Abstract
Genetic selection is a major force shaping life on earth. In classical genetic theory, response to selection is the product of the strength of selection and the additive genetic variance in a trait. The additive genetic variance reflects a population’s intrinsic potential to respond to selection. The ordinary additive genetic variance, however, ignores the social organization of life. With social interactions among individuals, individual trait values may depend on genes in others, a phenomenon known as indirect genetic effects. Models accounting for indirect genetic effects, however, lack a general definition of heritable variation. Here I propose a general definition of the heritable variation that determines the potential of a population to respond to selection. This generalizes the concept of heritable variance to any inheritance model and level of organization. The result shows that heritable variance determining potential response to selection is the variance among individuals in the heritable quantity that determines the population mean trait value, rather than the usual additive genetic component of phenotypic variance. It follows, therefore, that heritable variance may exceed phenotypic variance among individuals, which is impossible in classical theory. This work also provides a measure of the utilization of heritable variation for response to selection and integrates two well-known models of maternal genetic effects. The result shows that relatedness between the focal individual and the individuals affecting its fitness is a key determinant of the utilization of heritable variance for response to selection.
GENETIC selection is both a major force shaping life and the principal human tool to improve agricultural populations. In nature, differences in fitness among individuals lead to the evolution of adaptive traits (Haldane 1932; Wright 1937), and in agriculture, breeders improve their populations by selecting the best individuals as parents of the next generation (Lush 1937). Populations respond to selection only if they contain heritable variation, meaning that individuals differ in the effects they transmit to their offspring. In classical quantitative genetics, heritable variation equals the additive genetic variance (Fisher 1918; Haldane 1932; Wright 1937; Robertson 1966; Price 1970). The heritable variation reflects the potential of a population to respond to selection (Robertson 1966; Price 1970; Lande and Arnold 1983), which is important for adaptive evolution in nature and for genetic improvement in agriculture. A clear definition of heritable variation allows one to investigate the mechanisms maintaining heritable variation in nature and to optimize artificial selection schemes in agriculture.
Here I propose a general definition of the heritable variation that determines the potential of a population to respond to genetic selection. Thus, throughout this work, “heritable variation” refers to the quantity that determines the intrinsic potential of a population to respond to genetic selection. The remainder of this Introduction summarizes the classical definition of heritable variation and discusses its limitations. Subsequently, I generalize the definition of heritable variation.
In classical quantitative genetics, heritable variation follows from partitioning individual trait values, z, into a heritable component, A, and a nonheritable residual, e:
| (1) |
(Fisher 1918; Haldane 1932; Wright 1937; Falconer and Mackay 1996; Lynch and Walsh 1998). The A is the additive genetic value, or breeding value, which is the sum of the average effects of the alleles carried by the individual, including the average contributions arising from dominance and/or epistasis (Fisher 1918; Falconer and Mackay 1996; Lynch and Walsh 1998). The residual, e, includes all nonheritable components, which may originate from nonadditive genetic effects and the environment. The heritable variation is defined as the variance of the breeding values among individuals, , which is known as the additive genetic variance. (Note that additive genetic variance excludes transient effects transmitted to offspring, such as additive-by-additive epistatic effects). The additive genetic variance is a component of phenotypic variance (Fisher 1918; Falconer and Mackay 1996; Lynch and Walsh 1998),
| (2) |
Response to selection, , is the additive genetic change in mean trait value from one generation to the next and depends on both the selection process and the additive genetic variance. Although the common expressions for response to selection differ between the fields of evolutionary quantitative genetics and artificial breeding, they are equivalent when traits and fitness follow a multivariate normal distribution (Appendix A). In evolutionary quantitative genetics, response is expressed as the product of the selection gradient, β, and the additive genetic variance,
| (3a) |
(Robertson 1966; Price 1970; Rausher 1992; Rice 2004), where β is the regression coefficient of an individual’s relative fitness on its breeding value. (For this work, it suffices to consider a single trait and discrete generations.) In artificial breeding, response is expressed as the product of the intensity of selection, ι, the accuracy of selection, ρ, and the additive genetic standard deviation (Falconer and Mackay 1996),
| (3b) |
The accuracy of selection is the correlation between an individual’s value for the selection criterion and its breeding value. The intensity of selection is the selection differential expressed in standard deviation units. (The symbol ι is used here rather than the usual i, to avoid confusion with the index for the focal individual).
In accordance with current belief, Equations 3a and 3b show that additive genetic standard deviation, , determines the potential of a population to respond to selection. In Equation 3b, intensity and accuracy of selection are standardized, scale free, parameters that depend on the breeding design; neither intensity nor accuracy includes any information on heritable variation in the trait. The additive genetic standard deviation, by contrast, expresses the heritable differences in the population in trait units and reflects the intrinsic potential of a population to respond to selection. Equation 3a may suggest that , rather than , is the relevant parameter. The selection gradient, however, is not scale free. A full separation of selection and heritable variation follows from expressing Equation 3a as
where is the standardized selection gradient, which expresses the selection differential in units and is scale free. Hence, this expression also identifies as the relevant parameter.
The above shows that a variance partitioning perspective (Equations 1 and 2) and a response to selection perspective (Equations 3a and 3b) yield the same result; both identify additive genetic standard deviation as the relevant parameter. In classical theory, therefore, the heritable variation that determines potential response to selection follows from partitioning the phenotypic variance into additive genetic and remaining components. Because additive genetic variance is a component of phenotypic variance, it cannot exceed phenotypic variance (Equation 2). In the classical perspective, therefore, phenotypic variance restricts a population’s potential to respond to selection.
While the classical quantitative genetic model has increased our understanding of inheritance and response to selection tremendously, it overlooks part of the heritable effects that may contribute to response. Specifically, it states that the heritable effects on the focal individual’s trait value originate solely from the focal individual itself, disregarding the social organization of life (Dawkins 1982; Frank 1998; West-Eberhard 2003). It ignores the effects of an individual’s genes on trait values of others, known as indirect genetic or associative effects (Griffing 1967; Kirkpatrick and Lande 1989; Moore et al. 1997; Wolf et al. 1998; McAdam et al. 2002; Muir 2005; Wilson et al. 2009; Chenoweth et al. 2010; McGlothlin et al. 2010). There is growing evidence that such indirect genetic effects are widespread (Frank 2007). Individual fitness, for example, depends on number and quality of offspring, which are affected not only by the genes in the focal individual, but also by those in its mate. Conspecifics often compete for access to mates, making individual fitness dependent on genes in competitors. Many species, moreover, live in groups or colonies, where individual trait values may depend on genes in group mates. Although indirect genetic effects are often associated with behavioral interactions, such as interference competition (e.g., Wilson et al. 2009) or social learning, they may also work via the environment through effects on resources or exposure to infectious disease. Not only animals, but also microorganisms and plants exhibit numerous social interactions, both in agriculture and in nature (Crespi 2001; Frank 2007; Griffin et al. 2004; Muir 2005; West et al. 2006; Karban 2008).
The phenomenon that trait values depend on multiple individuals is not restricted to indirect genetic effects. Some traits cannot be attributed to single individuals, but emerge only at a higher level of organization, such as growth rate of a colony in social insects (Wheeler 1933) or the number of prey caught by a hunting pack. For such cases, current quantitative genetic theory does not provide a measure of heritable variation. Nevertheless, the individuals involved are members of the same population, and response to selection is the result of changes in a single gene pool, which suggests that it should be possible to define heritable variation also for such cases.
An example of an emergent trait in agriculture is the output of a farm, which may depend on different types of individuals. In pig production, for example, the meat produced per sow is the product of the number of offspring of that sow and average meat yield of her offspring (see example below). Hence, the heritable variation in meat yield per sow will depend on both genes for reproduction in the sow and genes for growth rate in her offspring. While meat yield per sow is not a fundamental biological parameter, the heritable variation in this parameter does reflect the potential for genetic improvement of pork production, which is highly relevant to breeders. Moreover, a comparison of realized rates of improvement to the heritable variation provides a measure of efficiency of breeding schemes. Hence, a definition of heritable variation for output parameters of production systems is relevant for breeders.
The importance of social organization is recognized in the biological sciences (Frank 2007), and appropriate quantitative genetic models of inheritance have been developed (Hamilton 1964; Griffing 1981; Moore et al. 1997). Genetic models accounting for indirect genetic effects, however, have yielded complex expressions for response to selection, involving multiple genetic variances and covariances (Griffing 1981; Kirkpatrick and Lande 1989; Moore et al. 1997; Wolf et al. 1998; McAdam et al. 2002; Muir 2005; Bijma and Wade 2008; Wilson et al. 2009; McGlothlin et al. 2010). In contrast to Equations 3a and 3b, those expressions do not partition the response into a parameter describing selection and a parameter reflecting heritable variation. Hence, while those expressions have identified important factors in response to selection, they do not reveal the intrinsic potential of a population to respond to selection or provide a measure of the utilization of that potential.
A more general definition of heritable variation has been proposed for the special case of populations structured into groups of equal size, with a single category of individuals (Bijma et al. 2007). The expression for response to selection in Bijma et al. (2007), however, does not distinguish heritable variation from selection and does not connect heritable variation to common expressions for response such as Equations 3a and 3b. Ellen et al. (2007) show that response in artificial sib selection schemes, with social interactions among pen mates, can be expressed in terms of Equation 3b (see also Wade et al. 2010). In none of those cases, however, has heritable variation been connected to the selection gradient expression for response to selection (Equation 3b), and heritable variation has not been defined for emergent traits. Thus there are special cases indicating that a general definition of heritable variation may exist, but it has not yet been identified.
In the following, I propose a general definition of the heritable variation that determines a population’s potential to respond to selection. The result will have the same simple form as the classical expressions (Equations 3a and 3b), separating selection from heritable variation, but will reveal that heritable variation is not restricted by the phenotypic variation among population members. I provide an approach to derive this heritable variation, which can be applied to any level of organization. Application of this approach is illustrated using examples for natural and agricultural populations and is used to integrate two common models of maternal genetic effects.
Heritable Variation
Individual trait values can always be partitioned into a heritable component and a nonheritable residual, using the method of least squares (Fisher 1918; Lynch and Walsh 1998). The heritable component may arise not only from the focal individual’s own genes, but also from genes in its conspecifics. We can, therefore, represent an individual’s trait value, , as the sum of all heritable effects, , and a nonheritable residual, ,
| (4) |
where i denotes the focal individual, j the individual contributing the heritable effect to the focal individual’s trait value, and k indexes the different categories of heritable effects underlying individual trait values. The summation is over all individuals affecting the focal individual’s trait value, including j = i, in which case k indicates direct effects and the classical (direct) additive genetic effect due to the focal individual’s own genes. In the following, it is assumed that the population structure remains the same over time, so that Equation 4 applies also in subsequent generations. (Note that this assumption is not required in the classical prediction of response to selection.)
For example, juvenile growth rate in a mammal may be modeled as the sum of a heritable direct effect of the focal individual’s genes, a heritable maternal effect of its mother’s genes, and a nonheritable residual (Willham 1963). Then, j = 1 denotes the focal individual, j = 2 its mother, k = 1 refers to direct genetic effects, k = 2 to maternal genetic effects, and the trait value is given by . (See Example 4 below for a treatment of maternal effects.)
In general, the in Equation 4 is the best predictor of from the focal individual’s own genes (j = i) and the genes in its population members, defined using least squares (Fisher 1918). Each is the sum of the so-called average effects of the genes in individual j on (Fisher 1918; Lynch and Walsh 1998). k indexes average effects of genes by category, for example direct vs. maternal effects, whereas j indexes the individuals carrying the genes. Thus is the additive genetic effect of the genes for category k in individual j and is expressed in the phenotype of individual i. In total, the includes the direct average effects of genes in i and the indirect average effects of the genes in all population members that affect zi.
Equation 4 does not imply additive gene action. Irrespective of the mode of inheritance, one can always partition the resulting phenotype into heritable effects that are additive by construction, using the method of least squares (Fisher 1918). This is fundamental to quantitative genetics (Lynch and Walsh 1998).
By virtue of least squares, in Equation 4, so that the mean trait value equals . Response to selection, i.e., the change in mean trait value due to a change in gene frequencies, therefore equals
| (5) |
which is determined by the genes passed on to the next generation. From the Robertson–Price theorem (Robertson 1966; Price 1970), the change in mean trait value ascribable to selection equals the covariance with relative fitness, . Because an individual passes on its own genes, which may differ in part from the genes affecting its trait value, this covariance is between an individual’s relative fitness, , and the (direct and indirect) average effects of its own genes; hence it includes , rather than the that affects the trait value of i. With a maternal effect, for example, refers to the genes for maternal effect carried by i, whereas the trait value of i depends on , which refers to the genes for maternal effect in its mother. The distinction between the heritable components of an individual’s trait value and the heritable effects that an individual passes on to the next generation is central to this work.
It follows from that an individual’s value for response to selection is measured by the average effects of its own genes, summed over all heritable categories underlying individual trait values,
| (6) |
so that . Since Hi reflects an individual’s value for response to selection, it has been called the “total breeding value” by Bijma et al. (2007), and I use that term in the remainder of this article. Hi is entirely a heritable property of individual i, irrespective of whether or not is a component of . With a maternal effect, for example, is the sum of the direct and maternal breeding value of i, , even though the maternal breeding value of i does not contribute to . (Hence, i may be a male.) Finally, from the definition of a selection gradient, , response to selection is
| (7a) |
where
| (7b) |
is the regression coefficient of an individual’s relative fitness on the summed average effects of its own genes, and the heritable variation, , is the variance among individuals in the summed average effects of their own genes. Thus, in contrast to phenotypic variance, cannot contain covariances between individuals. Equation 7a has the same form as Equation 3a, but is not restricted to classical average effects of genes. Note that the selection gradient in Equation 7a differs from the classical selection gradient, which is discussed further in the section on utilization of heritable variation below.
While Equations 7a and 7b have been derived for individual trait values, they apply also to traits that cannot be attributed to a single individual, such as the number of prey caught by a hunting pack. In that case, the i index is omitted from zi in Equation 4, but the still specifies the heritable effects on the trait.
Equation 7a predicts the ultimate response attributable to a selection. When social interactions act across generations, this response may not surface immediately in the next generation. Maternal effects, for example, result in time lags in the response, causing populations to continue evolving after selection ceases and creating dynamic patterns over time (Kirkpatrick and Lande 1989). Equation 7a, however, predicts the permanent response due to the allele frequency change created by a selection, after transient effects have decayed away (see Example 4); it is not intended to capture transient effects. Hence, the heritable variance in Equation 7b reflects the potential of a population for a permanent genetic change in trait value due to selection.
The expression for response to selection common in breeding, Equation 3b, can be generalized in the same way. Consider selection for a criterion x. Response to selection follows from regressing H on x, = = , where is the regression coefficient of H on x, and is the selection differential. Substituting , using = , and multiplying both numerator and denominator by yield
| (7c) |
Just as Equation 2b, this expression is an approximation when H and x do not follow a bivariate normal distribution.
Equations 7a and 7c are generalizations of Equations 3a and 3b. They have the same simple form, separating selection from heritable variation, but capture the full heritable variation in a trait, including the component originating from effects of genes on trait values of others. The generality of the derivation suggests that Equation 7a applies to any trait and level of organization. The classical expression appears as a special case, obtained when trait values depend on direct genetic effects only. This work, therefore, shows that the classical expressions for heritable variation and response to selection can be generalized to include traits affected by genes in multiple individuals, which have so far been treated as special cases (Griffing 1981; Kirkpatrick and Lande 1989; Moore et al. 1997; Wolf et al. 1998; McAdam et al. 2002; Muir 2005; Bijma and Wade 2008; Wilson et al. 2009; McGlothlin et al. 2010).
Equation 7b represents a general definition of the heritable variation that determines a population’s potential to respond to selection, , which is the variance among individuals in the average effects of their genes, summed over all heritable categories underlying individual trait values. The selection gradient, , in turn, expresses the extent to which selection acts on this full heritable variation. The following shows examples of application of Equations 7a–7c, followed by a section on mechanisms determining the selection gradient.
Examples
The following examples serve to illustrate the meaning of Equations 7a–7c and demonstrate their application; they are not intended to accurately capture all biological detail of the cases considered. Example 4 also integrates the maternal-effect model of Willham (1963) with that of Falconer (1965) and Kirkpatrick and Lande (1989).
Example 1—interactions among trees
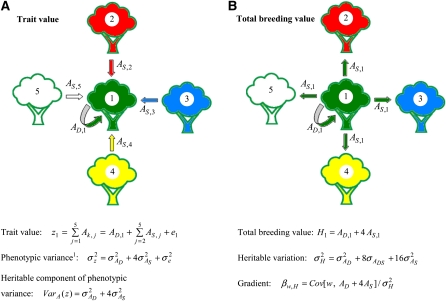
In Figure 1a, the trait value of focal tree 1 depends on the direct genetic effect of the focal tree itself and the indirect genetic effects of its neighbors, trees 2–5, , where k = D refers to direct genetic effects, and k = S to indirect genetic effects (“social effects”) (after Muir 2005). When trees are unrelated, phenotypic variance equals , so that the additive genetic component of phenotypic variance equals . When each tree has four neighbors, this model applies to all trees and response to selection equals .
Figure 1.
(A and B) Heritable variation with interactions among trees. Phenotypic variance1: phenotypic variance for a population with unrelated neighbors.
The total breeding value of the focal tree equals its total heritable effect, summed over all heritable categories (Figure 1b). Since each tree has four neighbors, this is the direct genetic effect of the focal individual on its own trait value plus its total indirect genetic effect on the trait values of its four neighbors, . Hence, response to selection equals the change in mean total breeding value of the population, . The heritable variation available for response to selection equals the variance in H among individuals, . differs from the additive genetic component of phenotypic variance, . This difference arises because H is entirely a genetic property of the focal individual, whereas the focal individual’s trait value depends on multiple individuals. Comparison of and clearly shows that indirect genetic effects contribute substantially more to the heritable variance that determines potential response to selection () than to phenotypic variance ().
The selection gradient is the regression coefficient of an individual’s relative fitness on its total breeding value, , and response to selection follows from Equations 7a–7c. The depends not only on the selection process, but also on relatedness between interacting trees (Equations 8a–8d below).
Example 2—adult body weight in the African wild dog
The African wild dog (Lycaon pictus) is a social carnivore living in packs of approximately seven adults. Only the dominant female usually breeds, while subordinates help to raise the pups. Packs hunt collectively, but the alpha female usually stays behind to guard her pups. The hunters share the prey with the pups and their mother. Adult body weight ranges from 18 to 28 kg, and relatedness among pack members averages ∼0.3 (Creel and Creel 2002).
On the basis of the social organization, an individual’s adult body weight may depend on a direct genetic effect of its own, , a maternal genetic effect of its mother, , and the indirect genetic effects its pack mates, , where j denotes an adult pack member, n adult pack size excluding the alpha female, and S indirect effects,
Maternal effects may, for example, relate to the maternal care and milk provided by the mother, whereas indirect effects of pack mates may relate to their hunting success and willingness to share prey.
The additive genetic component of phenotypic variance equals = + + . Using a relatedness of 0.5 between mother and offspring and of 0.3 between pack members yields
From the genetic mean trait value, it follows that response to selection equals . Thus the total breeding value equals
The heritable variance determining the population’s potential to respond to selection, therefore, equals,
Comparison of the expressions for and clearly shows that indirect genetic effects of pack mates contribute substantially more to heritable variation for response than to additive genetic variance: = ≫ = .
Estimated genetic parameters for direct, maternal, and indirect effects on adult body weight in the African wild dog are not available. An indication of the difference between and may, however, come from domestic pigs for which estimates have been published, yielding = 0.21 and = 0.007 (Bergsma et al. 2008). Estimates for maternal effects are commonly ∼ = 0.04 in domestic pigs (Johnson et al. 2002). Using these values, genetic covariances of zero, and a pack size of 7 yields = 0.59, whereas = 0.39, and ordinary heritability, = 0.21. This suggests that the heritable variation for response to selection may be ∼50% greater than the additive genetic component of phenotypic variance, and ∼180% greater than the ordinary (direct) additive genetic variance. This number is probably conservative, since social interactions are much more critical in African wild dogs than in domestic pigs. No unaided pair of wild dogs has, for example, been observed to raise pups (Creel and Creel 2002), whereas nursing sows are usually kept individually in pig herds. Moreover, there is clear opportunity for heritable variance to exceed phenotypic variance. For packs of 14 adults and genetic parameters as above, for example, = 1.62. The selection gradient for this example is a complex expression and is not given here, but follows from Equation 10 below.
Example 3—genetic improvement in pig breeding
Livestock genetic improvement aims to increase the efficiency of food production for human consumption. The prospects for genetic improvement are reflected by the heritable standard deviation in output parameters of agricultural production systems. Consider, for example, a sow line in an integrated pork production system. (In pig breeding, the mothers of fattening pigs come from a breeding line specialized for reproduction traits, known as a sow line, whereas the fathers come from a line specialized for growth traits.) Because farm size is usually measured by the number of sows, interest is in the total amount of meat produced from a sow, which is the product of offspring number (n) and offspring meat yield (y),
Offspring number is genetically determined by the sow, whereas offspring meat yield has both a direct and a maternal genetic component (Rothschild and Ruvinsky 1998). Meat yield per sow, therefore, is an emergent trait that cannot be attributed to a single individual.
Linearization at the current average trait values, using a first-order bivariate Taylor series, yields
Thus response due to selection in the sow line equals
where D and M indicate direct and maternal effects on meat yield, respectively, and the indicates that the sow line contributes only half of the genes in the fattening pigs. (The other half comes from the father, who comes from another breeding line.) Thus the heritable quantity determining the response in fattening pigs due to selection in the sow line equals
and response equals . Because individuals transmit their own genes to the next generation, refer to genes in a single individual, even though meat yield depends on multiple individuals (the sow and her offspring). Heritable variation equals the variance in ,
where variances and covariances are ordinary additive genetic (co)variances. Finally, response to selection equals , where the accuracy, ρ, is the correlation between the selection criterion and the H-values in the candidates for selection, which measures the quality of the selection criterion.
With an additive genetic relatedness of 0.5 between mother and offspring, the additive genetic component of phenotypic variance equals
which differs from . Hence, also this example illustrates that partitioning phenotypic variance into an additive genetic and residual component may not yield the heritable variance for response to selection, as already observed by Willham (1963).
Example 4—maternal effects and time dynamics of response
Kirkpatrick and Lande (1989) showed that maternal inheritance creates time lags in response to selection, causing dynamic response patterns over time. The following illustrates that Equation 7b provides the heritable variance also for such traits. This heritable variation refers to the ultimate response attributable to a selection, excluding transient effects and temporary dynamics. As a second objective, this example integrates the maternal-effect model of Willham (1963) with that Falconer (1965) and Kirkpatrick and Lande (1989).
Following Falconer (1965), Kirkpatrick and Lande (1989) considered a maternal effect that is a simple linear regression on maternal trait value,
where is the trait value of individual i, the ordinary direct breeding value of i, the (direct) environmental component of , m the partial regression coefficient of offspring trait value on maternal trait value, with , and the trait value of the mother of i. As above, the follows from identifying the heritable component of the trait mean and treating this as a property of a single individual. Repeatedly substituting by the expression for , ignoring nongenetic effects, and assuming that genetic parameters remain constant over time yield
Thus the heritable standard deviation determining potential response to selection equals
This result shows that positive feedback of maternal trait value on offspring trait value (m > 0) increases the heritable variation available for response to selection.
Whether m > 0 increases the actual response to selection will depend also on the selection process. Falconer (1965) and Kirkpatrick and Lande (1989) considered direct selection on offspring trait value,
where is the ordinary gradient of individual fitness on individual trait value in generation t. The standardized total gradient in generation t follows from regression of on . Using and a mother–offspring relatedness of 0.5 yields
This result shows that positive feedback of maternal trait value on offspring trait value (m > 0) increases the strength of direct selection. The denominator of this expression actually equals , indicating that a mother–offspring relatedness >0.5 inflates the effect of feedback (i.e., ) on the strength of selection. This occurs, for example, when mating is preferentially between kin.
The ultimate response due to selection in generation t, excluding transient effects and temporary dynamics, equals , giving
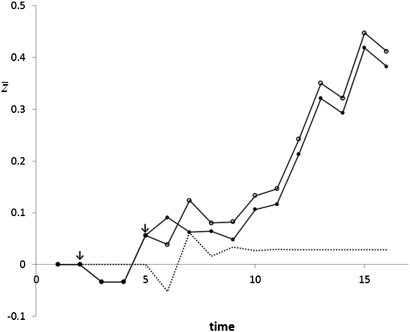
This result shows that m > 0 increases response to direct selection, which is partly due to increased heritable variation and partly due to increased strength of selection. This expression is identical to the result of repeatedly applying Equation 3 of Kirkpatrick and Lande (1989) to remove transient effects (Appendix B). Figure 2 illustrates that the response of a single selection in generation t asymptotes to this value, irrespective of selection in later generations.
Figure 2.
Evolution of mean trait value with a maternal effect. Selection starts at generation 2 for both populations. The selection gradients are the same for both populations and are randomly sampled from a uniform distribution between 0 and 0.4. For the population indicated with solid circles, selection ceases in generation 5 () and continues in generation 6. The dotted line is the difference in mean trait value between both populations and represents the response of the selection in generation 5 for the population indicated with open circles. Response initially fluctuates due to transient effects, but stabilizes at a value of 0.0289. Inputs are , , m = –0.4, and for the population indicated with open circles. Predicted response equals = = 0.0289, which is identical to the stabilized value.
The relationship between Willham’s and Falconer’s models:
In the main results of this work, I have used Willham’s (1963) maternal-effects model to illustrate the meaning of expressions. Willham’s model ignores the time dynamics of response, but predicts the same asymptotic response as Falconer’s (1965) model. Both models are well known and widely used (Lynch and Walsh 1998) and are related as follows (using W63 and F65 to indicate parameters referring to Willham’s model and Falconer’s model, respectively):
Depending on the sign of m, Falconer’s model corresponds to a direct-maternal genetic correlation of either +1 or –1 in Willham’s model. When individual fitness is determined by individual trait value, , then the standardized total selection gradient is given by
and the permanent response to selection by
Utilization of Heritable Variation
As shown above, the standardized selection gradient of relative fitness on total breeding value, , measures the utilization of heritable variation. This standardized gradient differs from the classical gradient of individual fitness on individual trait value. Previous work on indirect genetic effects has shown that relatedness between individuals and selection between groups rather than individuals may considerably increase response to selection (Griffing 1967, 1976, 1977; Wade 1977; Muir 1996, 2005; Craig and Muir 1996). This suggests that relatedness and multilevel selection are important determinants of . The following first illustrates the effects of relatedness and multilevel selection on in populations structured into groups, and subsequently considers the general case.
Natural selection
Consider a population structured into a large number of groups of n individuals each, where indirect genetic effects of group mates affect individual trait values,
as in Example 1. Moreover, suppose that selection is a function of individual trait value and the summed trait values of all n – 1 group mates,
The g represents the degree of between-group selection relative to individual selection; it is the ratio of the selection gradient on the summed trait values of group mates over the selection gradient on individual trait value. A g = 0 represents individual selection, and a g = 1 full between-group selection (Bijma and Wade 2008). Combining Equation 15 of Bijma and Wade (2008) with Equation 7a, using r to denote additive genetic relatedness between group members, yields the following expressions for the standardized total selection gradient. For individual selection with unrelated group members (g, r = 0),
| (8a) |
For individual selection (g = 0) with related group members, ,
| (8b) |
For multilevel selection, , with unrelated group members (r = 0),
| (8c) |
and for multilevel selection with related group members,
| (8d) |
In those expressions, is the variance of direct genetic effects, the covariance between direct and indirect genetic effects, and β the ordinary selection gradient.
The effects of relatedness and/or multilevel selection on the utilization of heritable variation follow from the partial derivatives of with respect to g or r. For example, the partial derivative of Equation 8b with respect to r equals
This derivative takes positive values whenever
where is the genetic correlation between direct and indirect genetic effects. This condition will probably be satisfied in most cases, except when group sizes are small, indirect genetic effects have little variance and are strongly negatively correlated with direct genetic effects. For example, when n = 4, , and , relatedness increases the standardized total gradient when .
Because Equation 8d is symmetric with respect to g and r, the effect of between-group selection on may seem to be identical to that of relatedness. However, at the same overall strength of selection, stronger between-group selection reduces the ordinary selection gradient β, which in turn reduces . This issue is investigated further in the next section.
Artificial selection
With artificial selection, the accuracy reflects the utilization of heritable variation by the breeder. (The intensity of selection merely reflects the overall strength of selection.) The above case may be investigated in the context of artificial selection by replacing fitness by a selection index of its own trait value and the summed trait values of group mates,
The accuracy of this index equals
| (9) |
Note that the second term in the numerator may take negative values when direct and indirect genetic effects are negatively correlated, whereas the first term is always positive. In the denominator, the increases with relatedness between group mates and particularly with the degree of between-group selection (Appendix C).
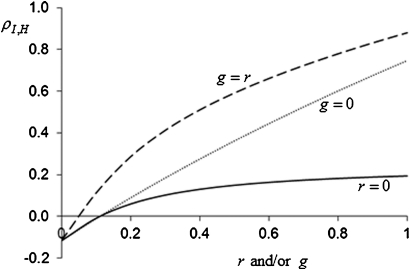
Figure 3 shows that accuracy increases much more with relatedness than with between-group selection. This occurs because an increase in g strongly increases the in Equation 9. This is the same phenomenon as the reducing effect of between-group selection on the magnitude of the ordinary selection gradient, mentioned above. In most cases, therefore, relatedness can contribute more to the utilization of heritable variation than multilevel selection.
Figure 3.
The effect of between-group selection (g) and/or relatedness (r) on the accuracy of selection (). Solid line: r = 0, . Dotted line: , g = 0. Dashed line: . Input values are n = 8, , , , and . Details of calculations are in Appendix C.
Generalization
The above considered the special case where population members interact within groups. The following investigates whether general statements about the utilization of heritable variance can be made.
Since any trait can be decomposed into additive effects using the method of least squares, this decomposition can be applied also to fitness,
where j′ denotes the individual contributing the jth heritable effect, , to the focal individual’s fitness, l indexes the different categories of heritable effects on fitness, and the summation is over all individuals affecting the focal individual’s fitness, including the focal individual itself. (For multilevel selection in group-structured populations, the relationship between the elements of wi and the selection parameters is given in Equation 11 and Table 2 of Bijma and Wade 2008). Because heritable effects on fitness may have a different origin from those on trait values, the indexing in the fitness model, j′ and l, differs from that in the trait model (j and k, Equation 4). From the Robertson–Price theorem, response to selection equals . Substituting by Equation 6, and using , yields
| (10) |
where is the additive genetic relatedness between the focal individual and j′, and is the ordinary additive genetic covariance (i.e., within individuals) between the kth heritable component of the trait value and the lth heritable component of fitness.
Equation 10 reveals two points of interest. First, response to selection depends on relatedness between the focal individual and the individuals affecting its fitness, , not on relatedness with the individuals affecting its trait value, . This makes sense in the light of the Robertson–Price equation. (Of course, those individuals may partly or entirely be the same.) Second, when relatedness takes positive values, the direction of response in a component of the trait value is the same as the sign of the within-individual covariance of that trait component with fitness. In other words, if > 0, then the response originating from the lth fitness component is positive when is positive and negative when is negative. Conversely, when < 0, then the response originating from the lth fitness component is negative when is positive and positive when is negative. [Negative relatedness may occur because relatedness is a measure of correlation, which is zero on average by definition. It merely means that both individuals have below average additive genetic similarity (Powell et al. 2010).] Hence, this result shows that relatedness works to change trait values in the direction of increased fitness, suggesting that relatedness between the focal individual and the individuals affecting its fitness causes an adaptive response to selection. This result agrees with the observation of Bijma (2010), who showed that relatedness contributes to a positive response in fitness when individuals interact.
Discussion
In this work a definition has been proposed of the heritable variance that determines the potential of a population to respond to selection. In this definition, heritable variance equals the variance among individuals in the heritable quantity that determines the mean trait value of the population, rather than the additive genetic component of phenotypic variance. This definition encompasses both traits affected by the focal individual’s genes only, in which case heritable variance equals the ordinary additive genetic variance and traits depending on heritable effects originating from multiple individuals. This work, therefore, generalizes the classical definition of heritable variance and the usual quantitative genetic expressions for response to selection to cases where trait values depend on genes in multiple individuals.
Because individuals transmit the genes they carry themselves, the heritable variance relevant for response to selection may differ from the additive genetic component of phenotypic variance, which may originate in part from genes in others. As a consequence, the heritable variance in traits that depend on genes in multiple individuals is not limited to phenotypic variance, which is a fundamental difference from classical theory (Fisher 1918; Lynch and Walsh 1998). For such traits, heritable variance may exceed the phenotypic variance among population members and has no theoretical upper bound. This result implies that social organization may allow populations to evolve faster by natural or artificial selection.
The partitioning of response to selection into contributions from heritable variation and selection facilitates research aiming to identify the mechanisms that determine the utilization of heritable variation by natural or artificial selection. The standardized total selection gradient, , is a scale-free measure of the utilization of heritable variation. The differs from the classical selection gradient of individual fitness on individual trait value and depends on relatedness between individuals and the levels of selection. Equation 10 shows that positive relatedness causes trait values to respond in the direction of increased fitness.
A partitioning of response into contributions from heritable variation and selection is also useful for breeders. Breeders want to know how much genetic improvement is possible in principle and to be able to assess the quality of their breeding programs. The heritable variation, as defined in Equation 7b, reflects the genetic improvement that is possible in principle. An efficient breeding scheme generates ∼1 unit response per generation. While heritable variation is a biological property of the population that is outside the breeder’s control, accuracy of selection depends on the breeding design and can be optimized. Hence, accuracy is an important quality criterion for a breeding scheme. This work has generalized the definition of accuracy, to include traits affected by genes in multiple individuals.
The definition of heritable variance provided here also explains in a natural way why certain heritable traits cannot respond to selection. Consider, for example, the rank of racing horses. The mean rank cannot respond to selection, because it is fully determined by the number of competitors. With eight competitors, for example, the mean rank is always 4.5. Racing ability nevertheless shows additive genetic variance (Langlois 1980). While it is obvious that rank cannot respond to selection, this case violates the ordinary quantitative genetic expression for response to selection (Equation 3). Equation 7b, however, reveals that heritable variance in rank is zero. This occurs because a 1-unit increase in an individual’s rank always decreases the average rank of its n − 1 competitors by an exact amount of . The variance in indirect genetic effects, therefore, equals times the variance in direct genetic effects, and the correlation between direct and indirect genetic effects equals –1. Substitution into Equation 7b shows that heritable variance equals zero: = – + = 0. While the mean rank cannot respond to selection, the model allows for response to selection in the underlying direct and indirect genetic effects; individuals can, for example, become more competitive. Wilson et al. (2011) applied a similar approach to genetically analyze dyadic interactions in Scottish deer. This approach may also be used to genetically analyze fitness in populations where mean fitness cannot respond to selection because population size is limited by the carrying capacity of the environment, which is very common.
Acknowledgments
I thank J. Bruce Walsh, Antti Kause, Duur K. Aanen, Johan A. M. van Arendonk, Martien A. M. Groenen, Michael Grossman, James H. Hunt, Hans Komen, Ole Madsen, Arie J. van Noordwijk, Elisabeth H. van der Waaij, and Michael J. Wade for reviewing drafts of this manuscript. This research was financially supported by the Dutch Science Council and was coordinated by the Netherlands Technology Foundation.
Appendix A
This Appendix shows that the common expressions for response to selection used in evolutionary genetics and artificial breeding are equivalent under multivariate normality (Equations 3a and 3b). Consider artificial selection for a criterion, say x, so that individual fitness is determined entirely by individual x. Thus the effect of breeding value for the trait on individual fitness must arise entirely via x. Moreover, under multivariate normality, regressions are linear and represent conditional expectations (Stuard and Ord 2004). Thus the regression coefficient of fitness on breeding value, , must equal the product of the regression coefficient of the selection criterion on breeding value, , and the regression coefficient of fitness on the selection criterion, ,
Next, in Equation 3b, , the intensity of selection is the ratio of the selection differential over the standard deviation of the selection criterion (Falconer and Mackay 1996); . From the Robertson–Price theorem, the selection differential equals the covariance with relative fitness, , giving . Moreover, from the definition of the correlation coefficient, the accuracy equals . Substituting these values into Equation 3b yields , which is Equation 3a.
Appendix B
This Appendix shows that the expression for response to selection derived in Example 4 is identical to the response from repeatedly applying Equation 3 of Kirkpatrick and Lande (1989),
| (B1) |
The last term in this expression is the phenotypic effect of the selection differential in the mothers on the trait value of their offspring and is therefore transient. The second term is partly transient because it includes . The first term reflects the change in breeding value and is permanent. Interest is in the ultimate response ascribable to selection in generation t, , after transient effects and temporary dynamics have decayed away. The follows from the difference in the ultimate mean trait value between a population where selection ceases after generation t and a population where selection ceases after generation t – 1,
| (B2) |
From Equation B1, the series of responses when selection ceases after generation t is
The series of responses when selection ceases after generation is
Calculating Equation B2 yields , giving
Substituting yields
which is identical to the result presented in Example 4.
Appendix C
This Appendix provides background information on Figure 3. Direct and indirect additive genetic (co)variances follow from , , and . Total heritable variance equals . Phenotypic variance equals . The variance of the index equals , in which denotes the phenotypic covariance among group mates, which equals . Substitution of the input values given in the Figure 3 legend into these expressions and using Equation 9 of the main text produces Figure 3.
Literature Cited
- Bergsma R., Kanis E., Knol E. F., Bijma P., 2008. The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics 178: 1559–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma P., 2010. Fisher’s fundamental theorem of inclusive fitness and the change in fitness due to natural selection when conspecifics interact. J. Evol. Biol. 23: 194–206 [DOI] [PubMed] [Google Scholar]
- Bijma P., Wade M. J., 2008. The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. J. Evol. Biol. 21: 1175–1188 [DOI] [PubMed] [Google Scholar]
- Bijma P., Muir W. M., Van Arendonk J. A. M., 2007. Multilevel selection 1: quantitative genetics of inheritance and response to selection. Genetics 175: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth S. F., Rundle H. D., Blows M. W., 2010. Experimental evidence for the evolution of indirect genetic effects: changes in the interaction effect coefficient, PSI (Ψ), due to sexual selection. Evolution 64: 1849–1856 [DOI] [PubMed] [Google Scholar]
- Craig J. V., Muir W. M., 1996. Group selection for adaptation to multiple-hen cages: beak-related mortality, feathering, and body weight responses. Poult. Sci. 75: 294–302 [DOI] [PubMed] [Google Scholar]
- Creel S., Creel N. M., 2002. The African Wild Dog: Behavior, Ecology and Conservation. Princeton University Press, Princeton, NJ [Google Scholar]
- Crespi B. J., 2001. The evolution of social behaviour in microorganisms. Trends Ecol. Evol. 16: 178–183 [DOI] [PubMed] [Google Scholar]
- Dawkins R., 1982. The Extended Phenotype. Oxford University Press, Oxford [Google Scholar]
- Ellen E. D., Muir W. M., Teuscher F., Bijma P., 2007. Genetic improvement of traits affected by interactions among individuals: sib selection schemes. Genetics 176: 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. S. 1965. Maternal effects and selection response, pp. 763–774 Genetics Today, Proceedings of the XIth International Congress on Genetics, Vol. 3, edited by Geerts S. J. Pergamon, New York [Google Scholar]
- Falconer D. S., Mackay T. F. C., 1996. Introduction to Quantitative Genetics, Ed. 4 Longman, London [Google Scholar]
- Fisher R. A., 1918. The correlation between relatives on the supposition of Mendelian inheritance. Trans. R. Soc. Edinb. 52: 399–433 [Google Scholar]
- Frank S. A., 1998. Foundations of Social Evolution. Princeton University Press, Princeton, NJ [Google Scholar]
- Frank S. A., 2007. All of life is social. Curr. Biol. 17(16): R650. [DOI] [PubMed] [Google Scholar]
- Griffin A. S., West S. A., Buckling A., 2004. Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027 [DOI] [PubMed] [Google Scholar]
- Griffing B., 1967. Selection in reference to biological groups. I. Individual and group selection applied to populations of unordered groups. Aust. J. Biol. Sci. 20: 127–142 [PubMed] [Google Scholar]
- Griffing B., 1976. Selection in reference to biological groups. VI. Analysis of full sib groups. Genetics 82: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffing B., 1977. Selection for populations of interacting genotypes, pp. 413–434 Proceedings of the International Conference on Quantitative Genetics, edited by Pollak E., Kempthorne O., Bailey T. B. Iowa State University Press, Ames, IA [Google Scholar]
- Griffing B., 1981. A theory of natural-selection incorporating interaction among individuals. 2. Use of related groups. J. Theor. Biol. 89: 659–677 [DOI] [PubMed] [Google Scholar]
- Haldane J. B. S., 1932. A mathematical theory of natural and artificial selection. Part IX. Rapid selection. Proc. Camb. Philos. Soc. 28: 244–248 [Google Scholar]
- Hamilton W. D., 1964. The genetical evolution of social behaviour. J. Theor. Biol. 7: 1–16 [DOI] [PubMed] [Google Scholar]
- Johnson Z. B., Chewning J. J., Nugent R. A., III, 2002. Maternal effects on traits measured during postweaning performance test of swine from four breeds. J. Anim. Sci. 80: 1470–1477 [DOI] [PubMed] [Google Scholar]
- Karban R., 2008. Plant behaviour and communication. Ecol. Lett. 11: 727–739 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M., Lande R., 1989. The evolution of maternal characters. Evolution 43: 485–503 [DOI] [PubMed] [Google Scholar]
- Lande R., Arnold S. J., 1983. The measurement of selection on correlated characters. Evolution 37: 1210–1226 [DOI] [PubMed] [Google Scholar]
- Langlois B., 1980. Heritability of racing ability in thoroughbreds: a review. Livest. Prod. Sci. 7: 591–605 [Google Scholar]
- Lush J. L., 1937. Animal Breeding Plans. Iowa State University Press, Ames, IA [Google Scholar]
- Lynch M., Walsh J. B., 1998. Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- McAdam A. G., Boutin S., Réale D., Berteaux D., 2002. Maternal effects and the potential for evolution in a natural population of animals. Evolution 56: 846–851 [DOI] [PubMed] [Google Scholar]
- McGlothlin J. W., Moore A. J., Wolf J. B., Brodie E. D., III, 2010. Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution 64: 2558–2574 [DOI] [PubMed] [Google Scholar]
- Moore A. J., Brodie E. D., III, Wolf J. B., 1997. Interacting phenotypes and the evolutionary process. I. Direct and indirect genetic effects of social interactions. Evolution 51: 1352–1362 [DOI] [PubMed] [Google Scholar]
- Muir W. M., 1996. Group selection for adaptation to multiple-hen cages: selection program and direct responses. Poult. Sci. 75: 447–458 [DOI] [PubMed] [Google Scholar]
- Muir W. M., 2005. Incorporation of competitive effects in forest tree or animal breeding programs. Genetics 170: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. E., Visscher P. M., Goddard M. E., 2010. Reconciling the analysis of IBD and IBS in complex trait studies. Nat. Rev. Genet. 11: 800–805 [DOI] [PubMed] [Google Scholar]
- Price G. R., 1970. Selection and covariance. Nature 227: 520–521 [DOI] [PubMed] [Google Scholar]
- Rausher M. D., 1992. The measurement of selection on quantitative traits: biases due to environmental covariances between traits and fitness. Evolution 46: 616–626 [DOI] [PubMed] [Google Scholar]
- Rice S. H., 2004. Evolutionary Theory: Mathematical and Conceptual Foundations. Sinauer Associates, Sunderland, MA [Google Scholar]
- Robertson A., 1966. A mathematical model of the culling process in dairy cattle. Anim. Prod. 8: 95–108 [Google Scholar]
- Rothschild M. F., Ruvinsky A., 1998. The Genetics of the Pig. CAB International, Wallingford, Oxon, UK [Google Scholar]
- Stuard A., Ord J. K., 2004. Kendall’s Advanced Theory of Statistics (Distribution Theory, Vol. 1) Hodder Education, London [Google Scholar]
- Wade M. J., 1977. An experimental study of group selection. Evolution 31: 134–153 [DOI] [PubMed] [Google Scholar]
- Wade M. J., Bijma P., Ellen E. D., Muir W. M., 2010. Group selection and social evolution in domesticated animals. Evol. Appl. 3: 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. A., Griffin A. S., Gardner A., Diggle S. P., 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4: 597–607 [DOI] [PubMed] [Google Scholar]
- West-Eberhard M. J., 2003. Developmental Plasticity and Evolution. Oxford University Press, Oxford [Google Scholar]
- Wheeler W. M., 1933. Colony Founding Among Ants, with an Account of Some Primitive Australian Species. Harvard University Press, Cambridge, MA [Google Scholar]
- Willham R. L., 1963. The covariance between relatives for characters composed of components contributed by related individuals. Biometrics 19: 18–27 [Google Scholar]
- Wilson A. J., Gelin U., Perron M.-C., Reale D., 2009. Indirect genetic effects and the evolution of aggression in a vertebrate system. Proc. Biol. Sci. 276: 533–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A. J., Morrissey M. B., Adams M. J., Walling C. A., Guinness F. E., et al. , 2011. Indirect genetics effects and evolutionary constraint: an analysis of social dominance in red deer, Cervus elaphus. J. Evol. Biol. 24: 772–783 [DOI] [PubMed] [Google Scholar]
- Wolf J. B., Brodie E. D., III, Cheverud J. M., Moore A. J., Wade M. J., 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13: 64–69 [DOI] [PubMed] [Google Scholar]
- Wright S., 1937. The distribution of gene frequencies in populations. Proc. Natl. Acad. Sci. USA 23: 307–320 [DOI] [PMC free article] [PubMed] [Google Scholar]