Abstract
The chloroplast atpB and atpE genes encode subunits β and ε of the ATP synthase, respectively. They are co-transcribed as dicistronic mRNAs in flowering plants. An unusual feature is an overlap (AUGA) of the atpB stop codon (UGA) with the atpE start codon (AUG). Hence, atpE translation has been believed to depend on atpB translation (i.e. translational coupling). Using an in vitro translation system from tobacco chloroplasts, we showed that both atpB and atpE cistrons are translated from the tobacco dicistronic mRNA, and that the efficiency of atpB translation is higher than that of atpE translation. When the atpB 5′-UTR was replaced with lower efficiency 5′-UTRs, atpE translation was higher than atpB translation. Removal of the entire atpB 5′-UTR arrested atpB translation but atpE translation still proceeded. Introduction of a premature stop codon in the atpB cistron did not abolish atpE translation. These results indicate that atpE translation is independent of atpB translation. Mutation analysis showed that the atpE cistron possesses its own cis-element(s) for translation, located ~25 nt upstream from the start codon.
INTRODUCTION
Chloroplasts carry their own genomes and gene expression systems. The chloroplast genome in flowering plants encodes about 80 proteins (1,2). Chloroplast gene expression is mainly regulated at post-transcriptional steps such as translation (3–10). Many protein-coding genes in flowering plant chloroplasts are clustered (11,12). These genes are co-transcribed to produce polycistronic transcripts that are then processed into mature mRNAs (1,13–17).
In the tobacco chloroplast genome, the protein-coding regions (hereafter cistrons) of eight genes partially overlap; these are psbD–psbC, ndhC–ndhK, atpB–atpE and rpl22–rps3 (18). This feature implies that translation of a downstream gene depends on that of its upstream gene, namely translational coupling or termination-dependent translation, as reported for some genes from Escherichia coli, its bacteriophages and some eukaryotic viruses [reviewed in (19)]. A typical case of translational coupling in partially overlapping cistrons is explained by the downstream cistron being translated exclusively by the ribosomes that completed translation of the upstream cistron. In this case, a small fraction of the ribosomes participates in translating the downstream cistron and the majority of the ribosomes are released at the stop codon of the upstream cistron (20,21). Hence, translation of the downstream cistron is usually very low when compared to that of the upstream cistron. When the distance between the stop codon of an upstream cistron and the start codon of the downstream cistron increases, for example, by insertion of a premature termination codon in the upstream cistron, it abolishes translation of the downstream cistron.
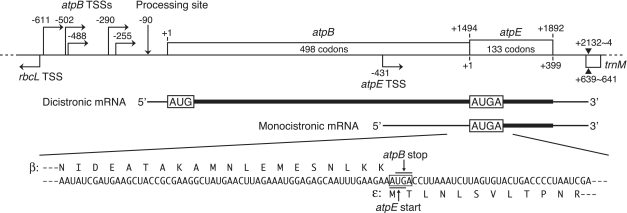
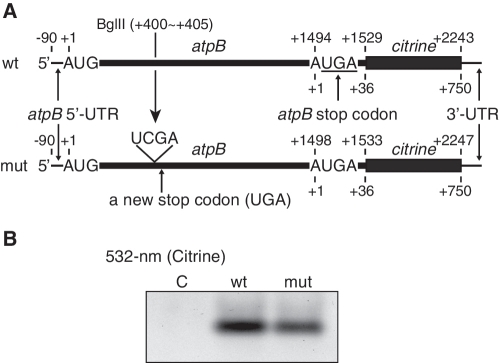
The chloroplast ATP synthase consists of nine distinct subunits that are encoded in both the nuclear and chloroplast genomes: three in the nucleus (atpC, atpD and atpG) and six in the chloroplast (atpA, atpB, atpE, atpF, atpH and atpI) (22,23). The six subunits synthesized within chloroplasts are encoded by two separate operons: the atpB–atpE (atpB/E) operon and the atpI–atpH–atpF–atpA cluster. The rps2 gene precedes the latter cluster and these five genes constitute an operon (24–27). In most flowering plants, the atpB stop codon (UGA) overlaps the atpE start codon (AUG), with the AUGA containing overlapping stop and start codons, and hence translation of these genes has been assumed to be coupled (28,29) (Figure 1). Expression of the maize chloroplast atpB/E genes in transformed E. coli and the cyanobacterium Synechocystis sp. PCC6803 was examined, and translation of the downstream atpE cistron was reported to depend on translation of the upstream atpB cistron in these heterologous systems (30). In tobacco, the atpE gene is transcribed from its own promoter located within the atpB cistron to provide monocistronic mRNA (31) (Figure 1). This monocistronic atpE mRNA is translatable in vitro (32,33). However, translation of the atpE cistron from the dicistronic atpB/E mRNA has not been investigated.
Figure 1.
The tobacco chloroplast atpB/atpE operon and its transcripts. Nucleotide positions relative to AUG start codons (A as +1) are shown above for atpB and below for atpE. Bent arrows indicate atpB and atpE transcription start sites (TSSs) (31,42–44). A vertical arrow indicates a processing site (−90) (42). The common 3′-end is designated by closed triangles (31). AUG, atpB start codon; AUGA, overlapping atpB stop-atpE start codons. A partial mRNA sequence around the atpE start codon is shown below. Deduced amino acid sequences from atpB and atpE genes (β- and ε-subunits) are shown above and below the mRNA sequence, respectively.
Here, we show that the atpE cistron can also be translated from a dicistronic transcript using a chloroplast in vitro translation system. Unexpectedly, the majority of the translation of the atpE cistron does not depend on the upstream cistron; the atpE cistron is translated via its own cis-element(s). An upstream sequence of ~25 nt is necessary for efficient atpE translation.
MATERIALS AND METHODS
Chloroplast extracts
Growth of Nicotiana tabacum (var. Bright Yellow 4), isolation of intact chloroplasts and preparation of chloroplast extracts (S30 fractions) for in vitro translation were as described (34). An RNase inhibitor (Takara Bio, Otsu, Japan) was added during preparation of S30 fractions.
Plasmid construction and RNA synthesis
The mGFP-coding region in plasmid pHK309 (34), derived from pKH6 (35), was modified to create coding regions for Citrine (36) and enhanced GFP (EGFP) (37). Necessary parts of the atpB/E, atpH and rbcL sequences were amplified by PCR from tobacco chloroplast DNA (38). Strategies to construct plasmid DNAs are shown in Supplementary Figure S1. Deletion and point mutations were performed as described (39). A frameshift was introduced in the atpB cistron by inserting the PCR product of a frameshifted atpB segment into the SpeI/BglII region (Supplementary Figure S2). The resulting DNA fragments were confirmed to be the ones wanted by DNA sequencing. Plasmid DNAs were linearized with XhoI at the end of the 3′-UTRs and purified with a Wizard SV DNA Purification Kit (Promega, Madison, WI, USA). Test mRNAs were synthesized from the linearized DNAs using an AmpliScribe T7 Flash Transcription kit (Epicentre, Madison, WI, USA). After purification using the SV Total RNA isolation kit (Promega), the mRNAs were precipitated with ethanol and finally dissolved in nuclease-free water.
In vitro translation assays
Assays were carried out basically as described (34). Briefly, the 20-µl reaction mixture contained 30 mM HEPES/KOH (pH 7.7), 50 mM potassium acetate, 9 mM magnesium acetate, 60 mM ammonium acetate, 5 mM dithiothreitol, 40 µM of each of the 20 common amino acids, 2 mM ATP (lithium salt), 0.2 mM GTP (lithium salt), 8 mM phosphocreatine, 8 mg/ml creatine kinase, 12 A280 U/ml S30 fraction and 4 pmol test mRNA. After incubation for 1 h at 28°C, the reaction mixture was mixed with 4 µl of sample buffer (375 mM Tris–HCl, pH 6.8, 6% Triton X-100, 600 mM dithiothreitol, 30% glycerol, 50 µg/ml chloramphenicol). Protein products were separated by 12.5% native polyacrylamide gel electrophoresis (PAGE). Fluorescent signals were detected and quantified with a Typhoon 9400 Imager (GE Healthcare, Little Chalfont, UK) using 520BP40 and 555BP20 filters when excited by 488 and 532-nm actinic light, respectively.
RESULTS
Translation of the atpE cistron from dicistronic atpB/E mRNA
The tobacco chloroplast atpE gene is mainly co-transcribed with its upstream atpB gene as multiple atpB/E transcripts varying in size from 2.3 to 2.7 kb (31,40). Although the dicistronic transcript is expected to be translated into the ε subunit (the atpE product), no direct evidence for this has been reported. The overlapping stop and start codons (AUGA) might hamper translation initiation of the downstream atpE cistron in tobacco chloroplasts.
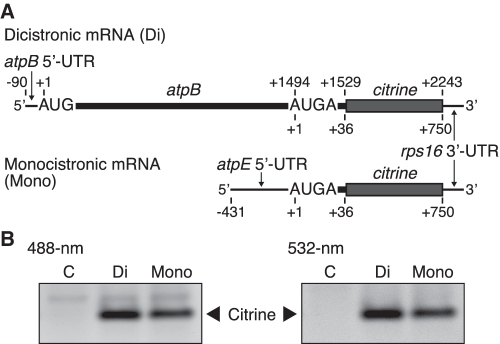
To examine whether the atpE cistron is translated from the dicistronic mRNA, we designed test mRNAs from which translation of only the atpE cistron can be monitored. A large portion of the atpE cistron (133 codons) was replaced with the coding region (citrine) for Citrine (an improved version of yellow fluorescent protein) (36). Specifically, a citrine-coding sequence (237 codons without start codon) was fused 36 nt downstream from the atpE start codon, and then the tobacco chloroplast rps16 3′-UTR (150 nt) was added to stabilize the mRNA (41) (Figure 2A). The tobacco atpB gene has multiple transcription initiation sites (TSSs) and one processing site at position −90 (11,40,42–44) (Figure 1). We used the processed 5′-UTR of 90 nt because it was much more efficiently translated in vitro than the primary 5′-UTR (34). Monocistronic atpE mRNA was also prepared as a control. We left the first 36 nt of the atpE cistron in the construct because sequences downstream of start codons are sometimes necessary for efficient translation (45).
Figure 2.
Translation of the atpE cistron from dicistronic and monocistronic mRNAs. (A) mRNA templates. A large portion of the atpE cistron was replaced by the Citrine-coding region (citrine). Positions are as in Figure 1. The full sequence is in Supplementary Figure S3. (B) Gel patterns of translation products. Synthesized Citrine products were detected by 488-nm light (left panel) and 532-nm light (right panel). Lanes labeled C, no-mRNA template control. Di and Mono indicate dicistronic and monocistronic mRNAs, respectively. Citrine bands represent atpE products (ε-subunit). Faint bands above the main Citrine bands are probably non-specific fluorescent substances in the S30 fraction.
In vitro translation reaction was performed under template limiting (4 pmol/20 µl) and linear progression (1 h at 28°C) conditions, which enable estimation of the relative rate of translation (34). Test mRNAs in the reaction were stable for at least 2 h, probably due to the presence of an RNase inhibitor (34). After reaction, translation products were separated by native PAGE and the fluorescence intensity of Citrine products was detected by irradiation with 488 or 532-nm actinic light. As shown in Figure 2B, 488 and 532-nm excitation gave single fluorescent bands from both the dicistronic mRNA and the control monocistronic mRNA. This result clearly indicates that atpE translation occurs from the dicistronic mRNA.
Test for translational coupling of atpB and atpE cistrons
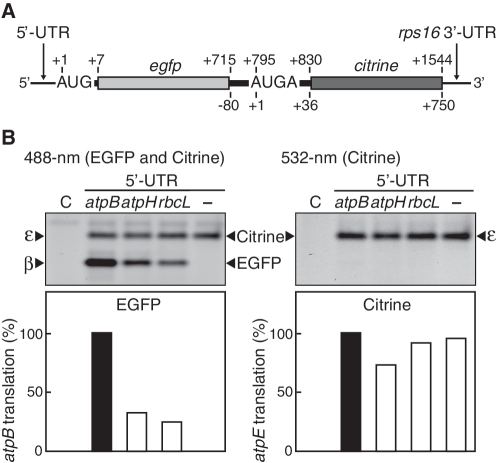
To examine whether translational coupling occurs, we devised an assay system to differentially monitor translation of the atpB and atpE cistrons. A large portion (from +4 to +1413, 470 codons) of the atpB cistron (498 codons) was replaced with a reading frame (236 codons without start and stop codons, egfp) for EGFP (37) in the dicistronic mRNA described above (Figure 3A). We left the last 81 nt of the atpB cistron in the construct because sequence upstream of the atpE start codon could be required for atpE translation initiation. Actinic light at 488 nm excites EGFP and Citrine, while 532-nm light excites only Citrine. After in vitro translation, two fluorescent bands were detected under 488-nm light while only the upper band was detected by 532-nm light (Figure 3B, lanes labeled atpB), indicating that the upper band corresponds to a Citrine product (from the atpE start codon) and the lower band to an EGFP product (from the atpB start codon). Hence, translation from atpB and atpE start codons can be differentially detected in the same gel. Based on the normalized intensity of EGFP and Citrine fluorescence, the efficiency of atpE translation was estimated to be ~80% of atpB translation (Supplementary Figure S4).
Figure 3.
Effect of atpB translation on atpE translation. (A) mRNA templates with atpB and atpE cistrons replaced by EGFP (egfp) and Citrine (citrine)-coding regions, respectively. Positions are as in Figure 1. The full sequence is in Supplementary Figure S5. (B) Gel patterns of translation products. Lanes labeled C, no-mRNA control. The 5′-UTR was either from atpB, atpH or rbcL, and lane ‘–‘ denotes no 5′-UTR or following AUG (Supplementary Figure S5). Synthesized EGFP and Citrine products were detected by 488-nm light (left panel) and 532-nm light (right panel). EGFP and Citrine bands represent atpB and atpE products (β- and ε-subunits), respectively. Quantification of translation products based on intensity of fluorescent bands (atpB value defined as 100%) is shown in bar graphs below. Faint bands above the main EGFP bands could be loosely folded EGFP.
If translational coupling occurs, the translation efficiency of the atpE cistron should depend on translation of the upstream atpB cistron. We replaced the atpB 5′-UTR with either the atpH 5′-UTR or the rbcL 5′-UTR, both of which have lower translation efficiencies than the atpB 5′-UTR (34). As expected, replacing the 5′-UTR resulted in significant reduction of atpB translation (EGFP bands) (Figure 3B, left panel). However, atpE translation (Citrine bands) decreased only slightly (Figure 3B, right panel). As a result, translation of the downstream atpE cistron became higher than that of the upstream atpB cistron. Furthermore, removal of the entire atpB 5′-UTR and the following AUG completely abolished atpB translation while no significant effect was observed on translation of the atpE cistron (Figure 3B, lanes ‘–‘). If translational coupling occurs, atpE translation should be absent or much less than atpB translation. Hence, the translational coupling model cannot explain our present observations.
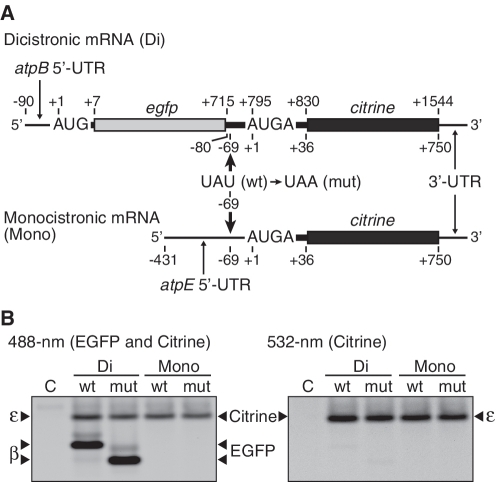
To confirm this result, we introduced a U to A point mutation at position −69 relative to the A (+1) of the atpE start codon, which created a premature stop codon, UAA, from UAU (Tyr), right after egfp (within the 3′-atpB cistron) (Figure 4A). In the mutant mRNA, the upstream atpB and the downstream atpE cistrons no longer overlap and are separated by a 69-nt spacer, which is enough to interfere with translational coupling (19). As shown in Figure 4B, no significant reduction in atpE translation was observed. Using the monocistronic mRNA as a control, the same point mutation hardly affected atpE translation (Figure 4B, lanes labeled ‘Mono’). Similarly, atpB translation was not influenced by premature termination, though the size of EGFP products was reduced (Figure 4B, left panel).
Figure 4.
Effect of premature termination of the upstream atpB cistron on atpE translation. (A) mRNA templates as in Figures 3A (Di) and 2A (Mono). Bold arrows indicate locations of premature termination codons. The last U (position −69) of UAU was replaced with A to create a stop codon (UAA) in the 3′ atpB cistron or in the atpE 5′-UTR. (B) Gel patterns of translation products. Lanes labeled C, no-mRNA control. Di and Mono indicate dicistronic and monocistronic mRNAs, respectively. ‘wt’, wild-type (UAU); ‘mut’, mutant (UAG). Synthesized EGFP and Citrine products were detected as in Figure 3. The EGFP product from the wt mRNA includes a β subunit fragment (27 amino acids) at the C-terminus. The premature termination deleted 24 amino acids from this extension, and therefore the band migrated faster. Faint bands above the main EGFP bands could be loosely folded EGFP.
To exclude the possibility that egfp interferes with translational coupling, we introduced a frameshift mutation in the middle of the original atpB cistron (insertion of UCGA between the 134th and the 135th codons at a BglII site), creating a premature stop codon (UGA) at the 151st codon (Figure 5A). Again, no significant inhibition of atpE translation was observed (Figure 5B). This mutation is identical to that applied to the maize chloroplast atpB/E operon expressed in E. coli cells, which caused almost complete prevention of expression of the atpE cistron (30). Taking this information together, we concluded that the downstream atpE cistron is translated almost independently from translation of the upstream atpB cistron (with little or no translational coupling) in the homologous chloroplast system. Therefore, the atpE cistron should be translated via its own cis-element.
Figure 5.
Effect of a frameshift mutation in the atpB cistron on atpE translation. (A) mRNA templates (as in Figure 2A) and location of the frameshift. Insertion of 4 nt at the first BglII site caused a frameshift, creating a stop codon within the atpB cistron. ‘wt’, wild type; ‘mut’, frameshift mutant. Sequences around the frameshift are in Supplementary Figure S2. (B) Gel patterns of translation products. Lanes labeled C, no-mRNA control. Synthesized Citrine products were detected by 532-nm light.
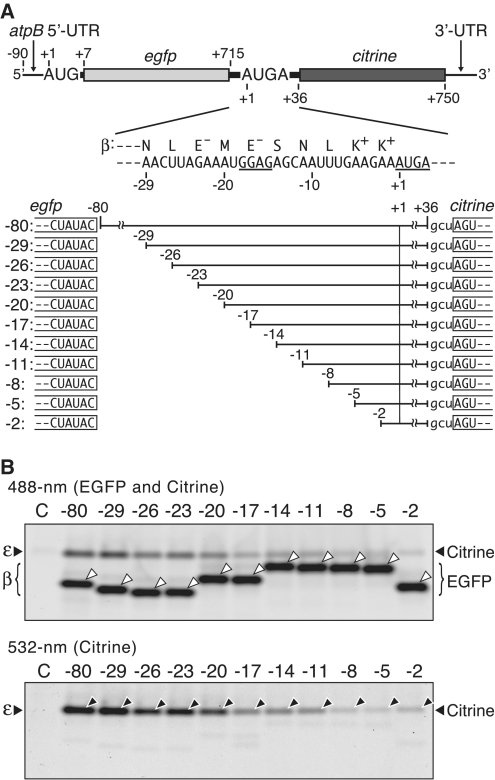
Cis-elements required for atpE translation
To define the sequences required for translation of the atpE cistron from dicistronic mRNAs, we prepared a series of dicistronic mRNAs with internal deletions and assayed their translation. Step-wise deletions (3 nt each) in the 5′ atpE cistron (from +6 to +36) hardly affected translation of either the atpB or atpE cistron (Supplementary Figure S6), indicating that the cis-element is localized upstream of the atpE start codon. Figure 6A shows parts deleted in the region upstream (from −80 to −3) of the atpE start codon (hereafter atpE 5′-UTR). As shown in Figure 6B (lower panel), no significant reduction of atpE translation was observed by deletion up to −20, while atpE translation dropped with a further 3-nt deletion (−17 mRNA), by which the SD-like sequence (from −18 to −15) was truncated (GGAG to cGAG, c from the egfp 3′-end) (Figure 6A). Further deletions to −11 led to no additional decrease in atpE translation, but deletions from −8 to −2 significantly inhibited its translation, though not completely. These observations indicate that the sequence, at least from −18 to −2, is important for atpE translation. Translation of the atpB cistron was hardly affected by these deletions (Figure 6B, bold bands in upper panel). Note that migration of EGFP products in native gels depends largely on their sizes and charges. 5′-Deletion analysis of the monocistronic atpE mRNA showed that a region of ~30 nt beginning 1 nt upstream from the start codon (−1) is also important for atpE translation (Supplementary Figure S7).
Figure 6.
Effect of internal deletions in the atpE 5′-UTR on atpE translation. (A) The mRNA (as in Figure 3A) and partial atpE 5′-UTR sequence. The SD-like sequence and the overlapping stop and start codons are underlined. mRNAs with internal deletions (indicated as blanks, from −80 to −3) are shown below. Terminal sequences of egfp and citrine are boxed. The gcu triplet after +36 is a ligation product. (B) Gel patterns of translation products. Synthesized EGFP and Citrine products were detected by 488-nm light (upper panel) and 532-nm light (lower panel). White arrowheads in the upper panel point to EGFP bands and black arrowheads in the lower panel point to Citrine bands. In the upper panel, migration of EGFP bands differed due to charge and size differences on native gels; i.e. deletion of one negatively charged glutamate (E−) caused slower migration (‘−23’ to ‘-20’ and ‘−17’ to ‘−14’) and deletion of positively charged lysine (K+) caused faster migration (‘−5’ to ‘−2’).
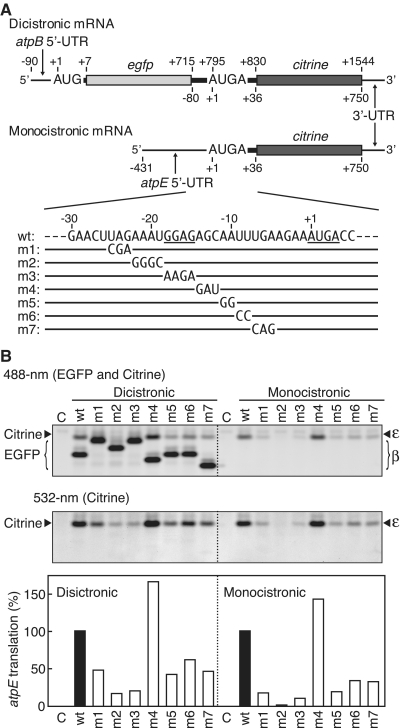
We then introduced mutations in the atpE 5′-UTR (positions −25 to −5) of both dicistronic and monocistronic mRNAs (Figure 7A) and assayed translation. Mutation from UAG to CGA (m1, −25 to −23) partly inhibited atpE translation and mutation from AAAU to GGGC (m2, −22 to −19) severely impaired translation from both dicistronic and monocistronic mRNAs (Figure 7B, lower panel, lanes m1 and m2). As expected, mutation of the SD-like sequence GGAG to AAGA (m3, −18 to −15) strongly inhibited atpE translation from both mRNAs (lanes m3; ~5-fold decrease in dicistronic and ~10-fold decrease in monocistronic mRNAs). Changes from AA to GG (m5, −11 to −10), UU to CC (m6, −9 to −8) and UGA to CAG (m7, −7 to −5) also inhibited atpE translation (lower panel, lanes m5, m6 and m7). Inhibitory effects of these mutations were less in the dicistronic mRNA than in the monocistronic mRNA (lower panel). These mutations caused no significant effect on atpB translation (Figure 7B, left upper panel). Migration of EGFP products (bold bands) differed largely due to charge differences in native gels (see Supplementary Figure S8 for altered amino acid sequences). Together with the results shown in Figures 6 and 7, this shows that the upstream sequence at least 25 nt from the atpE start codon plays an important role in efficient translation initiation of the atpE cistron in both dicistronic and monocistronic mRNAs.
Figure 7.
Effect of mutations in the atpE 5′-UTR on atpE translation. (A) mRNA templates as in Figures 3A (Dicistronic) and 2A (Monocistronic) and partial atpE 5′-UTR sequence. The SD-like sequence and the overlapping stop and start codons are underlined. Nucleotide and amino acid sequences are in Supplementary Figure S8. (B) Gel patterns of translation products. Lanes labeled C, no-mRNA control. Synthesized EGFP and Citrine products were detected as in Figure 6. In the upper left panel, migration of EGFP bands differed due to charge differences on native gels. The m1 and m3 dense bands overlap Citrine and EGFP products. Quantification of Citrine products based on intensity of fluorescent bands (wt value defined as 100%) is shown in bar graphs below.
Internal deletion up to −20 caused no significant inhibition of atpE translation (Figure 6B, lower panel, lanes −29 to −20), whereas mutations between −25 and −20 caused inhibition (Figure 7, lanes m1 and m2). The internal deletion resulted in fusion of the 3′-end of egfp (UAUAC, −85 to −81 in Figure 6A) to position −20, which could mimic the original sequence (UAGAA, −25 to −21), as these two sequences are similar. Mutation from AGC to GAU (−14 to −12, lane m4) caused no apparent inhibition from either dicistronic or monocistronic mRNAs. These observations suggest that either some nucleotide changes within the 25 nt sequence are tolerable as a regulatory sequence for atpE translation initiation or there are two separate cis-elements.
Deletion from positions −80 to −3 in the atpE 5′-UTR did not completely inhibit atpE translation (Figure 6B, lanes −5 and −2). Furthermore, mutation m2 abolished atpE translation from monocistronic mRNA whereas slight atpE translation was observed from dicistronic mRNA (Figure 7B, lanes m2). The residual translation of the atpE cistron from the dicistronic mRNA may be dependent on the upstream atpB cistron (see ‘Discussion’ section).
DISCUSSION
The chloroplast genes for ATP synthase subunits β and ε in many flowering plants possess overlapping start and stop codons. Based on this structure, it has been assumed that translation of these genes is coupled. Gatenby et al. (30) reported that the maize chloroplast atpB and atpE genes are translationally coupled in transformed E. coli and Synechocystis cells. They used these heterologous systems because no in vitro translation system from chloroplasts existed. We have established a highly active in vitro translation system from tobacco chloroplasts (34). Using this system, we showed that translational coupling does occur in the partially overlapping tobacco ndhC–ndhK transcripts (46) and in the overlapping tobacco psbD–psbC mRNA (Y. Adachi, H. Kuroda, Y. Yukawa and M. Sugiura).
However, unexpectedly, we concluded that the tobacco atpB/E mRNAs are not translationally coupled (except perhaps for a minor fraction) in the homologous tobacco chloroplast translation system. To arrive at this conclusion, we first devised an assay to detect translation from two cistrons differentially using two different fluorescent proteins. Results obtained by exchanging 5′-UTRs (Figure 3) and introduction of premature stop codons (Figures 4 and 5) indicated that no translational coupling occurs between the two cistrons.
Translation of the tobacco atpE cistron should occur via its own cis-element. Deletion and mutation analyses showed that the cis-element(s) resides within 25 nt or so from the atpE start codon. We previously reported that the SD-like sequence is required for translation of the monocistronic atpE mRNA because changing the GGAG to CCUC reduced atpE translation to <5% of the original mRNA (33). This was confirmed by our present study (see Figure 6, lane −17 and Figure 7, lane m3). However, our present study showed that the SD-like sequence is not the sole determinant. A sequence of at least 25 nt (up to −29) encompassing the SD-like sequence is necessary for efficient atpE translation from both dicistronic and monocistronic mRNAs (Figures 6 and 7). Therefore, the GGAG may not function similarly to the SD sequence found in most E. coli mRNAs. Baecker et al. (47) analyzed translation of chimeric uidA with the tobacco chloroplast atpI 5′-UTR in transplastomic tobacco plants. They found that efficient translation of uidA requires element(s) upstream of the putative SD sequence. Our results were similar. Translation of the tobacco atpE cistron may also require two or more cis-elements. In spinach chloroplasts, the atpB/E transcript lacking its 5′-UTR was found in crude polysomal fractions (48). Our study has clearly indicated that removal of the atpB 5′-UTR completely arrests translation of the atpB cistron but not the atpE cistron (see Figure 3, lanes ‘–‘). Hence, this leaderless transcript is likely to support only atpE translation.
Chloroplast translation is usually regulated by positive trans-acting regulatory proteins (8,10,49,50). The maize nuclear atp1 gene product is required for translation of the maize atpB/E mRNA (51). The tobacco chloroplast atpB mRNA contains no SD-like sequence but does contain a U-rich sequence in the 5′-UTR (−25 to −1), and this 25 nt upstream sequence was essential for translation of the atpB cistron (32). A 50-kDa protein (p50), detected by UV-crosslinking, specifically bound the 25 nt sequence, suggesting that p50 is a trans-acting factor or a component of such a factor for atpB translation initiation. One possibility is that p50 is the homolog of the maize atp1 gene product. Specific chloroplast proteins were reported to bind to the atpE 5′-UTR as well as to the atpB 5′-UTR from spinach chloroplast mRNAs (52). Therefore, it is possible that a trans-acting factor(s) is also necessary for atpE translation. If this is the case, this factor binds to the atpE 5′-UTR, namely the 3′ side of the atpB coding region, which would hamper the elongation of atpB translation. It may be that a fraction of atpB/E mRNAs produces solely subunit β and the other fraction provides subunit ε.
Our study showed that the residual atpE translation from the dicistronic mRNA that may be due to translational coupling is <10% of atpB translation (Figures 6 and 7). The SD sequence is necessary to translate downstream cistrons by translational coupling in E. coli (19). However, residual translation was still observed after deletion and mutation of the SD-like sequence in the atpE 5′-UTR. Thus, we cannot rule out the possibility that an unknown mechanism supports this residual atpE translation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Nagoya City University Funds; New Energy and Industrial Technology Development Organization; Grants-in-Aid for Scientific Research (15370025, 17370020 to M.S., 20570043 to H.K.). Funding for open access charge: Grant-in-Aid for Scientific Research (1737002).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEGEMENTS
The authors thank their laboratory members for help and discussion.
REFERENCES
- 1.Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 2.Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol. 19. Potsdam-Golm: Springer; 2007. pp. 29–63. [Google Scholar]
- 3.Deng XW, Gruissem W. Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell. 1987;49:379–387. doi: 10.1016/0092-8674(87)90290-x. [DOI] [PubMed] [Google Scholar]
- 4.Mullet JE. Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1988;39:475–502. [Google Scholar]
- 5.Marín-Navarro J, Manuell AL, Wu J, Mayfield SP. Chloroplast translation regulation. Photosynth. Res. 2007;94:359–374. doi: 10.1007/s11120-007-9183-z. [DOI] [PubMed] [Google Scholar]
- 6.Wobbe L, Schwarz C, Nickelsen J, Kruse O. Translational control of photosynthetic gene expression in phototrophic eukaryotes. Physiol. Plant. 2008;133:507–515. doi: 10.1111/j.1399-3054.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 7.Sugiura M, Hirose T, Sugita M. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 1998;32:437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 8.Peled-Zehavi H, Danon A. Translation and translational regulation in chloroplasts. In: Bock R, editor. Cell and Molecular Biology of Plastids. Vol. 19. Berlin Heidelberg: Springer; 2007. pp. 248–281. [Google Scholar]
- 9.Gruissem W, Tonkyn JC. Control mechanisms of plastid gene expression. Crit. Rev. Plant Sci. 1993;12:19–55. [Google Scholar]
- 10.Rochaix J-D. The role of nucleus- and chloroplast-encoded factors in the synthesis of the photosynthetic apparatus. In: Wise RR, Hoober JK, editors. The Structure and Function of Plastids. Vol. 23. Dordrecht: Springer; 2006. pp. 145–165. [Google Scholar]
- 11.Shinozaki K, Sugiura M. Sequence of the intercistronic region between the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit and coupling factor β subunit gene. Nucleic Acids Res. 1982;10:4923–4934. doi: 10.1093/nar/10.16.4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiratsuka J, Shimada H, Whittier R, Ishibashi T, Sakamoto M, Mori M, Kondo C, Honji Y, Sun CR, Meng BY, et al. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 1989;217:185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Obokata J, Chunwongse J, Shinozaki K, Sugiura M. Rapid splicing and stepwise processing of a transcript from the psbB operon in tobacco chloroplasts: determination of the intron sites in petB and petD. Mol. Gen. Genet. 1987;209:427–431. doi: 10.1007/BF00331145. [DOI] [PubMed] [Google Scholar]
- 14.Matsubayashi T, Wakasugi T, Shinozaki K, Yamaguchi-Shinozaki K, Zaita N, Hidaka T, Meng BY, Ohto C, Tanaka M, Kato A, et al. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol. Gen. Genet. 1987;210:385–393. doi: 10.1007/BF00327187. [DOI] [PubMed] [Google Scholar]
- 15.Barkan A. Proteins encoded by a complex chloroplast transcription unit are each translated from both monocistronic and polycistronic mRNAs. EMBO J. 1988;7:2637–2644. doi: 10.1002/j.1460-2075.1988.tb03116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westhoff P, Herrmann RG. Complex RNA maturation in chloroplasts. The psbB operon from spinach. Eur. J. Biochem. 1988;171:551–564. doi: 10.1111/j.1432-1033.1988.tb13824.x. [DOI] [PubMed] [Google Scholar]
- 17.Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 18.Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson RJ, Kaminski A, Pöyry TAA. Coupled termination-reinitiation events in mRNA translation. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. New York: Cold Spring Harbor Laboratory Press; 2007. pp. 197–223. [Google Scholar]
- 20.Inokuchi Y, Hirashima A, Sekine Y, Janosi L, Kaji A. Role of ribosome recycling factor (RRF) in translational coupling. EMBO J. 2000;19:3788–3798. doi: 10.1093/emboj/19.14.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powell ML, Napthine S, Jackson RJ, Brierley I, Brown TD. Characterization of the termination-reinitiation strategy employed in the expression of influenza B virus BM2 protein. RNA. 2008;14:2394–2406. doi: 10.1261/rna.1231008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hennig J, Herrmann RG. Chloroplast ATP synthase of spinach contains nine nonidentical subunit species, six of which are encoded by plastid chromosomes in two operons in a phylogenetically conserved arrangement. Mol. Gen. Genet. 1986;203:117–128. [Google Scholar]
- 23.Wollman F-A, Minai L, Nechushtai R. The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim. Biophys. Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- 24.Hudson GS, Mason JG, Holton TA, Koller B, Cox GB, Whitfeld PR, Bottomley W. A gene cluster in the spinach and pea chloroplast genomes encoding one CF1 and three CF0 subunits of the H+-ATP synthase complex and the ribosomal protein S2. J. Mol. Biol. 1987;196:283–298. doi: 10.1016/0022-2836(87)90690-5. [DOI] [PubMed] [Google Scholar]
- 25.Stollar NN, Kim J-K, Hollingsworth MJ. Ribosomes pause during the expression of the large ATP synthase gene cluster in spinach chloroplasts. Plant Physiol. 1994;105:1167–1177. doi: 10.1104/pp.105.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyagi T, Kapoor S, Sugita M, Sugiura M. Transcript analysis of the tobacco plastid operon rps2/atpI/H/F/A reveals the existence of a non-consensus type II (NCII) promoter upstream of the atpI coding sequence. Mol. Gen. Genet. 1998;257:299–307. doi: 10.1007/s004380050651. [DOI] [PubMed] [Google Scholar]
- 27.Green CD, Hollingsworth MJ. Expression of the large ATP synthase gene cluster in spinach plastids during light-induced development. Plant Physiol. 1992;100:1164–1170. doi: 10.1104/pp.100.3.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zurawski G, Bottomley W, Whitfeld PR. Structures of the genes for the β and ε subunits of spinach chloroplast ATPase indicate a dicistronic mRNA and an overlapping translation stop/start signal. Proc. Natl Acad. Sci. USA. 1982;79:6260–6264. doi: 10.1073/pnas.79.20.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krebbers ET, Larrinua IM, McIntosh L, Bogorad L. The maize chloroplast genes for the β and ε subunits of the photosynthetic coupling factor CF1 are fused. Nucleic Acids Res. 1982;10:4985–5002. doi: 10.1093/nar/10.16.4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gatenby AA, Rothstein SJ, Nomura M. Translational coupling of the maize chloroplast atpB and atpE genes. Proc. Natl Acad. Sci. USA. 1989;86:4066–4070. doi: 10.1073/pnas.86.11.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kapoor S, Wakasugi T, Deno H, Sugiura M. An atpE-specific promoter within the coding region of the atpB gene in tobacco chloroplast DNA. Curr. Genet. 1994;26:263–268. doi: 10.1007/BF00309558. [DOI] [PubMed] [Google Scholar]
- 32.Hirose T, Sugiura M. Multiple elements required for translation of plastid atpB mRNA lacking the Shine-Dalgarno sequence. Nucleic Acids Res. 2004;32:3503–3510. doi: 10.1093/nar/gkh682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirose T, Sugiura M. Functional Shine-Dalgarno-like sequences for translational initiation of chloroplast mRNAs. Plant Cell Physiol. 2004;45:114–117. doi: 10.1093/pcp/pch002. [DOI] [PubMed] [Google Scholar]
- 34.Yukawa M, Kuroda H, Sugiura M. A new in vitro translation system for non-radioactive assay from tobacco chloroplasts: effect of pre-mRNA processing on translation in vitro. Plant J. 2007;49:367–376. doi: 10.1111/j.1365-313X.2006.02948.x. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi K, Shiina T, Ishii N, Iwai K, Ishizaki Y, Morikawa K, Toyoshima Y. A role of the -35 element in the initiation of transcription at psbA promoter in tobacco plastids. Plant Cell Physiol. 2003;44:334–341. doi: 10.1093/pcp/pcg041. [DOI] [PubMed] [Google Scholar]
- 36.Heikal AA, Hess ST, Baird GS, Tsien RY, Webb WW. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: coral red (dsRed) and yellow (Citrine) Proc. Natl Acad. Sci. USA. 2000;97:11996–12001. doi: 10.1073/pnas.97.22.11996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 38.Yukawa M, Tsudzuki T, Sugiura M. The 2005 version of the chloroplast DNA sequence from tobacco (Nicotiana tabacum) Plant Mol. Biol. Rep. 2005;23:1–7. [Google Scholar]
- 39.Kuroda H, Suzuki H, Kusumegi T, Hirose T, Yukawa Y, Sugiura M. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol. 2007;48:1374–1378. doi: 10.1093/pcp/pcm097. [DOI] [PubMed] [Google Scholar]
- 40.Shinozaki K, Deno H, Kato A, Sugiura M. Overlap and cotranscription of the genes for the beta and epsilon subunits of tobacco chloroplast ATPase. Gene. 1983;24:147–155. doi: 10.1016/0378-1119(83)90074-4. [DOI] [PubMed] [Google Scholar]
- 41.Shiina T, Allison L, Maliga P. rbcL transcript levels in tobacco plastids are independent of light: reduced dark transcription rate is compensated by increased mRNA stability. Plant Cell. 1998;10:1713–1722. doi: 10.1105/tpc.10.10.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orozco EMJ, Chen LJ, Eilers RJ. The divergently transcribed rbcL and atpB genes of tobacco plastid DNA are separated by nineteen base pairs. Curr. Genet. 1990;17:65–71. doi: 10.1007/BF00313250. [DOI] [PubMed] [Google Scholar]
- 43.Kapoor S, Suzuki JY, Sugiura M. Identification and functional significance of a new class of non-consensus-type plastid promoters. Plant J. 1997;11:327–337. doi: 10.1046/j.1365-313x.1997.11020327.x. [DOI] [PubMed] [Google Scholar]
- 44.Hajdukiewicz PTJ, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuroda H, Maliga P. Sequences downstream of the translation initiation codon are important determinants of translation efficiency in chloroplasts. Plant Physiol. 2001;125:430–436. doi: 10.1104/pp.125.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yukawa M, Sugiura M. Termination codon-dependent translation of partially overlapping ndhC-ndhK transcripts in chloroplasts. Proc. Natl Acad. Sci. USA. 2008;105:19550–19554. doi: 10.1073/pnas.0809240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baecker JJ, Sneddon JC, Hollingsworth MJ. Efficient translation in chloroplasts requires element(s) upstream of the putative ribosome binding site from atpI. Am. J. Bot. 2009;96:627–636. doi: 10.3732/ajb.0800259. [DOI] [PubMed] [Google Scholar]
- 48.Bennett DC, Rogers SA, Chen L-J, Orozco EM., Jr A primary transcript in spinach chloroplasts that completely lacks a 5′ untranslated leader region. Plant Mol. Biol. 1990;15:111–119. doi: 10.1007/BF00017728. [DOI] [PubMed] [Google Scholar]
- 49.Goldschmidt-Clermont M. Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 1998;177:115–180. doi: 10.1016/s0074-7696(08)62232-9. [DOI] [PubMed] [Google Scholar]
- 50.Nickelsen J. Chloroplast RNA-binding proteins. Curr. Genet. 2003;43:392–399. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 51.McCormac DJ, Barkan A. A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell. 1999;11:1709–1716. doi: 10.1105/tpc.11.9.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robida MD, Merhige PM, Hollingsworth MJ. Proteins are shared among RNA-protein complexes that form in the 5′ untranslated regions of spinach chloroplast mRNAs. Curr. Genet. 2002;41:53–62. doi: 10.1007/s00294-002-0283-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.