 mM imidazole (Figure S2, lane 12). To determine if glycine substitutions in CcmF could be corrected for function, we replaced transmembrane His491 or His261 with glycine and assayed for cytochrome c4 assembly in the absence and presence of imidazole. Substitution of either His261 or His491 with glycine abolished or severely impaired cytochrome c4 assembly in vivo (quantified in Figure 6A and 6B; representative heme stains for cytochrome c4 are in Figure S3). Addition of 10 mM imidazole restored function to CcmF His261Gly and to His491Gly (Figure 6A and 6B). We propose that the CcmF His261Gly and His491Ala/Gly proteins form “cavities” in which imidazole can bind and serve as a ligand to the b-heme in CcmF. Based on these results, we predicted that amino acids with bulkier side chains (that are unable to function as b-heme ligands) would not accommodate imidazole binding in the “cavity”, and thus would not permit functional correction. We tested this by engineering additional substitutions at His261 and His491. As mentioned above, His261Cys was roughly 70 % functional relative to wild-type in the absence of imidazole (Figure 6A). However, neither His261Tyr nor His261Met were functional above background levels, nor were these substitutions corrected substantially by imidazole (Figure 6A). The same was true for Cys and Tyr substitutions at His491 (Figure 6B). His491Arg showed a low level of function relative to WT (approximately 18 %), but imidazole did not improve the function of this mutant (Figure 6B). These results are consistent with His261 and His491 acting as heme ligands, such that when these His residues are substituted with amino acids containing small side chains (Gly or Ala), a cavity is formed in which imidazole can bind and serve as a surrogate heme axial ligand. The inability of imidazole to correct the cytochrome c assembly defect of CcmF His261Ala could be the result of the methyl group of Ala occluding the “cavity” such that imidazole is unable to bind. We have not investigated this further.
Figure 6.
In vivo heme attachment to cytochrome c4 (i.e., holocytochrome c formation) by the indicated CcmF His261 and His491 mutants in the presence or absence of 10 mM imidazole. The function of each of the indicated substitutions at CcmF His261 (A) or His491 (B) is reported as a percentage of wild-type CcmF (WT) function. Holocytochrome c4 was quantified by measuring the chemiluminescent heme stain signal of 100 μg of BPER-isolated proteins from three independent experiments. Representative heme stains are provided in Supporting Information (Figure S3). Error bars denote standard deviation. “C” denotes control, which represents chemiluminescent signal in the absence of CcmF (any signal lower than the control is considered background).
Conservation of the b-heme in CcmF
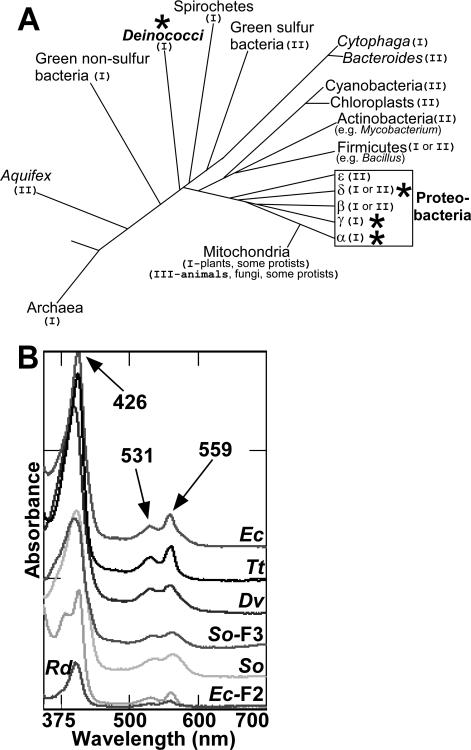
To determine if the b-heme in CcmF is conserved, we engineered CcmF proteins from a phylogenetically diverse group of prokaryotes (Figure 7A; gamma proteobacteria- Escherichia coli and Shewanella oneidensis; delta proteobacteria-Desulfovibrio vulgaris, alpha proteobacteria-Roseobacter denitrificans; deinococci-Thermus thermophilus). CcmF proteins from E. coli (NrfE, here called CcmF2) and Shewanella (here called CcmF3) that recognize and mature cytochromes c with alternate heme binding sites (Cys-Xxx-Xxx-Cys-Lys in E. coli; and, putatively, Cys-Xxx15-Cys-His in Shewanella) were also engineered with hexahistidine tags and purified (Figure S4). Each of these CcmF homologs possesses the four conserved histidine residues studied above in the E. coli CcmF. Heme analysis by UV/Vis absorption spectroscopy (Figure 7B) and heme stain (Figure S4) revealed that each purified CcmF protein contained b-heme in approximately equimolar stoichiometry.
Figure 7.
Phylogenetic distribution and spectral analysis of diverse CcmF proteins. (A) Adapted from reference (23). Representative distribution of systems I, II and III among the bacteria and archaea. The system number is noted in parentheses after each group name, and stars indicate groups containing organisms from which recombinant hexahistidine-tagged CcmF was analyzed in the present study. (B) Reduced UV/Vis absorption spectra of CcmF from E. coli (Ec), Thermus thermophilus (Tt; Deinococcus group), Desulfovibrio vulgaris (Dv), Roseobacter denitrificans (Rd), and Shewanella oneidensis (So); Shewanella oneidensis CcmF-3 (So-F3), and E. coli CcmF-2 (Ec-F2). Absorption maxima are indicated with arrows. Spectra have been offset for clarity. Approximately 20 μM of each purified CcmF protein was analyzed for which Coomassie and heme stains are provided in Supporting Information (Figure S4).
Reduction potential of the b-heme in CcmF
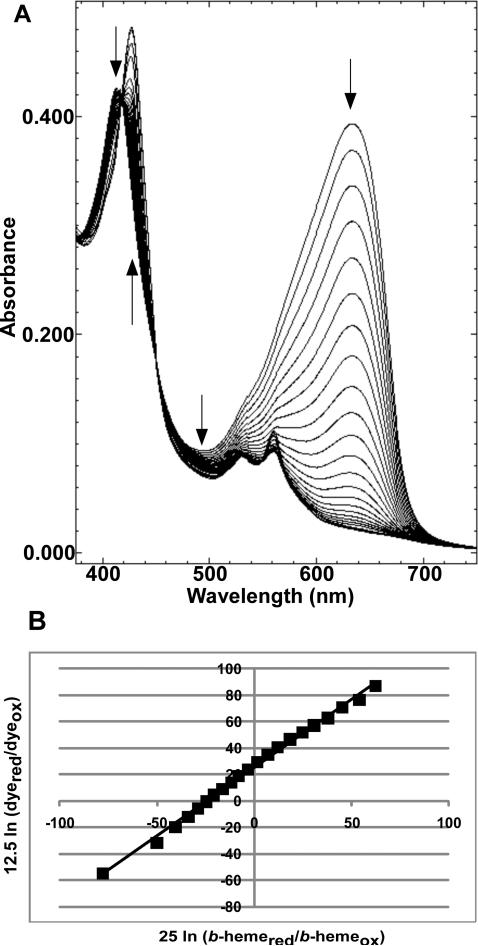
We determined the reduction potential for the CcmF b-heme using the purified CcmF from E. coli (Figure 8). Using a modification of the method of Massey (with xanthine/xanthine oxidase to generate reductant) (25), values for the midpoint potential of the CcmF b-heme (Fe3+/Fe2+) were determined against nile blue chloride (Em= -116 mV) at pH 7 (26). The spectral results obtained from a typical reduction run with CcmF and nile blue chloride are shown in Figure 8A. The decreases in absorbance at 412 nm and 480 nm and the increase in absorbance at 426 nm are indicative of reduction of the CcmF b-heme. The decrease in absorbance at 632 nm is due to reduction of nile blue chloride. The linear Nernst plot for reduction of the b-heme and nile blue chloride produced the expected slope of 1 (Figure 8B), indicating that the reduced and oxidized forms of the dye and the heme were at their equilibrium concentrations at the time each spectrum was recorded. This behavior ensures that there is no overpotential contribution to the midpoint potentials determined from these measurements. The reduction potential of the b-heme in CcmF was found to be -147 ± 2 mV. This value was confirmed using resorufin (Em = -50 mV) (26) or safranin O (Em = -280 mV) (26) as alternate redox dyes (Figure S5). It has been observed previously that the type of detergent (50, 51) and detergent concentration (50) can alter reduction potentials of b-type hemes in membrane proteins. To assess the effects of DDM, we determined the reduction potential of the CcmF b-heme in solutions containing 0.02 % (Figure 8) or 1.2 % DDM. The reduction potential in 1.2 % DDM was found to be substantially more positive (Em = -110 ± 4 mV) than in 0.02 % DDM (Figure S6). This is likely attributable to the same DDM-dependent speciation of the ferric heme-b between HS and LS states that was revealed by the rR spectra (vide supra).
Figure 8.
Redox titration of the CcmF b-heme. Spectra collected during a typical reductive titration of CcmF b-heme with nile blue chloride (A) and the corresponding linear Nernst plot (B). Arrows in (A) indicate the direction of changes in absorption during the course of the titration. In (B), [25 mV ln (b-hemered/b-hemeox)] was used for the one-electron reduction of heme and [12.5 mV ln (dyered/dyeox)] was used for the two-electron reduction of dye, where bhemered/b-hemeox and dyered/dyeox represent ratios of the molar concentrations of the reduced and oxidized forms of the b-heme and the dye, respectively. Conditions: 20 mM Tris-HCl, pH 7, 100 mM NaCl, 0.02 % DDM.
DISCUSSION
Here, we present several lines of evidence that TMD His261 and His491 are the ligands to the CcmF b-heme: (i) Gly or Ala substitutions at either His result in approximately 80 % less heme as compared to WT CcmF (Figure 4), (ii) substitution of His261 with Cys, a known heme ligand, yields a functional CcmF that shows improved heme binding relative to substitutions with non-ligand residues (Ala or Gly) and shows a perturbed UV/Vis spectrum (Figure 5), and (iii) exogeneous imidazole chemically corrects the in vivo cytochrome c synthetase function of small residue substitutions at CcmF His261 and His491, but not substitutions with bulkier residues (Figure 6). The results of rR spectroscopy confirm the presence of hexacoordinate, low-spin b-heme (Figure 2), and rR analysis of the heme-carbonyl complex suggests axial His coordination of the b-heme (Figure 3). Additionally, we show that the CcmF b-heme is widely conserved by analyzing recombinant CcmF proteins from diverse bacteria (Figure 7).
Implications of His261 and His491 as the b-heme axial ligands in CcmF
His261 in CcmF is located in TMD5, and His491 is located near the C-terminus in TMD12 (see Figure 1B). In order for these residues to coordinate the iron of the b-heme, we propose that TMD5 and TMD12 are positioned close to each other in the three-dimensional structure of CcmF. These findings suggest that the most likely position for the b-heme is directly below the periplasmic WWD domain in CcmF (refer to Figure 1B). The WWD domain is a highly conserved tryptophan-rich motif that is located at the outer leaflet of the inner membrane and has been shown to interact directly with heme in the WWD family member CcmC (7). In CcmC, heme axial ligands are provided by two His residues located in periplasmic loops that flank the WWD domain (7, 8). Similar to CcmC, we predict that the WWD domain of CcmF forms a heme binding site for heme from holoCcmE, with periplasmic His173 and His303 of CcmF serving as heme axial ligands (Figure 1B). Thus, the positioning of the b-heme directly below the CcmF WWD domain is consistent with our hypothesized role for CcmF in reducing heme from holoCcmE prior to attachment to the apocytochrome (see below).
The function of CcmF b-heme as a holoCcmE reductase
Reduction of the heme in holoCcmE is required for covalent attachment of this heme to the apocytochrome c. We have previously discussed that reduction of the holoCcmE heme could also favor ejection of the CcmE His130 covalent linkage (5). To address our hypothesis that the b-heme in CcmF may be involved in electron transfer to holoCcmE (5, 8), we determined the reduction potential of the CcmF b-heme. We report here that the CcmF b-heme has a midpoint potential of -147 mV at pH 7 (Figure 8). Harvat et al. have reported a redox potential of -121 mV at pH 7.3 for holoCcmE′, a soluble version of CcmE without the single TMD (52). Despite the differences in the buffer conditions used in determining the reduction potentials for holoCcmE′ and here for CcmF (e.g., the presence of DDM), it is certainly feasible that CcmF functions as a holoCcmE reductase. However, the reduction potential of isolated, purified holoCcmE′ may not reflect the physiological potential of this heme prior to attachment to the apocytochrome c. Recall that, en route to attachment to apocytochrome c, the holoCcmE heme likely enters the WWD domain of CcmF, where heme axial ligands are provided by His173 and His303 of CcmF (see Figure 1B). In purified holoCcmE, Tyr134 of CcmE has been shown to serve as an axial ligand to the heme (12, 53). It is well-established that heme reduction potential is sensitive to changes in axial ligation (38, 41, 54), and tyrosine axial ligation is typically associated with a lowering of the reduction potential (i.e., more negative) relative to ligation by histidine (38, 55, 56). Thus, the heme ligand “switch” from CcmE Tyr134 to CcmF His173/His303 upon binding of holoCcmE in the CcmF WWD domain could result in an increase in the reduction potential (i.e., more positive) of the heme in holoCcmE. Although the identity of the physiological electron donor to the CcmF b-heme is unknown, we previously showed that CcmF could be reduced by select quinone species in vitro in the presence of dithiothreitol (8). Further studies are needed to investigate these features of cytochrome c assembly.
Interaction between CcmF and holoCcmE
Although it is widely believed that the heme from holoCcmE moves onto the apocytochrome c via CcmF, to our knowledge, an interaction between CcmF and holoCcmE has never been detected. Thony-Meyer and colleagues have suggested that apoCcmE and CcmF interact (16). However, in that study, because the genes encoding CcmABCD were absent, all of the CcmE was in the apo- form, so the physiological relevance of this finding is unclear. Furthermore, while Ren et al. (16) showed that antisera to CcmF co-immunoprecipitated apoCcmE, the reverse co-immunoprecipitation with anti-sera to CcmE yielded “non-specific signals” (16). In our hands, the only protein that co-purifies with CcmF is CcmH (for example, see Figure 4 in ref. (8)). We have been unable to detect apo- or holoCcmE above background levels in purifications of hexahistidine-tagged CcmF. We suggest that holoCcmE may not co-purify with CcmF (or the CcmF/H complex) unless the apocytochrome c acceptor (Cys-Xxx-Xxx-Cys-His) is bound. This proposal is in part based on our recent findings on the “acceptor-dependent” nature of the CcmC-heme-CcmE complex of system I (7). CcmC does not bind detectable levels of heme in its WWD domain in the absence of CcmE (Figure 1A), indicating that heme binding in CcmC requires the presence of the heme acceptor, CcmE. By analogy, heme in holoCcmE may not enter the WWD domain of CcmF unless the heme acceptor protein, apocytochrome c, is present. Future work will be dedicated to better understanding the interaction of holoCcmE with CcmF; specifically, investigating if the docking of holoCcmE in the CcmF WWD domain requires the presence of the apocytochrome c acceptor, and, if the holoCcmE/CcmF/CcmH complex can be “trapped,” determining the reduction potential of the heme in holoCcmE when (and if) it is bound in the CcmF WWD domain.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Robert Blankenship, Dr. Emma Raven, and Dr. Igor Efimov for helpful discussions on reduction potentials. We thank Dr. Cynthia Richard-Fogal for helpful discussions and manuscript comments. We also thank Dr. Huifen Zhu for construction of the CcmF His303Ala mutant (pRGK405).
Abbreviations
- Ccm
cytochrome c maturation
- Cyt
cytochrome
- TMD
transmembrane domain
- BPER
bacterial protein extraction reagent
- WT
wild-type
- rR
resonance Raman
- DDM
n-Dodecyl-β-D-Maltopyranoside
Footnotes
This work was supported by National Institutes of Health grants GM47909 to R. G. K. and AI072719 to K. R. R.
SUPPORTING INFORMATION AVAILABLE
A complete description of the plasmids and strains used in this research along with other data are available as Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Barker PD, Ferrer JC, Mylrajan M, Loehr TM, Feng R, Konishi Y, Funk WD, MacGillivray RT, Mauk AG. Transmutation of a heme protein. Proc Natl Acad Sci U S A. 1993;90:6542–6546. doi: 10.1073/pnas.90.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicholson DW, Neupert W. Import of cytochrome c into mitochondria: reduction of heme, mediated by NADH and flavin nucleotides, is obligatory for its covalent linkage to apocytochrome c. Proc Natl Acad Sci U S A. 1989;86:4340–4344. doi: 10.1073/pnas.86.12.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferguson SJ, Stevens JM, Allen JW, Robertson IB. Cytochrome c assembly: a tale of ever increasing variation and mystery? Biochim Biophys Acta. 2008;1777:980–984. doi: 10.1016/j.bbabio.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Hamel P, Corvest V, Giege P, Bonnard G. Biochemical requirements for the maturation of mitochondrial c-type cytochromes. Biochim Biophys Acta. 2009;1793:125–138. doi: 10.1016/j.bbamcr.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard-Fogal C, Kranz RG. The CcmC:heme:CcmE complex in heme trafficking and cytochrome c biosynthesis. J Mol Biol. 2010;401:350–362. doi: 10.1016/j.jmb.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richard-Fogal CL, Frawley ER, Bonner ER, Zhu H, San Francisco B, Kranz RG. A conserved haem redox and trafficking pathway for cofactor attachment. Embo J. 2009;28:2349–2359. doi: 10.1038/emboj.2009.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulz H, Fabianek RA, Pellicioli EC, Hennecke H, Thony-Meyer L. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc Natl Acad Sci U S A. 1999;96:6462–6467. doi: 10.1073/pnas.96.11.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee D, Pervushin K, Bischof D, Braun M, Thony-Meyer L. Unusual heme-histidine bond in the active site of a chaperone. J Am Chem Soc. 2005;127:3716–3717. doi: 10.1021/ja044658e. [DOI] [PubMed] [Google Scholar]
- 11.Stevens JM, Daltrop O, Higham CW, Ferguson SJ. Interaction of heme with variants of the heme chaperone CcmE carrying active site mutations and a cleavable N-terminal His tag. J Biol Chem. 2003;278:20500–20506. doi: 10.1074/jbc.M212925200. [DOI] [PubMed] [Google Scholar]
- 12.Uchida T, Stevens JM, Daltrop O, Harvat EM, Hong L, Ferguson SJ, Kitagawa T. The interaction of covalently bound heme with the cytochrome c maturation protein CcmE. J Biol Chem. 2004;279:51981–51988. doi: 10.1074/jbc.M408963200. [DOI] [PubMed] [Google Scholar]
- 13.Christensen O, Harvat EM, Thony-Meyer L, Ferguson SJ, Stevens JM. Loss of ATP hydrolysis activity by CcmAB results in loss of c-type cytochrome synthesis and incomplete processing of CcmE. Febs J. 2007;274:2322–2332. doi: 10.1111/j.1742-4658.2007.05769.x. [DOI] [PubMed] [Google Scholar]
- 14.Feissner RE, Richard-Fogal CL, Frawley ER, Kranz RG. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol Microbiol. 2006;61:219–231. doi: 10.1111/j.1365-2958.2006.05221.x. [DOI] [PubMed] [Google Scholar]
- 15.Goldman BS, Beckman DL, Bali A, Monika EM, Gabbert KK, Kranz RG. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J Mol Biol. 1997;268:724–738. doi: 10.1006/jmbi.1997.0992. [DOI] [PubMed] [Google Scholar]
- 16.Ren Q, Ahuja U, Thony-Meyer L. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J Biol Chem. 2002;277:7657–7663. doi: 10.1074/jbc.M110979200. [DOI] [PubMed] [Google Scholar]
- 17.Feissner RE, Richard-Fogal CL, Frawley ER, Loughman JA, Earley KW, Kranz RG. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol Microbiol. 2006;60:563–577. doi: 10.1111/j.1365-2958.2006.05132.x. [DOI] [PubMed] [Google Scholar]
- 18.Sanders C, Turkarslan S, Lee DW, Onder O, Kranz RG, Daldal F. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J Biol Chem. 2008;283:29715–29722. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman BS, Beck DL, Monika EM, Kranz RG. Transmembrane heme delivery systems. Proc Natl Acad Sci U S A. 1998;95:5003–5008. doi: 10.1073/pnas.95.9.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feissner R, Xiang Y, Kranz RG. Chemiluminescent-based methods to detect subpicomole levels of c-type cytochromes. Anal Biochem. 2003;315:90–94. doi: 10.1016/s0003-2697(02)00658-9. [DOI] [PubMed] [Google Scholar]
- 21.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 22.Richard-Fogal CL, Frawley ER, Feissner RE, Kranz RG. Heme concentration dependence and metalloporphyrin inhibition of the system I and II cytochrome c assembly pathways. J Bacteriol. 2007;189:455–463. doi: 10.1128/JB.01388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frawley ER, Kranz RG. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc Natl Acad Sci U S A. 2009;106:10201–10206. doi: 10.1073/pnas.0903132106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massey V. In: Flavins and Flavoproteins. Curti B, Ronchi S, Zanetti G, editors. Walter de Gruyter & Co.; New York: 1991. pp. 59–66. [Google Scholar]
- 25.Efimov I, Papadopoulou ND, McLean KJ, Badyal SK, Macdonald IK, Munro AW, Moody PC, Raven EL. The redox properties of ascorbate peroxidase. Biochemistry. 2007;46:8017–8023. doi: 10.1021/bi7006492. [DOI] [PubMed] [Google Scholar]
- 26.Clark WM. Oxidation-Reduction Potentials of Organic Systems. Waverly Press; Baltimore, MD: 1960. [Google Scholar]
- 27.Li XY, Czernuszewicz RS, Kincaid JR, Stein P, Stein TG. Consistent porphyrin force field. 2. Nickel octaethylporphyrin skeletal and substituent mode assignments from nitrogen-15, meso-d4, and methylene-d16 Raman and infrared isotope shifts. J Phys Chem. 1990;94:47–61. [Google Scholar]
- 28.Smulevich G, Hu S, Rodgers KR, Goodin DB, Smith KM, Spiro TG. Heme-protein interactions in cytochrome c peroxidase revealed by site-directed mutagenesis and resonance Raman spectra of isotopically labeled hemes. Biospectroscopy. 1996;2:365–376. [Google Scholar]
- 29.Spiro TG, editor. Biological Applications of Raman Spectroscopy, Vol 3: Resonance Raman Spectra of Heme and Metalloproteins. John Wiley and Sons; 1988. [Google Scholar]
- 30.Spiro TG, Czernuszewicz RS, Li XY. Metalloporphyrin structure and dynamics from Resonance Raman spectroscopy. Coord Chem Rev. 1990;100:541–571. [Google Scholar]
- 31.Rodgers KR, Lukat-Rodgers GS, Barron JA. Structural basis for ligand discrimination and response initiation in the heme-based oxygen sensor FixL. Biochemistry. 1996;35:9539–9548. doi: 10.1021/bi9530853. [DOI] [PubMed] [Google Scholar]
- 32.Bangcharoenpaurpong O, Schomacker KT, Champion PM. Resonance Raman investigation of myoglobin and hemoglobin. J Am Chem Soc. 1984;106:5688–5698. [Google Scholar]
- 33.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel heme ligand displacement by CO in the soluble hemophore HasA and its proximal ligand mutants: implications for heme uptake and release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 34.Ray GB, Li XY, Ibers JA, Sessler JL, Spiro TG. How far can proteins bend the FeCO unit? Distal polar and steric effects in heme proteins and models. J Am Chem Soc. 1994;116:162–176. [Google Scholar]
- 35.Smulevich G, Evangelista-Kirkup R, English A, Spiro TG. Raman and infrared spectra of cytochrome c peroxidase-carbon monoxide adducts in alternative conformational states. Biochemistry. 1986;25:4426–4430. doi: 10.1021/bi00363a038. [DOI] [PubMed] [Google Scholar]
- 36.Spiro TG, Wasbotten IH. CO as a vibrational probe of heme protein active sites. J Inorg Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 37.Adachi S, Nagano S, Ishimori K, Watanabe Y, Morishima I, Egawa T, Kitagawa T, Makino R. Roles of proximal ligand in heme proteins: replacement of proximal histidine of human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry. 1993;32:241–252. doi: 10.1021/bi00052a031. [DOI] [PubMed] [Google Scholar]
- 38.Adachi S, Nagano S, Watanabe Y, Ishimori K, Morishima I. Alteration of human myoglobin proximal histidine to cysteine or tyrosine by site-directed mutagenesis: characterization and their catalytic activities. Biochem Biophys Res Commun. 1991;180:138–144. doi: 10.1016/s0006-291x(05)81266-5. [DOI] [PubMed] [Google Scholar]
- 39.Hildebrand DP, Ferrer JC, Tang HL, Smith M, Mauk AG. Trans effects on cysteine ligation in the proximal His93Cys variant of horse heart myoglobin. Biochemistry. 1995;34:11598–11605. doi: 10.1021/bi00036a036. [DOI] [PubMed] [Google Scholar]
- 40.Wang WH, Lu JX, Yao P, Xie Y, Huang ZX. The distinct heme coordination environments and heme-binding stabilities of His39Ser and His39Cys mutants of cytochrome b5. Protein Eng. 2003;16:1047–1054. doi: 10.1093/protein/gzg134. [DOI] [PubMed] [Google Scholar]
- 41.Mowat CG, Miles CS, Munro AW, Cheesman MR, Quaroni LG, Reid GA, Chapman SK. Changing the heme ligation in flavocytochrome b2: substitution of histidine-66 by cysteine. J Biol Inorg Chem. 2000;5:584–592. doi: 10.1007/s007750000141. [DOI] [PubMed] [Google Scholar]
- 42.Dawson JH, Andersson LA, Sono M. Spectroscopic investigations of ferric cytochrome P-450-CAM ligand complexes. Identification of the ligand trans to cysteinate in the native enzyme. J Biol Chem. 1982;257:3606–3617. [PubMed] [Google Scholar]
- 43.Shelver D, Kerby RL, He Y, Roberts GP. CooA, a CO-sensing transcription factor from Rhodospirillum rubrum, is a CO-binding heme protein. Proc Natl Acad Sci U S A. 1997;94:11216–11220. doi: 10.1073/pnas.94.21.11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dawson JH, Andersson LA, Sono M. The diverse spectroscopic properties of ferrous cytochrome P-450-CAM ligand complexes. J Biol Chem. 1983;258:13637–13645. [PubMed] [Google Scholar]
- 45.Reynolds MF, Shelver D, Kerby RL, Parks RB, Roberts GP, Burstyn JN. EPR and Electronic Absorption Spectroscopies of the CO-Sensing CooA Protein Reveal a Cysteine-Ligated Low-Spin Ferric Heme. J Am Chem Soc. 1998;120:9080–9081. [Google Scholar]
- 46.Vetter SW, Terentis AC, Osborne RL, Dawson JH, Goodin DB. Replacement of the axial histidine heme ligand with cysteine in nitrophorin 1: spectroscopic and crystallographic characterization. J Biol Inorg Chem. 2009;14:179–191. doi: 10.1007/s00775-008-0436-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrick D. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93-->Gly. Biochemistry. 1994;33:6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- 48.Meek L, Arp DJ. The hydrogenase cytochrome b heme ligands of Azotobacter vinelandii are required for full H(2) oxidation capability. J Bacteriol. 2000;182:3429–3436. doi: 10.1128/jb.182.12.3429-3436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kern M, Scheithauer J, Kranz RG, Simon J. Essential histidine pairs indicate conserved haem binding in epsilonproteobacterial cytochrome c haem lyases. Microbiology. 2010;156:3773–3781. doi: 10.1099/mic.0.042838-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorence RM, Miller MJ, Borochov A, Faiman-Weinberg R, Gennis RB. Effects of pH and detergent on the kinetic and electrochemical properties of the purified cytochrome d terminal oxidase complex of Escherichia coli. Biochim Biophys Acta. 1984;790:148–153. doi: 10.1016/0167-4838(84)90218-8. [DOI] [PubMed] [Google Scholar]
- 51.Matsuda H, Butler WL. Restoration of high-potential cytochrome b-559 in liposomes. Biochim Biophys Acta. 1983;724:123–127. [Google Scholar]
- 52.Harvat EM, Redfield C, Stevens JM, Ferguson SJ. Probing the heme-binding site of the cytochrome c maturation protein CcmE. Biochemistry. 2009;48:1820–1828. doi: 10.1021/bi801609a. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Rubio I, Braun M, Gromov I, Thony-Meyer L, Schweiger A. Axial coordination of heme in ferric CcmE chaperone characterized by EPR spectroscopy. Biophys J. 2007;92:1361–1373. doi: 10.1529/biophysj.106.098277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sligar SG, Egeberg KD, Sage JT, Morikis D, Champion PM. Alteration of Heme Axial LIgands by Site-Directed Mutagenesis: A Cytochrome Becomes a Catalytic Demethylase. J Am Chem Soc. 1987;109:7896–7897. [Google Scholar]
- 55.de Lacroix de Lavalette A, Barucq L, Alric J, Rappaport F, Zito F. Is the redox state of the ci heme of the cytochrome b6f complex dependent on the occupation and structure of the Qi site and vice versa? J Biol Chem. 2009;284:20822–20829. doi: 10.1074/jbc.M109.016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hildebrand DP, Burk DL, Maurus R, Ferrer JC, Brayer GD, Mauk AG. The proximal ligand variant His93Tyr of horse heart myoglobin. Biochemistry. 1995;34:1997–2005. doi: 10.1021/bi00006a021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.